Abstract
Africa is the birthplace of modern humans, and is the source of the geographic expansion of ancestral populations into other regions of the world. Indigenous Africans are characterized by high levels of genetic diversity within and between populations. The pattern of genetic variation in these populations has been shaped by demographic events occurring over the last 200,000 years. The dramatic variation in climate, diet, and exposure to infectious disease across the continent has also resulted in novel genetic and phenotypic adaptations in extant Africans. This review summarizes some recent advances in our understanding of the demographic history and selective pressures that have influenced levels and patterns of diversity in African populations.
Introduction
Modern humans evolved in Africa around 200 kya (thousand years ago), and have lived continuously on the African continent longer than in any other geographic region. Africa not only has the highest levels of human genetic variation in the world but also contains a considerable amount of linguistic, environmental and cultural diversity. For example, more than 2,000 distinct ethno-linguistic groups, representing nearly a third of the world’s languages, currently exist in Africa (http://www.ethnologue.com/) (Figure 1). Africans live in a wide range of environments, such as deserts, tropical rainforests, savannas, swamps, and mountain highlands [1, 2]. Furthermore, some of these environments have undergone dramatic changes over the course of modern human evolution [1, 3, 4]. African populations also practice a wide array of subsistence strategies, including various forms of hunting-gathering, agriculture and pastoralism, across the continent perhaps in response to this environmental variability over time and geographic space.
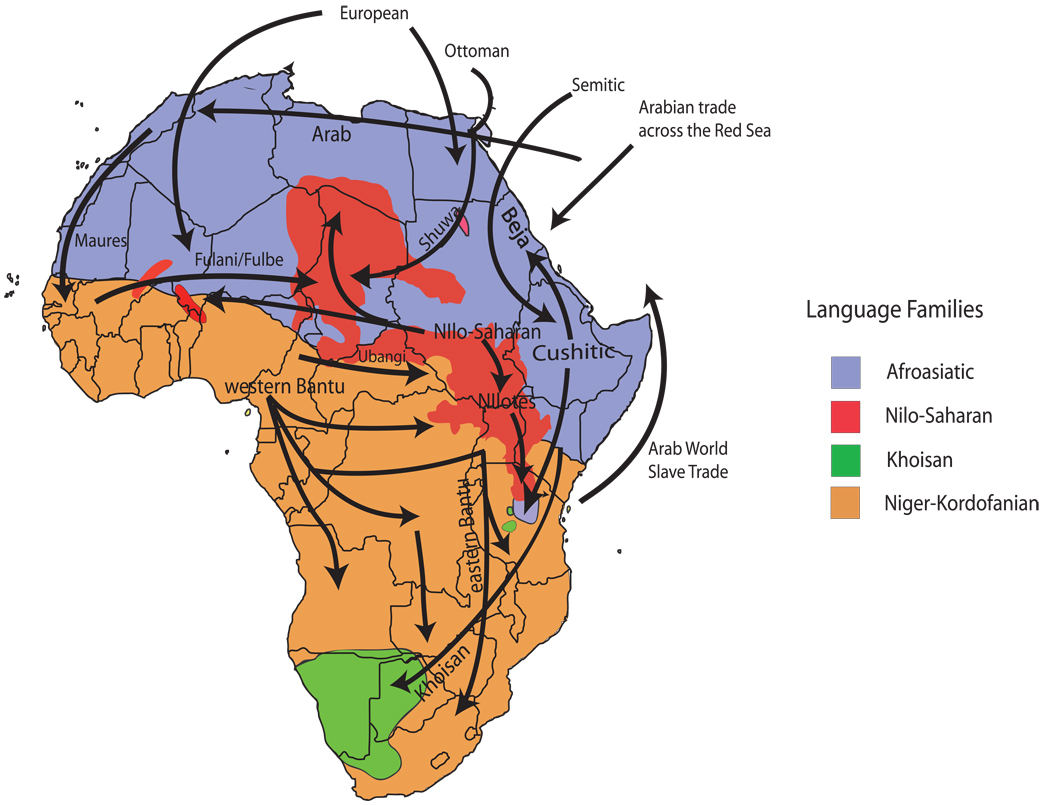
Figure 1. Classification of major language families and proposed historic and prehistoric migration events within Africa.
African languages have been classified into four major linguistic families as follows: Niger-Kordofanian (spoken predominantly by agriculturalist populations across a broad geographic distribution in Africa), Afroasiatic (spoken predominantly by northern and eastern African pastoralists and agropastoralists), Nilo-Saharan (spoken predominantly by central and eastern African pastoralists) and Khoesan (a language containing click-consonants, spoken by eastern and southern African hunter-gatherers). This map also shows a number of key migration events that occurred in Africa, most notably the expansion of Bantu-speakers from a homeland in Nigeria and Cameroon first into the equatorial rainforests and then into eastern and southern Africa, as well as the migration of Nilo-Saharan-speakers both westward and eastward from a Sudanese homeland. Additionally, Cushitic-speakers from the Ethiopian highlands migrated northward to the Red Sea coast of Sudan (where their modern-day descendants, the Beja, presently live) as well as southward into Kenya and Tanzania [6]. (based on Figure 1 from reference [2]).
African demographic history has consisted of fluctuations in population size, short- and long-range migration, admixture and extensive population structure which have resulted in complex patterns of variation in modern populations [1, 5]. The timing and duration of some of these demographic events were often correlated with known major environmental changes and/or cultural developments in Africa [6]. A number of novel genetic and phenotypic adaptations have also evolved in Africans in response to dramatic variation in environment, diet, and exposure to infectious disease across the continent. In some cases, these adaptations have occurred in the last several thousand years, exemplifying the ongoing evolution of human populations. Thus, present-day patterns of variation in African genomes are a product of both demographic and selective events.
The characterization of extant genetic diversity in Africa will be critical for reconstructing modern human origins and African demographic history. In addition, this genetic information, together with phenotype data on variable traits, will be informative for identifying population-specific variants that play a role in gene function, phenotypic adaptation and complex disease susceptibility in Africans and populations of African descent.
Evolutionary History of Modern Humans in Africa
Current paleontological data suggest that the transition to anatomically modern Homo sapiens occurred in Africa, supporting the ‘Recent African Origin’ model of human evolution (Figure 2). The earliest known suite of derived traits associated with anatomically modern humans was identified in fossil remains from East Africa dating to around 195–150 kya [7–9]. Thus, the basic morphology of modern humans was established in Africa about 200 kya [10]. Other early anatomically modern humans, with a more full set of modern features, also appear in Africa before 100 kya and in the Near East around 100 kya [11–13], followed by the more recent expansion of anatomically modern humans into Eurasia within the past 40,000–80,000 years [1, 2](Figure 2). Although the mode of evolution is still unclear, it has been suggested that the emergence of modern humans was not a sudden event, but rather a continuous process of gradual morphological change from archaic to modern H. sapiens [11]. However, it has also been argued that modern human origins likely involved episodes of sudden morphological change, leading to the appearance of anatomically modern H. sapiens as a species distinct from archaic humans [14]. Regardless of the mode of evolution, current fossil and chronological evidence indicate that modern humans existed in Africa for a relatively long period of time before their migration across much of the globe.
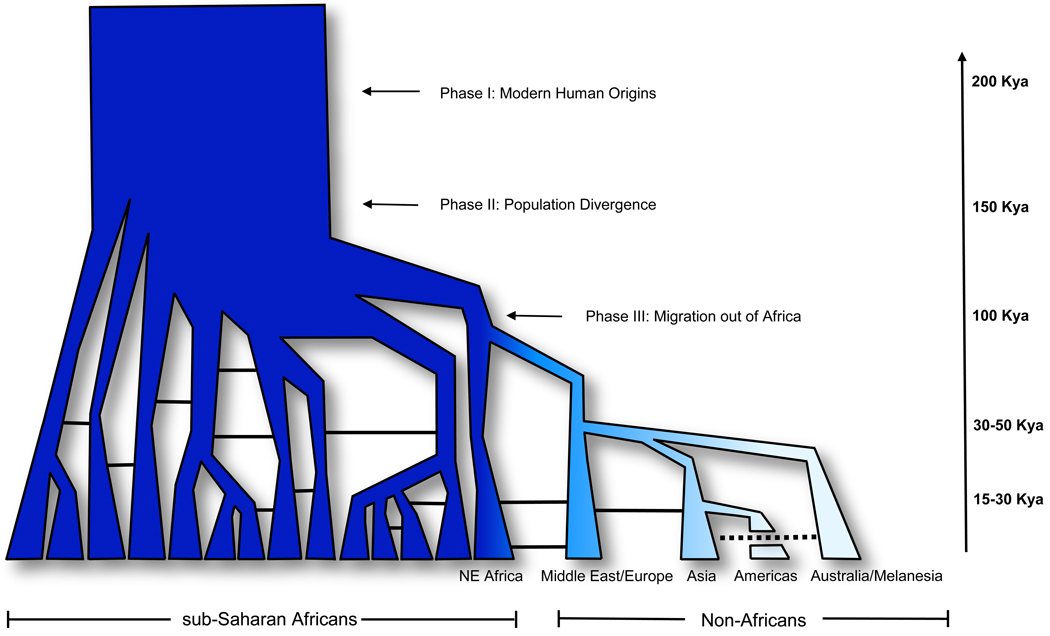
Figure 2. The ‘Recent African Origin’ model of modern humans and population substructure in Africa.
A study of genome-wide polymorphic markers in 121 ethnically diverse African populations indicated the presence of 14 genetically distinct ancestral population clusters in Africa [18]. According to the Recent African Origin model of modern human origins, anatomically modern humans evolved in Africa around 200 kya, migrated to Eurasia within the last 40,000–80,000 years and then migrated to the Americas within the last 15,000–30,000 years. The geographic expansion from Africa is thought to have been accompanied by a population bottleneck and a concomitant loss of genetic diversity. Studies have also suggested that a serial founder model of migration occurred in the history of non-Africans in which the migration of populations across much of the globe occurred in many small steps, with each migration event involving a sampling of variation from the previous population [25, 29, 33, 46]. In this figure, decreasing intensity of color represents the concomitant loss of genetic diversity as populations migrated in an eastward direction from Africa. Solid horizontal lines indicate gene-flow between ancestral human populations and the dashed horizontal line indicates recent gene-flow between Asian and Australian/Melanesian populations (based on Figure 2 from reference [1]).
Two main migratory routes out of Africa have been hypothesized for anatomically modern humans. The traditionally favored model involves a northern route of migration via North Africa and the Nile valley into the Levant with subsequent dispersal into both Europe and Asia [15]. Alternatively, an earlier southern coastal route has also been proposed in which modern humans first left Africa by crossing the Bab-el-Mandeb strait at the mouth of the Red Sea and then rapidly migrated along the South Asia coastline to Australia/Melanesia where evidence of human settlement dating to around 55 Kya can be found [15–17]. A recent genetic study that correlated levels of microsatellite diversity and the geographic position of sampled populations inferred a waypoint of dispersal of anatomically modern humans out of Africa centered on the Red Sea, strongly supporting an East African origin of migration of modern humans [18]. Although this study was not able to rule out the possibility of multiple migrations out of Africa, prior analysis of autosomal haplotype variability suggests that migration events originating from multiple genetically distinct source populations in Africa are unlikely [19].
The geographic expansion of a small number of anatomically modern humans out of Africa resulted in a population bottleneck. The size of the ancestral population(s) that left Africa is estimated to be around 1000 effective founding males and females based on autosomal microsatellite loci [20] or around 1500 effective founding males and females based on combined mtDNA, Y-chromosome, and X-chromosome resequencing data [21]. Recent studies comparing X-chromosome and autosomal diversity also suggest that the effective population sizes of founding males and females were not equal due to sex-biased migration. However, whether the sex-ratio of migrating individuals was male-biased or female-biased is currently under debate [22–24].
Patterns of Genetic Variation in Africa
Genome-wide data have indicated higher levels of genetic diversity in Africans compared to non-Africans, confirming the results of previous mtDNA, X-chromosome, and Y-chromosome studies [1, 25–33]. For example, a survey of 1327 nuclear microsatellite, insertion/deletion (INDEL) and single nucleotide polymorphism (SNP) markers showed that African populations, as well as African Americans, have the highest levels of within population genetic diversity relative to non-Africans [18]. In addition, more private alleles are present in Africa than in other geographic regions. The high level of genetic diversity in Africa is consistent with a larger long-term effective population size (Ne) which is estimated to be ~15,000 for Africans (and ~7,500 for non-Africans) based on resequencing data from a 10-kb autosomal non-coding region of the genome[34]. However, a recent study of 98-kb of sequence data from 20 loci on the X-chromosome indicated that effective population sizes of individual populations may be smaller (ranging from 2,300 to 9,000 for African populations and from 300 to 3,300 in non-African populations) [35]. Non-African populations also appear to have fewer private alleles and a subset of the genetic diversity present in sub-Saharan Africa, as expected under the ‘Recent African Origin’ model [1] (Figure 2).
A number of recent studies have also identified a considerable amount of structural variation in the human genome. ‘Structural variation’ commonly refers to genomic alterations that may be as small as a single nucleotide (excluding SNPs) or as large as millions of nucleotides in size, such as INDELs, copy number variation, translocations and inversions [36, 37]. Although the population genetics of structural variation are still in their infancy, information regarding the distribution of such variation within and between populations is emerging [1, 29, 38–40]. For example, a recent phylogenetic analysis based on 396 non-singleton copy number variants demonstrated that globally diverse populations clustered roughly by geographic region [29], and a study of 67 common copy number variants in the HapMap populations also noted that average FST, a classical measure of population divergence, of these loci was 0.11, comparable to previous estimates based on nucleotide and haplotype data [1, 38, 40]. Overall, results from these different studies suggest that levels and patterns of copy number variation in global populations are influenced by demographic history. However, technical limitations associated with a number of methods commonly used for structural variation detection [36, 37], including bias in structural variation ascertainment, can possibly distort estimates of demographic parameters. Advances in next-generation sequencing and other technologies, as well as the identification of structural variation in ethnically diverse Africans, will be needed to better determine patterns of structural variation in Africa and their possible role in phenotypic variability.
Population Structure
Several studies have indicated that ancestral populations were geographically structured before modern humans migrated out of Africa [1]. For example, it has been suggested that the deep coalescence times of mtDNA [41, 42] and X-chromosome [43, 44] lineages are consistent with a demographic scenario of ancient population structure in Africa. A recent analysis of cranial shape variability in anatomically modern human fossils (dating to 200–60 kya) from Africa and the Middle East also reported a high level of morphological divergence among these fossil hominids which was interpreted as evidence for ancestral population structure in Pleistocene Africa [45]. Thus, arguably, a considerable amount of genetic and phenotypic diversity may have been present at an early stage of modern human evolution.
Studies of global population structure in samples from the Human Genome Diversity Panel (HGDP-CEPH) also identified substructure in Africa, particularly between hunter-gatherers and other Africans [29, 46]. However, because the HGDP-CEPH contains a small number of African populations, some of which likely share recent common ancestry, these results do not reflect the full extent of population structure in Africa [1]. A recent genome-wide study of a much larger set of diverse Africans detected more extensive population structure within Africa than had been previously observed. Specifically, an analysis of 848 short tandem repeat polymorphisms (STRPs), 476 INDELs and 3 SNPs genotyped in ~2,400 individuals from 121 geographically diverse populations revealed the presence of 14 genetically distinct ancestral population clusters in Africa [18]. Each cluster consisted of populations that shared genetic similarity, as well as cultural and/or linguistic properties (for example, Pygmies, Khoesan-speaking hunter-gatherers, Bantu-speakers, Cushitic-speakers). Thus, there is a general correlation between genetic relatedness and linguistic/cultural similarity in Africa, although some exceptions exist. This study [18] also observed fine-scale genetic structure among populations speaking languages that belong to the same linguistic family, such as between western African and East African Bantu Niger-Kordofanian-speakers, among other examples [18]. East Africa was characterized by high levels of population differentiation, likely resulting from the historical migration of linguistically distinct populations into this geographic region and the long-term presence of indigenous East African Khoesan-speakers, namely the Hadza and Sandawe [18]. A pattern of strong genetic differentiation was also observed in East Africa based on mtDNA data [47]. Additionally, the above genome-wide analysis [18]in geographically diverse Africans showed that Central African Pygmies share common ancestry with several southern and eastern African Khoesan-speaking populations, who speak a language with click consonants, suggesting that these contemporary hunter-gatherers may have descended from a proto-Khoesan-Pygmy population of hunter-gatherers that diverged more than 35 kya [18]. This finding also raises the intriguing possibility that the original language spoken by African Pygmies, who are known to have lost their indigenous language, may have contained click consonants [18].
Several studies have also identified genetic structure within Central Africa, particularly between Pygmy and non-pygmy populations [18, 31, 48–51], as well as subtle substructure among Pygmies [18, 48, 50, 51]. Specifically, data have shown that the inferred ancestors of modern Pygmy hunter-gatherers and Bantu-speaking agriculturalists could have diverged as long as 70 kya [49], and that ancestral western and eastern Pygmy populations separated more than 18 kya [48] with subsequent genetic differentiation among the western Pygmies within the past 2,800 years [50]. The subtle structure among western Pygmies may be due to recent geographic isolation, genetic drift, and differential levels of admixture between Pygmies and neighboring Bantu-speaking populations [18, 49, 50, 52–54]. Overall, these patterns of genetic and phenotypic variation suggest that African populations have maintained a large and subdivided population structure throughout much of their evolutionary history. This population subdivision may have been facilitated by a number of factors, including ethnicity, culture, language, geography, as well as past fluctuations in geology and climate which may have affected population growth, contraction, fragmentation, and gene-flow in Africa [18, 55].
Migration and Admixture
Both short- and long- range migration events, and subsequent admixture between migrating and indigenous populations have also influenced the current genetic landscape of sub-Saharan Africa. One of the most significant migration events in recent African history was the geographic expansion of the Bantu Niger-Kordofanian-speakers from Nigeria and Cameroon first into the rainforests of equatorial Africa, and then into eastern and southern Africa within the past 5,000 years [18, 56] (Figure 1). Indeed, the presence of Bantu Niger-Kordofanian ancestry in many African populations and the widespread distribution of Bantu-related languages are signatures of the historical migration of Bantu-speakers across Africa and their subsequent admixture with other indigenous populations [18, 57]. Genetic evidence also indicates that independent waves of migration of western African and East African Bantu-speakers into southern Africa occurred, in agreement with previous linguistic and archaeological studies [18, 58]. Although the reasons for the radiation of Bantu-speakers across sub-Saharan Africa are not entirely known, it has been suggested that a shift from a humid to a drier climate around 5,000 years ago and the adoption of new crops suited to this drier environment may have contributed to the widespread movement of Bantu farmers throughout Africa [58, 59].
Genetic signatures of both historic and prehistoric migration events are also observed in other regions of Africa [18, 42, 57, 60–62]. For example, an analysis of microsatellite, INDEL and SNP polymorphisms in the nuclear genome showed that populations from central/southern Sudan, such as the Nuer and Dinka, have the highest proportion of Nilo-Saharan ancestry, with decreasing frequency observed in populations from northern Kenya to northern Tanzania in East Africa. These data suggest a Sudanese origin of Nilo-Saharan-speaking populations, with subsequent migration(s) southeastward to East Africa [18]. In addition, Nilo-Saharan-speakers from Sudan, Tanzania, Kenya and Chad also clustered closely with Afroasiatic Chadic-speaking populations from the southern Lake Chad Basin in genetic structure analyses, suggesting that these Chadic-speakers of Nilo-Saharan ancestry likely migrated westward from a Sudanese homeland to Lake Chad and adopted an Afroasiatic language at some point in their history without significant genetic exchange [18]. This shift in language may have occurred through interactions with proto-Chadic Afroasiatic-speakers who migrated from the central Sahara to the Lake Chad Basin around 8 kya [6, 56, 58, 63]. These genetic data are in general agreement with archaeological and linguistic studies that advocate a common origin of Nilo-Saharan populations in eastern Sudan, with subsequent migration events northward to the eastern Sahara, westward to the Chad Basin, and southeastward into Kenya and Tanzania [6, 64].
The migration of Nilo-Saharan-speakers may have been associated with past changes in environmental conditions. For example, archeological data suggest that following a climatic shift from dry to more humid conditions in the early Holocene around 10.5 kya, several Nilo-Saharan-speaking populations expanded westward from the Middle Nile Basin to Lake Chad and southeastward to northern Kenya to exploit newly created aquatic food resources [6, 65]. However, a small number of Nilo-Saharan-speakers, collectively referred to as Northern Sudanians, migrated northward to the eastern Sahara where they engaged in cattle domestication [6, 65] (Figure 3). During the Mid-Holocene arid phase around 8,500-7,500 years ago, aquatic food collecting Nilo-Saharans continued their mode of subsistence mainly along permanent rivers, such as the middle Niger and the Nile, and remaining lakes such as Lake Chad. However, during the return of more humid conditions after 7,500 years ago, Northern Sudanic cattle raisers expanded westward across the Sahara [6, 56, 65] (Figure 3). A subset of Nilo-Saharans, namely Nilotic pastoralists, originating from eastern Sudan is also known to have migrated southeastward to Kenya and northern Tanzania within the past 3,000 years [64].
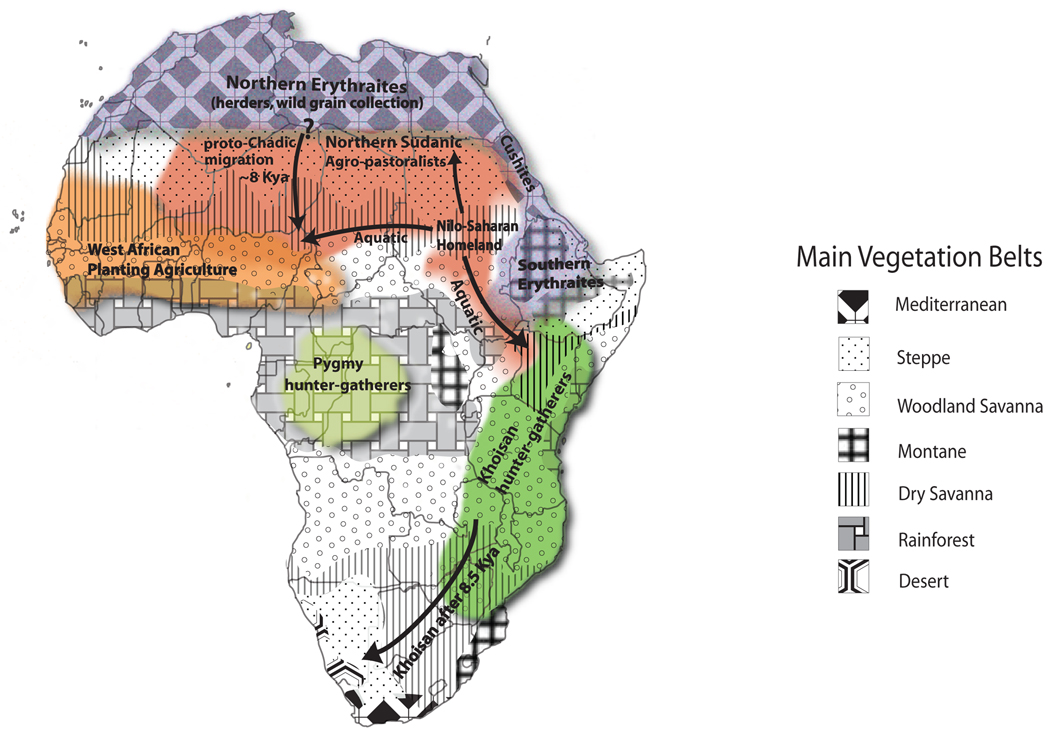
Figure 3. Map of African climate, vegetation and culture in the early Holocene (adapted from Maps 5 and 6 in reference [6]).
Following a period of dramatic increase in rainfall ~10.5 kya, the expansion of both rainforest vegetation in the Congo Basin and woodland savanna farther north and south of the rainforest belt, as well as the expansion of tropical steppe and grassland vegetation into the Sahara region occurred. New lakes also appeared, while old lakes grew in size along the southern edges of the Sahara [6]. Between 10.5 – 8.5 kya, several key demographic events occurred among culturally distinct ancestral African populations, including the migration of Nilo-Saharan-speakers from their homeland in Sudan northward to the dry tropical steppe vegetation of the eastern Sahara where they practiced agropastoralism [6]. Northern Erythraites (Afroasiatic-speakers) also lived mainly in the Mediterranean climate of the northern Sahara where they engaged in herding and wild grain collection[6]. The distributions of these cultural groups, among others depicted on this map, roughly correspond to the present-day distribution of major linguistic groups in Africa (Refer to Figure 1). The shift from dry to wetter climatic conditions in the early Holocene likely provided an opportunity for ancestral Africans to develop cultural traditions, such as agriculture and herding, in a more favorable environment leading to further technological developments in Africa [6].
Additionally, many Nilo-Saharan-speaking populations from East Africa have high levels of Afroasiatic Cushitic ancestry (likely of Ethiopian origin [64]), suggesting a long history of gene-flow between Nilo-Saharans and Cushites [18]. Archaeological studies have indicated a shared cultural practice of cattle herding between Nilo-Saharans and Cushites in northern Kenya which likely brought these linguistically distinct populations into repeated contact with one another over the past several thousand years [6, 56]. Cushitic agropastoralists are also thought to have expanded into southern Kenya and central northern Tanzania, engaging in cultural exchange and inter-marriage with southern and eastern Nilotic pastoralists [6] (Figure 1). This archaeological and genetic evidence of gene-flow between Nilo-Saharans and Cushites is consistent with other genetic data that showed the shared presence of an East African-specific mutation associated with lactose tolerance in these linguistically distinct populations [66].
The reverse migration of non-Africans into Africa was also shown to contribute to the gene-pool of modern African populations. For example, high levels of both Middle Eastern/European and eastern African Cushitic ancestry were detected in the Saharan African Beja, indicative of possible gene-flow from non-African populations [18]. These genetic patterns correlate well with linguistic and archaeological data that suggest that modern-day Beja pastoralists descended from northern Cushitic-speakers who migrated from Ethiopia to the Red Sea coast of Sudan [56]. Furthermore, the Beja have also had more intensive contact with the Middle East through commercial trade across the Red Sea as early as the 9th century A.D. and with nomadic camel herders of Arab Bedouin origin who settled in Sudan beginning in the 14th century A.D. [6] (Figure 1). These studies demonstrate that migration in Africa occurred at different points in time and over a range of geographic areas, resulting in complex patterns of genetic variation.
Dietary Adaptations
Natural selection, the process by which favorable heritable traits become more common in successive generations and unfavorable heritable traits become less common [67], operates to either increase or decrease the frequency of mutations that have an effect on an individual’s fitness. Selectively advantageous mutations associated with diet have evolved in populations which likely enabled ancestral humans to adapt to their environment, including changes in cultural practices. This section will focus on a few case studies of dietary adaptation primarily in African populations (for a detailed review of additional genetic adaptations in Africa, see reference [1]).
Lactase persistence, the ability to digest fresh milk and other dairy products into adulthood, varies in frequency in different human populations. However, the lactase persistence trait is common in pastoralist and dairying populations, such as northern Europeans, and certain African and Arabic nomadic groups [68]. Previous studies identified the T allele at a C/T SNP located 13910 bp upstream of the lactase gene (LCT) as the likely causal mutation of lactase persistence in Europeans and a few West African pastoralist populations, such as the Fulani. However, this mutation was not found to be a strong predictor of lactase persistence in other African populations that practice pastoralism [69, 70]. A recent study [66] of 43 populations from Tanzania, Kenya, and the Sudan identified three polymorphisms (G/C-14010, common in Tanzania and Kenya, T/G-13915 and C/G-13907, common in northern Sudan and Kenya) located ~14 kb upstream of LCT that are significantly associated with lactase persistence in East African populations. Further analysis revealed evidence consistent with a recent ongoing selective sweep over the past 3,000–7,000 years [66]. Recent resequencing studies have also found additional variants within this genomic region upstream of LCT in other African populations [71, 72]. However, the functional impact of these polymorphisms on lactose tolerance has not been firmly established.
Genetic data have indicated that past migration events may have resulted in the presence of shared genetic variation among pastoralist populations in different regions of Africa. For example, a survey of SNPs associated with lactase persistence in Africa found the C-14010 mutation at low frequency in several southern African Bantu-speaking pastoralists [73]This study argued for the spread of this mutation into southern Africa by migrating Khoe pastoralists who may have admixed with migrating Nilotic or Cushitic herders from East Africa, with subsequent admixture occurring between Khoe and Bantu-speakers in southern Angola around 2 kya [73]. Similar results have been observed in other East African and southern African populations (A. Ranciaro & S.A. Tishkoff, unpublished data, [72]). Recent Y-chromosome evidence has also suggested that eastern African Nilotic pastoralists migrated to southern-central Africa and admixed with local populations within the last few thousand years, supporting the hypothesis of an East African origin of southern African pastoralism [60]. Overall, these findings are a striking example of convergent evolution, local adaptation due to strong selective pressure resulting from shared cultural practices in Europeans and Africans, and gene-culture co-evolution [66, 74].
Similarly, starch consumption is variable among populations with different subsistence patterns, but is more notably elevated in agricultural and certain hunter-gatherer groups [75]. It has been suggested that a genetic adaptation arose in distinct populations in response to an increase in dietary starch intake. Perry and colleagues [75] examined the number of copies of the salivary amylase gene (AMY1), which plays a role in starch hydrolysis, in distinct populations with contrasting levels of starch consumption, including several African populations. This study observed a greater number of AMY1 gene copies in populations with a high starch diet (consisting of European Americans, Japanese, Hadza (African)) compared to populations with a low-starch diet (consisting of Biaka and Mbuti Pygmies (African), Datog (African), and Yakut (Asian)), arguing that dietary differences rather than ancestry influenced gene copy number [75]. Perry and coworkers [75] suggest that positive selection has been the dominant force affecting the divergence in gene copy number among populations at the AMY1 locus. However, targeted resequencing and long- range haplotype analyses of the AMY1 gene in additional populations with distinct subsistence patterns, including culturally diverse Africans, might be informative for confirming signatures of recent selection, providing further evidence for adaptive evolution at this locus.
Another important dietary adaptation is bitter taste perception which may have evolved in humans to prevent the ingestion of plant toxins. The ability to taste the bitter synthetic compound phenylthiocarbamide (PTC) is a highly variable trait in humans, and is correlated with both the ability to taste naturally bitter substances in food and food preference [76, 77]. A large proportion of the phenotypic variance in PTC sensitivity has been attributed to genetic variability at TAS2R38, a bitter taste receptor gene, located on chromosome 7 [78]. Specifically, three amino acid substitutions at TAS2R38 that form two common amino acid haplotypes have been shown to influence PTC sensitivity: a dominant taster haplotype, PAV and a non-taster haplotype, AVI [1]. A genetic analysis of TAS2R38 in geographically diverse populations, including a small number of Africans, uncovered evidence of balancing selection at this locus, such as an excess of intermediate-frequency variants and an ancient divergence between the major taster and nontaster haplotypes (estimated to be 1.5 million years ago) [79]. These patterns of diversity suggest that variants at this locus are selectively advantageous, likely playing a role in food choice [77, 79–81], and may represent long-term genetic adaptations in humans. More recently, genetic data have also shown that additional haplotypes associated with a broader range of PTC sensitivity are present in culturally and linguistically distinct African populations (M.C. Campbell and S.A. Tishkoff, unpublished data), raising the possibility that a wider range of sensitivity to naturally bitter compounds among Africans might be advantageous in Africa. These studies provide further information regarding the evolution of genetic variation associated with bitter taste perception and the role that genetic/phenotypic variability may play in dietary preference in human populations.
Future Directions
To date, only a fraction of the many ethno-linguistic groups in Africa has been extensively studied for genome-wide variation. Analyses of genetic diversity in more geographically and ethnically distinct African populations will be critical for testing models of modern human origins and dispersal out of Africa, as well as for inferring African demographic history. With the recent development of both “next generation” sequencing technology and methods for targeted sequencing, together with their rapidly decreasing costs, it may be feasible to resequence large portions of the genome, and conceivably the entire genome[82], in diverse African populations to more accurately test evolutionary models and to infer demographic history. The integration of archaeological, linguistic, paleoclimatic, and geographical information with these genetic data will be particularly useful in this reconstruction, providing a more unified account of historic and prehistoric events in Africa. Indeed, the “1000 genomes” project, which aims to discover novel variants through targeted and whole-genome resequencing in a subset of the extended HapMap populations, represents a first step in identifying novel genome-wide variation in more diverse Africans. However, given the extensive population structure present in Africa, additional linguistically and culturally distinct Africans will likely be needed to more fully characterize levels and patterns of variation which can then be used to infer population history.
Finally, African populations have been severely underrepresented in studies of genetic adaptation. Given that Africans possess high levels of genetic and phenotypic diversity, and live in distinct environments, it is likely that ethnically and geographically diverse populations in Africa have undergone local adaptation. To gain a better understanding of genetic and phenotypic adaptations in human populations, it is imperative to include a wider range of ethnically diverse African populations living in distinct environments in genetic studies. Moreover, genome-wide resequencing in a number of African populations is likely to lead to the identification of novel population-specific variation associated with variable traits. The development of statistical and computational methods for detecting selection across the genome that distinguish the effects of selection and demography will also be critical for identifying the genetic basis of adaptation in Africa. Overall, studies of adaptation combined with information on African demographic history will provide a more robust and accurate view of the processes that have shaped the genomes of African populations.
Acknowledgments
We thank A. Ranciaro, J. Hirbo, F. Gomez, and S. Soi for critical review of the manuscript. We are especially grateful to C. Ehret for his careful review of the manuscript and figures, and for his suggestions regarding African population history. The authors are funded by the U.S. National Science Foundation (NSF) grant BCS-0552486, U.S. NSF grant BCS-0827436, U.S. National Institutes of Health (NIH) grant R01GM076637, U.S. NIH Director's Pioneer Award Program DP1-OD-006445, and a David and Lucile Packard Career Award to S.A.T.
Contributor Information
Michael C. Campbell, Email: mcam@mail.med.upenn.edu.
Sarah A. Tishkoff, Email: tishkoff@mail.med.upenn.edu.
References
- 1.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed FA, Tishkoff SA. African human diversity, origins and migrations. Curr Opin Genet Dev. 2006;16:597–605. doi: 10.1016/j.gde.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Scholz CA, Johnson TC, Cohen AS, King JW, Peck JA, Overpeck JT, Talbot MR, Brown ET, Kalindekafe L, Amoako PY, et al. East African megadroughts between 135 and 75 thousand years ago and bearing on early-modern human origins. Proc Natl Acad Sci U S A. 2007;104:16416–16421. doi: 10.1073/pnas.0703874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trauth MH, Maslin MA, Deino AL, Strecker MR, Bergner AG, Duhnforth M. High- and low-latitude forcing of Plio-Pleistocene East African climate and human evolution. J Hum Evol. 2007;53:475–486. doi: 10.1016/j.jhevol.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Manica A, Amos W, Balloux F, Hanihara T. The effect of ancient population bottlenecks on human phenotypic variation. Nature. 2007;448:346–348. doi: 10.1038/nature05951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehret C. The Civilizations of Africa: A History to 1800. Charlottesville: University of Virginia Press; 2002. [Google Scholar]
- 7.McDougall I BF, Fleagle JG. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433:733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 8.Stringer C. Human evolution: Out of Ethiopia. Nature. 2003;423:692–693. 695. doi: 10.1038/423692a. [DOI] [PubMed] [Google Scholar]
- 9.White T, Asfaw B, DeGusta D, Gilbert H, Richards G, Suwa G, Howell F. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- 10.Tattersall I. Becoming Modern Homo sapiens. Evolution: Education and Outreach. 2009 Epub Ahead of print. [Google Scholar]
- 11.Brauer G. The Origin of Modern Anatomy: By Speciation or Intraspecific Evolution. Evolutionary Anthropology. 2008;17:22–37. [Google Scholar]
- 12.Niewoehner WA. Behavioral inferences from the Skhul/Qafzeh early modern human hand remains. Proc Natl Acad Sci U S A. 2001;98:2979–2984. doi: 10.1073/pnas.041588898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rightmire GP. Out of Africa: Modern Human Origins Special Feature: Middle and later Pleistocene hominins in Africa and Southwest Asia. Proc Natl Acad Sci U S A. 2009;106:16046–16050. doi: 10.1073/pnas.0903930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberman DE, McBratney BM, Krovitz G. The evolution and development of cranial form in Homosapiens. Proc Natl Acad Sci U S A. 2002;99:1134–1139. doi: 10.1073/pnas.022440799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mellars P. Why did modern human populations disperse from Africa ca. 60,000 years ago? A new model. Proc Natl Acad Sci U S A. 2006;103:9381–9386. doi: 10.1073/pnas.0510792103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanyon R, Sazzini M, Luiselli D. Timing the first human migration into eastern Asia. J Biol. 2009;8:18. doi: 10.1186/jbiol115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet. 1999;23:437–441. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- 18.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ, Kidd JR, Cheung K, Bonne-Tamir B, Santachiara-Benerecetti AS, Moral P, Krings M. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am J Hum Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrigan D, Kingan SB, Pilkington MM, Wilder JA, Cox MP, Soodyall H, Strassmann B, Destro-Bisol G, de Knijff P, Novelletto A, et al. Inferring human population sizes, divergence times and rates of gene flow from mitochondrial, X and Y chromosome resequencing data. Genetics. 2007;177:2195–2207. doi: 10.1534/genetics.107.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustamante CD, Ramachandran S. Evaluating signatures of sex-specific processes in the human genome. Nat Genet. 2009;41:8–10. doi: 10.1038/ng0109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammer MF, Mendez FL, Cox MP, Woerner AE, Wall JD. Sex-biased evolutionary forces shape genomic patterns of human diversity. Plos Genet. 2008;4 doi: 10.1371/journal.pgen.1000202. e1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat Genet. 2009;41:66–70. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- 26.Guthery SL, Salisbury BA, Pungliya MS, Stephens JC, Bamshad M. The structure of common genetic variation in United States populations. Am J Hum Genet. 2007;81:1221–1231. doi: 10.1086/522239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 28.Hughes AL, Welch R, Puri V, Matthews C, Haque K, Chanock SJ, Yeager M. Genome-wide SNP typing reveals signatures of population history. Genomics. 2008;92:1–8. doi: 10.1016/j.ygeno.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 30.Wall JD, Cox MP, Mendez FL, Woerner A, Severson T, Hammer MF. A novel DNA sequence database for analyzing human demographic history. Genome Res. 2008;18:1354–1361. doi: 10.1101/gr.075630.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long JC, Li J, Healy ME. Human DNA sequences: more variation and less race. Am J Phys Anthropol. 2009;139:23–34. doi: 10.1002/ajpa.21011. [DOI] [PubMed] [Google Scholar]
- 32.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 33.Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci U S A. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Z, Yu N, Fu YX, Li WH. Nucleotide variation and haplotype diversity in a 10-kb noncoding region in three continental human populations. Genetics. 2006;174:399–409. doi: 10.1534/genetics.106.060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox MP, Woerner AE, Wall JD, Hammer MF. Intergenic DNA sequences from the human X chromosome reveal high rates of global gene flow. BMC Genet. 2008;9:76. doi: 10.1186/1471-2156-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu X, Xiao H. Progress in the detection of human genome structural variations. Sci China C Life Sci. 2009;52:560–567. doi: 10.1007/s11427-009-0078-4. [DOI] [PubMed] [Google Scholar]
- 38.Consortium TIH. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 40.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen WW, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behar DM, Villems R, Soodyall H, Blue-Smith J, Pereira L, Metspalu E, Scozzari R, Makkan H, Tzur S, Comas D, et al. The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol. 2007;24:757–768. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- 43.Shimada MK, Panchapakesan K, Tishkoff SA, Nato AQ, Jr, Hey J. Divergent haplotypes and human history as revealed in a worldwide survey of X-linked DNA sequence variation. Mol Biol Evol. 2007;24:687–698. doi: 10.1093/molbev/msl196. [DOI] [PubMed] [Google Scholar]
- 44.Yotova V, Lefebvre JF, Kohany O, Jurka J, Michalski R, Modiano D, Utermann G, Williams SM, Labuda D. Tracing genetic history of modern humans using X-chromosome lineages. Hum Genet. 2007;122:431–443. doi: 10.1007/s00439-007-0413-4. [DOI] [PubMed] [Google Scholar]
- 45.Gunz P, Bookstein FL, Mitteroecker P, Stadlmayr A, Seidler H, Weber GW. Early modern human diversity suggests subdivided population structure and a complex out-of-Africa scenario. Proc Natl Acad Sci U S A. 2009;106:6094–6098. doi: 10.1073/pnas.0808160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 47.Poloni E, Naciri Y, Bucho R, Niba R, Kervaire B, Excoffier L, Langaney A, Sanchez-Mazas A. Genetic Evidence for Complexity in Ethnic Differentiation and History in East Africa. Annals of Human Genetics. 2009;73:582–600. doi: 10.1111/j.1469-1809.2009.00541.x. [DOI] [PubMed] [Google Scholar]
- 48.Patin E, Laval G, Barreiro LB, Salas A, Semino O, Santachiara-Benerecetti S, Kidd KK, Kidd JR, Van der Veen L, Hombert JM, et al. Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set. Plos Genet. 2009;5 doi: 10.1371/journal.pgen.1000448. e1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintana-Murci L, Quach H, Harmant C, Luca F, Massonnet B, Patin E, Sica L, Mouguiama-Daouda P, Comas D, Tzur S, et al. Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers. Proc Natl Acad Sci U S A. 2008;105:1596–1601. doi: 10.1073/pnas.0711467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verdu P, Austerlitz F, Estoup A, Vitalis R, Georges M, Thery S, Froment A, Le Bomin S, Gessain A, Hombert JM, et al. Origins and genetic diversity of pygmy hunter-gatherers from Western Central Africa. Curr Biol. 2009;19:312–318. doi: 10.1016/j.cub.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 51.Batini C, Coia V, Battaggia C, Rocha J, Pilkington MM, Spedini G, Comas D, Destro-Bisol G, Calafell F. Phylogeography of the human mitochondrial L1c haplogroup: genetic signatures of the prehistory of Central Africa. Mol Phylogenet Evol. 2007;43:635–644. doi: 10.1016/j.ympev.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Coia V, Caglia A, Arredi B, Donati F, Santos FR, Pandya A, Taglioli L, Paoli G, Pascali V, Spedini G, et al. Binary and microsatellite polymorphisms of the Y-chromosome in the Mbenzele pygmies from the Central African Republic. Am J Hum Biol. 2004;16:57–67. doi: 10.1002/ajhb.10236. [DOI] [PubMed] [Google Scholar]
- 53.Destro-Bisol G, Coia V, Boschi I, Verginelli F, Caglia A, Pascali V, Spedini G, Calafell F. The analysis of variation of mtDNA hypervariable region 1 suggests that Eastern and Western Pygmies diverged before the Bantu expansion. Am Nat. 2004;163:212–226. doi: 10.1086/381405. [DOI] [PubMed] [Google Scholar]
- 54.Destro-Bisol G, Donati F, Coia V, Boschi I, Verginelli F, Caglia A, Tofanelli S, Spedini G, Capelli C. Variation of female and male lineages in sub-Saharan populations: the importance of sociocultural factors. Mol Biol Evol. 2004;21:1673–1682. doi: 10.1093/molbev/msh186. [DOI] [PubMed] [Google Scholar]
- 55.Premo LS, Hublin JJ. Culture, population structure, and low genetic diversity in Pleistocene hominins. P Natl Acad Sci USA. 2009;106:33–37. doi: 10.1073/pnas.0809194105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blench R. Archaeology, Language, and the African Past. New York: Rowman & Littlefield Publishers, Inc; 2006. [Google Scholar]
- 57.Berniell-Lee G, Calafell F, Bosch E, Heyer E, Sica L, Mouguiama-Daouda P, van der Veen L, Hombert JM, Quintana-Murci L, Comas D. Genetic and demographic implications of the Bantu expansion: insights from human paternal lineages. Mol Biol Evol. 2009;26:1581–1589. doi: 10.1093/molbev/msp069. [DOI] [PubMed] [Google Scholar]
- 58.Ehret C. An African Classical Age: Eastern and Southern Africa in World History, 1000 B.C. to A.D. 400. Charlottesville: University Press of Virginia; 1998. [Google Scholar]
- 59.Marret F, Maley J, Scourse J. Climatic Instability in West Equatorial Africa during the Mid- and Late Holocene Quaternary International. 2006;150:71–81. [Google Scholar]
- 60.Henn BM, Gignoux C, Lin AA, Oefner PJ, Shen P, Scozzari R, Cruciani F, Tishkoff SA, Mountain JL, Underhill PA. Y-chromosomal evidence of a pastoralist migration through Tanzania to southern Africa. Proc Natl Acad Sci U S A. 2008;105:10693–10698. doi: 10.1073/pnas.0801184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tishkoff SA, Gonder MK, Henn BM, Mortensen H, Knight A, Gignoux C, Fernandopulle N, Lema G, Nyambo TB, Ramakrishnan U, et al. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol Biol Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 62.Wood ET, Stover DA, Ehret C, Destro-Bisol G, Spedini G, McLeod H, Louie L, Bamshad M, Strassmann BI, Soodyall H, et al. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur J Hum Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- 63.Cerny V, Fernandes V, Costa MD, Hajek M, Mulligan CJ, Pereira L. Migration of Chadic speaking pastoralists within Africa based on population structure of Chad Basin and phylogeography of mitochondrial L3f haplogroup. BMC Evol Biol. 2009;9:63. doi: 10.1186/1471-2148-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ehret C. Population Movement and Culture Contact in the Southern Sudan, c. 3000 B.C. to A.D. 1000. Nairobi: British Institute in Eastern Africa; 1982. [Google Scholar]
- 65.Ehret C. Linguistic Stratigraphies and Holocene History in Northeastern Africa. Posnan: Posnan Archaeological Museum; 2006. [Google Scholar]
- 66.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hancock A, DiRienzo A. Detecting the Genetic Signature of Natural Selection in Human Populations: Models, Methods, and Data. Annual Review of Anthropology. 2008;37:197–217. doi: 10.1146/annurev.anthro.37.081407.085141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hollox EJ, Poulter M, Zvarik M, Ferak V, Krause A, Jenkins T, Saha N, Kozlov AI, Swallow DM. Lactase haplotype diversity in the Old World. Am J Hum Genet. 2001;68:160–172. doi: 10.1086/316924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 70.Poulter M, Hollox E, Harvey CB, Mulcare C, Peuhkuri K, Kajander K, Sarner M, Korpela R, Swallow DM. The causal element for the lactase persistence/non-persistence polymorphism is located in a 1 Mb region of linkage disequilibrium in Europeans. Ann Hum Genet. 2003;67:298–311. doi: 10.1046/j.1469-1809.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 71.Ingram CJ, Raga TO, Tarekegn A, Browning SL, Elamin MF, Bekele E, Thomas MG, Weale ME, Bradman N, Swallow DM. Multiple Rare Variants as a Cause of a Common Phenotype: Several Different Lactase Persistence Associated Alleles in a Single Ethnic Group. J Mol Evola. 2009 doi: 10.1007/s00239-009-9301-y. [DOI] [PubMed] [Google Scholar]
- 72.Torniainen S, Parker MI, Holmberg V, Lahtela E, Dandara C, Jarvela I. Screening of variants for lactase persistence/non-persistence in populations from South Africa and Ghana. BMC Genet. 2009;10:31. doi: 10.1186/1471-2156-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coelho M, Sequeira F, Luiselli D, Beleza S, Rocha J. On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola. BMC Evol Biol. 2009;9:80. doi: 10.1186/1471-2148-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gerbault P, Moret C, Currat M, Sanchez-Mazas A. Impact of selection and demography on the diffusion of lactase persistence. PLoS One. 2009;4:e6369. doi: 10.1371/journal.pone.0006369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garcia-Bailo B, Toguri C, Eny KM, El-Sohemy A. Genetic variation in taste and its influence on food selection. Omics. 2009;13:69–80. doi: 10.1089/omi.2008.0031. [DOI] [PubMed] [Google Scholar]
- 77.Sandell MA, Breslin PA. Variability in a taste-receptor gene determines whether we taste toxins in food. Curr Biol. 2006;16:R792–R794. doi: 10.1016/j.cub.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 78.Drayna D. Human taste genetics. Annu Rev Genomics Hum Genet. 2005;6:217–235. doi: 10.1146/annurev.genom.6.080604.162340. [DOI] [PubMed] [Google Scholar]
- 79.Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim U, Drayna D. Genetics of Individual Differences in Bitter Taste Perception: Lessons from the PTC Gene. Clinical Genetics. 2004;67:275–280. doi: 10.1111/j.1399-0004.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 81.des Gachons CP, Beauchamp GK, Breslin PA. The genetics of bitterness and pungency detection and its impact on phytonutrient evaluation. Ann N Y Acad Sci. 2009;1170:140–144. doi: 10.1111/j.1749-6632.2009.04594.x. [DOI] [PubMed] [Google Scholar]
- 82.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]