Abstract
With the notable exception of humans, uric acid is degraded to (S)-allantoin in a biochemical pathway catalyzed by urate oxidase, 5-hydroxyisourate (HIU) hydrolase, and 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase in most vertebrate species. A point mutation in the gene encoding mouse HIU hydrolase, Urah, that perturbed uric acid metabolism within the liver was discovered during a mutagenesis screen in mice. The predicted substitution of cysteine for tyrosine in a conserved helical region of the mutant-encoded HIU hydrolase resulted in undetectable protein expression. Mice homozygous for this mutation developed elevated platelet counts secondary to excess thrombopoietin production and hepatomegaly. The majority of homozygous mutant mice also developed hepatocellular carcinoma, and tumor development was accelerated by exposure to radiation. The development of hepatomegaly and liver tumors in mice lacking Urah suggests that uric acid metabolites may be toxic and that urate oxidase activity without HIU hydrolase function may affect liver growth and transformation. The absence of HIU hydrolase in humans predicts slowed metabolism of HIU after clinical administration of exogenous urate oxidase in conditions of uric acid-related pathology. The data suggest that prolonged urate oxidase therapy should be combined with careful assessment of toxicity associated with extrahepatic production of uric acid metabolites.
Keywords: N-ethyl-N-nitrosourea mutagenesis, uric acid, 5-hydroxyisourate hydrolase, hepatocellular carcinoma, thrombopoietin
Uric acid is the product of purine metabolism. In most mammals, uric acid, or urate, the form that predominates in vivo, is further metabolized to (S)-allantoin (1, 2). In humans and some higher primates, the urate oxidase gene is dysfunctional because of the introduction of premature stop codons during recent evolution (3). A nonfunctional urate oxidase enzyme causes a relative excess of uric acid in humans compared with most other vertebrate species. Because uric acid is relatively insoluble, humans are susceptible to diseases resulting from precipitation of uric acid such as gout and kidney stones as well as urate nephropathy associated with the tumor lysis syndrome. Chronic hyperuricemia in humans has also been linked to the development of cardiovascular disease and hypertension (4, 5).
It is unclear why humans have evolved this block in uric acid metabolism during evolution, but suggested theoretical advantages of increased levels of circulating uric acid include its role as a potent antioxidant offering protection from cancer and aging (6) and its potential function in maintenance of adequate blood pressure during periods of low salt intake (7).
The components of the full metabolic pathway that converts urate to allantoin in lower mammals have only recently been identified (1). Although it was originally thought that urate oxidase was the sole enzymatic component of this pathway, recent studies have established that conversion of uric acid to allantoin occurs by sequential chemical modifications catalyzed by urate oxidase, 5-hydroxyisourate (HIU) hydrolase, and 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) decarboxylase. Given the recent elucidation of this pathway, relatively little is known of the detailed actions of the enzymes that regulate the intermediates in the uric acid degradation pathway or the relative importance and extent of their roles in physiology.
Forward genetic screens in mice afford a powerful unbiased approach to identification of genes controlling diverse biological pathways. In this report, we describe a loss-of-function allele of the gene encoding the second enzyme in the uric acid degradation pathway, HIU hydrolase, termed UrahPlt2, which emerged from an N-ethyl-N-nitrosourea (ENU) mutagenesis screen in mice. Young homozygous UrahPlt2/Plt2 mice exhibited hepatomegaly and increased circulating platelet counts resulting from excess hepatic production of thrombopoietin (TPO), the major cytokine regulator of platelet number. By the age of 2 y, the majority of UrahPlt2/Plt2 mice spontaneously developed hepatocellular carcinoma, suggesting that in the presence of urate oxidase activity, HIU hydrolase is necessary to metabolize further the toxic intermediates that predispose to liver cancer.
Results
platelet 2, an ENU-Induced Mutation Causing Thrombocytosis and Hepatomegaly.
A forward genetic screen was performed by treating male C57BL/6 mice with ENU; after recovery of spermatogenesis, they were mated with isogenic C57BL/6 mice to produce first-generation (G1) progeny. G1 male and female mice were then interbred, and their offspring were brother-sister mated to produce families of G3 animals. In this third generation, a pedigree termed platelet 2 (plt2) was identified in which 6 of 16 G3 mice displayed thrombocytosis, consistent with segregation of a recessive mutation causing elevated platelet number. A plt2/plt2 homozygous pedigree was then established by breeding animals with the highest platelet counts.
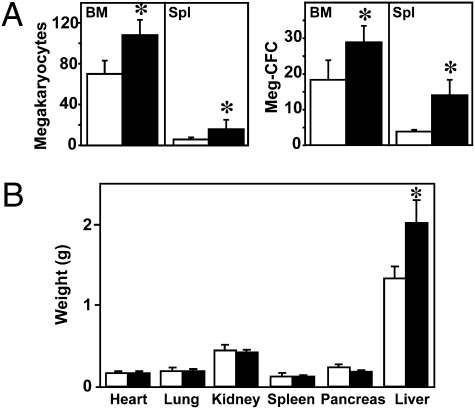
The plt2/plt2 mice appeared overtly normal and were fertile. The plt2/plt2 mice consistently had circulating platelet numbers 1.5-fold higher than WT controls, whereas the numbers of red and white blood cells were normal (Table S1). Excessive cellular production appeared to account for the thrombocytosis, because the numbers of megakaryocytes in the bone marrow and spleen, as well as in their progenitors, were also moderately elevated (Fig. 1A). The plt2/plt2 mice were similar in size to WT animals at 7–10 wk of age (body weight: plt2/plt2, 22 ± 2 g, n = 35; WT, 23 ± 3 g, n = 24; P = 0.2); however, plt2/plt2 mice invariably displayed significant enlargement of the liver without an accompanying increase in the size of any other organ (Fig. 1B). Liver weight, expressed as a proportion of total body weight, was 0.074 ± 0.005 for plt2/plt2 mice (n = 35) compared with 0.054 ± 0.004 for WT mice (n = 24; P < 0.001). Heterozygous plt2/+ animals were indistinguishable from the WT population, demonstrating the recessive nature of this phenotype.
Fig. 1.
Urahplt2/plt2 mice develop megakaryocytosis and hepatomegaly. (A) Numbers of megakaryocytes per 10 high-power microscope fields (hpf, 200×) and megakaryocyte progenitors per 2.5 × 104 bone marrow (BM) cells or 5 × 104 spleen (Spl) cells in WT (open bars) and Urahplt2/plt2 (filled bars) mice. Data are from six to seven mice of each genotype. (B) Absolute weight of internal organs from seven male Urahplt2/plt2 (filled bars) and seven male WT (open bars) animals. *P < 0.05 for pairwise comparison of data from Urahplt2/plt2 and WT mice.
Point Mutation in Urah Is Responsible for the plt2 Mutant Phenotype.
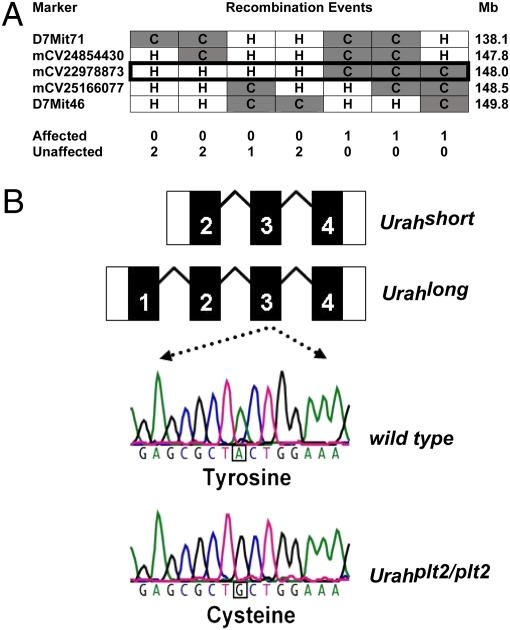
A positional cloning approach was used to identify the plt2 locus within the mouse genome. C57BL/6 plt2/plt2 mice with high platelet counts were mated with BALB/c mice, and the offspring were bred to C57BL/6 plt2/plt2 mice to produce a second generation backcross cohort (N2) or interbred to generate second generation intercross (F2) mice. A genome-wide scan was performed using 162 simple sequence length polymorphisms (SSLPs) in 89 N2 backcross mice. Linkage to platelet phenotype was then assessed using quantitative trait analysis in the statistical package R/qtl (8). Genetic linkage for thrombocytosis was observed on chromosome 7 at D7Mit189 with a peak logarithm of the odds (LOD) score of 20. Additional polymorphic markers were analyzed in a total of 634 N2 and F2 animals, refining the location of the plt2 locus to a 0.6-Mb interval (Fig. 2A). Fifteen genes were selected and sequenced within this interval, and an A-to-G point mutation was identified in plt2/plt2 mice in the Urah gene that was not present in WT samples (Fig. 2B). When mice from the mapping crosses were genotyped for the presence or absence of this mutation, thrombocytosis segregated with the mutant allele.
Fig. 2.
plt2 is a mutation in the Urah gene encoding mouse HIU hydrolase. (A) Informative mouse haplotypes were studied on chromosome 7 to localize the site of the plt2 mutation. Three mice with thrombocytosis (affected) were homozygous for a C57BL/6 (C) polymorphism (mCV22978873) at 148.0 Mb on chromosome 7. Mice with a normal platelet count (unaffected, n = 7) were heterozygous (H) for C57BL/6 and BALB/c at the same marker. (B) In plt2/plt2 mice, there is an A-to-G point mutation in exon 3 of the Urah transcript that predicts a tyrosine-to-cysteine substitution in the HIU hydrolase protein.
Urah is the mouse homolog of the bacterial UraH gene, which encodes an HIU hydrolase. HIU hydrolase is a member of the transthyretin-related protein (TRP) family, which displays high sequence conservation throughout evolution (9). Current evidence suggests that TRP family members act as HIU hydrolases in bacteria and that the product of the Urah gene performs this biochemical function in mice (1, 10, 11), catalyzing the second of three steps in the conversion of uric acid to allantoin (Fig. S1A). Expressed sequence tags covering this genomic region identify two alternative transcripts with different 5′-untranslated regions: a short three-exon transcript and a longer four-exon transcript with an additional 5′ exon. Thus, two potential HIU hydrolase isoforms could be translated, with the longer containing a putative peroxisomal targeting signal (1, 12). The mutation in plt2/plt2 mice occurs within the third exon of the long Urah transcript with a tyrosine-to-cysteine amino acid substitution at residue 98 of the predicted longer translation product (Fig. 2B).
Hepatocytes from Urahplt2/plt2 Mutant Mice Lack HIU Hydrolase.
Tyrosine-98 of HIU hydrolase is conserved from bacteria to mice (Fig. S1B), suggesting that this residue may be important for normal protein function. Crystal structures of HIU hydrolase from prokaryotes as well as zebra fish (13–15) predict that the tyrosine substituted in Urahplt2/plt2 mice (Y98) normally exists in a highly structured helical region and forms a strong set of hydrophobic interactions between the helix and adjacent β-sheet strands (Fig. S1C). Although not part of the predicted active site at the interface between the subunits of the HIU hydrolase tetramer (13–15), replacement of the aromatic tyrosine residue with a sulfur-containing cysteine in the UrahPlt2 protein is likely to disrupt this highly ordered hydrophobic core and lead to protein instability.
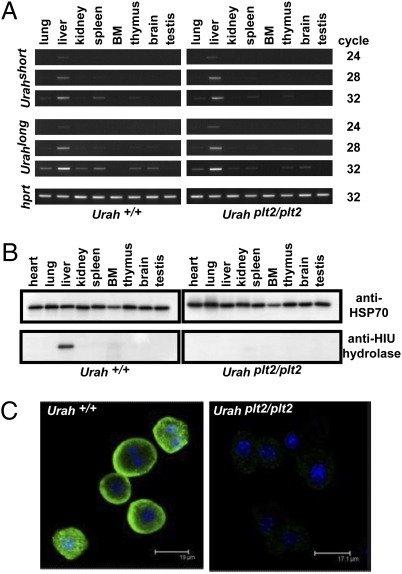
To examine the expression of HIU hydrolase, Urah mRNA was assayed by semiquantitative reverse transcriptase PCR. In both mutant and WT mice, the liver was the most abundant source of the short and long Urah transcripts. There was no difference in expression pattern between mutant and WT mice (Fig. 3A).
Fig. 3.
Urahplt2/plt2 hepatocytes are deficient in HIU hydrolase. (A) Reverse transcriptase PCR was performed on cDNA from a panel of tissues from WT and Urahplt2/plt2 mice. Urahshort and Urahlong transcripts were present in samples derived from liver at a relatively low PCR cycle number. (B) Polyclonal antisera raised against murine HIU hydrolase recognize a single 15-kDa band in WT liver that was absent in Urahplt2/plt2 liver. (C) Isolated hepatocytes were stained with HIU hydrolase antisera and then Alexa Fluor 488-conjugated anti-rabbit IgG (green), and cell nuclei were stained with DAPI (blue). Images captured by confocal microscopy demonstrate that murine HIU hydrolase is normally present in hepatocyte cytoplasm but is deficient in Urahplt2/plt2 hepatocytes.
A rabbit anti-mouse HIU hydrolase polyclonal antiserum was derived using a mixed peptide immunogen corresponding to the first and second exons of the long transcript. Specificity of the antisera was confirmed in 293T cells transiently transfected with vectors containing the short or long form of the murine Urah cDNA, including an N-terminal FLAG epitope tag: Western blotting revealed a band of 15 kDa that was detected by both anti-FLAG antibody and anti-HIU hydrolase antisera but was not detected in untransfected cells (Fig. S2). The organ distribution of endogenous HIU hydrolase in mice was then examined. In WT mice, HIU hydrolase was detected in the liver. No band was appreciable in lysates from any other organ examined or from protein derived from serum (Fig. 3B). In Urahplt2/plt2 mutant mice, there was no detectable HIU hydrolase protein in the liver or any other organ, suggesting that translation of the plt2 mutant form of Urah RNA may be aberrant or that the protein is unstable in these mutant mice. To confirm this finding and establish the specific cell type expressing HIU hydrolase, hepatocytes were isolated from WT and Urahplt2/plt2 mice and the presence of HIU hydrolase was determined by confocal microscopy (Fig. 3C). In WT hepatocytes, HIU hydrolase was present and located predominantly in the cytoplasm. In contrast, hepatocytes isolated from Urahplt2/plt2 mice demonstrated no fluorescent signal, indicating that the translated product of the mutant UrahPlt2 gene was markedly reduced or absent from these cells.
Uric acid metabolism does not occur in humans as a result of the evolution of nonsense mutations in the urate oxidase gene (3). Alignment of the mouse Urah nucleotide sequence with the human genome suggested similar loss of HIU hydrolase function in humans, with the presence of a 14-nt deletion in the homolog of exon 3 predicting premature termination of the HIU hydrolase peptide. Sequencing of cDNA from pooled human tissues confirmed the presence of this 14-nt deletion. Consistent with the absence of HIU hydrolase in humans, no protein was detected in Western blots of HEP-G2 human hepatoma cell line lysates (Fig. S2).
Transgenic Rescue of the UrahPlt2/Plt2 Phenotype.
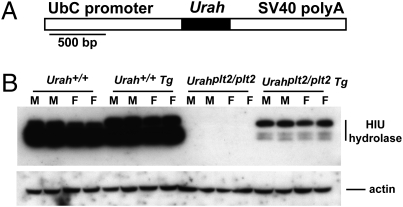
To confirm that the Y98C mutation was responsible for the phenotype in UrahPlt2/Plt2 mice, transgenic rescue was performed. Mice carrying a transgene in which the long form of Urah cDNA was linked to the ubiquitin C promoter (Fig. 4A) were bred onto the UrahPlt2/Plt2 background. Western blot analysis confirmed expression of the transgene. Although no HIU hydrolase was detectable in nontransgenic UrahPlt2/Plt2 livers, a prominent protein band of ∼16 kDa was detected in homozygous mutants carrying the transgene, consistent with the predicted size of the longer HIU hydrolase isoform (Fig. 4B). Less abundant and smaller HIU hydrolase species were also detected, which were presumably degradation products of the longer form. On a WT background, a smaller HIU hydrolase species was most abundant; however, increased amounts of the larger species were evident in longer exposures of Western blots from livers of mice carrying the transgene (Fig. 4B). Expression of the WT Urah transgene reverted the phenotype in UrahPlt2/Plt2 mice. Platelet numbers and liver weight in UrahPlt2/Plt2 transgenic mice were indistinguishable from those of WT mice, whereas thrombocytosis and hepatomegaly were observed as expected in a cohort of contemporaneously generated UrahPlt2/Plt2 transgene-negative littermates (Table 1). This observation provides strong confirmatory evidence that the phenotype in UrahPlt2/Plt2 mice is caused by the mutation in Urah.
Fig. 4.
Transgenic expression of WT HIU hydrolase in Urahplt2/plt2 mice. (A) Structure of the Urah transgene, with the ubiquitin C (UbC) promoter and SV40 polyadenylation (polyA) signal regulatory regions and Urah cDNA sequence (shaded) indicated. (B) Expression of WT transgenic (Tg) HIU hydrolase in liver extracts from individual male (M) and female (F) WT (+/+) or Urahplt2/plt2 mice.
Table 1.
Transgenic rescue of thrombocytosis and hepatomegaly in Urahplt2/plt2 mice
| Genotype | n | Body weight, g | Liver weight, g | Liver/body weight ratio | Platelets, 109/L |
| plt2/plt2 | 15 | 22.3 ± 2.8 | 1.47 ± 0.24 | 0.066 ± 0.007* | 1,913 ± 309* |
| plt2/plt2 Tg | 14 | 22.3 ± 3.5 | 1.15 ± 0.19 | 0.052 ± 0.004† | 1,201 ± 138† |
| plt2/+ | 11 | 21.9 ± 3.7 | 1.08 ± 0.23 | 0.049 ± 0.005 | 1,257 ± 176 |
| plt2/+ Tg | 10 | 21.6 ± 2.7 | 1.08 ± 0.18 | 0.050 ± 0.007 | 1,164 ± 278 |
| +/+ | 13 | 21.6 ± 3.7 | 1.08 ± 0.22 | 0.050 ± 0.007 | 1,113 ± 197 |
| +/+ Tg | 8 | 20.0 ± 3.2 | 0.98 ± 0.14 | 0.049 ± 0.003 | 1,109 ± 124 |
Tg, presence of the WT Urahlong transgene; n, number of mice analyzed.
*P < 10−4 for comparison of plt2/plt2 with +/+ mice.
†P < 10−4 for comparison of plt2/plt2 Tg with plt2/plt2 mice.
Thrombocytosis in Urahplt2/plt2 Mice Is Caused by Increased TPO Secretion from the Liver.
TPO is the principal hematopoietic cytokine responsible for platelet production. Because TPO is produced predominantly in the liver, the role of TPO in producing the thrombocytosis in Urahplt2/plt2 mice was investigated. Serum TPO was elevated by 64% in Urahplt2/plt2 mice (mean: 4,930 ± 1,309 pg/L, n = 18) compared with WT mice (mean: 3,010 ± 1,016 pg/L, n = 22; P < 0.001). When Urahplt2/plt2 mice were generated on a genetic background lacking the TPO receptor (Mpl−/−) (16), the platelet count was indistinguishable from that of Mpl−/− mice, consistent with the thrombocytosis in Urahplt2/plt2 mice being driven by TPO signaling.
Given the prominent hepatomegaly demonstrated in Urahplt2/plt2 mice, we hypothesized that increased production of TPO protein by the liver was responsible for the increased serum TPO. In liver lysates, TPO protein per gram of liver was increased in Urahplt2/plt2 mice by 51% (259 ± 87 absorbance units per gram of liver weight, n = 8) compared with WT animals (173 ± 56 absorbance units per gram of liver weight, n = 8; P = 0.03), although no general increase in protein content per gram of liver weight was observed. TPO production was then examined on a per-cell basis by measurement of TPO secretion by a defined number of primary hepatocytes in culture. Urahplt2/plt2 hepatocytes secreted more TPO (1.15 ± 0.19 absorbance units per 106 hepatocytes, n = 4) than hepatocytes isolated from WT mice (0.63 ± 0.12 absorbance units per 106 hepatocytes, n = 4; P < 0.01).
Urah Is a Liver-Specific Tumor Suppressor Gene.
Urahplt2/plt2 mice invariably displayed liver enlargement from an early age. No histological basis for the enlarged liver was found in mice at 7–10 wk of age. The number of hepatocyte nuclei was the same between Urahplt2/plt2 (18 ± 2 per high-power field, n = 5) and Urah+/+ mice (20 ± 2 per high-power field, n = 5), suggesting that increased liver size was the result of excessive cellular production rather than increased cell size. There was no cellular infiltrate or abnormal frequency of mitoses, and the hepatic architecture was normal. Serum biochemical markers of liver function were also indistinguishable in young Urahplt2/plt2 mice compared with controls (Table S1).
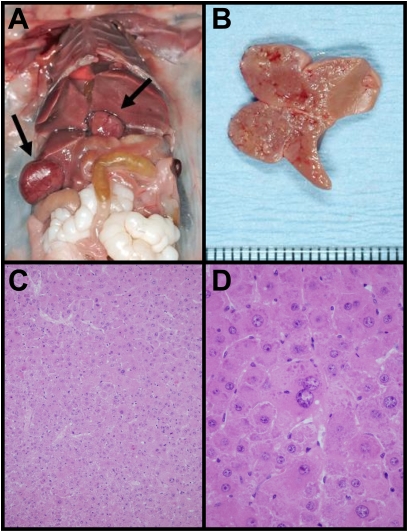
The natural history of the altered liver growth was examined by observing a cohort of mice for up to 2 y. The majority of Urahplt2/plt2 mice developed abdominal distention with aging, and 54% of animals (n = 37) were found to have hepatic tumors at autopsy (Fig. 5 A and B). In contrast, no case of hepatic tumor was identified in a parallel cohort of WT mice by 600 d of age (n = 18). The incidence of hepatic tumors was similar between male and female Urahplt2/plt2 mice. The histology of the tumors indicated that they were composed of hepatocytes but showed an absence of portal tracts, suggesting a neoplastic proliferation. In addition to moderate cytological atypia, the normal slender trabecular architecture of the liver was replaced by thicker trabeculae with some nested and pseudoglandular arrangements of cells (Fig. 5 C and D). There was neovascularization of the sinusoids and reticulin depletion. The combined cytological and architectural features were most consistent with hepatocellular carcinoma (17). This neoplastic disease was typically multifocal within the liver, and there was no evidence of metastasis outside that organ. Tumor predisposition in Urahplt2/plt2 mice was liver-specific, because occult malignancies (predominantly sarcoma and lymphoid leukemia) were identified at a similar frequency in nonhepatic organs within WT and Urahplt2/plt2 aging cohorts.
Fig. 5.
Urahplt2/plt2 mice develop multifocal hepatic tumors with aging. (A) Mouse abdominal cavity demonstrating multifocal hepatic tumor formation (arrows). (B) Hepatic tumor extending from the cut surface of the liver. (C) Representative histology of a liver tumor that consists of hepatocytes only without portal tracts. The tumor architecture demonstrates some slender trabeculae and other hepatocytes forming nests of up to eight cells (H&E stain, magnification 400×). (D) High-power photomicrograph of hepatocellular carcinoma that displays nuclear atypia which is easily discernible (H&E, magnification 600×).
High-dose total body radiation was administered in combination with bone marrow reinfusion to rescue the hematopoietic system. Hepatocellular carcinoma was identified in 89% of Urahplt2/plt2 mice (n = 9) by 14 mo after radiation exposure compared with 1 of 9 WT mice (11%). Multifocal neoplastic disease was present in the irradiated Urahplt2/plt2 mice, whereas there was only one focus of tumor in the affected WT mouse. The histopathology of liver tumors after radiation was the same as that observed with aging, indicating that high-dose radiation appeared to accelerate the malignant transformation of hepatocytes in animals lacking HIU hydrolase.
Discussion
Members of the TRP family of proteins (9) are considered to function as HIU hydrolases (1), converting HIU to OHCU as the second step in the pathway converting uric acid to allantoin. Urah, the gene encoding the mouse TRP family member HIU hydrolase, encodes two alternative transcripts that are both predominantly expressed in the liver. Experimental evidence suggests that the HIU hydrolase catalytic function is contained within exons 2–4 of this murine gene (1, 11). The long Urah transcript contains an additional exon that is thought to contain a type II peroxisome targeting signal based on a consensus sequence pattern (1, 12). This targeting signal predicts colocalization of HIU hydrolase with urate oxidase, because this first enzyme in the uric acid degradation pathway is known to be located in the peroxisomes of the hepatic parenchymal cells (18, 19). Our protein expression studies confirm the localization of HIU hydrolase in the cytoplasm of mouse hepatocytes.
The plt2/plt2 mutant mouse contains a point mutation in Urah rendering hepatocytes deficient in HIU hydrolase. Absent or markedly reduced HIU hydrolase function at this normal site of uric acid metabolism produced the unexpected hepatic phenotype of abnormal growth, propensity to tumor formation, and abnormal hepatic metabolism. Anomalous hepatic metabolism in Urahplt2/plt2 mutant mice was demonstrated with the increased secretion of the liver-derived protein TPO. Current understanding of the regulation of platelet number via TPO focuses on clearance of the cytokine by the platelet and megakaryocyte mass (20). This mutant mouse model suggests that intrahepatic factors may also be significant. An increase in TPO per gram of liver weight and increased TPO secretion by individual hepatocytes in culture from Urahplt2/plt2 animals suggest that the hepatocytes present in these mutant mice produce excess amounts of this hematopoietic cytokine. Thus, under some circumstances, regulation of hepatic production may also be a mechanism for controlling TPO levels. Thrombocytosis in humans is observed in hospitalized patients with infection, cancer, and chronic inflammation (21, 22), and this is frequently associated with increased circulating TPO (23). The liver mediates many of the protein changes recognized during the acute-phase response, and pathways present in the liver that produce reactive changes in TPO may provide a mechanistic explanation for these clinical observations.
Spontaneous liver tumor formation occurred in more than half of the Urahplt2/plt2 mutant mice after 600 d and increased to 89% of mutant animals after irradiation. Tumors were typically multifocal in Urahplt2/plt2 mice, indicating a field effect with the entire dysfunctional liver being prone to tumor development. Alteration in uric acid metabolism has not previously been demonstrated to promote neoplastic growth in the liver, and biochemical studies of this pathway have only recently appreciated the complexity of this breakdown process (1, 2). The product of urate oxidase, HIU, is unstable and undergoes spontaneous degradation to a racemic mix of allantoin (1). This degradation process is accelerated by the presence of HIU hydrolase and OHCU decarboxylase, and only (S)-allantoin is produced in the presence of these additional enzymes. The evolution of enzymes that accelerate this degradation suggests that these uric acid degradation intermediates may be toxic, a notion supported by current bioinformatic data. The uric acid degradation pathway appears to have been lost or gained independently several times throughout evolution. When urate oxidase is functionally absent in an organism, the other members of the biochemical pathway (HIU hydrolase and OHCU decarboxylase) are also usually absent or mutated. Conversely, when urate oxidase is functional, HIU hydrolase and OHCU decarboxylase are also present. This correlation between the combined presence or absence of all three enzymes throughout different species occurs despite in vitro evidence of slow spontaneous degradation of HIU to allantoin (1). Presumably, this slow degradation is not preferred, because the evolution of vertebrate species with urate oxidase function only has not occurred. These observations suggest that HIU hydrolase and perhaps OHCU decarboxylase limit the presence of biologically active chemical intermediates that appear to be growth-promoting and oncogenic in the mouse. The specific identity and biological function of these chemical intermediates remain to be determined.
Humans have relatively high levels of uric acid because of the absence of the urate degradation pathway. This may present advantages, because urate is considered to be an antioxidant; however, it also predisposes humans to diseases associated with uric acid excess. The health problems related to hyperuricemia have been approached therapeutically by suppressing production of uric acid using the xanthine oxidase inhibitor allopurinol or by introducing recombinant urate oxidase to promote uric acid breakdown. The data presented here suggest that the second therapeutical approach may produce unexpected consequences. Administration of recombinant urate oxidase produces HIU in an extrahepatic site without the addition of downstream enzymes (HIU hydrolase and OHCU decarboxylase) that promote the rapid degradation of the uric acid breakdown products. Within the liver of the mouse, where these enzymes normally reside, this appears to promote dysregulated cell growth and tumor formation. At present, most of the clinical data concerning recombinant urate oxidase administration in humans pertain to the prevention of urate nephropathy associated with the tumor lysis syndrome. In this setting, administration of the recombinant protein is limited to a few days and excessive unexpected clinical outcomes have not been reported (24, 25). During cytotoxic treatment of patients with high-grade malignancy at risk for tumor lysis syndrome, urate oxidase is more effective at reducing uric acid levels than allopurinol (25) and the theoretical disadvantages associated with HIU metabolism may be relatively unimportant. Nevertheless, adverse events are common in clinical trials studying patient populations with high-grade malignancy, and long-term safety of urate oxidase should be monitored.
Urate oxidase therapy is also being explored in tophaceous gout (23, 26) and in patients with gout refractory to standard therapies (27, 28). It has also been administered to a small number of patients after stroke (29) and in an individual with the Lesch–Nyhan syndrome (30). In patients with gout, recombinant urate oxidase (26) and chemically modified preparations of recombinant urate oxidase with a prolonged half-life (27, 31) have been used for up to 6 mo in clinical trials. These patients demonstrate prolonged biochemical evidence of uric acid breakdown, and some patients appeared to derive clinical benefit from this therapy (23). Animal model data presented here relate urate oxidase activity without HIU hydrolase function to abnormal growth and tumor development; thus, prolonged exposure to urate oxidase therapy in patients with nonmalignant medical conditions should be closely monitored with surveillance for adverse outcomes.
Materials and Methods
Mice.
Male C57BL/6 mice were treated with two doses of ENU (70 mg/kg of weight) administered i.p. (32) and were mated with isogenic female mice to produce G1 progeny. After further breeding, individuals in a G3 pedigree (called plt2) were recognized to have thrombocytosis. Most experiments were performed on mice genotyped for the presence of the Urahplt2 point mutation; for serum biochemistry and organ weights, the plt2/plt2 cohorts were chosen based on distinctly high platelet counts. Peripheral blood was collected from the retroorbital plexus, and the platelet count was determined using an Advia120 automated hematological analyzer (Bayer). Mouse serum was analyzed for biochemical markers of liver function using an Olympus AU400 Autoanalyser. The plt2 mutation was established on a Mpl−/− background by mating plt2/plt2 male mice with Mpl−/− female mice on a C57BL/6 background (16) to produce offspring that were obligate heterozygotes for the plt2 mutation and the Mpl null allele. These mice were then intercrossed.
All experiments performed on animals were approved by animal ethics committees of the Melbourne Health Research Directorate or The Walter and Eliza Hall Institute.
Genetic Mapping.
The plt2/plt2 male mice on a C57BL/6 background were mated with WT BALB/c female mice to produce F1 mice that, in turn, were either mated with plt2/plt2 male mice to produce N2 backcross mice or brother-sister mated to produce an F2 cohort. A genome-wide scan was performed on DNA prepared from liver using 162 SSLPs spaced evenly throughout the genome with markers derived from the Mouse Genetic Mapping Project of the Broad Institute (www.broad.mit.edu) (33). SSLPs were amplified by PCR using fluorescent dye-labeled oligonucleotides, and the sequence length of the PCR product was determined on an ABI 3700 DNA sequence analyzer. Once the position of the plt2 allele was established, animals with informative recombinations were examined for the presence of SNPs identified in DNA sequence databases. For sequencing, genomic DNA was amplified by PCR, treated with ExoSAP-IT (USB Corporation), and sequenced with a Big Dye Terminator 3.1 (Applied Biosystems).
Cloning and Expression of Murine Urah.
RNA was isolated from organs after they were snap-frozen in liquid nitrogen and then homogenized in TRIzol reagent (Invitrogen). First-strand cDNA synthesis was performed using SuperScript II Reverse Transcriptase (Invitrogen). Full-length Urah was amplified with PCR using primers specific for the predicted short and long transcripts of Urah (short: 5′-ACGGACTGGCTGATCACTCT-3′, 5′-CAAAGCCCATGATTTGTGTG-3′; long: 5′-TGCACAGACCAGAGCTTCAG-3′, 5′-CAGGCAGATAGATGGCTTTCTT-3′) and was cloned into the pEF-BOS plasmid (34), which was modified to encode a FLAG epitope. NIH 3T3 cells were transiently transfected with 1 μg of pEF-BOS-FLAG-Urah with FuGENE reagent (Roche). For tissue expression studies, the HPRT gene was amplified in parallel using 5′-TCCCTGGTTAAGCAGTAC-3′ and 5′-GATGGCCACAGGACTAGAACA-3′ as primers.
Production of HIU Hydrolase Antiserum and Protein Expression Analysis.
A rabbit was injected with 100 μg of peptide (MSSRTAPRLMTLQC and TTHVLDTASGLPAC) conjugated to keyhole limpet hemocyanin (KLH) in complete Freund's adjuvant. At 6-wk intervals, two additional injections were given containing 100 μg of peptide-KLH conjugate in incomplete Freund's adjuvant, followed by a last injection of peptide-KLH conjugate in normal saline before serum collection. Protein was isolated from mouse organs that were snap-frozen and then mechanically disrupted in a Dounce homogenizer (Wheaton Science Products) in KALB lysis buffer [150 mM NaCl, 50 mM Tris (pH 7.5), 1% Triton X-100, 1 mM EDTA] with protease inhibitors (Complete mixture tablets; Roche) on ice. Total protein content of lysates was calculated using the BCA protein assay kit (Pierce). For Western blotting, proteins were resolved by SDS/PAGE and transferred onto PVDF-plus filters (GE Waters and Process Technologies) and blotted using HIU hydrolase antiserum or anti-HSP-70 (1:1,000; Santa Cruz Biotechnology) and then sheep anti-rabbit IgG (1:5,000; Chemicon) or sheep anti-mouse IgG (1:5,000; Chemicon) as the secondary antibody. For confocal microscopy, purified hepatocytes were fixed with 4% (wt/vol) paraformaldehyde and permeabilized with methanol, and HIU hydrolase antiserum (1:500) was added. A secondary incubation was performed with Alexa Fluor 488-conjugated anti-rabbit IgG (Molecular Probes), and cells were then counterstained with DAPI (0.5 μg/mL). Images were captured with a Leica TCS4 SP2 spectral confocal scanner.
Statistical Analysis.
All data are presented as the mean ± 1 SD, and all experimental animals were on a C57BL/6 genetic background. The statistical significance of differences observed between Urahplt2/plt2 and WT mice was assessed using two-sided Student's t tests.
Supplementary Material
Acknowledgments
We thank Jason Corbin and Janelle Lochland for technical assistance and David De Sousa and Malcolm McConville for helpful discussions and input. This work was supported by Program Grant 461219, fellowships (to D.J.H., W.S.A., N.A.N., A.W.R., and M.B.), a scholarship (to W.S.S.), and Independent Research Institutes Support Scheme Grant 361646 from the Australian National Health and Medical Research Council of Australia; a Fellowship from the Cancer Council, Victoria (to D.M.); the Australian Cancer Research Fund; a Victorian State Government Operational Infrastructure Support grant; and MuriGen Pty. Ltd.
Footnotes
Conflict of interest statement: N.A.N., D.J.H., and W.S.A. hold shares in MuriGen Pty. Ltd., which funded part of this work.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010390107/-/DCSupplemental.
References
- 1.Ramazzina I, Folli C, Secchi A, Berni R, Percudani R. Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat Chem Biol. 2006;2:144–148. doi: 10.1038/nchembio768. [DOI] [PubMed] [Google Scholar]
- 2.Tipton PA. Urate to allantoin, specifically (S)-allantoin. Nat Chem Biol. 2006;2:124–125. doi: 10.1038/nchembio0306-124. [DOI] [PubMed] [Google Scholar]
- 3.Wu XW, Lee CC, Muzny DM, Caskey CT. Urate oxidase: Primary structure and evolutionary implications. Proc Natl Acad Sci USA. 1989;86:9412–9416. doi: 10.1073/pnas.86.23.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alderman MH, Cohen H, Madhavan S, Kivlighn S. Serum uric acid and cardiovascular events in successfully treated hypertensive patients. Hypertension. 1999;34:144–150. doi: 10.1161/01.hyp.34.1.144. [DOI] [PubMed] [Google Scholar]
- 5.Verdecchia P, et al. Relation between serum uric acid and risk of cardiovascular disease in essential hypertension. The PIUMA study. Hypertension. 2000;36:1072–1078. doi: 10.1161/01.hyp.36.6.1072. [DOI] [PubMed] [Google Scholar]
- 6.Ames BN, Cathcart R, Schwiers E, Hochstein P. Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: A hypothesis. Proc Natl Acad Sci USA. 1981;78:6858–6862. doi: 10.1073/pnas.78.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe S, et al. Uric acid, hominoid evolution, and the pathogenesis of salt-sensitivity. Hypertension. 2002;40:355–360. doi: 10.1161/01.hyp.0000028589.66335.aa. [DOI] [PubMed] [Google Scholar]
- 8.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 9.Eneqvist T, Lundberg E, Nilsson L, Abagyan R, Sauer-Eriksson AE. The transthyretin-related protein family. Eur J Biochem. 2003;270:518–532. doi: 10.1046/j.1432-1033.2003.03408.x. [DOI] [PubMed] [Google Scholar]
- 10.Hennebry SC. Evolutionary changes to transthyretin: Structure and function of a transthyretin-like ancestral protein. FEBS J. 2009;276:5367–5379. doi: 10.1111/j.1742-4658.2009.07246.x. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y, et al. Mouse transthyretin-related protein is a hydrolase which degrades 5-hydroxyisourate, the end product of the uricase reaction. Mol Cells. 2006;22:141–145. [PubMed] [Google Scholar]
- 12.Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hennebry SC, Law RH, Richardson SJ, Buckle AM, Whisstock JC. The crystal structure of the transthyretin-like protein from Salmonella dublin, a prokaryote 5-hydroxyisourate hydrolase. J Mol Biol. 2006;359:1389–1399. doi: 10.1016/j.jmb.2006.04.057. [DOI] [PubMed] [Google Scholar]
- 14.Jung DK, et al. Structural and functional analysis of PucM, a hydrolase in the ureide pathway and a member of the transthyretin-related protein family. Proc Natl Acad Sci USA. 2006;103:9790–9795. doi: 10.1073/pnas.0600523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanotti G, et al. Structure of zebra fish HIUase: Insights into evolution of an enzyme to a hormone transporter. J Mol Biol. 2006;363:1–9. doi: 10.1016/j.jmb.2006.07.079. [DOI] [PubMed] [Google Scholar]
- 16.Alexander WS, Roberts AW, Nicola NA, Li R, Metcalf D. Deficiencies in progenitor cells of multiple hematopoietic lineages and defective megakaryocytopoiesis in mice lacking the thrombopoietic receptor c-Mpl. Blood. 1996;87:2162–2170. [PubMed] [Google Scholar]
- 17.Hirohashi S, Ishak K, Kojiro M. Hepatocellular carcinoma. In: Hamilton S, Aaltonen L, editors. Pathology and Genetics of Tumours of the Digestive System. Lyon, France: International Agency for Research on Cancer; 2000. pp. 159–172. [Google Scholar]
- 18.Reddy PG, et al. Isolation and sequence determination of a cDNA clone for rat peroxisomal urate oxidase: Liver-specific expression in the rat. Proc Natl Acad Sci USA. 1988;85:9081–9085. doi: 10.1073/pnas.85.23.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Usuda N, Reddy MK, Hashimoto T, Rao MS, Reddy JK. Tissue specificity and species differences in the distribution of urate oxidase in peroxisomes. Lab Invest. 1988;58:100–111. [PubMed] [Google Scholar]
- 20.Kuter DJ. The physiology of platelet production. Stem Cells. 1996;14(Suppl 1):88–101. doi: 10.1002/stem.5530140711. [DOI] [PubMed] [Google Scholar]
- 21.Griesshammer M, et al. Aetiology and clinical significance of thrombocytosis: Analysis of 732 patients with an elevated platelet count. J Intern Med. 1999;245:295–300. doi: 10.1046/j.1365-2796.1999.00452.x. [DOI] [PubMed] [Google Scholar]
- 22.Schafer AI. Thrombocytosis. N Engl J Med. 2004;350:1211–1219. doi: 10.1056/NEJMra035363. [DOI] [PubMed] [Google Scholar]
- 23.Baraf HS, Matsumoto AK, Maroli AN, Waltrip RW., II Resolution of gouty tophi after twelve weeks of pegloticase treatment. Arthritis Rheum. 2008;58:3632–3634. doi: 10.1002/art.23993. [DOI] [PubMed] [Google Scholar]
- 24.Coiffier B, et al. Groupe d'Etude des Lymphomes de l'Adulte Trial on Rasburicase Activity in Adult Lymphoma. Efficacy and safety of rasburicase (recombinant urate oxidase) for the prevention and treatment of hyperuricemia during induction chemotherapy of aggressive non-Hodgkin's lymphoma: Results of the GRAAL1 (Groupe d'Etude des Lymphomes de l'Adulte Trial on Rasburicase Activity in Adult Lymphoma) study. J Clin Oncol. 2003;21:4402–4406. doi: 10.1200/JCO.2003.04.115. [DOI] [PubMed] [Google Scholar]
- 25.Goldman SC, et al. A randomized comparison between rasburicase and allopurinol in children with lymphoma or leukemia at high risk for tumor lysis. Blood. 2001;97:2998–3003. doi: 10.1182/blood.v97.10.2998. [DOI] [PubMed] [Google Scholar]
- 26.Richette P, Brière C, Hoenen-Clavert V, Loeuille D, Bardin T. Rasburicase for tophaceous gout not treatable with allopurinol: An exploratory study. J Rheumatol. 2007;34:2093–2098. [PubMed] [Google Scholar]
- 27.Sundy JS, et al. Pegloticase Phase 2 Study Investigators. Reduction of plasma urate levels following treatment with multiple doses of pegloticase (polyethylene glycol-conjugated uricase) in patients with treatment-failure gout: Results of a phase II randomized study. Arthritis Rheum. 2008;58:2882–2891. doi: 10.1002/art.23810. [DOI] [PubMed] [Google Scholar]
- 28.Sundy JS, et al. Pharmacokinetics and pharmacodynamics of intravenous PEGylated recombinant mammalian urate oxidase in patients with refractory gout. Arthritis Rheum. 2007;56:1021–1028. doi: 10.1002/art.22403. [DOI] [PubMed] [Google Scholar]
- 29.Amaro S, et al. A pilot study of dual treatment with recombinant tissue plasminogen activator and uric acid in acute ischemic stroke. Stroke. 2007;38:2173–2175. doi: 10.1161/STROKEAHA.106.480699. [DOI] [PubMed] [Google Scholar]
- 30.Roche A, et al. Efficacy of rasburicase in hyperuricemia secondary to Lesch-Nyhan syndrome. Am J Kidney Dis. 2009;53:677–680. doi: 10.1053/j.ajkd.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Yue CS, et al. Population pharmacokinetic and pharmacodynamic analysis of pegloticase in subjects with hyperuricemia and treatment-failure gout. J Clin Pharmacol. 2008;48:708–718. doi: 10.1177/0091270008317589. [DOI] [PubMed] [Google Scholar]
- 32.Bode VC. Ethylnitrosourea mutagenesis and the isolation of mutant alleles for specific genes located in the T region of mouse chromosome 17. Genetics. 1984;108:457–470. doi: 10.1093/genetics/108.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dietrich W, et al. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.