Abstract
Cyclic diguanylate (c-di-GMP) is a ubiquitous second messenger regulating diverse cellular functions including motility, biofilm formation, cell cycle progression and virulence in bacteria. In the cell, degradation of c-di-GMP is catalyzed by highly specific EAL domain phosphodiesterases whose catalytic mechanism is still unclear. Here, we purified 13 EAL domain proteins from various organisms and demonstrated that their catalytic activity is associated with the presence of 10 conserved EAL domain residues. The crystal structure of the TDB1265 EAL domain was determined in a free state (1.8 Å) and in complex with c-di-GMP (2.35 Å) and unveiled the role of the conserved residues in substrate binding and catalysis. The structure revealed the presence of two metal ions directly coordinated by six conserved residues, two oxygens of the c-di-GMP phosphate, and potential catalytic water molecule. Our results support a two-metal-ion catalytic mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
Keywords: EAL domain, cyclic di-GMP, phosphodiesterase, X-ray crystallography, Thiobacillus denitrificans
INTRODUCTION
Cyclic diguanylate (or bis-(3`–5`) cyclic dimeric guanosine monophosphate, c-di-GMP) is a global second messenger which controls a range of different cellular functions in bacteria on transcriptional, translational, and post-translational levels 1; 2. It was discovered over 20 years ago as an allosteric regulator of cellulose synthesis in Gluconacetobacter xylinus 3. Presently, it has been established that c-di-GMP binds to various receptor proteins or riboswitches and regulates biofilm formation, motility, virulence, antibiotic production, progression through the cell cycle and other cellular functions in a wide variety of organisms 4; 5; 6; 7; 8; 9.
The intracellular level of c-di-GMP is controlled by the opposing action of two different groups of enzymes. The diguanylate cyclases containing the GGDEF domain produce c-di-GMP from two molecules of GTP, whereas specific phosphodiesterases (PDEs) associated with EAL or HD-GYP domains break c-di-GMP to 5`-phosphoguanylyl-(3`–5`)-guanosine, which is subsequently hydrolyzed into two GMP molecules 10; 11; 12; 13; 14; 15. Most organisms have multiple proteins with these domains, which can exist as stand-alone proteins or are fused to various signalling or signal output domains (PAS, GAF, MASE, CHASE, REC, HTH) 9; 10. For example, Escherichia coli encodes 29 proteins with 19 GGDEF and/or 17 EAL domains, whereas Shewanella oneidensis has 57 such proteins 6; 16. The remarkable multiplicity of GGDEF and EAL domains in bacterial genomes and their association with numerous proteins suggest that many different signals can be integrated into the cellular c-di-GMP pool and diverse processes can be targeted and regulated in parallel. Numerous proteins contain both GGDEF and EAL domains, as well as different sensory domains, indicating that complex signal integration and domain interaction may occur in single proteins. There are three classes of these enzymes: GGDEF-domain proteins, EAL-domain proteins and proteins containing both domains in tandem. Presently, it is not clear how the opposing activities of the fused EAL and GGDEF domains are regulated or coordinated. A hypothesis has been proposed that in proteins containing both EAL and GGDEF domains, one of these domains might be catalytically inactive 14. This model was supported by work on BifA protein (PA4367) from Pseudomonas aeruginosa 17. It is also corroborated by the observation that many organisms possess GGDEF and EAL domain proteins with degenerate catalytic motifs and for some of them the absence of catalytic activity was experimentally demonstrated 1; 9; 18; 19. Interestingly, some degenerate GGDEF and EAL domains retain the ability to bind c-di-GMP and function as c-di-GMP receptors, regulating the associated protein domains and biological processes 20; 21. Therefore, it has been suggested that degenerate GGDEF and EAL domains function through c-di-GMP binding and/or macromolecular interactions with partner proteins 9.
Recently, PDE activity of EAL domains (EC 3.1.4.52) has been demonstrated in vitro, and several proteins have been biochemically investigated including the E. coli YahA and Dos, Pseudomonas aeruginosa RocR, Vibrio cholerae VieA and Caulobacter crescentus CC3396 13; 14; 22; 23. These enzymes were found to be strictly specific to c-di-GMP and required the addition of Mg2+ or Mn2+ for activity, whereas Ca2+, Ni2+, Fe2+ and Zn2+ did not support the PDE activity 14; 22; 24. The first two crystal structures of the EAL domain were determined for the apo-forms of the Bacillus subtilis YkuI in 2005 (PDB code 2bas; 25) and Thiobacillus denitrificans TBD1265 in 2007 (PDB code 2r6o; our work). Both EAL domains have a triose-phosphate isomerase (TIM) barrel-like fold implying that, as in other TIM barrel enzymes, the EAL domain active site is located at the C-terminal part of the β-barrel. The TBD1265 structure (PDB code 2r6o) revealed the presence of one or two Mg2+ ions in protein monomers suggesting that the one- or two-metal cation mechanisms might be employed by the EAL domain phosphodiesterases 23. However, the mutational analysis of the conserved residues of the P. aeruginosa RocR suggested that EAL domains are more likely to use a one-metal cation mechanism 23. Recently, the structures of YkuI, Pseudomonas aeruginosa FimX, and Klebsiella pneumoniae BlrP1 were determined in complex with c-di-GMP and presented the real view of the active site of EAL domain PDEs 21; 24; 25. However, FimX has a degenerate (inactive) EAL domain and functions as a c-di-GMP sensor, whereas YkuI was found to be catalytically inactive in vitro leaving the catalytic mechanism of the EAL domain uncertain 21; 25. The only structural insight into the catalytic mechanism of the EAL domain was provided by the crystal structure of the BlrP1/c-di-GMP complex 24. The BlrP1 structure revealed the presence of two bound metal ions (Mn2+) coordinated by several conserved carboxylates and one asparagine of the EAL domain. In contrast to earlier work 23, this study proposed a catalytic mechanism for EAL domains based on two metal cations coordinating a nucleophilic water molecule 24.
In this work, we present the results of the structural, biochemical, and mutational analysis of PDE activity of the EAL domain of TBD1265 from T. denitrificans. We determined the crystal structure of the TBD1265 EAL domain in the apo-form (2r6o) and in complex with c-di-GMP and two Mg2+ ions (3n3t). Alanine replacement mutagenesis of the TBD1265 EAL domain identified eight residues absolutely required for activity, whereas sequence-activity analysis of 13 purified EAL domains from various organisms pointed to an important role of these conserved residues in the activity of EAL domains. We propose that EAL domain PDEs use a two-metal-ion catalytic mechanism for c-di-GMP hydrolysis.
RESULTS and DISCUSSION
Phosphodiesterase activity of purified EAL domains
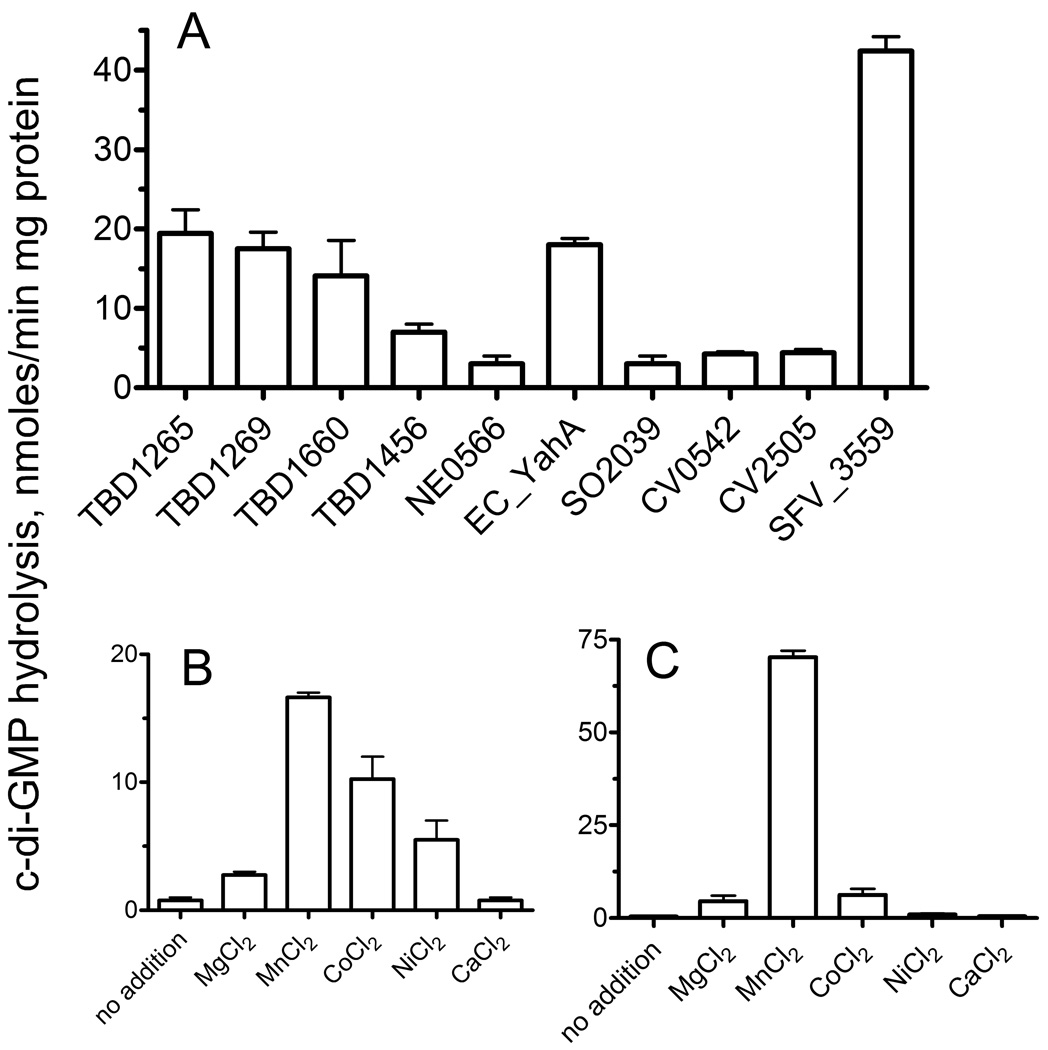
To characterize the relationship between the sequence and enzymatic activity of EAL domain proteins, we recombinantly expressed and purified 13 EAL domains from several bacteria: T. denitrificans (TBD1265, TBD1269, TBD1456, and TBD1660), E. coli (YahA and YdiV), Chromobacterium violaceum (CV0542 and CV2505), Nitrosomonas europaea (NE0566), Shigella flexneri (SFV_3559), Salmonella typhimurium (STM1344), Shewanella oneidensis (SO2039), and B. subtilis (BSU14090 or YkuI) (Table 1). Ten purified EAL proteins exhibited significant or detectable hydrolysis of c-di-GMP in the presence of Mn2+ (5 to 45 nmoles/min mg protein), whereas three other proteins showed no detectable activity (<0.1 nmol/min mg protein) (Table 1; Fig. 1A). The highest c-di-GMP PDE activity was observed in the purified EAL domains from Shi. flexneri SFV_3559 (aa 395–649) and T. denitrificans TBD1265 (aa 487–758). The presence or absence of catalytic activity has been previously demonstrated for the E. coli YahA and B. subtilis BSU14090 (YkuI) respectively 14; 25. The EAL domain proteins which were active against c-di-GMP were also active toward the general PDE substrate bis-p-nitrophenyl phosphate (bis-p-NPP) with the E. coli YahA and T. denitrificans TBD1660 showing the highest activities (Suppl. Fig. 1). PDE activity of an EAL domain against bis-p-NPP was previously demonstrated in crude extracts containing BifA from P. aeruginosa and in the purified HmsP from Yersinia pestis 17; 26. Thus, enzymatic activity of the EAL domain proteins could be assessed using the inexpensive model substrate bis-p-NPP.
Table 1.
Conserved residues of purified EAL domains and their ability to hydrolyze c-di-GMP
| Proteinsa | Conserved residues | Activityb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | #10 | ||
| 1. TBD1265 | E523 | R527 | E546 | N584 | E616 | D646 | D647 | K667 | E703 | Q723 | Yes |
| 2. TBD1269 | E | R | E | N | E | D | D | K | E | Q | Yes |
| 3. TBD1660 | E | R | E | N | E | D | D | K | E | Q | Yes |
| 4. TBD1456 | E | R | E | N | E | D | D | K | E | Q | Yes |
| 5. EC YahA | E | R | E | N | E | D | D | K | E | Q | Yes |
| 6. SFV_3559 | E | R | E | N | E | D | D | K | E | Q | Yes |
| 7. CV0542 | E | R | E | N | E | D | D | K | E | Q | Yes |
| 8. CV2505 | E | R | E | N | E | D | D | K | E | Q | Yes |
| 9. SO2039 | E | R | E | N | E | D | D | K | E | Q | Yes |
| 10. NE0566 | E | R | E | N | E | D | D | K | E | Q | Yes |
| 11. EC YdiV | E | H | E | Q | L | G | N | M | G | Q | No |
| 12. STM1344 | E | T | V | A | A | A | N | M | A | Q | No |
| 13. BS YkuI | E | R | I | D | E | D | N | K | E | Q | No |
|
Residue role (interactionc) |
Me1 | P2 | G1 | Me1, P1 |
Me1 | Me1, Me2 |
Me2 | Water-1, E616 |
Me2 | E523, K667 |
|
The EAL domains of the following proteins were cloned and purified in this study: T. denitrificans TBD1265 (Q3SJE6; aa 487–758), TBD1269 (Q3SJE2; aa 442–705), TBD1660 (Q3SIB5; aa 443–698), TBD1456 (Q3SIW6; aa 955–1212), E. coli YahA (P21514; aa 73–362), S. flexneri SFV_3559 (YhjK; Q83J53; aa 395–649), C. violaceum CV0542 (Q7P0M4; aa 362–612), CV2505 (Q7NV41; aa 146–401), S. oneidensis SO2039 (Q8EFE2; aa 1–233), N. europaea NE0566 (Q82WU5; aa 1–415), E. coli YdiV (P76204; aa 1–237), S. typhimurium STM1344 (CdgR; Q8ZPS6; aa 1–237), and B. subtilis YkuI (BSU14090; O35014; aa 1–407).
Hydrolysis of c-di-GMP.
Me1: metal ion-1; P2: c-di-GMP phosphate-2; G1: c-di-GMP guanine base-1; P1: c-di-GMP phosphate-1; Water-1: catalytic water molecule-1.
Fig. 1.
PDE activity of purified EAL domains. (A), c-di-GMP hydrolysis. Effect of different metals on the hydrolysis of c-di-GMP by the purified EAL domains from TBD1265 (B) and SFV_3559 (C).
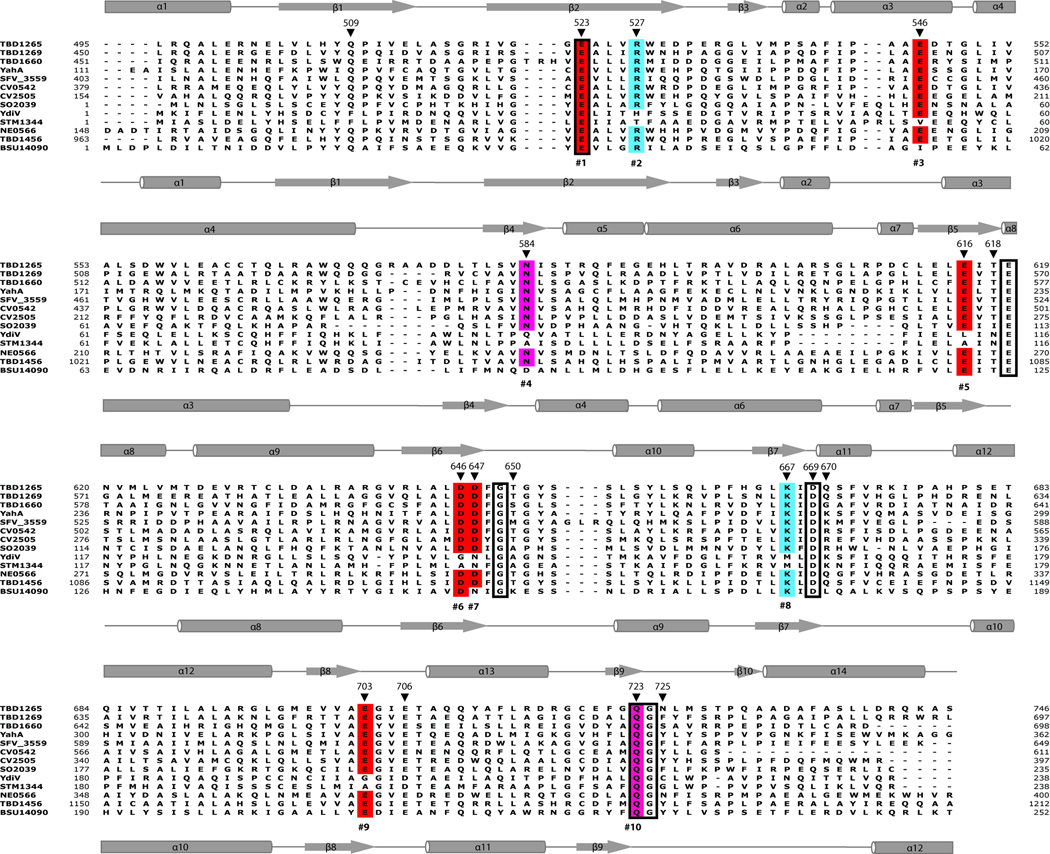
Sequence alignment of the 13 purified EAL domains (Fig. 2), as well as over 3,700 EAL domain sequences from sequenced genomes (Suppl. Fig. 2), revealed the presence of 10 conserved charged or polar residues which were identical in all active proteins. In the catalytically active EAL domain PDE BlrP1, four conserved residues (#1, 4, 5, and 6 (including the Glu residue of the EAL signature sequence) bind the metal ion, whereas the conserved E359 (residue #9) functions as a general base, coordinating the catalytic water molecule 24. The functional role of the conserved residues #2, 3, 8 and 10 is presently unknown. The sequences of the three inactive EAL domains found in our work show “normal” (active) residue #10, but the other residues were modified. Two inactive EAL domains (E. coli YdiV and S. typhimurium STM1344) have modified residues #2, 3, 4, 5, 6, 7, 8 and 9, and these proteins are likely unable to coordinate the metal ion (Fig. 2; Table 1). BSU14090 (YkuI) from B. subtilis has changes in conserved residues #3, 4, and 7 (Fig. 2; Table 1). In the purified EAL domain proteins RocR and PA2567 from P. aeruginosa, the conserved loop-6 (DFG[T/A]GYSS) has been shown to be essential for activity and for binding of Mg2+ and c-di-GMP, as well as for protein dimerization 27. However, the homologous loop-6 sequences in the proteins from our work showed many variations and no correlation with their activity. Thus, the inactive EAL domains found in our work display various changes in their conserved residues and therefore are likely affected in different steps of substrate binding and/or catalysis.
Fig. 2.
Structure-based sequence alignment of the EAL domains from TBD1265 and other proteins used in this study. Absolutely conserved residues are boxed, and 10 conserved EAL domain residues are colored and numbered below the alignment. The secondary structure elements of TBD1265 (2r6o) and YkuI (BSU14090, 2bas) are shown above and below the alignment, respectively. The TBD1265 residues mutated to Ala in this work are marked with the black triangles above the alignment. The compared proteins are listed in the legend of Table 1. The conserved motif residues are colored as in the WebLogo diagram (Suppl. Fig. 2): acidic residues (D, E), red; positively charged residues (K, R, H), blue; small residues (G, S, C), green; polar uncharged residues (N, Q, T), purple; and hydrophobic residues (A, F, L, M, I, P, V, W,Y), dark yellow.
Activity of EAL domains from TBD1265 and SFV_3559
The reaction requirements of EAL PDEs were characterized using purified EAL domains of TBD1265 (aa 487–758) and SFV_3559 (aa 395–649). TBD1265 is an uncharacterized putative diguanylate cyclase/PDE (758 aa) containing three domains: the N-terminal SBP (Solute Binding Protein) domain (aa 1–234), the central GGDEF domain (aa 316–473) and the C-terminal EAL domain (aa 493–730), whereas the uncharacterized protein CFV_3559 (YhjK, 649 aa) has the N-terminal signal transduction HAMP domain (aa 1–220), the central GGDEF domain (aa 229–381), and the C-terminal EAL domain (aa 401–635). Both proteins showed a broad pH optimum at alkaline pH (8.0–9.3) and required the addition of a divalent metal cation for hydrolysis of both c-di-GMP and bis-pNPP. With c-di-GMP as substrate, Mn2+ was the best metal ion for both proteins followed by Co2+, Ni2+, and Mg2+, whereas Ca2+ did not support activity (Fig. 1B, 1C). This is similar to other known EAL domain PDEs, most of which have showed higher activity in the presence of Mn2+ and can also be activated by Mg2+ or Co2+ 14; 22; 23; 24; 26. With bis-pNPP as a substrate, the TBD1265 EAL domain showed no saturation kinetics with Mn2+ (up to 15 mM), whereas SFV_3559 was saturated at approximately 10 mM concentration (Km 3.8 mM) (Table 2). In the presence of c-di-GMP, TBD1265 exhibited higher affinity to Mn2+ (Km 0.4 mM). Both TBD1265 and SFV_3559 demonstrated high affinity to c-di-GMP in vitro and their Km’s to c-diGMP (13.6 nM and 15.4 nM) (Table 2) were significantly lower than those reported for other EAL domains (from 60 nM to 3.2 µM) 9; 22; 23. The intracellular concentrations of c-di-GMP in various organisms have been reported to be in the submicromolar range and up to 10 µM 13; 28; 29; 30, suggesting that in vivo, the EAL domains of both TBD1265 and SFV_3559 are likely to function under saturation by c-di-GMP. Like other c-di-GMP PDEs 13; 23; 24, purified EAL domains from TBD1265 and SFV_3559 showed rather low rates of c-di-GMP hydrolysis in vitro (kcat 0.03–0.04 s−1), but the catalytic efficiency was high because of the high substrate affinity of both proteins to c-di-GMP (Table 2).
Table 2.
Kinetic parameters of the EAL domains of TBD1265 and SFV_3559
| EAL domain | Variable substrate |
Km, |
Vmax, U/mga |
kcat, s−1 |
kcat/Km, M−1, s−1 |
|---|---|---|---|---|---|
| TBD1265 | bis-p-NPP | 20.0 ± 1.3 mM | 0.1 ± 0.02 | 0.06 | 0.3×10 |
| MnCl2b | 0.4 ± 0.1 mM | 0.03 ± 0.003 | 0.02 | 0.5×102 | |
| c-diGMP | 13.6 ± 1.5 nM | 0.05 ± 0.002 | 0.03 | 2.3×106 | |
| SFV_3559 | bis-p-NPP | 14.1 ± 4.2 mM | 2.1 ± 0.6 | 8 | 0.6×103 |
| MnCl2c | 3.8 ± 0.4 mM | 0.4 ± 0.02 | 0.2 | 0.5×102 | |
| c-diGMP | 15.4 ± 2.3 nM | 0.07 ± 0.01 | 0.04 | 2.7×106 |
U/mg, µmoles/min mg protein
c-diGMP was used as a substrate
bis-p-NPP was used as a substrate
Crystal structure of the TBD1265 EAL domain with bound Mg2+
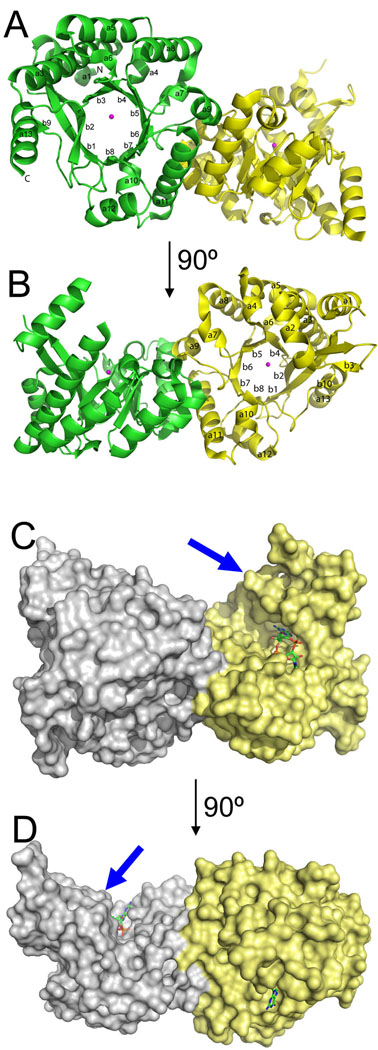
The Se-Met substituted EAL domain of TDB1265 was crystallized in the presence of Mg2+ and the structure was solved to 1.8 Å resolution using the single wavelength anomalous diffraction (SAD) method (PDB code 2r6o; Table 3). The asymmetric unit of the apo-structure shows a protein dimer with two monomers (A and B) connected through interactions between two antiparallel α-helices (α9 and α11) and β6-α9 loop from each monomer (Fig. 3). Accordingly, size-exclusion chromatography of the purified TBD1265 EAL domain showed that it forms a stable homodimer in solution (78 ± 5 kDa; the predicted molecular mass of the dimer is 66 kDa). Similar dimerization patterns were also observed in the full-length EAL domain proteins YkuI from B. subtilis and BlrP1 from K. pneumoniae 24; 25, suggesting that the full-length TBD1265 might also form a dimer through interactions between its EAL domains. In contrast, a tetrameric state was found for the P. aeruginosa RocR 23 and a monomeric state for the E. coli YahA 14.
Table 3.
Crystallographic data collection and model refinement statistics
| Apo-TBD1265 (2r6o) | TBD1265 + c-di-GMP (3n3t) | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9786 | 0.9794 |
| Space group | P212121 | P212121 |
| Cell parameters(Å) | a=50.05, b=63.14, c=173.28 | a=51.56, b=63.20, c=173.99 |
| Resolution (Å) | 50 - 1.8 (1.86 – 1.8) | 50 - 2.35 (2.37 – 2.35) |
| Total reflections | 535,897 | 169,249 |
| Unique reflections | 50,452 | 24,433 |
| Completeness (%) | 97.3 (90.1) | 99.9 (99.9) |
| I/sigma | 39.7 (3.3) | 34.6 (5.2) |
| Rmerge(%) | 6.0 (45.4) | 10.0 (49.9) |
| Refinement | ||
| Rcryst (%) | 19.0 | 18.4 |
| Rfree (%) | 21.8 | 25.4 |
| R.m.s. deviations from ideality | ||
| Bond lengths (Å) | 0.012 | 0.014 |
| Angles (°) | 1.21 | 1.54 |
| No. of protein atoms | 2054+1908 | 2033+2011 |
| No. of solvent atoms | 202+176 | 225 |
| No. of ligand atoms | 6 | 97 |
| Average B-factor | ||
| main-chain atoms (Å2) | 28.8 | 29.1 |
| Side-chain atoms (Å2) | 30.8 | 30.3 |
| Protein atoms (Å2) | 29.8 | 29.5 |
| Solvent atoms (Å2) | 41.6 | 40.0 |
| Ligand atoms (Å2) | 27.1 | 52.4 |
Values in parentheses are for the highest resolution shell.
Fig. 3.
Crystal structure of the TDB1265 EAL domain. (A,B), Overall structure of the apo-protein dimer (PDB code 2r6o): two views related by a 90° rotation. Protein subunits are shown as ribbons and colored in different colors with the labelled secondary structure elements. The position of the potential active site is indicated by the metal ion (shown as magenta-colored spheres). (C,D), Surface presentation of the TBD1265 EAL domain dimer with bound c-di-GMP (PDB code 3n3t): two views related by a 90° rotation. Two monomers are shown in different colors, and c-di-GMP is shown as sticks. Note that c-di-GMP is bound in the large pocket between the lobe (indicated by blue arrows) and TIM barrel domain.
The TBD1265 EAL domain has a modified TIM-barrel-like fold, in which the β1 strands runs antiparallel to other seven strands (Fig. 3A, 3B). TIM barrel is the most common and versatile structural fold in the protein universe and most of its members have hydrolase, oxidoreductase, transferase or isomerase activities 31. The antiparallel orientation of the β1 strand was also found in structures of the EAL domains from B. subtilis YkuI, K. pneumoniae BlrP1, and P. aeruginosa FimX 21; 24; 25 and therefore appears to be a characteristic feature of TIM barrels of the EAL domain. In addition, the structure of the TBD1265 EAL domain revealed the presence of a lobe positioned on the side of the TIM barrel core and formed by four helices (α1, α2, α3, and α5) (Fig. 3). A Dali search for structural homologues identified three EAL domain structures as the best hits: FimX (PDB 3hv8, Z-score 28.7), BlrP1 (PDB 3gg1, Z-score 25.6), and YkuI (PDB 2w27, Z-score 23.1). The second group of structural homologues includes proteins with a classical TIM-barrel: the copper homeostasis protein CutC (PDB 2bdg, Z-score 9.6), pyruvate kinase (PDB 2e28, Z-score 9.4), and aldose reductase (PDB 2bgs; Z-score 8.8).
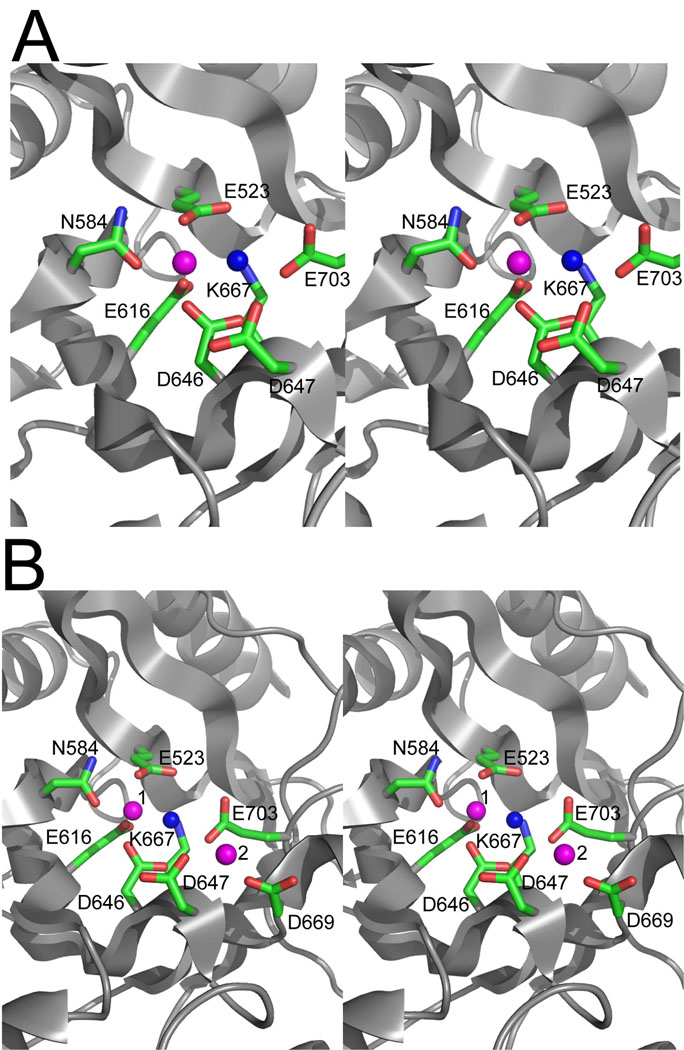
The structure of the TBD1265 EAL domain has also revealed the presence of a single bound metal ion in molecule A and two metal ions in molecule B (Fig. 4). These metal ions were interpreted as Mg2+ (Mg1 and Mg2) since MgCl2 was present in the crystallization solution and because of the characteristic coordination sphere and metal-oxygen distances (~2.2 Å). Since Mg2+ and Mn2+ share a similar size (0.86 Å and 0.97 Å, respectively), coordination number, octahedral geometry, and ligand preference 32, the Mg2+-bound structure of the TBD1265 EAL domain represents an adequate model for the characterization of the protein active site. Both metal ions are located at the C-terminal end of the TIM barrel (Fig. 3A, 3B), the usual location of the active site in known TIM-barrel enzymes 31. In both molecules, the Mg1 ion is coordinated by the side chains of four conserved residues: E523 (conserved residue #1, 2.10 Å), N584 (residue #4, 2.1 Å), E616 (residue #5, 2.1 Å), and D646 (residue #6, 2.1 Å) (Fig. 4A, 4B). Similar coordination was observed for metal ion Me1 in the apo-structure of BlrP1 and for the Ca2+ ion in the YkuI structure 24; 25. In molecule B, the Mg2 ion is located on the protein surface (8Å from Mg1) and is coordinated by the side chain carboxylates of the conserved D647 (conserved residue #7, 2.4 Å) and D669 (2.1 Å) residues (Fig. 4B). The position of Mg2 in the TBD1265 structure is different from that of the second metal ion in the structure of BlrP1 (4 Å further away from the metal ion 1). In TBD1265, Mg2 was not found in all molecules, and it has a temperature factor (38 Å3), which is two times higher than that of Mg1 (21–23 Å3) suggesting that the Mg2 site is likely to be a low affinity site.
Fig. 4.
Close-up stereo view of the active site of the TDB1265 EAL domain with the bound Mg2+ (2r6o) in the molecules A (A) and B (B). The metal ions (Mg2+) are denoted by the magenta spheres and numbered in the molecule B. The potential catalytic water molecule is shown as a blue sphere, whereas the amino acid residues in contact with the metal ions and water are shown as sticks along a TBD1265 ribbon (grey).
Structure of the TBD1265 EAL domain in complex with c-di-GMP
The structure of the TBD1265 EAL domain in complex with c-di-GMP was solved using the SAD method after soaking the protein crystals in a solution of c-di-GMP (2.5 mM) (Table 3). The structure revealed a dimer with each monomer containing two metal ions (interpreted as Mg2+) and c-di-GMP bound to the predicted active site and covering both Mg2+ ions (Fig. 5A, 5B). One of the metal ions (Me1) corresponds to the Mg1 ion of the apo-structure (Fig. 4), whereas the second Mg2+ ion (Me2) is located close to Me1 (4.1 Å) and does not match the position of the second metal ion of the apo-structure (which is 8 Å away from Mg1). However, the position of the Me2 ion in the TBD1265 complex with c-di-GMP is equivalent to that of the second Mn2+ ion in the BlrP1 structure (PDB codes 3gg0 and 3gfz; 3.7 – 4.3 Å between two metals). In TBD1265 complex structure, the two metal ions are bridged by the side chain oxygens of the conserved D646 (2.2 Å), by the potential catalytic water molecule-1 (2.1 Å and 3.1 Å), and by two oxygens of the c-di-GMP phosphate-1 (1.9 Å and 2.2 Å) (Fig. 5C). Similar to the TBD1265 apo-structure, the Me1 ion is additionally coordinated by the conserved E523 (2.0 Å), N584 (1.9 Å), and E616 (2.2 Å) (Fig. 5C). The Me2 is close to the side chains of the conserved D647 (2.5 Å) and E703 (2.1 Å). Thus, in the structures of the BlrP1 and TBD1265 EAL domains two metal ions show similar coordination, which generally resembles the classical two-metal ion model proposed for nucleases 33. However, in EAL domains the two metal ions are bound to the two different oxygens of the phosphate, whereas in nucleases both metal ions are bound to the same phosphate oxygen.
Fig. 5.
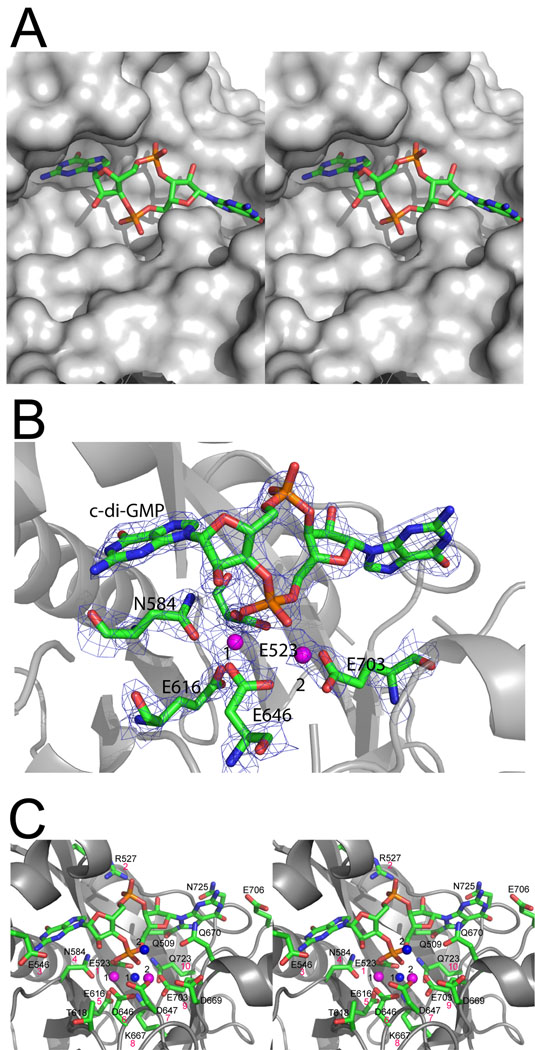
Close-up view of the active site of TBD1265 EAL domain in complex with c-di-GMP and Mg2+ (3n3t). (A), Stereo view of the active site showing the bound c-di-GMP. The protein is shown as surface presentation (gray), and c-di-GMP is shown as colored sticks. Note that one of the c-di-GMP bases (G1 in Fig. 7) is completely buried inside the large cavity formed between the lobe and TIM barrel domains. (B), Difference electron density observed for c-di-GMP bound to the TBD1265 EAL domain active site. The omit map was generated by removing c-di-GMP from the model followed by several rounds of refinement using Refmac5 (the resulting 2F0-Fc map was contoured at 2 σ). (C), Stereo view of the active site showing the residues coordinating c-di-GMP, two Mg2+ ions, and two water molecules. Two metal ions are denoted by magenta spheres, the water molecules by blue spheres (1 and 2), and c-di-GMP is shown as sticks along with the protein ribbon (gray). The magenta-colored numbers (below the residue labels) designate the number of the conserved EAL motif residue (as shown in Table 1).
The c-di-GMP molecule is bound in an extended conformation with two of the oxygen atoms of the phosphate-1 (P1; makes the scissile O3′-P bond) coordinated by the bound Mg2+ ions (1.8 – 2.2 Å) and the side chains of the conserved N584 (2.9 Å), E523 (2.8 Å), D646 (3.1 Å), and D647 (3.3 Å) (Fig. 5C). Another phosphate (P2) of c-di-GMP interacts through its non-bridging oxygen atom with the guanidium group of the conserved R527 (2.4 Å and 2.9 Å) (Fig. 5C). The guanine base G1 of c-di-GMP (on the left in Fig. 5) is positioned inside the mostly hydrophobic cavity formed by the lobe (α2 and α3), β2 strand and short loop β4-α4 (V526, I542, I551, L554, and V558) and makes no specific contacts with the protein side or main chain atoms (Fig. 5A, 5C). In contrast, the second guanine base (G2) of c-di-GMP interacts electrostatically with the side chains of the conserved Q509 (N7, 3 Å), E706 (N1, 2.7 Å; C2-NH2, 3.1 Å), and with the main-chain amide of N725 (2.5 Å). Although the general orientation of c-di-GMP in the active site of the TBD1265 EAL domain is the same as in the structures of BlrP1, YkuI and FimX, the residues involved in the binding of nucleotide bases are not conserved in these proteins. For example, in TBD1265 there is no carboxylate residue coordinating the G1 base (D215 in BlrP1), and the aromatic side chain coordinating the G2 base (F381 in BlrP1) is replaced by N725.
A limited number of specific contacts between c-di-GMP and the active site of the TBD1265 EAL domain are the likely reason for the ability of this protein and several other EAL domains to hydrolyze the model PDE substrate bis-pNPP (Suppl. Fig. 1). Recently, structural and biochemical characterization of the unknown domain (DUF147) of the DNA integrity scanning protein A (DisA) from B. subtilis revealed the presence of cyclic dimeric AMP (c-di-AMP) bound to the protein, as well as the presence of diadenylate cyclase activity 34. It is possible that c-di-AMP represents another bacterial second messenger, the level of which (like for c-di-GMP) might be regulated by its synthesis and degradation. Since the structure of the TBD1265 EAL domain suggests that c-di-AMP would fit into the protein active site, we wanted to check if the EAL domain was capable of hydrolyzing c-di-AMP. When five EAL domain proteins were tested for the ability to hydrolyze c-di-AMP, only E. coli YahA showed low activity (<4 nmoles/min mg protein) whereas EAL domains of the other four proteins (TBD1265, TBD1269, TBD1660, and SFV_3559) were inactive. Thus, EAL domain proteins seem to be incapable of hydrolysis of c-di-AMP, which is probably hydrolyzed by another presently unknown PDE.
Mutational studies of TBD1265 and implications for the catalytic mechanism of EAL domains
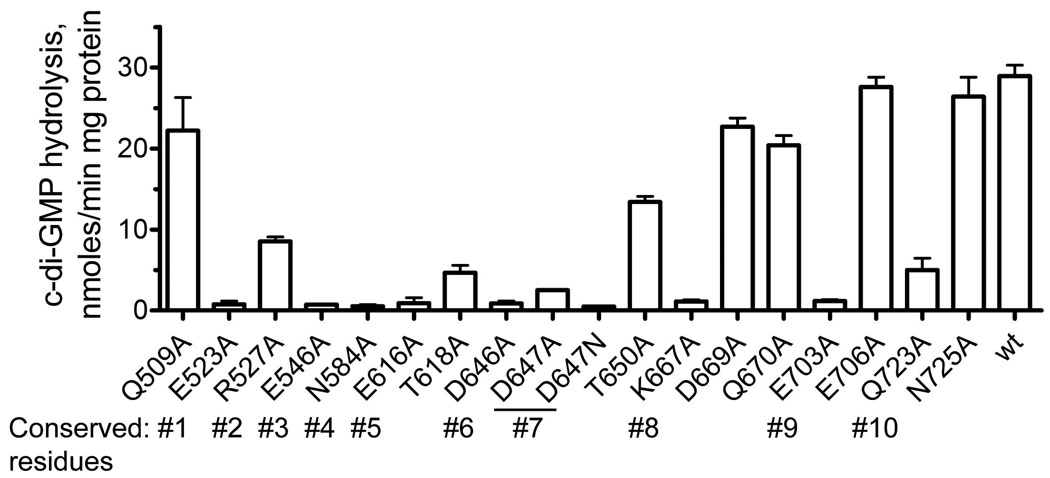
To characterize the role of conserved and semi-conserved residues of the EAL domain PDEs in the hydrolysis of c-di-GMP, we have mutated 17 residues of the TBD1265 EAL domain to Ala (or Asn) and assayed the activity of purified mutant proteins (Fig. 6). Eight mutant proteins exhibited greatly reduced catalytic activity, four proteins showed lower activity (R527A, T618A, T650A, and Q723A), and five proteins had wild-type level activity (Q509A, D669A, Q670A, E706A, and N725A). All mutations of the metal-binding residues produced inactive proteins (E523A, N584A, E616A, D646A, D647A, and E703) indicating that both metal ions play a crucial role in the catalytic mechanism of EAL domains. The replacement of D647 by Asn (as in YkuI) produced inactive TBD1265 EAL domain (D647N) (Fig. 6) explaining the absence of activity in YkuI and supporting the important role of this residue in catalysis. The side chain of E546 (conserved residue #3) might be involved in the binding of the substrate G1 base, whereas K667 (residue #8) interacts with the metal-coordinating residues E523 (2.9 Å) and E616 (2.6 Å) (Fig. 5C). Mutant proteins with wild-type or slightly reduced activity had mutations in residues interacting with the substrate G2 base (Q509, Q670, E706, and N725), P2 oxygens (R527), or in residues located outside of the active site (T650 and D669) (Fig. 6).
Fig. 6.
Alanine replacement mutagenesis of the TBD1265 EAL domain: hydrolysis of c-di-GMP by purified mutant proteins. The conserved EAL motif residues are indicated below the graph.
In recent years, the catalytic mechanism of EAL domain PDEs has been the subject of much discussion 23; 24; 25. Initially, a one metal ion mechanism was proposed based on modeling and mutagenesis of the P. aeruginosa RocR and on the structure of the B. subtilis YkuI 23; 25. In this model, a single divalent cation is coordinated by four conserved residues, a c-di-GMP phosphate oxygen, and a catalytic water molecule, which is also bonded to a general base carboxylate (E209 in YkuI). However, the recent work on the EAL PDE BlrP1 from K. pneumoniae has proposed a two metal-assisted catalytic mechanism, which is based on the conserved Glu-His tandem and bimetal system in the active site of the PDEs hydrolyzing 3′,5′-cyclic nucleoside monophosphates (cAMP, cGMP) 24; 35. These PDEs have a highly conserved catalytic pocket with several residues which are absolutely conserved across the 11 PDE families 36. In the crystal structure of PDE4B, two different metal ions (Zn2+ and Mg2+) are bridged by a conserved Asp (Asp275) and the catalytic hydroxide, while two conserved His residues (His238 and His274) and another Asp (Asp392) coordinate one of the metals 36. During the reaction, the catalytic hydroxide performs a nucleophilic attack on the phosphorous atom, whereas the conserved Glu-His tandem (Glu413 and His234) donates a proton to O3′ 36; 37; 38. While the two metals in the structures of BlrP1 and TBD1265 are positioned at similar distances and are bridged by a conserved Asp and catalytic water molecule as the two metals in PDE4B (4.2–4.3 Å), the EAL PDEs have no His residues in the active site and use two identical metal ions, which are bound to two different phosphate oxygens and are coordinated in a different manner. In the classical model of the two-metal mechanism, two metal ions interact with the same phosphate oxygen, but proposed to play different roles in reaction 39. The metal-A activates a catalytic water molecule, whereas the metal-B stabilizes the pentacovalent intermediate and facilitates leaving of the 3′ oxyanion group 39. In EAL domain PDEs, both metal ions appear to play similar roles in catalysis, and they are both involved in the activation of a water nucleophile and potentially in the stabilization of the pentacovalent intermediate.
Although binuclear metal centers are well characterized in hydrolytic enzymes, there is also extensive experimental data to support a one-metal-ion model as the general mechanistic scheme in considerations of metal-mediated hydrolysis of phosphodiester bonds 33. For example, the 3′–5′ Klenow exonuclease from E. coli is a paradigm for the two-metal mechanism 40. However, the calorimetric and kinetics experiments indicate the presence of only one metal ion in the active site of this enzyme 41. Moreover, recent works on restriction endonucleases argue that a single metal ion can perform the catalysis and that the second metal ion is not critical 42; 43; 44.
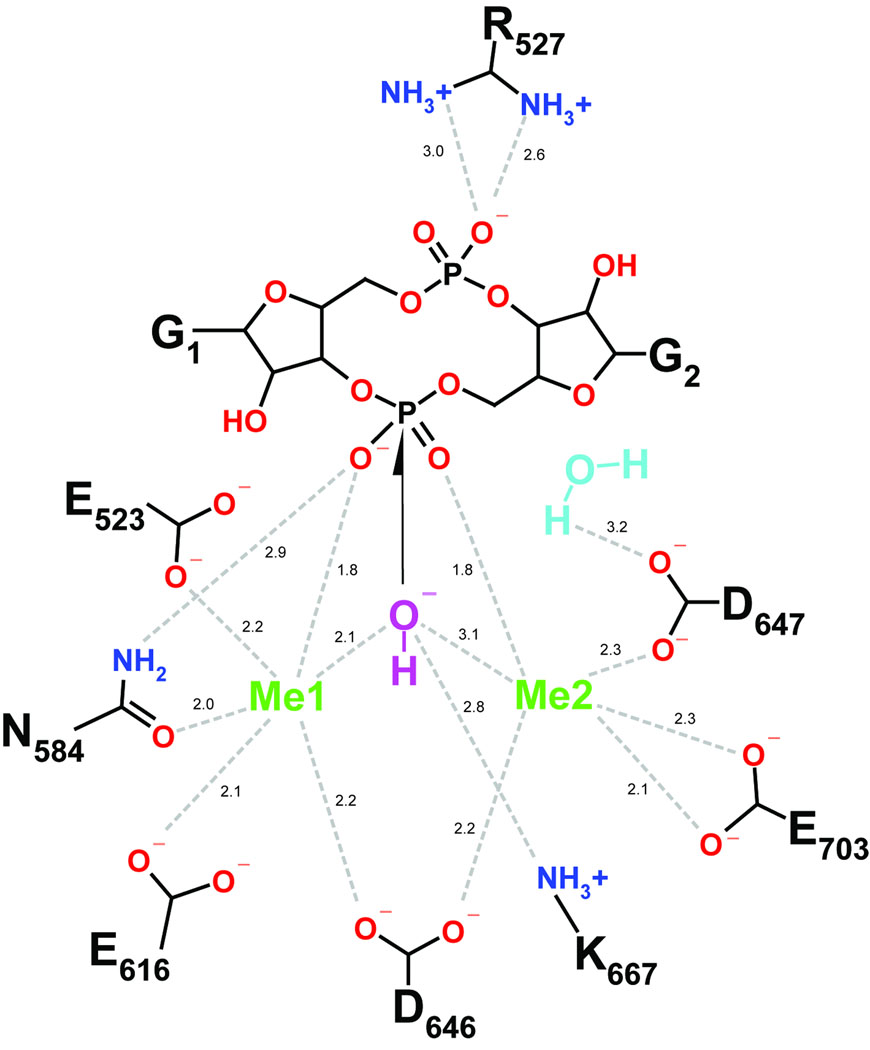
Our structural and mutational studies of the TBD1265 EAL domain together with the previous work on the K. pneumonia BlrP1 24 support a two metal-based mechanism for the hydrolysis of c-di-GMP by EAL domain PDEs (Fig. 7). In this model, two divalent metal cations (Me1 and Me2; Mn2+ or Mg2+) are directly coordinated by the side chains of six conserved residues #1, 4, 5, 6, and 9 (E523, N584, E616, D646, D647, and E703 in TBD1265), the two P1 phosphate oxygens of c-di-GMP, and by the catalytic water molecule (water 1 in Fig. 5C). As in nucleases 33; 45, these metal ions are likely to be involved in several steps including the P1 phosphate coordination (Lewis acid activation), water nucleophile activation (by reducing the pKa and inducing the deprotonation), and in transition state stabilization. It has been proposed that the higher activity of BlrP1 in the presence of Mn2+ (compared with Mg2+) can be explained by the higher Lewis acidity of Mn2+ facilitating the deprotonation of the catalytic water molecule 24. Similar to nucleases 45, the direct coordination of the P1 phosphate oxygens of c-di-GMP by two metal ions may also change the O-P-O angle helping to stabilize the transition state. The side chains of E523 and E616 also interact with the conserved K667 (conserved residue #8) and Q723 (residue #10) (2.6–2.9 Å) and form a network of interacting residues (EEKQ) (Fig. 5C, 7). These interactions are likely to stabilize the unprotonated states of E523 and E616 (by reducing their pKa) which are important for the metal coordination. The important role of these network interactions is supported by the low activities of K667A and Q723A mutant proteins (Fig. 6).
Fig. 7.
A diagram showing the coordination of c-di-GMP and Mg2+ in the active site of the TBD1265 EAL domain and the proposed reaction mechanism of c-di-GMP hydrolysis. Two metal ions (Me1 and Me2) together with the conserved K667 are proposed to be responsible for the activation of the catalytic water molecule and production of the nucleophilic hydroxyl. Another water molecule (coordinated by D647) is proposed to function as a proton donor for the leaving O3′ group. The numbers represent inter-atomic distances in Å.
In the TBD1265 EAL/c-di-GMP structure, the potential catalytic water molecule (#1) is located between two metal ions and is 3.2 Å away from the P1 phosphorus atom (Fig. 5C). A number of nuclease structures show the presence of a highly conserved Lys side chain in their active sites, which is positioned to interact with the water nucleophile and proposed to help in nucleophile activation 46; 47. In the TBD1265 EAL domain structure (Fig. 5C), the side chain of the conserved K667 (conserved residue #8) is positioned 2.8 Å from the catalytic water molecule suggesting that this residue might also contribute to nucleophile activation. We propose that K667 together with two metal cations activate the bound water molecule generating the catalytic hydroxide which performs a nucleophilic attack on the P1 phosphorus atom (Fig. 7). After the reaction, the leaving group O3′ is likely to be protonated by the water molecule (#2) located close to O3′ (2.8 Å) and coordinated by the conserved D647 (2.3 Å; conserved residue #7) (Fig. 5C). This is supported by the crucial role of D647 in the activity of the TBD1265 EAL domain (Fig. 6). Another conserved Asp residue (D669) is positioned in the vicinity of D647 (3.5 Å) and might facilitate its protonated state, which is important for the coordination of the water proton donor molecule. In the catalytically inactive EAL domain of YkuI, the D647 residue is replaced by Asn (N152) suggesting that this replacement produced an inactive protein. A coordinating role for a water proton donor molecule was also proposed for the homologous D303 in BlrP1 24.
A large number of metal-dependent PDEs have been crystallized with one, two, or three metal ions in their active sites and one-, two-, and three-metal-based mechanisms have been proposed fuelling debates about the roles of metal ions in the phosphodiester bond cleavage 33; 47; 48. Although the well-established two-metal-ion mechanism dominates the field 40, recent developments based on the application of novel experimental tools indicate that there is probably more than one way to hydrolyze phosphodiester bonds, and that some enzymes can use both one-metal-ion and two-metal-ions mechanisms 33; 42; 43. Our work suggests that c-di-GMP hydrolysis mechanism involving two metal ions per active site can be operative in EAL domain PDEs. However, a one-metal mechanism is also possible and can occur under different (solution or regulatory) conditions. The active site of EAL domain PDEs shows no obvious similarity to known PDEs or nucleases with the one- or two-metal-ion mechanisms expanding the diversity of catalytic mechanisms of phosphodiester hydrolyzing enzymes.
MATERIALS and METHODS
Gene cloning, protein expression and purification
The EAL domains of TBD1265 and other proteins (Table 1) were PCR-amplified using genomic DNAs from these organisms and cloned into a modified pET15b expression vector (Novagen) 49. The overexpression plasmid was transformed into the E. coli BL21 (DE3) Gold strain (Stratagene), and the proteins were purified using metal-chelate affinity chromatography on nickel affinity resin (Qiagen) as described previously 49. Gel-filtration analysis of the oligomeric state of the TBD1265 EAL domain was performed with a Superdex-75 16/60 column (Amersham Biosciences) equilibrated with 10 mM HEPES-K (pH 7.5) and 0.2 M NaCl using AKTA FPLC (Amersham Biosciences).
Site-directed mutagenesis of the TBD1265 EAL domain
Alanine replacement mutagenesis was performed using the QuikChange™ site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol. The residues selected to be mutated were changed to Ala (or to Asn). DNA encoding the wild-type TBD1265 EAL domain cloned into the modified pET15b was used as a template for mutagenesis. Plasmids were purified using the QIAprep Spin mini prep kit (Qiagen), and all mutations were verified by DNA sequencing. Verified plasmids containing the desired mutations were transformed into the E. coli BL21 (DE3) strain, and the mutant proteins were purified using the same protocol as for the wild-type protein.
Enzymatic assays
PDE activity of purified proteins against c-di-GMP was determined spectrophotomerically by measuring Pi release using the Malachite Green reagent as described previously 50. The reactions were performed in 80 µl assays in 96-well microplates containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 0.1 mM c-di-GMP and 5 µg of protein. After 25 min incubation at 37°C, the reaction was stopped by the addition of 80 µl of alkaline phosphatase buffer (0.2 M CHES buffer, pH 9.0, and 10 mM MgCl2) and incubated with 1 unit of shrimp alkaline phosphatase (SAP) for 10 min at 37°C. The SAP reaction was terminated by the addition of 40 µl of Malachite Green, and the production of Pi was measured at 630 nm. The hydrolysis of c-di-AMP was measured using similar reaction conditions. Cyclic di-GMP was from BioLog Life Science Institute (www.biolog.de), cyclic di-AMP was from Axxora LLC (www.axxora.com), and all other substrates and chemicals were from Sigma. PDE activity against the general PDE substrate bis-p-nitrophenyl phosphate (bis-pNPP) was measured spectrophotometrically in 200 microliter assays containing 50 mM HEPES (pH 7.5), 2 mM MnCl2, 10 mM bis-pNPP, and 5–10 µg of enzyme. After 25 min incubation at 37°C, the absorbance at 410 nm was determined. The pH dependence of PDE activity toward c-di-GMP was characterized using three buffers: MES-K (pH 5.5 to 6.5), Tris-HCl (pH 7.0 to 9.0), and CAPS-K (pH 9.4 to 11.0). The metal dependence of c-di-GMP hydrolysis was determined at optimal pH using various metals (2 mM MnCl2, 5 mM MgCl2, or 0.5 mM of other metals), 0.1 mM c-di-GMP and 5 µg of enzyme. Kinetic parameters were determined by non-linear curve fitting from the Lineweaver-Burk plot using GraphPad Prism software (version 4.00 for Windows, GraphPad Software, San Diego, CA).
Protein crystallization and data collection
Protein crystals were grown at 22°C by the sitting-drop vapour-diffusion method, with 2 µl of protein sample (23 mg/ml) mixed with an equal volume of reservoir buffer. The crystals of the TBD1265 EAL domain apo-protein grew after 2–3 weeks in the presence of 0.2 M MgCl2 and 20% PEG-3350 (pH 5.8). For the TBD1265/c-di-GMP complex, crystals of the apo-protein were obtained with a reservoir buffer containing 0.2 M MgCl2, 25% PEG-3350 and 0.1 M Bis-Tris (pH 6.5). These crystals were washed in 2 µl of reservoir solution and transferred to the freshly made reservoir solution containing 2.5 mM c-di-GMP for overnight soaking. All crystals were cryoprotected in Paratone-N oil (Hampton Research, Cat.# HR2-643) prior to flash-freezing in liquid nitrogen for data collection.
Structure determination
The structure of the TBD1265 apo-protein was solved with the single-wavelength anomalous dispersion (SAD) method using the anomalous signal from selenium atoms. Data was collected to 1.8 Å resolution on beamline 19-BM (Structural Biology Center, Advanced Photon Source, Argonne National Laboratory). The data was integrated and scaled, the positions of anomalous scatterers were found, and a density-modified electron density map was calculated using the HKL-3000 software package 51. ARP/wARP 52 within the HKL-3000 package was used to build a nearly complete model, followed by cycles of manual building using COOT 53 and restrained refinement against a maximum-likelihood target 54 with 5% of the reflections randomly excluded as an Rfree test set. TLS refinement 55; 56 was also used in the final stages of refinement. The structure of the TBD1265/c-di-GMP complex was solved to 2.35 Å on beamline 19-BM, data was integrated and scaled, and the structure was solved in an analogous manner to apo-TBD1265 using HKL-3000.
Protein Data Bank Accession Codes
Coordinates and structure factors have been deposited with accession codes 2r6o (wild-type, apo-structure) and 3n3t (complex with c-di-GMP).
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Ontario Centre for Structural Proteomics in Toronto (SPiT) for help in conducting experiments and discussions. The authors acknowledge the support of Genome Canada (through the Ontario Genomics Institute) and the Protein Structure Initiative of the National Institutes of Health (Midwest Center for Structural Genomics, NIH grant GM074942, A.J.). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, and the use Structural Biology Center beamlines was supported by Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. M.Y.G. is supported by the Intramural Research Program of the NLM, NIH.
Abbreviations used
- bis-pNPP
bis-para-nitrophenyl phosphate
- c-di-AMP
cyclic dimeric adenosine monophosphate
- c-di-GMP
cyclic diguanylate (or bis-(3`–5`) cyclic dimeric guanosine monophosphate)
- PDE
phosphodiesterase
- SAD
single wavelength anomalous diffraction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 2.Ryan RP, Fouhy Y, Lucey JF, Dow JM. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J Bacteriol. 2006;188:8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 4.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 7.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 10.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 11.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Wolfe AJ, Visick KL. Get the message out: cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sommerfeldt N, Possling A, Becker G, Pesavento C, Tschowri N, Hengge R. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology. 2009;155:1318–1331. doi: 10.1099/mic.0.024257-0. [DOI] [PubMed] [Google Scholar]
- 19.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3'–5')-cyclic-GMP in virulence. Proc Natl Acad Sci U S A. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland LM, O'Donnell ST, Ryjenkov DA, Gomelsky L, Slater SR, Fey PD, Gomelsky M, O'Gara JP. A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J Bacteriol. 2008;190:5178–5189. doi: 10.1128/JB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarro MV, De N, Bae N, Wang Q, Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure. 2009;17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamayo R, Tischler AD, Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J Biol Chem. 2005;280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao F, Yang Y, Qi Y, Liang ZX. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol. 2008;190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 25.Minasov G, Padavattan S, Shuvalova L, Brunzelle JS, Miller DJ, Basle A, Massa C, Collart FR, Schirmer T, Anderson WF. Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J Biol Chem. 2009;284:13174–13184. doi: 10.1074/jbc.M808221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bobrov AG, Kirillina O, Perry RD. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol Lett. 2005;247:123–130. doi: 10.1016/j.femsle.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 27.Rao F, Qi Y, Chong HS, Kotaka M, Li B, Li J, Lescar J, Tang K, Liang ZX. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J Bacteriol. 2009;191:4722–4731. doi: 10.1128/JB.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinhouse H, Sapir S, Amikam D, Shilo Y, Volman G, Ohana P, Benziman M. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1997;416:207–211. doi: 10.1016/s0014-5793(97)01202-7. [DOI] [PubMed] [Google Scholar]
- 29.Simm R, Morr M, Remminghorst U, Andersson M, Romling U. Quantitative determination of cyclic diguanosine monophosphate concentrations in nucleotide extracts of bacteria by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal Biochem. 2009;386:53–58. doi: 10.1016/j.ab.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 30.Spangler C, Bohm A, Jenal U, Seifert R, Kaever V. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods. 81:226–231. doi: 10.1016/j.mimet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 31.Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 32.Shannon RD. Acta Crystallogr. 1976;A32:751–767. [Google Scholar]
- 33.Dupureur CM. Roles of metal ions in nucleases. Curr Opin Chem Biol. 2008;12:250–255. doi: 10.1016/j.cbpa.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 35.Salter EA, Wierzbicki A. The mechanism of cyclic nucleotide hydrolysis in the phosphodiesterase catalytic site. J Phys Chem B. 2007;111:4547–4552. doi: 10.1021/jp066582+. [DOI] [PubMed] [Google Scholar]
- 36.Xu RX, Hassell AM, Vanderwall D, Lambert MH, Holmes WD, Luther MA, Rocque WJ, Milburn MV, Zhao Y, Ke H, Nolte RT. Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]
- 37.Xu RX, Rocque WJ, Lambert MH, Vanderwall DE, Luther MA, Nolte RT. Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram. J Mol Biol. 2004;337:355–365. doi: 10.1016/j.jmb.2004.01.040. [DOI] [PubMed] [Google Scholar]
- 38.Huai Q, Colicelli J, Ke H. The crystal structure of AMP-bound PDE4 suggests a mechanism for phosphodiesterase catalysis. Biochemistry. 2003;42:13220–13226. doi: 10.1021/bi034653e. [DOI] [PubMed] [Google Scholar]
- 39.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beese LS, Steitz TA. Structural basis for the 3'–5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Black CB, Cowan JA. A critical evaluation of metal-promoted Klenow 3`–5` exonuclease activity: calorimetric and kinetic analyses support a one-metal-ion mechanism. J. Biol. Inorg. Chem. 1998;3:292–299. [Google Scholar]
- 42.Dupureur CM. An integrated look at metallonuclease mechanism. Curr Chem Biol. 2008;2:159–173. [Google Scholar]
- 43.Xie F, Qureshi SH, Papadakos GA, Dupureur CM. One- and two-metal ion catalysis: global single-turnover kinetic analysis of the PvuII endonuclease mechanism. Biochemistry. 2008;47:12540–12550. doi: 10.1021/bi801027k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mones L, Kulhanek P, Florian J, Simon I, Fuxreiter M. Probing the two-metal ion mechanism in the restriction endonuclease BamHI. Biochemistry. 2007;46:14514–14523. doi: 10.1021/bi701630s. [DOI] [PubMed] [Google Scholar]
- 45.Horton NC, Perona JJ. Making the most of metal ions. Nat Struct Biol. 2001;8:290–293. doi: 10.1038/86149. [DOI] [PubMed] [Google Scholar]
- 46.Horton NC, Newberry KJ, Perona JJ. Metal ion-mediated substrate-assisted catalysis in type II restriction endonucleases. Proc Natl Acad Sci U S A. 1998;95:13489–13494. doi: 10.1073/pnas.95.23.13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sissi C, Palumbo M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009;37:702–711. doi: 10.1093/nar/gkp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang RG, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Savchenko A. Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase. Structure. 2001;9:1095–1106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savchenko A, Proudfoot M, Skarina T, Singer A, Litvinova O, Sanishvili R, Brown G, Chirgadze N, Yakunin AF. Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli. J Mol Biol. 2007;374:1091–1103. doi: 10.1016/j.jmb.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 51.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 52.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 55.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 56.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.