Abstract
Hyper-IgM syndrome and Common Variable Immunodeficiency are heterogeneous disorders characterized by predisposition to serious infection and impaired or absent neutralizing antibody responses. While a number of single gene defects have been associated with these immune deficiency disorders, the genetic basis of many cases is not known. To facilitate mutation screening in patients with these syndromes, we have developed a custom 300-kb resequencing array, the Hyper-IgM/CVID chip, which interrogates 1576 coding exons and intron-exon junction regions from 148 genes implicated in B cell development and immunoglobulin isotype switching. Genomic DNAs extracted from patients were hybridized to the array using a high-throughput protocol for target sequence amplification, pooling, and hybridization. A web-based application, SNP Explorer, was developed to directly analyze and visualize the single nucleotide polymorphism annotation and for quality filtering. Several mutations in known disease-susceptibility genes such as CD40LG, TNFRSF13B, IKBKG, AICDA, as well as rare nucleotide changes in other genes such as TRAF3IP2 were identified in patient DNA samples and validated by direct sequencing. We conclude that the Hyper-IgM/CVID chip combined with SNP Explorer may provide a cost-effective tool for high-throughput discovery of novel mutations among hundreds of disease-relevant genes in patients with inherited antibody deficiency.
Keywords: Hyper-IgM, CVID, B cell, somatic hypermutation, resequencing microarray, SNP
Introduction
Hyper-IgM syndrome is a rare primary immune deficiency disorder that is characterized by low or absent serum IgG, IgA, or IgE antibodies and predisposition to recurrent infection in infancy and early adulthood [Conley, et al., 2009]. Most Hyper-IgM patients have single-gene defects in genes that are involved in the CD40-CD40LG signaling pathways, such as CD40LG (MIM# 300386) [Allen, et al., 1993; Aruffo, et al., 1993], CD40 (MIM# 109535) [Ferrari, et al., 2001], AICDA (MIM# 605257) [Revy, et al., 2000], UNG (MIM# 191525) [Imai, et al., 2003b], and IKBKG (NEMO; MIM# 300248) [Jain, et al., 2001]. However, additional disease susceptibility genes responsible for this disorder await identification [Imai, et al., 2003a].
In contrast, common variable immunodeficiency (CVID; MIM #240500) is a relatively prevalent disease (1:25,000) that is usually diagnosed in youth or adulthood [Conley, et al., 2009]. CVID patients usually present with recurrent infections in mucosal tissues as consequence of marked hypogammaglobulinemia. Mutations in T cell co-stimulators such as ICOS (MIM# 604558) [Grimbacher, et al., 2003], or B-cell receptors such as CD19 (MIM# 107265) [van Zelm, et al., 2006], TNFRSF13C (BAFFR; MIM# 606269) [Warnatz, et al., 2009], and TNFRSF13B (TACI; MIM# 604907) [Castigli, et al., 2005; Salzer, et al., 2005] were found to be associated with CVID; however, the genetic basis of the majority of CVID cases remains unknown.
Due to large genetic diversity and sporadic occurrence, direct mutation detection among a number of candidate genes in clinically relevant gene sets and pathways is necessary to pinpoint exact genetic defects for each Hyper-IgM/CVID patient. A common approach is to re-sequence all candidate genes through traditional Sanger sequencing. However, despite technological improvements the Sanger dideoxy sequencing remains laborious and costly even for sequencing a limited number of genes [Pettersson, et al., 2009]. Although next-generation sequencing technology may allow large-scale de novo sequencing due to its capability of performing millions of parallel sequencing in one single run [Bentley, et al., 2009], at the present time it requires substantial computational resources for sequence analysis, and is therefore not a cost-effective way for mutation screening in a clinical setting.
Compared with the two sequencing technologies described above, a resequencing array is able to detect sequence variations in multiple genes simultaneously with high accuracy and reproducibility, provided the reference sequence is available and tiled on the chip [Hacia, 1999]. Since data generated by a resequencing array do not require post-sequencing assembly, post-array data analysis is relatively straightforward. So far, resequencing arrays have been evaluated as diagnostics tools in many complex genetic diseases such as amyotrophic lateral sclerosis (MIM# 105400) [Takahashi, et al., 2008], severe combined immunodeficiency (SCID; MIM# 602450) [Lebet, et al., 2008], and hypertrophic cardiomyopathy [Fokstuen, et al., 2008]. However, most of these studies were limited to a simultaneous test of 30 genes or fewer. Cost-effective high-throughput protocols for resequencing assays or user-friendly tools for resequencing and SNP (single nucleotide polymorphism) data analysis are currently lacking, and their absence prevents the widespread use of this technology for mutation screening in the diploid genome.
Based on the 300-kb array platform, we developed a custom DNA resequencing array called the Hyper-IgM/CVID chip, which covers 148 genes implicated in T cell - B cell interaction, NF-κB activation, class-switch initiation, DNA repair, somatic hypermutation (SHM), or gene set enrichment analysis from previous microarray studies in NEMO-deficient Hyper-IgM patients [Jain, et al., 2004]. To facilitate mutation detection in a robust and high-throughout manner, we developed a resequencing array assay protocol and a new web-based data analysis tool – SNP Explorer.
Materials and Methods
Patient DNA sample preparation
Patients with Hyper-IgM or CVID were enrolled in NIAID IRB-approved protocol 006-I-0049 with informed consent. Genomic DNA was extracted from patient blood using the QIAamp DNA Blood Midi Kit (Qiagen, Valencia, CA). All patient DNA samples were first screened for mutations in known disease-susceptibility genes including IKBKG, CD40LG, and TNFRSF13B, at their original DNA collection sites. Thirty-four patient samples from unrelated families with no detectable mutation in these genes were used in the resequencing chip study. Seven DNA samples from healthy volunteers were also assayed on the chip to evaluate array performance.
Hyper-IgM/CVID chip design
A 49-format Affymetrix resequencing array platform was used to construct the Hyper-IgM/CVID chip (Table 1, full gene list in Supp. Table S1). The CustomSeq® Resequencing Array Design Guide (www.affymetrix.com/support/) was strictly followed. In brief, the “sequence file” was prepared by downloading FASTA format reference sequences (genome assembly: NCBI36/hg18) containing only coding exons and 18 bp of flanking sequences on either side of the exons from the UCSC genome browser (http://genome.ucsc.edu). All repeats and cross-hybridized sequences were identified by either the RepeatMasker program (www.repeatmasker.org) or BLAST search between the reference sequences and PCR amplicons. An “instruction file” was then compiled with exclusion of any repeat sequence and cross-hybridizing sequence larger than 9 bp. The final design of the Hyper-IgM/CVID chip interrogates 1576 coding exons and 6 bp of intron-exon splicing sites from 148 genes and is capable of sequencing 262,703 bases in double-strands (Supp. Table S1). The Hyper-IgM/CVID chip is manufactured by Affymetrix (Santa Clara, CA).
Table 1.
Classification of Candidate Genes on Hyper-IgM/CVID Chip
| Pathways | Number of Genes | Representative Genes |
|---|---|---|
| NF-κB Pathway | 66 | RELA, NFKB1, IKBKG, BCL3, CYLD, MAP2K1 |
| Class-switch, SHM/DNA repair | 33 | CD40, CD40LG, XRCC4, ADA, AICDA, UNG, NBN |
| Immunodeficiency | 13 | ICOS, TNFRSF13B, TNFSF13, MSH4, XBP1, CD79A |
| B-cell development | 9 | PAX5, BLNK, CD70, FOXP1, TNFRSF13C, CD19, TNFSF13B |
| Others | 27 | SIAH1, CRTC2, PELI2, etc |
| Total | 148 | |
Target DNA amplification, quantitation, pooling, and fragmentation
All primers were designed using Primer3 (http://frodo.wi.mit.edu/primer3/) with the Tm generally set at 60°C and length at 25 nt. Two PCR conditions (for detail see Supp. Table S2) were adopted to produce approximately 800 amplicons ranging from 198 bp to 5 kb with LA Taq polymerase (Takara Bio, Madison, WI) on either a Veriti 96-well Thermocycler or GeneAmp 9700 (Applied Biosystems, CA). Multiplex PCR involving two primer pairs were also arranged between amplicons with size < 3 kb and with similar GC content, size, and amplification efficiency. Approximately 10–20 μg of genomic DNA is required to amplify all target exons in six 96-well plates with a 7-μl reaction volume. All primer sequences along with amplicon sizes and PCR conditions are included in Supp. Table S3.
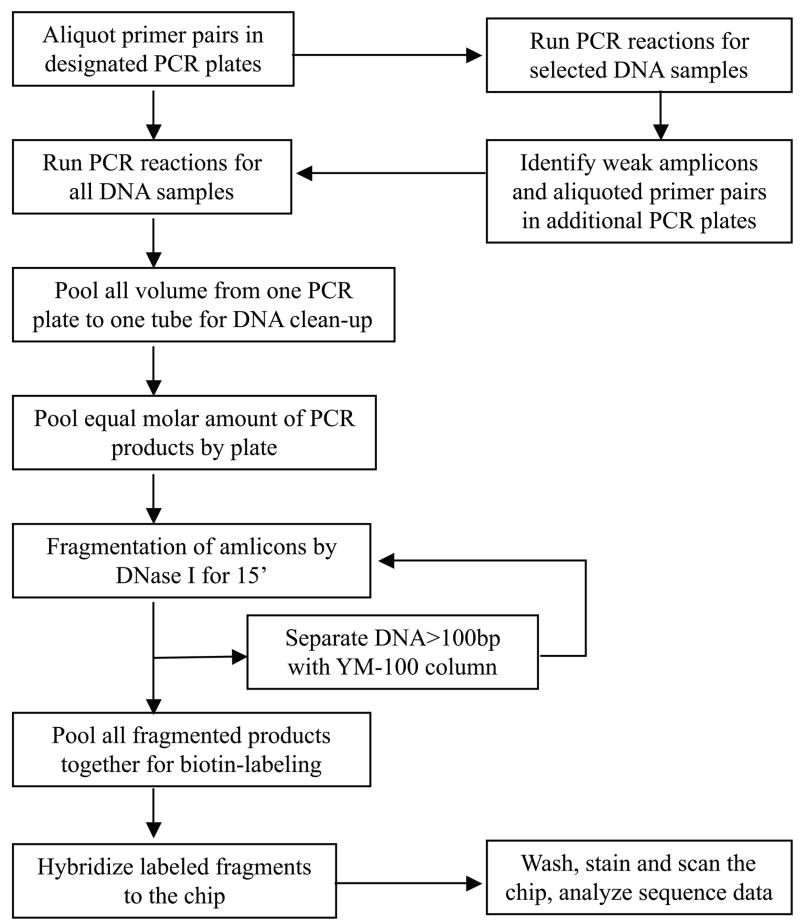
The complete schematic workflow of target sequence amplification, DNA pooling, fragmentation, labeling, and hybridization is shown in Fig. 1. In detail, for high-throughput DNA amplification, all primer pairs were pre-aliquoted by a Biomek FX robot (Beckman Coulter, Inc., Brea, CA) into PCR plates. The PCR amplification efficiency of each amplicon was tested for at least three different genomic DNA samples and determined by electrophoresing 0.5 μl of each PCR product in a 1–2% agarose gel. Any amplicon showing weak amplification was identified and re-amplified in supplemental primer plates with an increased amount of the template DNA. Thus, for all weak amplicons PCR amplification was conducted in both supplemental primer plates and along with the original set of primer plates; The PCR success rate of each DNA sample was assessed by analyzing 0.5 μl of 25% of all amplicons from each PCR plate on agarose gels. Plates with < 90% PCR success rate were re-amplified with an increased amount of the template DNA. After PCR, the entire volume of each PCR plate was pooled into one tube.
Figure 1.
The Schematic Workflow of Target Sequence Amplification, Pooling, and Hybridization for the Resequencing Chip Assay. Note that a two-round amplification strategy was applied for all weak PCR products and two-round fragmentation steps were applied to target sequences with variable amplicon lengths to improve fragmentation efficiency and uniformity.
PCR products from each plate were purified using the QIAquick PCR purification kit (Qiagen) and quantitated by a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). Equal molar amounts of DNA from each pool were combined based on total base pairs amplified per PCR plate and subjected to DNA fragmentation using DNase I (Roche, Indianapolis, IN). The fragmentation reaction was conducted at 37°C for 15 minutes with 0.08 U DNase I per μg DNA and inactivated at 95°C for 10 minutes. The efficacy of fragmentation was checked on a 2% agarose gel. Under-fragmented DNAs (>100 bp) were retrieved from the fragmented mix using YM-100 column (Millipore, Billerica, MA) and a second round of fragmentation was conducted. The properly fragmented DNAs were then labeled with Terminal Transferase from the GeneChip Resequencing Assay Kit (Affymetrix) following the GeneChip® CustomSeq® Resequencing Array Protocol (www.affymetrix.com/support/).
Resequencing chip hybridization and initial data analysis
Chip hybridization, washing, staining, and scanning were carried out following the GeneChip® CustomSeq® Resequencing Array Protocol using GeneChip® Fluidics Station 450 and the GeneChip® Scanner 3000 (Affymetrix). The Affymetrix GCOS 1.4 and GSEQ 4.0 software were used for image and initial sequence analysis with default settings under diploid mode except for quality score threshold, which was set at 0. Chip data from all DNA samples were analyzed in a batch mode as a requirement of the resequencing array ABACUS algorithm to produce reliable base calls. After that, the SNP sequences of each DNA sample and base call assignment plus quality score value at each base position for all samples were downloaded from the GSEQ for later SNP quality filtering and annotation.
SNP Explorer program design
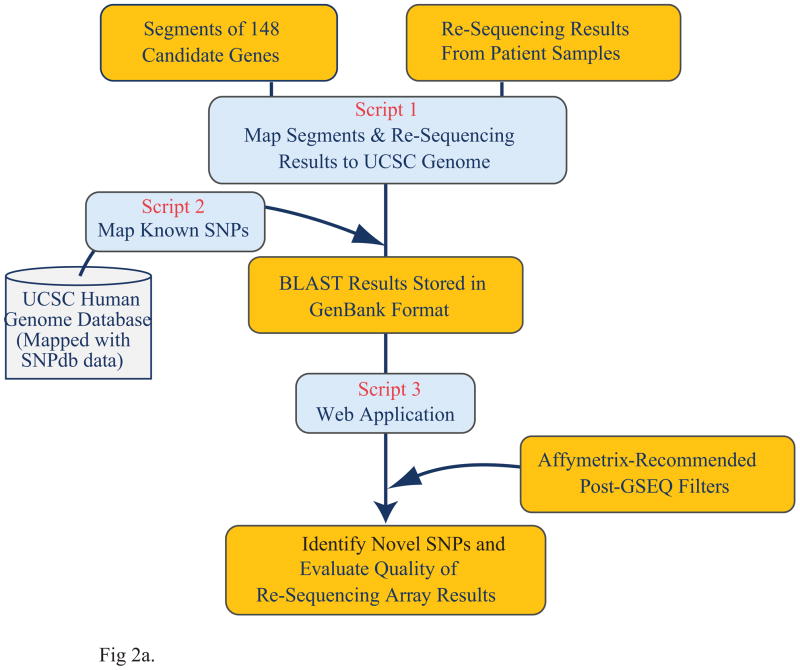
This web-based visualization and analysis system was developed for post-GSEQ resequencing data annotation and SNP data quality filtering (http://exon.niaid.nih.gov/SNPexplorer.html). Several Perl scripts were written to handle each task (Fig. 2a) using BioPerl and BioGraphics libraries. In detail, all coding sequences (CDS) and intron/exon junction regions of the 148 genes, as well as known SNPs present in these regions, were obtained from the NCBI36/hg18 version of the human genome and dbSNP annotations (build 130) in the UCSC genome database; The raw SNP sequence data or raw sequence data with quality score downloaded from Affymetrix GSEQ software were automatically aligned to the reference sequences based on the instruction file prepared during the chip design phase and the header information provided for each SNP base; Unmatched bases (except no call or “n” call) were identified as SNPs and displayed in the SNP Explorer graphic interface together with cDNA position number, protein position number, and amino acid substitution information. Nucleotide numbering throughout this manuscript reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, according to the guidelines of the Human Genome Variation Society (www.hgvs.org/mutnomen). The initiation codon is codon 1.
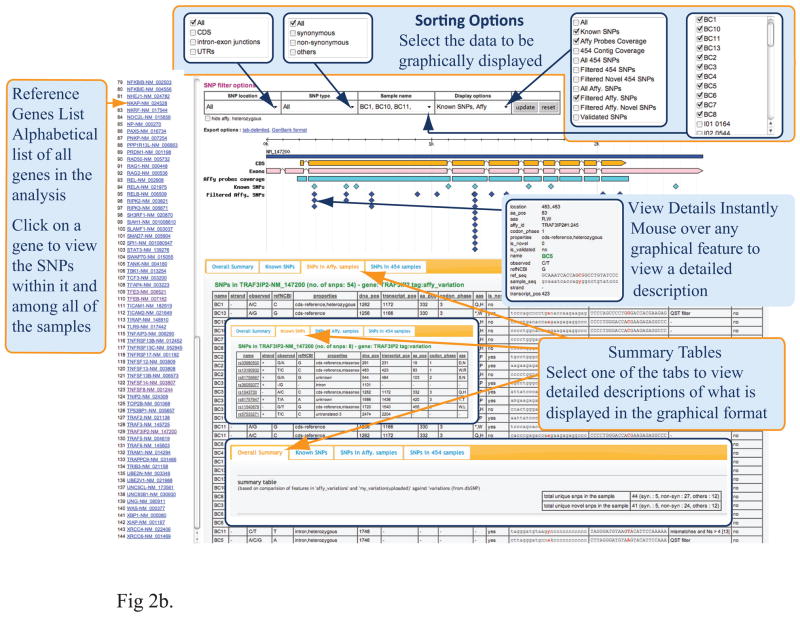
Figure 2.
Overview of the Web-Based Tool, SNP Explorer for Resequencing Array Data Analysis. (a) The data process workflow of SNP Explorer. (b) Graphic presentation of SNP Explorer.
Additional SNP filters with pre-loaded quality control parameters based on GeneChip® CustomSeq® Resequencing Array Base Calling Algorithm Version 2.0 (www.affymetrix.com/support/) were run at the same time to remove poor quality SNPs resulting from PCR failure, cross-hybridization, or SNP footprint effects. In addition, all novel SNP calls occurring in more than 12 samples (≥30%) were removed to decrease the false-positive rate. A simple search interface has been provided to select unfiltered or filtered SNPs from all samples or partial samples. The selected SNP data are displayed in both graphic presentation and table view (Fig. 2b). The SNP data can be exported into an Excel file or as a GenBank format file for further analysis.
Dideoxy sequencing analysis
Selected DNA samples that harbor non-synonymous nucleotide changes in certain exons identified by the chip were subjected to validation by direct sequencing. Each exon was amplified from the same DNA sample, purified using the QIAquick PCR purification kit, and sequenced through a contract facility at SAIC (Frederick, MD). The sequence trace files and their NCBI reference sequences were uploaded into Sequencher (V4.7, Gene Codes Corp, Ann Arbor, MI) for alignment. All trace profiles at the base positions of suspected SNP calls were visually inspected.
SNP effect analysis
For all validated non-synonymous variant calls identified in patient samples, the predicted SNP functional effects were calculated through an online program, Polyphen (http://genetics.bwh.harvard.edu/pph/). In addition, for any variant call occurring in multiple patient samples, the allele frequency was calculated by dividing the number of mutant alleles by the total number of alleles in all patient samples and compared to the allele frequency in a European population obtained from NCBI dbSNP build 131. The estimated odds ratio, confidence interval, and P-value were also calculated through an online tool, JavaStat (http://statpages.org/ctab2x2.html).
Results
Improvement of assay throughput and reduction of assay cost by multiplex PCR and multiple PCR and fragmentation steps
Enrichment of target sequences from genomic DNA is a necessary but time-limiting factor for a large-scale resequencing project. A widely accepted method is to amplify target sequences by PCR using specific primer pairs. However, highly variable amplification efficiency due to sequence secondary structure, primer design, or template quality limits this enrichment method and requires normalization of each amplicon before sequence detection. Previous protocols for amplicon normalization require quantitation by a reliable dsDNA quantitation assay and subsequent pooling of a desired volume of each amplicon by a programmable robot. This step has proved to be time-consuming and potentially inaccurate due to the presence of non-specific and primer-dimer bands in any PCR reaction.
To reduce the number of PCR reactions and associated cost of reagents and primers, we developed both short-range and long-range PCR conditions to amplify 1576 coding exons in 148 genes. Since some of our DNA samples had limited quantity, to further reduce reagent cost and template DNA usage, multiplex PCR reactions involving two more primer pairs were set up for certain amplicons with similar amplification efficiency (Supp. Table S3 and Supp. Figure S1).
To eliminate the target amplicon normalization process and thereby reduce the overall labor time of the resequencing chip assay, all weak amplicons identified during the initial QC process were amplified twice in all samples to increase their percentage in the final target DNA pool. Generally, up to 99% of target sequences were amplified in sufficient quantity with this two-round PCR amplification scheme. Additional quantitation and normalization of individual amplicons prior to array hybridization was not needed.
On the other hand, properly fragmented DNA is required for high-quality array hybridization as under- or over-fragmented DNA may lead to weak or non-specific hybridization signal. To ensure equal cutting of DNA fragments of different sizes while avoiding over-fragmentation, a two-round DNA fragmentation strategy each operating at sub-optimal conditions was employed. Under-fragmented DNA was separated after the first-round of fragmentation and went through the second-round of DNA fragmentation reaction. To sum up, with this improved amplification, fragmentation and hybridization protocol (Fig. 1), we achieved a base call rate > 99% for most of our DNA samples (Supp. Table S4).
Development of SNP Explorer tool and resequencing data analysis
To efficiently retrieve good quality SNPs from the large amount of resequencing array data and to provide annotated information for each identified SNP, we developed the SNP Explorer web application to perform all the post-GSEQ analysis.
Unlike other sequence alignment and annotation tools such as Sequencher and Lasagene, SNP Explorer is specifically designed to handle the noisy sequence data produced by the Affymetrix gene chips, which may contain too many “n” calls that prevent proper alignment with reference sequences, and directly compares the SNP calls with known SNPs present in the dbSNP database. The SNP Explorer includes implementation of a series of data quality filters based on the GeneChip® CustomSeq® Resequencing Array Base Calling Algorithm V2.0 (www.affymetrix.com/support/). In addition, we found that many novel SNP calls were identified in multiple DNA samples but most of these could not be confirmed. This probably indicates regions of poor hybridization or non-specific hybridization, or system errors resulting from the chip fabrication. We therefore implemented another filter in the SNP Explorer to remove novel SNP calls occurring in more than 30% of all test DNA samples. After filtering, >90% bad-quality SNP calls resulting from PCR failure, cross-hybridization, SNP footprint effects, or system errors, were automatically filtered out (Supp. Table S4), allowing us to retrieve reliable SNP calls for dideoxy sequencing confirmation or other downstream analysis. In summary, the SNP Explorer processes raw Affymetrix resequencing data and provides general users with a simple graphic interface to visualize SNP results before or after filtering and in one or more samples (Fig 2b).
Analysis of Hyper-IgM/CVID chip resequencing data from 41 DNA samples through GSEQ and SNP Explorer resulted in the identification of an average of 300 sequence changes per sample after data filtering (Supp. Table S4). This accounts for one SNP call per 900 bp, of which 58% were novel SNP calls thus a higher sensitivity of novel SNP discovery is achieved. Due to resource and time constraints, we initially selected 272 variant calls (SNP calls or “n” calls) identified mainly in patient DNA samples for dideoxy sequencing validation. We focused on novel non-synonymous nucleotide changes with significant amino acid substitutions, e.g., charged to non-charged or oppositely charged. In addition, if a SNP call was validated in one sample, other samples with no call at the same base position were also sequenced to avoid potentially missed SNP calls by the ABACUS algorithm [Lebet, et al., 2008]. By this validation strategy, the false-negative variant rate was reduced to 6.4% within the validated PCR amplicons (excluding “n” calls) (Table 2). This result was comparable to other similar reports regarding the performance of resequencing array [Kothiyal, et al., 2010]. Upon further examination of these false-negative SNP calls, we found that about two-thirds were low-quality reference calls (Quality Score < 3) and about 89% of them matched known dbSNPs. On the other hand, about 51% of variant calls were confirmed to be false positive. Interestingly, 92.8% of known dbSNPs identified by the resequencing chips were confirmed to be true; in contrast, only 10.9% of novel SNP calls were correct (Table 2). This indicates that for any novel variant call identified by the resequencing array, validation by an independent technology is essential. The higher false-positive rate of a resequencing chip assay is likely due to DNA sample quality, hybridization noises, or data analysis related issues.
Table 2.
Distribution of Known And Novel SNPs in Dideoxy-validated Variant Calls
| Variant Call Category | Total Validated Calls | SNP Calls Confirmed | Calls Not-confirmed | Confirmed SNP Calls% | Missed SNP Calls (called as reference base) |
|---|---|---|---|---|---|
| Match known dbSNPs | 125 | 116 | 9 | 92.80% | 8 |
| Novel or rare SNPs | 147 | 16 | 131 | 10.90% | 1 |
| Total | 272 | 132 | 140 | 9 | |
| Variant false-positive rate | 51.47% | ||||
| Variant false-negative rate | 6.38% | ||||
Selected variant calls (SNP or no calls) identified mainly in the patient group by resequencing chip assays were subjected to dideoxy sequencing validation. Many of these lead to non-synonymous amino acid changes.
Identification of disease-susceptibility genes and disease-associated SNPs by resequencing chip assays
Based on dideoxy sequencing results prior to resequencing chip assays, all of the samples tested on the Hyper-IgM/CVID chip were considered negative for mutations in known disease-susceptibility genes including IKBKG, CD40LG, and TNFRSF13B. However, we still identified several causative mutations and disease-associated SNPs that involved these three genes in seven patients (Table 3), suggesting a significant rate of mis-diagnosis by routine sequencing tests. Interestingly, we found that three of these seven patients demonstrated a combination of rare nucleotide changes occurring in two known disease-susceptibility genes, e.g., [c.62C>G, p.P21R] in TNFRSF13C and [c.655G>A, p.G217R] in CD40LG. Although possessing one of these variants was not confirmed to be the causative mutation of Hyper-IgM or CVID [Lee, et al., 2005; Losi, et al., 2005], it is still possible that the presence of concurrent mutations in both genes may eventually block the activation of B cell class-switch recombination pathways, a hypothesis that awaits future experimental confirmation. Other mutations or disease-associated SNPs in known disease-susceptibility genes such as AICDA and TNFRSF13C were identified in six other patients and subsequently confirmed by dideoxy sequencing (Table 3). In addition, we found that one patient’s DNA sample consistently failed hybridization for the AICDA gene, which was later found to be due to a large genomic deletion disrupting the whole AICDA gene (data not shown). In total, we identified disease-causative or contributory variants for antibody deficiency in 41% (14/34) of patients.
Table 3.
Selected Disease-Associated SNPs or Mutations Identified in Patient DNA Samples By Hyper-IgM/CVID chip
| Gene Symbol (Name, Refseq) |
Nucleotide Change |
Protein Change |
SNP ID | PolyPhen Score |
Predicted SNP Effect |
Number of Samples |
Disease Association |
Note |
|---|---|---|---|---|---|---|---|---|
| CD40LG (NM_000074.2) | c.346+1G>A | 1 | Hyper-IgM [Lee, et al., 2005] | Splicing site mutation | ||||
| c.655G>A | p.G219R | 1.442 | Benign | 3* | Hyper-IgM [Lee, et al., 2005] | SNP; two are heterozygotes | ||
| AICDA (NM_020661.1) | c.334C>T | p.R112C | 3.361 | Probably damaging | 1 | Hyper-IgM [Lee, et al., 2005] | Mutation; compound heterozygotes | |
| c.374G>A | p.G125E | 2.185 | Probably damaging | |||||
| IKBKG (NEMO, NM_003639.3) | c.337G>A | p.D113N | 1.082 | Benign | 1 | Hyper-IgM [Salt, et al., 2008] | Mutation; heterozygote in female | |
| TNFRSF13C (BAFFR, NM_052945.2) | c.475C>T | p.H159Y | rs61756766 | 2.082 | Probably damaging | 2+ | CVID [Losi, et al., 2005] | SNP; all are heterozygotes; +: compound heterozygotes |
| c.62C>G | p.P21R | 1.376 | Benign | 8 | ||||
| TNFRSF13B (TACI, NM_012452.2) | c.310T>C | p.C104R | rs34557412 | 3.571 | Probably damaging | 1 | CVID [Salzer, et al., 2005] | Mutation; heterozygote |
| c.58C>T | p.R20C | 1.8 | Possibly damaging | 1 | CVID [Salzer, et al., 2007] | SNP; Heterozygote | ||
| Total patients with causative mutations or disease-associated SNPs: | 13 | |||||||
The PolyPhen score and predicted SNP effect were obtained from PolyPhen (http://genetics.bwh.harvard.edu/pph/). Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence. All mutations reported here are present in the Locus Specific Mutation Databases (http://www.hgvs.org/dblist/glsdb.html).
Samples with * also contained [c.62C>G; p.P21R] variant in TNFRSF13C.
To identify additional disease-associated sequence variants, we evaluated the allele frequencies of validated SNP calls identified only in the patient group but not in the seven healthy controls, and compared them to the frequencies of an European population reported in dbSNP, since most of our patients are Caucasians. We found that the allele frequencies of two SNPs (rs33980500:G>A, p.D19N; rs13190932:C>T, p.R83W) of the TRAF3IP2 gene (MIM# 607043) were very low (< 0.1) in the reported European population, but were significantly increased in our patient group with estimated odds ratio >2 (Table 4). TRAF3IP2 has been shown to be a negative regulator of CD40LG and TNFSF13B (BAFF; MIM# 603969) mediated signaling pathways [Qian, et al., 2004] but a key positive regulator in IL-17 signaling pathways [Liu, et al., 2009]. The amino acid changes (D19N and R83W) are both located in the N-terminal part of TRAF3IP2 protein. Although this region was not shown to bind to IKK complex, it is required for TRAF3IP2-induced NF-κB activation [Mauro, et al., 2003] and therefore mutation in this region could affect the activity of TRAF3IP2. Future large-scale screening of these potential disease-associated SNPs in a larger cohort of patients with antibody deficiency, combined with functional studies, may reveal the disease mechanism for certain types of Hyper-IgM/CVID.
Table 4.
Known SNPs With Increased Allele Frequency In Hyper-IgM/CVID Patients
| SNP ID | Gene Symbol | Protein change | PolyPhen Score | Predicted SNP Effect | Allele Freq. in Europe population | Allele Freq. in patient group | Estimate Odds Ratio | 95% confidence Interval | P-value |
|---|---|---|---|---|---|---|---|---|---|
| rs13190932:C>T | TRAF3IP2 | p.R83W | 2.257 | probably damaging | 3.3% | 13.3% | 4.495 | from 1.379 to 14.55 | 0.003 |
| rs33980500:G>A | TRAF3IP2 | p.D19N | 1.614 | possibly damaging | 9.5% | 21.4% | 2.594 | from 1.157 to 5.803 | 0.008 |
Variant allele frequencies in European population were obtained from NCBI dbSNP build 131. Allele frequency in patient group was calculated by dividing the number of mutant allele by the total number of alleles in all patient samples. The PolyPhen score and predicted SNP effect were obtained from PolyPhen (http://genetics.bwh.harvard.edu/pph/). The odds ratio, confidence interval, and P-value were calculated through an online tool (http://statpages.org/ctab2x2.html).
Discussion
To date, we have successfully applied the Hyper-IgM/CVID chip to screen 34 patients and seven healthy donors. A total of 10,651,393 bp from 41 DNA samples was sequenced during the course of this study with a mean nucleotide call rate of 99%. As a first step toward our goal of discovering novel mutations, we have developed a high-throughput microarray-resequencing assay through the stages of array design, assay development, and effective data analysis by GSEQ and SNP Explorer.
The new resequencing assay protocol is more robust and of higher throughput than the standard resequencing array protocol but still retains sufficient sensitivity and specificity for 99% of target sequences. Generally, the complete assay of 148 genes for one patient sample from PCR amplification to data reporting takes only 3 to 4 days. Several DNA samples could be processed together depending on the number of available PCR instruments, which further reduces assay cost. Although there is a one-time mask fee of ~ $35,000 required by the manufacturer to design a new resequencing chip, once the photolithographic mask for the chip is made, any subsequent chip order costs ~ $300 to $500 depending on the order quantity. We calculate that < $1000 ($300 chip + $400 reagent costs) is sufficient to conduct one resequencing assay in 3.5 days, whereas dideoxy sequencing requires at least 30 plates to sequence 1576 exons in 2x coverage, which may easily add up to > $5,000 per patient (based on the current sequencing price of at least $150 per plate and $500 for PCR reagents and clean-up per sample). Therefore the resequencing chip assay represents a cost reduction of at least 4 to 5-fold, even including the cost of final sequencing confirmation.
Since the current PCR protocols did not eliminate the sequencing bias associated with primer design such as rare mutations within the primer region and variable amplification efficiencies for different templates, improvement of the sequence enrichment protocol may increase the robustness and accuracy of resequencing assays. Several groups [Albert, et al., 2007; Bau, et al., 2009; Okou, et al., 2007] reported a universal sequence enrichment method involving sequence-capture microarray in which thousands of array-fixed sequence-specific probes were used to retrieve targets from a pool of fragmented genomic DNA. The retained target sequences were then amplified by universal PCR and detected on either gene chip [Okou, et al., 2007] or next-generation sequencing platforms [Albert, et al., 2007; Bau, et al., 2009]. We are currently evaluating several sequence capture technologies and may incorporate one of these into our next resequencing assay system.
Compared with data generation by chip hybridization, data interpretation without an appropriate annotation tool could be very time consuming. The development of SNP Explorer dramatically reduced the data analysis time previously associated with resequencing chip assays. Since the structure of the SNP Explorer is flexible and open, other data filters for the resequencing assay could be easily built in once a better data analysis algorithm for the resequencing assay has been developed. In addition, although the SNP Explorer was initially designed for the Hyper-IgM/CVID chip study, it can be quickly modified for other resequencing array platforms or even next-generation sequencing platforms. In fact, the integration of next-generation data into the SNP Explorer is currently underway in our laboratory. Upon completion, the SNP Explorer will undoubtedly facilitate sequence analysis in any large-scale mutation screening studies.
Besides identification of several known causative mutations in known disease-susceptibility genes, we have also validated rare nucleotide changes in other genes not previously associated with Hyper-IgM/CVID in some patients (data not shown). We are currently evaluating the pathogenic significance of these sequence variants in other studies. Thus, our findings indicate that resequencing chip assays can be used efficiently to identify both known and novel mutations in a large number of candidate genes.
Nevertheless, the recent rapid advancement of next-generation sequencing technology poses a significant challenge to resequencing applications on an array platform. To take advantage of this technology trend, we have conducted a test run of mutation screening by the 454-platform using five amplified DNA products that were previously screened by the Hyper-IgM/CVID chip. The final results were not encouraging due to the following factors: 1) more sophisticated target sequence enrichment and normalization protocols beyond the ones practiced in this paper are required for effective pyrosequencing as unbalanced target representation and sequencing bias toward 3’ end were found for many target regions; 2) the average sequence coverage was 10x, therefore fewer reliable SNP calls were identified since coverage above 20x is required [Bentley, et al., 2009]; 3) there is a lack of user-friendly tools for mapping and SNP identification, therefore the vast amount of sequencing data generated per run requires significant further bioinformatics analysis; 4) the cost of one sequencing run in a Roche/454 Instrument ($9k per run) was much higher than that of the resequencing chip. Further reduction in the cost of target amplification and sequencing chemistries, as well as improvements in data analysis tools are necessary for next-generation resequencing to become widely adopted for molecular diagnosis.
Taken together, our data show that mutation screening by resequencing microarray technology is an important tool for the molecular delineation of B cell immune deficiency. As mentioned, the basic defect inherent in these disorders is an inability to generate neutralizing antibody responses that predisposes the host to infection. However, as is the case with other monogenetic disorders, there is significant clinical variability among siblings and other affected family members with a defined gene defect, thus clearly other genetic loci must be involved. The Hyper-IgM chip, combined with the SNP Explorer program, may offer a cost-effective mutation-screening tool to identify additional genetic modifiers that contribute to disease severity. In addition, our Hyper-IgM/CVID chip contains a significant number of genes involved in the activation of NF-κB pathways. Some mutations in these genes may result in a gain of function or loss of function that actively regulates cancer development and progression. For example, gain-of function mutation in REL [Pfeifer, et al., 2007] and loss of function in TRAF3 [Keats, et al., 2007], which result in increased transcription of NF-κB targeted genes, are associated with chronic lymphocytic leukemia and myeloma, respectively. Thus the tools we have created could also be useful in the molecular delineation of B cell malignancies or other related diseases. We expect that evaluation of resequencing assays with Hyper-IgM/CVID chip and the SNP Explorer tool for such applications will be conducted in the near future.
Supplementary Material
Acknowledgments
We thank Dr. Elaine Remmers, Ms. Julie Le, and other members of NIAMS Genomic Section for their technical assistance and Mary Derry for editorial assistance. We also like to thank the NIAID Research Technologies Section and NIAID Office of Cyber Infrastructure and Computational Biology (OCICB) for their support and valuable discussions. This research was supported by the Intramural Research Program of the NIH, NIAID.
Footnotes
Methods
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
Refer to Online publication for Supporting Information. The SNP Explorer is a free online program available at http://exon.niaid.nih.gov/SNPexplorer.html. As per federal government regulations, request of the source code should be made through the NIAID Office of Technology Development (OTD). For Hyper-IgM/CVID chip, interested group are invited to contact Dr. Ashish Jain for permission to purchase the chip from Affymertrix for non-profit research.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4(11):903–5. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Allen RC, Armitage RJ, Conley ME, Rosenblatt H, Jenkins NA, Copeland NG, Bedell MA, Edelhoff S, Disteche CM, Simoneaux DK, et al. CD40 ligand gene defects responsible for X-linked hyper-IgM syndrome. Science. 1993;259(5097):990–3. doi: 10.1126/science.7679801. [DOI] [PubMed] [Google Scholar]
- Aruffo A, Farrington M, Hollenbaugh D, Li X, Milatovich A, Nonoyama S, Bajorath J, Grosmaire LS, Stenkamp R, Neubauer M, et al. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 1993;72(2):291–300. doi: 10.1016/0092-8674(93)90668-g. [DOI] [PubMed] [Google Scholar]
- Bau S, Schracke N, Kranzle M, Wu H, Stahler PF, Hoheisel JD, Beier M, Summerer D. Targeted next-generation sequencing by specific capture of multiple genomic loci using low-volume microfluidic DNA arrays. Anal Bioanal Chem. 2009;393(1):171–5. doi: 10.1007/s00216-008-2460-7. [DOI] [PubMed] [Google Scholar]
- Bentley G, Higuchi R, Hoglund B, Goodridge D, Sayer D, Trachtenberg EA, Erlich HA. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens. 2009;74(5):393–403. doi: 10.1111/j.1399-0039.2009.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castigli E, Wilson SA, Garibyan L, Rachid R, Bonilla F, Schneider L, Geha RS. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat Genet. 2005;37(8):829–34. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- Conley ME, Dobbs AK, Farmer DM, Kilic S, Paris K, Grigoriadou S, Coustan-Smith E, Howard V, Campana D. Primary B cell immunodeficiencies: comparisons and contrasts. Annu Rev Immunol. 2009;27:199–227. doi: 10.1146/annurev.immunol.021908.132649. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Giliani S, Insalaco A, Al-Ghonaium A, Soresina AR, Loubser M, Avanzini MA, Marconi M, Badolato R, Ugazio AG, et al. Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc Natl Acad Sci U S A. 2001;98(22):12614–9. doi: 10.1073/pnas.221456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokstuen S, Lyle R, Munoz A, Gehrig C, Lerch R, Perrot A, Osterziel KJ, Geier C, Beghetti M, Mach F, et al. A DNA resequencing array for pathogenic mutation detection in hypertrophic cardiomyopathy. Hum Mutat. 2008;29(6):879–85. doi: 10.1002/humu.20749. [DOI] [PubMed] [Google Scholar]
- Grimbacher B, Warnatz K, Peter HH. The immunological synapse for B-cell memory: the role of the ICOS and its ligand for the longevity of humoral immunity. Curr Opin Allergy Clin Immunol. 2003;3(6):409–19. doi: 10.1097/00130832-200312000-00001. [DOI] [PubMed] [Google Scholar]
- Hacia JG. Resequencing and mutational analysis using oligonucleotide microarrays. Nat Genet. 1999;21(1 Suppl):42–7. doi: 10.1038/4469. [DOI] [PubMed] [Google Scholar]
- Imai K, Catalan N, Plebani A, Marodi L, Sanal O, Kumaki S, Nagendran V, Wood P, Glastre C, Sarrot-Reynauld F, et al. Hyper-IgM syndrome type 4 with a B lymphocyte-intrinsic selective deficiency in Ig class-switch recombination. J Clin Invest. 2003a;112(1):136–42. doi: 10.1172/JCI18161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003b;4(10):1023–8. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- Jain A, Ma CA, Liu S, Brown M, Cohen J, Strober W. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2(3):223–8. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- Jain A, Ma CA, Lopez-Granados E, Means G, Brady W, Orange JS, Liu S, Holland S, Derry JM. Specific NEMO mutations impair CD40-mediated c-Rel activation and B cell terminal differentiation. J Clin Invest. 2004;114(11):1593–602. doi: 10.1172/JCI21345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12(2):131–44. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothiyal P, Cox S, Ebert J, Husami A, Kenna MA, Greinwald JH, Aronow BJ, Rehm HL. High-throughput detection of mutations responsible for childhood hearing loss using resequencing microarrays. BMC Biotechnol. 2010;10:10. doi: 10.1186/1472-6750-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebet T, Chiles R, Hsu AP, Mansfield ES, Warrington JA, Puck JM. Mutations causing severe combined immunodeficiency: detection with a custom resequencing microarray. Genet Med. 2008;10(8):575–85. doi: 10.1097/gim.0b013e31818063bc. [DOI] [PubMed] [Google Scholar]
- Lee WI, Torgerson TR, Schumacher MJ, Yel L, Zhu Q, Ochs HD. Molecular analysis of a large cohort of patients with the hyper immunoglobulin M (IgM) syndrome. Blood. 2005;105(5):1881–90. doi: 10.1182/blood-2003-12-4420. [DOI] [PubMed] [Google Scholar]
- Liu C, Qian W, Qian Y, Giltiay NV, Lu Y, Swaidani S, Misra S, Deng L, Chen ZJ, Li X. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Sci Signal. 2009;2(92):ra63. doi: 10.1126/scisignal.2000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losi CG, Silini A, Fiorini C, Soresina A, Meini A, Ferrari S, Notarangelo LD, Lougaris V, Plebani A. Mutational analysis of human BAFF receptor TNFRSF13C (BAFF-R) in patients with common variable immunodeficiency. J Clin Immunol. 2005;25(5):496–502. doi: 10.1007/s10875-005-5637-2. [DOI] [PubMed] [Google Scholar]
- Mauro C, Vito P, Mellone S, Pacifico F, Chariot A, Formisano S, Leonardi A. Role of the adaptor protein CIKS in the activation of the IKK complex. Biochem Biophys Res Commun. 2003;309(1):84–90. doi: 10.1016/s0006-291x(03)01532-8. [DOI] [PubMed] [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4(11):907–9. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- Pettersson E, Lundeberg J, Ahmadian A. Generations of sequencing technologies. Genomics. 2009;93(2):105–11. doi: 10.1016/j.ygeno.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Pfeifer D, Pantic M, Skatulla I, Rawluk J, Kreutz C, Martens UM, Fisch P, Timmer J, Veelken H. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109(3):1202–10. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- Qian Y, Qin J, Cui G, Naramura M, Snow EC, Ware CF, Fairchild RL, Omori SA, Rickert RC, Scott M, et al. Act1, a negative regulator in CD40- and BAFF-mediated B cell survival. Immunity. 2004;21(4):575–87. doi: 10.1016/j.immuni.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102(5):565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Salt BH, Niemela JE, Pandey R, Hanson EP, Deering RP, Quinones R, Jain A, Orange JS, Gelfand EW. IKBKG (nuclear factor-kappa B essential modulator) mutation can be associated with opportunistic infection without impairing Toll-like receptor function. J Allergy Clin Immunol. 2008;121(4):976–82. doi: 10.1016/j.jaci.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer U, Birmelin J, Bacchelli C, Witte T, Buchegger-Podbielski U, Buckridge S, Rzepka R, Gaspar HB, Thrasher AJ, Schmidt RE, et al. Sequence analysis of TNFRSF13b, encoding TACI, in patients with systemic lupus erythematosus. J Clin Immunol. 2007;27(4):372–7. doi: 10.1007/s10875-007-9094-y. [DOI] [PubMed] [Google Scholar]
- Salzer U, Chapel HM, Webster AD, Pan-Hammarstrom Q, Schmitt-Graeff A, Schlesier M, Peter HH, Rockstroh JK, Schneider P, Schaffer AA, et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat Genet. 2005;37(8):820–8. doi: 10.1038/ng1600. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Seki N, Ishiura H, Mitsui J, Matsukawa T, Kishino A, Onodera O, Aoki M, Shimozawa N, Murayama S, et al. Development of a high-throughput microarray-based resequencing system for neurological disorders and its application to molecular genetics of amyotrophic lateral sclerosis. Arch Neurol. 2008;65(10):1326–32. doi: 10.1001/archneur.65.10.1326. [DOI] [PubMed] [Google Scholar]
- van Zelm MC, Reisli I, van der Burg M, Castano D, van Noesel CJ, van Tol MJ, Woellner C, Grimbacher B, Patino PJ, van Dongen JJ, et al. An antibody-deficiency syndrome due to mutations in the CD19 gene. N Engl J Med. 2006;354(18):1901–12. doi: 10.1056/NEJMoa051568. [DOI] [PubMed] [Google Scholar]
- Warnatz K, Salzer U, Rizzi M, Fischer B, Gutenberger S, Bohm J, Kienzler AK, Pan-Hammarstrom Q, Hammarstrom L, Rakhmanov M, et al. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proc Natl Acad Sci U S A. 2009;106(33):13945–50. doi: 10.1073/pnas.0903543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.