Abstract
Rtt109p, a histone acetyltransferase, associates with active genes and acetylates lysine 56 on histone H3 in Saccharomyces cerevisiae. However, the functional role of Rtt109p or H3 Lys56 acetylation in chromatin assembly/disassembly (and hence gene expression) immediately switching transcription on or off has not been clearly elucidated in vivo. Here, we show that Rtt109p promotes the eviction of histone H3 from a fast inducible yeast gene, GAL1, following transcriptional initiation via histone H3 Lys56 acetylation. Conversely, the deposition of histone H3 to GAL1 is significantly decreased in the presence of Rtt109p following transcriptional termination. Intriguingly, we also find that the deposition of histone H2B on preexisting non-acetylated histone H3 Lys56 at GAL1 in Δrtt109 is significantly increased independently of histone H3 deposition immediately following transcriptional termination subsequent to a short induction. Consistently, histone H2B is not efficiently evicted from GAL1 in the absence of Rtt109p immediately following transcriptional induction. Furthermore, we show that the stimulated eviction or reduced deposition of histones by Rtt109p promotes the association of RNA polymerase II with GAL1 and hence the synthesis of GAL1 mRNA. These results, taken together, support the fact that Rtt109p regulates the deposition/eviction of histone H2B in addition to its role in stimulating histone H3 eviction, thus providing insight into chromatin assembly/disassembly and hence gene expression in vivo.
Keywords: Chromatin, Gene Regulation, Histone Modification, RNA Polymerase II, Transcription, GAL1, Histone, Rtt109p
Introduction
The post-translational covalent modifications of histones play important roles in DNA transacting processes, such as transcription, replication, and DNA repair (1–7). Most of the covalent modifications occur at the N-terminal tails of histones with the exception of ubiquitylation that occurs at the C-terminal tails of histones H2A and H2B. However, covalent modifications also occur in the globular core domain. One such modification is acetylation of lysine 56 on histone H3 (8, 9). This modification has been speculated to play an important role in the regulation of histone H3-DNA interaction within the nucleosome because it is present at the entry and exit sites of nucleosomal DNA (8, 10, 11). Histone H3 Lys56 acetylation functions in promoting cell survival following exposure to genotoxic agents (9). Further, this covalent modification has been implicated in error-free replication (12, 13), gene activation (8, 11), RNA polymerase II transcript elongation through heterochromatin (14), and the pathogenesis of Candida albicans (15). Thus, histone H3 Lys56 acetylation is an important covalent modification that regulates transcription, replication, DNA repair, and pathogenesis (8, 9, 11–19).
To identify the factor(s) or enzyme(s) responsible for histone H3 Lys56 acetylation, Schneider et al. (16) performed the GPS (global proteomic analysis in Saccharomyces cerevisiae) screen, which revealed that Rtt109p is required for acetylation of Lys56 on histone H3. Consequently, several other studies (17–19) also demonstrated the role of Rtt109p in histone H3 Lys56 acetylation. Rtt109p is a fungal histone acetyltransferase (16–20) and shares structural homology with metazoan p300/CREB4-binding protein (21–23). In fact, Das et al. (24) have recently demonstrated the role of human p300/CREB-binding protein in histone H3 Lys56 acetylation. Rtt109p requires either of two chaperones, namely Asf1p and Vps75p, for stimulation of its enzymatic activity (18, 19, 25–27). Han et al. (26) have shown that Rtt109p acetylates core but not nucleosomal histone H3 in vitro. However, acetylated Lys56 on histone H3 has also been observed at the active genes in vivo (8, 16). The presence of such histone H3 Lys56 acetylation at the active genes has been attributed to incorporation of Lys56-acetylated histone H3 into nucleosomes (9, 19, 27–29). However, it remains to be addressed whether Rtt109p can also acetylate nucleosomal histone H3 in vivo.
Although Rtt109p associates with transcriptionally active genes and is involved in histone H3 Lys56 acetylation (16), its function in chromatin assembly/disassembly and transcription was not known. Recently, Williams et al. (11) have demonstrated that histone H3 Lys56 acetylation is essential for histone H3 eviction from the promoter of a slowly induced PHO5 gene during transcriptional activation. However, what happens at the early time periods of transcriptional induction remains unknown. Further, the role of histone H3 Lys56 acetylation or Rtt109p in the nucleosomal assembly/disassembly of histone H2B (or histone H2A-H2B dimer) is not clearly understood. In view of this, we have analyzed the eviction/deposition of histones H3 and H2B at a fast inducible gene, GAL1, in S. cerevisiae immediately following transcriptional induction/repression in the RTT109 deletion mutant and its isogenic wild type equivalent. Our data reveal significant functional insight of Rtt109p or histone H3 Lys56 acetylation in chromatin assembly/disassembly during transcription in vivo, as presented below.
EXPERIMENTAL PROCEDURES
Plasmids
The plasmid pRS406 was used in the PCR-based gene disruption of the RTT109 gene in the strain bearing FLAG-tagged histone H2B.
Strains
The yeast (S. cerevisiae) strain bearing FLAG-tagged histone H2B (YTT31) was obtained from the Osley laboratory (30). The RTT109 gene was deleted in the YTT31 strain using a PCR-based gene disruption method to generate SBY1 (Δrtt109, URA1).
Growth Media
For the transcriptional induction of GAL1, yeast strains were initially grown in YPR (yeast extract, peptone plus raffinose) up to an A600 of 0.9 and then switched to YPG (yeast extract, peptone plus galactose) for different induction time periods prior to formaldehyde-based in vivo cross-linking. For transcriptional repression of GAL1, the yeast cells were first grown in YPR up to an A600 of 0.9 and then switched to YPG for 90 min prior to transfer to YPD (yeast extract, peptone plus dextrose). Alternatively, the transcriptional repression of GAL1 was also performed by growing the cells initially in YPG up to an A600 of 0.9 prior to transfer to YPD.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed as described previously (31–34). Briefly, yeast cells were treated with 1% formaldehyde, collected, and resuspended in lysis buffer. Following sonication, cell lysate (400 μl of lysate from 50 ml of yeast culture) was precleared by centrifugation, and then 100 μl of lysate was used for each immunoprecipitation. Immunoprecipitated protein-DNA complexes were treated with proteinase K, the cross-links were reversed, and DNA was purified. Immunoprecipitated (IP) DNA was dissolved in 20 μl of TE 8.0 (10 mm Tris-HCl, pH 8.0, and 1 mm EDTA) and was analyzed by PCR using [α-32P]dATP. PCR products were detected by autoradiography after separation on a 6% polyacrylamide gel. As a control, “input” DNA was used in the PCR analysis. The input DNA was isolated from 5 μl of lysate without going through the immunoprecipitation step and dissolved in 100 μl of TE 8.0. Serial dilutions of input and IP DNAs were used to assess the linear range of PCR amplification as described previously (31). All of the PCR data presented in this article are within the linear range of the PCR analysis. For the ChIP analysis of histone H3, we modified the above ChIP protocol as described previously (33, 35). The primer pairs used for PCR analysis are as follows: GAL1 (core), 5′-ATAGGATGATAATGCGATTAGTTTTTTAGCCTT-3′and 5′-GAAAATGTTGAAAGTATTAGTTAAAGTGGTTATGCA-3′; GAL1 (ORF), 5′-CAGTGGATTGTCTTCTTCGGCCGC-3′ and 5′-GGCAGCCTGATCCATACCGCCATT-3′; ADH1 (ORF), 5′-CGGTAACAGAGCTGACACCAGAGA-3′ and 5′-ACGTATCTACCAACGATTTGACCC-3′. Autoradiograms were scanned and quantitated by the National Institutes of Health Image 1.62 program. IP DNAs were quantitated as the ratio of IP to input.
For eviction analysis of histones H2B and H3 from GAL1 in YPG following growth in YPR, the ChIP signal of histone H2B (or histone H3) at 0 min of induction was set to 100 for both the wild type and RTT109 deletion mutant strains. The ChIP signals for histone H2B (or histone H3) at GAL1 in the RTT109 wild type and deletion mutant strains at 0 min of induction are not significantly different (supplemental Fig. S1, A and B). The ChIP signals at later induction time periods were normalized with respect to 100. Similar normalization was also performed for deposition of histones H2B and H3 in YPD following growth in YPG as discussed under “Results and Discussion.”
RT-PCR Assay
The RT-PCR assay was performed as described previously (36). Briefly, total mRNA was prepared from 10 ml of yeast culture grown to an A600 of 1.0. Ten micrograms of total mRNA was used in the reverse transcription assay. mRNA was treated with RNase-free DNase (M610A, Promega) and then reverse-transcribed to cDNA using oligo(dT) as described in the protocol supplied by Promega (A3800, Promega). PCR was performed using synthesized first strand as template and the primer pairs targeted to the GAL1 and ADH1 coding sequences: GAL1, 5′-CAGAGGGCTAAGCATGTGTATTCT-3′ and 5′-GTCAATCTCTGGACAAGAACATTC-3′; ADH1, 5′-CGGTAACAGAGCTGACACCAGAGA-3′ and 5′-ACGTATCTACCAACGATTTGACCC-3′. RT-PCR products were separated by 2.2% agarose gel electrophoresis and visualized by ethidium bromide staining.
RESULTS AND DISCUSSION
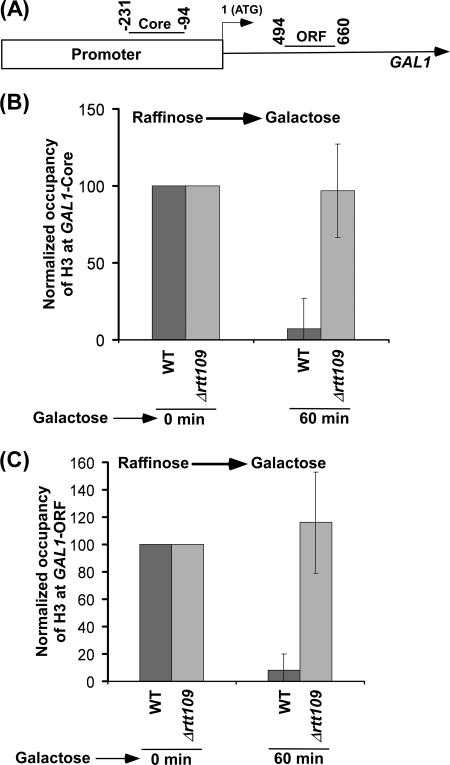
We have demonstrated recently that Rtt109p associates with active genes, including GAL1 (16), and is involved in acetylation of Lys56 on histone H3 (16) (supplemental Fig. S2). Because histone H3 Lys56 is located at/near the DNA entry and exit sites within the nucleosome (8, 10, 11), acetylation of Lys56 on histone H3 is expected to regulate the interaction of histone H3 with DNA (and hence chromatin assembly/disassembly). In view of this, we analyzed the eviction of histone H3 at the core promoter and ORF of the GAL1 gene (Fig. 1A) following transcriptional induction (upon switching the growth medium containing raffinose to galactose). We found that histone H3 was evicted from GAL1 in the wild type cells following transcriptional induction in galactose-containing growth medium (Fig. 1, B and C, and supplemental Fig. S3A), consistent with previous studies (37–46). However, such an eviction of histone H3 was dramatically decreased in the absence of Rtt109p (Fig. 1, B and C). Further, histone H3 Lys56 acetylation is observed at GAL1 in the wild type strain (16). Such acetylation is absent in the RTT109 deletion mutant strain (16) (supplemental Fig. S2), whereas a significantly high level of non-acetylated histone H3 Lys56 was present at GAL1 in the absence of Rtt109p (Fig. 1, B and C). These results support the fact that Rtt109p promotes the eviction of histone H3 through the destabilization of the interaction between DNA and histone H3 via histone H3 Lys56 acetylation. Because histones H3 and H4 form a tetramer, the eviction of histone H4 would thus be presumably facilitated by histone H3 Lys56 acetylation. Consistent with our observations, Williams et al. (11) have also demonstrated recently that histone H3 Lys56 acetylation promotes the eviction of histone H3 from the PHO5 promoter at ∼5 h following transcriptional induction. Although the transcriptional induction of PHO5 was much slower than GAL1, both studies support the role of histone H3 Lys56 acetylation in promoting the eviction of histone H3. Further, Schwabish and Struhl (46) have previously demonstrated that the eviction of histone H3 from GAL1 is significantly decreased in the absence of a histone H3 chaperone Asf1p that stimulates the enzymatic activity of Rtt109p for histone H3 Lys56 acetylation. These studies, together, implicate a role of Rtt109p or histone H3 Lys56 acetylation in promoting the eviction of histone H3 (and hence histone H3-H4 tetramer) during transcription.
FIGURE 1.
Histone H3 is not evicted from the core promoter and coding sequence of GAL1 following transcriptional induction in galactose-containing growth medium in the RTT109 deletion mutant strain. A, the PCR primer pairs located at the core promoter and coding sequence of GAL1 are schematically shown. Core, core promoter. B, Rtt109p is required for histone H3 eviction from the GAL1 core promoter. Both the wild type and RTT109 deletion mutant strains were grown in YPR up to an A600 of 0.9 and then switched to YPG for transcriptional induction prior to formaldehyde-based in vivo cross-linking. Immunoprecipitations were performed using an anti-H3 antibody (Ab-1791, Abcam) against histone H3. Immunoprecipitated DNA was analyzed by PCR, using the primer pair targeted to the core promoter of GAL1. C, Rtt109p is required for histone H3 eviction from the GAL1 coding sequence. Immunoprecipitated DNA was analyzed by PCR, using the primer pair targeted to the coding sequence of GAL1.
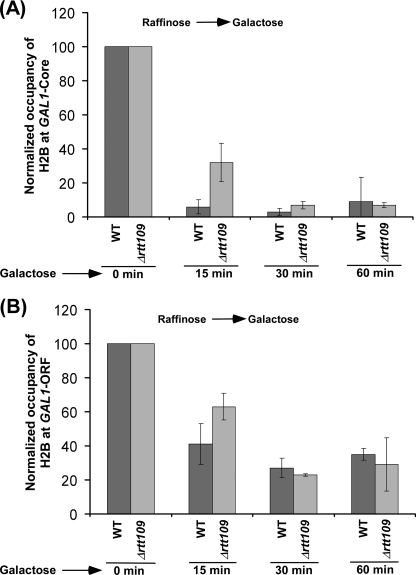
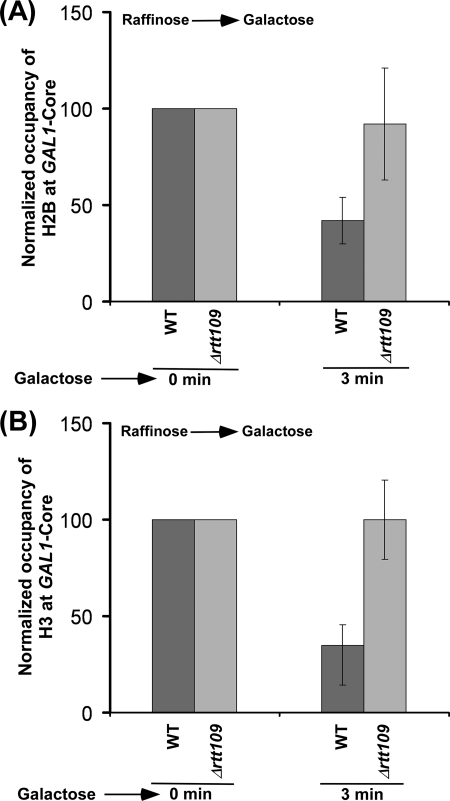
Next, we analyzed the effect of histone H3 Lys56 acetylation on the eviction of histone H2A-H2B dimer from GAL1 following transcriptional induction in galactose-containing growth medium. With this view, we determined the occupancy of histone H2B (as a representative component of histone H2A-H2B dimer) at the core promoter and coding sequence of GAL1 following transcriptional induction in the presence and absence of Rtt109p. We found that, like histone H3, histone H2B was evicted from GAL1 following transcriptional induction in the wild type cells (Fig. 2, A and B, and supplemental Fig. S3A), consistent with previous studies (37–46). Intriguingly, we found that histone H2B was evicted in the absence of Rtt109p at 60 min following transcriptional induction (Fig. 2, A and B, and supplemental Fig. S3B), even when histone H3 was not evicted (Fig. 1, B and C). These observations clearly demonstrate that histone H2B is evicted independently of histone H3. Thus, our data support the fact that the eviction of histone H3-H4 tetramer and histone H2A-H2B dimer occurs separately in vivo, as opposed to the eviction of the whole histone octamer. However, like histone H3, histone H2B was not efficiently evicted from GAL1 in the absence of Rtt109p at 3 min following transcriptional induction (Fig. 3, A and B), whereas significant eviction of histone H2B (as well as histone H3) occurred in the wild type cells (Fig. 3, A and B). Similarly, the eviction of histone H2B from GAL1 was also decreased at 15 min following transcriptional induction in Δrtt109 (Fig. 2). However, at later time points of transcriptional induction, histone H2B was evicted normally in Δrtt109 as compared with the wild type strain (Fig. 2, A and B). Such an eviction of histone H2B in Δrtt109 at later induction time points could be facilitated by the association of the FACT complex (which plays important roles in histone H2A-H2B dimer eviction/deposition) with GAL1 as demonstrated previously (47–54). The decreased eviction of histone H2B in the absence of Rtt109p immediately following transcriptional induction supports the fact that an impaired histone H3 eviction delays the eviction of histone H2B in vivo. Taken together, our data demonstrate that the eviction of histones H2B and H3 is significantly decreased immediately following transcriptional induction, and histone H2B, but not histone H3, is evicted normally as compared with the wild type equivalent at later time points of transcriptional induction in the absence of Rtt109p or histone H3 Lys56 acetylation.
FIGURE 2.
Analysis of histone H2B eviction from the core promoter (A) and coding sequence (B) of GAL1 following transcriptional induction in galactose-containing growth medium in the RTT109 deletion mutant and its isogenic wild type equivalent. Both the wild type and mutant strains carrying FLAG-tagged histone H2B were grown and cross-linked as in Fig. 1B. Immunoprecipitations were performed using an anti-FLAG antibody (F1804, Sigma) against the FLAG epitope attached to histone H2B.
FIGURE 3.
The eviction analysis of histones H2B (A) and H3 (B) from the GAL1 core promoter immediately following transcriptional induction in the wild type and Δrtt109 strains. Both the wild type and mutant strains were grown as in Fig. 1B.
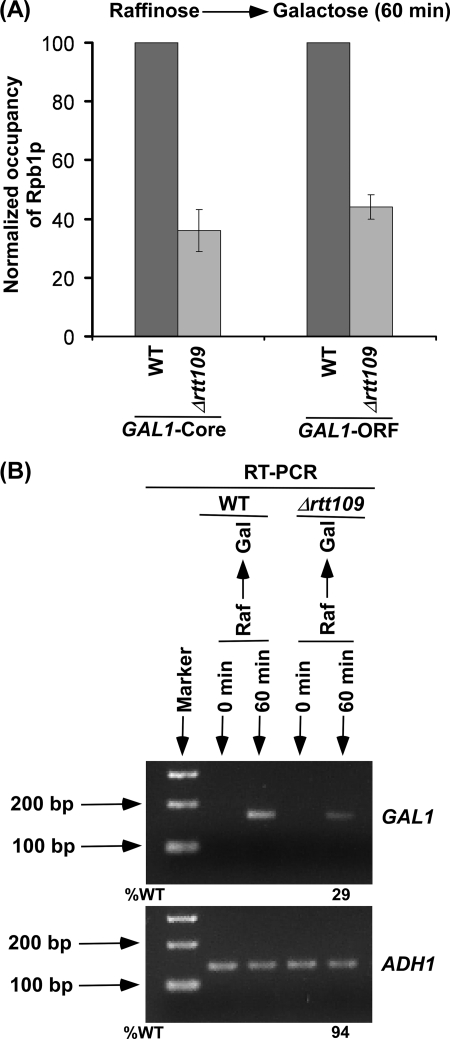
The nucleosomes present within the coding sequence of the gene would inhibit the passage of RNA polymerase II during transcriptional elongation. Thus, the association of RNA polymerase II with the active GAL1 would be decreased in the absence of Rtt109p as eviction of histone H3 is significantly diminished. To test this possibility, we analyzed the level of RNA polymerase II at the GAL1 coding sequence in the presence and absence of Rtt109p at 60 min following transcriptional induction. We found that the association of RNA polymerase II with the active GAL1 coding sequence was significantly decreased in the absence of Rtt109p (Fig. 4A). Thus, the reduced eviction of histone H3 in the absence of Rtt109p lowers the association of RNA polymerase II with the GAL1 coding sequence following transcriptional induction. Similarly, the recruitment of RNA polymerase II to the GAL1 core promoter was significantly decreased in the absence of Rtt109p (Fig. 4A) because histone H3 was not efficiently evicted from the GAL1 core promoter in Δrtt109 (Fig. 1B). Because the association of RNA polymerase II with active GAL1 is decreased in the absence of Rtt109p, the synthesis of mRNA would thus be significantly impaired. In view of this, we analyzed the level of GAL1 mRNA in the wild type and RTT109 deletion mutant strains using an RT-PCR assay. Our RT-PCR analysis revealed that the synthesis of GAL1 mRNA was significantly decreased in the RTT109 deletion mutant strain (Fig. 4B), consistent with the decrease in the association of RNA polymerase II with active GAL1 in response to reduced histone H3 eviction in the absence of Rtt109p. As an internal control, we have used a constitutively active gene, ADH1, because the association of RNA polymerase II with ADH1 was not altered in the absence of Rtt109p (supplemental Fig. S4). This is consistent with the fact that the steady-state level of RNA polymerase II at GAL1 after long induction in galactose-containing growth medium was not changed in Δrtt109 (supplemental Fig. S4).
FIGURE 4.
Rtt109p stimulates GAL1 transcription. A, Rtt109p promotes the association of RNA polymerase II with the GAL1 core promoter and coding sequence. Both the wild type and RTT109 deletion mutant strains were grown as in Fig. 1B. Immunoprecipitations were performed using 8WG16 antibody (Covance, Inc.) against the C-terminal domain of the Rpb1p subunit of RNA polymerase II. B, RT-PCR analysis. Both the wild type and mutant strains were grown as in A. Total RNA was prepared and analyzed for transcription. Oligo(dT) primer was used for cDNA synthesis. Raf, raffinose.
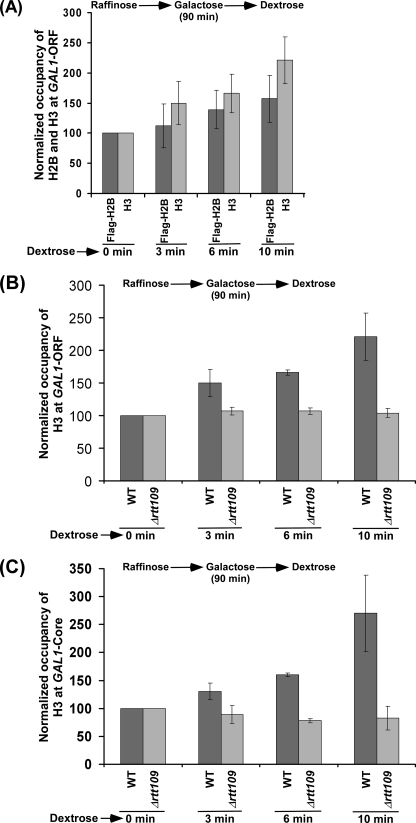
Because Rtt109p stimulates the eviction of histone H3 during transcriptional induction of GAL1, it is likely that the deposition of histone H3 to GAL1 would be decreased in the presence of Rtt109p. To test this hypothesis, we carried out the ChIP experiments to analyze the role of Rtt109p in deposition of histones H3 and H2B to GAL1 after switching off transcription. In this direction, we first analyzed the deposition of histones H3 and H2B to the coding sequence of GAL1 following transcriptional termination in dextrose-containing growth medium in the wild type strain (that was previously grown in raffinose-containing growth medium up to an A600 of 0.9 and then switched to galactose-containing growth medium for 90 min prior to transfer to dextrose-containing growth medium). We found that both histones H3 and H2B deposited to the GAL1 coding sequence in the wild type strain following transcriptional termination (Fig. 5A), and consistently, the association of RNA polymerase II was impaired (supplemental Fig. S5A). Next, we asked how Rtt109p regulates the deposition of histones H3 and H2B to the GAL1 coding sequence following its transcriptional termination. In view of this, we first analyzed the level of histone H3 at the coding sequence of GAL1 in the RTT109 deletion mutant and wild type strains under similar growth conditions. We found that histone H3 was gradually deposited to the GAL1 coding sequence in the wild type strain (Fig. 5B). However, the deposition of histone H3 was not observed in the RTT109 deletion mutant strain (Fig. 5B). Similar results were also obtained at the GAL1 core promoter (Fig. 5C and supplemental Fig. S5B). Such an absence of histone H3 deposition at the core promoter and coding sequence of the GAL1 gene can be attributed to the impaired eviction of histone H3 in the absence of Rtt109p following a short transcriptional induction in galactose-containing growth medium (Fig. 1, B and C, and supplemental Fig. S6 (left). Although a high level of histone H3 is found at 0 min in dextrose-containing growth medium in Δrtt109 as compared with wild type cells, it was set to 100 to compare the relative level of deposition of histone H3 at later time points of transcriptional repression. To overcome this problem, we induced both the wild type and RTT109 deletion mutant cells for a long time in galactose-containing growth medium (basically, yeast cells were grown in galactose-containing growth medium up to an A600 of 0.9) to allow eviction of histone H3 in the absence of Rtt109p (supplemental Fig. S6, right), and subsequently, the cells were transferred to dextrose-containing growth medium for transcriptional termination of GAL1. We found that histone H3 was deposited to the GAL1 core promoter in both the wild type and mutant strains following transcriptional termination in dextrose-containing growth medium (Fig. 6A). Importantly, histone H3 was deposited more efficiently to the GAL1 core promoter in the RTT109 deletion mutant strain as compared with the wild type equivalent (Fig. 6A). Thus, Rtt109p appears to prevent histone H3 deposition and hence nucleosomal assembly of the histone H3-H4 tetramer.
FIGURE 5.
Analysis of histone H3 deposition to GAL1 in the RTT109 wild type and deletion mutant strains following transcriptional termination. A, deposition of histones H3 and H2B to the GAL1 coding sequence in the wild type strain carrying FLAG-tagged histone H2B. The yeast strain was initially grown in YPR up to an A600 of 0.9 and then switched to YPG for 90 min prior to transfer to YPD. B, histone H3 is not deposited to the GAL1 coding sequence following transcriptional termination in Δrtt109 under the growth conditions as mentioned in A. C, histone H3 is not deposited to the GAL1 core promoter following transcriptional termination in Δrtt109 under the growth conditions mentioned for A.
FIGURE 6.
The deposition of histones H3 and H2B to GAL1 is significantly increased in the absence of Rtt109p following transcriptional termination. A, histone H3 is deposited more efficiently to the GAL1 core promoter in Δrtt109. Both the wild type and RTT109 deletion mutant strains were initially grown in YPG up to an A600 of 0.9 and then transferred to YPD prior to formaldehyde-based in vivo cross-linking. B, histone H2B is deposited more efficiently to the GAL1 core promoter in Δrtt109.
Nucleosomal assembly of histone H2A-H2B dimer follows histone H3-H4 tetramer. Thus, Rtt109p would lower the deposition of histone H2B because histone H3 was deposited more efficiently to GAL1 in Δrtt109 (Fig. 6A). To test this possibility, we analyzed the deposition of histone H2B at the GAL1 core promoter in the presence and absence of Rtt109p in dextrose-containing growth medium following long induction in galactose-containing growth medium. We found significant deposition of histone H2B to the GAL1 core promoter in both the wild type and RTT109 deletion mutant strains following transcriptional termination (Fig. 6B). However, histone H2B was deposited more efficiently to the GAL1 core promoter in the RTT109 deletion mutant strain as compared with the wild type equivalent (Fig. 6B). Thus, the inhibition of histone H3 deposition in the presence of Rtt109p appears to decrease the deposition of histone H2B, supporting the fact that the deposition of histone H3-H4 tetramer promotes the nucleosomal assembly of histone H2A-H2B dimer in vivo.
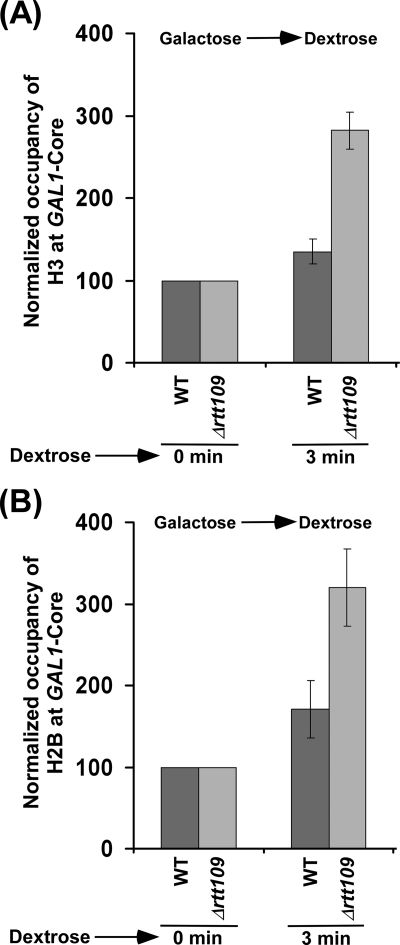
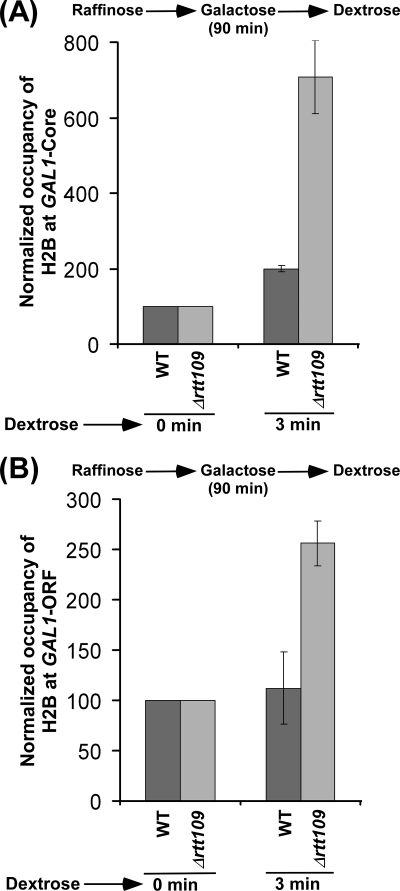
Next, we asked whether Rtt109p or histone H3 Lys56 acetylation regulates the nucleosomal assembly of histone H2A-H2B dimer independently of histone H3 deposition. With this view, we analyzed the level of histone H2B at the core promoter and coding sequence of the GAL1 gene following the transfer of the wild type and RTT109 deletion mutant cells (that were initially grown in raffinose-containing growth medium up to an A600 of 0.9 and then switched to galactose-containing growth medium) to dextrose-containing growth medium. Under these growth conditions, histone H3 was not deposited to GAL1 in dextrose-containing growth medium in the absence of Rtt109p (Fig. 5, B and C) because histone H3 was not efficiently evicted within a short period of induction in galactose-containing growth medium (Fig. 1, B and C, and supplemental Fig. S6, left). However, histone H3 was deposited to the core promoter and coding sequence of GAL1 in the wild type strain (Fig. 5, B and C). In this situation, where non-acetylated histone H3 Lys56 was not evicted from GAL1 (Fig. 1, B and C, and supplemental Fig. S2) (16) or non-acetylated histone H3 Lys56 was not deposited to GAL1 (Fig. 5, B and C, and supplemental Fig. S6, left) (16) in the absence of Rtt109p, we analyzed the deposition of histone H2B to the core promoter and coding sequence of GAL1 in the RTT109 deletion mutant and its isogenic wild type equivalent. We found that histone H2B was deposited to GAL1 in the wild type and RTT109 deletion mutant strains (Fig. 7, A and B). Intriguingly, we found that histone H2B was deposited more efficiently to GAL1 in Δrtt109 as compared with the wild type equivalent (Fig. 7, A and B), even when histone H3 was not deposited in the absence of Rtt109p following transcriptional termination (Fig. 5, B and C). Further, under these growth conditions, a significantly high level of non-acetylated histone H3 Lys56 was present at GAL1 in Δrtt109 (16) (Fig. 1, B and C, and supplemental Figs. S2 and S6, left). Thus, preexisting non-acetylated histone H3 at GAL1 in Δrtt109 enhances the deposition of histone H2B (and hence nucleosomal assembly of histone H2A-H2B dimer) independently of histone H3 deposition. Therefore, Rtt109p or histone H3 Lys56 acetylation appears to repress the association of histone H2A-H2B dimer with histone H3-H4 tetramer and hence chromatin assembly in vivo. Consistently, we also found that histone H2B was not efficiently evicted from the GAL1 core promoter in the absence of Rtt109p immediately following transcriptional induction (Figs. 2A and 3A) when non-acetylated histone H3 Lys56 was present at GAL1 in Δrtt109 (16) (Figs. 1, B and C, and 3B, and supplemental Fig. S2). However, at later induction time points, histone H2B was evicted in the absence of Rtt109p (Fig. 2A), whereas the eviction of non-acetylated histone H3 Lys56 was dramatically decreased in Δrtt109 following transcriptional induction (Figs. 1, B and C, and 3B and supplemental Fig. S6, left). This could be due to more accumulation of the FACT complex to GAL1 at later time points of the transcriptional induction for nucleosomal disassembly/assembly of histone H2A-H2B dimer, as demonstrated previously (47–54).
FIGURE 7.
Histone H2B is deposited more efficiently to the GAL1 core promoter (A) and coding sequence (B) following transcriptional termination in Δrtt109 as compared with the wild type equivalent. Both the wild and mutant strains were grown as in Fig. 5A.
In summary, our data support the fact that histone H3-H4 tetramer and histone H2A-H2B dimer are assembled/disassembled discretely in vivo, and such a process is intimately regulated by Rtt109p or histone H3 Lys56 acetylation. We demonstrate the role of Rtt109p in promoting eviction or repressing deposition of histone H3 (and hence histone H3-H4 tetramer) at GAL1 via histone H3 Lys56 acetylation. Further, we find that nucleosomal assembly/disassembly of the histone H2A-H2B dimer at GAL1 is regulated by Rtt109p through the acetylation status of histone H3 at Lys56 in vivo. Such modulation of nucleosomal assembly/disassembly of histone H3-H4 tetramer and histone H2A-H2B dimer by Rtt109p orchestrates the association of RNA polymerase II with GAL1 and hence its expression. These results, together, provide significant functional insights of Rtt109p or histone H3 Lys56 acetylation in regulation of chromatin assembly/disassembly and hence transcription and other DNA transacting processes in vivo.
Supplementary Material
Acknowledgments
We thank Mary Ann Osley for yeast strains and Shruti Bagla for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R15GM088798-01. This work was also supported by American Heart Association National Scientist Development Grant 0635008N, American Cancer Society Research Scholar Grant 06-52, a Mallinckrodt Foundation award, and several internal grants from Southern Illinois University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- CREB
- cAMP-response element-binding protein
- IP
- immunoprecipitated.
REFERENCES
- 1.Bhaumik S. R., Smith E., Shilatifard A. (2007) Nat. Struct. Mol. Biol. 14, 1008–1016 [DOI] [PubMed] [Google Scholar]
- 2.Shukla A., Chaurasia P., Bhaumik S. R. (2009) Cell Mol. Life Sci. 66, 1419–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malik S., Bhaumik S. R. (2010) FEBS J. 277, 1805–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg A. D., Allis C. D., Bernstein E. (2007) Cell 128, 635–638 [DOI] [PubMed] [Google Scholar]
- 5.Lin W., Dent S. Y. (2006) Curr. Opin. Genet. Dev. 16, 137–142 [DOI] [PubMed] [Google Scholar]
- 6.Peterson C. L., Laniel M. A. (2004) Curr. Biol. 14, R546–R551 [DOI] [PubMed] [Google Scholar]
- 7.Millar C. B., Grunstein M. (2006) Nat. Rev. Mol. Cell Biol. 7, 657–666 [DOI] [PubMed] [Google Scholar]
- 8.Xu F., Zhang K., Grunstein M. (2005) Cell 121, 375–385 [DOI] [PubMed] [Google Scholar]
- 9.Masumoto H., Hawke D., Kobayashi R., Verreault A. (2005) Nature 436, 294–298 [DOI] [PubMed] [Google Scholar]
- 10.Watanabe S., Resch M., Lilyestrom W., Clark N., Hansen J. C., Peterson C., Luger K. (2010) Biochim. Biophys. Acta 1799, 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams S. K., Truong D., Tyler J. K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9000–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J., Zhou H., Li Z., Xu R. M., Zhang Z. (2007) J. Biol. Chem. 282, 28587–28596 [DOI] [PubMed] [Google Scholar]
- 13.Yang J. H., Freudenreich C. H. (2010) DNA Repair 9, 414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Värv S., Kristjuhan K., Peil K., Lõoke M., Mahlakõiv T., Paapsi K., Kristjuhan A. (2010) Mol. Cell. Biol. 30, 1467–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopes da Rosa J., Boyartchuk V. L., Zhu L. J., Kaufman P. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1594–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider J., Bajwa P., Johnson F. C., Bhaumik S. R., Shilatifard A. (2006) J. Biol. Chem. 281, 37270–37274 [DOI] [PubMed] [Google Scholar]
- 17.Han J., Zhou H., Horazdovsky B., Zhang K., Xu R. M., Zhang Z. (2007) Science 315, 653–655 [DOI] [PubMed] [Google Scholar]
- 18.Driscoll R., Hudson A., Jackson S. P. (2007) Science 315, 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsubota T., Berndsen C. E., Erkmann J. A., Smith C. L., Yang L., Freitas M. A., Denu J. M., Kaufman P. D. (2007) Mol. Cell 25, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xhemalce B., Miller K. M., Driscoll R., Masumoto H., Jackson S. P., Kouzarides T., Verreault A., Arcangioli B. (2007) J. Biol. Chem. 282, 15040–15047 [DOI] [PubMed] [Google Scholar]
- 21.Stavropoulos P., Nagy V., Blobel G., Hoelz A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12236–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin C., Yuan Y. A. (2008) Structure 16, 1503–1510 [DOI] [PubMed] [Google Scholar]
- 23.Tang Y., Holbert M. A., Wurtele H., Meeth K., Rocha W., Gharib M., Jiang E., Thibault P., Verreault A., Cole P. A., Marmorstein R. (2008) Nat. Struct. Mol. Biol. 15, 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das C., Lucia M. S., Hansen K. C., Tyler J. K. (2009) Nature 459, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., Chu C. S., Schuldiner M., Gebbia M., Recht J., Shales M., Ding H., Xu H., Han J., Ingvarsdottir K., Cheng B., Andrews B., Boone C., Berger S. L., Hieter P., Zhang Z., Brown G. W., Ingles C. J., Emili A., Allis C. D., Toczyski D. P., Weissman J. S., Greenblatt J. F., Krogan N. J. (2007) Nature 446, 806–810 [DOI] [PubMed] [Google Scholar]
- 26.Han J., Zhou H., Li Z., Xu R. M., Zhang Z. (2007) J. Biol. Chem. 282, 14158–14164 [DOI] [PubMed] [Google Scholar]
- 27.Adkins M. W., Carson J. J., English C. M., Ramey C. J., Tyler J. K. (2007) J. Biol. Chem. 282, 1334–1340 [DOI] [PubMed] [Google Scholar]
- 28.Rufiange A., Jacques P. E., Bhat W., Robert F., Nourani A. (2007) Mol. Cell 27, 393–405 [DOI] [PubMed] [Google Scholar]
- 29.Kaplan T., Liu C. L., Erkmann J. A., Holik J., Grunstein M., Kaufman P. D., Friedman N., Rando O. J. (2008) PLoS Genet. 4, p. e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsukuda T., Fleming A. B., Nickoloff J. A., Osley M. A. (2005) Nature 438, 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhaumik S. R., Green M. R. (2002) Mol. Cell. Biol. 22, 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhaumik S. R., Green M. R. (2003) Methods Enzymol. 370, 445–454 [DOI] [PubMed] [Google Scholar]
- 33.Shukla A., Stanojevic N., Duan Z., Sen P., Bhaumik S. R. (2006) Mol. Cell. Biol. 26, 3339–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhaumik S. R., Raha T., Aiello D. P., Green M. R. (2004) Genes Dev. 18, 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malik S., Shukla A., Sen P., Bhaumik S. R. (2009) J. Biol. Chem. 284, 35714–35724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukla A., Durairaj G., Schneider J., Duan Z., Shadle T., Bhaumik S. R. (2009) J. Mol. Biol. 389, 238–247 [DOI] [PubMed] [Google Scholar]
- 37.Ohsawa R., Adkins M., Tyler J. K. (2009) Epigenetics Chromatin 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korber P., Luckenbach T., Blaschke D., Hörz W. (2004) Mol. Cell. Biol. 24, 10965–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorch Y., Maier-Davis B., Kornberg R. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3090–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boeger H., Griesenbeck J., Strattan J. S., Kornberg R. D. (2004) Mol. Cell 14, 667–673 [DOI] [PubMed] [Google Scholar]
- 41.Williams S. K., Tyler J. K. (2007) Curr. Opin. Genet. Dev. 17, 88–93 [DOI] [PubMed] [Google Scholar]
- 42.Adkins M. W., Williams S. K., Linger J., Tyler J. K. (2007) Mol. Cell. Biol. 27, 6372–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwabish M. A., Struhl K. (2007) Mol. Cell. Biol. 27, 6987–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C. C., Carson J. J., Feser J., Tamburini B., Zabaronick S., Linger J., Tyler J. K. (2008) Cell 134, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jamai A., Puglisi A., Strubin M. (2009) Mol. Cell 35, 377–383 [DOI] [PubMed] [Google Scholar]
- 46.Schwabish M. A., Struhl K. (2006) Mol. Cell 22, 415–422 [DOI] [PubMed] [Google Scholar]
- 47.Orphanides G., LeRoy G., Chang C. H., Luse D. S., Reinberg D. (1998) Cell 92, 105–116 [DOI] [PubMed] [Google Scholar]
- 48.Orphanides G., Wu W. H., Lane W. S., Hampsey M., Reinberg D. (1999) Nature 400, 284–288 [DOI] [PubMed] [Google Scholar]
- 49.Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., Reinberg D. (2003) Science 301, 1090–1093 [DOI] [PubMed] [Google Scholar]
- 50.Schwabish M. A., Struhl K. (2004) Mol. Cell. Biol. 24, 10111–10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mason P. B., Struhl K. (2003) Mol. Cell. Biol. 23, 8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mason P. B., Struhl K. (2005) Mol. Cell 17, 831–840 [DOI] [PubMed] [Google Scholar]
- 53.Ransom M., Williams S. K., Dechassa M. L., Das C., Linger J., Adkins M., Liu C., Bartholomew B., Tyler J. K. (2009) J. Biol. Chem. 284, 23461–23471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reinberg D., Sims R. J., 3rd (2006) J. Biol. Chem. 281, 23297–23301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.