Abstract
The functional relationship and cross-regulation between autophagy and apoptosis is complex. Here we show that high-mobility group box 1 protein (HMGB1) is a redox-sensitive regulator of the balance between autophagy and apoptosis. In cancer cells, anti-cancer agents enhanced autophagy and apoptosis as well as HMGB1 release. HMGB1 release may be a pro-survival signal for residual cells following various cytotoxic cancer treatments. Diminished HMGB1 by shRNA transfection or inhibition of HMGB1 release by ethyl pyruvate or other small molecules led to predominantly apoptosis and decreased autophagy in stressed cancer cells. In this setting, reducible HMGB1 binds to the receptor for advanced glycation end products (RAGE) but not Toll-like receptor 4 (TLR4), induces Beclin1-dependent autophagy, and promotes tumor resistance to alkylators (melphalan), tubulin disrupting agents (paclitaxel), DNA crosslinkers (ultraviolet light) and DNA-intercalators (oxaliplatin or adriamycin). Oxidized HMGB1 conversely increases the cytotoxicity of these agents and induces apoptosis mediated by the caspase-9/-3 intrinsic pathway. HMGB1 release as well as its redox state thus link autophagy and apoptosis, representing a suitable target when coupled with conventional tumor treatments.
INTRODUCTION
Broadly, stress can simultaneously provoke both an adaptive and apoptotic response within cells. Integration of these programmed survival and death signals determines the fate of the cell. Multiple and rather consistent defects in pathways that control apoptosis are found in virtually all human epithelial and lymphoid tumors. Removal or functional inhibition of proteins essential for the apoptotic machinery can promote a cellular stress response characterized by decreased apoptosis and increased autophagy (Han et al., 2008; Lum et al., 2005) and when adenosine triphosphate (ATP) sources are depleted, necrosis. Understanding this alternative, adaptive pathway known as autophagy, has thus become increasingly important (Amaravadi and Thompson, 2007). Autophagy presently is viewed as a “doubled-edged sword” whereby downregulation of this process promotes tumorigenesis and upregulation in an established tumor promotes cell survival (White and DiPaola, 2009). Autophagy allows tumor cells to survive bioenergetic stress via clearance of damaged organelles and mutant or unfolded proteins and generation of glycolytic substrates (Degenhardt et al., 2006; Levine, 2007). However excessive autophagy promotes programmed cell death under some specific condition (Kroemer and Levine, 2008). Although most cancer therapies such as radiation and cytotoxic drugs, activate apoptosis these treatments also induce autophagy (Apel et al., 2008; Levine, 2007). A thorough understanding of the functional relationship and cross-regulation between these paired processes, apoptosis and autophagy will reveal mechanisms of resistance and identify novel targets for cancer treatment (Maiuri et al., 2007).
High-mobility group box 1 protein (HMGB1) is a highly conserved nuclear protein which acts as a chromatin-binding factor that bends DNA and promotes access to transcriptional protein assemblies on specific DNA targets (Lotze and Tracey, 2005; Muller et al., 2001). In addition to its intra-nuclear role, HMGB1 also functions as an extracellular signaling molecule (Lotze and Tracey, 2005; Muller et al., 2001). HMGB1 is passively released from necrotic cells and is actively secreted by inflammatory cells. Released HMGB1 mediates the response to infection and injury by binding with high affinity to several receptors including the receptor for advanced glycation end products (RAGE), and Toll-like receptors (TLR)-2 and -4, thereby promoting inflammation (Lotze and Tracey, 2005; Scaffidi et al., 2002; Tang et al., 2010; Wang et al., 1999).
The pathogenic role of HMGB1 release in patients undergoing cancer treatment remains largely unexplored (Tang et al., 2010). HMGB1 release has been identified as a means by which acute immune responses are initiated against tumor cells undergoing chemotherapy-induced necrosis (Apetoh et al., 2007). Here we demonstrate that HMGB1 release is a critical regulator of the response to various forms of metabolic stress. Inhibition of autophagy limited HMGB1 release and promoted apoptosis in cancer cells. We demonstrate that in apoptosis-defective tumor cells, autophagy is upregulated and HMGB1 release is limited, suggesting that HMGB1 release is associated with sustained autophagy. Depletion of HMGB1 by RNAi or inhibition of HMGB1 release with small-molecule inhibitors increases tumor cell sensitivity to several clinically useful agents. Moreover, the redox state of HMGB1 is critical as exogenous delivery of reduced HMGB1 protein promotes autophagy and oxidized HMGB1 promotes apoptosis in cancer cells. HMGB1-mediated autophagy is dependent on RAGE/Beclin1, but not TLR4. These findings define new molecular mechanisms for HMGB1 action and support the notion that inhibition of HMGB1 release could decrease autophagy and thus limit resistance to treatment.
RESULTS
Cytotoxic agents induce cellular stress and HMGB1 translocation and release
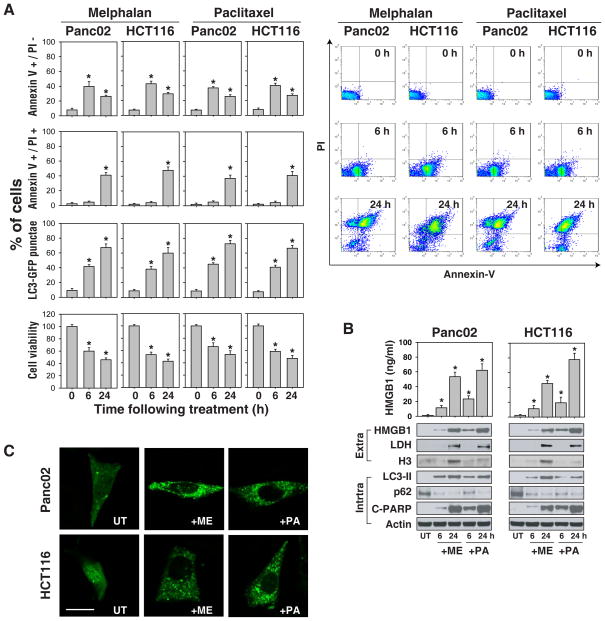
A number of DNA-damaging agents including the alkylator, melphalan (“ME”) and the tubulin depolymerizing agent, paclitaxel (“PA”) induce cell death in a time dependent manner. We have confirmed this finding using a CCK8 cell viability assay (Figure 1A), and a trypan blue dye assay (data not shown). With prolonged treatment, there was a decrease in early apoptotic cells (Annexin V positive and PI negative) and an increase in late apoptotic and necrotic cells (both Annexin V-PE and PI positive) (Figure 1A). Western blotting analysis confirmed PARP cleavage, a marker of cells undergoing apoptosis, following chemotherapuetic treatment (Figure 1B). Microtubule-associated protein light chain 3 (LC3) is now widely used as a marker to monitor autophagy (Mizushima and Yoshimori, 2007). When autophagy is upregulated, a cleaved cytosolic form of LC3 (LC3-I) is conjugated to phosphatidylethanolamine to form LC3-phosphatidylethanolamine (LC3-II), which is recruited to autophagosomal membranes. Melphalan and paclitaxel treatment triggered a time-dependent accumulation of GFP-LC3 punctae (Figure 1A and C) and induction of LC3-II (Figure 1B). The addition of the lysosomal protease inhibitors pepstatin and E64D led to a further increase in the amount of LC3-II (data not shown), consistent with increased autophagic flux. An alternative method for detecting autophagic flux is measuring enhanced degradation of p62 (SQSTM1/sequestosome), a long lived scaffolding protein involved in the transport of ubiquitinylated proteins destined for proteosomal digestion (Pankiv et al., 2007). Consistently, cells treated with melphalan and paclitaxel had reduced expression of p62 (Figure 1B) and increased levels of extracellular HMGB1 demonstrated by western blot analysis of the cell culture supernatants. Treatment with melphalan and paclitaxel for 6 h induced HMGB1 release unaccompanied by measurable lactate dehydrogenase (LDH, a marker of necrosis) or histone 3 (H3) release (Figure 1B). This suggests that early HMGB1 release is an active process. Taken together, these data suggest that in tumor cells treated with a variety of cytotoxic agents, HMGB1 release is a widespread phenomenon regardless of the type of tumor cell death.
Figure 1. Cell injury/stress promotes HMGB1 release from cancer cell lines.
(A) Small molecule anticancer agents decreased cell viability and induced both apoptotic and autophagic pathways. Panc02 and HCT116 cancer cells were treated with either a DNA alkylating agent or a tubulin depolymerization inhibitor (melphalan, “ME”, 160 μg/ml; paclitaxel, “PA”, 10 μg/ml respectively) for 0–24 h, and then assayed for cell viability using measures of NADH dehydrogenases [CCK8], apoptosis by flow cytometric analaysis (right panel) using Annexin V/PI stain and autophagy by quantification of the percentage of cells with GFP-LC3 punctae as described in methods (N=3, * p<0.05 versus control group). (B) The anticancer agents indicated in (A) induced HMGB1 release by ELISA assay (N=3, * p<0.01 versus untreated “UT” group, top), and western blot analysis (bottom), A representative western blot analysis of the protein levels indicated is included (LDH and H3 were both used as controls for protein leakage from damaged cells). (C) GFP-LC3 punctae are induced by melphalan and paclitaxel following 6 h treatment in Panc2.03 cells transfected with a GFP-LC3 reporter plasmid. “UT”: untreated. The percentage of cells showing accumulation of GFP-LC3 in punctae is reported in panel (A). Bar=20 μm.
Inhibition of autophagy diminishes HMGB1 release and enhances selective apoptosis
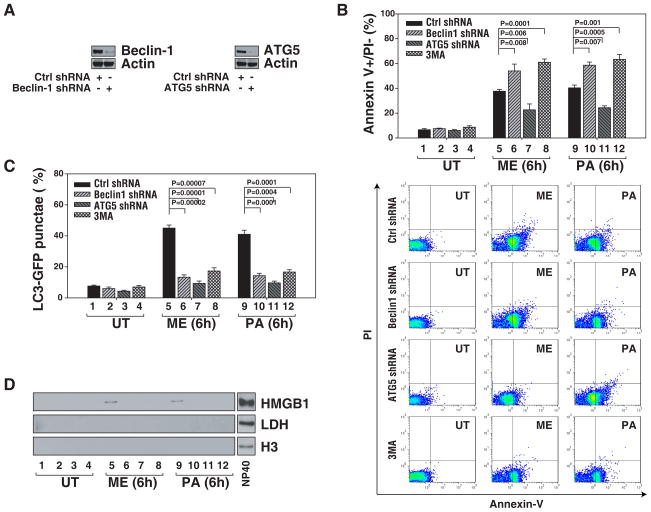
To explore the relationship between HMGB1 release and autophagy/apoptosis, we inhibited the autophagy regulator Beclin1 and ATG5 (Maiuri et al., 2007) by RNAi in Panc02 cells. Knockdown of Beclin1 and ATG5 by shRNA (Figure 2A) inhibited stress-induced accumulation of GFP-LC3 punctae (Figure 2C) and HMGB1 release (Figure 2D). Neither histone 3 (H3) nor LDH were detected in the cellular supernatants following short treatment for 6 h, indicating that HMGB1 release did not depend on conventionally measured early necrosis. As reported previously (Han et al., 2008; Sy et al., 2008), knockdown of Beclin1 promoted apoptosis as assessed by flow cytometry (Figure 2B). However, knockdown of ATG5 in vitro also inhibited apoptosis (Figure 2B) (Yousefi et al., 2006). To further confirm that HMGB1 release is associated with autophagy, we treated cells with phosphatidylinositol-3 kinase (PI-3K) inhibitors including 3-methyladenine (3MA), a wide-used autophagy inhibitor. Indeed, 3MA limited autophagy as well as HMGB1 release and increased apoptosis (Figure 2B–D). Our findings suggest that autophagic stimuli regulate HMGB1 release.
Figure 2. Inhibition of autophagy diminishes HMGB1 release and enhances selective apoptosis.
(A) Immunoblots are shown for Beclin1 and ATG5 knockdown performed in Panc02 cells. (B–D) Panc02 cells as indicated were treated with the anticancer agents (melphalan, “ME”, 160 μg/ml; paclitaxel, “PA”, 10 μg/ml) for 6 h. and then assayed for early apoptosis (annexin V+/PI−) by flow cytometry (B), autophagy by quantification of the percentage of cells with GFP-LC3 punctae (C) and HMGB1 release, by western blotting analysis (LDH and H3 were both used as controls for protein leakage from damaged cells) (D). PI3-kinase inhibitor 3-methyladenine (3MA, 5mm) was used as a nominal autophagy inhibitor. Representative western blots of the indicated proteins are presented.
Anticancer agents promote HMGB1 release and autophagy and limit apoptosis
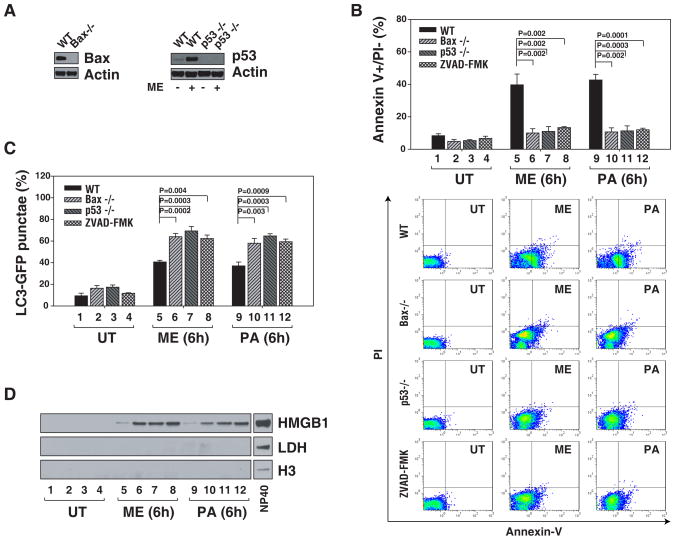
Bax and p53 are key proteins in the regulation of apoptosis following stress (Zhang et al., 2000). To further characterize the relationship between HMGB1 release and cell death, we treated apoptosis-defective Bax−/− and p53−/− HCT116 cancer cells with anticancer agents. The absence of Bax or p53 in the well-characterized HCT116 colorectal cancer cell line (Figure 3A) completely abolished the apoptotic response to melphalan or paclitaxel (Figure 3B). However, HMGB1 release and GFP-LC3 punctae were increased in Bax−/− and p53−/− HCT116 cells following treatment (Figure 3C and D). A necrotic marker, LDH was not detected in supernatants when cells were treated for 6 h, suggesting that there is an alternative mechanism responsible for HMGB1 release. Furthermore, pretreatment with the pancaspase inhibitor ZVAD-FMK promoted accumulation of GFP-LC3 punctae (Figure 3C) and associated HMGB1 release (Figure 3D).
Figure 3. HMGB1 release and autophagy is detected in the absence of measurable apoptosis.
(A) Immunoblots are shown for Bax and p53 knockout in HCT116 cells. (B-D) WT, Bax knock out, p53 knockout or pan-caspase inhibitor treated (ZVAD-FMK, 20 μm) HCT116 cells were treated with melphalan, “ME”, 160 μg/ml or paclitaxel, “PA”, 10 μg/ml for 6 h. and then assayed for measures of early apoptosis (annexin V+/PI−) by flow cytometry (B), autophagy by quantification of the percentage of cells with GFP-LC3 punctae (C) and HMGB1 release by western blot analysis (LDH and H3 were both used as controls for protein leakage from damaged cells) (D). Representative western blots of the indicated proteins are presented.
Inhibition of HMGB1 release promotes apoptosis and limits autophagic flux
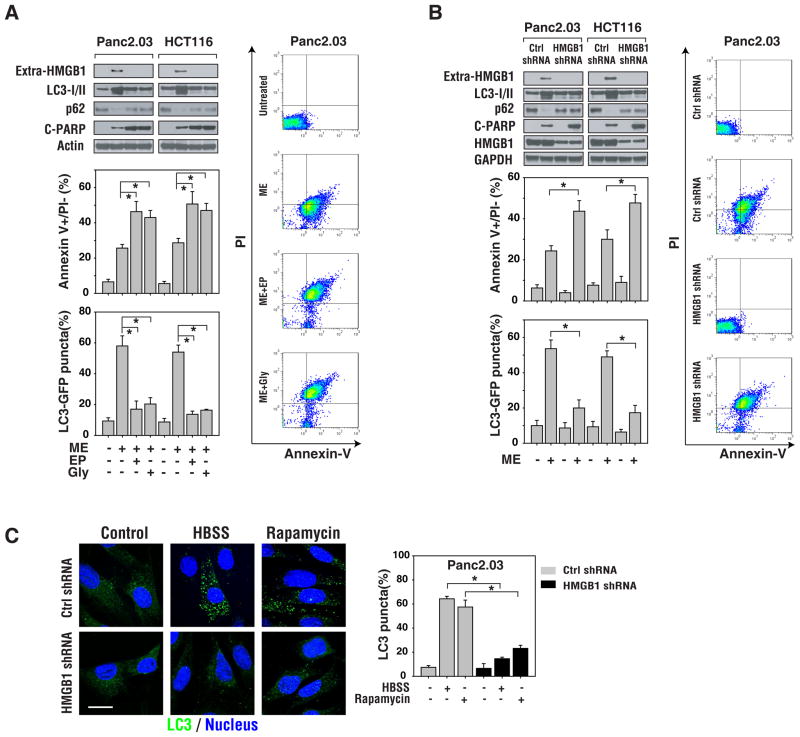
Ethyl pyruvate (“EP”) and glycyrrhizin (“Gly”) were identified as experimental inhibitors of HMGB1 release and activity (Mollica et al., 2007; Ulloa et al., 2002). To examine whether inhibition of HMGB1 release sensitizes cancer cells to cytotoxic agents, we pretreated cells with ethyl pyruvate or glycyrrhizin. These inhibitors attenuated HMGB1 release induced by melphalan (Figure 4A). Interestingly, inhibition of HMGB1 release in Panc2.03 and HCT-116 tumor cells treated with chemotherapy decreased autophagy as assessed by GFP-LC3 punctae and LC3-II/p62 western blot analysis and increased apoptosis as determined by flow cytometry and cleaved PARP assays (Figure 4A).
Figure 4. Inhibition of HMGB1 release increases tumor cell sensitivity to anticancer agents.
(A) Inhibition of HMGB1 release with small molecule drugs increases tumor cell sensitivity to anticancer agents. Panc2.03 and HCT116 cells were pretreated with the HMGB1-release inhibitors ethyl pyruvate (EP, 10 mm) or glycyrrhizin (Gly, 500 μm) for 2 h and then cultured in the presence of melphalan for an additional 24 h. Representative western blotting analysis of protein levels are presented. In parallel, measures of apoptosis (annexin V+/PI−) were assayed by flow cytometry and autophagy by quantifying the percentage of cells with GFP-LC3 punctae. (B) Panc2.03 and HCT116 cells were knocked down for HMGB1 using shRNA for 48 h, and then stimulated with melphalan for 24 h. Representative western blotting analysis of protein levels is presented. In parallel, apoptosis (annexin V+/PI−) was assayed by flow cytometry (right panel) and autophagy by quantifying the percentage of cells with GFP-LC3 punctae (N=3, p<0.01). Representative FACS plots are presented. (C) HMGB1 was knocked down in Panc2.03 using shRNA for 48 h, and then these cells were stimulated with starvation (HBSS, 3 h) and rapamycin (1 μm, 6 h). Autophagy was evaluated using the percentage of cells with LC3 punctae (N=3, p<0.01). Representative image are presented. Bar=20 μm.
To further characterize the role of HMGB1 release in cancer cells following chemotherapy, a target-specific HMGB1 shRNA (Figure 4B) or HMGB1 neutralizing antibody (data not shown) was used in Panc2.03 and HCT116 tumor cells. Inhibition of HMGB1 release by shRNA or antibody in these cells rendered them significantly more sensitive to melphalan-induced apoptotic cell death, which was also associated with lower levels of autophagy (Figure 4B). Moreover, knockdown of HMGB1 in Panc2.03 cells inhibited serum starvation (e.g. HBSS) and rapamycin-induced accumulation of LC3 punctae (Figure 4C), supporting a critical role for HMGB1 in the regulation of autophagy.
Provision of exogenous reduced HMGB1 increases autophagy
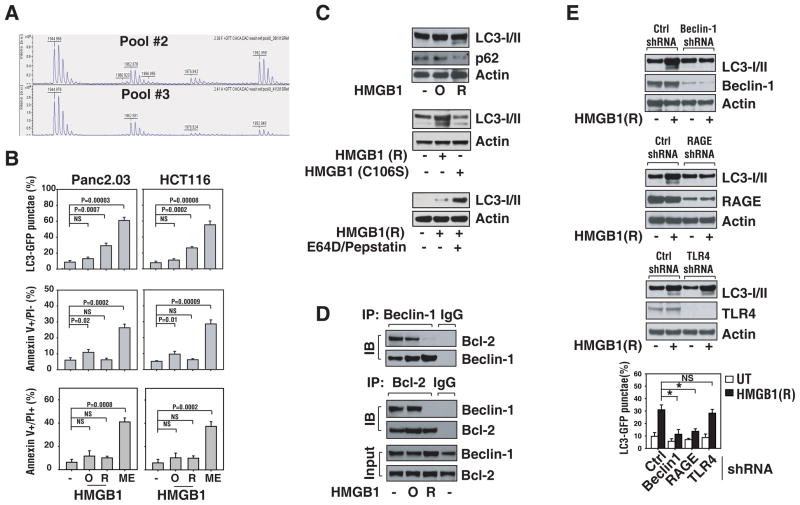
To further characterize the role of HMGB1 release in the setting of autophagy, we treated Panc2.03 and HCT116 cancer cells with recombinant HMGB1 proteins, which has been assessed for the oxidation state of the Cysteine residues in HMGB1 pools #2 and #3. HMGB1 contains three cysteine residues at positions 23, 45 and 106. To determine the relative oxidation states of these residues, tyrptic fragments were prepared from a solution digest of HMGB1 pool #2 and pool #3 preparations. A portion of the digest was subjected to reduction with DTT and was compared to the non-reduced digest and analyzed by MALDI-TOF mass spectrometry (Figure 5A). In the absence of DTT, both HMGB1 pool #2 and pool #3 digests yielded fragments indicative of Cys 23–45 crosslinks which were capable of being reduced with DTT (not shown). In addition, both pool #2 and pool #3 preparations displayed essentially the same relative ratios of Cys 23 involved in disulfide bonding as compared to the total (disulfide-linked + free sulfhydryl). One major difference between HMGB1 pool #2 and pool #3 fractions was evident in the oxidation state of Cys 106. Nearly half of the Cys 106 containing fragments (m/z 1944.9, residues 97–112, free sulfhydryl) in the HMGB1 pool #2 preparation were oxidized to the irreversible sulfonic acid (m/z, 1992.9) and were not able to be reduced in the presence of DTT (Figure 5A). In contrast, only 17% of the Cys 106 residues were oxidized to the sulfonic acid form in the pool #3 preparation. Oxidation to the sulfonic acid was confirmed by MS/MS analysis of the m/z 1992.9 mass ion (see materials and methods). Cys 106 has been shown to be critical for HMGB1 nuclear-cytoplasmic shuttling (Hoppe et al., 2006) and thus may account, at least in part, for the observed differences in biological activity between these two preparations.
Figure 5. Provision of exogenous reduced HMGB1 increases autophagy in cancer cells.
(A) Relative amounts of oxidized Cys106 (as Cys sulfonic acid) in Lilly Pool #2 and #3. MALDI-TOF Mass Spectrum of tryptic fragments of Lilly Pool #2 (top) and Pool #3 (bottom). The Cys106 containing fragment is amino acids 97–112. The free sulfhydryl (-SH) of total reducible cysteine is at a mass of 1944.9 Da. The monoxide is faintly seen at a mass of 1960.9 Da. The di- and tri- oxides are at masses of 1976.9 Da and 1992.9 Da, respectively. The peak at 1962.9 Da is the free sulfhydryl of the 13–28 fragments, used as an internal standard to verify the DTT reduction went to completion. (B) Reduced HMGB1 protein induces autophagy and oxidized HMGB1 mildly induces apoptosis. Panc2.03 and HCT116 cancer cells were treated with oxidized HMGB1 (“O”, 10 μg/ml) or reduced HMGB1 (“R”, 10 μg/ml) for 24 h, and then assayed for apoptosis by FACS using Annexin V/PI stain and autophagy by quantification of the percentage of cells with GFP-LC3 dots as described in methods. (C) Western analysis of LC3 processing in the presence or absence of lysosomal protease inhibitors pepstatin A (10 μg/ml) and E64D (10 μg/ml) and degradation of p62 by autophagy after HMGB1 or HMGB1 C106S mutant treatment. (D) Reduced HMGB1 protein regulates Beclin1/Bcl-2 complex formation in autophagy. Panc2.03 cells were treated with oxidized HMGB1 (“O”, 10 μg/ml) or reduced HMGB1 (“R”, 10 μg/ml), for 6 h, then cell lysates were prepared for IP with anti-Beclin1/-Bcl-2 or IgG. The resulting immune complexes and inputs were analyzed by western blotting as indicated. Representative western blotting analysis of protein levels is presented. (E) RAGE/Beclin1 but not TLR4 is required for HMGB1 mediated autophagy. Cells were transfected with the indicated shRNA for 48 h and then were treated with reduced HMGB1 (“R”, 10 μg/ml) for 24 h. Representative western blotting analysis of protein levels is presented. In parallel, autophagy was assayed by the percentage of cells with GFP-LC3 dots (N=3, * p<0.001).
Treatment with reducible (“R”) but not oxidized HMGB1 (“O”) increased accumulation of GFP-LC3 punctae (Figure 5B), induced LC3-II formation (Figure 5C) and reduced expression of p62 (Figure 5C). Moreover, HMGB1 C106S mutant protein significantly decreased autophagy compared with wild-type reduced HMGB1 protein (Figure 5C). There is further accumulation of LC3-II in the presence of the lysosomal protease inhibitors pepstatin and E64D (Figure 5C), indicating the enhancement of autophagic flux. Conversely, oxidized HMGB1 led to a modest increased apoptosis but not necrosis in cancer cells assessed by flow cytometry (Figure 5B). Pretreatment with inhibitors of caspase-3 and -9 [mitochondrial pathway], but not −8 [non-mitochondrial pathway] inhibited oxidized HMGB1-induced apoptosis in Panc2.03 cells (Figure 6C), suggesting that oxidized HMGB1 actives the mitochondrial apoptosis pathway.
Figure 6. Redox of HMGB1 regulates chemotherapy effectiveness.
(A) Cell viability and apoptosis assay. Panc2.03 and HCT116 cells were treated with oxaliplatin (160 μg/ml), melphalan (320 μg/ml), adriamycin (1.6 μg/ml), paclitaxel (20 μg/ml) with or without oxidized HMGB1 (“O”, 10 μg/ml) or reduced HMGB1 (“R”, 10 μg/ml). Cell death was analysis at indicated time by CCK-8 cell viability assay (n=3, * and #, p< 0.05 versus no HMGB1 group, left panel). In parallel, cell death was assayed by Annexin-V/PI using flow cytometry when Panc2.03 and HCT116 cells were exposed to paclitaxel (20 μg/ml) for 48 h (right panel). (B) Colony formation assay. Panc2.03 and HCT116 cells were treated with oxaliplatin (160 μg/ml), melphalan (320 μg/ml), adriamycin (1.6 μg/ml), paclitaxel (20 μg/ml) with or without oxidized HMGB1 (“O”, 10 μg/ml) or reduced HMGB1 (“R”, 10 μg/ml) for 24 h or 72 h, then 1, 000 cells were plated into 24 well plates. Colonies were visualized by crystal violet staining 3 weeks later. (C) Effects of caspase inhibitors on oxidized HMGB1-induced caspase 3 activity. Panc2.03 cells were treated with HMGB1 (“O”, 10 μg/ml) with or without a pan-caspase inhibitor (ZVAD-FMK, 20 μM), caspase-3 inhibitor (Z-DEVD-FMK, 20 μM), caspase-8 inhibitor (Z-IETD-FMK, 20 μM) or caspase-9 inhibitor (Z-LEHD-FMK, 20 μM) for 24 h, and then analyzed Caspase 3 activity. (n=3, ** p<0.001, *** p<0.0001). (D) The relationship between HMGB1 release and autophagy. Anticancer agents such as melphalan and paclitaxel promote HMGB1 release. Redox status of the tumor microenvironment and internal environment decides the activity and function of HMGB1. Reduced extracellular HMGB1 binds to the RAGE receptor but not TLR4 and induces Beclin1-dependent autophagy, which in turn promotes tumor survival. In addition, oxidized HMGB1 increases the cytotoxicity of anticancer agents and induces apoptosis via activation of caspase-3 and -9. HMGB1 is involved in the cross-regulation between autophagy and apoptosis.
Another important molecular event in autophagosome formation is the disassociation of the Bcl-2/Beclin1 complex (Pattingre et al., 2005). Only reduced HMGB1 suppressed interaction of Beclin1/Bcl-2 (Figure 5D). Moreover, knockout of Beclin1 by shRNA inhibited reduced HMGB1-induced autophagy (Figure 5E), suggesting that reduced HMGB1 promotes the Beclin1 dependent autophagic pathway.
To determine whether the receptor RAGE and/or TLR4 mediate HMGB1-induced autophagy, a target-specific shRNA against these receptors was transfected into tumor cells. Knockdown of RAGE in cancer cells diminished HMGB1-induced autophagy (Figure 5E). In contrast, there was no affect on HMGB1-induced autophagy in the TLR4 knockdown, suggesting that RAGE is required for HMGB1 promotion of Beclin1-dependent autophagy.
Redox status of HMGB1 regulates the effect of chemotherapy
To examine whether the redox state of the exogenous or released HMGB1 protein modifies the response to chemotherapy, we performed cell viability assays on cancer cells treated with various cytotoxic agents. The reduced form of HMGB1 decreased the effectiveness of many anticancer agents including oxaliplatin, melphalan, adriamycin, and paclitaxel against Panc2.03 and HCT116 cancer cell lines at 12–48 h by CCK8 cell viability assay (Figure 6A). However, the oxidized form of HMGB1 increased cell death following treatment with chemotherapeutic agents. Similarly, oxidized HMGB1 but not reduced HMGB1 increased paclitaxel-induced apoptosis by flow cytometry (Figure 6A). To confirm the role of HMGB1 release during therapy-induced stress in cancer cells, we evaluated colony forming assays. Consistent with the cell viability assays, treatment for 24 h and 72 h with reduced HMGB1 but not oxidized HMGB1 increased long term cellular viability (Figure 6B). Taken together, these data suggest that reduced HMGB1 increases resistance to a variety of cytotoxic agents.
DISCUSSION
Autophagy is a process associated with the degradation of intracellular organelles following sequestration within double-membrane delimited vacuoles. At present, the role of autophagy in tumor cells is not well characterized, particularly when subjected to various stressors including chemotherapy. Here we demonstrate that HMGB1 release and its redox state critically regulates the autophagic response to anticancer agents and influences antitumor efficacy.
Autophagy regulates HMGB1 release in tumor cells. Inhibition of autophagy by genetic manipulation or small molecule inhibitors minimized HMGB1 release. Similarly, autophagy is upregulated in apoptosis-defective tumor cells, which results in greater amounts of HMGB1 release in response to treatment in vitro. Furthermore, inhibition of HMGB1 release by genetic manipulation or small molecule inhibitors limited autophagy and increased the apparent efficacy of anticancer agent-induced cell death. Our previous studies have demonstrated that quercetin and wortmannin inhibit LPS-induced markers of autophagy (LC3-II production and punctae) as well as HMGB1 translocation and release in sepsis (Tang et al., 2009). A diphtheria toxin targeted to the EGF receptor (DT-EGF) kills glioblastoma cells through a caspase-independent mechanism that is associated with high levels of autophagy and HMGB1 release (Thorburn et al., 2009). These findings suggest that HMGB1 is released during autophagy and is a rather universal finding in the cellular response to stress.
Is HMGB1 released during apoptotic cell death? This has been considered possible (Tesniere et al., 2008) but it appears that in normal cells, HMGB1 is released during necrosis, but not apoptosis (Ohndorf et al., 1999; Scaffidi et al., 2002). Two findings have challenged this notion. First, nuclear DNA is released in a time-dependent manner following induction of apoptosis (Choi et al., 2004). Second, during apoptosis there is increased binding of HMGB1 to DNA, consistent with the notion that uningested, late-stage apoptotic cells can release both DNA and HMGB1. Recent studies (Bell et al., 2006; Kazama et al., 2008; Tian et al., 2007) have confirmed that HMGB1 can also be released from apoptotic tumor cells, at least at later stages of dissolution. Although necrotic cells release HMGB1, signaling tissue injury and initiating inflammatory responses (Scaffidi et al., 2002), apoptotic cells produce reactive oxygen species and oxidized HMGB1 released from apoptotic cells promotes tolerance (Kazama et al., 2008). Virtually all stressful stimuli not only induce apoptosis but also autophagy at early stages and promote necrosis at late stages in cancer cells with intrinsic apoptotic defects or with ATP depletion. Early inhibition of apoptosis in Bax−/− and p53−/− HCT116 cells promotes drug induced autophagy as well as HMGB1 release. Importantly, knockdown of ATG5, which decreases autophagy and apoptosis, inhibits HMGB1 release, suggesting that autophagy is a major regulator of HMGB1 localization.
When considering application of anticancer agents, it is important not only to consider the lethal effectiveness but also the characteristics of tumor cell death which we believe determines the long-term effectiveness of the treatment. At present the effect of the HMGB1 protein released from tumor cells and its microenvironment on tumor cell persistence and survival is not well characterized. Here we demonstrate that reducible exogenous HMGB1 protein regulates cell death and survival in tumor cells. Reducible HMGB1 decreases cell injury/death in tumor cells by increasing Beclin1-dependent autophagy whereas provision of oxidized HMGB1 enhanced cell injury/death in response to anticancer agents. This suggests that the local redox state controls HMGB1’s function (Figure 6D).
Oxidative stress occurs when the generation of reactive oxygen species (ROS) in a system exceeds its ability to neutralize and eliminate them. Compared with normal cells, both ROS and autophagy are altered in cancer cells. On one hand, ROS can induce autophagy through several distinct mechanisms involving autophagy-related gene 4 (ATG4), catalase and the mitochondrial electron transport chain (Azad et al., 2009). On the other hand, defective autophagy can increases oxidative stress in tumor cells (Mathew et al., 2009). Moreover, suppressing ROS or p62 accumulation prevents damage resulting from autophagy defects (Mathew et al., 2009). This suggests that autophagy defects may increase the proportion of oxidized HMGB1.
Once present in the extracellular space, HMGB1 can bind to a range of receptors, including RAGE and TLR4 (Kang et al., 2010a; Lotze and Tracey, 2005; Sparvero et al., 2009). For example, if dying tumor cells release the immune modulator HMGB1 following treatment, they can activate a TLR4-dependent tumor-specific immune response that enhances the effectiveness of the initial treatment (Apetoh et al., 2007). We found that RAGE RNAi abolished HMGB1-induced autophagy in cancer cells while TLR4 RNAi had no effect, suggesting that there are separable roles for RAGE and TLR4 in cancer and immune cells in response to anticancer chemotherapy or radiotherapy. Moreover, the involvement of HMGB1/RAGE in the NF-κB pathway has been demonstrated in many studies (Bierhaus et al., 2001; Liliensiek et al., 2004), although the precise mechanism is unknown. Interestingly, a recent study suggests that the IKK complex has a role in the induction of autophagy by physiological and pharmacological stimuli (Criollo et al.), suggesting that IKK has a novel function in regulating autophagy. Thus HMGB1 may active IKK and promote autophagy through RAGE.
In summary, these studies suggest that release of HMGB1 by autophagic cells promotes local cancer cell survival following administration of chemotherapy. Reducible HMGB1 protein induces autophagy in cancer cells that is RAGE/Beclin1-dependent. These findings serve as the basis for a novel therapeutic approach to potentiate cancer treatment efficacy via inhibition of HMGB1 release and resultant autophagy or by enhancing the aerobic denaturation of HMGB1. Future studies to elucidate the molecular mechanisms and impact of HMGB1-mediated autophagy regulating both tumor and immune cells within the tumor microenvironment are areas of great interest and worthy of further study.
MATERIALS AND METHODS
Antibodies
The antibody to HMGB1 was generated as previous described (Ito et al., 2007); The antibodies to cleaved-PARP, Bcl-2, Bax and H3 were obtained from Cell Signaling Technology (Danvers, MA, USA); The antibodies to GAPDH, actin and RAGE were from Sigma (St. Louis, MO, USA); The antibodies to LC3-I/II and Beclin1 were from Novus (Littleton, CO, USA); The antibodies to TLR4 and LDH were from Abcam (Cambridge, MA, USA); Anti-p62 antibody was from Santa Cruz (Santa Cruz, CA, USA); Caspase inhibitors were purchased from Calbiochem (Gibbstown, NJ, USA). Other anticancer agents and inhibitors were from Sigma.
HMGB1
Oxidized (Pool #2) and reducible (Pool #3) recombinant HMGB1 proteins were from Eli Lilly and Company (Indianapolis, IN, USA); the endotoxin content was 1.9 EU/ml for Pool #2 and 3.1 EU/ml for Pool #3. Authentic full length HMGB1 was transiently expressed in human embryonic kidney 293 cells. Cell lysates were collected and passed first over a DEAE sepharose then over a heparin-sepharose Pharmacia XK16 23ml column. The samples were loaded at 10ml/minute and eluted at 8 ml/min with a fraction size of one minute. The two buffers used were A] 30 mM tris, 1 mM CaCl2, pH = 8.0 and B] 30 mM tris, 1 mM CaCl2, 1M NaCl, pH=8.0. For the first 60 minutes, a linear gradient up to 50% B was used, ramping up to 100% B over the next ten minutes, which was maintained for the next five minutes. Pool #2 (fractions 53–60) 387 μg/ml, and Pool #3 (fractions 42–45 and 61–68) 258 μg/ml demonstrated a band with the predicted molecular weight of 27,987.1 Daltons as well as a higher smaller band collapsing to single bands under reducing conditions on a 4–20% Tris-Gly gel. We obtained 60.8 mls of Pool #2 and 92.7 mls of Pool #3. All pools were aliquoted and frozen at −80°C until use. MALDI mass spectrometry revealed that Pool #2 was primarily (>70%) oxidized based on analysis of peptide fragments and Pool #3 was primarily (>70%) reduced. HMGB1 C106S mutant protein was a gift from Dr. Helena Erlandsson Harris (Karolinska Institutet, Sweden).
Measurements of HMGB1 release
HMGB1 released into cell culture supernatants was evaluated using Western blotting as previously described (Scaffidi et al., 2002; Tang et al., 2008; Tang et al., 2007b; Wang et al., 1999) or enzyme-linked immunoabsorbent assay (ELISA) kits from the Shino-Test Corporation (Sagamihara-shi, Kanagawa, Japan) according to the manufacturer’s instructions.
MALDI-TOF mass spectrometry
For MALDI-TOF Mass Spectrometry sample preparation, solid-phase extraction pipette tips (C18 ZipTip) from Millipore were used. Protein sequencing grade trifluoroacetic acid (TFA) was purchased from Fisher Scientific. Mass Spectrometry grade acetonitrile (MeCN) and alpha-cyano-4-hydroxycinnamic acid (CHCA) were purchased from Sigma-Aldrich. ACS reagent grade dithiothreitol (DTT), ammonium citrate and ammonium bicarbonate were purchased from Sigma-Aldrich. Modified trypsin was purchased from Promega as Gold grade (proteomics quality). The UPCI Clinical Proteomics Facility Bruker Ultraflex II Mass Spectrometer (nitrogen laser, 337nm) was used in either reflector-positive or LIFT (MS/MS) mode as appropriate.
To determine the oxidation state of HMGB1, three samples each of HMGB1 Pool #2 and #3 were diluted to 25ng/uL in 25mM ammonium bicarbonate, pH7.4, and digested at 37’C overnight with trypsin (100:1 ratio protein:trypsin). Two 50uL aliquots from each digest where acidified with 5% TFA and passed through ZipTips as per the manufacturer’s directions to bind tryptic peptides. Each tip was washed once with 20uL water to remove residual salts. One tip from each digest was incubated (100mM DTT in ammonium bicarbonate) at 56’C for 1hr to reduce the available cysteines entirely to sulfhydryls. Both tips from each digest were washed four times with 0.1% TFA and eluted with 3uL 50/0.1 MeCN/TFA. Each elutant was mixed 1:1 with CHCA solution (10mg/mL) in the same solvent, centrifuged very briefly, then applied to a MALDI target plate in 0.75uL drops and allowed to air-dry. Once dry, they were on-plate washed with 2uL of ice cold 5mM ammonium citrate.
The MALDI-TOF Mass Spectrometer was recalibrated with a peptide standard before each analysis. Each MS sample was acquired as the sum of 200 shots from each five different regions within the same spot. Tryptic peptides were identified with the assistance of the Protein Prospector software suite (Baker, P.R. and Clauser, K.R., http://prospector.ucsf.edu). MS/MS spectra were acquired with varying laser intensities depending on fragmentation pattern and a sum total of 12,000 shots.
Cell culture
Human pancreatic cancer Panc2.03 cells were from American Type Culture Collection (USA) and human colon cancer HCT116 cells were a kind gift of Dr. Bert Vogelstein [Baltimore]. Mouse pancreatic cancer Panc02 cells were from National Cancer Institute (USA). HCT116 Bax−/− and p53−/− cells were kind gifts of Dr. Lin Zhang (Department of Pathology, University of Pittsburgh). These cells were cultured in RPMI 1640 or McCoy’s 5a medium supplemented with 10% heat-inactivated FBS, 2 mM glutamine and antibiotic-antifungal mix in a humidified incubator with 5% CO2 and 95% air.
Western blotting
Whole cell lysate were resolved on 4–12% Criterion XT Bis-Tris gels (Bio-Rad, Hercules, CA, USA) and transferred to a nitrocellulose membrane as previously described (Tang et al., 2007a; Tang et al., 2007b). After blocking, the membrane was incubated for 2 h at 25°C or overnight at 4°C with various primary antibodies respectively. After incubation with peroxidase-conjugated secondary antibodies for 1 h at 25°C, the signals were visualized by enhanced chemiluminescence (Pierce, Rockford, IL, USA) according to the manufacturer’s instruction. Levels of HMGB1 in the culture medium were determined by Western blotting analysis as previously described (Tang et al., 2007a; Tang et al., 2007b; Tang et al., 2005; Tang et al., 2007c; Wang et al., 1999).
RNAi by shRNA
RAGE-shRNA, Beclin1-shRNA, ATG-5 shRNA, HMGB1-shRNA, TLR4 shRNA, and control shRNA (from Sigma, USA) were transfected into cells using Lipofectamine 2000 reagent (Invitrogen, San Diego, CA, USA) according to the manufacturer’s instructions. At the end of the shRNA treatment (48–72 h), the medium over the cells was change before the addition of a chemotherapy agent.
Immunoprecipitation analysis
Cells were lysed at 4°C in ice-cold RIPA lysis buffer (Millipore, Billerica, MA, USA), and cell lysates were cleared by a brief centrifugation (12,000 g, 10 min). Concentrations of proteins in the supernatant were determined by BCA assay. Prior to immunoprecipitation, samples containing equal amounts of proteins were pre-cleared with Protein A or protein G agarose/sepharose (Millipore) (4°C, 3 h) and subsequently incubated with various irrelevant IgG or specific antibodies (5 μg/mL) in the presence of protein A or G agarose/sepharose beads for 2 h or overnight at 4°C with gently shaking (Kang et al., 2010b; Tang et al., 2007a; Tang et al., 2007b). Following incubation, agarose/sepharose beads were washed extensively with PBS and proteins eluted by boiling in 2 × SDS sample buffer before SDS-PAGE electrophoresis.
Apoptosis assays
Apoptosis in cells was assessed using the BD Pharmingen (San Jose, CA, USA) FITC Annexin V Apoptosis Detection Kit (Annexin V-FITC, Propidium Iodide (PI) solution and Annexin V binding buffer). This assay involves staining cancer cells with Annexin V-FITC (a phospholipid-binding protein binding to disrupted cell membranes) in combination with propidium iodide (PI, a vital dye binding to DNA penetrating into apoptotic cells). Flow cytometric analysis was performed on cancer cells that were in early apoptosis (annexin V+/PI−) or late apoptosis/necrosis (annexin V+/PI+) phase (Geft et al., 2008). Caspase-3 activity assays were performed using a Caspase-3 Colorimetric Assay Kit from Calbiochem. Cleaved-PARP was measured by western blotting analysis.
Autophagy assays
Formation of autophagic vesicles was monitored by GFP-LC3 (gifts of Dr. Xiao-Ming Yin, Department of Pathology, University of Pittsburgh) or endogenous LC3 aggregation in cell lines. The percentage of cells with LC3 dots was quantified by assessing 50 randomly chosen cells from three separate experiments. Autophagic flux assays were performed by western blotting for LC3-II formation and p62 expression, after treatment with HMGB1 protein in the presence or absence of lysosomal protease inhibitors (E64d/pepstatin A) (Mizushima and Yoshimori, 2007).
Cell viability and cell survival assay
Cells were plated at a density of 2×104 cells/well on 96-well plates in 100 μl RPMI. Cell viability was determined by WST-8 (2-(2-methoxy-4-nitrophenyl) - 3 - (4-nitrophenyl) - 5 - (2, 4-disulfophenyl) - 2 H - tetrazolium, monosodium salt), assay using a Cell Counting Kit - 8 (CCK-8) from Dojindo Laboratories (Tokyo, Japan) according to the manufacturer’s instructions (Hamamoto et al., 2004). In parallel, trypan blue exclusion test of cell viability was also used (data not included). Long term cell survival was monitored by colony formation assay. In brief, 1000 cells treated with chemotherapeutic drugs for 24 h were plated into 24 well plates. Colonies were visualized by crystal violet staining 3 weeks later (Wang et al., 2007).
Statistical analysis
Data are expressed as means ± SEM of three independent experiments performed in triplicate. One-way ANOVA was used for comparison among the different groups. When the ANOVA was significant, post hoc testing of differences between groups was performed using LSD test. In general, a p-value < 0.05 was considered significant.
Acknowledgments
This project was funded by a grant from the NIH 1 P01 CA 101944-04 (Michael T. Lotze) Integrating NK and DC into Cancer Therapy from National Cancer Institute. Thoughtful discussions and review of this work with. Timothy Billiar and Sarah Berman at the University of Pittsburgh and external colleagues, Guido Kroemer, Douglas Green, Matthew Albert, and Beth Levine are much appreciated.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- Amaravadi RK, Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res. 2007;13:7271–9. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–94. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–9. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- Azad MB, Chen Y, Gibson SB. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxid Redox Signal. 2009;11:777–90. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- Bell CW, Jiang W, Reich CF, 3rd, Pisetsky DS. The extracellular release of HMGB1 during apoptotic cell death. Am J Physiol Cell Physiol. 2006;291:C1318–25. doi: 10.1152/ajpcell.00616.2005. [DOI] [PubMed] [Google Scholar]
- Bierhaus A, Schiekofer S, Schwaninger M, Andrassy M, Humpert PM, Chen J, et al. Diabetes-associated sustained activation of the transcription factor nuclear factor-kappaB. Diabetes. 2001;50:2792–808. doi: 10.2337/diabetes.50.12.2792. [DOI] [PubMed] [Google Scholar]
- Choi JJ, Reich CF, 3rd, Pisetsky DS. Release of DNA from dead and dying lymphocyte and monocyte cell lines in vitro. Scand J Immunol. 2004;60:159–66. doi: 10.1111/j.0300-9475.2004.01470.x. [DOI] [PubMed] [Google Scholar]
- Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, et al. The IKK complex contributes to the induction of autophagy. Embo J. 29:619–31. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geft D, Schwartzenberg S, Rogowsky O, Finkelstein A, Ablin J, Maysel-Auslender S, et al. Circulating apoptotic progenitor cells in patients with congestive heart failure. PLoS ONE. 2008;3:e3238. doi: 10.1371/journal.pone.0003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–40. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- Han J, Hou W, Goldstein LA, Lu C, Stolz DB, Yin XM, et al. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J Biol Chem. 2008;283:19665–77. doi: 10.1074/jbc.M710169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe G, Talcott KE, Bhattacharya SK, Crabb JW, Sears JE. Molecular basis for the redox control of nuclear transport of the structural chromatin protein Hmgb1. Exp Cell Res. 2006;312:3526–38. doi: 10.1016/j.yexcr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Ito N, Demarco RA, Mailliard RB, Han J, Rabinowich H, Kalinski P, et al. Cytolytic cells induce HMGB1 release from melanoma cell lines. J Leukoc Biol. 2007;81:75–83. doi: 10.1189/jlb.0306169. [DOI] [PubMed] [Google Scholar]
- Kang R, Tang D, Schapiro NE, Livesey KM, Farkas A, Loughran P, et al. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death Differ. 2010a;17:666–76. doi: 10.1038/cdd.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Tang D, Yu Y, Wang Z, Hu T, Wang H, et al. WAVE1 regulates Bcl-2 localization and phosphorylation in leukemia cells. Leukemia. 2010b;24:177–86. doi: 10.1038/leu.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama H, Ricci JE, Herndon JM, Hoppe G, Green DR, Ferguson TA. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity. 2008;29:21–32. doi: 10.1016/j.immuni.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–10. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–7. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- Liliensiek B, Weigand MA, Bierhaus A, Nicklas W, Kasper M, Hofer S, et al. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J Clin Invest. 2004;113:1641–50. doi: 10.1172/JCI18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–42. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Mollica L, De Marchis F, Spitaleri A, Dallacosta C, Pennacchini D, Zamai M, et al. Glycyrrhizin binds to high-mobility group box 1 protein and inhibits its cytokine activities. Chem Biol. 2007;14:431–41. doi: 10.1016/j.chembiol.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Muller S, Scaffidi P, Degryse B, Bonaldi T, Ronfani L, Agresti A, et al. New EMBO members’ review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. Embo J. 2001;20:4337–40. doi: 10.1093/emboj/20.16.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–12. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–5. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- Sparvero LJ, Asafu-Adjei D, Kang R, Tang D, Amin N, Im J, et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J Transl Med. 2009;7:17. doi: 10.1186/1479-5876-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy LK, Yan SC, Lok CN, Man RY, Che CM. Timosaponin A-III induces autophagy preceding mitochondria-mediated apoptosis in HeLa cancer cells. Cancer Res. 2008;68:10229–37. doi: 10.1158/0008-5472.CAN-08-1983. [DOI] [PubMed] [Google Scholar]
- Tang D, Kang R, Cao L, Zhang G, Yu Y, Xiao W, et al. A pilot study to detect high mobility group box 1 and heat shock protein 72 in cerebrospinal fluid of pediatric patients with meningitis. Crit Care Med. 2008;36:291–295. doi: 10.1097/01.CCM.0000295316.86942.CE. [DOI] [PubMed] [Google Scholar]
- Tang D, Kang R, Xiao W, Jiang L, Liu M, Shi Y, et al. Nuclear Heat Shock Protein 72 as a Negative Regulator of Oxidative Stress (Hydrogen Peroxide)-Induced HMGB1 Cytoplasmic Translocation and Release. J Immunol. 2007a;178:7376–7384. doi: 10.4049/jimmunol.178.11.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Xiao W, Wang H, Calderwood SK, Xiao X. The Anti-inflammatory Effects of Heat Shock Protein 72 Involve Inhibition of High-Mobility-Group Box 1 Release and Proinflammatory Function in Macrophages. J Immunol. 2007b;179:1236–1244. doi: 10.4049/jimmunol.179.2.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Xiao W, Zhang H, Lotze MT, Wang H, et al. Quercetin prevents LPS-induced high-mobility group box 1 release and proinflammatory function. Am J Respir Cell Mol Biol. 2009;41:651–60. doi: 10.1165/rcmb.2008-0119OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Zeh HJ, 3rd, Lotze MT. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799:131–140. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Shi Y, Jang L, Wang K, Xiao W, Xiao X. Heat shock response inhibits release of high mobility group box 1 protein induced by endotoxin in murine macrophages. Shock. 2005;23:434–40. doi: 10.1097/01.shk.0000159556.95285.df. [DOI] [PubMed] [Google Scholar]
- Tang D, Shi Y, Kang R, Li T, Xiao W, Wang H, et al. Hydrogen peroxide stimulates macrophages and monocytes to actively release HMGB1. J Leukoc Biol. 2007c;81:741–7. doi: 10.1189/jlb.0806540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesniere A, Panaretakis T, Kepp O, Apetoh L, Ghiringhelli F, Zitvogel L, et al. Molecular characteristics of immunogenic cancer cell death. Cell Death Differ. 2008;15:3–12. doi: 10.1038/sj.cdd.4402269. [DOI] [PubMed] [Google Scholar]
- Thorburn J, Horita H, Redzic J, Hansen K, Frankel AE, Thorburn A. Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death Differ. 2009;16:175–83. doi: 10.1038/cdd.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–96. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- Ulloa L, Ochani M, Yang H, Tanovic M, Halperin D, Yang R, et al. Ethyl pyruvate prevents lethality in mice with established lethal sepsis and systemic inflammation. Proc Natl Acad Sci U S A. 2002;99:12351–6. doi: 10.1073/pnas.192222999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285:248–51. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- Wang P, Yu J, Zhang L. The nuclear function of p53 is required for PUMA-mediated apoptosis induced by DNA damage. Proc Natl Acad Sci U S A. 2007;104:4054–9. doi: 10.1073/pnas.0700020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–16. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu J, Park BH, Kinzler KW, Vogelstein B. Role of BAX in the apoptotic response to anticancer agents. Science. 2000;290:989–92. doi: 10.1126/science.290.5493.989. [DOI] [PubMed] [Google Scholar]