Abstract
Gram-negative bacteria, such as Escherichia coli, frequently utilize tripartite efflux complexes in the resistance-nodulation-cell division (RND) family to expel diverse toxic compounds from the cell.1,2 The efflux system CusCBA is responsible for extruding biocidal Cu(I) and Ag(I) ions.3,4 No prior structural information was available for the heavy-metal efflux (HME) subfamily of the RND efflux pumps. Here we describe the crystal structures of the inner membrane transporter CusA in the absence and presence of bound Cu(I) or Ag(I). These CusA structures provide important new structural information about the HME sub-family of RND efflux pumps. The structures suggest that the metal binding sites, formed by a three-methionine cluster, are located within the cleft region of the periplasmic domain. Intriguingly, this cleft is closed in the apo-CusA form but open in the CusA-Cu(I) and CusA-Ag(I) structures, which directly suggests a plausible pathway for ion export. Binding of Cu(I) and Ag(I) triggers significant conformational changes in both the periplasmic and transmembrane domains. The crystal structure indicates that CusA has, in addition to the three-methionine metal binding site, four methionine pairs - three located in the transmembrane region and one in the periplasmic domain. Genetic analysis and transport assays suggest that CusA is capable of actively picking up metal ions from the cytosol, utilizing these methionine pairs/clusters to bind and export metal ions. These structures suggest a stepwise shuttle mechanism for transport between these sites.
In Gram-negative bacteria, efflux systems of the resistance-nodulation-cell division (RND) superfamily play major roles in the intrinsic and acquired tolerance of antimicrobials, including silver and copper ions.1,2 The Gram-negative bacterium Escherichia coli harbors seven RND efflux transporters. These transporters can be categorized into two distinct sub-families, the hydrophobic and amphiphilic efflux RND (HAE-RND) and heavy-metal efflux RND (HME-RND) families.1,2 Six of these transporters, including AcrB, AcrD, AcrF, MdtB, MdtC and YhiV, are multidrug efflux pumps, which belong to the HAE-RND protein family.1 E. coli also contains one HME-RND transporter, CusA, which specifically confers resistance to Cu(I)/Ag(I).3,4 The two sub-families of these RND transporters share relatively low sequence homology. For example, alignment shows that CusA and AcrB have only 19% identity. CusA works in conjunction with the membrane fusion protein CusB,3,4 and the outer membrane channel CusC.3,4 Presumably, these three components form a tripartite efflux complex CusCBA, which spans the entire cell envelope of E. coli to export Cu(I)/Ag(I).
At present, only two crystal structures of the RND pumps are available. These pumps, belonging to the HAE-RND sub-family, are the E. coli AcrB5–11 and Pseudomonas aeruginosa MexB12 multidrug transporters. In addition, the structures of other components of these tripartite systems have also been determined. These include the outer membrane channels, E. coli TolC13 and P. aeruginosa OprM,14 as well as the membrane fusion proteins, E. coli AcrA15 and P. aeruginosa MexA.16–18 Currently, no structural information has yet been available for any HME-RND pumps. To elucidate the mechanisms used by the CusCBA system for Cu(I)/Ag(I) recognition and extrusion, we previously reported the crystal structure of CusB,19 and now describe the crystal structures of the HME-RND transporter CusA, both in the absence and presence of Cu(I)/Ag(I). The structures suggest that CusA relies upon methionine residues to bind and export the metal ions, as was proposed.4
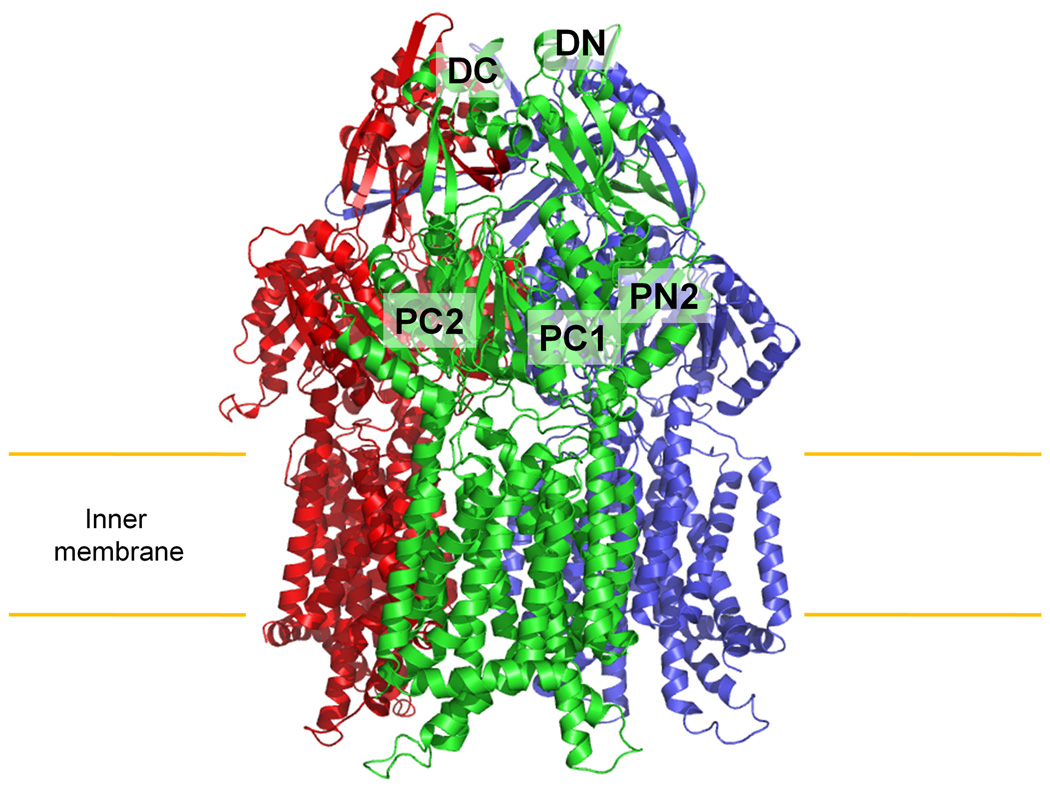
Multiple-isomorphous replacement with anomalous scattering (MIRAS) was used to determine the crystal structure (Fig. S1; Tables S1 and S2), which suggests that CusA exists as a homotrimer. Each subunit of CusA has12 transmembrane helices (TM1–TM12) and a large periplasmic domain formed by two loops between TM1 and TM2, and between TM7 and TM8 (Fig. 1 and Fig. S2). In the transmembrane region, TM1–TM6 are related to TM7–TM12 by pseudo-twofold symmetry. Differently from AcrB and MexB, four helices, TM4, TM5, TM10 and TM11, extend into the cytoplasm, forming the cytoplasmic domain of the pump. Two other helices, TM2 and TM8, protrude into the periplasm and contribute part of the periplasmic domain.
Figure 1.
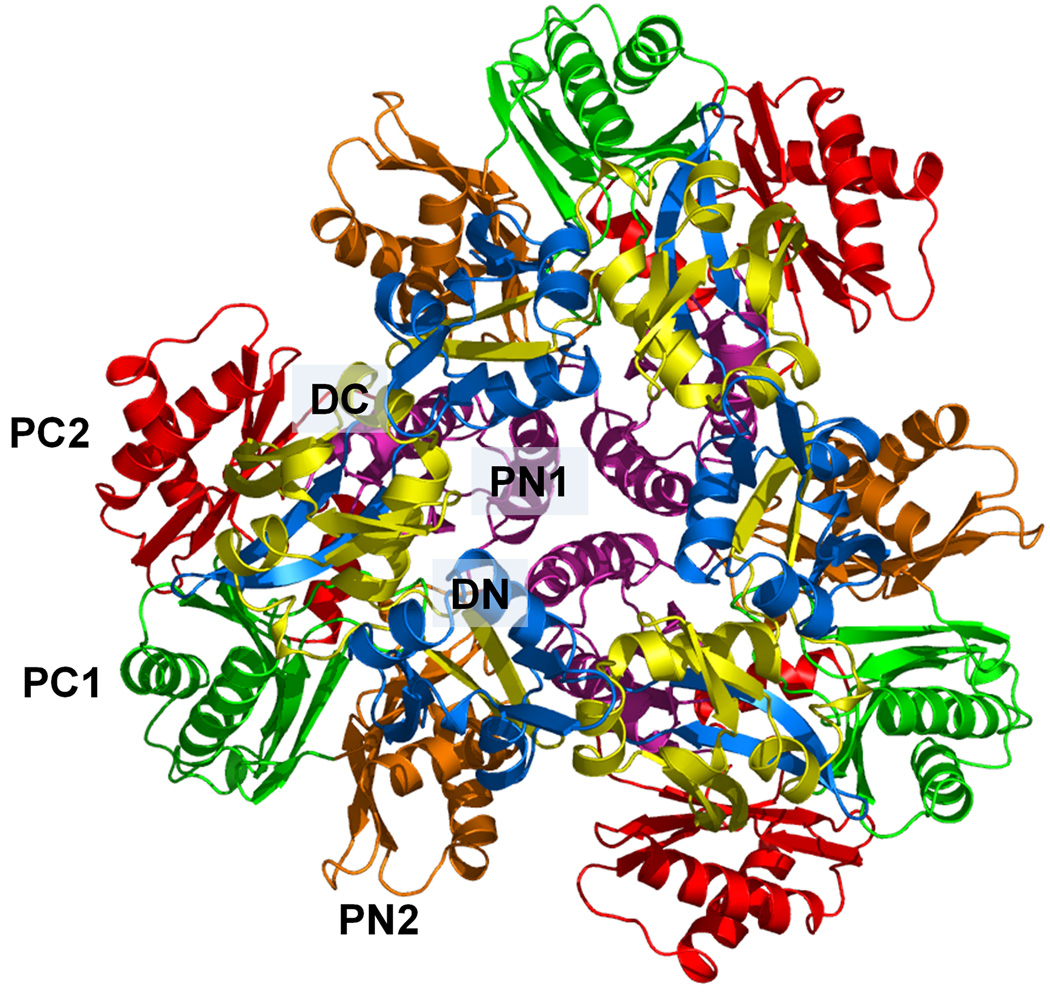
Structure of the apo CusA efflux pump. (a) Ribbon diagram of the CusA homotrimer viewed in the membrane plane. Each subunit of CusA is labeled with a different color. Sub-domains DN, DC, PN2, PC1 and PC2 are labeled on the front protomer (green). The location of PN1 in this protomer is behind PN2, PC1 and PC2 (see text). (b) Top view of the CusA trimer. The six sub-domains are labeled blue (DN), yellow (DC), pink (PN1), orange (PN2), green (PC1) and red (PC2). In the apo-CusA structure, the cleft between PC1 and PC2 is closed.
Like AcrB and MexB, the periplasmic domain of CusA can be divided into six sub-domains, PN1, PN2, PC1, PC2, DN and DC (Fig. 1). Sub-domains PN1, PN2, PC1 and PC2 form the pore domain, with PN1 making up the central pore stabilizing the trimeric organization. Sub-domains DN and DC, however, contribute to the docking domain, presumably interacting with the CusC channel. The trimeric CusA structure suggests that sub-domains PN2, PC1 and PC2 are located at the outermost core of the periplasmic domain, facing the periplasm. PC1 and PC2 also form an external cleft, and this cleft is closed in the apo-CusA structure (Figs. 1b and 2a).
Figure 2.
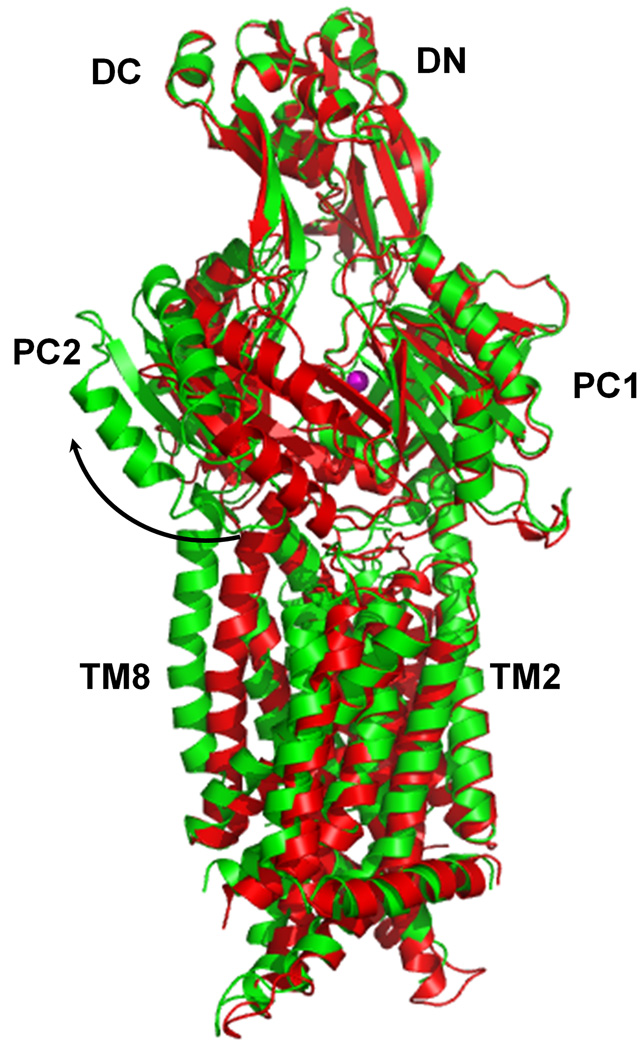
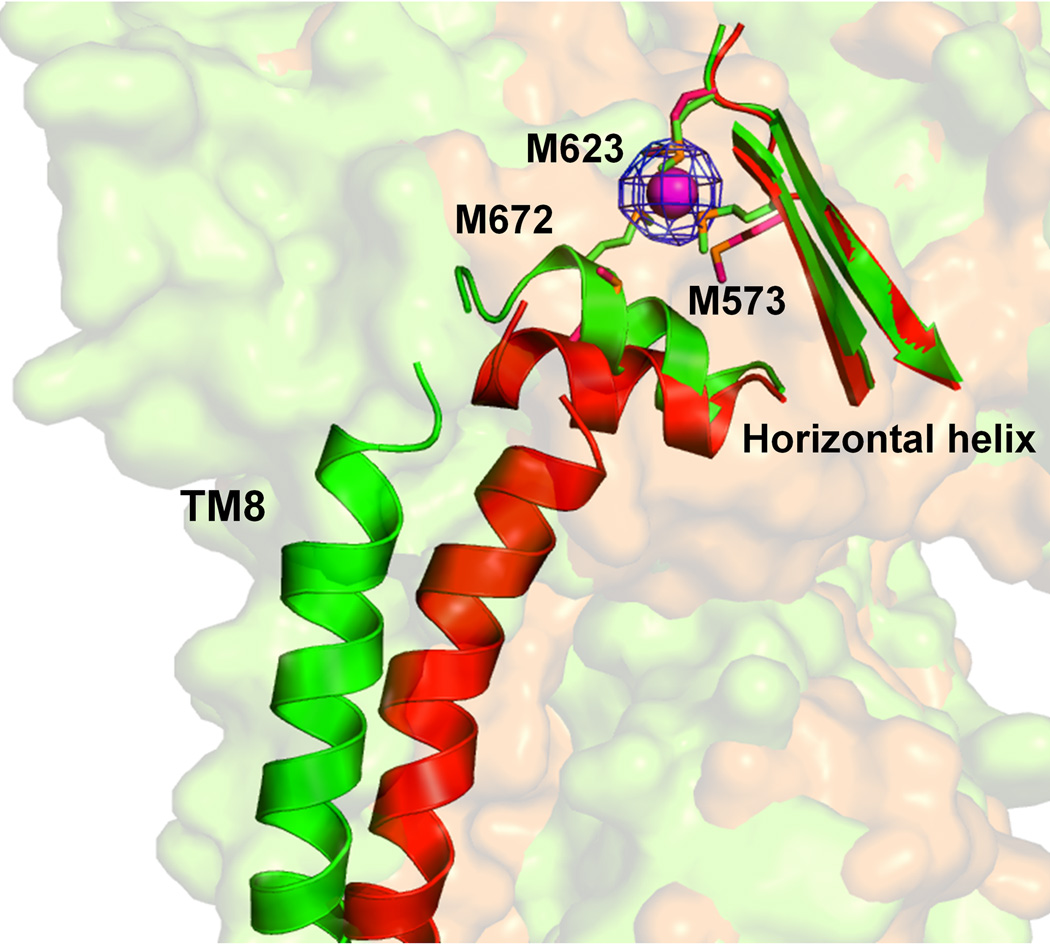
Comparison of the apo and metal-bound structures of CusA. (a) Superposition of apo-CusA (red) onto CusA-Cu(I) (green). The bound Cu(I) is pink. Arrow indicates the shift of PC2 when comparing these two structures. (b) Conformational changes of the periplasmic domain of CusA. The conformation of each sub-domain of CusA before (left) and after (right) Cu(I) binding is shown. The cleft formed between PC1 and PC2 is opened after Cu(I) binding. This occurs by the horizontal helix moving to bind the ion, freeing TM8 to move and open. The bound coppers are blue. The arrows indicate the locations of the clefts.
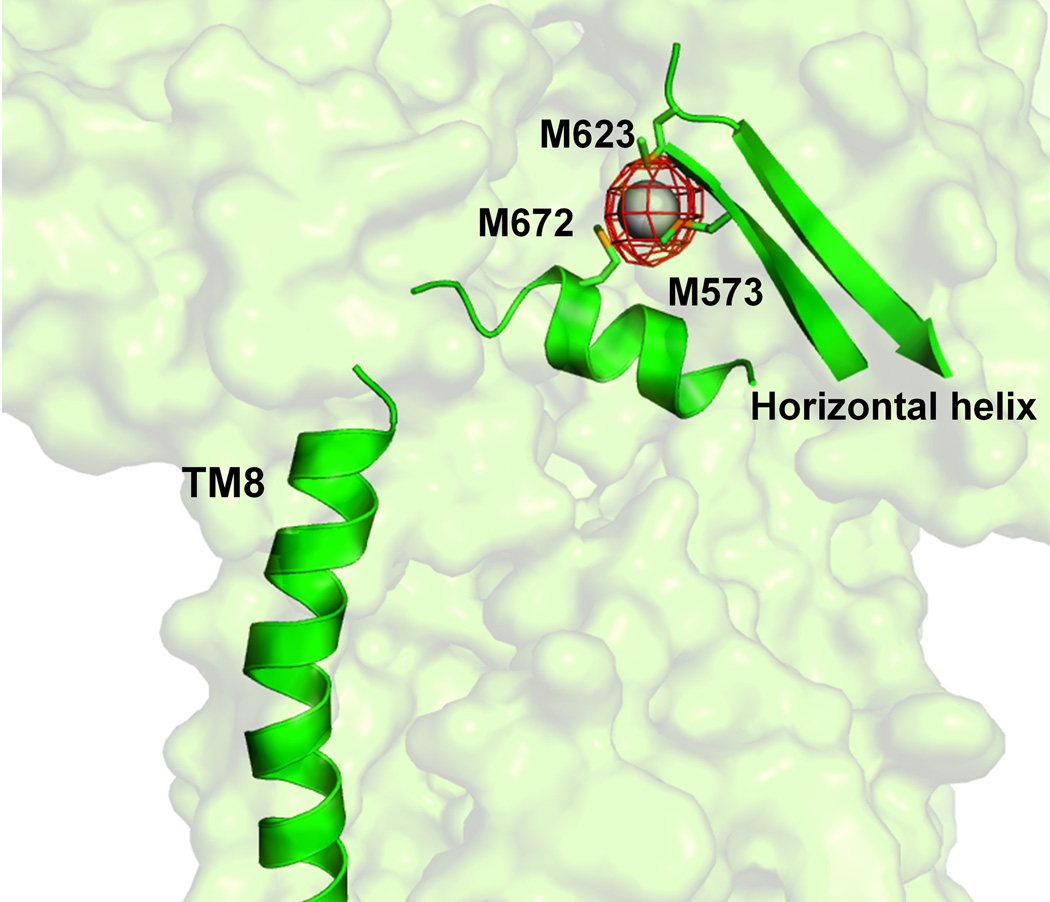
A most interesting feature appears in the cleft of the periplasmic domain with residues located on the left side of the wall (Fig. 2b), formed by one α-helix (residues 690–706) and three β-sheets (residues 681–687, 711–716 and 821–827), tilt into the cleft to close the opening. Surprisingly, residues 665–675, located at the bottom of the cleft, form an α-helix. This structural feature, not seen in AcrB or MexB, likely governs the specificity of CusA. The α-helix orients horizontally and roughly divides the transmembrane and periplasmic domains into two regions. Three proximal methionines, M573, M623 and M672, presumably creating a three-methionine specific binding site20,21 are found above this horizontal helix.
Intriguingly, the overall structure of CusA-Cu(I) is quite distinct from that of apo-CusA (Fig. 2). Superimposition of these two structures gives a RMSD of 3.9 Å (for 1,006 Cα atoms). The bound Cu(I) is found to coordinate residues M573, M623 and M672. Binding of Cu(I) seems to initiate significant conformational changes in the periplasmic as well as transmembrane domains of CusA. The most noticeable difference between the apo and ion-bound structures appears in the PC2 region (Fig. 2 and Fig. S3). A shift in position of the entire PC2 sub-domain is found in the Cu(I)-bound form. This shift results in formation of the cleft between sub-domains PC1 and PC2. The shift can be interpreted as a 30° swing of the PC2 sub-domain. The hinge for this movement appears to be at the junction between sub-domains PC2 and DC with G721 and P810 forming the hinge. This cleft presumably creates an entrance for metal ions from the periplasm. The horizontal helix, residues 665–675, located inside the cleft also makes a substantial movement. The C-terminal end of this helix is found to tilt upward by 21° in the Cu(I)-bound form in comparison with the apo structure (Fig. 3a). This tilting motion allows M672 to move closer to M573 and M623, forming a transient three-sulfur coordination site.22 Coupled with this movement, TM8 also shifts in position to a more vertical orientation while retaining its α-helical structure. Overall, the N-terminal end of TM8 is found to shift away from the core by 10 Å after Cu(I) binding.
Figure 3.
Anomalous maps of the bound metal ions. (a) The changes in conformation of the horizontal helix and TM8, are shown in a superimposition of the structures of apo (red) and Cu(I)-bound (green) CusA. The bound Cu(I) is shown as a pink sphere. Anomalous map of the bound Cu(I), contoured at 8σ, is in blue. M573, M623 and M672 are shown as sticks. (b) The Ag(I) binding site. The bound Ag(I) is shown as a gray sphere. Anomalous map of the bound Ag(I) , contoured at 10σ, is in red.
The overall conformational changes triggered by Cu(I) and by Ag(I) binding are nearly identical (Fig. 3b). Superimposition of the CusA-Cu(I) and CusA-Ag(I) structures gives a RMSD of 1.0 Å (for 1,021 Cαs). Based on these structures, the horizontal helix in the cleft directly interacts with the N-terminal end of TM8. The movement of TM8 may relate to transmembrane signaling and could initiate proton translocation across the membrane.
It appears that the binding of Cu(I)/Ag(I) also triggers significant conformational changes in the other transmembrane helices of the pump. In addition to the movement of TM8, all other transmembrane helices except TM2 shift horizontally by as much as 4 Å, mimicking the motion of TM8. Further, TM1, TM3 and TM6 readjust in an approximately 3 Å upward shift with respect to the inner membrane surface. The net result is that all three of these transmembrane helices move towards the periplasm by one turn. Coupled with the above conformational changes, PN1 also shifts upward by 3 Å. This moves the central pore helix upward by one turn upon metal binding. Mutagenesis studies indicated the conserved charged residue D405 of TM4 to be essential for transporter function.4 This acidic residue is found to interact with E939 and K984, possibly forming the proton-relay network of the pump (Fig S4).
Flexible docking was then carried out to assemble the CusBA complex. The resulting CusBA structure suggests that Domains 1 and 2 of CusB contact CusA at the interface between PN2 and PC1 (Fig. S5).
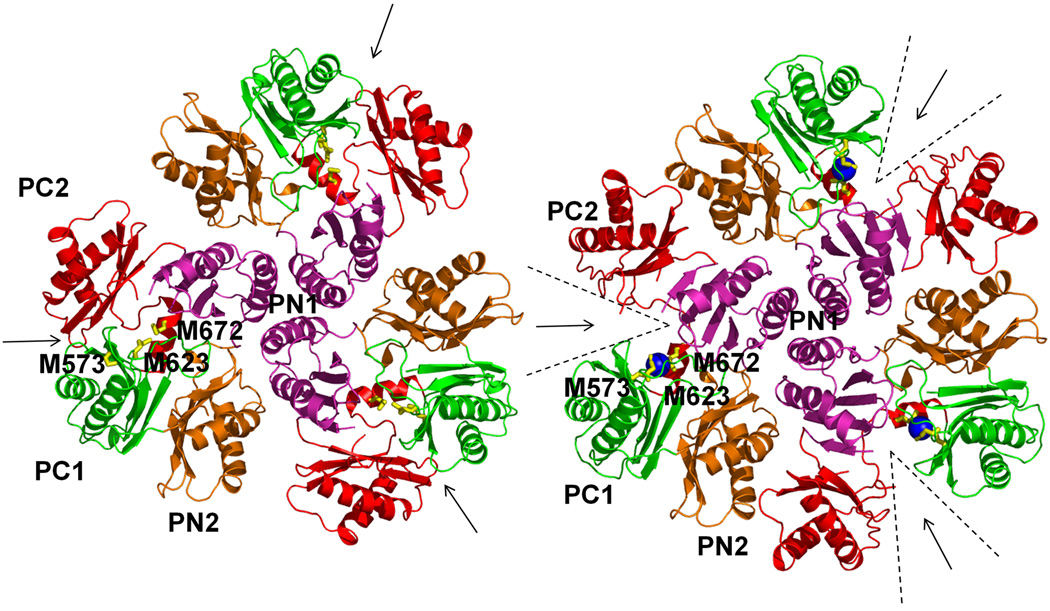
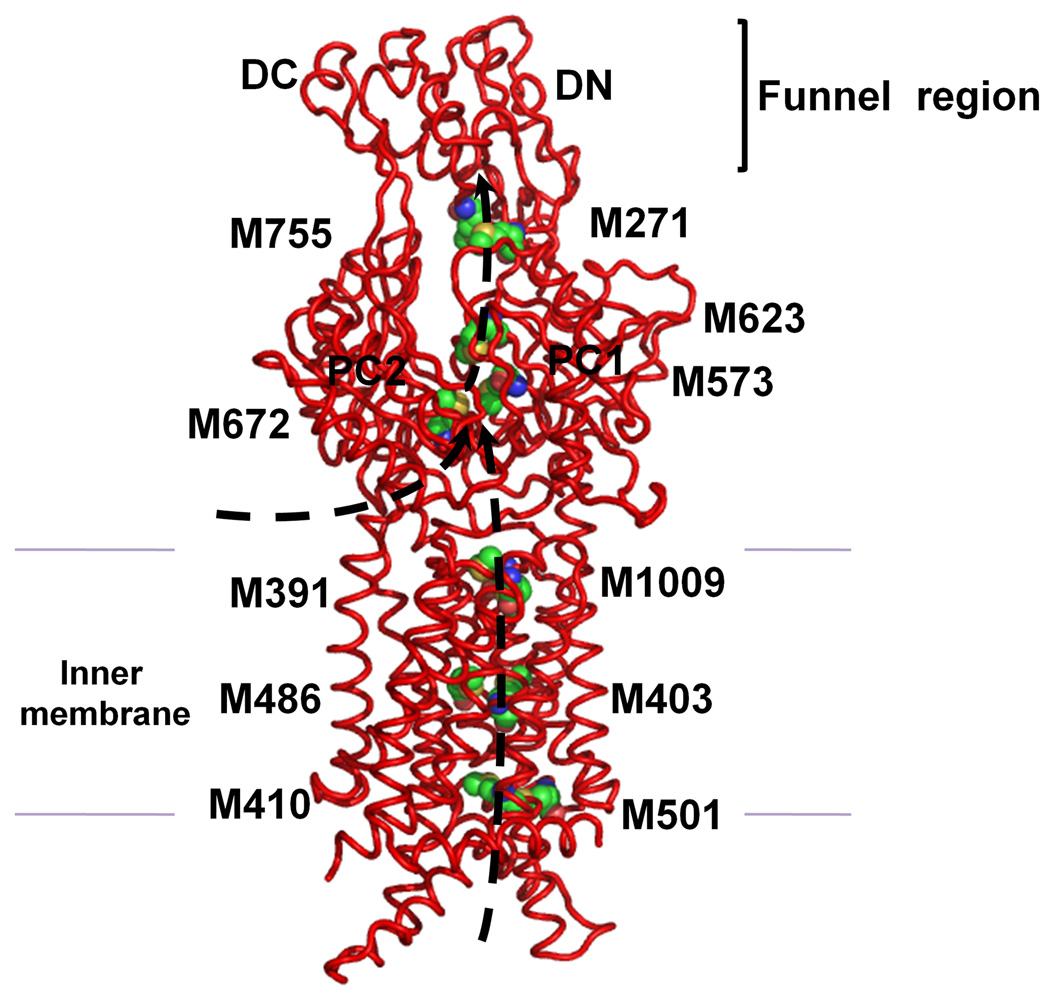
In the transmembrane domain, six methionines are paired up to form three distinct pairs (Fig. S1b). These methionine pairs are M410–M501, M403–M486, and M391–M1009 (Figs. 4, S6 and S7). Copper tolerance proteins, such as CusF,23,24 CueR25 and Atx1,26 have been found to utilize two methionines or two cysteines to carry their Cu(I)/Ag(I) cargos. Thus, these methionine pairs could potentially form binding sites for Cu(I)/Ag(I) in the transmembrane region. So, then CusA could relay these metal ions from the cytoplasm along these methionine pairs. In the periplasmic domain of CusA, we have found another pair (M271–M755) in addition to the three-methionine metal binding site (Fig. 4). These five methionine pairs/clusters are deemed to be important for binding and transport of metals.
Figure 4.
Proposed metal transport pathway of the CusA efflux pump. The five methionine pairs/clusters of the apo-CusA transporter form a pathway for metal export. These methionines are shown as spheres (C, green; O, orange; N, blue; S, yellow). The pair M271–M755 is located at the bottom of the periplasmic funnel where the metal ion could then be released for final extrusion. The paths for metal transport through the periplasmic cleft and transmembrane region are illustrated with black arrows.
The program CAVER (http://loschmidt.chemi.muni.cz/caver) indicates that each protomer of CusA forms a channel spanning the entire transmembrane region up to the bottom of the periplasmic funnel (Fig. S8). Intriguingly, this channel includes the four methionine-pairs (M410–M501, M403–M486, M391–M1009, and M271–M755) as well as the three-methionine binding site (M573, M623 and M672). Taken together these five methionine clusters are likely to form a relay network facilitating metal transport. We made an E. coli knock-out strain BL21(DE3)ΔcueOΔcusA that lacks both cueO and cusA. M573, M623 and M672 were mutated into isoleucines (M573I, M623I and M672I). These three mutants were expressed in BL21(DE3)ΔcueOΔcusA, and tested their ability to confer copper and silver resistance in vivo. Our studies show that the CusA mutants, M573I, M623I and M672I, are unable to relieve the copper or silver sensitivity of strain BL21(DE3)ΔcueOΔcusA (Tables S3 and S4), thus agreeing with the work of Franke et al.4 in which M573, M623 and M672 were shown to be essential. Further mutants M391I, M410I, M486I and M755I were prepared to disrupt the methionine pairs formed by the pump. Expression of these four mutants in BL21(DE3)ΔcueOΔcusA showed a significant decrease in the levels of copper/silver tolerance (Tables S3 and S4). Together, these results strongly support the importance of these methionine pairs/clusters for the methionine-residue ion relay channel. Also the charged residues D405, E939 and K984 are expected to be important for the proton-relay network, and these in turn were replaced with alanines (D405A, E939A and K984A). Indeed, cells expressing this mutant were unable to tolerate copper and silver, demonstrating that these charged residues are essential for the transport function.
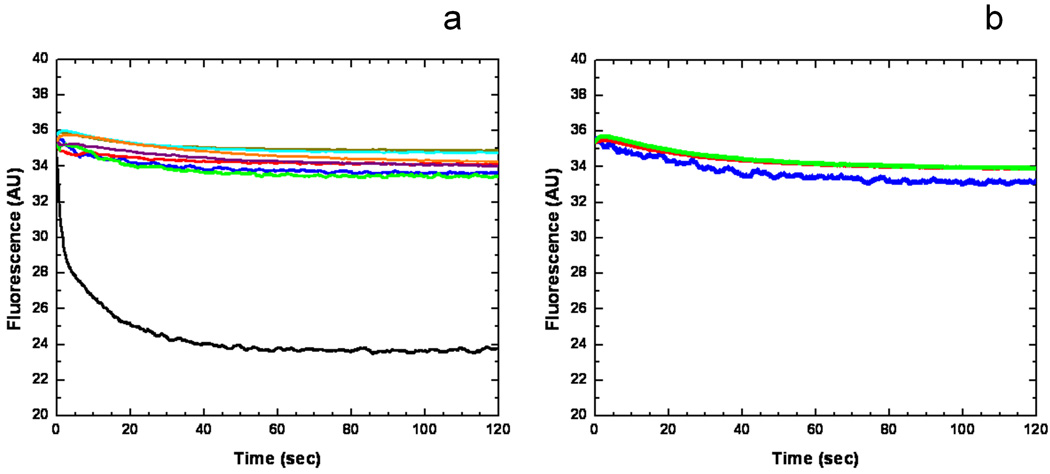
To investigate whether CusA can transport metal ions from the cytoplasm, the purified CusA protein was reconstituted into liposomes containing the fluorescent indicator Phen Green SK (PGSK) in the intravesicular space (Fig. S9). A stopped-flow transport assay was employed to determin whether these proteoliposomes can capture metal ions from the extravesicular medium. When Ag+ ions were added into the extravesicular medium, the quenching of the fluorescence signal was detected, suggesting the uptake of Ag+ into the intravesicular space (Fig. 5a).
Figure 5.
Stopped-flow transport assay of reconstituted CusA with extravesicular Ag+ ion. (a) Mutants of the methionine-residue relay network. The decrease in fluorescence signal of PGSK mediated by proteoliposomes of wild-type CusA indicates the transport of Ag+ across the membrane. The stopped-flow traces are the cumulative average of four successive recordings (wild-type CusA, black curve; M486I, green curve; M391I, brown curve; M573I, red curve; M623I, purple curve; M672I, orange curve; M755I, cyan curve; control liposome, blue curve). (b) Mutants of the proton-relay network. The traces are the cumulative average of four successive recordings (D405A, blue curve; E939A, red curve; K984A, green curve).
In addition, we investigated the methionine residues that were shown to be important for copper/silver tolerances with the above transport assay. We reconstituted the purified CusA mutants, M391I, M486I, M573I, M623I, M672I and M755I, into liposomes encapsulated with the same indicator. Surprisingly, these mutants do not take up Ag+ from the extravesicular medium of the proteoliposomes (Fig. 5a), suggesting that the mutant transporters are unable to actively transport Ag+ across the membrane.
The importance of the charged residues D405, E939 and K984 for proton translocation was investigated further. When reconstituted into liposomes, mutant transporters (D405A, E939A and K984A) do not take up Ag+ into the intravesicular space (Fig. 5b), thus confirming the importance of D405, E939 and K984.
It is likely that CusA operates through an alternating-access mechanism.7–9 The dynamics simulation of the trimeric CusA pump using elastic network model27 indeed suggests that CusA functions through three coupled motions in which the periplasmic cleft formed by sub-domains PC1 and PC2 alternately open and close (Figs. S10 and S11).
Our CusA structures suggest that this pump is able to take up metals from both the periplasm and cytoplasm. In fact, it has been demonstrated that both the CzcA28 and AcrD29 pumps are capable of transporting substrates from the cytosol. Thus, we hypothesize that CusA can pick up metal ions from both the cytoplasm and periplasm and utilizes methionine pairs/clusters to export Cu(I)/Ag(I). The periplasmic cleft of CusA presumably remains closed when there is no Cu(I)/Ag(I). In the presence of Cu(I)/Ag(I), the periplasmic cleft opens. This occurs by the horizontal helix moving to bind the ion tightly and TM8 then being freed to move and open the cleft. Metal ions could bind strongly in the open form to the three-methionine binding site directly through the periplasmic cleft or via the methionine pairs within the transmembrane region (Fig. 4). Thus, transport of a metal ion within the membrane is likely to involve a stepwise process that shuttles the metal ion from one methionine pair to another. The locations of these methionine pairs/clusters allow us to depict a direct pathway for metal transport. The metal ion bound at the three-methionine binding site of the periplasmic cleft could be transferred to the nearest methionine pair, M277–M755, following which the ion could then be released into the central funnel to reach the CusC channel for final extrusion.
Methods Summary
Protein preparation
The full-length CusA membrane protein containing a 6xHis tag at the N-terminus was overproduced in E. coli BL21(DE3)ΔacrB cells possessing pET15bΩcusA.19 Cells were grown in LB medium with 100 µg/ml ampicillin at 37°C. When the OD600 reached 0.5, the culture was induced with 1 mM IPTG, and cells were harvested within 3 h. The collected bacteria were resuspended in buffer containing 100 mM sodium phosphate (pH 7.2), 10 % glycerol, 1 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM phenylmethanesulfonyl fluoride (PMSF), and then disrupted with a French pressure cell. The membrane fraction was collected and washed twice with buffer containing 20 mM sodium phosphate (pH 7.2), 2 M KCl, 10 % glycerol, 1 mM EDTA and 1 mM PMSF, and once with 20 mM HEPES-NaOH buffer (pH 7.5) containing 1 mM PMSF. The protein was solubilized in 1 % (w/v) Cymal-6. Insoluble material was removed by ultracentrifugation at 100,000 × g. The extracted protein was purified with a Ni2+-affinity column. The purified protein was then dialyzed and concentrated to 20 mg/ml in a buffer containing 20 mM Na-HEPES (pH 7.5) and 0.05% CYMAL-6.
Protein crystallization
Crystals of the 6xHis CusA were obtained using sitting-drop vapor diffusion. A 2 µl protein solution containing 20 mg/ml CusA protein in 20 mM Na-HEPES (pH 7.5) and 0.05% (w/v) CYMAL-6 was mixed with 2 µl of reservoir solution containing 10% PEG 3350, 0.1 M Na-MES (pH 6.5), 0.4 M (NH4)2SO4, 1% JM 600 and 10% glycerol. The resultant mixture was equilibrated against 500 µl of the reservoir solution. Crystals grew to full size within a month. Cryoprotection was achieved by raising the glycerol concentration stepwise to 30% in 5% increments.
Phasing, refinements, statistics and details of the experimental procedures are provided in the Methods.
Methods
Preparation of the SeMet-CusA protein
For expression of the 6xHis SeMet-CusA protein, a 10 ml LB broth overnight culture containing E. coli BL21(DE3)ΔacrB/pET15bΩcusA cells was transferred into 120 ml of LB broth containing 100 µg/ml ampicillin and grown at 37°C. When the OD600 value reached 1.2, cells were harvested by centrifugation at 6000 rev/min for 10 min, and then washed two times with 20 ml of M9 minimal salts solution. The cells were re-suspended in 120 ml of M9 media and then transferred into a 12 L pre-warmed M9 solution containing 100 µg/ml ampicillin. The cell culture was incubated at 37°C with shaking. When the OD600 reached 0.4, 100 mg/l of lysine, phenylalanine and threonine, 50 mg/l isoleucine, leucine and valine, and 60 mg/l of L-selenomethionine were added. The culture was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) after 15 min. Cells were then harvested within 3 h after induction. The procedures for purifying the 6xHis SeMet-CusA were identical to those used for the native protein.
Crystals of the CusA derivatives
The crystallization conditions for SeMet-CusA were the same as those used for the native CusA protein. Crystals of the gold or tantalum derivatives were prepared by incubating the crystals of apo-CusA in a solution containing 10% PEG 3350, 0.1 M Na-MES (pH 6.5), 0.4 M (NH4)2SO4, 1% JM 600, 10% glycerol, 0.05% (w/v) CYMAL-6, and 1 mM KAuCl4 or 5 mM Ta6Br12 2Br for 1.5 hours at 25°C.
The CusA-Cu(I) complex crystals were prepared by incubating crystals of apo-CusA in solution containing 10% PEG 3350, 0.1 M Na-MES (pH 6.5), 0.4 M (NH4)2SO4, 1% JM 600, 10% glycerol, 2 mM [Cu(CH3CN)4]PF6, 2 mM tris(2-carboxyethyl)phosphine (TCEP) and 0.05% (w/v) CYMAL-6 for 1 hour at 25°C. Crystals of the CusA-Ag(I) complex were prepared using a similar method in a solution containing 10% PEG 3350, 0.1 M Na-MES (pH 6.5), 0.4 M (NH4)2SO4, 10% glycerol, 1 mM AgNO3 and 0.05% (w/v) CYMAL-6 for 1 hour at 25°C.
Data collection, structural determination and refinement
All diffraction data were collected at 100K at the beamline 24ID-C located at the Advanced Photon Source, using an ADSC Quantum 315 CCD-based detector. Diffraction data were processed using DENZO and scaled using SCALEPACK.30 The crystals belong to space group R32 (Tables S1 and S2). Based on the molecular weight of CusA (115.72 kDa), a single molecule per asymmetric unit with a solvent content of 67.5% is expected. Two heavy-atom derivatives (gold and Ta6Br12 cluster) and the selenomethionyl-substituted (SeMet) crystal were isomorphous with the native crystal (Table S1). Ten heavy-atom sites for the gold derivative were identified using SHELXC and SHELXD31 as implemented in the HKL2MAP package.32 These heavy-atom sites were refined using the program MLPHARE.33,34 The resulting phases were subjected to density modification by RESOLVE35 using the native structure factor amplitudes. Density modified phases were good enough to allow us to visualize the secondary structure features of the molecule. These phases were then used to locate three Ta6 clusters and 31 selenium sites using the corresponding data sets. The final experimental phases were generated using 10 Au, three Ta6 and 33 Se sites. The Ta6 sites were treated as super-atoms containing all six tantalum atoms at a given site. These phases were then subjected to density modification and phase extension to 3.52 Å-resolution using the program RESOLVE.35 The resulting phases were of excellent quality that enabled tracing of most of the molecule. The full-length CusA protein includes 34 methionine residues. The SeMet data not only augmented the experimental phases, but also helped in tracing the molecules by anomalous difference Fourier maps where we could ascertain the proper registry of SeMet residues. After tracing the initial model manually using the program Coot,36 the model was refined against the native data at 3.52 Å-resolution using TLS refinement techniques adopting a single TLS body as implemented in PHENIX37 leaving 5% of reflections in the Free-R set. Iterations of refinement using PHENIX37 and CNS38 and model building in Coot36 lead to the current model, which consists of 1,025 residues with excellent geometrical characteristics (Table S1).
While the structure of the CusA-Cu(I) complex could be determined using molecular replacement, the structure resulted in a relatively high refinement R-factor of 48.7%. Upon inspection of the electron density maps, it was found that several regions, especially the PC2 sub-domain, of CusA-Cu(I) have undergone significant conformational changes as discussed in the main text. To aid the modeling of these conformational changes in the least biased way, SAD phasing using the program PHASER39 was employed to obtain experimental phases in addition to the phases from the structural model of apo-CusA which had sub-domains PC2 and PN1 removed. Phases were then improved using RESOLVE.35 Even though the data resolution is low (4.10 Å), the phases were of sufficient quality to permit modeling of the conformational change (Fig. S3) using the program COOT.35 At a later stage, higher resolution (3.88 Å) data of the CusA-Cu(I) were collected (Table S2). To reduce radiation damage, the data was collected away from the copper absorption edge, at 1.033 Å, where Cu still has ~2.4 anomalous electrons. The model was refined using PHENIX37 and CNS.38
Since the X-ray absorption edge of Ag was not reachable at the synchrotron beamline (L-edges below 4 KeV and K-edge above 25 KeV), the data were collected at the copper edge where Ag has an anomalous contribution of ~3.5e−. The structure of the CusA-Ag(I) complex was phased using molecular replacement by using CusA-Cu(I) as the search model. Structural refinement was then performed using PHENIX37 and CNS38 by refining the model against the 4.35 Å-resolution data from the CusA-Ag(I) crystal (Table S2).
Docking structures of CusB onto CusA
Initial rigid-body docking of the crystal structures of CusA and CusB did not reveal any feasible complexes that satisfied the CusA-K150/CusB-K95 crosslinking constraint,19 indicating that docking most likely require a conformational change in one or both molecules. As previously mentioned it has been hypothesized that the PN2 and PC1 sub-domains may move to accommodate CusB. For CusA, the normal modes from the Anisotropic-Network-Models (ANMs)27 were calculated, but did not reveal any appropriate structures. For CusB, ANM revealed significant hinge motions between Domains 1 and 2. And, following this lead for a dominant transition, these two domains were docked individually onto CusA. Steered molecular dynamics simulations were then used to drive the CusB into a docked form agreeing with the individually docked fragments, thus allowing for the flexibility of the PC1, PN2 and TM2 domains of CusA.
Data-driven docking of Domains 1 and 2 of CusB
Rigid-body docking was performed using the 3D-dock software suite,40 which utilizes the Katchalski-Katzir algorithm.41 For all runs, 10,000 conformations were generated and filtered using the following data-driven constraints: transmembrane helices were masked from docking interactions; and side-chain amines of CusA-K150 and CusB-K95 were within 25 Å;19 CusB was oriented so that its domains were free to interact with CusC and its N-terminus was directed towards the inner membrane.
Steered Molecular Dynamics and Energy minimization
An initial conformation was calculated by aligning CusB with the two individually docked fragments. To relax CusB from this conformation, we utilized steered molecular dynamics to guide the center of mass of domains 1 and 2 into the corresponding docked position. Energy minimization was employed to alleviate steric clashes. During the simulation CusA was taken to be rigid with the exception of TM2, PN2, and PC1, which make up most of the CusB binding region. All calculations were performed using NAMD42 and the CHARMM27 force field.43
Double knocked-out strain and susceptibility assays
The double knocked-out E. coli strain BL21(DE3)ΔcueOΔcusA was produced from the BL21(DE3) strain using an RED disruption system as described by Datsenko and Wanner.44 The ΔcueO∷kan cassette, which was used to replace the chromosomal cueO gene, was produced by PCR, and then introduced into pKD46/BL21(DE3) by electroporation. The knocked-out BL21(DE3)ΔcueO∷kan strain was selected on LB plate containing 30 µg/ml kanamycin, and verified by PCR. The kanamycin resistant gene was then released to generate the BL21(DE3)ΔcueO knocked-out strain. The deletion of cusA from BL21(DE3)ΔcueO was performed with procedures similar to those described above to generate the final BL21(DE3)ΔcueOΔcusA double knocked-out strain.
The susceptibility to copper of E. coli BL21(DE3)ΔcueOΔcusA harboring pET15bΩcusA expressing the wild-type or mutant transporters, or the pET15b empty vector was tested on agar plates. Cells were grown in Luria Broth (LB) medium with 100 µg/ml ampicillin at 37 °C. When the OD600 reached 0.5, the cultures were induced with 0.5 mM IPTG and harvested within two hours after induction. The minimum growth inhibitory concentrations (MICs) to copper of E. coli BL21(DE3)ΔcueOΔcusA (incoculum, 500 cells/ml) harboring these vectors were then determined using LB agar containing 50 µg/ml ampicillin, 0.1 mM IPTG and different concentrations of CuSO4 (0.25 to 2.75 mM in steps of 0.25 mM).
For the susceptibility to silver of E. coli BL21(DE3)ΔcueOΔcusA harboring these vectors, cells were grown in Mueller Hinton (MH) medium containing 100 µg/ml ampicillin with the same procedures. The MICs to silver of E. coli BL21(DE3)ΔcueOΔcusA (incoculum, 500 cells/ml) harboring these vectors were then performed in MH agar containing 50 µg/ml ampicillin, 0.1 mM IPTG and different concentrations of AgNO3 (5.0 to 50.0 µM in steps of 2.5 µM). It should be noted that MH medium does not contain chloride to complex the silver and that the MIC determination for silver was done in the dark. The expression level of each mutant in BL21(DE3)ΔcueOΔcusA is similar to that of the wild-type transporter as indicated by Western blot analysis (Fig. S12). Bacterial growth in the LB or MH agar was recorded after 24 h of incubation at 37°C. Each assay was repeated at least four times to ensure the reproducibility of the results.
Reconstitution and stop-flow transport assay
The purified wild-type CusA, M391I, M486I, M573I, M623I, M672I, M722I, D405A, E939A or K984A was reconstituted into liposomes made of E. coli polar lipid and egg-yolk phosphatidylcholine (Avanti Polar Lipids) in a molar ratio of 3:1. Briefly, 50 µg of the purified protein (wild-type CusA or its mutant) was added to 4.5 mg of lipids dispersed in the reconstitution buffer containing 20 mM HEPES-KOH, pH 6.6. The protein samples were completely incorporated into liposomes as judged by using 10% SDS-PAGE stained with Coomassie Brilliant Blue. Control liposomes were prepared following the same procedure without adding protein. 200 µM fluorescence indicator Phen Green SK (PGSK) (Invitrogen) was encapsulated by two freeze-thaw cycles, followed by gel filtration to remove the untrapped dye. Transport experiments were performed at 25°C on a stopped-flow apparatus (Hi-Tech Scientific) connected to a spectrofluorometer (PerkinElmer LS55). Proteoliposomes and a transport assay buffer (20 mM HEPES-KOH pH 7.0 and 1 mM AgNO3) were loaded into two separate syringes of equal volume. Transport reactions were initiated by pushing 400 µl fresh reactants at a 1:1 ratio through the 90 µl mixing cell at a flow rate of 2 ml/s. Stopped-flow traces are the cumulative average of four successive recordings at 530 nm with the excitation wavelength at 480 nm.
For experiments in the absence of a pH gradient between the inside and outside of proteoliposomes containing the wild-type CusA transporter, the reconstitution buffer (20 mM HEPES-KOH, pH 6.6) and transport assay buffer (20 mM HEPES-KOH pH 6.6 and 1 mM AgNO3) were used to adjust the pH at 6.6 (intravesicular pH = extravesicular pH = 6.6). We also used the reconstitution buffer (20 mM HEPES-KOH, pH 7.0) and transport assay buffer (20 mM HEPES-KOH pH 7.0 and 1 mM AgNO3) to maintain the intravesicular pH and extravesicular pH at 7.0 for these experiments (Fig. S13). In addition, we performed the stopped-flow transport assay for the wild-type pump with a reverse pH gradient (intravesicular pH = 7.0 and extravesicular pH = 6.6) (Fig. S13). In this experiment, the reconstitution buffer (20 mM HEPES-KOH, pH 7.0) and transport assay buffer (20 mM HEPES-KOH pH 6.6 and 1 mM AgNO3) were used. The procedures for these transport experiments were the same as described above.
Dynamics Simulations
The observed conformational changes between the Cu(I) bound and the apo forms, displayed in Fig. 2, are shown to be intrinsically favored by the new structure through the use of normal mode analysis (Fig. S10). A coarse-grained elastic network model27 was investigated that is comprised of the Cα atom coordinates and a portion of coarse-grained lipid bilayer between the trans-membrane domains. The opening and closing of the space between PC1 and PC2 coupled with a shift of TM8 is reproduced by this simple model (Fig. S11). The model does not show simultaneous opening and closing of all three periplasmic heavy metal entrance sites, but rather has a more complex opening/closing motion in which each pair of two adjacent groups is alternately open and closed. All three combinations of these coupled motions are obtained by using just the two low energy motions.
Supplementary Material
Acknowledgements
This work is supported by NIH Grants GM 074027 (E.W.Y.), GM 086431 (E.W.Y.), GM 081680 (R.L.J.) and GM 072014 (R.L.J.). We thank Mathew D. Routh for critical reading of the manuscript. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by NIH award RR-15301 from the National Center for Research Resources. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This paper is dedicated to the memory of Prof. Victor S. Lin, a colleague and a friend.
Footnotes
Author Contributions
F.L., C.-C.S., and E.W.Y. designed research; F.L., and C.-C.S. performed experiments; M.T.Z., and S.E.B. performed simulations; C.-C.S., F.L., K.R.R., and E.W.Y. performed model building and refinement; and F.L., C.-C.S., R.L.J., and E.W.Y. wrote the paper.
Author Information
Atomic coordinates and structure factors have been deposited with the Protein Data Bank as codes 3KO7 (apo-CusA), 3KSS (CusA-Cu(I)) and 3KSO (CusA-Ag(I)).
References
- 1.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development protein. J. Mol. Microbiol. Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 2.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 3.Franke S, Grass G, Nies DH. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiol. 2001;147:965–972. doi: 10.1099/00221287-147-4-965. [DOI] [PubMed] [Google Scholar]
- 4.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 6.Yu EW, McDermott G, Zgruskaya HI, Nikaido H, Koshland DE., Jr Structural basis of multiple drug binding capacity of the AcrB multidrug efflux pump. Science. 2003;300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 7.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 8.Seeger MA, Schiefner A, Eicher T, Verrey F, Dietrichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 9.Sennhauser G, Amstutz P, Briand C, Storchengegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic-drug binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su C-C, Li M, Gu R, Takatsuka Y, McDermott G, Nikaido H, Yu EW. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 2006;188:7290–7296. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sennhauser G, Bukowska MA, Briand C, Grütter MG. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 2009;389:134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 14.Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwag T, Yoneyama H, Narita S, Nakagawa A, Nakae T. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:52816–52819. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 15.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA. 2004;101:9994–9999. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akama H, Matsuura T, Kashiwag S, Yoneyama H, Narita S, Tsukihara T, Nakagawa A, Nakae T. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 18.Symmons M, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. USA. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su C-C, Yang F, Long F, Reyon D, Routh MD, Kuo DW, Mokhtari AK, Van Ornam JD, Rabe KL, Hoy JA, Lee YJ, Rajashankar KR, Yu EW. Crystal structure of the membrane fusion protein CusB from Escherichia coli. J. Mol. Biol. 2009;393:342–355. doi: 10.1016/j.jmb.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H, Thiele DJ. Identification of a novel high affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 2001;276:20529–20535. doi: 10.1074/jbc.M102004200. [DOI] [PubMed] [Google Scholar]
- 21.Jiang J, Nadas IA, Kim MA, Franz KJ. A Mets motif peptide found in copper transport proteins selectively binds Cu(I) with methionine-only coordination. Inorg. Chem. 2005;44:9787–9794. doi: 10.1021/ic051180m. [DOI] [PubMed] [Google Scholar]
- 22.Banci L, Bertini I, Cantini F, Felli IC, Gonnelli L, Hadjiliadis N, Pierattelli R, Rosato A, Voulgaris P. The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nature Chem. Biol. 2006;2:367–368. doi: 10.1038/nchembio797. [DOI] [PubMed] [Google Scholar]
- 23.Xue Y, Davis AV, Balakrishnan G, Stasser JP, Staehlin BM, Focia P, Spiro TG, Penner-Hahn JE, O'Halloran TV. Cu(I) recognition via cation- and methionine interactions in CusF. Nature Chem. Biol. 2008;4:107–109. doi: 10.1038/nchembio.2007.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loftin IR, Franke S, Blackburn NJ, McEvoy MM. Unusual Cu(I)/Ag(I) coordination of Escherichia coli CusF as revealed by atomic resolution crystallography and x-ray absorption spectroscopy. Protein Sci. 2007;16:2287–2293. doi: 10.1110/ps.073021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O'Halloran TV, Mondragón A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 26.Arnesano F, Banci L, Bertini I, Huffman DL, O'Halloran TV. Solution structure of the Cu(I) and apo-forms of the yeast metallochaperone, Atx1. Biochemistry. 2001;40:1528–1539. doi: 10.1021/bi0014711. [DOI] [PubMed] [Google Scholar]
- 27.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldberg M, Pribyl T, Juhuke S, Nies DH. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J. Biol. Chem. 1999;274:26065–26070. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 29.Aires JR, Nikaido H. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 2005;187:1923–1929. doi: 10.1128/JB.187.6.1923-1929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor M. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 32.Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- 33.Otwinowski Z. MLPHARE, CCP4 Proc. 80. Warrington, UK: Daresbury Laboratory; 1991. [Google Scholar]
- 34.Collaborative Computational Project No. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Terwilliger TC. Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Cryst. 2001;D57:1755–1762. doi: 10.1107/S0907444901013737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCroy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 38.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabb HA, Jackson RM, Sternberg MJE. Modelling protein docking using shape complementarity, electrostatics, and biochemical information. J. Mol Biol. 1997;272:106–120. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 41.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aalo C, Wodak SJ. Molecular surface recognition: Determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Nat. Acad. Sci. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feller SE, MacKerell AD., Jr An improved empirical potential energy for molecular simulations of phospholipids. J. Phys. Chem. B. 2000;104:7510–7515. [Google Scholar]
- 44.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Nat. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.