Abstract
The catalytic domain of metalloelastase (matrix metalloproteinase-12 or MMP-12) is unique among MMPs in exerting high proteolytic activity upon fibrils that resist hydrolysis, especially elastin from lungs afflicted with chronic obstructive pulmonary disease or arteries with aneurysms. How does the MMP-12 catalytic domain achieve this specificity? NMR interface mapping suggests that α-elastin species cover the primed subsites, a strip across the β-sheet from β-strand IV to the II–III loop, and a broad bowl from helix A to helix C. The many contacts may account for the comparatively high affinity, as well as embedding of MMP-12 in damaged elastin fibrils in vivo. We developed a strategy called BINDSIght, for bioinformatics and NMR discovery of specificity of interactions, to evaluate MMP-12 specificity without a structure of a complex. BINDSIght integration of the interface mapping with other ambiguous information from sequences guided choice mutations in binding regions nearer the active site. Single substitutions at each of ten locations impair specific activity toward solubilized elastin. Five of them impair release of peptides from intact elastin fibrils. Eight lesions also impair specific activity toward triple helices from collagen IV or V. Eight sites map to the “primed” side in the III–IV, V–B, and S1′ specificity loops. Two map to the “unprimed” side in the IV–V and B–C loops. The ten key residues circumscribe the catalytic cleft, form an exosite, and are distinctive features available for targeting by new diagnostics or therapeutics.
Keywords: Collagen, Elastin, Matrix Metalloproteinase, NMR, Protein-Protein Interactions, Proteolytic Enzymes, Biomolecular Recognition, Catalytic Efficiency, Specificity
Introduction
Although protein-protein interactions are universally important, mechanistic understanding of their specificity is often poor (1). An impediment to detailed understanding of proteolytic attack of proteins is the transience and potential heterogeneity of the interactions, which interfere in capturing the structure of a substrate complex by crystallography or other methods. These complications affect characterization of matrix metalloproteinase-12 (MMP-12),3 the metalloelastase secreted by human macrophages at sites of inflammation. To investigate how MMP-12 achieves specificity for protein fibrils from lungs and arteries, we developed an approach designated BINDSIght, for its combination of bioinformatics and NMR discovery of specificity of interactions.
In lungs, arteries, skin, and basement membranes, elastin provides elastic recoil, is heavily cross-linked, and is difficult to digest. Collagens are ubiquitous and comprise ∼25% of the protein mass of the body. Damage to fibrils of the extracellular matrix by proteases such as MMP-12 contributes to the inflammation and chronic disease states of chronic obstructive pulmonary disease (2–4), atherosclerosis (5–6), abdominal aortic aneurysm (7), multiple sclerosis (8), ulcerative colitis (9), asthma (4), and rheumatoid arthritis (10). The progression of chronic obstructive pulmonary disease/emphysema and (abdominal aortic aneurysm) in smokers depends in large part on MMP-12 expression (11) and its degradation of elastin (7, 12). Release of elastin fragments provides crucial stimulation thought to amplify and prolong chronic inflammation (13). Minute quantities of 10- to 50-kDa fragments of elastin are potent chemoattractants of monocytes to the lung in pulmonary emphysema where they differentiate into alveolar macrophages (14) that secrete MMP-12 (15). Elastin degradation products are released largely by neutrophil elastase in concert with MMP-12 (12). 36 sites of MMP-12 digestion of elastin fibrils have been identified (16).
Mature elastin fibrils are insoluble, extensible, and intimately mingled with collagen, fibulin, other glycoproteins, and polysaccharides such as chondroitin sulfate (17–18). Mature elastin chains are cross-linked by desmosine linkages formed among most lysine residues (18, 19). Elastin is “rubber-like,” amorphous by x-ray fiber diffraction, and does not crystallize (20). It is heterogeneous from time of secretion as tropoelastin (21) through its maturation with progressively more cross-linking by lysyl oxidase (22) and when solubilized into α-elastin (23). Tropoelastin contains 700 residues that are mainly glycine, alanine, valine, and proline in repetitive sequences (rendering it unfit for NMR structure) and organized into domains that are alternately hydrophobic (Gly-rich or Pro-rich) or for cross-linking (Lys-containing and enriched in Ala or Pro) (20). Its structure largely appears to be extended polyproline II conformation in equilibrium with disorder, plus β-turns thought to shift in lending entropy gain and elasticity (20). Tropoelastin is susceptible to cross-linking in domains 12 and 19–25 (24). Investigations have focused on soluble derivatives of mature elastin such as α-elastin and peptide models of repeating sequences (20). When warmed toward 37 °C, α-elastin reversibly phase separates into a viscous, milky liquid (17) with increased structure (25) and formation of 50-Å filaments and 700-Å fibrils that resemble elastin (26). The reductionist approach has been validated by the self-assembly intrinsic to α-elastin and peptide fragments as small as 3.4 kDa (27, 28).
Ability to digest elastin, collagen IV, fibrillar collagens I and V, and the triple helical peptide (THP) mimic of the cleavage site in collagen V is shared among MMPs 2, 9, and 12 (29–34). Collagen V and this THP are not susceptible to cleavage by other MMPs (34). Collagen V is a component of ubiquitous collagen I fibrils that regulates their diameter (35). The catalytic domain of MMP-12 hydrolyzes skin collagen types I and III at several sites (32). Insight into the unique ability of its catalytic domain to digest collagens V and I may have broader relevance to the classic question of how homologous catalytic domains of collagenases engage and attack the triple helix. MMP-2 and -9 require their insertions of fibronectin-like modules to digest elastin, collagens, and THPs (36–40). By contrast, the activated form of MMP-12 in vivo is simply its catalytic domain (15). Why is the catalytic domain of MMP-12 sufficient for high activity in cleaving protease-resistant fibrils such as elastin and collagen V? Addressing this question may lend insight into specificity of MMPs more generally and into strategies for selective and clinical recognition and inhibition.
Past investigations of specificity focused on interactions of medicinal compounds with the S1′ specificity pocket (41–43) in a quest for selectivity to diminish side effects. MMP-12 and its close homologues of MMP-3, -8, and -13 possess similar S1′ specificity pockets (42). The side chains of protein substrates are, however, too short to sample the depths of the S1′ pocket. Peptides from elastin were recently simulated to extend across the active site cleft of MMP-12 (44). Given the high conservation of the central active site cleft, what might imbue the MMP-12 catalytic domain with its specificities? Elastins and collagens should spill out of the cleft. Consistent with this, the triple helical peptidase activity of MMP-1, -8, and -12 involves the V–B loop preceding the active-site helix B (33, 45–47). A triple helical peptide substrate derived from collagen V contacts not only this exosite but also appears to cover other sites in loops and atop the β-sheet even more remote from the active site cleft of MMP-12 (33).
To probe subtle features that tune the specificities, we employed: (i) NMR to locate physical contacts between inactivated enzyme and mini-protein substrates, (ii) sequence positions that distinguish enzyme subfamilies, and (iii) subtle, targeted lesions on the surface that aim to impair only specific activities. This BINDSIght combination of experimental interface mapping with distinctiveness in sequence reliably identified less than obvious residues as conferring advantages to MMP-12 in specific activity for substrates from elastin and collagen fibrils, relative to MMP-3. The catalytic domain of MMP-3 is useful for comparison due to its relative lack of the specific activities, its 60% sequence identity with MMP-12, and sharing almost indistinguishable backbone structural coordinates (48). The BINDSIght approach builds on NMR evidence that 20-kDa species of solubilized elastin protect the surface of MMP-12 from its active-site cleft across flanking loops and a strip across the β-sheet to a broad surface on the most remote side of the catalytic domain. Point mutations of interfacial residues at positions on the periphery of the active site that distinguish MMP-12 from other subfamilies preserve a wild-type level of general MMP peptidase activity while diminishing the specific activities toward much bulkier elastin and collagen triple helical substrates. The evidences suggest that optimization of MMP-12 for digestion of solubilized elastin and some triple helices can involve the perimeter of the active site and at least one exosite.
EXPERIMENTAL PROCEDURES
MMP Samples
The catalytic domain of human MMP-12 (EC 3.4.24.65) extends from Phe-100 to Gly-263. Samples of it were prepared as described (49), but with an optimized folding procedure (50). A final SP-Sepharose step (pH 7.5) removed any misfolded MMP-12. The catalytic domain of human MMP-3 (EC 3.4.24.17) was also prepared from inclusion bodies (51). Samples for activity analyses were stored in small aliquots at −80 °C at 10 μm in 20 mm Tris-HCl (pH 7.2), 10 mm CaCl2, 0.1 mm ZnCl2, 50% w/v glycerol. NMR samples of MMP-12(E219A) were typically 0.3–0.4 mm in 20 mm imidazole (pH 6.6), 10 mm CaCl2, and 20 μm ZnCl2.
Elastin Samples
Insoluble elastin fibrils conjugated with fluorescein were from Elastin Products Co. (Owensville, MO). Soluble α-elastin from bovine neck ligament (Elastin Products Co.), a mixture of fragments with a broad continuum of masses, was fractionated with size exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare) operated with a Bio-Rad BioLogic Duo Flow chromatography system. Desmosine cross-linking was quantified in the α-elastin species of nearly 20 kDa and nearly full length (∼125 kDa by SDS-PAGE and ∼100 kDa by MALDI-TOF). Each fraction was mixed with an equal volume 12 n HCl and hydrolyzed at 100 °C for 24 h. The acid was evaporated, and the sample was redissolved in water and assayed for desmosine (52) and protein (53).
Three of ∼32 lysines of the elastin sequence (54, 55) plus α-amino groups (17) were expected to be available for condensation with fluorophores close enough to quench one another. α-Elastin was conjugated with amine-reactive BODIPY FL (Invitrogen). 1 mg of the dimethyl-BODIPY was dissolved in 200 μl of anhydrous DMSO and added dropwise to 5 ml of a 10 mg/ml solution of α-elastin. The reaction was stirred in darkness at 25 °C for 1 h and size fractionated in darkness using the Superdex 200 column. The ∼100- and 20-kDa fractions were designated fEln-100 and fEln-20, respectively.
Proteolysis Substrates
The fEln-100 and fEln-20 were used in measuring and comparing apparent kcat and Km of proteolysis in solution. Proteolysis relieves the self quenching of the BODIPY FL to increase the fluorescence emission, which peaks at ∼515–516 nm (supplemental Fig. S1b), where progress curves were monitored. The assays were performed at 25 °C in TNC buffer (50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 10 mm CaCl2, 0.1 mm ZnCl2, and 0.035% Brij-35).
DQTM-collagens I and IV and the soluble Knight's substrate FS-6 were purchased from Invitrogen. The α1(V) 436–437 fTHP is selective for MMP-2, -9, and -12 and derived from the MMP-2 and -9 cleavage site in type V collagen (33–34). The proteolytic assays were performed at 25 °C in TNC buffer.
Kinetic Assays of MMPs and Variants
For accuracy in fits of steady-state kinetics, the concentrations of enzyme active sites in samples of all enzyme variants were carefully titrated with known concentrations of the tight-binding inhibitor galardin (GM6001, EMD) and fitted as described (56). The kcat/Km for each FRET substrate and enzyme variant was evaluated under pseudo first-order conditions of [S] ≪ Km (57). Individual kcat and Km parameters were determined for the substrates of better defined mass (fEln-100, fEln-20, α1(V) 436–447 fTHP, and FS-6) by the expedited method of fitting a few progress curves (58). This approach minimizes concerns about fluorescence non-linearity, expense of substrate, and substrate solubility. Progress curves for fEln-100, fEln-20, α1(V) 436–447 fTHP, and DQ-collagen I and IV substrates were fitted through the plateau of the first kinetic phase, corresponding to 1–3 h of the progress curves, prior to the subsequent kinetic phase.4 A lag of 10–15 min was observed prior to fluorescence increase of the fEln-100 substrate. Example fits of progress curves are illustrated for fEln-100 and fEln-20 and the DQ-collagens in supplemental Figs. S2 and S3, respectively.
The kinetics of fluorescence increase were monitored using an SLM-Aminco 8100 spectrofluorometer upgraded with photon counting and Peltier thermal control (ISS, Champaign-Urbana, IL) with excitation λ = 490 nm and detection λ = 512 nm. Kinetics of digestion of the fEln-100 substrate (and fEln-20) were also monitored with a BioTek Synergy MX plate reader reading from the bottom of 96-well plates covered to prevent evaporation during progress curves 3–6 h long (for WT-, S230A-, V243S-, G178N-, and I180S-substituted MMP-12).
NMR Assays of Elastin Interactions
The soluble α-elastin species of ∼20 kDa were designated Eln-20 and were used primarily for assays of interactions with inactivated MMP-12(E219A) monitored by NMR. TROSY spectra of 15N-labeled MMP-12(E219A) were acquired at 26 °C using a Bruker Avance III 800-MHz spectrometer fitted with a TCI cryogenic probe. The resulting high sensitivity and resolution enabled quantitative comparison of free and Eln-20-bound states of E219A-inactivated MMP-12. NMR spectra were processed with NMRPipe (59) or TopSpin 2.1 and interpreted using SPARKY (60). Stock solutions of Eln-20 were 10-fold more concentrated than MMP-12(E219A). Chemical shift perturbations of backbone amide 1H and 15N frequencies of MMP-12(E219A) upon additions of Eln-20 up to 1.5-fold molar excess were monitored in 800-MHz TROSY spectra (Fig. 1a).
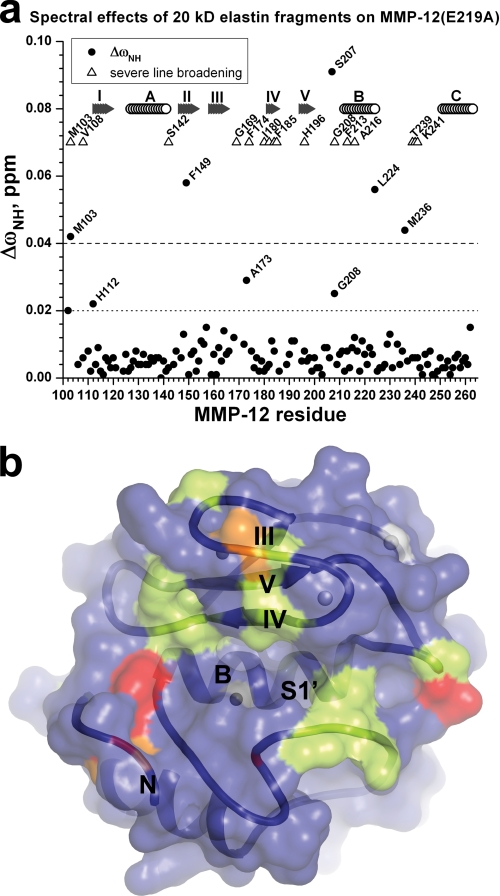
FIGURE 1.
Fragments of soluble elastin exert NMR spectral effects around the periphery of the active site cleft of inactivated MMP-12. Eln-20 was added at 1.5-fold molar excess over E219A-inactivated MMP-12 (0.3 mm). The 10% of amide TROSY peaks most broadened by the addition are marked with triangles in a and light green coloring of the NMR structure (49) in b. The backbone ribbon and transparent molecular surface in standard orientation in b displays unprimed subsites at left and primed subsites at right. Radial changes in chemical shift of amide NMR peaks in a, ΔωHN = [ΔωH2 + (ΔωN/5)2]1/2, were measured at 800 MHz, 26 °C, and pH 6.6. Residues with Eln-20-induced ΔωHN ≥ 0.04 ppm are red and those with 0.04 > ΔωHN ≥ 0.02 ppm are orange in b. β-Strands are labeled with Roman numerals, and helices are labeled with letters a and b.
Surfaces of MMP-12(E219A) that Eln-20 protects from the NMR line-broadening effects of Gd(III)·EDTA were mapped by the method of previous studies (33, 61–62). 800-MHz TROSY spectra were collected for 15N MMP-12(E219A), free and in the presence of 1.5-molar equivalents of Eln-20, each without and with the addition of 0.8 mm Gd(III)·EDTA. To allow the water and amide signals to recover a similar degree in the absence of the paramagnetic Gd(III)·EDTA as they do rapidly in its presence, the delay for recovery between transients was increased to 3 s. (Normalized peak heights, i.e. with red circles > 1.0 in Fig. 2a, suggest that the recovery was still not quite complete without Gd(III)·EDTA.) The broadening and exposure to the Gd(III)·EDTA probe in either the free or bound state is represented by the peak height in the presence of Gd(III)·EDTA divided by the peak height in its absence (Fig. 2a). The protection of the MMP-12 surface conferred by Eln-20 is represented by plotting the normalized peak heights of Gd(III)·EDTA-probed Eln-20-saturated state minus the normalized peak heights of Gd(III)·EDTA-probed free state (see Fig. 2b). A standard deviation of uncertainty in NMR peak height is estimated to be 1.5/(S/N), using the S/N reported by SPARKY, where noise N is root mean square noise. Standard rules of error propagation were used to estimate uncertainties of the ratios and differences of ratios.
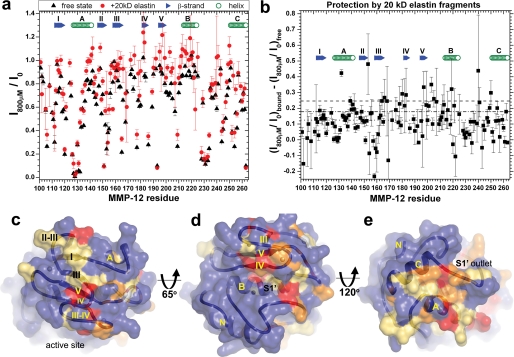
FIGURE 2.
Elastin fragments protect extensive surfaces of MMP-12(E219A) from a Gd(III)·EDTA probe. a, exposure of amide protons to NMR line broadening by 0.8 mm Gd(III)·EDTA. Peak heights are from 800-MHz TROSY spectra of 0.3 mm MMP-12(E219A) (pH 6.6, 26 °C) without or with addition of 1.5-fold molar excess of Eln-20. Peak heights with 0.8 mm Gd(III)·EDTA present, I800μM, are normalized by peak heights without it, I0, for the free state (black triangles) or Eln-20-bound state (red circles). Higher ratios in a indicate greater burial while low ratios toward zero indicate greater exposure to the soluble probe. b, plot of differences in I800μM/I0 between bound and free states. Large, positive differences in b indicate greater apparent burial by Eln-20, whereas smaller differences indicate less apparent burial. Overall line broadening in the presence of Eln-20 increases uncertainties in peak heights. The apparent protection in b is mapped onto the enzyme surface in c–e. Residues among the 10% most protected by Eln-20 from broadening by Gd(III)·EDTA (>0.246 and dashed line in b are red in c–e). Residues among the next 15%, most protected by Eln-20 (>0.178 and dash-dotted line), are orange. Residues in the third tier of protection (>0.125 and dotted line in b) are tan. d, standard orientation. c, rotated to view the β-sheet. e, rotated to view the “back” or “bottom” of the catalytic domain.
Distinctive Sequence Features by Evolutionary Trace Analysis
MMP sequences from diverse organisms aligned at the HSSP database were downloaded by Evolutionary Trace (ET) Report Maker (63), which deleted truncated and duplicate sequences of the catalytic domains. 286 unique sequences were submitted to the ET server at Cambridge (UK) for separation into subfamilies and construction of their class consensus sequences. Variable sequence positions were designated class-specific when conserved within ≥ 17 of 18 (51 of 54) subfamilies and nearly class-specific (64) when conserved within ≥ 15 of 18 (45 of 54) subfamilies (supplemental Fig. S4).
BINDSIght Prioritization of Mutagenesis
A residue's likelihood of contributing specificity was scored for evidences of distinctiveness in sequence (supplemental Figs. S4 and S5), burial within a binding interface with the protein partner/substrate (Fig. 2) and amide NMR peak being shifted or broadened (Fig. 1). The outcome of ET was assigned a score of 1 when a residue is class-specific (distinctive) or 0.5 when nearly class-specific (supplemental Figs. S4 and S5). A fully conserved residue was penalized by −3 and almost fully conserved by −2. Scores of 2, 1, or 0.5 were added in Fig. S5 for residues that had the most, medium, or moderate protection within interfaces with a mini-protein partner of either Eln-20 (Fig. 2) or α1(V) 436–450 THP (Fig. S5 of Ref. 33). Where the mini-protein partner introduces a significant perturbation in NMR chemical shift or line broadening, 1 was added to the score of the affected residue, or 0.5 for a modest peak shift (Fig. S5). The thresholds of protection within the interfaces and peak perturbations by the Eln-20 or α1(V) 436–450 THP partners are listed in the legend of Fig. S5. Mutagenesis avoided invariant or buried residues and instead focused on residues with high BINDSIght scores (supplemental Fig. S5) that lie within 22 Å of the catalytic zinc ion.
Site-directed Mutagenesis
Site-directed mutagenesis was performed using the QuikChange multisite-directed mutagenesis kit or Easy-A High-Fidelity PCR Master Mix (Stratagene) with only 12–15 cycles of PCR to avoid a second site mutation. DNA sequencing ensured the desired mutation and none others in the coding frame. Enzymes were expressed in Escherichia coli BL21 DE3.
Peptide Release from Insoluble Elastin Fibrils
The MMP variants were compared in terms of release of fluorescent peptides (65) from elastin-fluorescein particles (75–37 μm, Elastin Products Co.) in incubations at 37 °C at 2 mg/ml in TNC buffer with gentle shaking. 80 nm MMP variant was incubated with the fibrils for 16 h. Excitation was at 490 nm and emission at 520 nm. The supernatants containing the elastin peptides released were diluted 10-fold for fluorescence measurements to mitigate any loss of linearity from the inner-filtering effect.
Double Mutant Cycle Analysis
The catalytic efficiency (kcat/Km) of combined mutations was appraised in terms of coupling energy (ΔGI) for the pair of lesions using the expression (66),
where ΔΔGmut is defined as the transition-state stabilization energy (67) in Equation 2.
RESULTS
The questions of what surfaces of MMP-12 make it such a good elastase (15) and type V triple helical peptidase (33) spurred us to develop both quantitative activity assays (33, 58)4 and the BINDSIght approach for recognizing which among a plenitude of candidate residues contribute to the specificities. We decided that understanding the specificity without structures of the complexes requires (i) binding sites mapped accurately, (ii) finding distinguishing side chains at these interfaces, and (iii) functional testing of subtle mutations of these positions to be quantitative and efficient. The BINDSIght strategy incorporates these three values. First, we describe quantitative specific activities as benchmarks for comparison. Second, we report the mapping of crucial sites of contact between MMP-12 and fragments of solubilized α-elastin using NMR. We proceed to consider residues that distinguish MMP subfamilies. We combine this with the experimental interface mapping to anticipate the strongest candidates for tuning specific activities. Finally, the importance of these residues to specificity is quantified from perturbations of mild mutations.
High Activities of MMP-12 toward Substrates from Fibrils from Extracellular Matrix
We surveyed the steady-state kinetics parameters of elastolysis, triple helical peptidase, and linear peptidase activities of human MMP-12 and MMP-3 catalytic domains.
Soluble α-elastin (from complete solubilization of bovine neck elastin using 0.25 m oxalic acid) (17) has been a favorite substrate for elastolysis due to its sensitivity, specificity, reproducibility (68), retention of physiological cross-linking (17), and ability to form fibrils at 37 °C that resemble elastin (26). α-Elastin is polydisperse and averages 17 cross-linked chains with a mass of 60–84 kDa (among species up to 130 kDa) (17). We used gel filtration to isolate fractions from the α-elastin mixture appearing to migrate around 125 and 20 kDa by SDS-PAGE (supplemental Fig. S1a), for use in proteolytic enzyme kinetics and NMR interface mapping, respectively. Radioimmunoassay (52) detected 46.5 and 48.3 nmol of desmosine per milligram of each fraction of α-elastin. This corresponds to 2.4% and 2.3% (w/w) desmosine, respectively, a heavy degree of cross-linking similar to reports of 1.9% (w/w) desmosine for solubilized bovine neck elastin (54) and 1.5% for insoluble bovine elastin (69). Variability in the amount of cross-linking is suggested by pI values of the larger fraction ranging from 4 to 7 with most species having pI from 4.7 to 5.7.4 The two fractions were mixtures of masses peaking at ∼100 and 17.8 kDa, respectively, in MALDI-TOF mass spectra (supplemental Fig. S1, c and d).
We conjugated a sample of α-elastin with BO-DIPY FL, isolated gel-filtration fractions of these masses from the reaction mixture, and designated them fEln-100 and fEln-20 (referring to approximate mass). The labeling is heavy enough for self-quenching. Proteolysis of the cross-linked chains releases fluors and fluorescence quenching to support FRET assays (supplemental Figs. S1b and S2) for quantitative comparisons of apparent kcat and Km of soluble elastin hydrolysis by MMP-12 variants. Addition of MMP-12 initially and temporarily quenches fluorescence emission (supplemental Fig. S1b), suggesting that it might draw α-elastin species together. fEln-100 and fEln-20 are subsequently hydrolyzed with by far the slowest apparent turnover numbers kcat that we have observed among substrates of MMPs (supplemental Fig. S2, a and b, and Table 1). This is compensated by their high apparent affinities (lowest apparent Km values of 1.0 μm), relative to other substrates. MMP-12 digests them with apparent second order rate constants kcat/Km of around 10,000 m−1 s−1 for fELN-100 (Table 1) and 1,500 m−1 s−1 for fEln-20 fragments. MMP-3 catalytic domain has 12-fold lower kcat/Km of 830 m−1 s−1 toward fELN-100, which is mainly a consequence of its 13.5-fold lower kcat (Table 1). Km values for small linear peptide substrates, such as soluble Knight's substrate (FS-6) (70), are two orders of magnitude weaker (Table 1). MMP-12 is highly active upon a triple helical peptide from type V collagen (α1(V) 436–447 fTHP (49)), whereas MMP-3 and other MMPs except MMP-2 and -9 are inactive toward this substrate (34) (Table 1). MMP-12 also exhibits 265-fold greater catalytic efficiency (kcat/Km) toward DQ-collagen IV compared with MMP-3 (supplemental Fig. S3a and Table 1). Thus, homologous MMP-3 catalytic domain can serve as a “negative” control in comparison with MMP-12.
TABLE 1.
Catalytic properties of MMP-12 variants (25 °C, pH 7.5) for substrates from α-elastin and collagens IV and V
| MMP-12 variant | General substrate FS-6 |
fELN-100 |
α1(V) 436–447 fTHP |
DQ-Col IV |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| kcat/Km | kcat | Km | kcat/Km | kcat | Km | kcat/Km | kcat | Km | kcat/Km | |
| m−1s−1 | s−1 | μm | m−1s−1 | s−1 | μm | m−1s−1 | s−1 | μm | m−1s−1 | |
| Controls | ||||||||||
| MMP-12a | 133,800b ± 6000 | 17.4 ± 0.6 | 130 ± 4 | 9,880b ± 480 | 0.0105 ± 0.0005 | 1.1 ± 0.1 | 55,400c ± 350 | 1.23c ± 0.1 | 22.2c ± 5.6 | 29,200b ± 190 |
| MMP-3 | 20,660b ± 30 | 3.2 ± 0.1 | 154 ± 3 | 830b ± 40 | 0.0008 ± 0.0001 | 0.9 ± 0.1 | <50d | NDd | NDd | 110b ± 10 |
| MMP-12/MMP-3 | 6.5b | 5.4 | 0.84 | 12b | 13.5 | 1.2 | >1,000 | 265b | ||
| S230A | 148,100 ± 1130 | 20.0 ± 1.7 | 135 ± 1 | 9,570 ± 480 | 0.0105 ± 0.0005 | 1.1 ± 0.1 | 55,730 ± 210 | 1.11 ± 0.05 | 20 ± 1 | 22,130 ± 30 |
| V243S | 140,400 ± 650 | 18.0 ± 1.2 | 128 ± 1 | 9,640 ± 480 | 0.0096 ± 0.0005 | 1.0 ± 0.1 | 55,840 ± 70 | 2.79 ± 0.08 | 50 ± 1 | 25,448 ± 34 |
| Specificity | ||||||||||
| G178N | 107,900 ± 400 | 13.6 ± 0.4 | 126 ± 1 | 6,460 ± 320 | 0.0066 ± 0.0003 | 1.0 ± 0.1 | 41,670 ± 20 | 0.83 ± 0.04 | 20 ± 1 | 18,360 ± 40 |
| I180S | 141,800 ± 400 | 24.1 ± 2.2 | 170 ± 2 | 2,860 ± 140 | 0.0037 ± 0.0006 | 1.3 ± 0.1 | 42,240 ± 60 | 0.42 ± 0.04 | 10 ± 1 | 20,270 ± 40 |
| F185Y | 138,300 ± 300 | 20.7 ± 1.4 | 150 ± 10 | 3,910 ± 200 | 0.0164 ± 0.0019 | 4.2 ± 0.5 | 2,950 ± 10 | 0.44 ± 0.03 | 150 ± 11 | 4,080 ± 20 |
| F202Q | 139,000 ± 200 | 18.0 ± 0.9 | 130 ± 7 | 5,500 ± 280 | 0.0176 ± 0.0020 | 3.2 ± 0.4 | 5,790 ± 40 | 0.38 ± 0.03 | 65 ± 5 | 10,990 ± 90 |
| T205K | 135,400 ± 200 | 18.9 ± 2.0 | 140 ± 15 | 3,910 ± 200 | 0.0096 ± 0.0006 | 2.4 ± 0.2 | 13,540 ± 1080 | 0.41 ± 0.03 | 30 ± 1 | 8,200 ± 40 |
| H206D | 126,500 ± 100 | 17.7 ± 0.3 | 140 ± 2 | 4,450 ± 220 | 0.0183 ± 0.0018 | 4.1 ± 0.4 | 8,590 ± 180 | 0.27 ± 0.01 | 31 ± 1 | 9,220 ± 30 |
| T210Y | 145,700 ± 200 | 17.7 ± 1.2 | 122 ± 7 | 6,040 ± 300 | 0.0263 ± 0.0016 | 4.4 ± 0.3 | 26,150 ± 160 | 0.78 ± 0.02 | 30 ± 1 | 30,510 ± 1440 |
| G227F | 124,600 ± 100 | 19.0 ± 4.0 | 150 ± 6 | 5,410 ± 270 | 0.0119 ± 0.0008 | 2.2 ± 0.1 | 10,880 ± 80 | 0.11 ± 0.01 | 10 ± 1 | 16,830 ± 60 |
| T239L | 122,700 ± 200 | 17.0 ± 0.5 | 136 ± 4 | 4,680 ± 230 | 0.0112 ± 0.0006 | 2.4 ± 0.1 | 5,210 ± 20 | 0.09 ± 0.01 | 18 ± 1 | 7,820 ± 80 |
| K241H | 115,500 ± 200 | 16.0 ± 0.3 | 137 ± 3 | 5,250 ± 540 | 0.0176 ± 0.0029 | 3.35 ± 0.2 | 4,450 ± 30 | 0.20 ± 0.02 | 46 ± 2 | 11,880 ± 230 |
Chemical Shift Mapping of Elastin Binding Sites on MMP-12
To locate potential binding sites for α-elastin using NMR, we implemented Eln-20 due to its mass and solubility amenable for quantitative NMR in solution, its physiological and mature cross-linking, potential for self assembly (28), and its sharing 1.0 μm Km with fEln-100 for MMP-12. Use of E219A-inactivated MMP-12 prevented digestion of Eln-20. The extent of this structural perturbation is localized to residue 219 in the active site (49, 71–72).
Addition of a 1.5-fold molar excess of Eln-20 to 15N-labeled MMP-12(E219A) broadened the amide peaks in its TROSY spectra to a median of 13% of the peak heights of its free state (after correction for modest dilution). This suggests that the elastin fragments slow the tumbling of the MMP-12(E219A) substantially by increasing its hydrodynamic radii in complex, more than expected of a binary complex. The amide groups of MMP-12 (E219A) among the 10% most broadened by addition of Eln-20 (Fig. 1a), and potentially in contact with the α-elastin species, surround the active site (light green in Fig. 1b). The clearly broadened residues are found at the N terminus: Ser-142 of a remote loop; the upper curve of the S shape of the III–IV loop; Ile-180, Ala-182, and Phe-185 in and flanking β-strand sIV; His-196 (zinc ligand from sV); the V–B loop; the N-terminal end of helix hB under the active site; and Thr-239 to Lys-241 of the S1′ specificity loop known to contact competitive inhibitors (Fig. 1). Interspersed among these are residues with backbone amide NMR peaks that are slightly shifted by the addition of Eln-20. They lie at the N terminus, sII, III–IV loop, V–B loop, C-terminal end of hB, and conserved Met-236. The side chains of some affected residues are buried in the core: Val-108, Phe-149, Phe-213, Ala-216, Leu-224, and Met-236.
Elastin Binding Sites Identified Near and Far from Active Site Using Surface Probe
We are circumspect about chemical shift mapping of binding sites in that it sometimes overestimates interfaces (61), misses interfaces (73), and responds to binding-linked conformational adjustment (49, 74). A spectral “footprinting” method, however, seems to render consistently accurate discrimination of residues within versus outside a biomolecular interface, for protein-protein and protein-DNA associations ranging in Kd from 2 nm to 30 μm (33, 61–62, 75–76). It uses NMR to locate binding-dependent burial from an inert paramagnetic probe (preferably Gd(III)·EDTA or Gd(III)·DTPA·BMA) of exposed surfaces. Eln-20 protects extensive surfaces of MMP-12(E219A) from NMR line broadening by Gd(III)·EDTA (Fig. 2), including surfaces around the periphery of the active site. Eln-20 protects the following loci from the line broadening effects of the probe: the III–IV loop, sIV, the V–B loop, hB, and the S1′ specificity loop (Fig. 2, a and b). Eln-20 appears to block access by Gd(III)·EDTA specifically to the amide groups of Gly-166, His-168, Asp-175, Gly-178, and Ile-180 from the III–IV loop; Ala-182 and His-183 of sIV; Glu-199, Glu-201, Trp-203, His-205, and Gly-207 of the V–B loop; Thr-210 to Phe-213, Thr-215, Ala-216, Ala-219, Gly-221, and Gly-225 in and near hB; and Tyr-240 and Lys-241 of the S1′ specificity loop (Fig. 2, b and d).
Eln-20 also protects two novel patches remote from the active site. The first is a swath across the β-sheet from sV and the III–IV loop across sIII to sI and the II–III loop (Fig. 2c). These protected residues are: Thr-115 to Asn-119 of sI; Asn-153, Gly-155, and Met-156 of the II–III loop projecting above the β-sheet; Val-162 to Ala-164 of sIII; Gly-166 and His-168 of the III–IV loop; and Phe-197 and Glu-199 of sV. Eln-20 protects a second and larger remote patch extending from hA to hC and including neighboring loops. This Eln20-protected cluster includes Tyr-132 to Ser-142 of hA; Val-144, Leu-147, and Lys-148 of the A-II loop and sII; Thr-247 to Leu-250 at the C-terminal end of the B-C loop; and Ile-255, Gln-259, and Tyr-262 on an exposed side of hC (Fig. 2, b and e).
Sequence Positions Distinguishing Groups of MMP Catalytic Domains
To identify sequence positions characteristic of subfamilies of MMPs that might influence specificities, we conducted evolutionary trace analysis (ET) (77) of 286 unique and diverse MMP sequences. A threshold of 87% sequence identity separated the phylogenetic tree into 54 branches. The class consensus sequences representing the conservation within each of these 54 subfamilies are shown in supplemental Fig. S4. ET identifies residues, termed class-specific, that can distinguish one or more subfamilies and possibly tune specificities for partners (77). Of ∼165 amino acids of an MMP catalytic domain, 34 are class-specific (supplemental Fig. S4), and 51 are nearly class-specific. The nearly class-specific set was created to highlight more residues around the active site that are affected in NMR assays.
BINDSIght Combination of Interfaces and Distinctive Sequences
The key idea of BINDSIght is to identify the distinctive residues in or near binding sites that may confer specificity to the interaction. This involves combining maps of binding sites, whose borders may be defined ambiguously (e.g. chemical shift mapping), with the distinguishing sequence information, whose relevance to the molecular associations of interest may be in question. To address ambiguity in chemical shift mapping of binding sites, we also considered enzyme surfaces covered by the substrate protein and protected from the line broadening probe, a mapping expected to be more accurate and complete. Fig. 3 illustrates the concept of combining the interfaces that occlude the probe molecule (blue) with locations having NMR peaks perturbed by protein substrate (magenta) with sequence positions that distinguish protein subfamilies (green). Residues identified in two of the three assays may represent candidates with better prospects of tuning specificity, because of the increased likelihood that they are distinguishing features in or near binding sites.
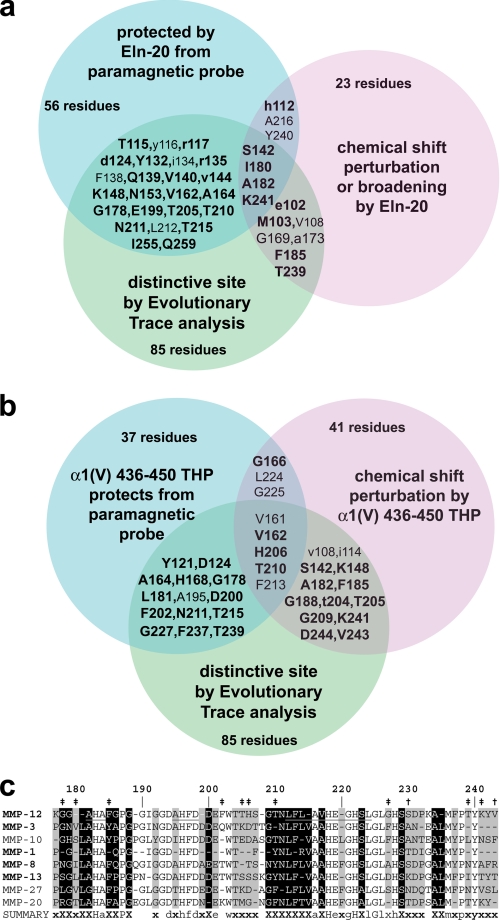
FIGURE 3.
BINDSIght combines maps of binding sites and distinctive sequence to suggest residues tuning specificity, which are enriched in MMP-12 loops flanking sIV and hB. a and b, Venn diagrams symbolize combination of NMR evidences of interfacial contacts (blue and lavender) with distinctive sequence positions (green; see supplemental Fig. S4). Side chains likely to influence specificity for elastin fragments or α1(V) 436–447 fTHP are listed in the intersections of the sets in a and b, respectively. The candidates in blue circles are protected by Eln-20 (a) (Fig. 2) or α1(V) 436–450 THP (b) (Figs. 4 and 5 of Ref. 33) from Gd(III)·EDTA. Candidates in lavender circles experience shifts or broadening of amide NMR peaks introduced by the partner of Eln-20 (a) (Fig. 1) or α1(V) 436–450 THP (b) (Fig. 2 of Ref. 33). Lowercase letters refer to effects being moderate in both assays. Residues with lighter font are buried or conserved. c, an excerpt from ET consensus sequences (of 54 subfamilies) listing some subfamilies close to MMP-12 in the phylogenetic tree and containing the human sequence listed. Class-specific positions are marked with a black background and an “X.” Nearly class specific positions are marked with a gray background and an “x” (see supplemental Fig. S4). β-Strands IV and V and helix B are marked by underlining of the MMP-12 consensus sequence. Sites chosen for mutagenesis are marked with “‡” or “†” where BINDSIght results forecast importance or little importance, respectively, to specificity.
The intersections of these sets narrowed the candidates to a number more manageable for site-directed mutagenesis, preparation of mutant enzymes, and functional comparison. Though the Eln-20 species appear to cover extensive surfaces of MMP-12 (Fig. 2, c–e), only 28 of these positions can distinguish sequences of MMP subfamilies (blue ∩ green in Fig. 3a). Eleven residues both have NMR peaks perturbed by Eln-20 and side chains that can distinguish MMP subfamilies (lavender ∩ green in Fig. 3a). The distinctive positions in sequence in these intersections map to the loops flanking sIV and hB (Fig. 3a) that form walls of the active site, and to remote sites.
Regarding interactions with the THP from collagen V, chemical shift mapping and this substrate's occlusion of the line broadening probe from the surface of MMP-12(E219A) were reported (33). This α1(V) 436–450 THP covers fewer surface residues (∼37) (Fig. 3b) than does Eln-20. Of these residues occluded by α1(V) 436–450 THP, 19 are distinctive positions in sequence (blue ∩ green in Fig. 3b). Eight of the THP-covered residues also have NMR peak positions perturbed by the THP (lavender ∩ blue in Fig. 3b). 18 residues are highlighted by chemical shift mapping and distinguishing side chains (lavender ∩ green in Fig. 3b). Thus, this BINDSIght result suggests at least 28 residues to be candidates for enhancing either the elastase activity of MMP-12 or triple helical peptidase activity toward α1(V) 436–447 fTHP (Fig. 3b). These are marked on the structure in supplemental Fig. S6.
BINDSIght-guided Choice of Mutations
In prioritizing residues listed in Fig. 3 for evaluation, we assigned a cumulative score to evidences of contributions to high activity toward fEln-100 or α1(V) 436–450 fTHP (supplemental Fig. S5). A residue's score increased if protected within a binding interface with Eln-20 (Fig. 2) or α1(V) 436–450 THP mini-proteins (Fig. 4 of Ref. 33), having sequence characteristic of its MMP subfamily (class-specific in supplemental Fig. S4), and/or having its NMR peak perturbed upon addition of Eln-20 (Fig. 1) or α1(V) 436–450 THP (Fig. 2 of Ref. 33). We deemed surface-exposed sites with net BINDSIght scores of 1.5 or greater (supplemental Fig. S5) more likely to enhance the activities of MMP-12 toward substrates derived from elastin or collagen triple helices. This criterion retained 28 or more candidate residues. We focused on candidates around the periphery of the active site. These are separated from the catalytic center but are near enough for potential contact with a protein fibril too large to be contained within the cleft. The BINDSIght plots of supplemental Fig. S5 raise the question of Gly-178, Phe-185, Thr-210, Thr-239, and Lys-241 enhancing specific activity for both substrates from elastin and collagen V.
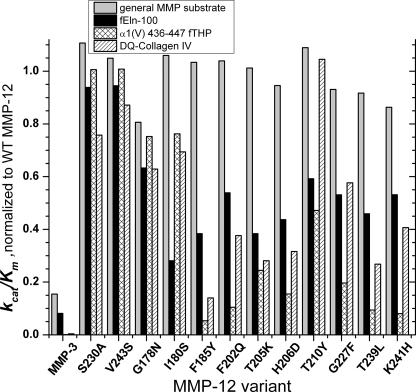
FIGURE 4.
The catalytic efficiency (kcat/Km) of point mutants relative to WT MMP-12 catalytic domain at 25 °C, pH 7.5. The general metalloproteinase substrate is the soluble form of Knight's substrate, FS-6 (70). S230A and V243S mutations test the ability of low BINDSIght scores to forecast inconsequential sites.
To evaluate candidates suggested by BINDSIght, we constructed single substitutions to the corresponding residue of a much less active MMP homologue, usually MMP-3 (stromelysin 1), but occasionally MMP-10 (stromelysin 2) or MMP-1 (collagenase 1): G178N, I180S, F185Y, F202Q, T205K, H206D, T210Y, G227F, T239L, and K241H. Natural selection has validated these side-chain substitutions as suitable in the corresponding environments of homologous MMPs to support their activity and very similar structure. Some are more subtle than alanine substitutions. To test the ability of BINDSIght to predict inconsequential mutations from lower BINDSIght scores of 0.5–1 (supplemental Fig. S5), we prepared S230A and V243S lesions at similar distance from the active-site cleft.
Peripheral Lesions Impair Activity for Substrates from Fibrils and Retain Activity upon a Peptide
We characterized the steady-state enzyme kinetics of each mutated enzyme with a control peptide substrate susceptible to metalloproteinases in general (soluble Knight's or FS-6 (70)), fEln-100, and α1(V) 436–447 fTHP (34) in terms of kcat and Km. The results with the general substrate FS-6 monitor catalytic consequences of any conformational adjustments from a mutation propagating to the active site. For testing relevance to other collagenase activities, we employed DQ-collagen IV (Table 1) and DQ-collagen I (supplemental Table S1). Control mutations S230A and V243S retain essentially wild-type activities toward the five substrates, except that S230A modestly diminishes activity toward DQ-collagen IV (Table 1). These results validate the BINDSIght prediction of inconsequential locations.
Each enzyme variant with a point mutation targeting specific activity for α-elastin or α1(V) 436–447 fTHP retained from 81% to 109% of WT general activity toward the small peptide substrate FS-6. This suggests a lack of catalytically important conformational adjustment from each mutation propagating to the active site. The catalytic efficiencies of the ten variants toward fEln-100 averaged around 2-fold down from WT, ranging from 28% of WT (I180S) to 63% of WT (G178N) (Table 1 and Fig. 4). To test how well these impaired activities might represent hydrolysis of intact insoluble elastin fibrils, activity in release of peptides from elastin fibrils was compared. The G178N, I180S, F202Q, T239L, and K241H lesions each impaired release of peptides from insoluble elastin in overnight digestions (Table 2).
TABLE 2.
Relative catalytic efficiencies of MMP-12 variants at pH 7.5
Transition-state stabilization energy, ΔΔG‡, can be calculated from the ratio according to eq. 2. The kinetic assays were each performed at 25 °C.
| MMP-12 variant | Distance, Cα to Zn2‡ | (kcat/Km)mut/(kcat/Km)WT |
RFUmut/RFUWT |
(kcat/Km)mut/(kcat/Km)WT |
||
|---|---|---|---|---|---|---|
| General substrate FS-6 | fEln-100 | Elastin-fluorescein peptide releasea | α1(V) 436–447 fTHP | DQ-collagen IV | ||
| Å | % | % | % | % | % | |
| Controls | ||||||
| MMP-12 | 100 | 100 | 100 | 100 | 100 | |
| MMP-3 | 15 | 8.4 | 7.5 ± 1 | <0.1 | 0.38 | |
| S230A | 10.9 | 111 | 94 | 131 ± 8 | 101 | 76 |
| V243S | 11.0 | 105 | 94 | 95 ± 5 | 101 | 87 |
| Tune specificity | ||||||
| G178N | 15.1 | 81 | 63 | 54 ± 5 | 75 | 63 |
| I180S | 10.0 | 106 | 28 | 49 ± 14 | 76 | 69 |
| F185Y | 10.2 | 103 | 38 | 110 ± 15 | 5.3 | 14 |
| F202Q | 18.4 | 104 | 54 | 73 ± 9 | 10 | 38 |
| T205K | 19.9 | 101 | 38 | 91 ± 2 | 24 | 28 |
| H206D | 20.7 | 94.5 | 44 | 105 ± 11 | 15.5 | 32 |
| T210Y | 14.5 | 109 | 59 | 121 ± 1 | 47 | 104 |
| G227F | 7.2 | 93 | 53 | 87 ± 11 | 20 | 58 |
| T239L | 7.8 | 92 | 46 | 54.5 ± 2 | 9.4 | 27 |
| K241H | 12.4 | 86 | 53 | 66 ± 17 | 8.0 | 41 |
a The elastin peptide release assays were performed at 37 °C for 16 h. The uncertainties are standard deviations from triplicate measurements.
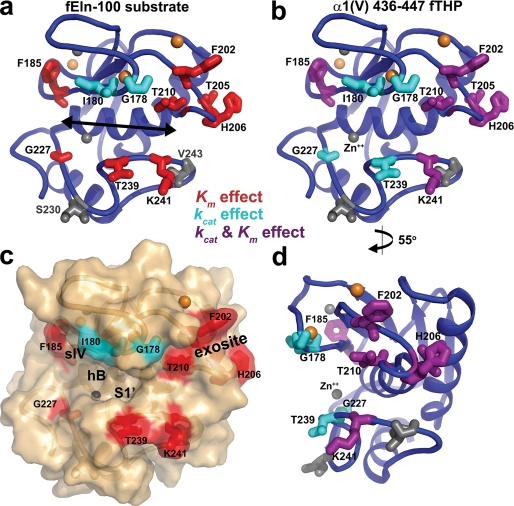
The lesions result in greater losses in activity toward α1(V) 436–447 fTHP that range from 19-fold for F185Y to 1.3-fold for G178N and I180S (Table 1 and Fig. 4). The impairments toward DQ-collagen IV are as much as 7.2-fold for F185Y. G178N or I180S only modestly diminish proteolytic turnover of either the α1(V) 436–447 fTHP or DQ-collagen IV (Table 1 and Fig. 4). All ten substitutions constructed diminish MMP-12 specific activity for fibril-derived substrates and map to sites encircling the active site cleft (Fig. 5). Only the side chains of T210Y and G227F are located within the catalytic cleft (Fig. 5).
FIGURE 5.
Sites of the lesions around the periphery of the active site cleft. The cleft is pointed out by the arrow in a. Panels a–c share near standard orientation. d, rotated 55° about the vertical axis places the cleft at the left, showing that most of the side chains probed lie outside the cleft. Catalytic consequences of substitutions upon activity toward fEln-100 are marked on the left panels and toward α1(V) 436–447 fTHP on the right panels. Sites of lesions that compromise Kmare colored red, or kcat cyan, or both purple. Sites of deliberately inconsequential substitutions are gray.
All ten point mutations targeting specificity preserve kcat and Km within 22% for the small peptide substrate FS-6 (Table 2). The I180S lesion increases kcat 39% and Km 31%, providing full catalytic efficiency toward FS-6 (Table 2). Eight mutations primarily increase Km for fEln-100, suggesting weaker affinity (Table 1 and Fig. 5a). The G178N and I180S lesions, in the lower curve of the S-shaped loop prior to sIV, instead diminish kcat for fEln-100 (Table 2 and Fig. 5a). The turnover rate constant kcat toward α1(V) 436–447 fTHP is slowed in each of the ten point mutants, but the decreases in kcat/Km of G178N or I180S are limited. Apparent affinity for the α1(V) 436–447 fTHP is impaired by lesions F185Y (IV–V loop), F202Q, T205K, H206D, T210Y (V–B loop), or K241H (S1′ specificity loop) (Km in Table 1 and Fig. 5b).
DISCUSSION
MMP-12 Specificity for Substrates from Fibrils Engages Periphery around Active Site
Structural studies searched the S1′ “specificity” pocket for features that might confer selectivity among MMPs by small inhibitory compounds (41–43, 78) (Thr-239 sits near this pocket; see Fig. 5). Yet MMP activity upon protein substrates is a much different and more physiological context for considering specificity. For example, the V–B loop influences triple helical peptidase activity of MMP-1, -8, and -12 (33, 45–47). About 30 of the distinctive sequence positions might contact 20-kDa α-elastin species (Eln-20) in the case of MMP-12 (Figs. 1, 2, 3a, S5a, and S6a). NMR-detected interactions (33) suggest that an overlapping subset of the distinctive positions could also interact with α1(V) 436–447 fTHP (Figs. 3 and S6). Among these candidates hypothesized using BINDSIght to contribute to MMP-12 specificities (Figs. 3 and S5), mutation of any of the ten tested around the periphery of the active site diminished catalytic efficiency toward fEln-100 and kcat toward α1(V) 436–447 fTHP, while preserving the activity toward the small peptide substrate (Tables 1 and 2 and Fig. 4). Thus, the ten residues encircling the active site of MMP-12 seem to enhance its uniquely efficient turnover of soluble α-elastin and substrates from collagen IV and V triple helices.
The importance of these peri-active site residues to digestion of fEln-100 α-elastin and triple helices from collagen IV and V suggests that they may interact with these bulky substrates directly, despite their distance from small peptide substrates that are confined within the cleft. These large substrates appear to cover similar surfaces around the primed subsites (at right in Figs. 1 and 5 (a–c)). The residues covered both by Eln-20 α-elastin and triple helical α1(V) 436–450 THP map to the N-terminal end of hB, the lower lobe of the S curve of the III–IV loop, sIV, Thr-239 to Lys-241 of the S1′ specificity loop, and V–B loop residues scattered from Glu-199 to Thr-210 (Figs. 2, S5, and S6, and in Fig. 4 of Ref. 33).
The impairment of peptide release from intact elastin fibrils by the G178N, I180S, T239L, and K241H lesions (Table 2) implies the importance of the region around the primed subsites of MMP-12. The mild impairment of elastin fibril digestion by the F202Q mutation suggests some involvement of the exosite beyond the primed subsites and the possibility of some elastin bending to reach the exosite. The results on peptide release from elastin fibrils should represent early digestion when the fibrils are intact and rigid in structure. Yet elastin digestion was observed to progress to clearance at sites of aneurysm in human aorta (7). The three dozen sites of cleavage by MMP-12 (16) may participate in the progressive damage. The progression of the elastolysis ought to damage the non-covalent packing of the filaments. Elastin molecules freed by digestion might revert toward the disordered-extended conformational equilibrium observed in tropoelastin (20). α-Elastin fractions fEln-100 and Eln-20 could share flexibility similar to elastin being digested. Thus the fEln-100 and Eln-20 substrates might have value for representing later stages of elastolysis, where peripheral and remote sites of MMP-12 could participate more.
Multiple Interactions with α-Elastin
Structural models in which peptides from elastin linearly extended only across the catalytic cleft (44) may need to be elaborated to account for Eln-20 coverage of remote surfaces of MMP-12 (Fig. 2), especially the best documented exosite where F202Q resides and modestly impairs hydrolysis of intact elastin fibrils (Table 2 and Fig. 5). One unexpected potential binding site is the narrow strip across the β-sheet from sV to sI and the II–III loop (Fig. 2c). The other is the broad bowl from hA and the B–C loop to hC of MMP-12 (Fig. 2e). These remote patches of hypothesized interaction (Fig. 2) suggest the possibility of more than one binding mode of Eln-20 and potentially complex interactions. Binding of a second molecule of Eln-20 is possible given the saturating 1.5-fold excess used to map-protected interfaces. The NMR line broadening of MMP-12 with Eln-20 present is larger than expected of a binary complex. It joins the fluorescence data (supplemental Fig. S1b) in suggesting that assemblies contain potentially more than one elastin molecule.
Could the NMR evidence of Eln-20 interactions with remote patches (Fig. 2) be artifacts of high concentrations in vitro and the sensitivity of NMR to weak biomolecular interactions? Weak protein-protein encounter complexes have been proposed to reload a specific, functional complex with physiological efficiency (79). Evidence that fEln-100 may interact with MMP-12 away from the active site at low concentrations is the initial fluorescence quenching of 100 nm fEln-100 upon addition of 10 nm MMP-12; in supplemental Fig. S1b, compare black and red spectra. This suggests that MMP-12 brings fluorophores of fEln-100 closer for more self-quenching, perhaps by bridging more than one fEln-100 molecule together. MMP-12 was observed embedded in digested elastin fibrils in human aorta (7) and colocalized with damaged elastin in UV-damaged human skin (80). The embedding of MMP-12 in damaged elastin fibrils implies locally high concentrations in which sites of MMP-12 with even weak affinity for elastin may contribute to interactions in vivo. Elastin binding sites located away from the active site could explain why MMP-12 bound to the inhibitor protein TIMP (across the active site) remained bound to elastin in arteries (7). Thus, some of the multiple sites of MMP-12 interaction with Eln-20 and fEln-100 could be relevant in vivo.
Multiple contact surfaces could account for the Km being 1 μm, suggestive of affinity that is at least 20-fold greater than for any other substrate measured. (The nearest Km is 22 μm for α1(V) 436–447 fTHP in Table 1.) Many contacts mooring MMP-12 to elastin could sum energetically to provide the favorable free energy of binding. Multiple remote contacts could enhance the affinity of MMP-12 found bound to damaged elastin fibrils in arteries (7) or UV-damaged skin (80). Perhaps the effect upon Km for fEln-100 of each point mutation of MMP-12 being limited to 4-fold or less (Table 1) results from it perturbing only a few of many contacts with elastin (Fig. 2).
Small increases in kcat of F185Y, F202Q, H206D, T210Y, and K241H mutants of MMP-12 acting upon fEln-100 compensate their increased Km (Table 2) to moderate the decreases of their catalytic efficiencies to 38 to 59% of WT (Table 1 and Fig. 4). The enhancements of kcat might result from slight alleviation of slow product release by off-rates enhanced with the 3- to 4-fold weakening of Km. The 2.8- and 1.6-fold decreases in kcat toward fEln-100 by I180S or G178N lesions, respectively, suggest the possibility of subtle adjustments of conformation of MMP-12 (their bulge in the III–IV loop), in the Michaelis complex, or in the positioning of MMP-12 around scissile bonds within elastin.
A caveat to the ability of the Km effects of the eight peripheral sequence positions (red in Fig. 5, a and c) to account for the high elastase activity of MMP-12 is that its advantage over MMP-3 is not in Km, but rather 13.5-fold in kcat (Table 1). The lower kcat of MMP-3 could be related to its greater extent of millisecond motions surrounding the active site (72).
Triple Helical Peptidase Perturbations
Eight of the ten lesions degrade activity toward the triple helical substrate from collagen V more than they do toward fEln-100. The mutants' bigger losses in activity upon α1(V) 436–447 fTHP coincide with the much larger advantage in activity of MMP-12 over MMP-3 of at least three orders of magnitude (Table 1). Generally, the mutations most detrimental to turnover of α1(V) 436–447 fTHP (Table 1) impair both kcat and Km (Table 1 and Fig. 5b). The G227F and T239L lesions simply impair kcat by at least 10-fold (Table 1). Gly-227 forms the base of the S2 subsite, whereas the side chain of Thr-239 points away from the S1′ subsite (Fig. 5). The G227F and T239L lesions are only 7–8 Å distant from the site of hydrolysis at the catalytic zinc (Table 2). These nearby lesions seem to interfere more directly in hydrolytic turnover of the triple helix. The smaller losses in rate of turnover of α1(V) 436–447 fTHP due to G178N or I180S lesions (Table 1) are mitigated by their preservation of Km. Despite the burial of Gly-178 in the interface with α1(V) 436–450 THP (33), addition of the asparagine side chain here seems not to affect the affinity (Table 1).
Adding the hydroxyl group to residue 185 decreases kcat/Km by a striking 19- and 7-fold for α1(V) 436–447 fTHP and DQ-collagen IV, respectively. Whether the phenyl ring of Phe-185 makes critical hydrophobic contacts with ligands of the structural zinc ion or with the triple helix is unclear. Judging from a model of peptide substrate bound to MMP-12 (48), Phe-185 is possibly near an S3 subsite. A direct interaction of Phe-185 with the triple helix could be more plausible when considering that the F185Y substitution has little indirect effect upon activity toward the peptide substrate (Tables 1 and 2 and Fig. 4). F202Q and H206D diminish triple helical peptidase activity the most among the substitutions in the V–B loop (Table 1 and Fig. 4). F202Q decreases kcat/Km 10-fold toward α1(V) 436–447 fTHP. The location of Phe-202 well outside the active site cleft (Fig. 5) supports the hypotheses that either α1(V) 436–447 fTHP bends around this “primed” side of MMP-12 to meet Phe-202, or that additional binding modes outside the catalytic cleft help establish an orientation susceptible to cleavage (33). The precedent of peptides bridging and bending between active site and a remote exosite is thoroughly established for thrombin (from the blood coagulation cascade) (81). Peptide substrates bending around other classes of proteases is also suggested by cathepsin G (a serine protease) and cathepsin K (a cysteine protease), each recognizing substrate positions out through P4′ and P5′ (82). Similarly, P4′ and even P5′ and P6′ positions influence peptide hydrolysis by MMP-2 (82), whereas MMP-1 triple helical peptidase activity is influenced by the P8–P13 and P12′–P17′ positions (83).
Nine of the lesions decrease catalytic efficiency toward DQ-collagen IV (Table 1 and Fig. 4). The high activity of MMP-12(T210Y) toward DQ-collagen IV is consistent with tyrosine occupying this position in type IV collagenases (MMP-2 and -9) and other collagenases (Fig. 3a and supplemental Fig. S4). The lesions' impact on specific activities for substrates from type IV and V collagen raises an important question: Do the corresponding positions in orthodox collagenolytic MMPs modulate their turnover of collagen triple helices? This possibility is suggested by MMP-1 and -8 relying on the V–B loop (45–47). The exosite of Fig. 5 corresponds in the structure of MMP-2 and -9 (84) to the channel between the FnII-like inserts and the catalytic domain where gelatin and type IV collagen might traverse between FnII-like module 2 and the active site (85).
Mutations were previously accumulated in one of the loops: T205K plus H206D and these two plus S207T and G208T of the V–B loop (33). MMP-12(T205K/H206D) has a weaker Km of 70 μm for α1(V) 436–447 fTHP than the 30 μm Km of either T205K or H206D alone. Nonetheless, MMP-12(T205K) and MMP-12(H206D) each possess around half the catalytic efficiency of MMP-12(T205K/H206D) (Tables 1 and 2, and S2), due to 3- to 4-fold losses in kcat. The non-additivity of T205K+H206D upon catalytic efficiencies has free energies of interaction ΔGint (Equation 1) between T205K and H206D substitutions for proteolysis of fEln-100, α1(V) 436–447 fTHP, DQ-collagen I and DQ-collagen IV of −0.7, −1.2, −1.0, and −1.4 kcal/mol, respectively. These are unfavorable and large compared with the transition state stabilization energies ΔΔG‡ of each individual mutation and substrate. This behavior is so non-additive as to be antagonistic. Non-additivity was correlated elegantly with the proximity of lesions within enzymes or within a protein-protein interface, in contrast with distantly separated mutations that are additive (86, 87). The unfavorable energetic coupling of T205K and H206D abides by the trend of non-additive effects of mutations clustered in structure and sequence in a protein-protein interface (86, 87).
Remote Mutations and Enzyme Function
Viewing MMP-12 as a product of evolution of elastase activity within its catalytic domain enables comparison with directed evolution of new activities in enzymes. Mutations that introduced new functions were found relatively far from key residues in catalytic sites, i.e. generally in loops on the periphery of the active site (88–92). The ten MMP-12 lesions tested herein are situated similarly. Random mutagenesis of enzymes identified many residues affecting activity that are remote from the active site, evidently for the simple reason that there are far more residues distant from active sites than lie within them (92). Mutations within 12 Å of the catalytic center appear more likely to enhance activity and selectivity upon small molecule substrates (92). However, for MMP-12 specificity for mini-protein substrates from fibrils, eight of ten residues identified by the BINDSIght process instead lie 10–21 Å distant from the catalytic zinc (Table 1). The effects of lesions at these distances seem related to the substrates' bulk, diminished apparent affinities (Table 1), and possible bending to reach the exosite (Fig. 5).
On BINDSIght
The strategy of identifying distinctive residues in or near confirmed binding sites provided much more guidance to consequential residues than using only either distinctive residues from ET or interfacial residues from NMR. If only class-specific residues (34 or more) or only NMR results (28 or more residues; Figs. 1–3) had been considered near the active site (from residue 178–242; Fig. 3c), testing ∼3-fold more mutational variants for relevance to specificity would have been warranted. The guidance by BINDSIght achieved the efficiency that all ten of ten candidate sites selected modulate specific activity toward fEln-100, and about eight of them toward triple helical substrates. Low BINDSIght scores also proved successful in suggesting deliberately inconsequential mutations. The functional comparison of the 16 enzyme variants with five substrates benefited from streamlined determination of steady-state enzyme kinetics parameters (58).
A caveat to expecting all residues conferring specificity to occupy binding interfaces is that some important electrostatic interactions lie outside interfaces. The observation of only seven or eight residues being both occluded in interfaces and perturbed in chemical shift mapping (blue ∩ lavender in Fig. 3) may seem surprising. Yet such differences have precedent in hyaluronan binding of CD44 introducing most of its chemical shift perturbations outside the interface (73). The chemical shift perturbations of some ligand binding events may be small for lack of aromatic ring current effects (73) or enough affinity or structural adjustment for larger peaks shifts.
A precursor to the BINDSIght approach successfully targeted mutagenesis to a functionally important interface between a DnaJ-like co-chaperone and mammalian Hsc70 chaperone, which had not been expected from the E. coli DnaJ–DnaK interface (62). Consequently, the BINDSIght approach could prove useful in elucidating the specificity of other transient protein-protein interactions. The interface mapping of BINDSIght could be expanded to other accurate solution methods such as NMR cross-saturation (73), intermolecular NOE, and hydrogen-deuterium exchange mass spectrometry.
Concluding Remarks
MMP-12 interactions with soluble derivatives of elastin appear to be multifaceted. A multiplicity of interactions could account for embedding of MMP-12 in human elastin fibrils to hasten the inherently slow elastolysis in chronic disease states of arteries (abdominal aortic aneurysm) and lungs (chronic obstructive pulmonary disease). Confirmation of eight residues as also modulating MMP-12 activity toward mimics of collagens IV or V begs the question of how important these sequence positions might also be in the triple helical peptidase activity of other MMPs. The work herein implicates five peripheral loops around the active site in the specificity of MMP-12. The peripheral loops represent a new frontier of distinctive features of MMPs that could be targeted by prospective therapeutic and diagnostic agents that bind them selectively (93) in the context of inflammatory diseases that damage the extracellular matrix.
Supplementary Material
Acknowledgments
We thank J. Lauer for THP, the Gehrke Proteomics Center for mass spectrometry, and B. Starcher for radioimmunoassay of desmosine, discussion of elastin cross-linking, and a critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM57289 (to S. R. V.). The 800-MHz NMR spectrometer was purchased with funding from NIH Grant RR022341 and the University of Missouri. THP development was supported by NIH Grant R01 CA098799 and the Robert A. Welch Foundation (to G. B. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
M. O. Palmier, Y. G. Fulcher, and S. R. Van Doren, unpublished observation.
- MMP
- matrix metalloproteinase
- BINDSIght
- bioinformatics and NMR discovery of specificity of interactions
- ET
- evolutionary trace analysis
- hA to hC
- helix A to helix C
- fEln-100
- fluorescently labeled 100-kDa fragments of cross-linked, solubilized elastin
- Eln-20
- 20-kDa fragments of solubilized elastin
- FnII
- fibronectin II
- sI to sV
- β-strands I to V
- THP
- triple helical peptide
- α1(V) THP
- THP derived from α1 chain of type V collagen
- TROSY
- transverse relaxation-optimized spectroscopy.
REFERENCES
- 1.Kortemme T., Baker D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14116–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hautamaki R. D., Kobayashi D. K., Senior R. M., Shapiro S. D. (1997) Science 277, 2002–2004 [DOI] [PubMed] [Google Scholar]
- 3.Churg A., Wang R. D., Tai H., Wang X., Xie C., Dai J., Shapiro S. D., Wright J. L. (2003) Am J. Respir Crit. Care. Med. 167, 1083–1089 [DOI] [PubMed] [Google Scholar]
- 4.Hunninghake G. M., Cho M. H., Tesfaigzi Y., Soto-Quiros M. E., Avila L., Lasky-Su J., Stidley C., Melen E., Söderhäll C., Hallberg J., Kull I., Kere J., Svartengren M., Pershagen G., Wickman M., Lange C., Demeo D. L., Hersh C. P., Klanderman B. J., Raby B. A., Sparrow D., Shapiro S. D., Silverman E. K., Litonjua A. A., Weiss S. T., Celedón J. C. (2009) N. Engl. J. Med. 361, 2599–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson J. L., George S. J., Newby A. C., Jackson C. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15575–15580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang J., Liu E., Yu Y., Kitajima S., Koike T., Jin Y., Morimoto M., Hatakeyama K., Asada Y., Watanabe T., Sasaguri Y., Watanabe S., Fan J. (2006) Circulation 113, 1993–2001 [DOI] [PubMed] [Google Scholar]
- 7.Curci J. A., Liao S., Huffman M. D., Shapiro S. D., Thompson R. W. (1998) J. Clin. Invest. 102, 1900–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vos C. M., van Haastert E. S., de Groot C. J., van der Valk P., de Vries H. E. (2003) J. Neuroimmunol 138, 106–114 [DOI] [PubMed] [Google Scholar]
- 9.Vaalamo M., Karjalainen-Lindsberg M. L., Puolakkainen P., Kere J., Saarialho-Kere U. (1998) Am J. Pathol. 152, 1005–1014 [PMC free article] [PubMed] [Google Scholar]
- 10.Liu M., Sun H., Wang X., Koike T., Mishima H., Ikeda K., Watanabe T., Ochiai N., Fan J. (2004) Arthritis Rheum. 50, 3112–3117 [DOI] [PubMed] [Google Scholar]
- 11.Valença S. S., da Hora K., Castro P., Moraes V. G., Carvalho L., Porto L. C. (2004) Toxicol Pathol. 32, 351–356 [DOI] [PubMed] [Google Scholar]
- 12.Houghton A. M., Quintero P. A., Perkins D. L., Kobayashi D. K., Kelley D. G., Marconcini L. A., Mecham R. P., Senior R. M., Shapiro S. D. (2006) J. Clin. Invest. 116, 753–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonicelli F., Bellon G., Debelle L., Hornebeck W. (2007) Curr Top Dev. Biol. 79, 99–155 [DOI] [PubMed] [Google Scholar]
- 14.Hunninghake G. W., Davidson J. M., Rennard S., Szapiel S., Gadek J. E., Crystal R. G. (1981) Science 212, 925–927 [DOI] [PubMed] [Google Scholar]
- 15.Shapiro S. D., Kobayashi D. K., Ley T. J. (1993) J. Biol. Chem. 268, 23824–23829 [PubMed] [Google Scholar]
- 16.Taddese S., Weiss A. S., Neubert R. H., Schmelzer C. E. (2008) Matrix Biol. 27, 420–428 [DOI] [PubMed] [Google Scholar]
- 17.Partridge S. M., Davis H. F., Adair G. S. (1955) Biochem. J. 61, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagenseil J. E., Mecham R. P. (2007) Birth Defects Res C Embryo Today. 81, 229–240 [DOI] [PubMed] [Google Scholar]
- 19.Partridge S. M., Elsden D. F., Thomas J., Dorfman A., Telser A., Ho P. L. (1966) Nature 209, 399–400 [DOI] [PubMed] [Google Scholar]
- 20.Tamburro A. M., Bochicchio B., Pepe A. (2005) Pathol. Biol. 53, 383–389 [DOI] [PubMed] [Google Scholar]
- 21.Parks W. C., Deak S. B. (1990) Am. J. Respir. Cell. Mol. Biol. 2, 399–406 [DOI] [PubMed] [Google Scholar]
- 22.O'Dell B. L., Elsden D. F., Thomas J., Partridge S. M., Smith R. H., Palmer R. (1966) Nature 209, 401–402 [DOI] [PubMed] [Google Scholar]
- 23.Podrazký V. (1968) Biochim. Biophys. Acta 160, 277–279 [DOI] [PubMed] [Google Scholar]
- 24.Wise S. G., Mithieux S. M., Raftery M. J., Weiss A. S. (2005) J. Struct. Biol. 149, 273–281 [DOI] [PubMed] [Google Scholar]
- 25.Urry D. W., Starcher B., Partridge S. M. (1969) Nature 222, 795–796 [DOI] [PubMed] [Google Scholar]
- 26.Cox B. A., Starcher B. C., Urry D. W. (1973) Biochim. Biophys. Acta 317, 209–213 [DOI] [PubMed] [Google Scholar]
- 27.Bellingham C. M., Lillie M. A., Gosline J. M., Wright G. M., Starcher B. C., Bailey A. J., Woodhouse K. A., Keeley F. W. (2003) Biopolymers 70, 445–455 [DOI] [PubMed] [Google Scholar]
- 28.Pepe A., Guerra D., Bochicchio B., Quaglino D., Gheduzzi D., Pasquali Ronchetti I., Tamburro A. M. (2005) Matrix Biol. 24, 96–109 [DOI] [PubMed] [Google Scholar]
- 29.Fu J. Y., Lyga A., Shi H., Blue M. L., Dixon B., Chen D. (2001) Protein. Expr. Purif. 21, 268–274 [DOI] [PubMed] [Google Scholar]
- 30.Liotta L. A., Lanzer W. L., Garbisa S. (1981) Biochem. Biophys. Res. Commun. 98, 184–190 [DOI] [PubMed] [Google Scholar]
- 31.Hibbs M. S., Hasty K. A., Seyer J. M., Kang A. H., Mainardi C. L. (1985) J. Biol. Chem. 260, 2493–2500 [PubMed] [Google Scholar]
- 32.Taddese S., Jung M. C., Ihling C., Heinz A., Neubert R. H., Schmelzer C. E. (2010) Biochim. Biophys. Acta 1804, 731–739 [DOI] [PubMed] [Google Scholar]
- 33.Bhaskaran R., Palmier M. O., Lauer-Fields J. L., Fields G. B., Van Doren S. R. (2008) J. Biol. Chem. 283, 21779–21788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauer-Fields J. L., Sritharan T., Stack M. S., Nagase H., Fields G. B. (2003) J. Biol. Chem. 278, 18140–18145 [DOI] [PubMed] [Google Scholar]
- 35.Linsenmayer T. F., Fitch J. M., Gordon M. K., Cai C. X., Igoe F., Marchant J. K., Birk D. E. (1998) Prog. Retin. Eye Res. 17, 231–265 [DOI] [PubMed] [Google Scholar]
- 36.Shipley J. M., Doyle G. A., Fliszar C. J., Ye Q. Z., Johnson L. L., Shapiro S. D., Welgus H. G., Senior R. M. (1996) J. Biol. Chem. 271, 4335–4341 [DOI] [PubMed] [Google Scholar]
- 37.O'Farrell T. J., Pourmotabbed T. (1998) Arch. Biochem. Biophys. 354, 24–30 [DOI] [PubMed] [Google Scholar]
- 38.Collier I. E., Krasnov P. A., Strongin A. Y., Birkedal-Hansen H., Goldberg G. I. (1992) J. Biol. Chem. 267, 6776–6781 [PubMed] [Google Scholar]
- 39.Tam E. M., Moore T. R., Butler G. S., Overall C. M. (2004) J. Biol. Chem. 279, 43336–43344 [DOI] [PubMed] [Google Scholar]
- 40.Lauer-Fields J. L., Whitehead J. K., Li S., Hammer R. P., Brew K., Fields G. B. (2008) J. Biol. Chem. 283, 20087–20095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovejoy B., Welch A. R., Carr S., Luong C., Broka C., Hendricks R. T., Campbell J. A., Walker K. A., Martin R., Van Wart H., Browner M. F. (1999) Nat. Struct. Biol. 6, 217–221 [DOI] [PubMed] [Google Scholar]
- 42.Nar H., Werle K., Bauer M. M., Dollinger H., Jung B. (2001) J. Mol. Biol. 312, 743–751 [DOI] [PubMed] [Google Scholar]
- 43.Moy F. J., Chanda P. K., Chen J., Cosmi S., Edris W., Levin J. I., Rush T. S., Wilhelm J., Powers R. (2002) J. Am. Chem Soc. 124, 12658–12659 [DOI] [PubMed] [Google Scholar]
- 44.Bertini I., Fragai M., Luchinat C., Melikian M., Venturi C. (2009) Chemistry 15, 7842–7845 [DOI] [PubMed] [Google Scholar]
- 45.Chung L., Shimokawa K., Dinakarpandian D., Grams F., Fields G. B., Nagase H. (2000) J. Biol. Chem. 275, 29610–29617 [DOI] [PubMed] [Google Scholar]
- 46.Pelman G. R., Morrison C. J., Overall C. M. (2005) J. Biol. Chem. 280, 2370–2377 [DOI] [PubMed] [Google Scholar]
- 47.Minond D., Lauer-Fields J. L., Cudic M., Overall C. M., Pei D., Brew K., Visse R., Nagase H., Fields G. B. (2006) J. Biol. Chem. 281, 38302–38313 [DOI] [PubMed] [Google Scholar]
- 48.Lang R., Kocourek A., Braun M., Tschesche H., Huber R., Bode W., Maskos K. (2001) J. Mol. Biol. 312, 731–742 [DOI] [PubMed] [Google Scholar]
- 49.Bhaskaran R., Palmier M. O., Bagegni N. A., Liang X., Van Doren S. R. (2007) J. Mol. Biol. 374, 1333–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng X., Ou L., Tong X., Zhu J., Wu H. (2007) Prot. Expr. Purif. 56, 160–166 [DOI] [PubMed] [Google Scholar]
- 51.Ye Q. Z., Johnson L. L., Hupe D. J., Baragi V. (1992) Biochemistry 31, 11231–11235 [DOI] [PubMed] [Google Scholar]
- 52.Starcher B., Conrad M. (1995) Connect. Tiss. Res. 31, 133–140 [DOI] [PubMed] [Google Scholar]
- 53.Starcher B. (2001) Anal. Biochem. 292, 125–129 [DOI] [PubMed] [Google Scholar]
- 54.Mecham R. P., Lange G. (1980) Connect. Tiss. Res. 7, 247–252 [DOI] [PubMed] [Google Scholar]
- 55.Daamen W. F., Hafmans T., Veerkamp J. H., Van Kuppevelt T. H. (2001) Biomaterials 22, 1997–2005 [DOI] [PubMed] [Google Scholar]
- 56.Williams J. W., Morrison J. F. (1979) Methods Enzymol. 63, 437–467 [DOI] [PubMed] [Google Scholar]
- 57.Niedzwiecki L., Teahan J., Harrison R. K., Stein R. L. (1992) Biochemistry 31, 12618–12623 [DOI] [PubMed] [Google Scholar]
- 58.Palmier M. O., Van Doren S. R. (2007) Anal. Biochem. 371, 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 60.Goddard T. D., Kneller D. G. (2000) SPARKY. University of California, San Francisco, San Francisco [Google Scholar]
- 61.Arumugam S., Hemme C. L., Yoshida N., Suzuki K., Nagase H., Berjanskii M., Wu B., Van Doren S. R. (1998) Biochemistry 37, 9650–9657 [DOI] [PubMed] [Google Scholar]
- 62.Garimella R., Liu X., Qiao W., Liang X., Zuiderweg E. R., Riley M. I., Van Doren S. R. (2006) Biochemistry 45, 6917–6929 [DOI] [PubMed] [Google Scholar]
- 63.Mihalek I., Res I., Lichtarge O. (2006) Bioinformatics 22, 1656–1657 [DOI] [PubMed] [Google Scholar]
- 64.Lee G. I., Ding Z., Walker J. C., Van Doren S. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11261–11266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huebner P. F. (1976) Anal. Biochem. 74, 419–429 [DOI] [PubMed] [Google Scholar]
- 66.Carter P. J., Winter G., Wilkinson A. J., Fersht A. R. (1984) Cell 38, 835–840 [DOI] [PubMed] [Google Scholar]
- 67.Wilkinson A. J., Fersht A. R., Blow D. M., Winter G. (1983) Biochemistry 22, 3581–3586 [DOI] [PubMed] [Google Scholar]
- 68.Keller S., Mandl I. (1971) Biochem. Med. 5, 342–347 [Google Scholar]
- 69.Starcher B. C., Galione M. J. (1976) Anal. Biochem. 74, 441–447 [DOI] [PubMed] [Google Scholar]
- 70.Neumann U., Kubota H., Frei K., Ganu V., Leppert D. (2004) Anal. Biochem. 328, 166–173 [DOI] [PubMed] [Google Scholar]
- 71.Steele D. L., El-Kabbani O., Dunten P., Windsor L. J., Kammlott R. U., Crowther R. L., Michoud C., Engler J. A., Birktoft J. J. (2000) Protein Eng. 13, 397–405 [DOI] [PubMed] [Google Scholar]
- 72.Liang X., Arunima A., Zhao Y., Bhaskaran R., Shende A., Byrne T. S., Fleeks J., Palmier M. O., Van Doren S. R. (2010) Biophys. J. 99, 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takeda M., Terasawa H., Sakakura M., Yamaguchi Y., Kajiwara M., Kawashima H., Miyasaka M., Shimada I. (2003) J. Biol. Chem. 278, 43550–43555 [DOI] [PubMed] [Google Scholar]
- 74.Tzeng S. R., Kalodimos C. G. (2009) Nature 462, 368–372 [DOI] [PubMed] [Google Scholar]
- 75.Arumugam S., Gao G., Patton B. L., Semenchenko V., Brew K., Van Doren S. R. (2003) J. Mol. Biol. 327, 719–734 [DOI] [PubMed] [Google Scholar]
- 76.Iwahara J., Zweckstetter M., Clore G. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15062–15067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lichtarge O., Bourne H. R., Cohen F. E. (1996) J. Mol. Biol. 257, 342–358 [DOI] [PubMed] [Google Scholar]
- 78.Verma R. P., Hansch C. (2007) Bioorg. Med. Chem. 15, 2223–2268 [DOI] [PubMed] [Google Scholar]
- 79.Fawzi N. L., Doucleff M., Suh J. Y., Clore G. M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1379–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saarialho-Kere U., Kerkelä E., Jeskanen L., Hasan T., Pierce R., Starcher B., Raudasoja R., Ranki A., Oikarinen A., Vaalamo M. (1999) J. Invest. Dermatol. 113, 664–672 [DOI] [PubMed] [Google Scholar]
- 81.Bode W. (2006) Blood Cells. Mol. Dis. 36, 122–130 [DOI] [PubMed] [Google Scholar]
- 82.Schilling O., Overall C. M. (2008) Nat. Biotechnol. 26, 685–694 [DOI] [PubMed] [Google Scholar]
- 83.Lauer-Fields J. L., Chalmers M. J., Busby S. A., Minond D., Griffin P. R., Fields G. B. (2009) J. Biol. Chem. 284, 24017–24024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morgunova E., Tuuttila A., Bergmann U., Isupov M., Lindqvist Y., Schneider G., Tryggvason K. (1999) Science 284, 1667–1670 [DOI] [PubMed] [Google Scholar]
- 85.Van Doren S. R. (2010) in Extracellular Matrix Degradation (Parks W. C., Mecham R. P. eds) Springer, Berlin, in press [Google Scholar]
- 86.Wells J. A. (1990) Biochemistry 29, 8509–8517 [DOI] [PubMed] [Google Scholar]
- 87.Reichmann D., Rahat O., Albeck S., Meged R., Dym O., Schreiber G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hedstrom L., Szilagyi L., Rutter W. J. (1992) Science 255, 1249–1253 [DOI] [PubMed] [Google Scholar]
- 89.Oue S., Okamoto A., Yano T., Kagamiyama H. (1999) J. Biol. Chem. 274, 2344–2349 [DOI] [PubMed] [Google Scholar]
- 90.Aharoni A., Gaidukov L., Khersonsky O., McQ Gould S., Roodveldt C., Tawfik D. S. (2005) Nat. Genet. 37, 73–76 [DOI] [PubMed] [Google Scholar]
- 91.Tomatis P. E., Rasia R. M., Segovia L., Vila A. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13761–13766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morley K. L., Kazlauskas R. J. (2005) Trends Biotechnol. 23, 231–237 [DOI] [PubMed] [Google Scholar]
- 93.Sela-Passwell N., Rosenblum G., Shoham T., Sagi I. (2010) Biochim. Biophys. Acta 1803, 29–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.