SUMMARY
Faithful translation of the genetic code depends on the GTPase EF-Tu delivering correctly charged aminoacyl-tRNAs to the ribosome for pairing with cognate codons. The accurate coupling of cognate amino acids and tRNAs by the aminoacyl-tRNA synthetases is achieved through a combination of substrate specificity and product editing. Once released by aminoacyl-tRNA synthetases, both cognate and near-cognate aminoacyl-tRNAs were considered to be committed to ribosomal protein synthesis through their association with EF-Tu. Here we show instead that aminoacyl-tRNAs in ternary complex with EF-Tu•GTP can readily dissociate and rebind to aminoacyl-tRNA synthetases. For mischarged species, this allows resampling by the product editing pathway, leading to a reduction in the overall error rate of aminoacyl-tRNA synthesis. Resampling of mischarged tRNAs was shown to increase the accuracy of translation over ten fold during in vitro protein synthesis, supporting the presence of an additional quality control step prior to translation elongation.
INTRODUCTION
Faithful translation of genetic information from mRNA to protein is critical for normal cellular functions. The protein synthesis machinery utilizes aminoacyl-tRNAs (aa-tRNAs), which are formed by aminoacyl-tRNA synthetases (aaRSs) and delivered to the ribosome by elongation factors (EF-Tu in bacteria and EF-1α in archaea and eukaryotes) (Ibba and Söll, 2000; Ogle and Ramakrishnan, 2005). The overall translation error rate of 10−3–10−4 is a net accumulation from several steps, including transcription (~10−4), aa-tRNA synthesis (~10−5), and ribosomal decoding (~10−4) (Loftfield and Vanderjagt, 1972; Rosenberger and Foskett, 1981; Ibba and Söll, 1999; Ogle and Ramakrishnan, 2005; Roy and Ibba, 2006). As the error rates from all the above steps are similar and additive, elevated mistakes during any step, such as aminoacylation, may limit the overall accuracy of protein synthesis. To maintain genetic code fidelity, aaRSs selectively pair the correct amino acids with their cognate tRNAs in a two-step aminoacylation reaction. AaRSs first activate the amino acid with ATP to form an aminoacyl-adenylate intermediate and then catalyze the esterification of the activated amino acid to the 2′ or 3′ hydroxyl at the 3′ end of tRNA. AaRSs are extremely selective for their cognate tRNAs due to highly specific binding and kinetic proofreading (Ibba and Söll, 1999; Guth and Francklyn, 2007). In contrast, several aaRSs lack sufficient discrimination against structurally similar near-cognate amino acids during activation. For example, phenylalanyl-tRNA synthetase (PheRS) misactivates Tyr at a level higher than the overall translation error rate (Lin et al., 1984; Roy et al., 2005). However, such mistakes by aaRSs are not directed to protein synthesis, due to a proofreading step (Baldwin and Berg, 1966). This step, called editing, occurs through hydrolysis of misactivated amino acids (pretransfer editing) or misacylated aminoacyl-tRNAs (posttransfer editing), while the correct products are excluded from the hydrolytic reaction. Editing activities have been shown to play critical roles in vivo (Döring et al., 2001; Roy et al., 2004; Lee et al., 2006) and are found in both aaRS structural classes (I and II) (Ibba and Söll, 2000; Hendrickson and Schimmel, 2003).
Proofreading aaRSs contain a hydrolytic editing site 30–40 Å away from the synthetic active site (Nureki et al., 1998; Fukai et al., 2000; Dock-Bregeon et al., 2000; Cusack et al., 2000; Kotik-Kogan et al., 2005; Crepin et al., 2006). How mischarged tRNA travels from the active to the editing site remains an open question. It has been suggested that the aminoacylated 3′ end of the tRNA translocates between the two sites while the rest of the tRNA molecule remains attached to the synthetase (Silvian et al., 1999; Dock-Bregeon et al., 2000; Fukunaga and Yokoyama, 2005; Tukalo et al., 2005). This model is consistent with several structural and modeling studies but fails to explain the posttransfer editing mechanism of alanyl-tRNA synthetase, which requires rearrangement of more substantial parts of the tRNA than the 3′ end alone (Musier-Forsyth et al., 1991; Beebe et al., 2008). An added complication is posed by EF-Tu, which would be expected to tightly bind any mischarged tRNAs that dissociate before editing is complete (Ling et al., 2007b). Using Tyr-tRNAPhe editing by PheRS as a model, we show here that a fraction of mischarged tRNA dissociates from the aaRS prior to entering the editing site. Rather than being sequestered directly by EF-Tu for protein synthesis, released mischarged tRNAs can rebind to the aaRS, leading to resampling by the editing site and a reduction in the aminoacylation error rate. The net effect of this resampling is to provide an additional quality control step before translation elongation.
RESULTS
Substrate Dissociation during Editing of Mischarged tRNAs
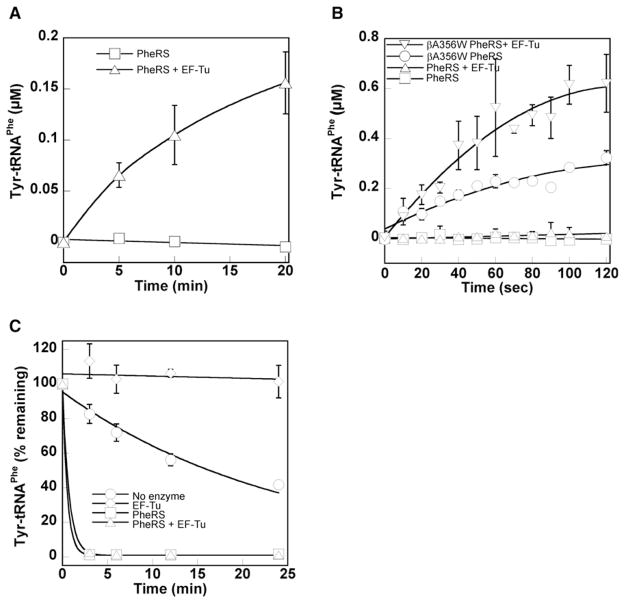
EF-Tu was used as a probe to investigate substrate movement during PheRS editing of Tyr-tRNAPhe. EF-Tu binds aa-tRNAs, including Tyr-tRNAPhe, with high affinity at low temperature and provides protection from both spontaneous and enzymatic hydrolysis (at 4°C, Kd = 5–50 nM) (LaRiviere et al., 2001; Ling et al., 2007b). Superimposition of the EF-Tu:Phe-tRNAPhe structure onto the PheRS:tRNAPhe complex by overlaying the tRNA backbones revealed extensive steric clashes (see Figure S1 available online) (Goldgur et al., 1997; Nissen et al., 1995). These structural constraints suggest that EF-Tu will not be able to protect Tyr-tRNAPhe from hydrolysis until after it is released by PheRS. This allowed us to use EF-Tu to trap and stabilize any Tyr-tRNAPhe that dissociates from PheRS before entering the editing site. An E. coli PheRS active site variant (αA294G) with enhanced Tyr activation activity was first tested (Ibba et al., 1994; Roy et al., 2004). This PheRS variant, which has wild-type editing activity, did not accumulate Tyr-tRNAPhe at 2°C in the absence of EF-Tu (Figures 1A and S2). Addition of EF-Tu led to accumulation of Tyr-tRNAPhe under the same conditions, indicating that at least a fraction of Tyr-tRNAPhe dissociates from PheRS before entering the editing site. This suggests that PheRS may also be able to compete with EF-Tu for mischarged tRNAs, which would constitute an additional quality control step during aminoacylation.
Figure 1. PheRS Editing Site Competes with EF-Tu for Tyr-tRNAPhe.
(A) Tyrosylation by E. coli PheRS (0.75 μM) at 2°C ± 10 μM activated E. coli EF-Tu.
(B) Tyrosylation by wild-type or editing-defective (βA356W) E. coli PheRS (0.25 μM) at 37°C ± 10 μM activated E. coli EF-Tu.
(C) Hydrolysis of Tyr-tRNAPhe. WT β subunit EcPheRS (0.75 μM) and activated E. coli EF-Tu (10 μM) were mixed and preincubated at 37°C for 3 min before the addition of 1 μM Tyr-tRNAPhe. Phe-tRNAPhe was previously shown to be stable under these conditions (Ling et al., 2007a).
Error bars correspond to the standard deviation from three independent experiments.
PheRS Competes Effectively with EF-Tu for Tyr-tRNAPhe
EF-Tu readily binds Tyr-tRNAPhe and can deliver it to the ribosome for translation elongation just as efficiently as it does Phe-tRNAPhe (Ling et al., 2007b). This observation, and the fact that EF-Tu is one of the most abundant proteins in E. coli, would seem to suggest that any mischarged tRNAs that escape PheRS will be sequestered by EF-Tu. However, PheRS with a wild-type editing site did not produce detectable levels of Tyr-tRNAPhe at 37°C even in the presence of excess EF-Tu, while an editing-defective PheRS variant (βA356W) stably accumulated the mischarged species (Figure 1B). These results suggest that, at physiological temperatures, little EF-Tu-bound Tyr-tRNAPhe is available for the ribosome, presumably due to the ability of PheRS to hydrolyze Tyr-tRNAPhe despite the presence of EF-Tu. To directly test the competition, purified Tyr-tRNAPhe was added to a mixture of activated EF-Tu and PheRS. Tyr-tRNAPhe was completely hydrolyzed by PheRS even though EF-Tu was present in excess (Figure 1C), confirming that the PheRS editing site effectively competes with EF-Tu for free Tyr-tRNAPhe under physiological conditions.
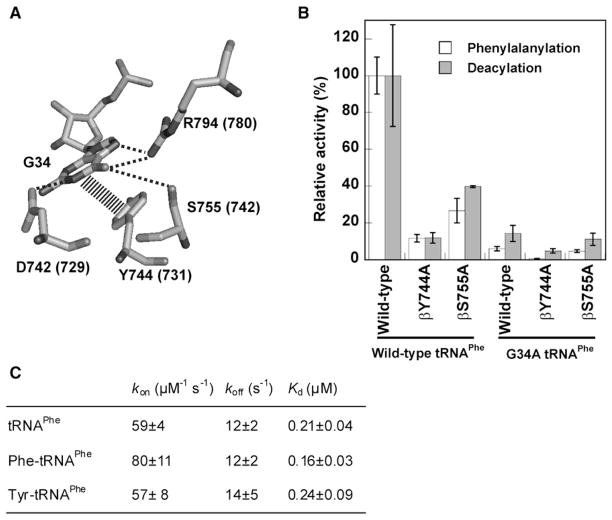
tRNAPhe Recognition Is Conserved between Aminoacylation and Editing
The recognition of tRNAPhe during editing was investigated in order to determine how PheRS rebinds Tyr-tRNAPhe after it initially dissociates. G34 in the anticodon of tRNAPhe is recognized by several conserved PheRS residues during aminoacylation, and this interaction was used to probe recognition (Figure 2A) (Peterson and Uhlenbeck, 1992; Goldgur et al., 1997). The G34A mutation in tRNAPhe decreased aminoacylation 17-fold, while the βY744A and βS755A replacements in PheRS reduced efficiency by 10-fold and 4-fold, respectively (Figure 2B and Table S1). Next we examined recognition of G34 by PheRS during editing of preformed Tyr-tRNAPhe. The G34A, βY744A, or βS755A replacements all showed reductions in editing activity comparable to the changes observed for aminoacylation (Figure 2B and Table S2). This indicates that at least some of the major protein-RNA recognition events are conserved between aminoacylation and editing in PheRS. This is consistent with earlier models that predicted that bending of the tRNAPhe 3′ end alone would be sufficient to switch the mischarged Tyr between the active and editing sites of PheRS (Roy et al., 2004).
Figure 2. Recognition of E. coli tRNAPhe during Aminoacylation and Editing.
(A) Recognition of G34 by PheRS in the aminoacylation conformation (Goldgur et al., 1997). E. coli PheRS numbering is shown (equivalent T. thermophilus residues in parentheses).
(B) Relative phenylalanylation and Tyr-tRNAPhe deacylation activities.
(C) Parameters for Phe-tRNAPhe and Tyr-tRNAPhe binding to βA356W PheRS.
Error bars correspond to the standard deviation from three independent experiments.
To further investigate aminoacyl-tRNA recognition during editing, the kinetics of Phe-tRNAPhe and Tyr-tRNAPhe binding were determined. To prevent hydrolysis of Tyr-tRNAPhe, an editing-defective PheRS variant (βA356W) was employed. Phe-tRNAPhe and Tyr-tRNAPhe showed no significant differences in any of the kinetic parameters measured (Figure 2C), and the constants were comparable to those previously measured for E. coli PheRS binding of Phe-tRNAPhe (Baltzinger and Holler, 1982).
Substrate Dissociation and Rebinding Is Specific for Class II-Type Editing
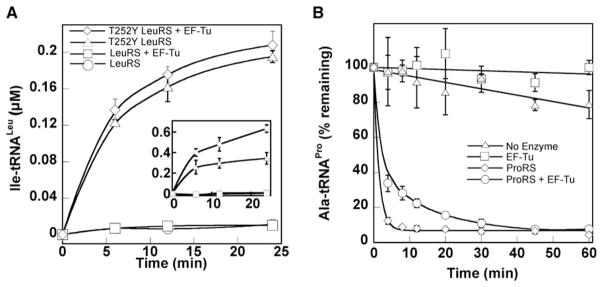
AaRSs are grouped into two mutually exclusive structural classes, I and II, based on differences in their active site topologies (Ibba and Söll, 2000). In class II aaRSs such as AlaRS, PheRS, and ProRS, ami-noacyl-tRNA release is rapid and not rate limiting during aminoacylation, while the rate of aminoacylation by class I enzymes is limited by product release (Baltzinger and Holler, 1982; Ibba et al., 1995; Zhang et al., 2006). To examine whether these class-specific differences in activity also extend to the dissociation and rebinding of mischarged tRNAs, we investigated the impact of EF-Tu on editing by LeuRS (class I) (Lincecum et al., 2003) and ProRS (class II) (Beuning and Musier-Forsyth, 2000).
EF-Tu did not stimulate accumulation of mischarged Ile-tRNALeu by wild-type LeuRS, irrespective of temperature (Figures 3A and S3). As expected, EF-Tu did stabilize Ile-tRNALeu in the presence of an editing-defective LeuRS variant (T252Y; Figure 3A, inset). Although Ala mischarging by ProRS is too weak to observe any reproducible effect of EF-Tu addition (data not shown), deacylation assays showed that ProRS competes with EF-Tu for mischarged Ala-tRNAPro, indicating that mischarged tRNAPro is rebound and edited after its initial synthesis and dissociation from ProRS (Figure 3B).
Figure 3. Misacylation by LeuRS and ProRS.
(A) Isoleucylation of total E. coli tRNAs by wild-type and editing-defective (T252Y) E. coli LeuRS variants at 2°C (8 μM enzyme) or 37°C (2.4 μM enzyme, inset) ± 10 μM activated E. coli EF-Tu.
(B) Hydrolysis of 0.5 μM Ala-tRNAPro at 37°C ± EF-Tu, 2.5 μM EcProRS, and 10 μM activated E. coli EF-Tu. Pro-tRNAPro was previously shown to be stable under these conditions (Beuning and Musier-Forsyth, 2000).
Error bars correspond to the standard deviation from three independent experiments.
Tyr-tRNAPhe Is Resampled Prior to Translation Elongation
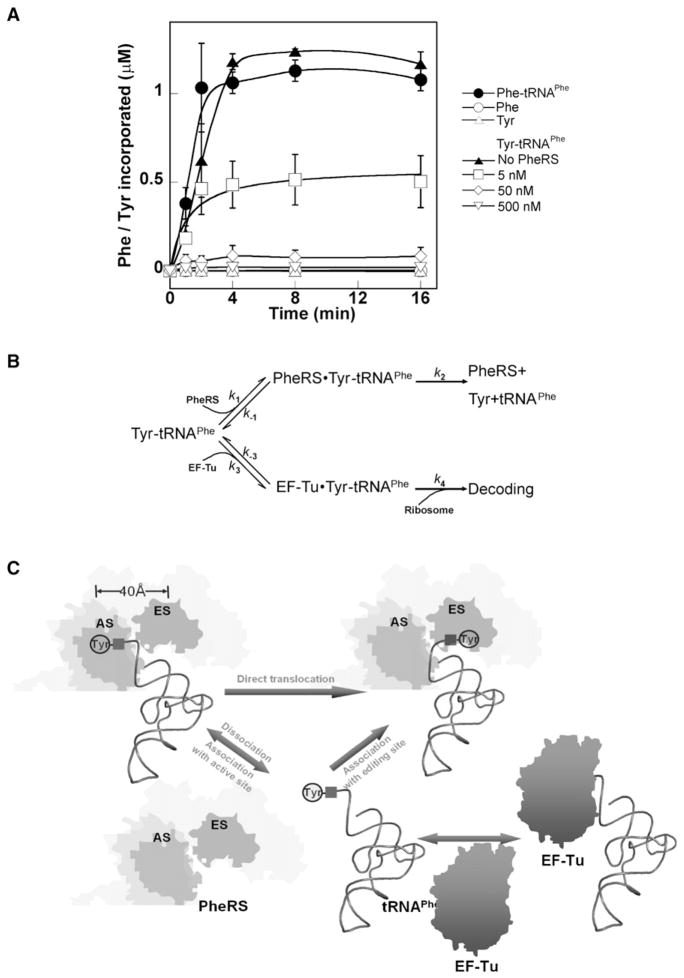
While our data showed that PheRS and other class II editing proteins could compete with EF-Tu to edit mischarged tRNA, it remained unclear how effective this competition would be compared to Tyr-tRNAPhe•EF-Tu•GTP binding by the ribosome. The effect of translating ribosomes on resampling and editing of mischarged tRNAs was investigated using poly(U)-directed ribosomal polypeptide synthesis. Previous studies showed that ribosomes utilize Tyr-tRNAPhe and Phe-tRNAPhe with equal efficiencies in the presence of poly(U) and purified EF-Tu and EF-G, but in the absence of PheRS (Ling et al., 2007b). These experiments were now repeated with the addition of increasing amounts of PheRS (Figure 4A). Upon addition of purified Phe-tRNAPhe, poly-Phe was rapidly synthesized with approximately 100% of the radiolabeled Phe finally incorporated into the polypeptide. When Tyr-tRNAPhe was used in the absence of PheRS, yields of poly-Tyr were obtained that were comparable to those for poly-Phe. If, instead, PheRS was included at 0.1% the level of EF-Tu, poly-Tyr synthesis was reduced by more than half, and at higher concentrations of the synthetase PheRS polypeptide synthesis was virtually eliminated. These data indicate that the majority of free Tyr-tRNAPhe is hydrolyzed before its utilization in translation despite the presence of both EF-Tu and ribosomes, a step that provides a substantial reduction in the error rate of protein synthesis.
Figure 4. Resampling and Editing of Misacylated tRNA.
(A) Poly(U)-directed poly-Phe and poly-Tyr synthesis at 37°C. As controls, 1 μM [14C] Phe or [3H] Tyr were added instead of [14C] Phe-tRNAPhe or [3H] Tyr-tRNAPhe. For poly-Tyr synthesis 0, 5, 50, or 500 nM PheRS was included as indicated. PheRS was also added to poly-Phe synthesis reactions at the same concentrations, but no change in poly-Phe synthesis was observed (data not shown).
(B) Scheme of competition for Tyr-tRNAPhe between PheRS and EF-Tu. As k1[PheRS] > k3 [EF-Tu] and k2 > > k−1, the majority of free-standing Tyr-tRNAPhe is bound by PheRS and hydrolyzed, whereas only a small fraction is utilized by the ribosome in protein synthesis.
(C) Model for concerted editing pathways. (Top) cis-editing pathway. Upon synthesis at the active site (AS), a fraction of Tyr-tRNAPhe directly translocates to the editing site for hydrolysis, which is not accessible to EF-Tu. (Bottom) trans-editing pathway. Tyr-tRNAPhe dissociates from PheRS and is competed for by EF-Tu and PheRS. Tyr-tRNAPhe bound to the editing site is rapidly hydrolyzed to yield a very low level of EF-Tu-bound Tyr-tRNAPhe in vivo.
Error bars correspond to the standard deviation from three independent experiments.
DISCUSSION
Resampling and Editing of Mischarged tRNA
In vitro poly-Tyr synthesis using Tyr-tRNAPhe was virtually eliminated upon addition of sufficient PheRS, suggesting that the majority of the mischarged tRNA was accessed and edited by the synthetase before it could be used for translation elongation (Figure 4A). While this contradicts the notion that aa-tRNAs are almost irreversibly bound to EF-Tu until GTP hydrolysis occurs on the ribosome, the data are consistent with substrate binding kinetics. EF-Tu binds aa-tRNAs very slowly, with an association rate constant (k3 in Figure 4B) of approximately 0.1 μM−1 s−1 at physiological temperatures (LaRiviere et al., 2001; Asahara and Uhlenbeck, 2002; Roy and Ibba, 2008). In contrast, the association rate of E. coli PheRS for Tyr-tRNAPhe is ~60 μM−1 s−1. The 500-fold difference in association rate constants ensures that PheRS can effectively compete with EF-Tu for Tyr-tRNAPhe even though EF-Tu is in about 100-fold excess over PheRS in E. coli (Furano, 1975; Neidhardt et al., 1977; Jakubowski and Goldman, 1984). Once bound by PheRS, Tyr-tRNAPhe is likely to be hydrolyzed rapidly.
The dissociation rate of Tyr-tRNAPhe from E. coli PheRS is about 14 s−1, so k2 in Figure 4B is estimated to be greater than 200 s−1 from the equation KM = (k−1 + k2)/k1 (KM of PheRS for Tyr-tRNAPhe hydrolysis exceeds 5 μM [Ling et al., 2007a]). This indicates that the association of Tyr-tRNAPhe to the PheRS editing site is nearly unidirectional. Conversely, the binding of aa-tRNAs by EF-Tu is predicted to be readily reversible, consistent with the recent descriptions of nonribosomal processes that rely on canonical elongator aa-tRNAs as substrates (Villet et al., 2007; Watanabe et al., 2007; Lloyd et al., 2008; Roy and Ibba, 2008).
A Concerted Translocation Model for Class II-Type Editing
In class II aaRS enzymes such as PheRS, ProRS, and ThrRS, mischarged tRNA is synthesized at the active site and hydrolyzed at the distinct editing site ~40 Å away. It had been believed that direct translocation of mischarged tRNA between these two sites on the aaRS was an absolute requirement for editing (Roy et al., 2004). Our data now show that, while this may hold true for class I aaRSs, class II type editing includes an additional resampling pathway (Figure 4C). Following synthesis at the active site, a portion of Tyr-tRNAPhe directly translocates to the editing site through the movement of the 3′ end and is hydrolyzed in cis, while the rest of the mischarged Tyr-tRNAPhe either partially (Yang et al., 2006) or completely dissociates from PheRS. At physiological temperatures, PheRS efficiently competes with EF-Tu to rebind released Tyr-tRNAPhe, with the Tyr moiety entering the editing site to be rapidly hydrolyzed in trans. In contrast to the reversible binding by EF-Tu, the binding and hydrolysis of Tyr-tRNAPhe by the PheRS editing site is unidirectional, promoting the equilibrium of EF-Tu binding to shift toward releasing Tyr-tRNAPhe. This maintains a very low level of EF-Tu-bound Tyr-tRNAPhe and restricts its use in protein synthesis. Nevertheless, the cis-editing pathway should not be excluded. Even in the presence of EF-Tu at 2°C, where aa-tRNAs are more tightly bound (Roy and Ibba, 2008), wild-type EcPheRS produces Tyr-tRNAPhe much more slowly than the editing-defective variant (data not shown), suggesting that a significant portion of the editing activity can not be sequestered by EF-Tu. Previously it was shown that in minimal medium enriched with Tyr and depleted for Phe, coexpressing wild-type and editing-defective PheRS variants resulted in Tyr misincorporation at Phe codons (Roy et al., 2004), indicating that Tyr-tRNAPhe overproduced by the editing-defective PheRS could not be completely hydrolyzed in trans. Although the relative contributions of cis and trans editing remain to be characterized, it is likely that PheRS trans editing complements cis editing by efficiently hydrolyzing misacylated Tyr-tRNAPhe that escapes from the enzyme, mimicking the function of autonomous trans-editing factors (Ahel et al., 2003; Wong et al., 2003; Korencic et al., 2004).
Trans Editing
Several autonomous trans-editing factors have been identified that hydrolyze mischarged tRNAs (Ahel et al., 2003; Wong et al., 2003; An and Musier-Forsyth, 2004; Korencic et al., 2004). One such protein, YbaK, was shown not to compete with EF-Tu for mischarged tRNA when free, but may compete more effectively when in complex with ProRS (An and Musier-Forsyth, 2005). The same may also be true for the trans-editing factor AlaX and AlaRS that compete with EF-1α to clear Ser-tRNAAla, accumulation of which can cause severe neurodegeneration (Lee et al., 2006). Like PheRS, AlaRS is a class II aaRS and is not likely to form a complex with EF-Tu and Ser-tRNAAla, indicating that Ser-tRNAAla must be released from AlaRS to be protected by EF-Tu. The notion that Ser-tRNAAla dissociates from wild-type AlaRS is also supported by studies of tRNA recognition during editing. The G3:U70 pair of tRNAAla is critical for both aminoacylation and posttransfer editing (Musier-Forsyth et al., 1991; Beebe et al., 2008) but is recognized by distinct domains during the two reactions (Beebe et al., 2008). The significant rearrangement of G3:U70 recognition during aminoacylation and editing correlates well with a trans-editing pathway, but not with cis editing alone.
Aminoacylation by class I aaRSs is suggested to be rate limited by product release for both editing (IleRS and ValRS) and nonediting (CysRS) enzymes (Eldred and Schimmel, 1972; Zhang et al., 2006). The dissociation rates of cognate aa-tRNAs from ValRS are estimated to be 3–4 s−1, comparable to the overall editing rate of ValRS (Nomanbhoy and Schimmel, 2000; Zhang et al., 2006). Structural modeling revealed that, in class I editing aaRSs, the CP1 domain sterically excludes EF-Tu from forming a complex with the aaRS and aa-tRNA (Zhang et al., 2006), consistent with the observation that bacterial EF-Tu does not increase aminoacylation or stimulate misacylation by LeuRS (Hausmann et al., 2007; Hausmann and Ibba, 2008). Such steric clashes may prevent EF-Tu from enhancing dissociation of misacylated aa-tRNAs, allowing editing to occur through the dominant cis pathway in class I aaRSs. All known autonomous trans-editing factors have evolved from class II aaRSs, consistent with the notion that class I enzymes do not release misacylated aa-tRNAs and so would not need to utilize trans editing for proofreading. These class-specific differences in proofreading may reflect early evolutionary constraints on accuracy that facilitated the introduction of particular amino acids into the genetic code.
EXPERIMENTAL PROCEDURES
General Methods
Protein and RNA preparation, aminoacylation, deacylation, determination of binding rates, and in vitro translation followed standard procedures as described in detail in the Supplemental Data.
2AP-tRNAPhe Synthesis and Aminoacylation
Blunt tRNAPhe sequence lacking the last four nucleotides at the 3′ end was cloned into pUC-18 vector, and E. coli XL1-Blue cells were transformed with this plasmid. The blunt tRNAPhe was in vitro transcribed and gel purified as described previously (Roy et al., 2004). A tetranucleotide oligo 5′p ACC-2APp 3′ where A76 on the tRNA 3′ end was replaced with 2-aminopurine was obtained commercially (TriLink BioTechnologies) and ligated to the blunt tRNAPhe to form an intact tRNAPhe using T4 RNA ligase (NEB Ltd). The ligated tRNA was phenol-chloroform extracted and gel filtered using sephadex G-25 (GEC). This tRNA was then dephosphorylated using antarctic phosphatase enzyme (NEB). A second phenol chloroform extraction was performed followed by gel filtration using sephadex G-50 (Pharmacia biotech). aa-tRNAPhe complexes were prepared as described previously (Ling et al., 2007a). In addition, gel filtration using sephadex G-25 was performed to remove any excess [3H]Tyr, [14C]Phe. The charging level and concentration of aa-tRNAPhe was measured by scintillation counting.
Supplementary Material
Acknowledgments
We thank Drs. B. Kraal (Leiden University, Leiden, the Netherlands), S. Martinis (University of Illinois, Urbana-Champaign), and D. Tirrell (California Institute of Technology, Pasadena, CA) for strains, plasmids, and enzymes. We thank C. Hausmann, N. Reynolds, and T. Rogers for critical reading of the manuscript; and D. Qin for materials. This work was supported by grants from the National Science Foundation (MCB-0344002 to M.I.) and the National Institutes of Health (GM49928 to K.M.-F. and GM72528 to K.F.), and by a predoctoral fellowship from the American Heart Association (to J.L.).
Footnotes
The Supplemental Data include Supplemental Experimental Procedures, two tables, and three figures and can be found with this article online at http://www.cell.com/molecular-cell/S1097-2765(09)00076-8.
References
- Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Musier-Forsyth K. Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J Biol Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- An S, Musier-Forsyth K. Cys-tRNAPro editing by Haemophilus influenzae YbaK via a novel synthetase. YbaK.tRNA ternary complex. J Biol Chem. 2005;280:34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- Asahara H, Uhlenbeck OC. The tRNA specificity of Thermus thermophilus EF-Tu. Proc Natl Acad Sci USA. 2002;99:3499–3504. doi: 10.1073/pnas.052028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AN, Berg P. Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J Biol Chem. 1966;241:839–845. [PubMed] [Google Scholar]
- Baltzinger M, Holler E. Kinetics of acyl transfer ribonucleic acid complexes of Escherichia coli phenylalanyl-tRNA synthetase. A conformational change is rate limiting in catalysis. Biochemistry. 1982;21:2460–2467. doi: 10.1021/bi00539a027. [DOI] [PubMed] [Google Scholar]
- Beebe K, Mock M, Merriman E, Schimmel P. Distinct domains of tRNA synthetase recognize the same base pair. Nature. 2008;451:90–93. doi: 10.1038/nature06454. [DOI] [PubMed] [Google Scholar]
- Beuning PJ, Musier-Forsyth K. Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc Natl Acad Sci USA. 2000;97:8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepin T, Yaremchuk A, Tukalo M, Cusack S. Structures of two bacterial prolyl-tRNA synthetases with and without a cis-editing domain. Structure. 2006;14:1511–1525. doi: 10.1016/j.str.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Cusack S, Yaremchuk A, Tukalo M. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dock-Bregeon A, Sankaranarayanan R, Romby P, Caillet J, Springer M, Rees B, Francklyn CS, Ehresmann C, Moras D. Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem. Cell. 2000;103:877–884. doi: 10.1016/s0092-8674(00)00191-4. [DOI] [PubMed] [Google Scholar]
- Döring V, Mootz HD, Nangle LA, Hendrickson TL, Crecy-Lagard V, Schimmel P, Marliere P. Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science. 2001;292:501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- Eldred EW, Schimmel PR. Investigation of the transfer of amino acid from a transfer ribonucleic acid synthetase-aminoacyl adenylate complex to transfer ribonucleic acid. Biochemistry. 1972;11:17–23. doi: 10.1021/bi00751a004. [DOI] [PubMed] [Google Scholar]
- Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev DG, Yokoyama S. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- Furano AV. Content of elongation factor Tu in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:4780–4784. doi: 10.1073/pnas.72.12.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgur Y, Mosyak L, Reshetnikova L, Ankilova V, Lavrik O, Khodyreva S, Safro M. The crystal structure of phenylalanyl-tRNA synthetase from thermus thermophilus complexed with cognate tRNAPhe. Structure. 1997;5:59–68. doi: 10.1016/s0969-2126(97)00166-4. [DOI] [PubMed] [Google Scholar]
- Guth EC, Francklyn CS. Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol Cell. 2007;25:531–542. doi: 10.1016/j.molcel.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann CD, Praetorius-Ibba M, Ibba M. An aminoacyl-tRNA synthetase:elongation factor complex for substrate channeling in archaeal translation. Nucleic Acids Res. 2007;35:6094–6102. doi: 10.1093/nar/gkm534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann CD, Ibba M. Structural and functional mapping of the archaeal multi-aminoacyl-tRNA synthetase complex. FEBS Lett. 2008;582:2178–2182. doi: 10.1016/j.febslet.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson TL, Schimmel P. In: Transfer RNA-dependent amino acid discrimination by aminoacyl-tRNA synthetases. Eurekah, Mechanisms T, Laponte J, Brakier-Gingras L, editors. Georgetown, TX: Landes Bioscience; 2003. pp. 34–64. [Google Scholar]
- Ibba M, Kast P, Hennecke H. Substrate specificity is determined by amino acid binding pocket size in Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry. 1994;33:7107–7112. doi: 10.1021/bi00189a013. [DOI] [PubMed] [Google Scholar]
- Ibba M, Johnson CM, Hennecke H, Fersht AR. Increased rates of tRNA charging through modification of the enzyme-aminoacyl-adenylate complex of phenylalanyl-tRNA synthetase. FEBS Lett. 1995;358:293–296. doi: 10.1016/0014-5793(94)01454-9. [DOI] [PubMed] [Google Scholar]
- Ibba M, Söll D. Quality control mechanisms during translation. Science. 1999;286:1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- Jakubowski H, Goldman E. Quantities of individual aminoacyl-tRNA families and their turnover in Escherichia coli. J Bacteriol. 1984;158:769–776. doi: 10.1128/jb.158.3.769-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korencic D, Ahel I, Schelert J, Sacher M, Ruan B, Stathopoulos C, Blum P, Ibba M, Söll D. A freestanding proofreading domain is required for protein synthesis quality control in Archaea. Proc Natl Acad Sci USA. 2004;101:10260–10265. doi: 10.1073/pnas.0403926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotik-Kogan O, Moor N, Tworowski D, Safro M. Structural basis for discrimination of L-phenylalanine from L-tyrosine by phenylalanyl-tRNA synthetase. Structure. 2005;13:1799–1807. doi: 10.1016/j.str.2005.08.013. [DOI] [PubMed] [Google Scholar]
- LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Lin SX, Baltzinger M, Remy P. Fast kinetic study of yeast phenylalanyl-tRNA synthetase: role of tRNAPhe in the discrimination between tyrosine and phenylalanine. Biochemistry. 1984;23:4109–4116. doi: 10.1021/bi00313a015. [DOI] [PubMed] [Google Scholar]
- Lincecum TL, Jr, Tukalo M, Yaremchuk A, Mursinna RS, Williams AM, Sproat BS, Van Den EW, Link A, Van Calenbergh S, Grotli M, et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- Ling J, Roy H, Ibba M. Mechanism of tRNA-dependent editing in translational quality control. Proc Natl Acad Sci USA. 2007a;104:72–77. doi: 10.1073/pnas.0606272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Yadavalli SS, Ibba M. Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. RNA. 2007b;13:1881–1886. doi: 10.1261/rna.684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AJ, Gilbey AM, Blewett AM, De Pascale G, El Zoeiby A, Levesque RC, Catherwood AC, Tomasz A, Bugg TD, Roper DI, Dowson CG. Characterization of tRNA-dependent peptide bond formation by MurM in the synthesis of Streptococcus pneumoniae peptidoglycan. J Biol Chem. 2008;283:6402–6417. doi: 10.1074/jbc.M708105200. [DOI] [PubMed] [Google Scholar]
- Loftfield RB, Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972;128:1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musier-Forsyth K, Usman N, Scaringe S, Doudna J, Green R, Schimmel P. Specificity for aminoacylation of an RNA helix: an unpaired, exocyclic amino group in the minor groove. Science. 1991;253:784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Pedersen S, Reeh S. Chemical measurement of steady-state levels of ten aminoacyl-transfer ribonucleic acid synthetases in Escherichia coli. J Bacteriol. 1977;129:378–387. doi: 10.1128/jb.129.1.378-387.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- Nomanbhoy TK, Schimmel PR. Misactivated amino acids translocate at similar rates across surface of a tRNA synthetase. Proc Natl Acad Sci USA. 2000;97:5119–5122. doi: 10.1073/pnas.090102197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nureki O, Vassylyev DG, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson TL, Schimmel P, Yokoyama S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Peterson ET, Uhlenbeck OC. Determination of recognition nucleotides for Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry. 1992;31:10380–10389. doi: 10.1021/bi00157a028. [DOI] [PubMed] [Google Scholar]
- Rosenberger RF, Foskett G. An estimate of the frequency of in vivo transcriptional errors at a nonsense codon in Escherichia coli. Mol Gen Genet. 1981;183:561–563. doi: 10.1007/BF00268784. [DOI] [PubMed] [Google Scholar]
- Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23:4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H, Ling J, Alfonzo J, Ibba M. Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 2005;280:38186–38192. doi: 10.1074/jbc.M508281200. [DOI] [PubMed] [Google Scholar]
- Roy H, Ibba M. Molecular biology: sticky end in protein synthesis. Nature. 2006;443:41–42. doi: 10.1038/nature05002. [DOI] [PubMed] [Google Scholar]
- Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci USA. 2008;105:4667–4672. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvian LF, Wang J, Steitz TA. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat Struct Mol Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- Villet R, Fonvielle M, Busca P, Chemama M, Maillard AP, Hugonnet JE, Dubost L, Marie A, Josseaume N, Mesnage S, et al. Idiosyncratic features in tRNAs participating in bacterial cell wall synthesis. Nucleic Acids Res. 2007;35:6870–6883. doi: 10.1093/nar/gkm778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Toh Y, Suto K, Shimizu Y, Oka N, Wada T, Tomita K. Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature. 2007;449:867–871. doi: 10.1038/nature06167. [DOI] [PubMed] [Google Scholar]
- Wong FC, Beuning PJ, Silvers C, Musier-Forsyth K. An isolated class II aminoacyl-tRNA synthetase insertion domain is functional in amino acid editing. J Biol Chem. 2003;278:52857–52864. doi: 10.1074/jbc.M309627200. [DOI] [PubMed] [Google Scholar]
- Yang XL, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Köhrer C, RajBhandary UL, Skene RJ, McRee DE, Schimmel P. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CM, Perona JJ, Ryu K, Francklyn C, Hou YM. Distinct kinetic mechanisms of the two classes of Aminoacyl-tRNA synthetases. J Mol Biol. 2006;361:300–311. doi: 10.1016/j.jmb.2006.06.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.