Abstract
Castellaniella (ex Alcaligenes) defragrans strain 65Phen mineralizes monoterpenes in the absence of oxygen. Soluble cell extracts anaerobically catalyzed the isomerization of geraniol to linalool and the dehydration of linalool to myrcene. The linalool dehydratase was present in cells grown on monoterpenes, but not if grown on acetate. We purified the novel enzyme ∼1800-fold to complete homogeneity. The native enzyme had a molecular mass of 160 kDa. Denaturing gel electrophoresis revealed one single protein band with a molecular mass of 40 kDa, which indicated a homotetramer as native conformation. The aerobically purified enzyme was anaerobically activated in the presence of 2 mm DTT. The linalool dehydratase catalyzed in vitro two reactions in both directions depending on the thermodynamic driving forces: a water secession from the tertiary alcohol linalool to the corresponding acyclic monoterpene myrcene and an isomerization of the primary allylalcohol geraniol in its stereoisomer linalool. The specific activities (Vmax) were 140 nanokatals mg−1 for the linalool dehydratase and 410 nanokatals mg−1 for the geraniol isomerase, with apparent Km values of 750 μm and 500 μm, respectively. The corresponding open reading frame was identified and revealed a precursor protein with a signal peptide for a periplasmatic location. The amino acid sequence did not affiliate with any described enzymes. We suggest naming the enzyme linalool dehydratase-isomerase according to its bifunctionality and placing it as a member of a new protein family within the hydrolyases (EC 4.2.1.X).
Keywords: Enzyme Catalysis, Enzyme Kinetics, Enzyme Purification, Membrane Enzymes, Metabolism, Alkene Hydratase, Allylalcohol Isomerase, Geraniol, Monoterpene, Myrcene
Introduction
Monoterpenes constitute a large and extremely diverse group of natural compounds within the isoprenoids (1–3). They are synthesized from two five-carbon units of isopentenyl pyrophosphate, a derivative of isoprene. This central intermediate is formed in two alternative pathways. In the mevalonate-dependent route, isopentenyl pyrophosphate is synthesized from acetyl-CoA via mevalonic acid. It represents an important cellular metabolic pathway in all higher eukaryotes, Archaea and many Bacteria. (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate is the central precursor in the nonmevalonate pathway utilized by algae, the plastids of higher plants and some Bacteria (4, 5).
The monoterpenes are divided into acyclic compounds, such as myrcene (7-methyl-3-methylene-1,6-octadiene) and ocimene, monocyclic monoterpenes, e.g. limonene and phellandrene, and bicyclic monoterpenes, e.g. pinene and sabinene. These unsaturated hydrocarbons are classified as highly volatile organic compounds. Plants as major producers emit more than 100 million tons/year to the atmosphere (6) where they are photooxidized and contribute to aerosol formation (7, 8). An example of physiological function is as defense against herbivores: plants often induce the synthesis of monoterpenes as repellents upon insect damage (9).
The mineralization of monoterpenes by aerobic microorganisms has been studied in detail with Pseudomonas species (10, 11). The aerobic metabolism depends on oxygenases that catalyze hydroxylation reactions with molecular oxygen as co-substrate (12). In the absence of oxygen, alternative biochemical pathways have been identified for hydrocarbon-mineralizing bacteria. Alkanes, e.g. n-hexane, and aromatic hydrocarbons with alkyl substituents, e.g. toluene, are anaerobically activated by glycine radical enzymes, and the radical intermediates add to fumarate, yielding methylalkylsuccinate and benzylsuccinate, respectively (13–15). Molybdenum-containing enzymes anaerobically hydroxylate ethylbenzene (16) and cholesterol (17).
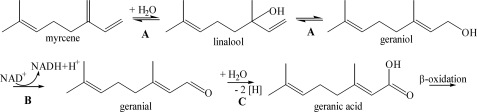
For monoterpenes, no pathway has been elucidated so far. The anaerobic mineralization of monoterpenes to carbon dioxide is frequently present in denitrifying bacteria (18). Cultivation approaches established the enrichment of monoterpene-mineralizing microorganisms (19) and the isolation of strains of Alcaligenes defragrans (20) and Thauera terpenica (21). A. defragrans was recently placed in the newly defined genus Castellaniella, as C. defragrans (22). Initial studies on potential metabolites of the degradation pathway identified isoterpinolene as metabolite that was apparently not further metabolized (23) and geranic acid as ionic intermediate present in nitrate-respiring cells that were grown on acyclic or cyclic monoterpenes, e.g. myrcene or limonene (24).
A simple pathway hypothesis is a hydration of myrcene, leading to geraniol and further to geranic acid (Fig. 1). We initiated biotransformation studies with soluble extracts of C. defragrans. In this article we report on the detection of novel enzyme activities and the isolation and characterization of an anaerobic linalool dehydratase-isomerase, a bifunctional enzyme that catalyzes the reversible dehydration and isomerization of linalool (3,7-dimethyl-1,6-octadien-3-ol) (Fig. 1).
FIGURE 1.
Proposed anaerobic transformation of myrcene in C. defragrans. A, linalool dehydratase-isomerase; B, geraniol dehydrogenase; C, geranial dehydrogenase.
EXPERIMENTAL PROCEDURES
Reagents
R-Limonene (95%), myrcene (90%), linalool (99%), and geraniol (98%) were purchased from Sigma-Aldrich. All other chemicals used were of the highest available purity and were purchased from Aldrich, Boehringer, Fluka (Neu-Ulm, Germany), Merck, Sigma, and Bio-Rad Laboratories. Gases (CO2 grade 4.8, N2 grade 5.0, and O2 grade 2.0) were supplied by Air Liquide (Düsseldorf, Germany). Chromatography media and instruments were from GE Healthcare.
Cell Growth and Preparation of Soluble Extracts
C. defragrans strain 65Phen was maintained as described (20). For biomass production, the strain was cultivated on 30 mm limonene and 100 mm nitrate (24). A 1-liter preculture was inoculated in a 10-liter vessel of carbonate-buffered mineral salt medium at pH 7.0. Filter-sterilized limonene and vitamins (25) were added after cooling, and the culture was incubated for 6–7 days with a CO2/N2 (10/90 (v/v)) gas stream of 24 ml h−1 at 28 °C. The stirrer frequency was initially 150 rpm and was increased during exponential growth phase of C. defragrans up to 250 rpm to ensure optimal substrate availability.
Cell harvest began after the addition of reducing agents, 50 μm Fe(II)Cl2 and 2 mm DTT. Cells in the late exponential growth phase (A600 ≈ 3) were transferred by gas pressure to centrifuge tubes and then collected by centrifugation for 15 min at 9000 × g at 4 °C. For the preparation of the soluble proteins, 40 g of wet or frozen cells were suspended in 60 ml of 25 mm sodium phosphate buffer, pH 8.0, containing 2 mm DTT and disintegrated in two passages through a French pressure cell press (Amincon, Rochester, NY) at 10.3 MPa. The soluble fraction was obtained by ultracentrifugation for 90 min at 150,000 × g at 4 °C to remove cell debris, unbroken cells, and membrane proteins.
Assays for Geraniol Isomerization and Linalool Dehydration
Salt or urea containing linalool dehydratase fractions were dialyzed three times against a 1000-fold volume of 80 mm Tris-HCl buffer, pH 9.0, for 20 min at 4 °C and under magnetic stirring. Purified and dialyzed linalool dehydratase fractions were stored under an anoxic gas phase at 4 °C.
Geraniol isomerization and linalool dehydration were assayed routinely in a two-phase system. Vials (17 × 38 mm; Zinsser Analytic, Frankfurt, Germany) were prewarmed at 35 °C. Anoxic protein solution was transferred into the vials, and DTT was added to 2 mm. The tests were sealed with a butyl septum, and the headspace was flushed with CO2/N2 (10/90 (v/v)). The reaction was started by adding a distinct linalool or geraniol concentration to investigate the reaction to myrcene. 10–100 mm organic substrate was dissolved in 2,2,4,4,6,8,8-heptamethylnonane (HMN).2 The organic phase was added in a 1:1 ratio to the aqueous protein solution. Kinetic parameters were determined in a one-phase system with 10% (v/v) DMSO. The reaction was started by adding monoterpene (0.1–10 mm) that was dissolved in anoxic 80 mm Tris-HCl buffer, pH 9.0, with 10% DMSO. In a third assay system, a pure myrcene phase (1/2 (v/v) myrcene/Tris-HCl buffer) was applied for measuring linalool and subsequently geraniol formation. The tubes were immediately transferred into a 35 °C shaking incubator. For kinetic analyses and the myrcene turnover, aqueous samples were taken at different time points and directly injected into the GC. In the two-phase system, 1 μl of the organic HMN carrier phase was injected to determine the substrate and product concentration.
To estimate the effect of temperature on linalool dehydratase activity, the two-phase assay was performed at temperatures between 4 °C and 45 °C. The pH optimum was tested by varying the buffer systems with pH values near the specific pKa values at 35 °C. The two-phase assay was also used to determine the influence of different effectors on enzyme activities.
The concentrations of the monoterpenes were analyzed by GC (Auto System XL; PerkinElmer Life Sciences) equipped with an Optima®-5 (0.25-μm film thickness, 50 m × 0.32-mm inner diameter; Macherey-Nagel, Düren, Germany) column and flame ionization detector. The following temperature program was applied: injection port temperature, 250 °C; column start temperature, 85 °C for 1 min, increasing to 120 °C at a rate of 5 °C min−1, 120 °C for 0.1 min, increasing to 290 °C at a rate of 45 °C min−1, 290 °C for 1 min; detection temperature, 350 °C. The split ratio was set to 1:25.
Purification of Linalool Dehydratase
The purification was performed with an Äkta system (GE Healthcare). All purification procedures were carried out at 4 °C with filtered (0.2 μm) and degassed buffers. 100 ml of soluble extract obtained from cells grown on limonene was applied to a Source 30Q column (5 × 30 cm) equilibrated in 50 mm sodium phosphate, pH 8.0 (AIE-Q1). The enzyme eluted at 200 mm NaCl in the aforementioned buffer during a stepwise gradient performed with 3.5 ml min−1. Fractions containing linalool dehydratase were pooled, and saturated ammonium sulfate solution was added to a final concentration of 15% (v/v). The protein solution was applied to a Butyl-Sepharose FF column (4.7 ml) preequilibrated with 15% (v/v) saturated ammonium sulfate in 80 mm Tris-HCl, pH 8.0. After a first elution with 80 mm Tris-HCl, pH 8.0, the target enzyme was eluted with 6 m urea. The urea fraction, typically 20 ml, was mixed with saturated ammonium sulfate solution to a final concentration of 40% (v/v). After centrifugation for 10 min at 20,000 × g and 20 °C, the supernatant was withdrawn, and the pellet was solved in 2 ml of 100 mm Tris-HCl, pH 8.0. The concentrated solution was passed through a SuperdexTM 200-pg column (120 ml) equilibrated with 10 mm Tris-HCl, pH 9.0. The active fractions from the gel filtration were applied to a second anion exchange chromatography with a different column material (ResourceQ, 1 ml) that was preequilibrated with 10 mm Tris-HCl, pH 7.0. The enzyme eluted at 120 mm NaCl in a step gradient performed with 2 ml min−1 (AIE-Q2).
Determination of Relative Molecular Mass
The apparent relative molecular mass of the native enzyme was determined by gel filtration on a SuperdexTM 200-pg column (120 ml) in 80 mm Tris-HCl buffer, pH 9.0, The standard proteins were: catalase (232 kDa), aldolase (158 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), and chymotrypsinogen A (25 kDa). The molecular mass of the monomeric enzyme was determined using a 12% SDS-polyacrylamide gel stained with Coomassie Blue R-250 (26). The protein ladder (Fermentas, St. Leon-Rot, Germany) covered a range molecular masses from 10 to 170 kDa.
UV-Visible Spectroscopy
Standards and purified linalool dehydratase fractions were dissolved in 80 mm Tris-HCl, pH 9.0, in the range between 10 and 100 μg ml−1. UV absorption spectra were obtained by using a DU 600 UV-visible spectrophotometer (Beckman Coulter, Krefeld, Germany).
Protein Determination
The protein content was measured by Coomassie Blue R-250 protein assay (27) and by using bovine serum albumin as the standard.
N-terminal Amino Acid Sequence Analysis
Purified linalool dehydratase was separated by 12% SDS-PAGE and electroblotted on a PVDF membrane (Sequi-Blot; Bio-Rad Laboratories) according to the method of Towbin et al. (28). The membrane was washed for 1 min with distilled water and stained with Coomassie Blue R-250 (0.025% (v/v) in 40% (v/v) methanol) for 30 s before destaining with a water/methanol/acetic acid mixture (50/45/5, v/v/v). The PVDF membrane was washed with distilled water and dried for 6 h. The protein was excised from the membrane, and Edman degradation of the N-terminal amino acid residues was performed by Toplab GmbH (Martinsried, Germany). The gene and the protein sequence were deposited at GenBank under accession no. FR669447.
Overexpression in Escherichia coli
Standard molecular biology methods were applied. In short, the ldi gene was amplified with the primers ldi_NdeI_fw (TGCGACATATGATGCGGTTCACATTG) and ldi_BglII_rw (CGCGAGATCTTTATTTCCCTGCGA) from genomic C. defragrans DNA and ligated into pCR4-TOPO (Invitrogen). The NdeI-BglII-flanked gene was transferred into pET-42a(+) (Novagen, Merck KGaA), and the gene was expressed in E. coli BL21 StarTM (DE3) (Invitrogen). The construct correctness was confirmed by sequencing. Cultures were induced with isopropyl 1-thio-β-d-galactopyranoside. Soluble extracts were assayed in the anaerobic two-phase system.
RESULTS
Soluble extracts of C. defragrans catalyzed the transformation of geraniol in two directions (Fig. 1). A NAD+-reducing activity showed the presence of a geraniol dehydrogenase.3 In the absence of an electron acceptor, the dialyzed soluble extract initially formed linalool, and then, after a certain linalool concentration was reached, myrcene appeared. Both compounds, linalool and myrcene, were detected and identified by GC and GC-MS (data not shown). In separate experiments, the dialyzed soluble extract transformed linalool to myrcene.
Biomass yields in a pH-controlled fermenter were lower on myrcene than on limonene. Hence, we grew C. defragrans on limonene. The crude extracts showed comparable specific linalool dehydratase activities. In contrast, the enzyme activity was not detected in cells grown on acetate. Addition of limonene (10 mm) to the culture growing on acetate resulted in induction of the enzyme activity after 10 h (data not shown).
Purification of Linalool Dehydratase from C. defragrans Strain 65Phen
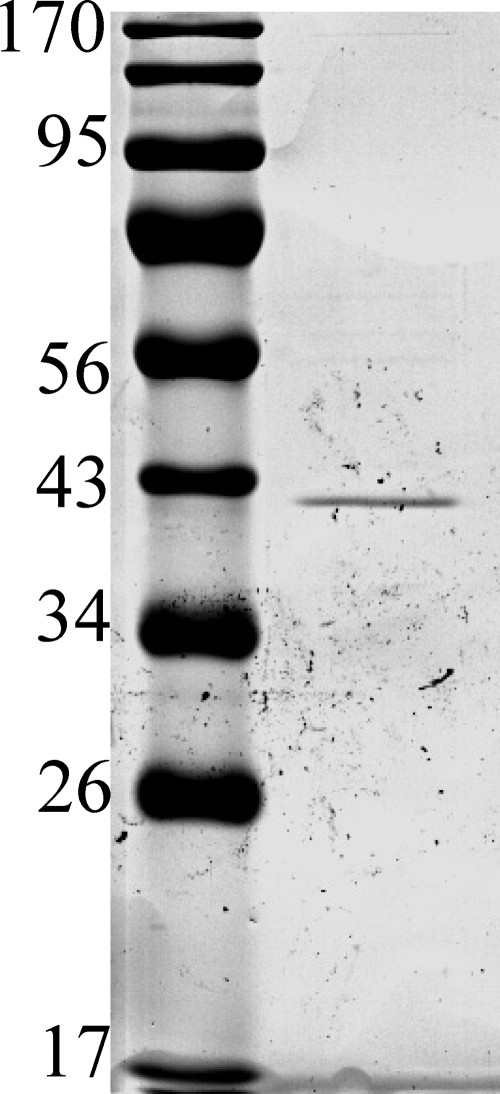
The purification of the linalool dehydratase initially yielded a preparation with several proteins (data not shown). The purification procedure was significantly improved by including a Butyl-Sepharose column. The protein eluted from this column with 6 m urea. In a five-step protocol, the enzyme activity was purified to a single protein band (Fig. 2). The linalool dehydratase protein yield was 0.02% of the initial protein, accompanied by a 1846-fold increase in the specific activity (Table 1). Gel filtration chromatography on a SuperdexTM 200-pg column gave a single peak of active protein after a retention volume of 62 ml. Based on a calibration with standard proteins, linalool dehydratase exhibited a native molecular mass of 160 kDa. SDS-PAGE of the purified enzyme showed a single band with a molecular mass of 40 kDa (Fig. 2). These observations suggested that the native form of linalool dehydratase from C. defragrans 65Phen is likely a homotetramer (α4). UV-visible absorbance spectra revealed the absence of chromophors between 300 and 850 nm, suggesting that there was no prosthetic group present.
FIGURE 2.
SDS-PAGE of the linalool dehydratase-isomerase after purification. The sizes of marker proteins are indicated in kDa. Gels with 7.5% acrylamide revealed the absence of smaller proteins (data not shown).
TABLE 1.
Purification of linalool dehydratase from C. defragrans 65Phen
| Purification step | Protein | Activity | Specific activity | Relative specific activity | Protein yield |
|---|---|---|---|---|---|
| mg | nanokatals | nanokatals/mg | % | ||
| Soluble extract | 2490.3 | 12.9 | 0.005 | 1 | 100 |
| Anion exchange (AIE-Q1) | 401.6 | 25.5 | 0.063 | 13 | 16.13 |
| Hydrophobic interaction | 33.3 | 19.2 | 0.575 | 115 | 1.34 |
| Ammonium sulfate precipitation | 15.3 | 33.9 | 2.213 | 443 | 0.61 |
| Size exclusion | 6.9 | 19.3 | 2.783 | 557 | 0.28 |
| Anion exchange (AIE-Q2) | 0.6 | 5.3 | 9.228 | 1846 | 0.02 |
Catalytic Properties of Purified Linalool Dehydratase
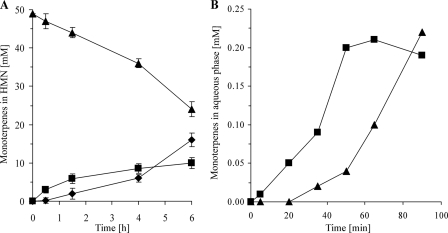
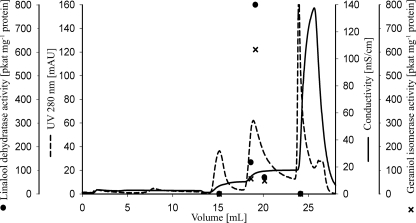
The purified protein catalyzed the dehydration of linalool to myrcene in the absence of molecular oxygen but required 2 mm DTT. Geraniol was isomerized initially to linalool, and subsequently myrcene appeared (Fig. 3A). Both activities occurred concurrently in all purification steps; e.g. Fig. 4 shows the final anion exchange purification. The purification of the geraniol isomerase activity was 1740-fold similar to the 1846-fold purification of the linalool dehydratase activity.
FIGURE 3.
Time course of monoterpene transformation by the purified linalool dehydratase-isomerase in a two-phase-system with HMN as organic carrier phase (A) and in the presence of a myrcene phase (B). ♦, myrcene; ■, linalool; ▴, geraniol.
FIGURE 4.
Enzyme purification by anion exchange chromatography (AIE-Q2). Linalool dehydratase and geraniol isomerase activities eluted together at 120 mm sodium chloride. The most active fraction contained a single protein on SDS-PAGE (Fig. 2).
The enzyme activity measurements were performed in a two-phase system with HMN as organic phase. Like other monoterpenes, myrcene is 100-fold less soluble in water than monoterpenoids, e.g. geraniol or linalool: myrcene has a solubility of 43 μm and a octanol/water partition coefficient of logP = 4.5 (29). The organic phase served also as reservoir for the monoterpenoids. This dilution influences the actual concentrations of geraniol and linalool in aqueous solution. In equilibrium with the organic phase, calculation revealed micromolar concentrations for geraniol and linalool in the aqueous phase. Observed rates under these conditions were low, 14.5 picokatals mg−1 for linalool dehydratase and 8.8 picokatals mg−1 for geraniol isomerase.
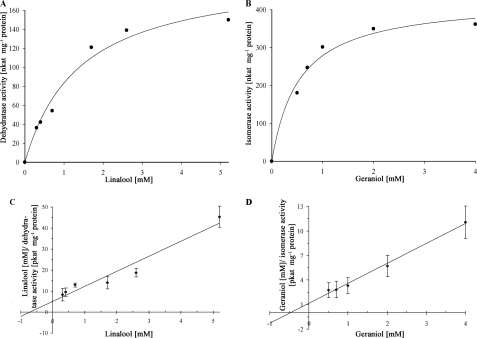
The enzyme activities were not inhibited by 10% (v/v) DMSO. Thus, we performed the kinetic characterization in a single-phase system with 10% (v/v) DMSO in water. The linalool dehydratase and the geraniol isomerase activities exhibited typical Michaelis-Menten kinetics with Vmax values of 140 and 410 nanokatals mg−1 protein, respectively (Fig. 5). The Km values for linalool and geraniol were 750 μm and 500 μm, respectively.
FIGURE 5.
A and B, Michaelis-Menten plots of linalool dehydratase activity (A) and geraniol isomerase activity (B). C and D, Hanes plots revealing the Km and Vmax values. pkat, picokatals.
Quantification of the reverse reactions, the hydration of myrcene to linalool and the isomerization of linalool to geraniol, were attempted with a myrcene-saturated aqueous phase that was maintained by a pure myrcene phase. The formation of linalool proceeded initially with a maximum specific activity of 133 picokatals mg−1 (Fig. 3B). After an accumulation of 0.2 mm linalool, geraniol was formed at a similar rate. This experiment revealed the reversibility of the enzyme activity. In systems without a pure myrcene phase, we never detected the formation of geraniol from linalool, neither in the two-phase system with an organic carrier nor in a DMSO-containing aqueous system. Myrcene was the only product detected in these experiments.
We tested other acyclic monoterpenes as substrate for the enzyme. Neither the monoterpenes α- and β-ocimene nor the monoterpenoids citronellol and nerol were transformed. A 3-methylene group is absent in the ocimenes that have a 3-methyl-1,3-diene structure. Of the cis-3-methyl-2-en-1-ol motif present in geraniol, citronellol lacks the double bond at the C2-carbon atom, and nerol is the trans-isomer to geraniol. This suggests a highly specific binding site for the substrates.
Effects of Various Compounds
The purified linalool dehydratase required only DTT as a reducing agent and an oxygen free microenvironment (<1% (v/v)) for the dehydration of linalool. The activity was not detectable in the presence of 1 mm Ti(III)citrate. Other inhibitors were molecular oxygen (Table 2) and high salt concentrations. NaCl, KCl, or MgCl2 at a concentration of 220 mm inhibited the enzyme activity completely. The metal-chelating agent EDTA (5 mm) did not affect the enzyme activity, suggesting that either the protein does not require metal ions for activity or the chelating molecule was not able to remove the metal ions under the assay conditions. Potassium nitrite or nitrate (20 mm) did not influence the enzyme activity. Coenzyme A was ineffective as a cofactor. However, phosphate as buffer or pyridoxal phosphate as well as S-adenosylmethionine modulated the enzyme activity. The enzyme is inhibited by urea: 20% activity remained at 3 m urea, and no activity was detected in 6 m urea.
TABLE 2.
Effectors on enzyme activity
The complete assay contained 150 μl of 100 mm monoterpene dissolved in HMN and 150 μl of protein solution (0.5 mg/ml) including 2 mm DTT.
| Assay | Linalool dehydratase | Geraniol isomerase |
|---|---|---|
| % | % | |
| Complete assay | 100 | 100 |
| −2 mm DTT | 10 | 10 |
| +1 mm Ti(III)citrate | 0 | 0 |
| +0.1% (v/v) O2 | 100 | 100 |
| +0.5% (v/v) O2 | 110 | 110 |
| +1% (v/v) O2 | 90 | 90 |
| +20% (v/v) O2 | 5 | 5 |
| +99.9% (v/v) O2 | 0 | 0 |
| +1 mm pyridoxal phosphate | 65 | 100 |
| +40 mmS-adenosylmethionine | 20 | 200 |
Optimal pH and Thermophilicity
The linalool dehydratase activity had an optimal temperature at 35 °C. The enzyme activity had a pH maximum at low alkaline conditions (supplemental Fig. S1), but there was a sharp decrease in linalool dehydratase activity beyond pH 9.0. The optimal pH was 9.0 with Tris-HCl buffer. The temperature dependence of the reaction showed a linear Arrhenius plot in the range from 22 °C to 35 °C (supplemental Fig. S2), with an activation energy of EA = 68.6 kJ/mol.
Identification of the Open Reading Frame
The N-terminal protein sequence was determined, and the corresponding open reading frame was found within a fosmid sequence obtained from C. defragrans 65Phen.4 The gene coded for a preprotein with 397 amino acids, including an N-terminal signal peptide sequence (MRFTLKTTAIVSAAALLAGFGPPPRAA) for transport into the periplasmatic space (supplemental Fig. S3). The alanine pair residues represent the cleavage motif. The experimentally determined N terminus of the purified protein started with the second alanine of the predicted cleavage motif (AELPPGRLATTE). Analyses with SignalP 3.0 (30) based on studies of signal-sequence cleavage sites (31) suggested a Sec-dependent membrane translocation mechanism for the preprotein into the periplasmatic space. Thus, the purified protein represents a mature protein. According to in silico mass calculation the precursor exhibits a molecular mass of 43 kDa.
Comparisons of the protein and of the gene sequences with nucleotide, microbial genome and environmental metagenomic datasets did not reveal significant relationships to known proteins and genes. TblastN (32) identified the closest relative as a hypothetical partial mRNA protein from the eukaryotic ascomycota Aspergillus oryzae RIB40 with an E value of 2E-08. The biotransformation potential of Aspergillus species on myrcene has been elucidated previously (33), although linalool was not detected as a transformation product. TblastP identified a hypothetical protein from another eukaryotic ascomycota, Nectria hematococca mpVI, as closest related protein, with an E value of 2E-12.
A ClustalW alignment of the linalool dehydratase-isomerase showed no relevant scores with characterized alkene hydratases, namely a γ-carotene 1,2-hydratase (CruF) from Deinococcus radiodurans R1 (34), a hydroxyneurosporene synthase from Rhodospirillum rubrum ATCC 11170, and a γ-carotene 1,2-hydroxylase from Synechococcus sp. PCC 7002. This was a further indication of the novel character of this enzyme and its catalytic activity.
Expression of Linalool Dehydratase-Isomerase in E. coli
The identified open reading frame was used to construct the expression vector pET-42a(+)-LDI. Isopropyl 1-thio-β-d-galactopyranoside-induced 3-ml cultures showed a linalool dehydratase activity of 380 nanokatals and a geraniol isomerase activity of 310 nanokatals in a 6-h assay. Control cultures with the vector lacking the ldi gene had no enzyme activity. Soluble extracts of the induced cells had a specific linalool dehydratase activity of 435 picokatals mg−1 protein and a geraniol isomerase activity of 116 picokatals mg−1 protein in the two-phase assay.
DISCUSSION
Myrcene is an acyclic C10-hydrocarbon and represents a large fraction (74%) of monoterpenes extracted from the essential oils of the hop plant Humulus lupulus (35). The transformation of this unsaturated hydrocarbon at the enzymatic level has never investigated under anaerobic conditions. Here, we describe a new initial reaction in the anaerobic degradation of hydrocarbons: the hydration of myrcene. First, a water molecule is added to the methylene double bond. Mechanistically, it may be equivalent to a chemical water addition catalyzed by acids, leading to linalool, a tertiary allylalcohol (3,7-dimethyl-1,6-octadien-3-ol). A subsequent isomerization yielded the primary allylalcohol geraniol (3,7-dimethylocta-2,6-dien-1-ol). These two reactions are catalyzed by a single bifunctional enzyme, the linalool dehydratase-isomerase.
The thermodynamic equilibrium favors the formation of myrcene from geraniol. Linalool is thermodynamically more stable than geraniol: according to experimental observations in a two-phase system with Thauera linaloolentis (36), linalool is 5.9-kJ mol−1 more stable than geraniol. Our observations confirmed this equilibrium with the enzyme from C. defragrans: the linalool dehydratase-isomerase catalyzes the formation of linalool from geraniol and subsequently of myrcene from linalool. In this thermodynamically favorable direction, the formation of myrcene from geraniol may be seen as detoxification process for the monoterpene alcohol. The monoterpene alcohols have a higher cell toxicity than the monoterpenes.
The observation of the reverse reactions, myrcene to linalool and linalool to geraniol, revealed that the Gibbs free energy change of the hydration is rather small. The steady-state equilibrium with 0.2 mm linalool in the presence of a myrcene phase (Fig. 3B) corresponds, considering a maximum water solubility of linalool of 10.1 mm (37), to a difference in free energy of −10.1 kJ mol−1 at 35 °C. Thus, the enzymatic reaction can provide a thermodynamically limited pool of geraniol for further metabolic reactions.
The in vivo monoterpene mineralization rate of 325 picokatals (milligrams of total protein)−1 (24) is higher than the in vitro formation rate of geranic acid (0.9 picokatals (milligrams of soluble protein)−1 (24)) and the “reverse” enzyme activity in soluble extracts (Table 1). The in vitro activity of the enzyme is too low to sustain the in vivo monoterpene turnover. This may suggest that the enzyme may contribute primarily to the monoterpene resistance. However, future studies on the genetic level, including the development of a genetic system for C. defragrans, are clearly required to reveal the importance of this enzyme in the monoterpene mineralization pathway(s).
The linalool dehydratase-isomerase seems to be a cofactor-free enzyme. The UV-visible absorption spectrum of the purified enzyme revealed only an absorption maximum at 280 nm, indicating the presence of aromatic amino acid residues (data not shown). The purification with an elution with 6 m urea suggests an unfolding and a spontaneous folding during the dialysis. This, together with the lack of inhibition by EDTA, argues for the lack of a nonpermanently bound cofactor. The expression in active form in E. coli can be interpreted as the absence of a complex posttranslational enzyme activation by metal cofactor integration. The only requirement for the enzyme activity of the purified protein was a mild reducing agent, DTT, and the absence of oxygen or a strong reducing agent, e.g. Ti(III)citrate.
The amino acid sequence analysis attested a Sec-dependent translocation of linalool dehydratase-isomerase. The process involves a cytosolic preprotein which, typical for these translocated proteins, has a short signal sequence with a two alanine motif at the end, representing the cleavage site (31). The preprotein is transported across the cytoplasmatic membrane in an unfolded state. During this process, the signal peptide is cleaved. A periplasmatic location for a hydrocarbon activation enzyme was already detected in the denitrifying Azoarcus strain EbN1 (38). A periplasmatic dehydrogenase oxidizes ethylbenzene to (S)-1-phenylethanol. Future experiments with protein labeling or specific antibodies may describe the translocation in more detail.
In summary, this work depicts the enzyme for the initial metabolism of myrcene, an acyclical monoterpene, under anaerobic conditions. It is a novel type of dehydratase-isomerase that acts on myrcene, linalool, and geraniol. We recommend the disposition of a new protein family with the EC number 4.2.1.X.
Supplementary Material
Acknowledgment
We thank Hannah Marchant for improvement of the language.
This work was supported by the Max Planck Society.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) FR669447.
J. Harder, unpublished results.
A. W̄ulfing, F. Germer, A. Meyerdierks, and J. Harder, unpublished results.
- HMN
- 2,2,4,4,6,8,8-heptamethylnonane
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1.Erman W. F. (1985) Chemistry of the Monoterpenes: An Encyclopedic Handbook, Marcel Dekker, New York [Google Scholar]
- 2.van der Werf M. J., Swarts H. J., de Bont J. A. M. (1999) Appl. Environ. Microbiol. 65, 2092–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guenther A., Hewitt C. N., Erickson D., Fall R., Geron C., Graedel T., Harley P., Klinger L., Lerdau M., Mckay W. A. P., Pierce T., Scholes B., Steinbrecher R., Tallamraju R., Taylor J., Zimmerman P. (1995) J. Geophys. Res. 100, 8873–8892 [Google Scholar]
- 4.Boucher Y., Doolittle W. F. (2000) Mol. Microbiol. 37, 703–716 [DOI] [PubMed] [Google Scholar]
- 5.Hunter W. N. (2007) J. Biol. Chem. 282, 21573–21577 [DOI] [PubMed] [Google Scholar]
- 6.Zimmerman P. R., Chatfield R. B., Fishman J., Crutzen P. J., Hanst P. L. (1978) Geophys. Res. Lett. 5, 679–682 [Google Scholar]
- 7.Kamens R., Jang M., Chien C., Leach K. (1999) Environ. Sci. Technol. 33, 1430–1438 [Google Scholar]
- 8.Kiendler-Scharr A., Wildt J., Dal Maso M., Hohaus T., Kleist E., Mentel T. F., Tillmann R., Uerlings R., Schurr U., Wahner A. (2009) Nature 461, 381–384 [DOI] [PubMed] [Google Scholar]
- 9.Pare P. W., Tumlinson J. H. (1997) Nature 385, 30–31 [Google Scholar]
- 10.Trudgill P. W. (1986) in Terpinoid Metabolism by Pseudomonas. The Bacteria: A Treatise on Structure and Function (Gunsalus I. C. ed) pp. 483–528, Academic Press, New York [Google Scholar]
- 11.Förster-Fromme K., Jendrossek D. (2008) FEMS Microbiol. 286, 78–84 [DOI] [PubMed] [Google Scholar]
- 12.Hartmans S., Weber F. J., Somhorst D. P., de Bont J. A. (1991) J. Gen. Microbiol. 137, 2555–2560 [DOI] [PubMed] [Google Scholar]
- 13.Evans P. J., Ling W., Goldschmidt B., Ritter E. R., Young L. Y. (1992) Appl. Environ. Microbiol. 58, 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabus R., Wilkes H., Behrends A., Armstroff A., Fischer T., Pierik A. J., Widdel F. (2001) J. Bacteriol. 183, 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heider J. (2007) Curr. Opin. Chem. Biol. 11, 188–194 [DOI] [PubMed] [Google Scholar]
- 16.Kloer D. P., Hagel C., Heider J., Schulz G. E. (2006) Structure 14, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 17.Chiang Y. R., Ismail W., Müller M., Fuchs G. (2007) J. Biol. Chem. 282, 13240–13249 [DOI] [PubMed] [Google Scholar]
- 18.Harder J., Heyen U., Probian C., Foss S. (2000) Biodegradation 11, 55–63 [DOI] [PubMed] [Google Scholar]
- 19.Harder J., Probian C. (1995) Appl. Environ. Microbiol. 61, 3804–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foss S., Heyden U., Harder J. (1998) System. Appl. Microbiol. 21, 237–244 [DOI] [PubMed] [Google Scholar]
- 21.Foss S., Harder J. (1998) System. Appl. Microbiol. 21, 365–373 [DOI] [PubMed] [Google Scholar]
- 22.Kämpfer P., Denger K., Cook A. M., Lee S. T., Jäckel U., Denner E. B., Busse H. J. (2006) Int. J. Syst. Evol. Microbiol. 56, 815–819 [DOI] [PubMed] [Google Scholar]
- 23.Heyen U., Harder J. (1998) FEMS Microbiol. Rev. 169, 67–71 [Google Scholar]
- 24.Heyen U., Harder J. (2000) Appl. Environ. Microbiol. 66, 3004–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aeckersberg F., Bak F., Widdel F. (1991) Arch. Microbiol. 156, 5–14 [Google Scholar]
- 26.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 27.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 28.Towbin H., Staehelin T., Gordon J. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid C., Steinbrecher R., Ziegler H. (1992) Trees 6, 32–36 [Google Scholar]
- 30.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 31.von Heijne G. (1983) Eur. J. Biochem. 133, 17–21 [DOI] [PubMed] [Google Scholar]
- 32.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farooq A. R., Rahman A., Choudhary A. I. (2004) Curr. Org. Chem. 8, 353–367 [Google Scholar]
- 34.Sun Z., Shen S., Wang C., Wang H., Hu Y., Jiao J., Ma T., Tian B., Hua Y. (2009) Microbiology 155, 2775–2783 [DOI] [PubMed] [Google Scholar]
- 35.Thompson M. L., Marriott R., Dowle A., Grogan G. (2010) Appl. Microbiol. Biotechnol. 85, 721–730 [DOI] [PubMed] [Google Scholar]
- 36.Foss S., Harder J. (1997) FEMS Microbiol. 149, 71–75 [Google Scholar]
- 37.Fichan I., Larroche C., Gros J. B. (1999) J. Chem. Eng. Data 44, 56–62 [Google Scholar]
- 38.Kniemeyer O., Heider J. (2001) J. Biol. Chem. 276, 21381–21386 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.