Abstract
We extend PRIME, an intermediate-resolution protein model previously used in simulations of the aggregation of polyalanine and polyglutamine, to the description of the geometry and energetics of peptides containing all twenty amino acid residues. The 20 amino acid side chains are classified into 14 groups according to their hydrophobicity, polarity, size, charge and potential for side chain hydrogen bonding. The parameters for extended PRIME, called PRIME 20, include hydrogen-bonding energies, side-chain interaction range and energy, and excluded volume. The parameters are obtained by applying a perceptron- learning algorithm and a modified stochastic learning algorithm that optimizes the energy gap between 711 known native states from the PDB and decoy structures generated by gapless threading. The number of independent pair-interaction parameters is chosen to be small enough to be physically meaningful yet large enough to give reasonably accurate results in discriminating decoys from native structures. The most physically meaningful results are obtained with 19 energy parameters.
INTRODUCTION
The purpose of this paper is to describe efforts to extend the force field for PRIME, an implicit-solvent intermediate- resolution protein model developed in our group, to proteins other than polyalanine, polyglutamine and polyglycine. PRIME was introduced in 20011 to enable simulation of the aggregation of simple homoproteins, particularly polyalanine, into fibrillar structures. By reducing the protein representation to four spheres per amino acid, treating solvent implicitly and modeling geometric constraints, hydrogen bonding and hydrophobic interactions through a combination of hard-sphere and square-well interactions, we were able to simulate the spontaneous formation of a system of 96 KA14K peptides into ordered fibrillar structures in hours on a fast workstation.2 PRIME has also been used to investigate the fibrillization of polyglutamine.3 The high speeds were achieved by using discontinuous molecular dynamics, a very fast alternative to traditional molecular dynamics, which is applicable to discontinuous potentials such as the hard sphere and square well potential.4 In this paper we describe efforts to extend the simple model used to describe polyalanine to all 20 amino acids.
PRIME is one of many coarse grained potentials currently being used to provide biophysical insights into protein folding and aggregation.2–12. Such models are particularly useful for studying the behavior of large systems of proteins over long time scales. This is because the detail and realism that makes atomistic force fields like Amber13 and CHARMM14, so useful for the study of specific proteins, comes with a computational price tag that limits the length of the simulations and hence the system sizes and time scales that can be accessed. Although the protein models developed in the early years of protein folding simulation could be classified as either coarse grained models or atomistic models, the spectrum between the two has filled in over time. In recent years major efforts have been underway to develop protein models of intermediate resolution whose simplified geometry and energetics are nevertheless capable of providing information about specific proteins.7,9,15 We aim to add PRIME to this group because it offers several advantages compared to other protein models that are particularly suitable for studying protein aggregation. These are: (1) the decomposition of forces between residues into discontinuous potentials allows us to take advantage of the high speeds possible in discontinuous molecular dynamics, (2) the simple 4-sphere-per-residue representation offers a good compromise between atomistic and pearl necklace treatments of protein geometry, (3) the use of discontinuous potentials, as opposed to harmonic potentials, to enforce bonding and angular restrictions speeds up the simulations, and (4) the simple yet accurate treatment of hydrogen bonding by a directional square-well potential enhances the formation of secondary structures like alpha helices and beta sheets.
The long term goal of this research is to develop the capability to simulate the various stages of protein aggregation, from the formation of low molecular weight oligomers early in the process to the assembly of ordered aggregates (fibrils or amyloid) later in the process. Protein aggregation is associated with serious and eventually-fatal neurodegenerative diseases including Alzheimer’s and Parkinson’s.16,17 Recent studies indicate that it is the early oligomers (and not the fully formed fibrils) that are likely responsible for toxicity in these diseases.17–20 This observation has stimulated computational researchers to examine the dynamical processes associated with the formation of small oligomers.21–23 However even though these events occur early in the aggregation process on time scales that might be accessible the limited number of peptides that can be simulated simultaneously makes atomistic simulations less than ideal for this purpose. Studies of oligomerization based on a small number of peptides must contend with the problem that the free energy barriers associated with oligomeric changes depend on the number of peptides.24 Coarse-grained simulations, however, allow us to simulate large systems having many peptides for long times, giving us the chance to examine more than one stage of the aggregation process.
The strengths of the effective interaction potentials in coarse-grained protein folding models have traditionally been obtained by one of the following two methods. The first method is the statistical extraction of energy parameters in which the residue-residue contact energies are estimated by relating the frequency of residue-residue contacts in the Protein Data Bank to those in a reference state, taken to be the quasichemical (Bethe) description of a random mixture of amino acids.25–36 Since the landmark work of Miyazawa and Jernigan29, which yielded 210 energy parameters for a Cα–based protein representation, a number of refinements of this approach have been made including: improved calculation of the reference state probabilities,29,36 more detailed descriptions of protein geometry and distance dependent forces,31,37 incorporation of protein-solvent interactions,36, and better optimization procedures including iterative methods.30,37,38
The second method is the energy gap optimization method which, following Anfinsen’s argument that the native state free energy in given physiological conditions is a global minima, searches for energy parameters that make the energy of the native structure less than those of a large set of decoy structures. In this method, the thermodynamic stabilities of a large number of native structures are optimized subject to a large number of constraints or inequalities. The optimized solutions have been found by various methods including linear programming, a mathematical technique which maximizes linear equations under constraints, or the perceptron learning algorithm, a neural network approach that iteratively adjusts weighting factors until reaching an expected or optimized output, etc.39–49 Most of the early methods for determining interaction energies (including those used in the statistical extraction of energy parameters) were based on a contact energy approach in which the protein was represented as a chain of united atoms and the range of interactions between two united atoms was assumed to be independent of the amino acid type.29,39 Later, methods were introduced in which the energy parameters depended on the distance between the united atoms; these approaches showed better performance in discriminating native structure from decoys.44,49 Other improvements include the development of new and better ways to generate decoys using a Monte Carlo algorithm, better energy minimization techniques for decoys, high resolution decoys,38,46,50 to include structure environments,47,48 and even to calculate pair interactions between side-chain centroids and backbone-backbone hydrogen bonds separately.51 Although most of the estimated parameter sets were used to discriminate native structures among decoys sets or for fold recognition, a few have been used in on- or off-lattice simplified polymer-like models for protein folding.28,52
Unlike most of the papers describing energy parameter estimation which seek 210 parameters, one for every possible amino acid pair, we seek to establish a smaller set of parameters that would be sufficiently detailed for protein aggregation simulations and also makes good sense physically. By “making sense physically” we mean that they should reflect the relative strengths of the types of forces thought to be operating between particular pairs of residues, e.g. hydrophobic/ hydrophilic, positive/negative charge, hydrogen bonding, etc. This is a challenging task since having more independent parameters generally gives a better performance in discriminating native structures from decoys.46,50 Furthermore, it is known that even 210 contact energy parameters can become insufficient for discriminating real native structures as the number of test structures grows.41,50 However, having too many parameters can lead to situations in which the parameter space of interaction energies becomes too complex and meta-stable, causing the estimated parameters to depend on the initial values or technical methods for estimating parameters. An additional type of problem that can be encountered when there are large numbers of parameters is that the energy parameters fitted for example for a particular type of interaction, for example hydrophobic interactions between V-V, L-L, I-L, M-I, F-I, can fluctuate and show no consistent trends compared to their hydrophobic scale.
The idea of having a small group of energy parameters to represent pair interactions in simplified protein folding models is, of course, not new. The simplest possible classification is the two-group HP classification introduced originally by Chan and Dill53 in which a protein is classified as either hydrophobic or polar. Another well known grouping is the five-group classification by Wang and Wang,54 which contains five representative amino acids (I, A, G, E, K) and was used successfully to build the SH3 domain in a protein engineering experiment.55 However this five-group classification is too simple to be applied to our extended PRIME model since it does not classify the charged and polar amino acids and it does not have any information about side-chain hydrogen-bonds. Other possible groupings include the 7-letter alphabet of Maiorov and Crippen,39 the four-letter alphabet of Betancourt and Thirumalai (based on a hydrophobicity scale),56 the 9 classes of Buchete and Thirumalai,57 and the hierarchical 2 to 14 group classification of Thomas and Dill.30
In this paper, we present procedures to estimate the side-chain/side-chain energy parameters (well depths) and hydrogen bonding energy parameters between backbone NH and CO, between side chain and side chain, and between side chain and backbone NH and CO for use in an extended version of the PRIME model, which we will call PRIME20. We also use PDB information to determine side chain diameters, Cα - Cβ bond distances, and square well diameters for all possible interacting pairs of sites. Our hope here is that the problems associated with having a small number of energy parameters may be compensated for by having physically meaningful energy parameters and relatively realistic protein geometry, including unique diameters and bond lengths for each side chain. The energy gap optimization approach is used. The contact energies from the native states of 711 proteins in the protein data bank (PDB) are compared with those of nearly two million decoy structures generated by gapless threading techniques.58 The optimum set of energy parameters is calculated using the perceptron learning algorithm.59 We also suggest a new modified version of the perceptron learning technique that includes a stochastic component. Our approach differs from other knowledge-based potential studies in that we tried to reduce the number of independent pair-interaction parameters to be small enough that our pair interaction energies are physically meaningful and yet large enough to give us reasonably accurate results in discriminating decoys from native structures in the PDB database
Highlights of our results are the following. The 20 amino acid side chains are classified into 14 groups according to their hydrophobicity, polarity, size, charge and potential for side chain hydrogen bonding. The latter property is further classified based on the type and number of polar atoms present on the side chain. The most physically meaningful results are obtained with 19 energy parameters. Addition of the stochastic algorithm to the perceptron learning scheme stabilizes the learning process by preventing dependence of the final solution on the initial guesses for the parameters. This is illustrated for the case of a simpler 7 group classification containing 12 energy parameters. Finally we consider a smaller database containing 585 PDBs and find that the types of proteins included in the PDB database have a significant effect on the quality of the results.
MODEL AND METHOD
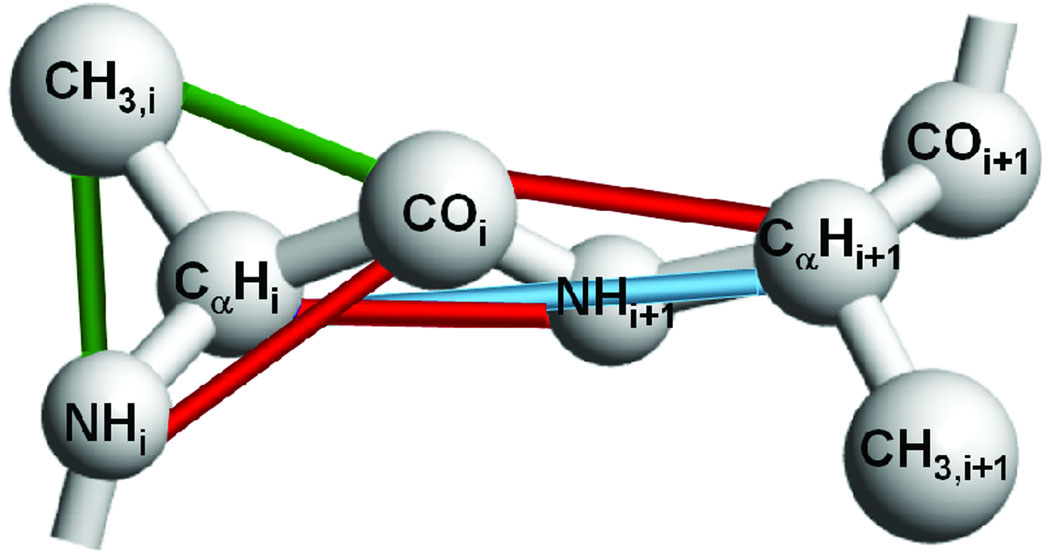
In the PRIME model, each amino acid residue is composed of a 3-sphere backbone comprised of united atoms NH, CαH, and CO, and a single-sphere side chain (CH3- for alanines).1–2 See Figure 1. Ideal backbone bond angles, Cα-Cα distances and residue L-isomerization are achieved by imposing pseudobonds. All forces are modeled by either hard-sphere or square-well potentials with realistic diameters. The solvent is modeled implicitly; its effect is factored into the energy function as a potential of mean force. Interactions between hydrophobic side chains are represented by a square-well potential. Hydrogen bonding between amide hydrogen atoms and carbonyl oxygen atoms is represented by a directionally-dependent square-well attraction between NH and CO united atoms.
Figure 1.
Geometry of PRIME. Covalent bonds are shown with gray lines connecting united atoms. At least one of each type of pseudobond is shown with a coloured line. Pseudobonds are used to maintain backbone bond angles, consecutive C distances, and residue L-isomerization. The united atoms are not shown full size for ease of viewing.
In order to obtain a realistic estimate of energy parameters it is necessary to have a good set of native structures and decoys. We downloaded 3693 PDBs having at most 25% sequence homology from PDB_select website.60 We eliminated membrane proteins and multi-domain proteins. We also eliminated PDBs with non-standard amino-acids, broken-chains, Cα-only representations, missing atoms, and ligands, small single helices and coiled peptides. Finally we set up the 711 PDBs listed in supplemental Table 1. Those PDBs were mapped onto the PRIME 4-sphere per residue geometry by assigning the centers of the PRIME backbone spheres, NH, Cα and CO, to lie at the same positions as the centers of the PDB backbone atoms N, Cα and C, and assigning the center of the PRIME side chain to lie at the same position as the center of mass of the PDB side-chain atoms.
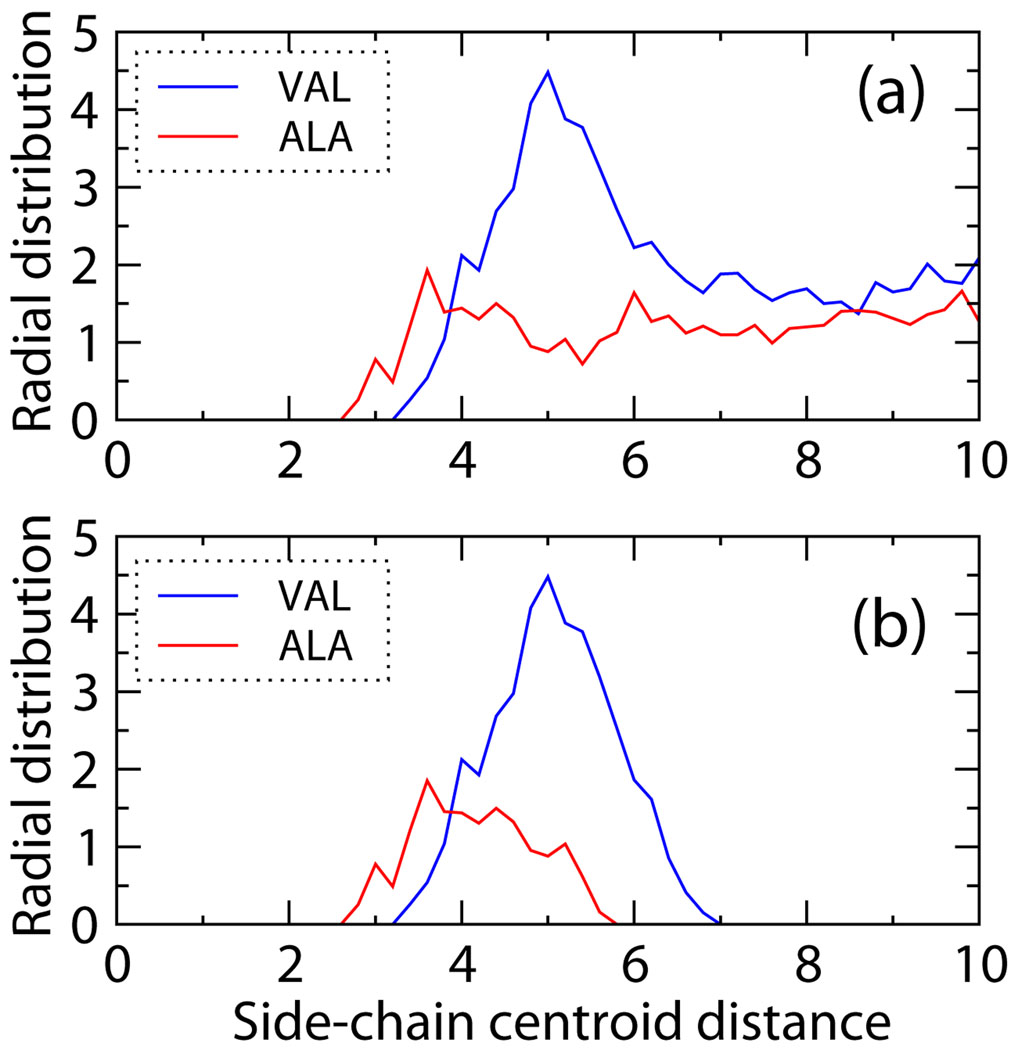
From those four united-atom model PDBs, radial distribution functions (RDF) for all pairs of united atoms were plotted to allow estimation of the side chain sphere and well diameters. Figure 2(a) is a sample plot for the radial distribution function between the Ala-Ala and Val-Val centroids. More precisely it is the population × 4πr2 versus the distance in Å between the side chain centroids estimated from 711 PDBs; bin sizes are set at 0.2. The sphere diameters are chosen to be the starting point of the distribution, i.e. the closest non-zero value of the RDF for each amino-acid pair. The well diameters are not as easy to choose since there is no obvious cutoff beyond which the RDF is again zero. Instead we plot an alternate distribution function, similar to the radial distribution function, which only counts pair interactions between united atoms when more than half of the distances between the heavy atoms on the first united atom and those on the second are less than 5.5Å. Sample distributions for Ala-Ala and Val-Val pair interactions with the 5.5Å criteria are shown in Figure 2(b). The well diameters are chosen to be the average values of the distribution plus 1.645σst, where σst is the standard deviation, which is a 90% criteria if the distribution follows the standard distribution. The sphere and well diameters are shown in supplemental Tables 2 and 3.
Figure 2.
(a) Radial distribution function for Ala-Ala and Val-Val pair interactions. (b) Pair distribution for Val-Val and Ala-Ala with heavy atom distance cutoff 5.5Å.
Approximately 1.6 to 1.9 million decoys (depending on the constraints for removing very compact decoys having more pair-interactions than the native structures), were generated using a gapless threading algorithm.58 Improved techniques for decoy generation using Monte Carlo or molecular dynamics simulation46,50 could have been applied but we do not need high resolution decoys since our goal is to find a small number of interaction parameters which can discriminate low-resolution decoys. Decoys are created by threading one native sequence onto the structures of other sequences. The Cα, N and C of the native sequence are placed in the same positions as the Cα, N and C on the other structures. The side-chain centroid positions of different amino acids on the threaded structures are adjusted so as to maintain the native distance associated with the original amino acids in the native sequence.
The generated decoys are, of course, not stable which means that their energies must be higher than native state energies. This gives 1.6 to 1.9 million inequalities which say that the difference between the energy of the kth decoy corresponding to native state j and the energy of native state j must be greater than a minimum energy gap parameter, Δ,
| (1) |
or equivalently
| (2) |
, where i is the type of interaction and ε (i) is the interaction parameter for the i-type pair interaction. Here, N(i,j,k) is the number of i-type pair interactions on the kth decoy structure corresponding to native state j, and N(i,j) is the number of i-type pair interactions on the jth native structure. N(i,j) and N(i,j,k) are calculated by using the sphere- and well-diameters described above.
The perceptron-learning algorithm was used to find the set of optimized contact energy parameters, ε(i) that satisfy the inequalities in eqn(2). The algorithm begins with an initial guess for ε(i). It then searches through all the proteins and their decoys for the (j, k) that yields the minimum energy difference between a decoy structure and its native structures (E(d − n)), i.e. the minimum scalar product (J⃗min · ε⃗). The search is subject to the following three constraints imposed in order to eliminate decoys (j, k) that are far more compact than native structures.
| (3) |
| (4) |
| (5) |
Here NHB is the number of hydrogen-bonds, NST is the number of strong interactions, and NWK is the number of weak interactions in the (j, k) decoy structures. These three constraints are needed because we do not have enough interaction parameters to satisfy the enormous number of inequalities. Once the (j, k) associated with the minimum scalar product is determined, the energy vector (ε⃗) is iterated by
| (6) |
, where t is the iteration step and δ is small value that controls convergence of the optimized solution. At the new iteration step, the (j,k) are searched at energy vector ε⃗(t + 1) until a new minimum energy difference and associated J⃗min is selected. This process is repeated until ε⃗ converges to a stable vector, which is the solution. Supplemental figure 1 illustrates the procedure. To satisfy all the inequalities, the angle between J⃗ and ε⃗ should be less than 90°. Therefore adding a small part of J⃗min to the next value of ε⃗ makes the angle between J⃗min and ε⃗ decrease, leading to improved likelihood of satisfying the inequalities after many iterations. If all the inequalities were satisfied, Δ would be zero or a small positive value and the learning process for this system would be called “learnable”. However given the small number of parameters used in this work it is impossible to satisfy the enormous number of inequalities. Instead we tried to find parameters to satisfy as many inequalities as possible, (i.e. to have Δ be the least possible negative value within the constraints of eqn (3) to (5).
Even within the framework of satisfying as many inequalities as possible we still face the barrier of the ruggedness of parameter space and the dependence on the initial values of the parameters. Problems associated with the dependence of our perceptron-learning-algorithm solution on the initial guesses for the energy parameters prompted us to introduce stochastic acceptance criteria into the learning algorithm iteration scheme.
In the perceptron learning plus stochastic process method, hereafter called the stochastic learning algorithm, new energy parameter vectors are updated either by the standard perceptron learning iteration scheme, Eqn (6), or by the addition of a random fluctuation, essentially a shuffling, to the old the energy vector based on a series of energy changes given by eqn(12) to (14) for a stochastic acceptance criteria. In order to set up the stochastic acceptance criteria, four energies are defined. The first energy is the average over all (j,k) of the normalized difference between the decoy and native energies divided by the product of the standard deviation (σ) for each protein and the normalization factor where Nres is the number of residues for each protein.
| (7) |
This corresponds to −Zscore. We divide by to even out the energy scale over the various protein sizes because the energies of the native structures are proportional to in our database. The second energy is the average over all (j,k) of the normalized difference between the decoy and native energies having negative values, again divided by the normalization factor .
| (8) |
The third energy is the minimum value over all possible (j,k) of the normalized decoy minus native energy and is given by
| (9) |
The fourth energy is a summation of weighted energy parameters given by
| (10) |
with
| (11) |
This fourth energy is a kind of statistical score function designed to account for and counterbalance any major differences in frequency of appearance of a particular type of interaction, i, in the native state and decoy structure databases. It has the effect of enhancing the strong attractive interactions and preventing repulsive interactions such as KK, EE from becoming too large.
The four types of energy changes associated with iterating between step t and t+1, ΔE1,ΔE2,ΔE3,ΔE4, can be evaluated as
| (12) |
| (13) |
| (14) |
| (15) |
The energy change ΔE that appears in our acceptance criteria for the stochastic update repeatedly cycles through the four energy changes given above; i. e.,
Finally our acceptance criterion for the iteration between step t and t+1 is given by the following. If ΔE ≤ 0 or a random number ≤ AR, where AR is the acceptance rate, then accept the original standard learning algorithm update iteration
| (16) |
Otherwise add random fluctuations to the parameter values.
| (17) |
where Ran is a random number.
The reason we add the random fluctuations to the energy vector in the second case is that without the addition of random fluctuations the energy would remain constant, and hence the next decoy selection would yield the same minimum scalar product as in the previous iteration. This would have rendered the process equivalent to the original perceptron learning algorithm. This somewhat complex modification of the perceptron learning algorithm yields unique and stable solutions for the enormous number of inequalities in our calculation without becoming trapped in local minima on the rugged parameter space landscape. During the learning process we need to normalize the parameters as the method proceeds; if we do not that, some of the parameters would grow to be very big and others will become zero. To prevent this we apply a normalization factor by resetting ε (i) at each iteration to be ε(i) = ε(i)/|ε(1)|.
RESULTS- Extended parameter set that accounts for side chain hydrogen-bonding
We have grouped the twenty amino acids into fourteen groups according to their physico-chemical characteristics. Our reasoning is the following. We start by noting that sixteen of the amino acids can be classified into groups based on whether or not they are hydrophobic {LVIMFYW}, negatively charged{ED}, positively charged {KR}, or polar{STNQH}. The remaining four amino acids, ALA, CYS, PRO, and GLY are fairly unique and for this reason are each classified into their own group. Thus the eight classifications are {LVIMFYW} {A} {C} {ED} {KR} {P} {STNQH} {G}. We further divide the hydrophobic residues into two groups, small hydrophobic residues {LVIM} and large hydrophobic residues {FYW}; the aromatic side chains in the latter group tend to enhance the hydrophobic effect. Thus so far we have identified nine groups. Next we break out those amino acids which are capable of side chain hydrogen bonding due to the presence of nitrogen, oxygen, or sulfur on their side chains. Thirteen of the amino acids have this capability (M,Y,W,C,D,E,K,R,T,S,N,Q,H); these can be classified based on the nature of their polar atoms. M is singled out from LVI due to its polar sulfur. Y with a oxygen and W with two nitrogens are distinguished from the other aromatic residues, and assigned to their own group due to their different polar atoms. S and T are grouped together because they both have one polar oxygen in their side chain. N and Q are distinguished from S, T and H since they have two polar atoms (nitrogen and oxygen). Finally H is also singled out due to its two nitrogens. From the above consideration, there are 14 amino acid groups: {LVI} {M} {F} {Y} {W} {A} {C} {ED} {KR} {P} {ST} {NQ} {H} {G}.
Given the 14 representative amino-acids, 91 pair interactions are possible, assuming that G does not interact. The number of pair interactions can be reduced by combining similar interaction types into a single group as is shown in Table 1. The top row and first column of Table 1 show the 13 groups (G is omitted).(The first column also lists the NH and CO backbone united atoms.) Each group is named according to its first amino acid so for example the group LVI is written in the table as L{VI} but in describing the interaction between this group and another group we will only indicate the first amino acid. So for example if we talk about the interaction “LE” we are referring to all possible interactions between the groups {LVI} and {ED}, i.e. LE, LD, VE, VD, IE, and ID. The second row indicates the type of polar atom, S, SH, O, OH or NH, on the side chain for each group. The third row indicates whether the side chain can act as an H- bond donor (D), or acceptor (A), or both (D&A). The numbers in the matrix indicate the interaction type—there are 23 types; these are described in Table 2. Examples that illustrate why certain pair interactions have been combined into the same group are the following. The interactions LL, LM, MM are assumed to have the same interaction strength since they are relatively small amino acids and they interact solely by hydrophobic interactions. The interactions YK, YQ, YH, WQ, WH are assumed to have the same interaction strength since they are between large aromatic hydrophobic and large polar amino acids with the possibility of hydrogen-bonding. Moreover, interactions involving hydrogen-bonding between polar atoms on side-chains (M,Y,W,C,E,K,S,Q,H) and NH, CO atoms on backbones are assumed to have the same interaction strength. In these cases the NH on the backbone can form hydrogen bonds with acceptors on the side chains of M, Y, C, E, S,Q, H, and the CO on the backbone can interact with donors on the side chains of Y,W,C,K,S,Q,H. Although the Pro-Pro interaction is likely different from the Pro-Polar interaction, it is difficult to estimate because the number of Pro-Pro interactions in the 711 native PDBs is very small. The absence of data to “learn” from can cause abnormal runaway behaviors in the learning process where the applicable energy parameter increases without bound upon parameter rescaling. Since both the Pro-Pro interaction and Pro-Polar interactions do not include any hydrophobic, hydrogen-bonding or charged interactions we have grouped them together for expediency.
Table 1.
Pair interaction parameters between the 14 groups of amino acids. The first row and first column show the 13 groups (glycine is assumed non-interacting). The first column also lists the NH and CO backbone united atoms. The second row shows the types of polar atom on the amino acid side chains. The third row indicates whether or not the polar atom in the second row is a donor (D) or acceptor (A) or both donor and acceptor (D & A). The indices (numbers 1–23) represent the type of interaction for 23 parameters. Also shown for each interacting pair is the possible hydrogen-bonding arrangement NHO, NHN, OHO, SHN, SHO.
| L[VI] | M | F | Y | W | A | C | E[D] | K[R] | P | S[T] | Q[N] | H | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polar Atom |
S | OH | NH | SH | O | NH | OH | NH O |
NH N |
||||
| DorA | A | D&A | D | D&A | A | D | D&A | D&A | D&A | ||||
| L[VI] | 2 | 2 | 3 | 3 | 3 | 12 | 13 | 15 | 16 | 15 | 15 | 16 | 16 |
| M | 2 | 3 | 4 SHO |
4 SHN |
12 | 13 | 15 | 17 S HN |
15 | 17 SHO |
17 S HN |
17 S HN |
|
| F | 5 | 5 | 5 | 12 | 13 | 15 | 16 | 15 | 15 | 16 | 16 | ||
| Y | 6 OHO |
6 OHN |
12 | 14 OHS |
18 OHO |
19 OHN |
15 | 18 OHO |
19 OHN OHO |
19 OHN |
|||
| W | 5 | 12 | 14 NHS |
18 NHO |
16 | 15 | 18 NHO |
19 NHN NHO |
19 NHN |
||||
| A | 7 | 13 | 20 | 20 | 20 | 20 | 20 | 20 | |||||
| C | 8 | 17 SH O |
17 SHN |
15 | 17 SHO |
17 SHN SHO |
17 SHN |
||||||
| E[D] | 9 | 10 | 21 | 18 OHO |
18 OHN |
18 OHN |
|||||||
| K[R] | 11 | 21 | 18 NHO |
18 NHN NHO |
18 NHN |
||||||||
| P | 21 | 21 | 21 | 21 | |||||||||
| S[T] | 18 OHO |
18 OHN OHO |
18 OHN |
||||||||||
| Q[N] | 22 NHO |
22 NHN OHN |
|||||||||||
| H | 22 NHN |
||||||||||||
| NH | 23 | 23 | 23 | 23 | 23 | 23 | 23 | ||||||
| CO | 23 | 23 | 23 | 23 | 23 | 23 | 23 |
Table 2.
Descriptions for 23(19) interaction parameters based on whether they have hydrophobic or charged interactions, the side chain size, and the possibility of hydrogen-bonding. Hydrogen bonding with sulfur(S) is considered separately because it is usually weaker than the hydrogen bonding via NH and O. The 22nd parameter (QQ,QH,HH) is separated out since those amino acids have two polar atoms in their side-chains.
| Parameter names and indices | Descriptions for Hydrophobic (HP) or Charge, Size or Types of Amino Acids (A.A.)Hydrogen-Bond (HB) types |
||
|---|---|---|---|
| Number of Parameters | 23 | 19 | |
| NHCO | 1 | 1 | Backbone HB |
| LL,LM,MM | 2 | 2 | Strong HP, Small-Small A.A. |
| L{FYW},MF | 3 | 3 | Strong HP, Small-Large A.A. HB with S |
| M{YW} | 4 | 4 | Strong HP, Small-Large A.A. |
| F{FYW},WW | 5 | 5 | Strong HP, Large-Large A.A. |
| Y{Y,W} | 6 | 6 | Strong HP, Large-Large A.A. Side HB |
| AA | 7 | 7 | Weak HP |
| CC(Covalent Bond) | 8 | 8 | Breakable disulfide-bond |
| EE | 9 | 9 | Charged(--) |
| EK | 10 | 10 | Charged(−+) |
| KK | 11 | 11 | Charged(++) |
| A{LMFYW} | 12 | 12 | Weak HP, Ala- HP sidechains |
| C{LMFAC} | 13 | 13 | Cys-HP sidechains |
| C{YW} | 14 | 15 | Cys-HP sidechains, Side HB with S |
| {LMF}{EP},{YWC}P,{LF}S | 15 | 14 | HP sidechains – Small Polar(P) |
| {LF}{KQH},WK | 16 | 14 | HP sidechains – Large P |
| {MC}{KSQH},CE | 17 | 15 | Side HB with S |
| {YW}{ES},{ES}{SQH},KS | 18 | 16 | Side HB |
| {YW}{QH},YK | 19 | 16 | HP sidechains – Large P, Side HB |
| A{EKPSQH} | 20 | 17 | Ala - P |
| P{EKPSQH} | 21 | 17 | Pro - P |
| QQ,QH,HH | 22 | 18 | Large P – Large P, Side HB |
| Backbone-SideChain HB | 23 | 19 | Backbone-Side HB |
Table 3(left) shows the interaction parameters obtained by the stochastic learning algorithm based on 711 PDBs with different R1,R2,R3 combinations under the constraint that the strong attractive hydrophobic interactions (parameter numbers 1,2,3,4,5,6,8) are more negative than weak attractive hydrophobic interactions (parameter number 12). In the calculation of R2 (Eqn 4) and Nst (the number of strong interactions), the strong interactions are taken to be backbone-backbone hydrogen bonding, strong hydrophobic interactions and disulfide bond interactions between C and C, i.e. parameter numbers (1,2,3,4,5,6,8). In the calculation of R3 (Eqn 5) and Nwk, the weak attractive interactions are taken to be the weak hydrophobic interactions, charged attractive interactions and hydrogen-bonding between side-chains, i.e. parameter numbers (10,12, 17,18,19,22). Even though the evaluated interaction parameters do not satisfy all of the inequalities in the learning process and depend on the values of R1, R2, and R3, the Z-score values of ~-2.3 signify that almost 98% of the inequalities are satisfied. Moreover each parameter set is very stable, meaning it does not depend on the initial parameters chosen in the learning algorithm. Even starting from very different initial values of the parameters gives results that are identical up to the third decimal digit. Thus the results are believed to be the global minimum in parameter space.
Table 3.
Twenty three (left) and nineteen (right) interaction parameters from the 711PDB database and the modified stochastic learning algorithm.
| 23 Parameters | 19 Parameters | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| R1= R2= R3= |
0.05 0.20 0.25 |
0.05 0.35 0.55 |
0.05 0.40 0.45 |
R1= R2= R3= |
0.05 0.15 0.15 |
0.05 0.35 0.35 |
0.05 0.35 0.40 |
0.05 0.40 0.40 |
Ave |
| 1.NHCO | −1.000 | −1.000 | −1.000 | 1.NHCO | −1.000 | −1.000 | −1.000 | −1.000 | −1.000 |
| 2.LL | −0.254 | −0.214 | −0.186 | 2.LL | −0.256 | −0.174 | −0.170 | −0.201 | −0.200 |
| 3.LF | −0.350 | −0.265 | −0.227 | 3.LF | −0.256 | −0.175 | −0.173 | −0.207 | −0.203 |
| 4.MY | −0.463 | −0.216 | −0.202 | 4.MY | −0.279 | −0.176 | −0.180 | −0.206 | −0.210 |
| 5.FF | −0.446 | −0.316 | −0.267 | 5.FF | −0.256 | −0.176 | −0.177 | −0.212 | −0.205 |
| 6.YY | −0.254 | −0.216 | −0.195 | 6.YY | −0.254 | −0.174 | −0.171 | −0.203 | −0.201 |
| 7.AA | −0.134 | −0.113 | −0.085 | 7.AA | −0.098 | −0.070 | −0.067 | −0.099 | −0.084 |
| 8.CC | −0.836 | −0.559 | −0.545 | 8.CC | −0.678 | −0.701 | −0.516 | −0.446 | −0.585 |
| 9.EE | 0.556 | 0.474 | 0.342 | 9.EE | 0.414 | 0.118 | 0.214 | 0.265 | 0.253 |
| 10.EK | −0.253 | −0.166 | −0.135 | 10.EK | −0.151 | −0.121 | −0.119 | −0.151 | −0.136 |
| 11.KK | 0.135 | 0.022 | 0.005 | 11.KK | 0.046 | 0.055 | 0.082 | 0.107 | 0.073 |
| 12.LA | −0.204 | −0.162 | −0.135 | 12.AL | −0.201 | −0.122 | −0.118 | −0.150 | −0.148 |
| 13.CL | −0.080 | −0.176 | −0.162 | 13.CL | −0.046 | −0.175 | −0.160 | −0.176 | −0.139 |
| 14.CY | 0.148 | −0.052 | 0.007 | 14.LE | 0.026 | 0.009 | 0.014 | 0.009 | 0.015 |
| 15.LE | 0.076 | −0.007 | 0.017 | 15.MK | −0.124 | −0.100 | −0.097 | −0.141 | −0.116 |
| 16.LK | 0.039 | 0.025 | 0.029 | 16.YE | −0.101 | −0.068 | −0.072 | −0.104 | −0.086 |
| 17.MK | −0.204 | −0.151 | −0.148 | 17.AE | 0.056 | 0.084 | 0.070 | 0.084 | 0.074 |
| 18.YE | −0.116 | −0.087 | −0.082 | 18.QQ | −0.033 | −0.104 | −0.043 | −0.141 | −0.080 |
| 19.YQ | −0.242 | −0.132 | −0.117 | 19.B-S | −0.012 | −0.018 | −0.015 | −0.016 | −0.015 |
| 20.AE | 0.148 | −0.014 | 0.003 | ||||||
| 21.PE | −0.054 | 0.110 | 0.084 | ||||||
| 22.QQ | −0.095 | −0.023 | −0.014 | ||||||
| 23.B-S | −0.011 | −0.024 | −0.019 | ||||||
| <En(d-n)> | 2.325 | 2.360 | 2.276 | 2.344 | 2.118 | 2.127 | 2.236 | ||
| <Enneg(d-n)> | −0.003 | −0.004 | −0.004 | −0.003 | −0.003 | −0.003 | −0.004 | ||
| Eminn | −0.013 | −0.017 | −0.015 | −0.009 | −0.013 | −0.013 | −0.015 | ||
| SWP | −8.346 | −7.004 | −6.510 | −7.000 | −6.086 | −5.874 | −6.307 | ||
Comparing the relative values of the different types of parameters in Table 3(left) indicates that our results are physically reasonable (with one exception to be discussed in the next paragraph). Values in Table 3(left) clearly show that the strong attractive interactions--- the hydrophobic interactions (LL,LF,FF,YY) and the CC disulfide bond --- have large negative values and the weak attractive interactions--- the hydrophobic (LA,AA), charged (EK), and side-chain hydrogen-bond interactions interaction numbers (17,18,19,22) --- have small negative values. Also the interactions between aromatic residues (5,6) are slightly stronger than the hydrophobic interactions between small residues(2). In calculating the C-C disulfide bond interaction, two parameters were determined: the first operates at close range (<2.8Å) to mimic the covalent bond and the second operates at longer range (>2.8Å) and is non-covalent. The first is interaction 8 and the second is included with interaction 13. The parameter ratio AA/NHCO is between 0.05 and 0.15, which is in good agreement with the ratio used in the PRIME model simulations of polyalanine. The parameter ratio CC/NHCO is between 0.5 and 0.8. The interaction 10(EK) between residues with unlike charges is negative and the interactions between residues with the same charge, 9(EE) and 11(KK), are positive although small; interaction 11(KK) is smaller than interaction 9(EE), probably due to the slight hydrophobic effect for large side-chains. The only value in Table 3 that is not reasonable is parameter 14(C{YW}), which shows a positive value, contradicting common sense. The explanation for this is that the (C{YW}) interaction is a relatively rare event in our database of native and decoy conformations.
To address the positive (C{YW}) problem we further reduce the number of independent parameters by grouping parameters (15,16) (14,17), (18,19), (20,21) into their own individual groups, now numbered 14,15,16, and 17 respectively, arriving at a set of 19 parameters whose results are shown in Table 3(right). (The correspondence between the interaction parameters indices in the 19 parameter set and those in the 23 parameter set is indicated in the 2nd and 3rd columns of Table 2.) All pair interactions are physically meaningful since their relative values are consistent with their physico-chemical nature, e.g. hydrophobic, charged, side-chain hydrogen-bonding, etc., without any unexpected values like CY in table 3(left). The ratios of the various pair-interactions to the backbone-backbone hydrogen bond (NHCO) are also sensible.
It is of interest to consider an even simpler classification in which the possibility of side chain hydrogen bonding is not accounted for, In this case we have 7 groups of amino acids: strongly hydrophobic (LIVFYWM), weakly hydrophobic (A), glycine, (G), covalent bonding (C), polar (PSTHNQ), negatively-charged (DE) and positively -charged (KR). For this set we designate 12 types of interactions: (NH-CO) hydrogen-bond, (LL) strong hydrophobic interaction, (LA) hydrophobic-alanine, (L{P,E,K}) hydrophobic-polar or charged, (AA) alanine-alanine, (A{P,E,K}) alanine- polar or charged, (CC) disulfide bond, (C{L,A,P,E,K}) cysteine-others, (P{P,E,K}) polar-polar or charged, (EE) negative charges repulsive interaction, (KK) positive charges repulsive interaction, (EK) opposite charges attractive interaction.
Supplemental tables 4 and 5 show the results for the 12 parameters estimated by perceptron learning without (table 4) and with (table 5) the stochastic algorithm starting from 4 different initial values (I.V.) of the interaction parameters. In these calculations the constraint parameters that appear in eqns (3)- (5) are chosen to be R1=0.2, R2=0.2, R3=0.3, since they tend to yield a ratio AA/NHCO= 0.15 which is close to 0.1. The interaction parameters in supplemental table 4 evaluated without using the stochastic procedure of eqn(17) have two different final values, depending on the initial guesses for the parameters. However when the stochastic procedure is added (Supplemental table 5) the parameter values no longer depend on the initial values, which means we are more likely to have arrived at the optimum parameter values. Therefore our complex stochastic procedure of eqn(17) based on the four energies of eqn(12) to eqn(15) is necessary if we are to obtain a unique solution to the enormous number of inequalities in eqns (1) and (2). One shortcoming of this set is that the value of interaction 3(LA) is rather large compared to those of LL and AA. The 12 parameter set could be useful as pair interactions in coarse-grained simulations but would be unable to account for the type of side chain hydrogen-bonding that plays an important role in fibril-forming peptide like GNNQQNY.61
It is also of interest to see how sensitive the results are to the size of the database. We set up another database of native structures with 585 PDBs. In this case we removed binding proteins and mutated PDBs from the 711 protein database but added in small helix and coiled peptides. The resulting values for the 19 parameters extracted from the 585 PDB are shown in supplemental table 6. The interactions determined using this 585 PDB database is not as physically reasonable as those determined using the 711 database. For example compare the first column in supplemental table 6 (with R1=0.05, R2= 0.15, R3=0.15 with the first column of table 3 (right) The weak attractive interactions (LA,MS,YS,QQ) are larger in supplemental table 6 than in table 3 (right), and QQ is larger than LL. Parameter values in the second, third and fourth columns are selected from among the many R1,R2,R3 combinations considered, since they have the attractive feature that AA/NHCO is close to 0.1. In addition the pair interactions (MS,YS,QQ) having side-chain hydrogen-bonding have large negative values and are even stronger than the hydrophobic interaction (LL). Therefore we can conclude that removing small proteins, which is how the 711 PDB database was constructed, yields a better database than removing binding and mutated proteins which is how the 585 PDB database was constructed for in estimating pair-interaction parameters.
The following question naturally arises. Can the parameters estimated in this paper, which are essentially intramolecular pair interactions since they are based on single chain information in the PDB, be applied to the aggregation of peptides, which is driven primarily by inter-chain interactions? The difference between intra- and intermolecular interactions has been addressed by Keskin et al.62 who concluded that the assumption that intra- and inter-molecular pair potentials are well correlated is valid only when the reference state used is solvent-mediated. Since no reference state is used in our work, an alternative way to answer this question is to compare our results with those of other potentials having different reference states. Pokarowski and coworkers analyzed 29 published contact-interactions., and classified them into two types.63 The first group includes the Miyazawa-Jernigan (based on solvent mediated reference state)64, Betancourt and Thirumalai65, Skolnick65, and Tobi -Bahar interactions51; these were all dominated by a one-body hydrophobicity scale similar to that of Wertz and Scheraga.66 The second group of contact potentials includes the Miyazawa-Jernigan potential (based on residue mediated reference state)64 and the contact energies of Baker’s group67; these were correlated with a residue-dependent factor called q as well as with the a one-body hydrophobicity scale close to the Kyte-Doolittle scale.68 We have calculated the correlations between the average 19 parameters in table 3 and the 29 different pairwise contact potentials selected by Pokarowski et al.63 by using their correlation definition given by <x −x̄,y− ȳ>/nσxσy, where x and y are parameter values, <…,…> represents a scalar product, the bar indicates an average, n is the number of parameters, and σx is a standard deviation. Only five types of potentials show a correlation value larger than 0.7 with our parameters. They are the BT(0.704), SKOb(0.732) SKOa(0.721), MJ3h(0.716), BFKV(0.732) which belong to the class where the solvent mediated reference state is employed.56,64,65,69 The correlations between our average 19 parameters and the potentials associated with the residue mediated reference state, MJ1(0.405), MJ3(0.447) and B2(0.340),64,67,70 are relatively low. Therefore our 19 pair-potential parameter set is closer to solvent mediated pair potentials than to residue mediated pair potentials. Thus our use of the pair potential parameters calculated here as inter-molecular potential parameters in simulations of protein aggregation is supported by Keskin et al’s discussion.
DISCUSSION
We developed a new stochastic procedure to overcome dependence on the initial values in the perceptron learning algorithm. The pair interaction parameters for use in the extended version of PRIME, called PRIME 20, are estimated using this modified stochastic perceptron learning algorithm. The aim here was to reduce the number of independent pair-interaction parameters (compared to the usual 210 parameters) to be small enough that our pair interaction energies would be physically meaningful and yet large enough to give us reasonably accurate results in discriminating decoys from native structures in the PDB database. The smallest number of parameters that gives reasonable interactions is 19; these account in a satisfactory way for hydrophobic, charged, side-chain hydrogen-bonding interactions. A 12 parameter set that does not account for side-chain hydrogen bonding was less successful at describing the relative values of the various types of interactions that one would expect on physico-chemical grounds. The types of proteins included in the learning algorithms; PDB database appear to be critical in estimating pairwise interaction parameters; in particular single helix and nearly unstructured coiled peptides should be removed in learning procedures.
Application of the newly constructed PRIME20 force field described here, with its heterogeneous values for the pair interaction energy strengths and ranges, mass, bond- and pseudo-bond lengths and, squeeze distances, to the aggregation of small peptides such as Aβ(16–22) and Aβ(25–35) are under way. Thus far PRIME 20 has been tested by applying DMD simulations to systems containing forty eight Abeta16–22(KLVFFAE) peptides. Spontaneous formation of twisted cross-beta structures or stacked beta-sheets is observed over a range of fixed protein concentrations and temperatures starting from random initial configurations.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health, USA under grant GM56766 and EB006006(MC,CKH) and National Creative Research Initiatives (Center for Proteome Biophysics) of National Research Foundation/Ministry of Education, Science and Technology, Korea (Grant No. 2008-0061984 to MC, IC)
REFERENCES
- 1.Smith AV, Hall CK. α-helix formation: discontinuous molecular dynamics on an intermediate resolution model. Proteins. 2001;44:376–391. doi: 10.1002/prot.1100. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen HD, Hall CK. Molecular dynamics simulations of spontaneous fibril formation by random-coil peptides. Proc Natl Acad Sci USA. 2004;101:16180–16184. doi: 10.1073/pnas.0407273101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchut A, Hall CK. Side-chain interactions determine amyloid formation by model polyglutamine peptides in molecular dynamics simulations. Biophys J. 2006;90:4574–4584. doi: 10.1529/biophysj.105.079269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alder BJ, Wainwright TE. Studies in molecular dynamics. I. General method. J Chem Phys. 1959;31:459–466. [Google Scholar]
- 5.Nguyen HD, Hall CK. Spontaneous fibril formation by polyalanines; discontinuous molecular dynamics simulations. J Am Chem Soc. 2006;128:1890–1901. doi: 10.1021/ja0539140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borreguero JM, Urbanc B, Lazo ND, Buldyrev SV, Teplow DB, Stanley HE. Folding events in the 21–30 region of amyloid beta-protein (Abeta) studied in silico. Proc Natl Acad Sci USA. 2005;102:6015–6020. doi: 10.1073/pnas.0502006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urbanc B, Cruz L, Ding F, Sammond D, Khare S, Buldyrev SV, Stanley HE, Dokholyan NV. Molecular dynamics simulation of amyloid beta dimer formation. Biophys J. 2004;87:2310–2321. doi: 10.1529/biophysj.104.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang H, Hall CK, Zhou Y. Assembly and kinetics folding pathways of a tetrameric beta-sheet complex: molecular dynamics simulations on simplified off-lattice models. Biophys J. 2004;86:31–49. doi: 10.1016/S0006-3495(04)74081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favrin G, Irback A, Mohanty S. Oligomerization of amyloid Abeta16–22 peptides using hydrogen bonds and hydrophobicity forces. Biophys J. 2004;87:3657–3664. doi: 10.1529/biophysj.104.046839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cellmer T, Bratko D, Prausnitz JM, Blanch HW. Protein aggregation in silico. TRENDS in Biochemistry. 2007;25:254–261. doi: 10.1016/j.tibtech.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheon M, Chang I, Mohanty S, Luheshi LM, Dobson CM, Vendruscolo M, Favrin G. Structural reorganization and potential toxicity of oligomeric species formed during the assembly of amyloid fibrils. Plos Comput Biol. 2007;3:1727–1738. doi: 10.1371/journal.pcbi.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auer S, Meersman F, Dobson CM, Vendruscolo M. A generic mechanism of emergence of amyoid protofilaments from disordered oligomeric aggregates. Plos Comput Biol. 2008;4:e1000222. doi: 10.1371/journal.pcbi.1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weiner P. A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc. 1984;106:765. [Google Scholar]
- 14.Brooks BR, Brooks CL, III, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma B, Nussinov R. Simulations as analytical tools to understand protein aggregation and predict amyloid conformation. Curr Opin Chem Biol. 2006;10:445–452. doi: 10.1016/j.cbpa.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Chiti F, Dobson CM. Protein misfolding, functional amyloid and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 17.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. 2003. [DOI] [PubMed] [Google Scholar]
- 18.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DL. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 19.Gong Y, Chang L, Viola KL, Lacor PN, Lambert MP, Finch CE, Krafft GA, Klein WL. Alzheimer’s disease-affected brain: presence of oligomeric A beta ligands (ADDLs) suggests a molecular basis for reversible memory loss. Proc Natl Acad Sci USA. 2003;100:10417–10422. doi: 10.1073/pnas.1834302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kayed R, Head E, Thompson JL, Mclntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen PH, Li MS, Stock G, Straub JE, Thirumalai D. Monomer adds to preformed structured oligomers of Abeta-peptides by a two-stage dock-lock mechanism. Proc Natl Acad Sci USA. 2007;104:111–116. doi: 10.1073/pnas.0607440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haspel N, Zanuy D, Ma B, Wolfson H, Nussinov R. A comparative study of amyloid fibril formation by residues 15–19 of the human calcitonin hormone: a single beta-sheet model with a small hydrophobic core. J Mol Biol. 2005;345:1213–1227. doi: 10.1016/j.jmb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Shen J, Luo X, Zhu W, Chen K, Ma J, Jiang H. Conformational transition of amyloid beta-peptide. Proc Natl Acad Sci USA. 2005;102:5403–5407. doi: 10.1073/pnas.0501218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheon M, Favrin G, Chang I, Dobson CM, Vendruscolo M. Calculation of the free energy barriers in the oligomerisation of A beta peptide fragments. Front Biosci. 2008;13:5614–5622. doi: 10.2741/3104. [DOI] [PubMed] [Google Scholar]
- 25.Bryant SH, Lawrence CE. The frequency of ion-pair substructures in proteins is quantitatively related to electrostatic potential. A statistical model for nonbonded interactions. Proteins. 1991;9:108–119. doi: 10.1002/prot.340090205. [DOI] [PubMed] [Google Scholar]
- 26.Sippl MJ. Knowledge based potentials for proteins. Curr Opin Struct Biol. 1995;5:229–235. doi: 10.1016/0959-440x(95)80081-6. [DOI] [PubMed] [Google Scholar]
- 27.Godzik A, Kolinski A, Skolnick J. Are proteins ideal mixtures of amino acids? Analysis of energy parameter sets. Prot Sci. 1995;4:2107–2117. doi: 10.1002/pro.5560041016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jernigan RL, Bahar I. Structure-derived potentials and protein simulations. Curr Opin Struct Biol. 1996;6:195–209. doi: 10.1016/s0959-440x(96)80075-3. [DOI] [PubMed] [Google Scholar]
- 29.Miyazawa S, Jernigan RL. Residue–residue potentials with a favorable contact pair term an unfavorable high packing density term, for simulation and threading. J Mol Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 30.Thomas PD, Dill KA. An iterative method for extracting energy-like quantities from protein structures. Proc Natl Acad Sci USA. 1996;93:11628–11633. doi: 10.1073/pnas.93.21.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park B, Levitt M. Energy Functions that Discriminate X-ray and Near-native Folds from Well-constructed Decoys. J Mol Biol. 1996;258:367–392. doi: 10.1006/jmbi.1996.0256. [DOI] [PubMed] [Google Scholar]
- 32.Skolnick J, Jaroszewski L, Kolinski A, Godzik A. Derivation and testing of pair potential for protein folding. When is quasichemical approximation correct? Prot Sci. 1997;6:676–688. doi: 10.1002/pro.5560060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melo F, Sanchez R, Sali A. Statistical potentials for fold assessment. Prot Sci. 2002;11:430–448. doi: 10.1002/pro.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Pro Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solis AD, Rackovsky S. Improvement of Statistical Potentials and Threading Score Functions Using Information Maximization. Proteins. 2006;62:892–908. doi: 10.1002/prot.20501. [DOI] [PubMed] [Google Scholar]
- 36.Dehouck Y, Gills D, Rooman M. A New Generation of Statistical Potentials for Proteins. Biophys J. 2006;90:4010–4017. doi: 10.1529/biophysj.105.079434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang SY, Zou X. An iterative knowledge-based scoring function for protein-protein recognition. Proteins. 2008;72:557–579. doi: 10.1002/prot.21949. [DOI] [PubMed] [Google Scholar]
- 38.Micheletti C, Seno F, Banavar JR, Maritan A. Learning Effective Amino Acid Interactions through Iterative Stochastic Techniques. Proteins. 2001;42:422–431. doi: 10.1002/1097-0134(20010215)42:3<422::aid-prot120>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Maiorov VN, Crippen GM. Contact potential that recognizes the correct folding of globular proteins. J Mol Biol. 1992;227:876–888. doi: 10.1016/0022-2836(92)90228-c. [DOI] [PubMed] [Google Scholar]
- 40.Mirny LA, Shakhnovich EI. How to Derive a Protein Folding Potential? A new approach to an old problem. J Mol Biol. 1996;264:1164–1179. doi: 10.1006/jmbi.1996.0704. [DOI] [PubMed] [Google Scholar]
- 41.Vendruscolo M, Domany E. Pairwise contact potentials are unsuitable for protein folding. J Chem Phys. 1999;109:11101–11108. [Google Scholar]
- 42.Dima R, Settanni G, Micheletti C, Banavar JR, Maritan A. Extraction of interaction potentials between amino acids from native protein structures. J Chem Phys. 2000;112:9151–9166. [Google Scholar]
- 43.Ohkubo YZ, Crippen GM. Potential energy functions for continuous state models of globular proteins. J Comput Biol. 2000;7:363–379. doi: 10.1089/106652700750050835. [DOI] [PubMed] [Google Scholar]
- 44.Tobi D, Elber R. Distance-dependent, pair potential for protein folding: Results from linear optimization. Proteins. 2000;41:40–46. [PubMed] [Google Scholar]
- 45.Meller J, Wagner M, Elber R. Maximum feasibility guideline in the design and analysis of protein folding potentials. J Comput Chem. 2002;23:111–118. doi: 10.1002/jcc.10014. [DOI] [PubMed] [Google Scholar]
- 46.Loose C, Klepeis JL, Floudas CA. A new pairwise folding potential based on improved decoy generation and side-chain packing. Proteins. 2004;54:303–314. doi: 10.1002/prot.10521. [DOI] [PubMed] [Google Scholar]
- 47.Chang I, Cieplak M, Dima RI, Maritan A, Banavar JR. Protein threading by learning. Proc Natl Sci USA. 2001;98:14350–14355. doi: 10.1073/pnas.241133698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heo M, Kim S, Moon EJ, Cheon M, Chung K, Chang I. Perceptron learning of pairwise contact energies for proteins incorporating the amino acid environment. Phys Rev E. 2005;72:011906. doi: 10.1103/PhysRevE.72.011906. [DOI] [PubMed] [Google Scholar]
- 49.Rajgaria R, McAllister SR, Floudas CA. Development of a novel high resolution Cα-Cα distance dependent force field using a high quality decoy set. Proteins. 2006;65:726–741. doi: 10.1002/prot.21149. [DOI] [PubMed] [Google Scholar]
- 50.Rajgaria R, McAllister SR, Floudas CA. Distance dependent centroid to centroid force fields using high resolution decoys. Proteins. 2008;70:950–970. doi: 10.1002/prot.21561. [DOI] [PubMed] [Google Scholar]
- 51.Tobi D, Bahar I. Optimal Design of Protein Docking Potentials: Efficiency and Limitations. Proteins. 2006;62:970–981. doi: 10.1002/prot.20859. [DOI] [PubMed] [Google Scholar]
- 52.Hinds DA, Levitt M. Exploring conformational space with a simple lattice model for protein structure. J Mol Biol. 1994;243:668–682. doi: 10.1016/0022-2836(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 53.Chan HS, Dill DA. Origins of structure in globular proteins. Proc Natl Acad Sci USA. 1990;87:6388–6392. doi: 10.1073/pnas.87.16.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Wang W. A computational approach to simplifying the protein folding alphabet. Nat Struct Biol. 1999;6:1033–1038. doi: 10.1038/14918. [DOI] [PubMed] [Google Scholar]
- 55.Riddle DS, Santiago JV, Bray-Hall ST, Doshi N, Grantcharova VP, Yi Q, Baker D. Functional rapidly folding proteins from simplified amino acid sequences. Nat Struct Biol. 1997;4:805–809. doi: 10.1038/nsb1097-805. [DOI] [PubMed] [Google Scholar]
- 56.Betancourt MR, Thirumalai D. Pair potentials for protein folding: Choice of reference states and sensitivity of predicted native states to variations in the interaction schemes. Prot Sci. 1999;8:361–369. doi: 10.1110/ps.8.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buchete N-V, Straub JE, Thirumalai D. Dissecting contact potentials for proteins: Relative contributions of individual amino acids. Proteins. 2008;70:119–130. doi: 10.1002/prot.21538. [DOI] [PubMed] [Google Scholar]
- 58.Jones DT, Taylor WR, Thorton JM. A new approach to protein fold recognition. Nature. 1992;358:86. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 59.Krauth W, Mezard M. Learning algorithms with optimal stability in neural networks. J Phys A. 1987;20:745–752. [Google Scholar]
- 60.Hobohm U, Scharf M, Schneider R, Sander C. Selection of a representative set of structures from the Brookhaven Protein Data Bank. Prot Sci. 1992;1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nelson R, Sawaya MR, Balbirnie M, Madsen A, Riekel C, Grothe R, Eisenberg D. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keskin O, Bahar I, Badretdinov AY, Ptitsyn OB, Jernigan RL. Empirical solvent-mediated potentials hold for both intra-molecular and inter-molecular inter-residue interactions. Prot Sci. 1998;7:2578–2586. doi: 10.1002/pro.5560071211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pokarowski P, Kloczkowski A, Jernigan RL, Kothari N, Pokarowska M, Kolinski A. Inferring ideal amino acid interaction forms from statistical protein contact potentials. Proteins. 2005;59:49–57. doi: 10.1002/prot.20380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyazawa S, Jernigan RL. Self-consistent estimation of inter-residue protein contact energies based on an equilibrium mixture approximation of residues. Proteins. 1999;34:49–68. doi: 10.1002/(sici)1097-0134(19990101)34:1<49::aid-prot5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 65.Skolnick J, Kolinski A, Ortiz A. Derivation of protein-specific pair potentials based on weak sequence fragment similarity. Proteins. 2000;38:3–16. [PubMed] [Google Scholar]
- 66.Wertz DH, Scheraga HA. Influence of water on protein structure. An analysis of the preferences of amino acid residues for the inside or outside and for specific conformations in a protein molecule. Macromolecules. 1978;11:9–15. doi: 10.1021/ma60061a002. [DOI] [PubMed] [Google Scholar]
- 67.Simons KT, Ruczinski I, Kooperberg C, Fox BA, Bystroff C, Baker D. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 68.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 69.Bastolla U, Farwer J, Knapp EW, Vendruscolo M. How to guarantee optimal stability for most representative structures in the Protein Data Bank. Proteins. 2001;44:79–96. doi: 10.1002/prot.1075. [DOI] [PubMed] [Google Scholar]
- 70.Miyazawa S, Jernigan RL. Estimation of effective interresidue contact energies from protein crystal structures—quasi-chemical approximation. Macromolecules. 1985;18:534–552. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.