Abstract
Producing polymeric or hybrid microfluidic devices operating at high temperatures with reduced or no water evaporation is a challenge for many on-chip applications including polymerase chain reaction (PCR). We study sample evaporation in polymeric and hybrid devices, realized by glass microchannels for avoiding water diffusion toward the elastomer used for chip fabrication. The method dramatically reduces water evaporation in PCR devices that are found to exhibit optimal stability and effective operation under oscillating-flow. This approach maintains the flexibility, ease of fabrication, and low cost of disposable chips, and can be extended to other high-temperature microfluidic biochemical reactors.
INTRODUCTION
The use of microfluidic-based integrated devices for biological, pharmaceutical, and medical applications traces back in the 1990s with the development of the first micrototal analysis system.1, 2 Thenceforth, several microfluidic devices for polymerase chain reaction (PCR) have been produced as miniaturized, compact alternative to conventional equipments and technologies, offering advantages in terms of reduced assay time, low consumption of biological samples, high portability and integration of multiple processing modules.3, 4, 5
PCR chips are mainly classified on the base of their operation in stationary or dynamic mode.4 Stationary devices are characterized by the use of single2, 6, 7, 8 or multiple reaction chambers or microchannel regions.9, 10, 11, 12 The PCR mixture is injected into microfabricated wells, and the system (including the biological sample) is heated and cooled according to a proper thermal cycling profile. In this architecture, the large involved mass generally determines high thermal inertia, with consequent long cycling times. Instead, in dynamic devices the PCR mixture flows continuously into a microchannel through regions at fixed temperature.13, 14 Only the sample mass undergoes repeated heating and cooling, thus reducing the relevant thermal inertia. The time between subsequent temperature transitions is therefore related to the sample flow rate and thermal equilibrium time, enabling shorter amplification processing.
Continuous-flow, dynamic PCR chips are realized by different designs, including oscillatory,15, 16, 17, 18 closed-loop,19, 20, 21 and fixed-loop devices,13, 14, 22, 23, 24 and by different fluidic geometries (linear, serpentines, rings, and spiral capillaries5). In the fixed-loop architecture, the microchannel length determines the final number of temperature cycles, making hardly changeable the PCR thermal profile. In oscillatory and closed-loop devices, the biological sample is moved back and forth (oscillatory chips) or around (closed-loop chips) the fluidic circuit, crossing the regions held at different temperatures. These devices are therefore generally more flexible in terms of adjustability of number of thermal cycles.
However, a few main issues need to be addressed in these chips, primarily related to the small sample volumes and large surface-to-volume ratios. In particular, the evaporation of sample solution during thermal cycles4 is especially significant during the PCR denaturation step, when the system temperature approaches 100 °C. This would result in loss of reagents and changes in concentration that are particularly critical for volumes within or below the microliter range. To date, sample evaporation still remains an important limitation to the applicability of many microfluidic devices using very small volumes, especially for plastic chips. Different proposed strategies include the use of valves11 and other elements for increasing the gas pressure around the sample.16
In addition, the inhibition of the amplification reaction, due to the interaction between the PCR mixture and microchannel surfaces,4 is more evident in systems working in dynamic mode. In fact, the PCR solution, flowing continuously through the reaction path, increases the overall surface and time of contact with the capillary walls. Coatings are realized onto capillary walls by means of silicon oxide deposition or silanization treatments,22, 25 otherwise by adding several passivation agents to the PCR mix solution, such as adjuvant-bovine serum albumin,26 polyethylene glycol,27 and polyvinylpyrrolidone.28
Here, we investigate the water solution evaporation in PCR microfluidic chips based on oscillating-flow, realized by polydimethylsiloxane (PDMS) and glass, and integrating microfabricated heaters and temperature sensors. We focus on avoiding sample evaporation during the PCR process by the choice of the device materials. In all-PDMS chips employing a continuous phase of oil, water volume reductions of about 60% in 120 min at 20 °C and of 100% in less than 4 min at 95 °C are measured because of evaporation. Glass capillaries, introduced as fluidic core of the PDMS device, allow us to dramatically reduce the evaporation making the elastomer material inaccessible to water diffusion in the device, while maintaining the flexibility, ease of fabrication, and low cost of disposable chips. Although PCR in glass capillaries or in oscillating conditions has been reported before, the here proposed combination of a glass core and a flexible, polymeric external architecture is particularly useful for the integration with external boards embedding microfabricated elements. Indeed, the PDMS embedment guarantees effective thermal exchange and structural flexibility, thus being easily coupled to heaters. The microfluidic element avoiding evaporation is therefore disposable, whereas boards and electrodes are used hundreds of times without any damage. Surface passivation to enable effective on-chip PCR is also performed, and the operation of hybrid devices is tested by a prototype amplification reaction on a 500-nucleotide target sequence. This approach guarantees optimal stability and effective operation of the device under oscillating-flow, and can be extended to other microfluidic, biochemical reactors working well above room-temperature.
EXPERIMENTAL
Our chip platform is made of (i) a board containing three independent Ti∕Pt heaters, with integrated temperature sensors, and (ii) three hydraulically connected reaction regions, placed in correspondence to the heaters, for sample amplification. The PCR mixture is moved into the microfluidic network back and forth in an oscillating way, by using external pumping systems. We produce both devices whose fluidic path is made by PDMS, and other chips in which the microchannel is a glass capillary.
Materials
The PDMS elastomer (Sylgard 184 Silicone Elastomer Kit) is purchased by Dow Corning (Midland, MI). The PDMS prepolymer is obtained by mixing the base and curing agent in 20:1 and 5:1 w∕w ratios. The negative-tone resist SU-8 2100, with OmniCoat primer and developer, from MicroChem Corp. (Newton, MA), is used for the production of master templates, and the positive-tone AZ5214E, with its primer and developer solution, from Clariant Corporation AZ Electronic Materials (Wiesbaden, Germany), are used for fabricating microheaters and temperature sensors. Round borosilicate capillaries, with internal diameter (i.d.) of 0.90 mm and external diameter (o.d.) of 1.10 mm, are purchased by Optics Planet Inc. (Northbrook, IL). Capillary tubing connections (Tygon), with i.d.=1 mm and o.d.=1.75 mm, are from Norton Performance Plastics (Akron, OH). PDMS oils with viscosities (η) of 350 and 500 cP, and the silanization solution, are purchased from Sigma Aldrich (St. Louis, MO). Disks of Ti and Pt for electrode fabrication are provided by Umicore Group (Brussels, Belgium). Primers for PCR are purchased from NBS Biotech INC (Milan, Italy), and bacteriophage λ DNA and deoxynucleotide triphosphates (dNTPs) are purchased from New England BioLabs (Ipswich, MA) and Euroclone (Milan, Italy), respectively. The Taq polymerase with the reaction buffer, including magnesium solution, is integrated in the EuroTaq mix (Euroclone).
Equipment
Photolithography is performed with an EVG620 mask aligner (EVGroup, St. Florian am Inn, Austria). Electron-beam evaporation (Temescal STIH-270-1CK, Boc Edwards, Livermore, CA) is used to deposit Ti and Pt for heaters and temperature sensors. A Keithley 2700 multimeter data acquisition system (Cleveland, OH) is used for the temperature versus voltage (T-Vb), and resistance versus voltage (R-Vb) calibrations of heaters and sensors. Dual dc power supplies (Iso-Tech IPS-3303, RS Components, Milan, Italy) are employed as voltage generators for heating. The temperature of the liquid phase into reaction regions of the chip is also measured by a Watlow mineral insulated thermocouple (St. Louis, MO), with external diameter of 0.25 mm. Two external microdiaphragm pumping systems (ThinXXS, Zweibrücken, Germany), working in infusion and withdraw mode, are employed for liquid injection and manipulation. The motion of the liquid into the microfluidic network is visualized by a Leica MZ16FA stereomicroscope (Wetzlar, Germany), equipped with a DFC490 camera. The Mastercycler ep gradient thermal cycler (Eppendorf, Wesseling-Berzdorf, Germany) is employed to control amplification reactions. The visualization of agarose gels is carried out by an UV transilluminator (EuroClone, Italy) equipped with a Canon digital camera.
Heaters and temperature sensors
Electrodes of Ti∕Pt (distanced by 4 mm) are fabricated on a 1 mm thick glass substrate by photolithography and subsequent metal evaporation and lift-off. Each electrode (area of 2.25 mm2) includes a heater, with 50 μm wide periodic lines, and a central temperature sensor having 10 μm wide periodic lines. For optical lithography, a film of AZ5214E:AZ EBR (1:1) is deposited on glass that provides thermal insulation between adjacent high-temperature regions during the device operation. This prevents heat loss and thermal interferences, allowing robust constant-temperature control. The pattern is exposed to UV light (wavelength of 295 nm) for 5 s at 350 W, and then developed for 60 s. A 20 nm thick Ti film and a 200 nm thick Pt film are deposited onto the photoresist pattern at an evaporation rate of ≈1 Å∕s. Lift-off in acetone completes the fabrication of heaters and sensors, which are then calibrated by their T-Vb and R-Vb characteristics, respectively. The heaters are found to work in a range of temperature between 20 and 150 °C. Each sensor is used to detect the temperature of the glass substrate in order to provide feedback to the input Vb value, guaranteeing constant-temperature control.
PDMS chips
All-plastic chips are realized by replicating master templates into PDMS by soft lithography [Fig. 1a].29 We first perform optical lithography using a photomask with two circular reservoirs (2 mm in diameter) for liquid inlet and outlet, connected to three elliptical reaction chambers (each having an area of 4.7 mm2) through a 500 μm wide channel. The total length of the network is 2.3 cm. After cleaning and drying (95 °C for 3 min) a Si substrate, we spin-cast a layer of OmniCoat (3000 rpm for 40 s), curing at 200 °C for 1 min. Then, a 150 μm thick layer of SU-8 2100 (4000 rpm for 40 s) is deposited, carrying out a soft bake process for 5 min at 65 °C and for 20 min at 95 °C guaranteeing complete evaporation of the solvent. The resist is exposed for 30 s to UV through the photomask, baked for 5 min at 65 °C, and for 15 min at 95 °C, developed for 10 min, and again baked for 10 min at 150 °C.
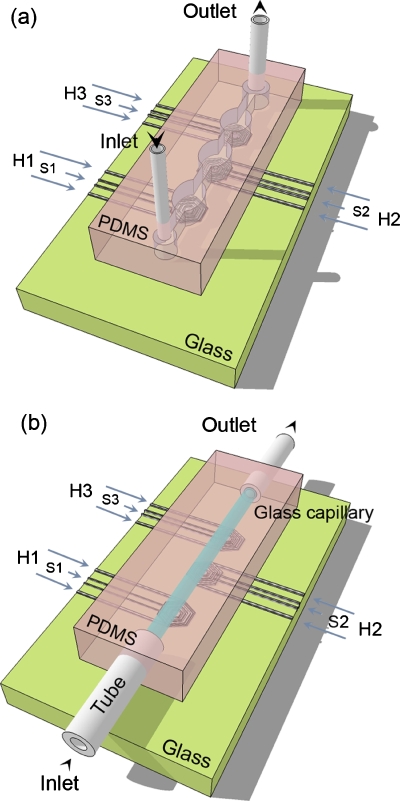
Figure 1.
Scheme of the (a) all-PDMS and (b) PDMS-glass devices with microfabricated heaters (H1–H3) and sensors (S1–S3). H1, H2, and H3 work at 66 °C (annealing temperature), 72 °C (extension), and 95 °C (denaturation), respectively. Features not in scale.
The patterns on the masters are transferred by replica molding,29 using a relative concentration of PDMS base and curing agent of 5:1 (w∕w). The elastomeric elements contain the network of microchannels and reaction chambers, and are used as structured layer for the final PDMS device. The second layer consists of a flat PDMS substrate, obtained from a 20:1 w∕w solution of base and curing agent. The polymerization of both the separated elements is carried out at 75 °C for 20 min. Afterward, calibrated holes are generated into the structured layer in correspondence with the circular reservoirs, to have stable and watertight connections between the PDMS chip and external pumping systems. Then, the structured element is placed in conformal contact with the flat PDMS layer, and cured at 75 °C for 2 h. The relative excess of base in one layer and of curing agent in the other promotes the cross-linking at the interface, and the irreversible bonding of the complete elastomeric device.
Hybrid PDMS-glass chips
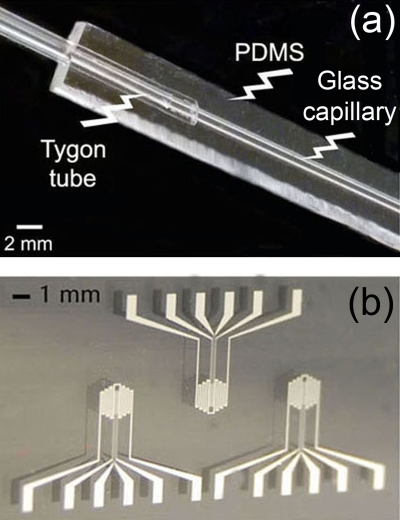
Figure 1b displays a schematics of our hybrid PDMS-glass device. Before chip assembly, the internal walls of borosilicate capillaries are treated with 2% v∕v of dimethyldichlorosilane in 1,1,1-trichloroethane.22, 30 Capillaries are filled with 20 μl of solution and placed on an oscillating shaker for 1.5 h in air. After solution evaporation, capillaries are dried at 60 °C for 30 min and connected to two plastic tubes as fluid inlet and outlet [Fig. 2a]. Then, prepolymeric liquid PDMS (20:1 w∕w base:curing agent) is poured onto the capillary and polymerized at 75 °C for 15 min, thus blocking the glass capillary and external connections into the elastomer. This stage is particularly useful for the subsequent integration with the board embedding the Ti∕Pt heaters [Fig. 2b]. The thickness of the PDMS layer underneath the glass capillary and in contact with the electrodes is below 500 μm, guaranteeing effective thermal exchange between the heaters and the microchannel. Moreover, the microfluidic element is flexible and able to be positioned by conformal contact since PDMS allows a reversible adhesion of the fluidic element to the glass substrate embedding the electrodes, without mutual misalignments and damages to the metal patterns. Typically, the microfluidic element is disposable, whereas the electrodes are used for more than 30 PCR experiments.
Figure 2.
(a) Photograph of the used glass capillary, with connections to plastic tubes, embedded into PDMS matrix. (b) Microfabricated Ti/Pt heaters and sensors.
PCR
A sequence of a 500-nucleotide segment (from 7131 to 7630) of the 48.5 kb bacteriophage λ genome is used as target DNA, and amplified with the forward primer 5′-GATGAGTTCGTGTCCGTACAACTGG-3′ and reverse primer 5′-GGTTATCGAAATCAGCCACAGCGCC-3′. The PCR mixture (1 μl) employed contains ultrapure water, reaction buffer [160 mM (NH4)2SO4, 670 mM tris∙HCl (pH 8.8), 0.1% v∕v Tween-20], 2 mM MgCl2, 200 μM of each dNTP, 1 μM of each primer, 0.02 unit of TaqDNA polymerase, and 0.02 μg of bacteriophage λ DNA. The thermal cycling profile, carried out both by a standard thermocycler as control and by the microfluidic chips, is as follows: (i) hot start at 95 °C for 2 min, (ii) 20 cycles, each composed by 30 s at 95 °C, 30 s at 66 °C, and 40 s at 72 °C, and (iii) final extension at 72 °C for 10 min. The PCR products are analyzed by electrophoresis on 1% agarose gel after ethidium bromide staining, and visualized by UV transillumination.
RESULTS AND DISCUSSION
Study of evaporation
The most used elastomer in microfluidics is PDMS due to its high flexibility, easy and low cost manufacturing, optical transparency, and biocompatibility. However, because of its large free volume and porosity, it allows significant vapor permeability,31 which may lead to concentration changes and ineffective DNA amplification in PCR applications.
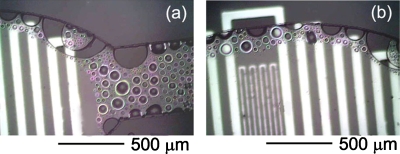
The contraction of a water volume because of losses due to thermal agitation can be described as a diffusional process, in which the distance traveled by molecules over time (t) is given by the length, .31 The diffusion coefficient, D, depends on temperature according to an Arrhenius behavior, D=D0 exp[−EA∕(RT)],32 where D0 is a pre-exponential factor, EA is the activation energy of diffusion, and R is the gas constant. For water through PDMS, D=2×10−9 m2∕s at 25 °C,33 from which one estimates D of the order of 10−9 m2∕s throughout the PCR thermal cycle, given an activation energy of 14 kJ∕mol.31 Over 5400 s (90 min), which is the characteristic duration of a PCR process, this results in a water diffusion length above 3 mm, i.e., much larger than the thickness of the PDMS layers used to fabricate our devices. In addition to such vapor escape from the device, other water molecules are directly adsorbed by the polymer (0.38% w∕w).31, 34 Overall, we can estimate that at least a volume of about 0.35 μl of water would be lost in 5400 s during the PCR thermal cycles, corresponding to a rate of about 6×10−5 μl∕s. Water loss to this extent severely limits the applicability of PDMS chips to PCR and to other applications using heated water solutions. The evaporation of water at 95 °C in our fully PDMS device is shown in Fig. 3. The process onset is revealed by air bubbles forming near the edges of the reaction chamber and of the microchannel, where water in contact with three walls of PDMS finds favorite paths to escape from the chip. The evaporation front proceeds then to the central regions of the fluidic network.
Figure 3.
[(a) and (b)] Micrographs of water evaporation at 95 °C into a microchannel and a reaction chamber of a fully PDMS device.
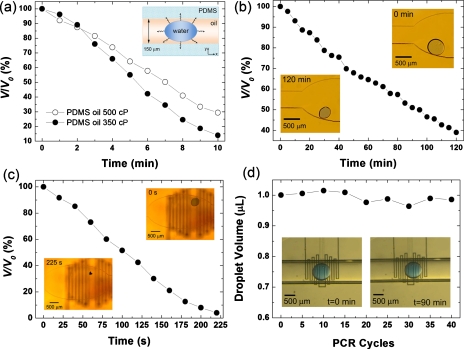
In order to quantify and reduce sample evaporation, we employ a two-phase liquid system, namely, a droplet of water-based biological solution in a continuous phase of silicone PDMS oil. We test two oils of different viscosities (η1=350 cP and η2=500 cP). Figure 4a shows the relative contraction of a water droplet of initial volume, V0=1 μl, due to evaporation at 95 °C. For the oil with low and high viscosity, the droplet volume (V) is reduced by about 85% and 70%, respectively, after 10 min. Being the chamber volume ≈1 μl and height=150 μm, one estimates that water fills the chamber almost completely, and that an oil film thickness of ≈1 μm can be present between the water-oil interface and the top PDMS layer of the device [inset of Fig. 4a]. Water molecules can diffuse into the oil phase and reach PDMS, through which diffusion and evaporation proceed further. The use of oils with higher η only weakly limits the evaporation, highlighting a weak dependence of water diffusion on the oil viscosity. The evaporation effects are even more evident for smaller droplets (V0=30 nl). The observed relative contraction at 20 and 95 °C is reported in Figs. 4b, 4c. We find a volume reduction of about 60% in 120 min at 20 °C, and an almost complete sample loss in 220 s at 95 °C.
Figure 4.
(a) Experimental data of volume (V) reduction of a water droplet (initial volume, V0=1 μl) in a PDMS chip, through a continuous phase of oil with η1=350 cP (dark circles) and η2=500 cP (empty circles), at 95 °C. The solid line is a guide for the eye. Inset: schematics of water diffusion. [(b) and (c)] Volume reduction of a water droplet (V0=30 nl) in a PDMS device at 20 and 95 °C, respectively. Oil viscosity is 500 cP. Insets: micrographs of the droplet during evaporation. (d) Volume of a droplet of PCR mix (V0=1 μl) during amplification cycles in a glass-PDMS device. Insets: micrographs of a water droplet (V0=0.3 μl) at the beginning (left) and the end (right) of a complete PCR process.
Importantly, the water evaporation can be dramatically reduced, making PDMS inaccessible to diffusion, by using glass capillaries. In this configuration, we monitor the volume evolution of a 1 μl droplet of PCR mix during 90 min reactions (40 thermal cycles), finding no appreciable sample reduction [Fig. 4d]. Therefore, using glass capillaries as fluidic cores and vapor barriers, one maintains constant concentration of PCR reagents in the mix, which is crucial for accomplishing effective amplification reactions. With respect to previously proposed vapor barrier methods based on organics such as parylene35 or polyethylene,31 this approach relying on glass has the advantage of using commercially available capillaries for producing simple linear microchannel geometries, and of exploiting already developed, reproducible passivation methods for treating the inner surfaces of the fluidic core, with no need of chemical vapor deposition35 or high-temperature curing and bonding of polymer layers.31
Device operation
Before performing PCR experiments, a thermocouple is inserted into the glass capillary to measure the temperature value of the oil phase in correspondence of the heaters. In this way, we calibrate each electrode, finding that voltages of 6.0, 4.8, and 18.8 V heat the fluid above heaters 1, 2, and 3 to 66, 72, and 95 °C, respectively. In fact, the sample temperature corresponding to each heater during PCR is not only given by the underlying electrodes but also by the hot neighboring regions of the substrate by thermal convection in the liquid.
A 1 μl droplet of PCR mix is then introduced into the capillary and moved in an oscillating way into the oil phase (η=500 cP). The droplet is first placed in the region at 95 °C (heater 3 in Fig. 1) for 2 min and 30 s (hot start and first denaturation step). Subsequently, it flows in correspondence of heater 1 with a velocity of 3×10−3 m∕s. After 30 s at 66 °C (annealing step), the fluid direction is inverted and the drop is moved, at the same velocity, toward heater 2, whose temperature is fixed at 72 °C. Here the extension step longs for 40 s. Afterward, the droplet is placed again on heater 3 for 30 s, starting the second thermal cycle. The fluid direction changes after each denaturation and annealing step. After 20 cycles, a final extension process is carried out at 72 °C for 10 min.
The capillary flow of a droplet in an immiscible liquid at low Reynolds numbers is governed by three parameters36 allowing one to foresee the interface deformation and breakup.37 These are the ratio of the undeformed radius of the drop to the capillary radius (Λ), the ratio of the viscosity of the drop liquid to that of the suspending fluid (k), and the capillary number (Ca) describing the competition between viscous forces and interfacial tensions. At fixed Λ and k, the deformation enhances upon increasing Ca, leading to instability and breaking above a critical value, Cac.
Controlling the fluid motion is therefore particularly important in chips based on oscillating-flow. The breakup of the PCR mix volume flowing through regions at different temperature may induce local changes in the mix reagents concentration. Moreover, imposing the same thermal cycling profile to droplets of different sizes would be quite hard during flow oscillations since the different volumes of water solution tend to reach different shapes and velocities. For our system (Λ≈1.38 and k≈1.8×10−3), we estimate Cac≈4.36 Using a fluid velocity of 3×10−3 m∕s, i.e., Ca≈3×10−2, well below the breaking threshold, the devices work with very stable droplets during the amplification process.
In general, due to their large surface-to-volume ratios, microfluidic devices may suffer from high non-specific adsorption of one or more components of the PCR mix (Taq DNA polymerase and template DNA) on microchannels and reaction chambers, causing significant changes in the reagents concentration.3 Methods relying on passivation agents directly added to the amplification mix generally require optimal relative concentrations of surface modifiers, reagents, and DNA, and potentially fail when used in microchannels of different volumes. To overcome Taq adsorption on the sidewalls of their oscillatory capillary PCR machine, Chiou et al.38, 39 reported on the use of high concentrations of Taq DNA polymerase (0.18−0.7 unit∕μl), flowing into a polytetrafluoroethylene microcapillary over 30 cycles. Here, a static silanization of glass is performed prior to the amplification experiments, allowing us to work with a standard quantity of Taq polymerase (0.02 unit∕μl). The PCR mixture also contains the biological detergent Tween-20; however, different from previous studies,38, 39 we could not note relevant stability issues for our droplets, which is likely related to the low fluid velocity (3×10−3 m∕s).
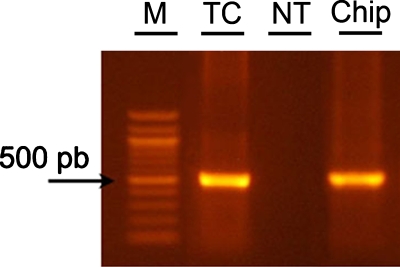
Glass capillaries treated by the coating solution are tested in the microfluidic devices using the same thermal profile. The method is found to enable on-chip PCR (Fig. 5), demonstrating the stability of the coatings under flow. Different from static conditions in thermal cycler, in the chip the biological sample continuously flows into the capillary, thus interacting with a large surface (≈35 mm2) during the process. The consequent possible surface adsorption of PCR mix components is therefore particularly critical to potentially determine an inhibition of amplification. Instead, our chips enable effective PCR, as evidenced by the product imaged in Fig. 5, whose quality and quantity are found to be comparable to results obtained by control experiments carried out in a thermal cycler.
Figure 5.
Gel analysis of PCR products. The different lanes show marker (M), positive control sample [in standard PCR tube in thermal cycler (TC)], results of experiments carried out in microfluidic hybrid PDMS-glass chips, with not-treated (NT) and passivated (Chip) glass capillary.
CONCLUSIONS
In conclusion, we report on a straightforward strategy to avoid sample evaporation in oscillating-flow microfluidic PCR devices. We investigate the evaporation of water solution droplets in PDMS and glass-PDMS chips. A drastic volume reduction (up to 100%) is found for water droplets in oil in PDMS chips during PCR heating due to the polymer low vapor barrier and high porosity, making these devices unsuitable for such applications. However, this problem can be overcome by making PDMS inaccessible to water diffusion by means of glass capillaries embedded in the elastomer. We believe that this method can be adapted to microfluidic networks and geometries of whatever complexity. This process would include the realization of elastomer devices by replica molding from microfabricated masters, and recently developed technologies for functionalizing the inner surfaces of polymer microchannels by glass layers.40 This concept is therefore useful to design and realize new low-volume, highly sensitive genomic and proteomic microfluidic components and devices. The device geometry proposed here, and its future developments, represent a general route to high-temperature (up to 100 °C) biochemical reactors relying on microflows.
ACKNOWLEDGMENTS
The authors are grateful to the support of the Apulia Regional Strategic Project PS_144, and of the Italian Ministry of University and Research through the FIRB Project Nos. RBIN045MNB and RBLA03ER38.
References
- Manz A., Graber N., and Widmer H. M., Sens. Actuators B 1, 244 (1990). 10.1016/0925-4005(90)80209-I [DOI] [Google Scholar]
- Northrup M. A., Ching M. T., White R. M., and Watson R. T., Proceedings of the Seventh International Conference on Solid-State Sensors and Actuators (Transducers ‘93), Yokohama, Japan, 7–10 June 1993, pp. 924–926.
- Zhang C., Xu J., Ma W., and Zheng W., Biotechnol. Adv. 24, 243 (2006). 10.1016/j.biotechadv.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Zhang C. and Xing D., Nucleic Acids Res. 35, 4223 (2007). 10.1093/nar/gkm389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. and Ozdemir P., Anal. Chim. Acta 638, 115 (2009). 10.1016/j.aca.2009.02.038 [DOI] [PubMed] [Google Scholar]
- Lee J. G., Cheong K. H., Huh N., Kim S., Choi J. W., and Ko C., Lab Chip 6, 886 (2006). 10.1039/b515876a [DOI] [PubMed] [Google Scholar]
- Legendre L. A., Bienvenue J. M., Roper M. G., Ferrance J. P., and Landers J. P., Anal. Chem. 78, 1444 (2006). 10.1021/ac0516988 [DOI] [PubMed] [Google Scholar]
- Chen Z., Wang J., Qian S., and Bau H. H., Lab Chip 5, 1277 (2005). 10.1039/b508275g [DOI] [PubMed] [Google Scholar]
- Matsubara Y., Kerman K., Kobayashi M., Yamanura S., Morita Y., and Tamiya E., Biosens. Bioelectron. 20, 1482 (2005). 10.1016/j.bios.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Oh K. W., Park C., Namkoong K., Kim J., Ock K. S., Kim S., Kim Y. A., Cho Y. K., and Ko C., Lab Chip 5, 845 (2005). 10.1039/b503437j [DOI] [PubMed] [Google Scholar]
- Toriello N. M., Liu C. N., and Mathies R. A., Anal. Chem. 78, 7997 (2006). 10.1021/ac061058k [DOI] [PubMed] [Google Scholar]
- Marcus J. S., Anderson W. F., and Quake S. R., Anal. Chem. 78, 956 (2006). 10.1021/ac0513865 [DOI] [PubMed] [Google Scholar]
- Kim J. A., Lee J. Y., Seong S., Cha S. H., Lee S. H., Kim J. J., and Park T. H., Biochem. Eng. J. 29, 91 (2006). 10.1016/j.bej.2005.02.032 [DOI] [Google Scholar]
- Tsai N. C. and Sue C. Y., Biosens. Bioelectron. 22, 313 (2006). 10.1016/j.bios.2005.12.023 [DOI] [PubMed] [Google Scholar]
- Wang W., Li Z. X., Luo R., Lu S. H., Xu A. D., and Yang Y. J., J. Micromech. Microeng. 15, 1369 (2005). 10.1088/0960-1317/15/8/001 [DOI] [Google Scholar]
- Cheng J. Y., Hsieh C. J., Chuang Y. C., and Hsieh J. R., Analyst (Cambridge, U.K.) 130, 931 (2005). 10.1039/b501061f [DOI] [PubMed] [Google Scholar]
- Bu M. Q., Tracy M., Ensell G., Wilkinson J. S., and Evans A. G. R., J. Micromech. Microeng. 13, S125 (2003). 10.1088/0960-1317/13/4/321 [DOI] [Google Scholar]
- Frey O., Bonneick S., Hierlemann A., and Lichtenberg J., Biomed. Microdevices 9, 711 (2007). 10.1007/s10544-007-9080-4 [DOI] [PubMed] [Google Scholar]
- Liu J., Enzelberger M., and Quake S., Electrophoresis 23, 1531 (2002). [DOI] [PubMed] [Google Scholar]
- West J., Karamata B., Lillis B., Gleeson J. P., Alderman J., Collins J. K., Lane W., Mathewson A., and Berney H., Lab Chip 2, 224 (2002). 10.1039/b206756k [DOI] [PubMed] [Google Scholar]
- Sadler D. J., Changrani R., Roberts P., Chou C. F., and Zenhausern F., IEEE Trans. Compon. Packag. Technol. 26, 309 (2003). 10.1109/TCAPT.2003.815093 [DOI] [Google Scholar]
- Kopp M. U., de Mello A. J., and Manz A., Science 280, 1046 (1998). 10.1126/science.280.5366.1046 [DOI] [PubMed] [Google Scholar]
- Wang H., Chen J., Zhu L., Shadpour H., Hupert M. L., and Soper S. A., Anal. Chem. 78, 6223 (2006). 10.1021/ac060568b [DOI] [PubMed] [Google Scholar]
- Crews N., Wittwer C., and Gale B., Biomed. Microdevices 10, 187 (2008). 10.1007/s10544-007-9124-9 [DOI] [PubMed] [Google Scholar]
- Shoffner M. A., Cheng J., Hvichia G. E., Kricka L. J., and Wilding P., Nucleic Acids Res. 24, 375 (1996). 10.1093/nar/24.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N. A. and Meldrum D. R., Anal. Chem. 70, 2997 (1998). 10.1021/ac971303n [DOI] [PubMed] [Google Scholar]
- Giordano B. C., Ferrance J., Swedberg S., Huhmer A. F. R., and Landers J. P., Anal. Biochem. 291, 124 (2001). 10.1006/abio.2000.4974 [DOI] [PubMed] [Google Scholar]
- Giordano B. C., Copeland E. R., and Landers J. P., Electrophoresis 22, 334 (2001). [DOI] [PubMed] [Google Scholar]
- Xia Y. and Whitesides G. M., Angew. Chem., Int. Ed. 37, 550 (1998). [DOI] [PubMed] [Google Scholar]
- Obeid P. J., Christopoulos T. K., Crabtree H. J., and Backhouse C. J., Anal. Chem. 75, 288 (2003). 10.1021/ac0260239 [DOI] [PubMed] [Google Scholar]
- Prakash A. R., Adamia S., Sieben V., Pilarski P., Pilarski L. M., and Backhouse C. J., Sens. Actuators B 113, 398 (2006). 10.1016/j.snb.2005.03.049 [DOI] [Google Scholar]
- George S. C. and Thomas S., Prog. Polym. Sci. 26, 985 (2001). 10.1016/S0079-6700(00)00036-8 [DOI] [Google Scholar]
- Watson J. M. and Baron M. G., J. Membr. Sci. 110, 47 (1996). 10.1016/0376-7388(95)00229-4 [DOI] [Google Scholar]
- Blume I., Schwering P. J. F., Mulder M. H. V., and Smolders C. A., J. Membr. Sci. 61, 85 (1991). 10.1016/0376-7388(91)80008-T [DOI] [Google Scholar]
- Shin Y. S., Cho K., Lim S. H., Chung S., Park S. J., Chung C., Han D. C., and Chang J. K., J. Micromech. Microeng. 13, 768 (2003). 10.1088/0960-1317/13/5/332 [DOI] [Google Scholar]
- Olbricht W. L. and Kung D. M., Phys. Fluids A 4, 1347 (1992). 10.1063/1.858412 [DOI] [Google Scholar]
- Borhan A. and Pallinti J., Phys. Fluids 11, 2846 (1999). 10.1063/1.870143 [DOI] [Google Scholar]
- Chiou J., Matsudaira P., Sonin A., and Ehrlich D., Anal. Chem. 73, 2018 (2001). 10.1021/ac001227f [DOI] [PubMed] [Google Scholar]
- Chiou J., Matsudaira P., and Ehrlich D., BioTechniques 33, 557 (2002). [DOI] [PubMed] [Google Scholar]
- Abate A. R., Lee D., Do T., Holtze C., and Weitz D. A., Lab Chip 8, 516 (2008). 10.1039/b800001h [DOI] [PubMed] [Google Scholar]