Abstract
NF-κB is an important transcription factor involved in various biological responses, including inflammation, cell differentiation, and tumorigenesis. κB-Ras was identified as an IκB-interacting small GTPase and is reported to disturb cytokine-induced NF-κB activation. In this study, we established that κB-Ras is a novel type of nuclear-cytoplasmic small GTPase that mainly binds to GTP, and its localization seemed to be regulated by its GTP/GDP-binding state. Unexpectedly, the GDP-binding form of the κB-Ras mutant exhibited a more potent inhibitory effect on NF-κB activation, and this inhibitory effect seemed to be due to suppression of the transactivation of a p65/RelA NF-κB subunit. κB-Ras suppressed phosphorylation at serine 276 on the p65/RelA subunit, resulting in decreased interaction between p65/RelA and the transcriptional coactivator p300. Interestingly, the GDP-bound κB-Ras mutant exhibited higher interactive affinity with p65/RelA and inhibited the phosphorylation of p65/RelA more potently than wild-type κB-Ras. Taken together, these findings suggest that the GDP-bound form of κB-Ras in cytoplasm suppresses NF-κB activation by inhibiting its transcriptional activation.
Keywords: G Proteins, NF-kappaB Transcription Factor, Nuclear Translocation, Ras, Transcription, Tumor Necrosis Factor (TNF), Ras Family Small GTP-binding Protein, p300/CBP
Introduction
Ras family GTP-binding proteins regulate cell growth, apoptosis, and cell differentiation in a wide variety of mammalian cells (1, 2). In various cell types, stimulation with several growth factors, including EGF, PDGF, NGF, erythropoietin, and various interleukins, elicits Ras activation, as evidenced by the accumulation of the GTP-bound form of the Ras family. Ras family proteins have significant roles in these growth factor-mediated signal transduction pathways (3–7) and interact with a variety of downstream effectors, including Raf family protein kinase and phosphatidylinositol 3-kinase (PI3K) (8). The activation of GTP-binding proteins occurs through the exchange of bound GDP and GTP, which is regulated by guanine nucleotide exchange factors and GTPase-activating proteins (9, 10). Among the GTPase-activating proteins for Ras, NF1 is well known to function as a tumor suppressor gene product. The amino acid replacement resulting from point mutations in codons 12, 13, or 61 of the ras genes converts these gene products into active oncogenes. In the case of H-Ras, substitution of glycine to valine at position 12 (G12V) or glutamine to leucine at position 61 (Q61L) is well known to decrease their GTPase activity, and these mutations cause transforming activity and have been found in various types of tumors (10, 11). On the other hand, serine at position 17 (Ser-17) and threonine at position 35 (Thr-35) are involved in the interaction with Mg2+ coordinated with guanine nucleotide (12). In particular, the substitution of Ser-17 by asparagine (called S17N) causes the Ras protein to exhibit higher binding affinity to GDP but not GTP, and the Ras (S17N) mutant behaves as a dominant negative mutant in the Ras signaling pathway (13).
The exchange reaction of GDP to GTP causes two regions, switch I (residues 32–40) and switch II (residues 60–76, to change their conformation (14). The conformational change of the switch I region is recognized by effector proteins, and this GTP binding-induced conformational change allows Ras to bind and activate downstream effectors (15). In addition, the C-terminal portion of the Ras family is modified by the farnesyl group, which is essential for their membrane localization and interaction with their target effectors. Recently, the number of Ras family proteins has increased, and their properties have diversified widely.
NF-κB2 is a family of dimeric transcription factors that activate the expression of a variety of genes involved in the immune response, inflammation, development, the regulation of apoptosis, and tumorigenesis (16–19). The five mammalian NF-κB subunits, p50, p52, p65/RelA, c-Rel, and RelB, exhibit homology in their N-terminal sequence of ∼300 amino acids in length, and this is known as the Rel homology domain, which is responsible not only for DNA binding but also for nuclear localization and dimerization. The C-terminal transcriptional activation domain is present only in p65/RelA, c-Rel, and RelB. In unstimulated cells, NF-κB forms an inactive complex in cytosol; for example, the p50 and p65/RelA heterodimer associates with the family of inhibitors of κB (IκB). To activate p50/p65 heterodimers, IκB kinase (IKK) phosphorylates IκBs, leading to the ubiquitination and proteasomal degradation of IκBs. The released p50/p65 heterodimer translocates into the nucleus and binds to promoter regions of its target genes. Furthermore, to exhibit the transcriptional activity of NF-κBs, their direct interaction with the transcriptional coactivator p300/CBP is required (20, 21). In the case of p65/RelA, phosphorylation of the serine residue at 276 by protein kinase A (PKA) accelerates the interaction between the NF-κB complex and p300/CBP (22, 23).
κB-Ras was originally identified as IκB-interacting Ras superfamily GTPases and includes two family proteins, κB-Ras1 and κB-Ras2. κB-Ras was reported to negatively regulate cytokine-induced NF-κB activation (24). Compared with other Ras family GTPases, κB-Ras1 and -2 harbor the well conserved switch I and switch II regions. Amino acid residues involved in the interaction between Ras family small GTPase and Mg2+ coordinated with guanine nucleotide are also conserved (Thr-18 and Thr-36), suggesting that modification of these amino acid residues could affect the function of κB-Ras. Fenwick et al. (24) found several structural differences between κB-Ras and other Ras families. First, κB-Ras harbors alanine or leucine at position 13 (instead of glycine) and leucine at position 65 (instead of glutamine), both of which are equivalent to positions 12 and 61, respectively, in H-Ras. Because these amino acids are known to be critical residues for exhibiting GTP hydrolysis activity, it can be simply speculated that κB-Ras may be constitutively present as a GTP-binding form in the cells. Second, κB-Ras lacks a C-terminal membrane attachment sequence (CAAX motif, where A is any aliphatic amino acid and X is any amino acid), suggesting that localization of κB-Ras is still uncertain. κB-Ras interacted with a complex, including IκBα and NF-κB or IκBβ alone, resulting in the suppression of their proteasomal degradation. Furthermore, another report showed that κB-Ras also inhibits the phosphorylation of IκBβ by the IKK complex in a manner dependent on both the GTP- and GDP-bound forms (25); however, the biochemical properties of κB-Ras and its detailed mechanism for inhibiting NF-κB activation remain to be elucidated.
Here, we found that κB-Ras is a novel type of nuclear-cytoplasmic small GTPase and that its localization may be regulated by its bound guanine nucleotides. We also propose a novel inhibitory mechanism of NF-κB mediated by interrupting the association between the NF-κB subunit, p65/RelA, and transcriptional coactivator p300.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 100 μg/ml penicillin and streptomycin at 37 °C and 5% CO2. KF-8 cells, a stable transfectant of NIH-3T3 cells harboring the NF-κB luciferase reporter gene, were maintained in medium, including 7.5 μg/ml puromycin (26). Transfection into HEK293T cells was performed by the calcium phosphate method or using Lipofectamine 2000 (Invitrogen). For RT-PCR and EMSA analysis, HEK293T cells were transfected by Lipofectamine 2000, and transfection efficiency was evaluated by flow cytometry analysis; efficiency was more than 80% in each experiment (data not shown).
Analysis of κB-Ras-bound GDP and GTP
HEK293T cells were transfected with plasmids encoding κB-Ras2 or κB-Ras2 (T18N) by the calcium phosphate method. Forty eight hours later, cells were incubated in phosphate-free DMEM containing 10% dialyzed FBS and [32P]orthophosphate (0.5 mCi/ml, carrier-free; GE Healthcare) for 120 min. Cells were then harvested and lysed in Ras·GTP immunoprecipitation buffer (4), including 50 mm Tris-HCl (pH 7.5), 20 mm MgCl2, 150 mm NaCl, 0.5% (v/v) Nonidet P-40, and 2 μg/ml aprotinin. κB-Ras proteins were immunoprecipitated with 0.5 μg of FLAG (M2) antibody (Sigma), and bound guanine nucleotides were eluted and developed by PEI-cellulose TLC. The radioactivities of κB-Ras-bound GDP and GTP were measured using an image analyzer BAS-2500 (Fuji Film, Tokyo, Japan).
RNA Interference
Annealed oligonucleotide coding short hairpin RNA (shRNA) for murine κB-Ras2 was inserted into pSUPER-retro-puro retroviral plasmid (Oligoengine, Seattle, WA). The retroviral plasmid and the plasmid encoding retroviral helper were transfected into HEK293T cells using Lipofectamine 2000. Thirty six hours after transfection, the retrovirus secreted into culture supernatant was collected. To infect KF-8 cells with retrovirus harboring shRNA against κB-Ras2, the collected retrovirus was mixed with 8 μg/ml Polybrene and then added to 60-mm culture dishes, including 1 × 105 of KF-8 cells. Three days later, knockdown efficiency was checked by immunoblot analysis for κB-Ras2, followed by reporter gene analysis. The sequences of oligonucleotides used for constructing the shRNA retroviral vector was 5′-GATCCCCGGGCCAAGTCCGAGAAGGTTTCAAGAGAACCTTCTCGGACTTGGCCCTTTTTA-3′ and 5′-AGCTTAAAAAGGGCCAAGTCCGAGAAGGTTCTCTTGAAACCTTCTCGGACTTGGCCCGGG-3′ (underlined sequences correspond to the sequence of murine κB-Ras2 (from 413 to 431 in ORF).
Reporter Gene Analysis
To perform reporter gene analysis, cells were plated in a 24-well plate. HEK293T cells transfected with the indicated combinations of plasmids, including κB-Ras2, κB-Ras2 (T18N), p65/RelA, and/or pNF-κB-luciferase, were stimulated with 10 ng/ml TNFα (PeproTech, Rocky Hill, NJ) for 12 h. Reporter gene analysis was performed using a luciferase assay kit (Promega, Madison, WI). Luciferase activity was normalized by the protein concentration of each lysate. For KF-8 cells, experiments were performed using the same procedure.
RT-PCR Analysis for IL-8
Cells were transfected with plasmids encoding κB-Ras2 or κB-Ras2 (T18N), and 48 h later, cells were stimulated with/without 10 ng/ml TNFα for 3 h. Total RNA was extracted with TRIzol (Sigma). Reverse transcription using oligo(dT)20 primer and PCR was performed by using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and rTaq DNA polymerase (Toyobo, Osaka, Japan). The PCR products were electrophoresed on 1% agarose gel and visualized by ethidium bromide. Then the intensity of the bands was measured and calculated by AE-9000 E-Graph and CS Analyzer 3 (ATTO, Tokyo, Japan), respectively. Primers used were as follows: human IL-8, 5′-GAGCCAGGAAGAAACCACCGGA-3′ (sense) and 5′-GCATCTGGCAACCCTACAACAGACC-3′ (antisense); human GAPDH, 5′-ACCACAGTCCATGCCATCAC-3′ (sense) and 5′-TCCACCACCCTGTTGCTGTA-3′ (antisense).
Preparation of Cytosolic and Nuclear Fraction and EMSA
Cells were transfected with plasmids encoding κB-Ras2 or κB-Ras2 (T18N), and 48 h later, cells were stimulated with 10 ng/ml TNFα for the indicated periods. Cells were lysed in buffer A (10 mm HEPES-KOH (pH 7.8), 10 mm KCl, 0.1 mm EDTA, 0.1% Nonidet P-40, 1 mm DTT, 0.5 mm PMSF, 2 μg/ml aprotinin, 2 μg/ml pepstatin, 2 μg/ml leupeptin). Nuclei were then isolated by centrifugation at 2,000 × g for 2 min. Isolated nuclei were resuspended in buffer C (50 mm HEPES-KOH (pH 7.8), 420 mm KCl, 0.1 mm EDTA, 5 mm MgCl2, 2% glycerol, 1 mm DTT, 0.5 mm PMSF, 2 μg/ml aprotinin, 2 μg/ml pepstatin, 2 μg/ml leupeptin) and rotated at 4 °C for 30 min. Nuclear extracts were prepared by centrifugation at 15,000 rpm for 15 min at 4 °C. The nuclear extracts were mixed with 3 μg of poly(dI-dC) (GE Healthcare) and the radioactively labeled probe (2 × 104 cpm) in a final volume of 25 μl of EMSA binding buffer (10 mm HEPES-KOH (pH 7.8), 1 mm EDTA, 5 mm MgCl2, 10% glycerol, 50 mm KCl) and incubated for 20 min at 25 °C. The protein-DNA complex was separated by 4.5% polyacrylamide gel using 0.5× TGE (12.5 mm Tris, 95 mm glycine, 0.5 mm EDTA) as a running buffer and detected by autoradiography. The annealed oligoprobe includes the κB-responsive element (probe sequence, 5′-AGTTGAGGGGACTTTCCCAGGC-3′). The oligoprobe was radioactively labeled with T4-polynucleotide kinase (Toyobo) using [γ-32P]ATP.
Purification of Recombinant Proteins
The cDNA of κB-Ras2 and T18N mutant was cloned into pCold-I-GST vector, with GST tag inserted into the original pCold-I bacterial expression vector (Takara, Shiga, Japan). pCold-I-GST vector was a gift from Dr. Chojiro-Kojima of Nara Institute of Science and Technology (45). The cDNA of p65/RelA, p65/RelA (S276A), and IκBα was cloned into pCold-I expression vector. Bacteria harboring each plasmid were cultured in LB medium at 37 °C until the A600 reached 0.5, and then the culture temperature was immediately lowered to 16 °C. After 1 h, 100 μm isopropyl 1-thio-β-d-galactopyranoside was added to each culture flask and incubated at 16 °C for 24 h. Bacterial pellets were harvested by centrifugation and suspended in TMNG buffer (50 mm Tris-HCl (pH 7.5), 1 mm MgCl2, 150 mm NaCl, 20% glycerol, and 5 units/ml aprotinin). The suspended bacteria were sonicated, and lysates were clarified by centrifugation at 15,000 rpm for 15 min at 4 °C. Each recombinant protein was collected with DEAE-Sepharose (GE Healthcare) and eluted using a NaCl gradient (0–300 mm). GST-tagged κB-Ras2 and T18N mutant were purified by GST-Sepharose (GE Healthcare). His-p65/RelA, His-p65/RelA (S276A), and His-IκBα were purified by nickel-nitrilotriacetic acid-agarose (Qiagen, Valencia, CA).
In Vitro Guanine Nucleotide Exchange Assays
Two picomoles of GST-κB-Ras2 were mixed with exchange buffer (20 mm HEPES-NaOH (pH 7.5), 5 mm MgCl2, 1 mm EDTA, 1 mm DTT, and 150 mm NaCl). The exchange reaction was started by adding 30 pmol of [35S]GTPγS, and the reactions were carried out for the indicated periods at 20 °C. Subsequently, the reactions were stopped by adding 1 ml of stop buffer (20 mm HEPES-NaOH (pH 7.5), 10 mm MgCl2, and 150 mm NaCl) and transferred to a nitrocellulose membrane. The membranes were washed three times with ice-cold stop buffer. The amount of [35S]GTPγS bound to membranes was counted with a scintillation counter. To compare the binding activity to GDP and GTP, GST-κB-Ras2 was incubated with the indicated concentration of [35S]GTPγS for 30 min at 20 °C. When GDP binding activity was tested, recombinant GST-κB-Ras2 was mixed with GDP exchange buffer (20 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 15 mm EDTA, 1 mm DTT, and 150 mm NaCl) and then incubated with the indicated concentration of [3H]GDP for 30 min at 20 °C.
In Vitro Binding Assay and Kinase Assay
One hundred picomoles of GST-κB-Ras2 or GST-T18N mutant were pretreated with 10 μm GTPγS or 10 μm GDP for 30 min on ice. Guanine nucleotide-loaded GST-κB-Ras2 or -T18N was then divided into 10, 30, 100, or 300 pmol for the following binding reaction. Twenty picomoles of His-p65/RelA were incubated with aliquots of GST-κB-Ras2 with GTPγS or GDP-loaded GST-T18N mutant in 200 μl of binding buffer (20 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 15 mm EDTA, 50 mm NaCl, 10% (v/v) glycerol, 1 mm DTT, 1 mm PMSF, 1 μg/ml aprotinin and leupeptin) for 30 min at 25 °C. To recover the protein complexes, glutathione-Sepharose CL-4B (GE Healthcare) was added to each reaction and gently rotated for 1 h at 4 °C. Recovered protein complexes were washed with binding buffer three times. The precipitates were resolved by SDS-PAGE and immunoblotted with anti-p65/RelA antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-GST antibody. For in vitro kinase assay, 20 pmol of p65/RelA or p65/RelA (S276A) was incubated with 40 units of PKA (New England Biolabs, Ipswich, MA) in kinase reaction buffer (50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 100 μm [γ-32P]ATP (37 TBq/mol) for the indicated periods at 37 °C. To test the effect of κB-Ras, guanine nucleotide-loaded GST-κB-Ras2 or T18N mutant (each 20 pmol) was added. The reaction was terminated by the addition of Laemmli sample buffer, and phosphorylated samples were then resolved by SDS-PAGE and analyzed by an image analyzer, BAS-2500.
Immunoprecipitation and Immunofluorescence Analysis
HEK293T cells were transfected with the indicated combinations of plasmids, including pCMV5-Myc-p65/RelA, pCMV5-FLAG-κB-Ras2, or pCMV5-FLAG-κB-Ras2 (T18N). Forty eight hours after transfection, cells were stimulated with 10 ng/ml TNFα for 60 min and then lysed in a lysis buffer (10 mm sodium phosphate (pH 7.2), 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, 1% sodium deoxycholate, 20 mm β-glycerophosphate, 1 mm NaF, 100 mm Na3VO4, 1 μg/ml aprotinin and leupeptin), and cell lysates were used for immunoprecipitation with 0.5 μg of anti-Myc antibody (9E10; Sigma) and protein-G-Sepharose (GE Healthcare), and the immunoprecipitates were analyzed by immunoblotting with anti-p65/RelA, anti-p300 (Santa Cruz Biotechnology), or anti-κB-Ras2 antibodies. For immunofluorescence analysis, transfected or untransfected HeLa cells were fixed with 4% formaldehyde in PBS for 20 min. After fixation, cells were permeabilized with 1% Triton X-100 in PBS and sequentially stained with anti-κB-Ras1 (Santa Cruz Biotechnology), anti-κB-Ras2, or FLAG (M2) antibodies and Alexa Fluor 488-conjugated goat anti-mouse antibody (Invitrogen). The nucleus was visualized by the addition of Hoechst 33258 (Invitrogen). In experiments testing the effect of leptomycin B (LMB), cells were treated with 10 nm LMB for 6 h and then fixed and stained as above.
Generation of Anti-κB-Ras2 Antibody
The cDNA of κB-Ras2 was cloned into pCold-I cold shock expression vector (Takara). The recombinant protein of His-tagged κB-Ras2 was sequentially purified by DEAE-Sepharose (GE Healthcare) and nickel-nitrilotriacetic acid-agarose (Qiagen). Five milligrams of recombinant protein were utilized as the antigen, and antiserum against κB-Ras2 was produced by Medical and Biological Laboratories (Nagoya, Japan). Anti-κB-Ras2 antibody was purified by κB-Ras2-conjugated agarose resin.
RESULTS
κB-Ras Is a Novel Type of Nuclear-Cytoplasmic Small GTPase
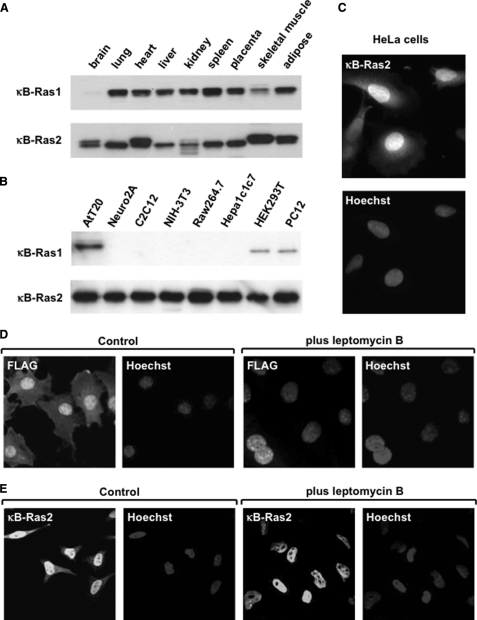
First, we examined the tissue distribution of κB-Ras, using lysates of various murine tissues, by immunoblot analysis. As shown in Fig. 1A, κB-Ras1 and κB-Ras2 were broadly expressed in various mouse tissues, except the brain, in which κB-Ras1 could not be detected; however, among the various cell lines, κB-Ras1 was detected in only AtT20, HEK293T, and PC12 cells (Fig. 1B). On the other hand, κB-Ras2 was broadly expressed in all cell lines examined, and these results encouraged us to focus on κB-Ras2 to test its inhibitory effect on the NF-κB signaling pathway. Next, we performed immunofluorescence analysis to investigate the localization of κB-Ras2. Surprisingly, κB-Ras2 was mainly localized in the nucleus, and slightly in the cytoplasm (Fig. 1C). As in a previous report (24), κB-Ras lacks a CAAX motif on its C terminus, the sequence for farnesylation, which is important for small GTPases to bind to the membrane. Our observation suggests that the absence of lipid modification may result in the nuclear localization of κB-Ras. To test whether κB-Ras shuttles between the nucleus and cytosol, we analyzed the localization of FLAG-κB-Ras2 in LMB-treated NIH-3T3 cells. LMB is known as an inhibitor of Crm1, and thus it inhibits the nuclear export of proteins harboring a nuclear export signal (NES). As shown in Fig. 1D, LMB treatment resulted in the nuclear accumulation of FLAG-κB-Ras2. Additionally, we tested the effect of LMB on the localization of endogenous κB-Ras2, and similar results were observed in HeLa cells (Fig. 1E). These results indicate that κB-Ras is a novel type nuclear cytoplasmic small GTPase and can shuttle between the nucleus and cytosol.
FIGURE 1.
κB-Ras is a nuclear-cytoplasm shuttling small GTPase. A, immunoblotting analysis of κB-Ras1 and κB-Ras2 in adult mouse tissues. Upper and lower photographs show the expression of κB-Ras1 and κB-Ras2. B, immunoblotting analysis of κB-Ras1 and κB-Ras2 in lysates of various human and mouse cell lines. C, immunostaining analysis of κB-Ras2 in HeLa cells. Endogenous κB-Ras2 was detected by anti-κB-Ras2 polyclonal antibody and Alexa594-conjugated secondary antibody. Nucleus was visualized by Hoechst 33258. D, confocal microscopy analysis of κB-Ras2 expressed in NIH-3T3 cells. NIH-3T3 cells expressing FLAG-κB-Ras2 were treated with LMB for 6 h. κB-Ras2 localization was detected by anti-FLAG (M2) antibody and anti-mouse IgG conjugated with Alexa594. Nucleus was visualized by Hoechst 33258. E, confocal microscopy analysis of endogenous κB-Ras2 in HeLa cells. Cells were treated with LMB as in D. κB-Ras2 localization was detected by anti-κB-Ras2 antibody and anti-mouse IgG conjugated with Alexa594. Nucleus was visualized by Hoechst 33258.
κB-Ras Changes Its Localization in a GTP/GDP-dependent Manner
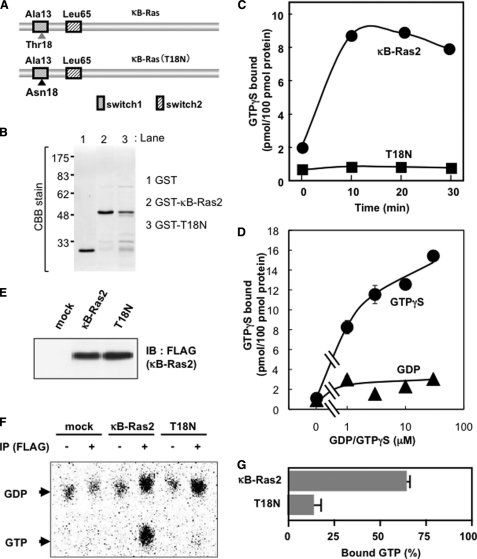
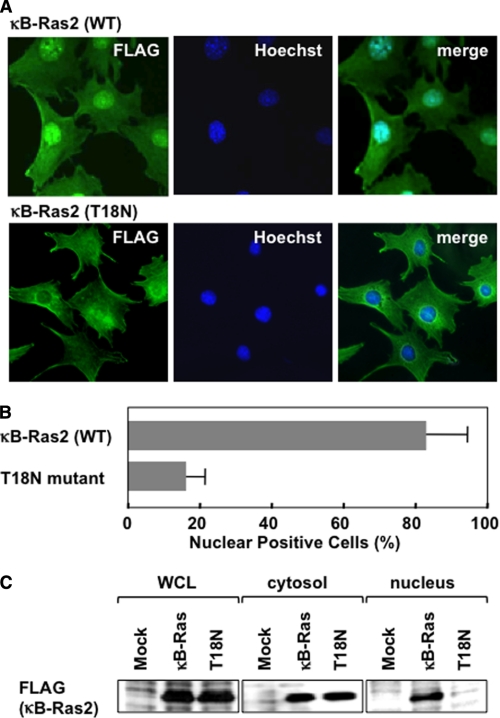
The structure of the Ras family has been analyzed in both the GDP- and GTP-bound forms, which showed marked conformational change confined to regions called switch I and II, as shown in Fig. 2A (14). To clarify the bound guanine nucleotide dependence on the function of κB-Ras, we introduced a point mutation in κB-Ras2. Ser-17 of H-Ras is essential for binding with GTP, and Ras (S17N) mutant behaves as a dominant negative mutant. According to the analogy with other Ras families, Thr-18 of κB-Ras could be important for binding with GTP, thus we substituted Thr-18 into asparagine and named this mutant κB-Ras (T18N) (Fig. 2A). First, we purified the recombinant proteins of κB-Ras2 and κB-Ras2 (T18N) (Fig. 2B), and we examined the GTP binding activity of both wild-type and mutated κB-Ras2 by the in vitro GTPγS binding assay. As shown in Fig. 2C, wild-type κB-Ras2 exhibited GTPγS binding activity; however, the T18N mutant failed to show binding activity. κB-Ras2 harbors alanine at position 13 and leucine at position 65 (24). Hydrophobic amino acids at these positions have been known to decrease the GTPase activity of small GTP-binding proteins, suggesting that κB-Ras may harbor low GTPase activity and exhibit the constitutively GTP-bound form. It is also striking that κB-Ras exhibited little binding ability to GDP, suggesting that κB-Ras has higher affinity to GTP rather than GDP (Fig. 2D). Thus, we next examined the κB-Ras-bound GDP/GTP state in HEK293T cells. As shown in Fig. 2E, wild-type and mutated κB-Ras2 (T18N) were equally expressed in transfected HEK293T cells. We metabolically labeled these transfected cells with [32P]phosphorus, and then guanine nucleotides bound to κB-Ras2 and κB-Ras2 (T18N) were evaluated by TLC using PEI-cellulose. As expected, κB-Ras2 dominantly bound to GTP (>60% total protein), suggesting that κB-Ras2 seems to be the constitutively GTP-bound form in cells (Fig. 2F). On the other hand, T18N mutant failed to show GTP binding activity and mainly bound with GDP (Fig. 2F). Interestingly, the T18N mutant was localized in the cytoplasm, but not the nucleus, although overexpressed wild-type κB-Ras2 was found in both the nucleus and cytoplasm (Fig. 3, A and B). As shown in Fig. 3C, cytosolic and nuclear fractions were prepared from HEK293T cells expressing FLAG-κB-Ras2 or T18N mutant, and localization of these proteins was analyzed by immunoblot analysis using anti-FLAG (M2) antibody. The results were well correlated with Fig. 3A and supported that T18N was specifically localized in the cytosol, whereas wild-type κB-Ras2 was localized in both the nucleus and cytoplasm. Wild-type κB-Ras2 exhibited binding activity to not only GTP but also GDP (Fig. 2F), suggesting that κB-Ras may be localized in a bound guanine nucleotide-dependent manner.
FIGURE 2.
T18N mutant of κB-Ras exhibits low affinity to GTP. A, molecular scheme of wild-type κB-Ras2 and T18N mutant. B, purified recombinant GST, GST-κB-Ras2, and GST-T18N mutant were analyzed by Coomassie Brilliant Blue (CBB) staining. C, GTPγS binding activity of κB-Ras2 and T18N mutant in vitro. Recombinant proteins of κB-Ras2 and T18N mutant were incubated with [35S]GTPγS during the indicated periods, and then protein-GTPγS complexes were collected on a nitrocellulose membrane. D, binding ability of κB-Ras2 to GTPγS and GDP was tested. Recombinant protein of κB-Ras2 was incubated with the indicated concentration of [35S]GTPγS or [3H]GDP for 30 min. Protein-GTPγS or -GDP complexes were collected on a nitrocellulose membrane, and radioactivity was measured. E, expressions of κB-Ras2 and T18N mutant in HEK293T cells. F, cells expressing FLAG-κB-Ras2 or T18N were metabolically labeled with [32P]phosphorus. Cell lysates were utilized for immunoprecipitation using anti-FLAG (M2) antibody. Bound guanine nucleotides were developed by TLC. G, percentage of GTP-bound form is shown on the graph. Error bars, S.D. (n = 3). IP, immunoprecipitation; IB, immunoblot.
FIGURE 3.
κB-Ras changes its localization in a GTP/GDP-dependent manner. A, immunostaining analysis of κB-Ras2 and T18N mutant in NIH-3T3 cells. Experiments were performed as in Fig. 1, C and D. κB-Ras2 localization was detected by anti-FLAG (M2) antibody and anti-mouse IgG conjugated with Alexa594 (green). Nucleus was visualized by Hoechst 33258 (blue). To confirm the localization of κB-Ras2, merged photographs are shown. B, percentage of positive cells with nuclear localization of κB-Ras2 and T18N mutant in NIH-3T3 cells were calculated and are shown in the graph. In the graph, error bars indicate S.D. (n = 3). C, to analyze the subcellular localization of the GDP- or GTP-bound form of κB-Ras, cytosolic and nuclear fractions were prepared from HEK293T cells expressing FLAG-κB-Ras2 or T18N. Each fraction was analyzed by immunoblot analysis with anti-FLAG (M2) antibody. WCL, whole cell lysate.
GDP-bound Form of κB-Ras Exhibits More Potent Inhibitory Effect on NF-κB Activation
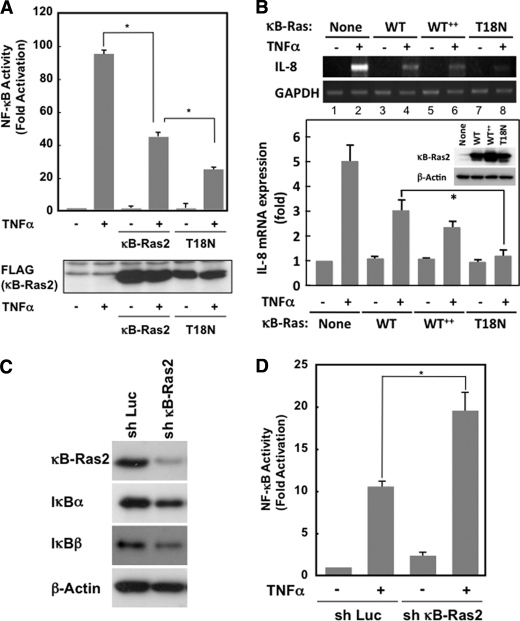
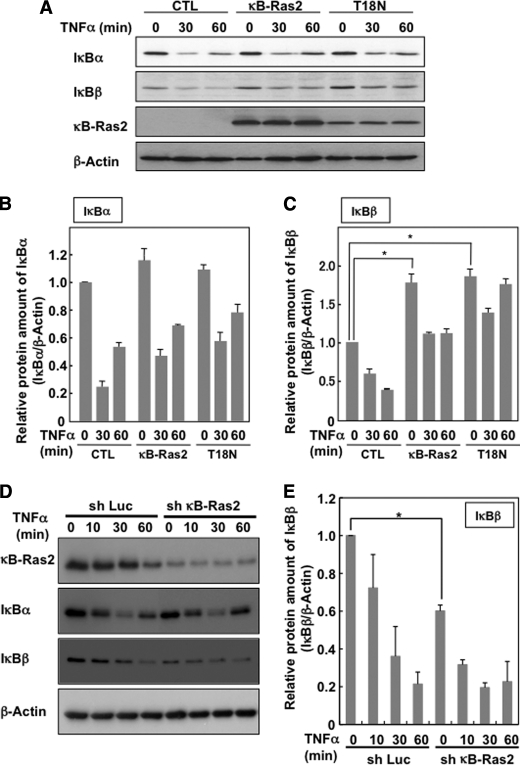
Next, we evaluated the inhibitory effect on NF-κB signal by κB-Ras and its mutant. The overexpression of wild-type κB-Ras2 effectively inhibited TNFα-induced NF-κB activation (Fig. 4A). Interestingly, T18N mutant exhibited a more potent inhibitory effect on NF-κB activation than wild-type κB-Ras2, suggesting that the GDP-bound form of κB-Ras may dominantly inhibit NF-κB activation. We next tested the effect of κB-Ras and its mutant on TNFα-induced interleukin (IL)-8 expression, because IL-8 is known as one of the target genes of NF-κB (27). As shown in Fig. 4B, in HEK293T cells, TNFα stimulation showed strong induction of IL-8 expression; however, κB-Ras2 and T18N mutant effectively diminished IL-8 expression. Wild type of κB-Ras2 exhibited the inhibitory effect on TNFα-induced IL-8 expression in a dose-dependent manner (compare lanes 2, 4, and 6 in RT-PCR, and Mock, WT, and WT++ in immunoblot analysis). When wild-type κB-Ras2 and T18N were equally expressed in the cells, T18N mutant exhibited a more potent inhibitory effect on NF-κB activation than wild-type κB-Ras2 (compare lanes 4 and 8 in RT-PCR and WT and T18N in immunoblot analysis). This result strongly supported the data of reporter gene analysis. Furthermore, we knocked down the expression of endogenous κB-Ras2 in KF-8 cells (26), which stably harbor the luciferase gene driven by κB-responsive element, by retrovirus, including shRNA against κB-Ras2, and then we evaluated the luciferase activity induced by TNFα stimulation. As shown in Fig. 4, C and D, shRNA infection effectively decreased the expression level of κB-Ras2, and this knockdown enhanced TNFα-induced NF-κB activation. When κB-Ras2 was knocked down, the expression of IκBβ but not IκBα was markedly decreased (Fig. 4C). This result was similar to the previous report describing the destabilization of IκBβ by knockdown of κB-Ras (25). Taken together, κB-Ras suppresses TNFα-induced NF-κB activation and the expression of the endogenous NF-κB target gene. This inhibitory effect was observed more potently in the GDP-bound form of the T18N mutant, suggesting that κB-Ras may exhibit the inhibitory effect on NF-κB, when it binds to GDP. These data also suggested that cytoplasmic κB-Ras may exhibit a more putative inhibitory effect on NF-κB activation.
FIGURE 4.
Effect of κB-Ras on cytokine-induced NF-κB activation. A, HEK293T cells transfected with pNF-κB-luciferase plus FLAG-κB-Ras2 or T18N were stimulated with TNFα, and luciferase activity was measured. To verify the protein expression, immunoblot analysis was performed for κB-Ras2 and T18N mutant. B, HEK293T cells seeded on a 60-mm dish were transfected with κB-Ras2 (0.1 or 0.3 μg) or T18N (1 μg). Cells expressing κB-Ras2 (0.1 or 0.3 mg) or T18N were stimulated with TNFα. RT-PCR analysis for IL-8 and GAPDH was performed. The expression level of κB-Ras2 and T18N mutant was evaluated by immunoblot analysis. In the experiments in C and D, NIH-3T3 cells that stably harbor κB-responsive luciferase gene (KF-8 cells) were utilized. C, KF-8 cells were infected with retrovirus harboring shRNA for luciferase (sh-Luc) or murine κB-Ras2 (sh-κB-Ras2). Six days later, cells were harvested for immunoblot analysis to test the expressions of κB-Ras2, IκBα, IκBβ, and β-actin. D, KF-8 cells stably harboring the κB-luciferase gene were infected with control virus (sh-luciferase) or sh-κB-Ras2 virus. Cells were stimulated with TNFα (10 ng/ml) for 12 h. Cells were harvested, and luciferase activity was measured. In each experiment, error bars = S.D. (n = 3; *, p < 0.005).
κB-Ras Inhibits Cytokine-induced IκBβ Degradation
Fenwick et al. (24) and Chen et al. (25) reported that κB-Ras interrupted the degradation of IκBβ. These reports and our current result seemed to support the idea that κB-Ras interrupted the activation of NF-κB mediated by the stabilization of IκBβ. We next tested the effect of κB-Ras2 and T18N mutant on the TNFα-induced degradation of IκB proteins. As shown in Fig. 5, A and B, neither κB-Ras2 nor T18N mutant affected the degradation of IκBα. On the other hand, the expression level of IκBβ in quiescent cells was increased by both κB-Ras2 and T18N mutant. In these cells, TNFα-induced degradation of IκBβ could still be observed; however, the amount of IκBβ protein remaining in TNFα-stimulated cells that expressed κB-Ras2 or T18N was comparable with that in unstimulated control cells (Fig. 5, A and C). Furthermore, we analyzed the TNFα-induced degradation of IκB in NIH-3T3 cells in which κB-Ras2 was knocked down. As shown in Fig. 5, D and E, knockdown of κB-Ras2 effectively decreased the expression of IκBβ and also moderately accelerated the degradation of IκBβ; however, no effect on IκBα was observed. These observations suggest that the suppression of IκBβ degradation seems to be part of the mechanism by which κB-Ras inhibits the activation of NF-κB, as reported previously (24, 25). However, previous reports and our data failed to explain why κB-Ras exhibited the potent inhibitory effect on NF-κB, shown in Fig. 4, because another main regulatory protein, IκBα, was well degraded by TNFα stimulation despite the presence of κB-Ras. These data encouraged us to consider another possible pathway, by which κB-Ras inhibits NF-κB in an IκB stabilization-independent manner.
FIGURE 5.
Effect of κB-Ras on TNFα-induced IκB degradation. A, FLAG-κB-Ras2 and T18N were ectopically expressed in HEK293T cells. Forty eight hours later, cells were stimulated with/without TNFα (10 ng/ml) for the indicated periods, and cells were then lysed with lysis buffer. Obtained lysates were analyzed by immunoblot analysis using anti-IκBα, IκBβ, and β-actin antibodies. CTL, control. To detect overexpressed κB-Ras2, anti-FLAG (M2) antibody was utilized. The degradation of IκBα and IκBβ was normalized with the protein of β-actin, and the quantified ratios of IκBα and IκBβ are shown in B and C, respectively. In each experiment, error bars = S.D. (n = 3; *, p < 0.005). D, NIH-3T3 cells were infected with control virus (sh-Luc) or sh-κB-Ras2 virus. Six days later, cells were stimulated with 10 ng/ml TNFα for the indicated periods and then harvested for immunoblot analysis to test the expressions of κB-Ras2, IκBα, IκBβ, and β-actin. E, relative protein of IκBα and IκBβ was normalized with the protein of β-actin, and the quantified ratios of IκBα and IκBβ are shown. Error bars, S.D. (n = 3; *, p < 0.005).
κB-Ras-induced Inhibition of NF-κB Does Not Require the Stabilization of IκBβ
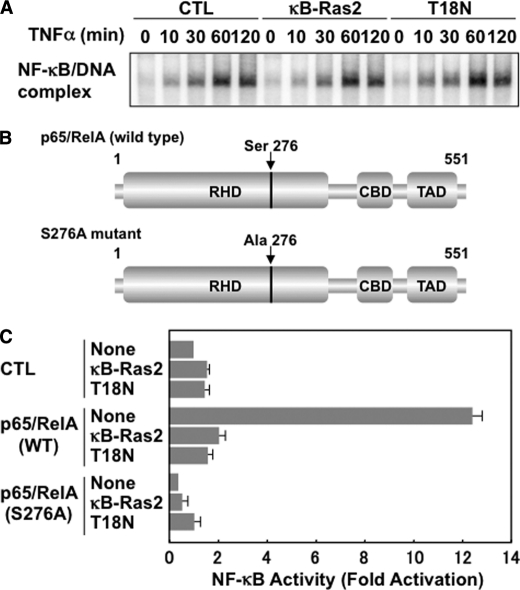
If the stabilization of IκBβ is the main mechanism of the κB-Ras-induced inhibition of NF-κB, it should be simple speculation that κB-Ras suppresses the cytokine-induced nuclear localization of NF-κB. To test this, we analyzed the effect of κB-Ras and T18N mutant on the nuclear localization and the DNA binding activity of NF-κB by EMSA. We stimulated the cells with TNFα for the indicated periods, and then their nuclear extracts were prepared and used for EMSA using an NF-κB-specific probe. In Fig. 6A, TNFα stimulation activated the DNA binding activity of p65/RelA; however, it was interesting that neither κB-Ras2 nor T18N mutant affected the TNFα-induced DNA binding activity of NF-κB. These data apparently abolished the possibility that κB-Ras-induced stabilization of IκBβ may contribute to NF-κB suppression. Furthermore, we next tested the effect of κB-Ras on the transcriptional activity of overexpressed p65/RelA. If κB-Ras inhibits NF-κB activation by inhibiting IκB degradation, κB-Ras should fail to affect the transcriptional activity of overexpressed p65/RelA. For this analysis, we generated an expression vector for wild-type p65/RelA and mutated p65/RelA (S276A) (Fig. 6B). As shown in Fig. 6C, overexpression of p65/RelA markedly induced luciferase expression. On the other hand, p65/RelA (S276A) failed to exhibit transcriptional ability. It is well known that Ser-276 is phosphorylated by several protein kinases, including PKA, and mitogen- and stress-activated protein kinase (MSK1). This phosphorylation is important for the transcriptional activation of p65/RelA (22, 23, 28). Interestingly, Fig. 6C also showed that the transcriptional activity of p65/RelA was markedly diminished by both κB-Ras and T18N mutant, indicating that the stabilization of IκBβ by κB-Ras is unlikely to be sufficient for the κB-Ras-induced potent inhibitory effect on NF-κB activation.
FIGURE 6.
κB-Ras inhibits p65/RelA activation in an IκBβ stabilization-independent manner. A, HEK293T cells expressing κB-Ras2 or T18N were stimulated with TNFα for the indicated periods, and nuclear extracts were then prepared and analyzed by EMSA using an NF-κB-specific probe. CTL indicates control cells transfected with empty vector. B, schemes of wild-type p65/RelA and p65/RelA (S276A) mutant were drawn. RHD, CBD, and TAD indicate Rel homology domain, CBP/p300-binding domain, and transcriptional activating domain, respectively. Numbers mean the position of amino acids in p65/RelA. C, HEK293T cells were transfected with pNF-κB-luciferase plus the indicated combination of plasmids encoding wild-type p65/RelA, p65/RelA (S276A), FLAG-κB-Ras2, and/or T18N mutant. Cells were harvested, and the luciferase activity in each transfected cell was measured. CTL indicates cells transfected without p65/RelA (WT) or p65/RelA (S276A) mutant. Error bars, S.D. (n = 3).
GDP-bound Form of κB-Ras Directly Binds to p65/RelA
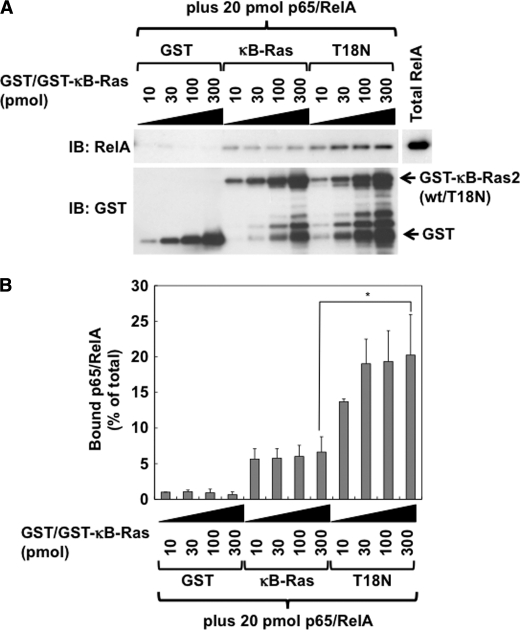
Next, to test whether κB-Ras directly binds to p65/RelA, we performed in vitro pulldown analysis. As shown in Fig. 2D, κB-Ras2 showed low binding ability to GDP. Thus, in the in vitro pulldown assay, T18N mutant was utilized as the GDP-bound form of κB-Ras. We purified GST-κB-Ras2 and GST-T18N mutant and tested their interaction ability with recombinant p65/RelA. It was observed that GDP-bound T18N mutant exhibited higher binding affinity with p65/RelA than wild-type κB-Ras2 (Fig. 7, A and B). These results seemed to support the results in Fig. 4 that the GDP-bound form of κB-Ras has a more potent inhibitory effect on NF-κB.
FIGURE 7.
GDP-bound form of κB-Ras directly interacts with p65/RelA. A, direct interaction between κB-Ras2/T18N and p65/RelA was analyzed by in vitro pulldown assay. Various amounts (10, 30, 100, and 300 pmol) of GST-κB-Ras2 (wild type) or T18N were incubated with 10 μm GTPγS or GDP for 30 min. Then 20 pmol of recombinant p65/RelA was added. Binding reaction was performed for 30 min at 25 °C, and then protein complexes were collected using glutathione-Sepharose. Protein complexes were analyzed by immunoblot (IB) analysis using anti-p65/RelA or anti-GST antibody. B, protein of myc-p65/RelA bound to GST-κB-Ras2 or T18N is shown on the graph. The amount of bound p65/RelA is shown as a percentage of the total amount of input p65/RelA. Error bars, S.D. (n = 3; *, p < 0.005).
κB-Ras Inhibits the Transcriptional Activation of NF-κB
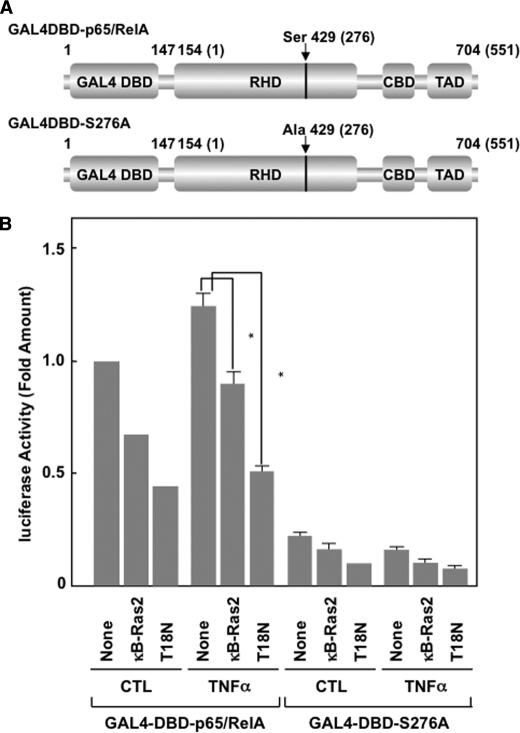
Next, we tested the effect of κB-Ras2 on the transcriptional activation of NF-κB by generating the expression plasmid of a fusion protein of p65/RelA, including the DNA-binding domain of GAL4 transcription factor on the N terminus of p65/RelA, named GAL4DBD-p65/RelA (see Fig. 8A) (29). We also generated an inactive mutant of GAL4DBD-p65/RelA, named GAL4DBD-p65/RelA (S276A), in which Ser at position 276 of p65/RelA was substituted into Ala. We transfected the expression vectors for these fusion proteins with a reporter plasmid harboring a GAL4-responsive element (pFR-luciferase) into HEK293T cells. As shown in Fig. 8B, GAL4DBD-p65/RelA exhibited high transcriptional activity, which was enhanced by TNFα stimulation. On the other hand, GAL4DBD-p65/RelA (S276A) failed to show transcriptional activity. Strikingly, the transcriptional activation of GAL4DBD-p65/RelA was markedly inhibited by κB-Ras2 and κB-Ras2 (T18N). In particular, T18N mutant exhibited a more potent inhibitory effect. These data strongly suggested that κB-Ras seems to stimulate a secondary mechanism that inhibits NF-κB activation mediated by suppression of the transcriptional activity of p65/RelA.
FIGURE 8.
κB-Ras specifically inhibits the transcriptional activation of p65/RelA. A, schemes of GAL4DBD-p65/RelA and GAL4DBD-p65/RelA (S276A) were drawn. RHD, CBD, and TAD indicate the Rel-homology domain, CBP/p300-binding domain, and transcriptional activating domain, respectively. Numbers in parentheses mean the original position of amino acids in p65/RelA. B, HEK293T cells were transfected with pFR-luciferase plus the indicated combination of plasmids encoding GAL4DBD-p65/RelA, GAL4DBD-p65/RelA (S276A), FLAG-κB-Ras2, and/or T18N mutant. Cells were stimulated with TNFα. Cells were harvested and their luciferase activity was measured. CTL indicates unstimulated control. Error bars, S.D. (n = 3; *, p < 0.005).
κB-Ras Inhibits the Interaction between p65/RelA and p300
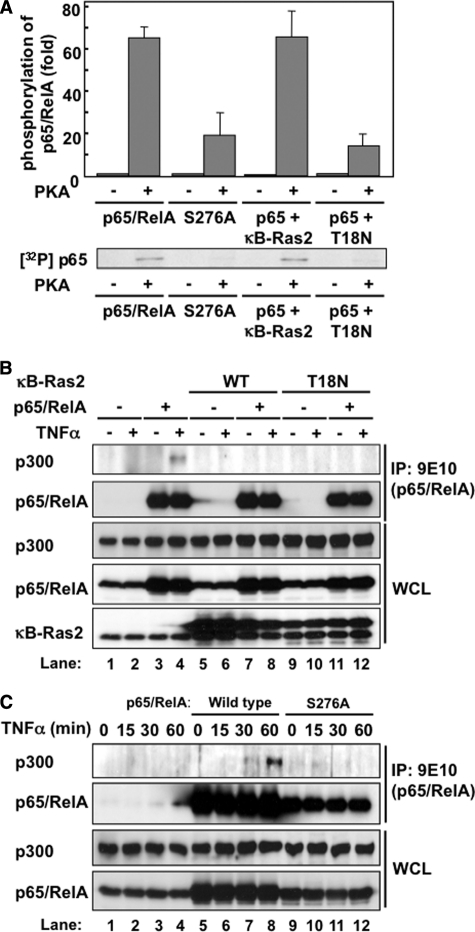
It has been well established that the phosphorylation of p65/RelA at Ser-276 is crucial for its transactivation. When Ser-276 was substituted into alanine in p65/RelA and GAL4DBD-p65/RelA (S276A mutant), these mutants exhibited no transcriptional activity (Figs. 6C and 8B). Zhong et al. reported that PKA phosphorylates p65/RelA at serine 276 (22). Thus, we tested whether κB-Ras inhibits the PKA-mediated phosphorylation of p65/RelA in vitro. To test this, we prepared recombinant proteins of p65/RelA and p65/RelA (S276A) mutant. PKA phosphorylated p65/RelA but failed to phosphorylate S276A mutant (data not shown), suggesting that PKA mainly phosphorylates p65/RelA at serine 276. Next, we tested the effect of κB-Ras2 on PKA-mediated phosphorylation of p65/RelA. As shown in Fig. 9A, wild-type κB-Ras2 with GTPγS showed no effect on p65/RelA phosphorylation, whereas T18N mutant with GDP effectively diminished the phosphorylation of p65/RelA. This result correlated well with the results in Fig. 7, A and B, in which the GDP-bound form of κB-Ras exhibited high binding affinity with p65/RelA and a potent inhibitory effect on p65/RelA phosphorylation. The phosphorylation of p65/RelA at Ser-276 is reported to be a trigger for the interaction between p65/RelA and transcriptional coactivator p300 (23). Thus, we next tested whether κB-Ras inhibits the interaction between p65/RelA and p300. As shown in Fig. 9B, TNFα stimulation accelerated the interaction of p65/RelA with p300 (compare lanes 3 and 4). Strikingly, both κB-Ras2 and κB-Ras2 (T18N) effectively diminished the binding of p65/RelA with p300 (Fig. 9B, compare lanes 4, 8, and 12). Additionally, we tested the importance of Ser-276 of p65/RelA for the interaction with p300. As shown in Fig. 9C, wild-type p65/RelA bound to p300 in a TNFα stimulation-dependent manner (lanes 7 and 8); however, p65/RelA (S276A) failed to interact with p300 when stimulated with TNFα (Fig. 9C, lanes 11 and 12). Taken together, our data strongly suggest a novel inhibitory mechanism of NF-κB transcriptional activation by κB-Ras though inhibition of the interaction between p65/RelA and p300, which may be due to the inhibition of p65/RelA phosphorylation by the GDP-bound form of κB-Ras (Fig. 10).
FIGURE 9.
κB-Ras interrupts the interaction between p65/RelA and p300. A, effect of κB-Ras on PKA-catalyzed phosphorylation of p65/RelA in vitro. Recombinant proteins of p65/RelA or p65/RelA (S276A) were incubated with recombinant PKA in kinase reaction buffer including [γ-32P]ATP for 2 h at 37 °C. Phosphorylated samples were resolved by SDS-PAGE and analyzed by autoradiography. B, HEK293T cells were transfected with the indicated combination of plasmids encoding myc-p65/RelA, FLAG-κB-Ras2, and T18N mutant. Cells were stimulated with TNFα for 30 min, and their cell lysates were then used for immunoprecipitation (IP) with anti-Myc (9E10) antibody. Immunoprecipitates and whole cell lysates (WCL) were analyzed by immunoblotting with anti-p65/RelA, anti-p300, and anti-κB-Ras2 antibodies. CTL indicates transfection without κB-Ras2 or T18N. C, HEK293T cells expressing myc-p65/RelA or myc-p65/RelA (S276A) mutant were stimulated with 10 ng/ml TNFα for the indicated periods, and then their cell lysates were used for immunoprecipitation with anti-Myc (9E10) antibody. Immunoprecipitated samples and WCL were analyzed by immunoblotting with anti-p65/RelA and anti-p300 antibodies.
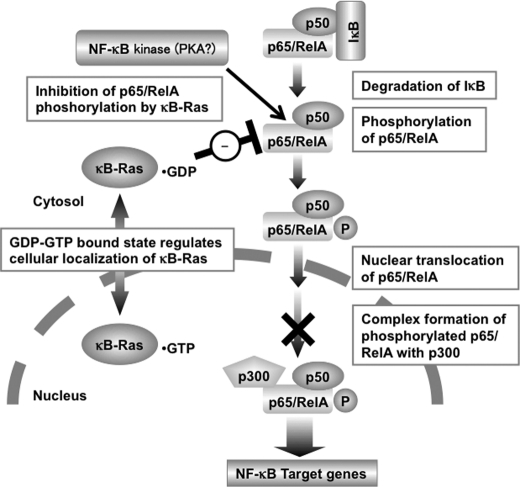
FIGURE 10.
Working hypothesis of κB-Ras-induced NF-κB inhibition. According to our current study, κB-Ras seems to inhibit NF-κB activation by interrupting the transcriptional activation of p65/RelA. The GDP-bound form of κB-Ras localized in cytosol mainly contributes to this suppressive mechanism.
DISCUSSION
p65/RelA is known to be phosphorylated by various protein kinases to exhibit its transcriptional activity (30). A previous report and our current data suggested that the phosphorylation of p65/RelA on serine 276 seems to be essential for its transcriptional activation (Figs. 6A and 7B) (18). Zhong et al. (23) also reported that the phosphorylation of the p65/RelA subunit at Ser-276 was catalyzed by PKA. The catalytic subunit of PKA is maintained in an inactive state by association with the NF-κB-IκB complex. Cytokine signals causing IκB degradation induce the activation of PKA in a cAMP-independent manner; however, the functional role of PKA in the NF-κB regulating signaling pathway is still controversial. Neumann et al. (31) reported that p65/RelA is one of the target molecules for PKA-mediated immunosuppression in T-lymphocytes. According to their study, PKA signal impairs the nuclear translocation and DNA binding activity of p65/RelA. Furthermore, Takahashi et al. (32) showed that PKA-activating reagents or the expression of the exogenous PKA catalytic subunit diminished TNFα-induced NF-κB activation. They suggested that the inhibitory action of PKA seems to be through the C-terminal transcription-activating domain of p65/RelA; however, the phosphorylation of p65/RelA at serine 276 is not involved in this action. As shown in Fig. 6A, the recombinant protein of p65/RelA (S276A) was barely phosphorylated by PKA in vitro, suggesting that serine 276 seems to be the main phosphorylation site by PKA. These observations highlight factors determining the directionality of PKA signaling on NF-κB activation. Recently, protein kinase A-interacting protein 1 (AKIP1) was identified as a binding partner of p65/RelA, and AKIP1 enhanced PKA-mediated phosphorylation at serine 276 and transcriptional activation of p65/RelA (33). This study showed that mainly the GDP-bound form of κB-Ras2 impaired the PKA-mediated phosphorylation of p65/RelA and its association with p300/CBP; however, it remains possible that the effect of κB-Ras on p65/RelA activation is indirect. Although we have no additional evidence, AKIP1 may be another targeting molecule of κB-Ras for NF-κB suppression.
In our study, we have proposed a new mechanism, by which κB-Ras2 inhibits cytokine-induced NF-κB activation. Fenwick et al. (24) reported that κB-Ras is associated with NF-κB-IκBβ complexes in cells and results in a slower rate of degradation of IκBβ than IκBα. Chen et al. (25) also provided supporting evidence that RNA interference with κB-Ras1 and κB-Ras2 accelerates the degradation of IκBβ but not IκBα. We also observed that κB-Ras2 and T18N mutant slowed down the degradation of IκBβ (Fig. 5, A and C); however, neither the nuclear translocation nor the DNA binding activity of p65/RelA was inhibited by κB-Ras2 and T18N mutant (Fig. 6A). This encouraged us to consider that κB-Ras may utilize a different pathway to inhibit NF-κB activation, in addition to the suppression of IκBβ degradation. According to our results, κB-Ras exhibits a putative inhibitory effect against NF-κB activation by interrupting the interaction between p65/RelA and p300/CBP through the suppression of p65/RelA phosphorylation.
Our results also suggested that κB-Ras might be a nucleo-cytoplasmic shuttling small GTPase and its localization seemed to change in a bound GTP/GDP-dependent manner (Fig. 3). Interestingly, the results of the in vitro kinase assay of p65/RelA suggested that the GDP-bound form of κB-Ras2 preferentially inhibited PKA-catalyzed p65/RelA phosphorylation (Fig. 9A); thus, it is suggested that κB-Ras-mediated inhibition of p65/RelA phosphorylation might occur in the cytoplasm. In a previous study, PKA associated with the NF-κB-IκB complex was reported to be activated via the degradation of IκB, which is induced in cytosol (22). This seemed to fit our proposed model that the cytoplasmic GDP-bound form of κB-Ras inhibits p65/RelA phosphorylation. On the other hand, another protein kinase, Msk1, is reported to phosphorylate p65/RelA at serine 276 in the nucleus (28). Msk1 exhibits structural similarity with the family protein of MAPKAP kinase, which functions downstream of the mitogen- and stress-activated protein kinase family (34). MSK1 was first identified as a nuclear CRAB and histone H3 kinase that responds to both ERKs and p38 MAPK (35, 36); however, it is still unclear if κB-Ras can affect Msk1-induced p65/RelA phosphorylation. During our study, we found several differences with previous reports. In Fig. 1A, Fenwick et al. (24) reported that κB-Ras1 was specifically expressed in the brain, heart, and skeletal muscle; however, our data showed a broad expression of κB-Ras1, except in the brain. There is no suitable explanation; however, this might have been due to the difference between RNA and protein expressions. Although we have no evidence for this, protein expression might be regulated by not only mRNA transcription but also other factors, including the activity of protein synthesis. In addition, we analyzed the lysates of mouse tissues, whereas Fenwick et al. (24) analyzed human RNA. Additionally, we could not clearly explain why T18N mutant exhibited a greater inhibitory effect on NF-κB activation than wild-type κB-Ras. As shown in Fig. 2F, T18N specifically bound to GDP, whereas wild-type κB-Ras could bind to both GDP and GTP. The differences in bound guanine nucleotide specificity may allow the T18N mutant to exhibit a greater inhibitory effect than wild-type κB-Ras.
On the other hand, our data exhibited some discrepancies that wild-type κB-Ras perfectly inhibited overexpressed p65/RelA-induced NF-κB-responsive promoter activation (Fig. 6C) and the interaction between p300 and p65/RelA (Fig. 9B), although wild-type κB-Ras failed to inhibit the PKA-induced phosphorylation of p65/RelA. We do not have additional data for explaining it. Activity of p300/CBP may be regulated by various signaling, although the functional modification of p300/CBP has been poorly understood except SUMOylation of p300/CBP (37). κB-Ras failed to induce the SUMOylation of p300/CBP (data not shown); however, it is still possible that κB-Ras may have the unknown functions and affect the activity of p300/CBP. Therefore, κB-Ras may affect the activities of various transcription factors. This problem would be a future project that should be clarified.
NF-κB complexes, including p65/RelA and c-Rel, are regulated by IκB proteins and can be activated by proteasomal degradation of IκBs; however, there have been no reports on whether κB-Ras affects the activation of other NF-κB complexes, including p50, p52, and RelB, which were regulated by the IκB-independent pathway. NF-κB complex of p50/p52 functions in B-cell receptor signaling, and this alterative pathway requires two protein kinases, NIK and IKKα but not IKKβ (38). In quiescent cells, RelB interacts with p100, a precursor of p52 subunit. Once cells are stimulated with NF-κB-activating signals, the associated p100 is digested to p52, and then RelB and p52 form a transcriptional active complex. RelB, unlike p65/RelA and c-Rel, includes a leucine zipper domain required for its full transcriptional activation (39). RelB-deficient mice did not show lethality but had altered lymph node development and defective splenic and thymic stroma cells (40). Our current observation not only suggested a new mechanism of NF-κB inhibition but also showed that κB-Ras may regulate the activation of this IκB-insensitive NF-κB family and its functions.
In this study, we reported that the GDP-bound form of κB-Ras mainly exhibits an inhibitory effect on the transcriptional activation of p65/RelA; however, this is a noncanonical case, because almost all types of GTP-binding proteins exhibit this ability when they bind to GTP but not GDP (41). Recently, the GDP-bound form of Arf6 was reported to recruit Rac guanine nucleotide exchange factor Kalirin to the plasma membrane and to be involved in Rac activation (42). Arf6 has been clarified to activate various signaling pathways, including phosphatidylinositol 5-phosphate kinase and an Arf guanine nucleotide exchange factor ARNO/cytohesin (43, 44). We showed that more than 60% of κB-Ras2 binds to GTP in cells (Fig. 2F). The κB-Ras-interacting protein harboring GTPase-activating protein activity for κB-Ras may have a pivotal role in the κB-Ras-mediated signaling pathway, including the suppression of NF-κB. To clarify these problems, it is important to identify κB-Ras-interacting proteins, including κB-Ras-specific GTPase-activating proteins.
Acknowledgment
We are grateful to Dr. Chojiro Kojima for providing the pCold-I-GST bacterial expression vector.
This work was supported by Grants-in-aid for Scientific Research 20770103 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
- NF-κB
- nuclear factor-κB
- IκB
- inhibitor of NF-κB
- IKK
- IκB kinase
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- LMB
- leptomycin B.
REFERENCES
- 1.Lowy D. R., Willumsen B. M. (1993) Annu. Rev. Biochem. 62, 851–891 [DOI] [PubMed] [Google Scholar]
- 2.Satoh T., Nakafuku M., Kaziro Y. (1992) J. Biol. Chem. 267, 24149–24152 [PubMed] [Google Scholar]
- 3.Satoh T., Endo M., Nakafuku M., Akiyama T., Yamamoto T., Kaziro Y. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 7926–7929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satoh T., Minami Y., Kono T., Yamada K., Kawahara A., Taniguchi T., Kaziro Y. (1992) J. Biol. Chem. 267, 25423–25427 [PubMed] [Google Scholar]
- 5.Gibbs J. B., Marshall M. S., Scolnick E. M., Dixon R. A., Vogel U. S. (1990) J. Biol. Chem. 265, 20437–20442 [PubMed] [Google Scholar]
- 6.Qiu M. S., Green S. H. (1991) Neuron 7, 937–946 [DOI] [PubMed] [Google Scholar]
- 7.Torti M., Marti K. B., Altschuler D., Yamamoto K., Lapetina E. G. (1992) J. Biol. Chem. 267, 8293–8298 [PubMed] [Google Scholar]
- 8.Cox A. D., Der C. J. (2002) Cancer Biol. Ther. 1, 599–606 [DOI] [PubMed] [Google Scholar]
- 9.Buday L., Downward J. (2008) Biochim. Biophys. Acta 1786, 178–187 [DOI] [PubMed] [Google Scholar]
- 10.Karnoub A. E., Weinberg R. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bos J. L. (1989) Cancer Res. 49, 4682–4689 [PubMed] [Google Scholar]
- 12.John J., Rensland H., Schlichting I., Vetter I., Borasio G. D., Goody R. S., Wittinghofer A. (1993) J. Biol. Chem. 268, 923–929 [PubMed] [Google Scholar]
- 13.Feig L. A., Cooper G. M. (1988) Mol. Cell. Biol. 8, 3235–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milburn M. V., Tong L., deVos A. M., Brünger A., Yamaizumi Z., Nishimura S., Kim S. H. (1990) Science 247, 939–945 [DOI] [PubMed] [Google Scholar]
- 15.Herrmann C. (2003) Curr. Opin. Struct. Biol. 13, 122–129 [DOI] [PubMed] [Google Scholar]
- 16.Hayden M. S., West A. P., Ghosh S. (2006) Oncogene 25, 6758–6780 [DOI] [PubMed] [Google Scholar]
- 17.Pacifico F., Leonardi A. (2006) Biochem. Pharmacol. 72, 1142–1152 [DOI] [PubMed] [Google Scholar]
- 18.Schulze-Luehrmann J., Ghosh S. (2006) Immunity 25, 701–715 [DOI] [PubMed] [Google Scholar]
- 19.Karin M. (2008) Cell Res. 18, 334–342 [DOI] [PubMed] [Google Scholar]
- 20.Gerritsen M. E., Williams A. J., Neish A. S., Moore S., Shi Y., Collins T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkins N. D., Felzien L. K., Betts J. C., Leung K., Beach D. H., Nabel G. J. (1997) Science 275, 523–527 [DOI] [PubMed] [Google Scholar]
- 22.Zhong H., SuYang H., Erdjument-Bromage H., Tempst P., Ghosh S. (1997) Cell 89, 413–424 [DOI] [PubMed] [Google Scholar]
- 23.Zhong H., Voll R. E., Ghosh S. (1998) Mol. Cell 1, 661–671 [DOI] [PubMed] [Google Scholar]
- 24.Fenwick C., Na S. Y., Voll R. E., Zhong H., Im S. Y., Lee J. W., Ghosh S. (2000) Science 287, 869–873 [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Vallee S., Wu J., Vu D., Sondek J., Ghosh G. (2004) Mol. Cell. Biol. 24, 3048–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonoda Y., Kasahara T., Yamaguchi Y., Kuno K., Matsushima K., Mukaida N. (1997) J. Biol. Chem. 272, 15366–15372 [DOI] [PubMed] [Google Scholar]
- 27.Funakoshi-Tago M., Shimizu T., Tago K., Nakamura M., Itoh H., Sonoda Y., Kasahara T. (2008) Biochem. Pharmacol. 76, 662–671 [DOI] [PubMed] [Google Scholar]
- 28.Vermeulen L., De Wilde G., Van Damme P., Vanden Berghe W., Haegeman G. (2003) EMBO J. 22, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furusawa J., Funakoshi-Tago M., Tago K., Mashino T., Inoue H., Sonoda Y., Kasahara T. (2009) Cell. Signal. 21, 778–785 [DOI] [PubMed] [Google Scholar]
- 30.Viatour P., Merville M. P., Bours V., Chariot A. (2005) Trends Biochem. Sci. 30, 43–52 [DOI] [PubMed] [Google Scholar]
- 31.Neumann M., Grieshammer T., Chuvpilo S., Kneitz B., Lohoff M., Schimpl A., Franza B. R., Jr., Serfling E. (1995) EMBO J. 14, 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi N., Tetsuka T., Uranishi H., Okamoto T. (2002) Eur. J. Biochem. 269, 4559–4565 [DOI] [PubMed] [Google Scholar]
- 33.Gao N., Asamitsu K., Hibi Y., Ueno T., Okamoto T. (2008) J. Biol. Chem. 283, 7834–7843 [DOI] [PubMed] [Google Scholar]
- 34.Gaestel M. (2006) Nat. Rev. Mol. Cell Biol. 7, 120–130 [DOI] [PubMed] [Google Scholar]
- 35.Deak M., Clifton A. D., Lucocq L. M., Alessi D. R. (1998) EMBO J. 17, 4426–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson S., Clayton A. L., Hazzalin C. A., Rose S., Barratt M. J., Mahadevan L. C. (1999) EMBO J. 18, 4779–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Girdwood D., Bumpass D., Vaughan O. A., Thain A., Anderson L. A., Snowden A. W., Garcia-Wilson E., Perkins N. D., Hay R. T. (2003) Mol. Cell 11, 1043–1054 [DOI] [PubMed] [Google Scholar]
- 38.Dejardin E. (2006) Biochem. Pharmacol. 72, 1161–1179 [DOI] [PubMed] [Google Scholar]
- 39.Dobrzanski P., Ryseck R. P., Bravo R. (1993) Mol. Cell. Biol. 13, 1572–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weih D. S., Yilmaz Z. B., Weih F. (2001) J. Immunol. 167, 1909–1919 [DOI] [PubMed] [Google Scholar]
- 41.Kaziro Y., Itoh H., Kozasa T., Nakafuku M., Satoh T. (1991) Annu. Rev. Biochem. 60, 349–400 [DOI] [PubMed] [Google Scholar]
- 42.Koo T. H., Eipper B. A., Donaldson J. G. (2007) BMC Cell Biol. 8, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honda A., Nogami M., Yokozeki T., Yamazaki M., Nakamura H., Watanabe H., Kawamoto K., Nakayama K., Morris A. J., Frohman M. A., Kanaho Y. (1999) Cell 99, 521–532 [DOI] [PubMed] [Google Scholar]
- 44.Cohen L. A., Honda A., Varnai P., Brown F. D., Balla T., Donaldson J. G. (2007) Mol. Biol. Cell 18, 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi K., Kojima C. (2008) Protein Expr. Purif. 62, 120–127 [DOI] [PubMed] [Google Scholar]