Abstract
Inhibin-α knockout (Inha−/−) female mice develop sex cord-stromal ovarian cancer with complete penetrance and previous studies demonstrate that the pituitary gonadotropins (FSH and LH) are influential modifiers of granulosa cell tumor development and progression in inhibin-deficient females. Recent studies have demonstrated that Inha−/− ovarian follicles develop precociously to the early antral stage in prepubertal mice without any increase in serum FSH. These studies suggest that in the absence of inhibins, granulosa cells differentiate abnormally and thus at sexual maturity may undergo an abnormal response to gonadotropin signaling contributing to tumor development. To test this hypothesis, we stimulated immature wild-type and Inha−/− female mice with gonadotropin analogs prior to tumor formation and subsequently examined gonadotropin-induced ovarian follicle development as well as preovulatory and human chorionic gonadotropin-induced gene expression changes in granulosa cells. We find that at 3 wk of age, inhibin-deficient ovaries do not show further antral development or undergo cumulus expansion. In addition, there are widespread alterations in the transcriptome of gonadotropin-treated Inha−/− granulosa cells, with significant changes in genes involved in extracellular matrix and cell-cell communication. These data indicate the gonadotropins initiate an improper program of cell differentiation prior to tumor formation in the absence of inhibins.
Inha null granulosa cells respond to gonadotropins with an abnormal gene expression pattern that is dissimilar to both wild-type mural and cumulus granulosa cells.
The pituitary gonadotropins FSH and LH are critical for the growth and development of postsecondary stage ovarian follicles as well as subsequent ovulation and luteinization (1,2,3). FSH receptors are expressed in granulosa cells of growing follicles, and FSH stimulates granulosa cell proliferation and follicular antrum formation and prevents apoptosis and follicular atresia. Subsequent stimulation via the LH surge leads to cumulus cell expansion, ovulation of the cumulus-oocyte complex through the ruptured follicle wall and ovarian surface, and terminal differentiation of the granulosa cells to form the corpus luteum. Accordingly, FSH-deficient (Fshb−/−) female mice have small ovaries and are infertile secondary to a block in folliculogenesis (2). Follicles in Fshb−/− ovaries arrest as multilayered preantral follicles and do not progress further to antral or preovulatory stages unless stimulated with exogenous FSH (2,4). LH-deficient (Lhb−/−) female mice have neither preovulatory follicles nor corpora lutea and also are infertile (3).
The balance of oocyte growth and somatic cell proliferation and differentiation during folliculogenesis is coordinated in part by functional interactions of the gonadotropins with inhibins and activins. TGFβ superfamily hormones were discovered for their respective roles as suppressors (i.e. inhibins) or stimulators (i.e. activins) of pituitary FSH synthesis and secretion (5). Deletion of the shared activin/inhibin-βΑ (Inhba) subunit results in perinatal lethality (6) or subfertility when conditionally null in the ovary (7), whereas deletion of the unique inhibin-α (Inha) subunit results in the development of sex cord-stromal tumors in adult mice as early as 4 wk of age (8).
Previous studies demonstrate that gonadotropins are influential modifiers of sex cord-stromal cancer development and progression in inhibin-deficient mice. Double-mutant mice lacking inhibins and GnRH (Inha−/− Gnrh1hpg/hpg) have suppressed FSH and LH and are free of gross ovarian tumors, although they develop abnormal foci of cells within the gonad (9). Adult Inha−/− mice with gross tumors have elevated serum FSH levels compared with wild-type (WT) mice (8), and removal of FSH by generating Inha−/− Fshb−/− double knockout mice leads to slower growing, less invasive ovarian tumors (9). Similarly, Inha−/− Lhb−/− females have delayed ovarian cancer progression correlated with increased tumor expression of the cell cycle inhibitors p15INK4b (Cdkn2b) and p27Kip1 (Cdkn1b) (10).
Reproductive function in Inha−/− females is compromised and our initial studies demonstrated that inhibin-deficient female mice are infertile (8). By 12 d of age, follicles in Inha−/− ovaries are more advanced than their WT counterparts; Inha−/− mice show development of large multilayered follicles within the ovary in the absence of elevated FSH but have not yet developed tumors or signs of abnormal cell foci (11). Preliminary studies with pharmacological superovulation before advanced tumor growth in prepubertal Inha−/− mice (i.e. 3–4 wk old) demonstrates reduced efficiency for ovulation compared with WT controls (12). Thus, the phenotypes of Inha−/−, Inha−/− Gnrh1hpg/hpg, Inha−/− Fshb−/−, and Inha−/− Lhb−/− females suggest that in the absence of inhibins, granulosa cells respond abnormally to gonadotropins with respect to subsequent follicle growth and gene expression, culminating in tumor growth. To understand the basis for how gonadotropins contribute to improper granulosa cell development without the variables of estrous cyclicity and the presence of gross tumors, we examined the response of sexually immature Inha−/− mice to stimulation by exogenous gonadotropins. Our experiments reveal widespread defects in granulosa cell differentiation in the absence of inhibins.
Materials and Methods
Experimental animals
Generation and genotyping of Inha mutant mice by PCR or Southern blot have been described (8,11,13). Mice were maintained in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Morphological and histological analysis
Twenty-one- to 23-d-old WT and Inha−/− female mice were injected ip with 5 IU pregnant mare serum gonadotropin (PMSG; Calbiochem, San Diego, CA) for 44–46 h alone or followed by ip injection with 5 IU human chorionic gonadotropin (hCG) (Novarel; Ferring Pharmaceuticals, Parsippany, NY) for 6 h. Ovaries were fixed in 10% neutral buffered formalin (Richard-Allan Scientific, Kalamazoo, MI) and subsequent processing, embedding, sectioning, and staining with periodic acid-Schiff/hematoxylin were performed by the Department of Pathology Core Services Laboratory (Baylor College of Medicine) or within the laboratory with standard techniques.
Granulosa cell collection
To collect granulosa cells, immature mice were stimulated for 44–46 h with PMSG. Additional mice were stimulated with 6 h of hCG as described above. Ovaries were collected, and a mixture of mural and cumulus granulosa cells were harvested by puncturing ovarian follicles in DMEM/F12 (Invitrogen, Carlsbad, CA) supplemented with 2.4 g NaHCO3, 0.3% BSA, 10 mm HEPES, and 1% penicillin-streptomycin (Invitrogen). Cells were filtered through a 40-μm nylon mesh (Nalgene, Rochester, NY) to remove tissue debris and oocytes. Granulosa cells were centrifuged at 1000 × g for 5 min, the supernatant was aspirated, and the pellet was frozen at −80 C until RNA isolation. In this study, we term preovulatory granulosa cells as those cells collected after 44–46 h of PMSG stimulation, regardless of genotype.
Microarray analysis
Total RNA was extracted from granulosa cells using the RNeasy Mini Kit (QIAGEN, Valencia, CA) with on-column deoxyribonuclease treatment (QIAGEN), and RNA quality was inspected on a 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA). Gene expression profiles were generated on GeneChip Mouse Genome 430 2.0 microarrays (Affymetrix, Santa Clara, CA; n = 3 chips for each of four genotype/treatment combinations). For each array, RNA was isolated from granulosa cells that were pooled from three to four mice. The array platform consists of more than 45,000 probe sets representing more than 39,000 mouse transcripts and variants. Expression data were analyzed with GeneSpring GX 10.0.2 software (Agilent). Probe level data were imported, background corrected, quantile normalized, and summarized using the GCRMA (GeneChip robust multiarray averaging) function. These raw expression values are available in the supplemental file entitled Array_ANOVA.xls, published on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org. The raw expression values were then subject to baseline transformation to the median of all samples. Differentially expressed genes (P < 0.02, fold change >2) were identified by one-way ANOVA across all four genotype/treatment combinations followed by Tukey’s honestly significant difference (HSD) test, with Benjamini-Hochberg multiple testing correction to control the false discovery rate. For Kyoto Encyclopedia of Genes and Genomes pathway analysis, the statistically significant genes found using Tukey’s HSD test between preovulatory (i.e. without hCG) WT and Inha−/− granulosa cells were uploaded into the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) and analyzed using the default settings. KEGG pathway categories were considered to be statistically significant at P < 0.05. All array data sets described in this study have been deposited into the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) as series GSE20466 (accession no. GSM513621-GSM513632).
Quantitative real-time PCR (qPCR)
qPCR validation of microarray data were performed on granulosa cells from PMSG-treated female mice independent of samples used in microarray experiments (n = 6 individual mice per genotype). Total RNA (200 ng) was reverse transcribed in a 20-μl reaction using the high capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA). cDNA was diluted 50-fold and 2 μl was used for each qPCR. qPCR was performed on an ABI Prism 7500 sequence detection system using TaqMan gene expression PCR master mix with TaqMan gene expression assays (Applied Biosystems) (Supplemental Table 4), or SYBR Green PCR master mix (Applied Biosystems) with custom primers designed using Primer Express software (Applied Biosystems) (Supplemental Table 5). The reaction volume was 20 μl and reaction conditions were 2 min at 50 C, 10 min at 95 C, 40 cycles of 15 sec at 95 C (denaturation), and 1 min at 60 C (annealing/extension). Each sample was analyzed in duplicate or triplicate. The relative quantity (RQ) of transcript was calculated using the 2−ΔΔCT method with the average ΔCT of the WT group as the calibrator and mouse Gapdh as an endogenous control (14). Statistical differences were tested using one-way ANOVA followed by Tukey’s HSD test or Fisher’s least significant difference (LSD) test as indicated in the text, with P < 0.05 considered statistically significant using JMP (SAS, Cary, NC) or SPSS version 17 (SPSS, Chicago, IL). There was good agreement between the microarray analysis and relative values by qPCR (Supplemental file Array_ANOVA.xls).
Results
Inha−/− ovaries do not develop large antral follicles after PMSG stimulation and do not undergo cumulus expansion after hCG
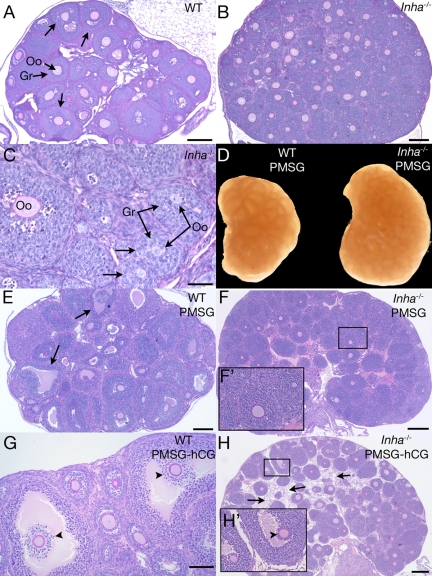
We recently showed that prepubertal follicle development occurs precociously in Inha−/− females, with small (<180 μm) antral follicles visible by 12 d of age without an increase in serum FSH (11). In this study, we observed that at 3 wk of age, Inha−/− ovaries are larger than WT but do not show any advanced antral follicle development beyond what is present at 12 d of age (Fig. 1, A–C) (11). Uninjected 3-wk Inha−/− ovaries also maintain the same follicle defects that have been described for the uninjected 12-d Inha−/− ovary (11), including a larger ovarian size, small oocytes within large follicles, and asymmetric growth of granulosa cells (Fig. 1C). In addition, as demonstrated in ovaries from 12-d WT and Inha−/− ovaries (11), at 21 d of age, FSH receptor (Fshr) is similarly expressed in ovaries of both genotypes (Supplemental Fig. 1). To determine whether exogenous FSH could induce subsequent follicle development, immature (3 wk old) mice were stimulated with PMSG for 44–46 h. Whereas PMSG-injected immature WT mice develop numerous preovulatory follicles (Fig. 1, D and E), Inha−/− ovaries show no antral follicle development, even though the ovaries are larger than WT ovaries (Fig. 1, D and F). As an attempt to prompt ovulation and cumulus cell expansion, mice were exposed to short-term hCG (6 h) after 44–46 h of PMSG injection. In contrast to WT mice (Fig. 1G), Inha−/− mice stimulated with hCG rarely demonstrated follicles at an advanced stage within the ovary (Fig. 1H). In the occasional Inha−/− antral follicle, visible cumulus expansion could not be seen (Fig. 1H, inset). Instead, hCG induced a widespread stromal cell or matrix-like component between follicles (three of five ovaries examined) (Fig. 1H).
Figure 1.
Histologic analysis of 3-wk-old WT and Inha−/− ovaries. Ovary from a 3-wk untreated WT mouse shows early antral follicle development (arrows) (A), whereas a 3-wk untreated Inha−/− ovary is larger and does not develop follicular antra to the same extent (B). C, Higher magnification of Inha−/− ovary, demonstrating uncoupled oocyte and granulosa cell growth (arrow) similar to studies of 12-d Inha−/− ovaries (11). D, Gross anatomy of 3-wk-old WT and Inha−/−ovaries injected for 44–46 h PMSG, demonstrating a larger size of the Inha−/− ovary. Three-week WT ovary 44–46 h after PMSG injection demonstrates preovulatory follicle development (E), whereas Inha−/− ovaries do not develop follicular antra (F). F′, Higher magnification of boxed area shown in F. G, Three-week-old WT ovary after 44–46 h PMSG and 6 h hCG containing cumulus oocyte complexes undergoing cumulus expansion (arrowheads). H, No cumulus expansion is visible in PMSG/hCG-treated Inha−/− ovaries (arrowhead). Boxed region in H is shown as a higher-magnification inset (H′). An unusual stromal component (arrows) between follicles is visible in PMSG/hCG-treated Inha−/− ovaries that is not seen in WT ovaries. Scale bars (A, B, E, F, and H), 200 μm. Scale bars (C), 50 μm. Scale bar (G), 100 μm. Oo, Oocyte; Gr, granulosa cells.
Preovulatory and hCG-primed Inha−/− granulosa cells demonstrate aberrant gene expression patterns
Because large antral (i.e. preovulatory) follicles did not develop in Inha−/− with PMSG treatment, we asked two questions: 1) What are the gene expression differences between WT and Inha−/− preovulatory granulosa cells (i.e. defined as those cells collected after 44–46 h of stimulation with PMSG); and 2) Do preovulatory granulosa cells from these genotypes respond similarly to an LH analog (i.e. hCG)? To answer these questions, we performed microarray analysis between WT and Inha−/− preovulatory granulosa cells as well as granulosa cells collected from mice treated additionally with 6 h of hCG. Statistical analysis by one-way ANOVA on all four genotype/treatment groups yielded 2763 differentially expressed probe sets (P < 0.02) (Supplemental file Array_ANOVA.xls). Post hoc analysis revealed 635 differentially expressed probe sets representing 235 unique genes up-regulated and 252 unique genes down-regulated (fold change >2) in preovulatory Inha−/− granulosa cells compared with WT before treatment with hCG. We used KEGG pathway analysis to identify statistically significant categories within the differentially expressed gene lists (Supplemental Table 1). In addition, we manually examined the differentially expressed gene list and selected those genes additionally relevant to reproduction and cancer development. These are presented in Tables 1–4: Extracellular matrix and cell adhesion (Table 1); Cytoskeleton (Table 2); Steroidogenesis (Table 3); and Cell cycle, proliferation, and apoptosis (Table 4).
Table 1.
Differentially expressed ECM and cell adhesion genes in preovulatory granulosa cells
| Gene | Description | Fold change (Inha−/−/WT) |
|---|---|---|
| Up in Inha−/− | ||
| Lamc3 | Laminin-γ3 | +16.6 |
| Bcan | Brevican | +16.2 (+12.3a) |
| Mmp2 | Matrix metallopeptidase 2 | +9.8 (+26.5a) |
| Wisp1 | WNT1 inducible signaling pathway protein 1 | +9.5 (+9.5a) |
| Boc | Biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein | +8.8 |
| Col1a2 | Collagen, type I, α2 | +7.0 |
| Adamtsl2 | ADAMTS-like 2 | +5.1 |
| Emilin2 | Elastin microfibril interfacer 2 | +5.0 |
| Col4a4 | Collagen, type IV, α4 | +3.9 |
| Col3a1 | Collagen, type III, α1 | +3.5 |
| Itga9 | Integrin α9 | +3.5 |
| Gjc1 | Gap junction protein, γ1 | +2.8 |
| Col6a1 | Collagen, type VI, α1 | +2.7 |
| Matn2 | Matrilin 2 | +2.5 |
| Jam2 | Junction adhesion molecule 2 | +2.4 |
| Down in Inha−/− | ||
| Coch | Coagulation factor C homolog (Limulus polyphemus) | −64.7 |
| Comp | Cartilage oligomeric matrix protein | −46.9 (−38.3a) |
| Cyr61 | Cysteine-rich protein 61 | −13.0 |
| Ctgf | Connective tissue growth factor | −8.2 (−6.1a) |
| Col4a6 | Collagen, type IV, α6 | −6.7 |
| Prss23 | Protease, serine, 23 | −4.7 |
| Col4a5 | Collagen, type IV, α5 | −3.7 |
| Itga3 | Integrin α3 | −3.3 |
Genes in bold were independently validated by qPCR with fold change in parentheses.
qPCR statistical significance at P < 0.05.
Table 2.
Differentially expressed cytoskeleton genes in preovulatory granulosa cells
| Gene | Description | Fold change (Inha−/−/WT) |
|---|---|---|
| Up in Inha−/− | ||
| Actg2 | Actin, γ 2, smooth muscle, enteric | +17.1 |
| Krt79 | Keratin 79 | +8.2 |
| Cnn1 | Calponin 1 | +7.3 |
| Myh11 | Myosin, heavy polypeptide 11, smooth muscle | +7.3 |
| Myl9 | Myosin, light polypeptide 9, regulatory | +6.6 |
| Tpm2 | Tropomyosin 2, β | +5.7 |
| Dnahc7b | Dynein, axonemal, heavy chain 7B | +5.1 |
| Acta2 | Actin, α2, smooth muscle, aorta | +4.6 |
| Myo18a | Myosin XVIIIA | +4.6 |
| Tube1 | ε-Tubulin 1 | +2.8 |
| Kif24 | Kinesin family member 24 | +2.1 |
| Lmna | Lamin A | +2.1 |
| Down in Inha−/− | ||
| Dnahc2 | Dynein, axonemal, heavy chain 2 | −24.3 |
| Obsl1 | Obscurin-like 1 | −18.9 |
| Krt8 | Keratin 8 | −3.0 |
| Krt18 | Keratin 18 | −2.8 |
| Krt19 | Keratin 19 | −2.8 |
| Myo7a | Myosin VIIA | −2.7 |
| Tbcel | Tubulin folding cofactor E-like | −2.2 |
| F11r | F11 receptor | −3.0 |
| Adamts1 | A disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 | −2.7 (−2.0a) |
| Pcdh7 | Protocadherin 7 | −2.7 |
| Col4a3bp | Collagen, type IV, α3 (Goodpasture antigen) binding protein | −2.1 |
| Ltbp1 | Latent TGFβ binding protein 1 | −2.1 |
Adamts1 is statistically different (P < 0.05) between preovulatory WT and Inha−/− granulosa cells by Fisher’s t test.
Table 3.
Differentially expressed steroidogenesis genes in preovulatory granulosa cells
| Gene | Description | Fold change (Inha−/−/WT) |
|---|---|---|
| Up in Inha−/− | ||
| Igfbp4 | IGF binding protein 4 | +4.8 |
| Cyp1b1 | Cytochrome P450, family 1, subfamily b, polypeptide 1 | +2.6 |
| Cyp51 | Cytochrome P450, family 51 | +2.2 |
| Down in Inha−/− | ||
| Lrp11 | Low-density lipoprotein receptor-related protein 11 | −32.2 (−11.7a) |
| Star | Steroidogenic acute regulatory protein | −24.2 (−3.6a) |
| Hsd17b2 | Hydroxysteroid (17β) dehydrogenase 2 | −15.7 |
| Cyp11a1 | Cytochrome P450, family 11, subfamily a, polypeptide 1; side chain cleavage enzyme | −9.4 (−3.9a) |
| Prlr | Prolactin receptor | −7.2 (−3.9a) |
| Lhcgr | LH/choriogonadotropin receptor | −7.1 (−6.2a) |
| Ephx1 | Epoxide hydrolase 1, microsomal | −6.1 |
| Il1r1 | IL-1 receptor, type I | −2.9 |
| Smarca1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 | −2.8 |
| Hsd17b11 | Hydroxysteroid (17β) dehydrogenase 11 | −2.5 |
| Cyp2d22 | Cytochrome P450, family 2, subfamily d, polypeptide 22 | −2.3 |
Genes in bold were independently validated by qPCR with fold change in parentheses.
qPCR statistical significance at P < 0.05.
Table 4.
Differentially expressed cell cycle, proliferation, and apoptosis genes in preovulatory granulosa cells
| Gene | Description | Fold change (Inha−/−/WT) |
|---|---|---|
| Up in Inha−/− | ||
| Fgfr4 | Fibroblast growth factor receptor 4 | +35.5 |
| Egfr | Epidermal growth factor receptor | +3.8 (+3.4a) |
| Fgfr1 | Fibroblast growth factor receptor 1 | +2.5 |
| Ccne1 | Cyclin E1 | +2.3 (+1.9a) |
| E2f1 | E2F transcription factor 1 | +2.2 (+1.9a) |
| Espl1 | Extra spindle poles-like 1 (Saccharomyces cerevisiae) | +2.1 |
| Down in Inha−/− | ||
| Sfrp4 | Secreted frizzled-related protein 4 | −23.2 (−6.4a) |
| Pik3r1 | Phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85α) | −21.0 |
| Pik3ip1 | Phosphoinositide-3-kinase interacting protein 1 | −12.2 |
| Sgk | Serum/glucocorticoid regulated kinase 1 | −4.1 |
| Trp53inp1 | Transformation related protein 53 inducible nuclear protein 1 | −3.9 |
| Myc | Myelocytomatosis oncogene | −3.7 |
| Cflar | CASP8 and Fas-associated death domain-like apoptosis regulator | −2.7 |
| Dhx32 | DEAH (Asp-Glu-Ala-His) box polypeptide 32 | −2.6 |
| Pik3cd | Phosphatidylinositol 3-kinase catalytic δ-polypeptide | −2.6 |
Genes in bold were independently validated by qPCR with fold change in parentheses.
qPCR statistical significance at P < 0.05.
Alterations in extracellular matrix (ECM), cell adhesion, cell communication, and cytoskeletal genes
Puncture of newly formed antral follicles after PMSG stimulation of immature mice is a common method of collecting preovulatory granulosa cells. In our initial studies, we noticed that PMSG-stimulated Inha−/− ovaries easily dissociated when isolated ovaries were punctured with needles during granulosa cell harvest, even though Inha−/− follicles do not show antral follicle formation like WT mice. Pathway analysis using the KEGG database on the complete set of 635 differentially expressed probe sets between preovulatory WT and Inha−/− cells demonstrated statistically significant differences in categories related to ECM, cell adhesion, and cell communication (Supplemental Table 1). Laminin-γ3 (Lamc3) was up-regulated approximately 17-fold, and we detected differential expression of multiple type IV and other collagen α-chains (Col1a2, Col3a1, Col4a4, Col6a1, Col4a5, Col4a6) (Table 1). In Inha−/− preovulatory granulosa cells, we also observed up-regulation of the collagenase Mmp2 (Table 1) and down-regulation of Adamts1, a metalloproteinase with critical roles in antrum formation and ovulation (15,16,17) (Table 2). There was elevation of Adamtsl2, an A disintegrin and metalloproteinase with thrombospondin-like repeats (ADAMTS)-like enzyme that regulates TGFβ signaling (Table 1), and up-regulation of the ADAMTS substrate Bcan (Table 1), a TGFβ-induced proteoglycan believed to modulate synaptogenesis in the central nervous system (18) but whose function in follicle development is unclear. Furthermore, there was underexpression of the ADAMTS substrate Comp (Table 1), a matrix protein implicated in skeletal disease (19).
In addition, three of six members of the CCN protein family [named for the founding members: cysteine rich 61 (CYR61), connective tissue growth factor (CTGF), and nephroblastoma overexpressed gene (NOV) are significantly altered in Inha−/− preovulatory granulosa cells (Table 1) (20). The CCN family are secreted factors that act as adhesive proteins between cells and the ECM as well as modulate growth factor activity (20). We detected up-regulation of Wisp1 and down-regulation of Cyr61 and Ctgf. Little is known about Wisp1 and Cyr61 in granulosa cells, but Ctgf is expressed in granulosa cells upon stimulation by TGFβ, activin, or growth and differentiation factor 9 (GDF9) and suppressed in antral follicles by FSH and LH (21,22).
Alterations in genes related to steroidogenesis, cell cycle, proliferation, and apoptosis
During the development to the preovulatory stage, granulosa cells undergo a number of gene expression changes, including changes in steroidogenic enzymes and acquisition of the LH receptor (Lhcgr). There were significant differences in genes necessary for ovulation and steroidogenesis between WT and Inha−/− preovulatory follicles (Table 3). Lhcgr is down-regulated in Inha−/− preovulatory granulosa cells, suggesting that Inha−/− granulosa cells are less differentiated than WT preovulatory granulosa cells. Inha−/− preovulatory granulosa cells also underexpressed genes encoding the steroidogenic enzymes Cyp11a1 and aromatase (Cyp19a1) as well as steroidogenic acute regulatory protein (Star). Interestingly, both Lhcgr and Cyp11a1 are known to have a differential expression in mural vs. cumulus cells, with mural granulosa cells demonstrating greater expression and cumulus cells showing lesser expression in preovulatory follicles (23).
Several growth factor receptors were found to be overexpressed in preovulatory granulosa cells from Inha−/− ovaries compared with WT (Table 4). Inha−/− preovulatory granulosa cells showed overexpression of Egfr and several fibroblast growth factor receptors (Fgfr4 and Fgfr1) and down-regulation of multiple genes involved in phosphatidylinositol 3-kinase signaling (Pik3cd, Pik3ip1, Pik3r1) (Table 4). Based on previous studies (24), we measured cyclin D2 (Ccnd2) levels and found that the gene was overexpressed in Inha−/− granulosa cells by 2.4-fold (Fig. 2E), although this was not initially identified in the microarray analysis.
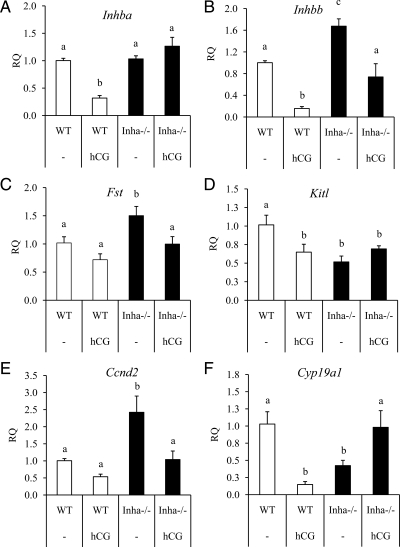
Figure 2.
qPCR analysis of genes related to inhibin/activin production and function in preovulatory and hCG-treated WT (white bars) and Inha−/− (black bars) granulosa cells. Preovulatory granulosa cells were collected 44–46 h after PMSG injection without (−) or with hCG for 6 h. qPCR results are expressed as RQ. hCG down-regulates Inhba (A) in WT preovulatory granulosa cells but not in hCG-treated Inha−/− preovulatory granulosa cells. B, Levels of Inhbb are increased in Inha−/− preovulatory granulosa cells compared with WT, and hCG treatment reduces those levels to that of preovulatory WT granulosa cells. C, Follistatin, an activin antagonist, is up-regulated in preovulatory Inha−/− and declines to WT preovulatory levels after hCG treatment. D, Kitl is suppressed in preovulatory Inha−/− granulosa cells compared with WT, and its levels are similar to hCG-treated WT cells. E, Ccnd2 is up-regulated in Inha−/− preovulatory granulosa cells compared with WT, although it is down-regulated by hCG to levels comparable with preovulatory WT cells. F, Cyp19a1 is down-regulated in Inha−/− preovulatory granulosa cells and shows an opposite regulation (i.e. shows an increase) to hCG than in WT cells (i.e. shows a decrease). Data are mean ± sem of a minimum of three independent samples with the control mean set to 1. Different letters above the bars indicate statistical significance at P < 0.05 by one-way ANOVA followed by Fisher LSD post hoc test.
Inha−/− granulosa cells have altered gene expression in response to hCG
Lhcgr is significantly reduced in Inha−/− granulosa cells, although it is not absent (Table 3 and Fig. 3A). This suggests that hCG may potentially affect gene expression in Inha−/− cells with altered consequences. Therefore, we were interested in identifying genes regulated by hCG in WT preovulatory granulosa cells that were misregulated by hCG in Inha−/− cells. We did this based on the following approach. From our 2763 differentially expressed probe sets (by ANOVA, P < 0.02), we first listed the probe sets that were regulated at a 2-fold or greater change by hCG in WT granulosa cells (1453 probe sets). Because of the large size of this list, we then selected for inclusion the 10% with the largest fold up-regulation in hCG-treated WT preovulatory cells vs. preovulatory WT granulosa cells (146 probe sets). Next, because we were interested in genes that showed changes only during hCG treatment, we excluded from the analysis probe sets that already demonstrated an initial baseline difference between preovulatory WT and Inha−/− granulosa cells (i.e. there were 22 probe sets that already had an initial difference in gene expression before hCG treatment). We then organized the remaining 124 probe sets based on their fold difference between hCG-treated WT and Inha−/− preovulatory granulosa cells as: 1) unchanged (i.e. granulosa cell genes that respond like WT to hCG in Inha−/− mutants), 2) not induced (induced by hCG in WT but not in Inha−/−), 3) underexpressed (i.e. lesser response to hCG in Inha−/− than WT), or 4) overexpressed (i.e. greater response to hCG in Inha−/− than WT) (Supplemental Table 2). A subset of these genes was verified by qPCR analysis of independent samples (Figs. 2–4).
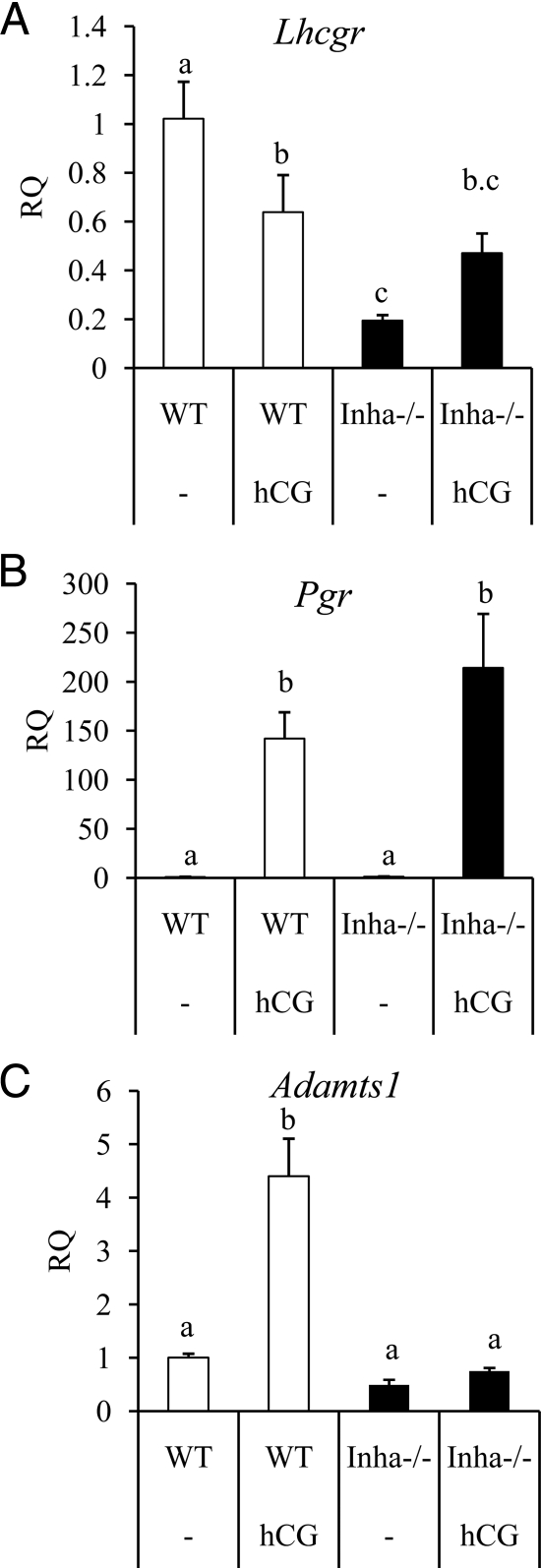
Figure 3.
qPCR analysis of genes related to ovulation and luteinization in WT (white bars) and Inha−/− (black bars) preovulatory granulosa cells collected from mice treated with and without hCG. Preovulatory granulosa cells were collected 44–46 h after PMSG injection without (−) or with hCG for 6 h. qPCR results are expressed as RQ. A, Lhcgr is suppressed in Inha−/− preovulatory granulosa cells with and without hCG. B, Pgr is up-regulated similarly between WT and Inha−/− preovulatory granulosa cells treated with hCG. C, Adamts1, regulated at ovulation by LH and progesterone receptor (56), is not up-regulated in Inha−/− cells. Data are mean ± sem of a minimum of three independent samples with the control mean set to 1. Different letters above the bars indicate statistical significance at P < 0.05 by one-way ANOVA and followed by Fisher LSD post hoc tests.
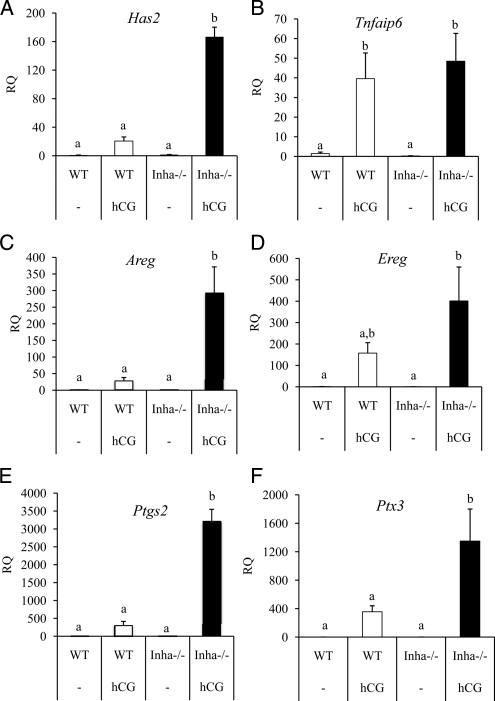
Figure 4.
qPCR analysis of genes related to cumulus expansion in WT (white bars) and Inha−/− (black bars) preovulatory granulosa cells collected from mice treated with and without hCG. Preovulatory granulosa cells were collected 44–46 h after PMSG injection without (−) or with hCG for 6 h. qPCR results are expressed as RQ. Has2 (A), Areg (C), Ereg (D), Ptgs2 (E), and Ptx3 (F) are more highly up-regulated in hCG-treated Inha−/− preovulatory granulosa cells compared with hCG-treated WT cells, whereas Tnfaip6 (B) shows a similar fold induction. Data are mean ± sem of a minimum of three independent samples with the control mean set to 1. Different letters above the bars indicate statistical significance at P < 0.05 by one-way ANOVA and followed by Fisher LSD post hoc tests.
Some genes regulated during ovulation are similarly expressed in Inha−/− granulosa cells compared with WT cells after 6 h of hCG and include progesterone receptor (Pgr) (Fig. 3B and Supplemental Table 2). However, some genes regulated by hCG show a lesser response to hCG in Inha−/− cells than WT cells, including Prkg2 (25) and Snap25 (Supplemental Table 2) (26). In contrast to these results, some genes found in the microarray analysis to be expressed during hCG treatment show greater induction in Inha−/− cells than WT cells, including amphiregulin (Areg), prostaglandin-endoperoxidase synthase 2 (Ptgs2), and TNF-α-induced protein 6 (Tnfaip6) (Supplemental Table 2). Because Areg, Ptgs2, and Tnfaip6 are typically associated with cumulus expansion, we examined other factors induced at this time including Has2, Ereg, Ptx3, and Adamts1. Similar to Areg and Ptgs2 expression patterns, Has2, Ereg, and Ptx3 are more highly up-regulated during hCG treatment of Inha−/− cells than WT cells (Fig. 4). In contrast, Adamts1 was not up-regulated in response to hCG in Inha−/− cells, whereas it is up-regulated by hCG in WT cells (Fig. 3C). Analysis of mural vs. cumulus cell markers (23) indicates that preovulatory granulosa cells from Inha−/− mice have not only reduced expression of the mural cell markers Cyp11a1 and Lhcgr but also increased expression of cumulus cell marker Ar compared with WT cells (Supplemental Table 3).
The LH surge down-regulates a number of genes in preparation for ovulation, including components of the inhibin and activin system (27,28,29). Because activins promote tumor development and disease progression in Inha−/− mice and are responsible for their cancer cachexia-induced death (30,31,32), we examined the behavior of the activin subunits, the activin antagonist follistatin, and activin downstream target genes in hCG-stimulated WT and Inha−/− preovulatory granulosa cells. Before hCG stimulation, the activin-βA (Inhba) subunit is expressed similarly in WT and Inha−/− preovulatory granulosa cells, and activin-βB (Inhbb) is expressed greater in Inha−/− preovulatory granulosa cells compared with WT (Fig. 2B). hCG treatment down-regulated Inhba and Inhbb in WT cells, but both genes continue to be expressed in hCG-treated Inha−/− cells at levels compared with untreated WT (Fig. 2, A and B). In contrast, the activin antagonist follistatin (Fst) is up-regulated in Inha−/− preovulatory granulosa cells but decreases to WT levels after hCG treatment (Fig. 2C). We also analyzed the expression of kit ligand (Kitl), cyclin D2 (Ccnd2), and aromatase (Cyp19a1) because they are target genes of FSH and activin signaling (7,33,34,35,36,37), all three of which are down-regulated by hCG in WT preovulatory granulosa cells. Treatment with hCG has no effect on Kitl levels, suggesting no further response in Inha−/− cells (Fig. 2D). Ccnd2 has a greater expression in Inha−/− preovulatory granulosa cells compared with WT preovulatory granulosa cells, but its levels are reduced by hCG to that of untreated WT preovulatory granulosa cells (Fig. 2E), similar to the pattern of Inhbb expression. Finally, as mentioned in Table 3, Cyp19a1 is down-regulated 2.4-fold in Inha−/− cells compared with WT (Fig. 2F). However, whereas hCG down-regulated Cyp19a1 (7-fold) in hCG-treated WT ovaries, this gene is up-regulated by hCG in Inha−/− preovulatory granulosa cells to levels comparable with preovulatory WT cells (Fig. 2F), demonstrating an opposite regulation to that found in WT preovulatory granulosa cells.
Discussion
In the present study, we investigated the consequence of genetic deletion of inhibin-α on gonadotropin-induced folliculogenesis in mice before sexual maturity and gross tumor formation. Inha−/− female mice are infertile (8), and we previously demonstrated that prepubertal (12-d-old) Inha−/− ovaries develop precociously to the early antral stage without increases in serum FSH and while maintaining expression of Fshr (11). However, little information is known regarding the Inha−/− phenotype between 12 d of age and the onset of puberty (i.e. 4–6 wk of age). This time frame is critical because the development of ovarian tumors in Inha−/− mice appears to correlate with the onset of sexual maturity (8). Three mouse models (Inha−/− Fshb−/−, Inha−/− Lhb−/−, and Inha−/− Gnrh1hpg/hpg) demonstrated that the pituitary gonadotropins modify the phenotype of Inha−/−, although the focus has been on the development of tumors in adult mice. Of these, only the Gnrh1hpg/hpg mouse crossed to Inha−/− failed to develop gross ovarian tumors (9), with the other two models showing delayed ovarian tumor progression and increased survival (10,38). In the current study, exogenous gonadotropin treatment of sexually immature Inha−/− female mice before tumor formation has uncovered abnormal ovarian follicle development to the preovulatory stage in mutant mice coupled with widespread changes in the transcriptome of gonadotropin-treated granulosa cells.
FSH signaling in granulosa cells is essential for the preantral to antral follicle transition (2,39,40). However, treatment with PMSG does not induce preovulatory follicles in Inha−/− ovaries, even though the FSH receptor is present in Inha−/− ovaries (11). The lack of preovulatory antral follicles in PMSG-treated Inha−/− ovaries indicates impaired follicle maturation in the absence of inhibins and suggests that cells respond to exogenous PMSG abnormally. Previous experiments have demonstrated that FSH is essential for follicle remodeling by inducing cytoskeletal rearrangements in granulosa cells (41,42,43) and causing retraction of transzonal projections and actin- and microtubule-based conduits that extend between granulosa cells and the oocyte to facilitate paracrine signaling (4,44). We found that preovulatory Inha−/− granulosa cells showed aberrant expression of many cytoskeleton genes compared with WT, including actin subunits (Acta2, Actg2) and tubulin-related genes (Tube1, Tbcel). The functional significance of these changes remains to be determined.
In this study, we were interested in the effects of signaling pathways in granulosa cell function before tumor formation. FSH activity on granulosa cells is modulated by multiple growth factors, including activin, which may act either synergistically or antagonistically with FSH. It has been hypothesized that because deletion of Inha results in a net gain of activin (as the β-subunits continue to be expressed in granulosa cells from Inha−/− mice), activin must be a major contributor to Inha−/− tumor development (45). In addition, data derived from mouse knockout models for various TGFβ family signaling components, including the Sma and mothers against decapentaplegic-related protein (SMAD) family, has led us to hypothesize that unbalanced signaling that favors activation of the activin/TGFβ pathway leads to granulosa cell tumor development (46,47,48). Thus, gene expression changes in granulosa cells collected from PMSG-treated Inha−/− ovaries should in part reflect continuous unopposed activin in the presence of FSH and reveal how activin may assist FSH in establishing the gene expression profile of Inha−/− granulosa cells and subsequent tumors. For example, in granulosa cell cultures, activin directly suppresses Kitl (7), and acts synergistically with FSH to increase Ccnd2 (37,49). Accordingly, in Inha−/− preovulatory granulosa cells, Kitl is significantly suppressed and Ccnd2 is significantly increased. However, other genes up-regulated by activin, or activin and FSH, in cell cultures of granulosa cells include Cyp19a1, Lhcgr, and Ctgf (21,36,37,50), but these genes are down-regulated in preovulatory Inha−/− cells and thus are inconsistent with high levels of activin or activin activity. It is possible that activin signaling in pretumor preovulatory granulosa cells is partly held in check by a concomitant up-regulation of follistatin that was also detected in Inha−/− preovulatory granulosa cells.
A different scenario for activin-β subunit and activin target gene expression occurs in preovulatory Inha−/− granulosa cells after treatment with hCG. In WT mice, activin-β subunit expression is suppressed by the LH surge (27,28,29), and this is recapitulated in hCG-treated preovulatory WT granulosa cells. Surprisingly, we found that hCG treatment did not suppress βA expression in Inha−/− mice, and both βΑ and βB continue to be expressed at levels similar to preovulatory WT granulosa cells. Furthermore, the continued expression of the activin-β subunits is accompanied by decreased levels of follistatin transcript detected in post-hCG treatment in Inha−/− granulosa cells. Overexpression of activin is a key component of the inhibin-α knockout phenotype because it is directly responsible for the development of the cancer cachexia that eventually results in death (8,32). Lack of regulation of activin subunit expression by hCG is accompanied by altered expression of two of activin’s proposed downstream target genes, Cyp19a1 and Lhcgr, when preovulatory Inha knockout mice are treated with hCG. Both Cyp19a1 and Lhcgr show an opposite response to hCG in Inha−/− preovulatory granulosa cells compared with WT cells: hCG treatment down-regulates both in WT cells but up-regulates both in Inha−/− cells, which would support a hypothesis of increased activin activity after hCG in Inha−/− granulosa cells. In addition, Lhcgr expression decreases in Inha knockout tumors (51) and Lhcgr is commonly down-regulated in human granulosa cell tumors compared with normal ovaries (52,53).
Several genes have been classified as mural or cumulus markers in preovulatory pre-LH surge follicles (23). Mural granulosa cells express Lhcgr, Cyp11a1, and Cd34, whereas cumulus cells express Slc38a3, Amh, and Ar (23). Inha−/− preovulatory granulosa cells have lower levels of the mural markers Lhcgr and Cyp11a1, similar to cumulus cells, but also express the mural marker Cd34 at levels comparable with WT preovulatory granulosa cells. Inha−/− granulosa cells are similar to WT mural cells for Amh expression, but Inha−/− granulosa cells express the cumulus marker Ar greater than WT. Inha−/− preovulatory granulosa cells also have significantly reduced levels of the luteal markers Sfrp4 and Prlr, the latter two indicating an early granulosa cell differentiation status (54). In total, these expression data suggest that Inha−/− preovulatory granulosa cells have a mixed phenotype with characteristics of both mural and cumulus cells, although Inha−/− preovulatory granulosa cells may be more similar to cumulus cells than mural cells. This hypothesis may explain the gene expression changes after hCG treatment, which indicate that Inha−/− granulosa cells behave similarly to cumulus cells at ovulation, as evidenced by the significant increases in cumulus expansion genes, Has2, Ptx3, and Tnfaip3. The majority of the cumulus expansion-related genes demonstrated a higher fold induction in Inha−/− cells than in WT cells. This may reflect a higher proportion of cells undergoing the expansion program (i.e. the majority of granulosa cells collected from PMSG/hCG-treated WT mice are mural cells, whereas granulosa cells from Inha−/− mice are mixed or potentially more cumulus-like).
Whereas Inha−/− preovulatory granulosa cells initiate the cumulus expansion at the transcriptional level, a critical component for full cumulus expansion appears to be missing, as the cells directly adjacent to the oocyte in antral follicles from Inha−/− mice do not show any morphological evidence of cumulus expansion (i.e. they do not become embedded in an extracellular matrix or move away from the oocyte). Interestingly, the protease Adamts1 was the only cumulus expansion-related gene that we tested that was not up-regulated in Inha−/− preovulatory cells in response to hCG. Adamts1 is known to be up-regulated after the LH surge in coordination with signaling via the progesterone receptor (Pgr) (55,56). The defect does not appear to be due to Pgr expression because Pgr shows normal regulation after treatment. Mice null for Adamts1 have ovulation defects but undergo partial cumulus expansion (16), suggesting that the cumulus expansion defect in Inha−/− mice cannot be attributed solely to loss of Adamts1. These data suggest that there are additional key components to cumulus expansion that are disrupted in Inha−/− granulosa cells and remain to be identified.
Unexpectedly, a large number of changes in extracellular matrix genes as well as genes that encode proteins that interact with the ECM were found when comparing preovulatory WT and Inha−/− granulosa cells. Significant changes were identified in the type of collagen being expressed (i.e. increases in Col1a2, Col4a4, Col3a1, and Col6a1 and decreases in Col4a6 and Col4a5) in preovulatory Inha−/− granulosa cells. It has been reported that collagen type IV chains-α3 to -α6 in the basal lamina decline during follicle growth (57,58), and in Inha−/−, these appear to be overexpressed compared with WT. How changes in collagen composition affect the phenotype of Inha−/− or contribute to alterations in Inha−/− gene expression is currently unknown. Interestingly, a previous study has shown a decrease in inhibin in follicular fluid in women with polycystic ovary syndrome (PCOS) (59), a disorder that results in premature arrest of the ovarian follicle as well as increased stromal collagen (60). A further investigation into these alterations may prove useful in understanding the role of the inhibin/activin system in changes in ovarian stroma in PCOS.
Changes in the expression of ECM genes were accompanied by significant changes in Inha−/− in genes of the related categories of ECM-receptor interactions, cell communication, and focal adhesion. Two members of the CCN family, Ctgf and Cyr61, were significantly down-regulated, and one member, Wisp1, was up-regulated. The CCN family modifies signaling of growth factors and cytokines and affects diverse processes such as proliferation, migration, and differentiation by mediating cellular adhesion and modulating the function of extracellular ligands (20). Little is known about this family in follicle growth, ovarian function, or granulosa cell tumor physiology. Wisp1 is downstream of the WNT1 signaling pathway and has been shown to be induced by β-catenin, and interestingly, overexpression of β-catenin in granulosa cells causes granulosa cell tumor formation (61). Wisp1 overexpression is also found in colorectal and breast cancer (62,63,64). The expression pattern and function of Cyr61 in the ovary is unknown. The dramatic changes in genes related to ECM production and function in Inha−/− granulosa cells are a novel and unexpected finding and may contribute importantly to both the loss of preovulatory follicle development and to subsequent tumor formation and growth.
In conclusion, our study demonstrates disrupted follicular development of gonadotropin-primed mouse ovaries lacking inhibin and highlights molecular changes associated with defects in granulosa cell development and differentiation in the absence of inhibin. The altered differentiation pattern of Inha−/− granulosa cells leads to aberrant responses of these cells to pituitary gonadotropins, which drives tumor growth and ultimately death. In part, this includes misregulation of the inhibin-βA and -βB subunits during the gonadotropin surges leading to production of excess activins, which are key contributors to the growth of Inha−/− ovarian tumors. Our studies may be additionally relevant to other diseases of the ovary such as PCOS because changes in the ovarian stroma, robust responses to FSH (65,66), and follicular arrest in PCOS appear to parallel some of the defects that we now demonstrate in our studies on inhibin-α KO granulosa cells. These data demonstrate that Inha expression is necessary for the appropriate response of granulosa cells during gonadotropin-dependent ovarian folliculogenesis and regulation of tumorigenesis and suggest that balanced regulation of ECM production by the inhibin/activin system may play an as-yet-undescribed role in granulosa cell tumor development.
Supplementary Material
Acknowledgments
We thank Dr. Claudia Andreu-Vieyra for helpful discussions and Dr. Joanne Richards for critical reading of the initial manuscript.
Footnotes
This work was supported by National Institutes of Health Grants R01CA60651 (to M.M.M.) and T32GM008307 (to A.K.N.); a National Cancer Institute administrative supplement from funds provided by the American Recovery and Reinvestment Act of 2009 supporting summer research experiences for students (to S.R.); the Joseph and Matilda Melnick Endowed Fund (to A.K.N.); Baylor Research Advocates for Student Scientists (to A.K.N.); a Burroughs Wellcome Career Award in the Biomedical Sciences grant (S.A.P.); a Dan L. Duncan Scholar Award from the Dan L. Duncan Cancer Center) (to S.A.P.); and a grant from the Caroline Wiess Law Fund for Molecular Medicine and L. E. and Josephine S. Gordy Memorial Cancer Research Fund (to S.A.P.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online August 25, 2010
Abbreviations: ADAMTS, A disintegrin-like and metalloproteinase with thrombospondin-like repeats; CCN, protein family named for CYR61, CTGF, and nephroblastoma overexpressed protein; CTGF, connective tissue growth factor; CYR61, cysteine rich 61; ECM, extracellular matrix; hCG, human chorionic gonadotropin; HSD, honestly significant difference; LSD, least significant difference; PCOS, polycystic ovary syndrome; PMSG, pregnant mare serum gonadotropin; qPCR, quantitative real-time PCR; RQ, relative quantity; WT, wild type.
References
- Edson MA, Nagaraja AK, Matzuk MM 2009 The mammalian ovary from genesis to revelation. Endocr Rev 30:624–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TR, Wang Y, Lu N, Matzuk MM 1997 Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet 15:201–204 [DOI] [PubMed] [Google Scholar]
- Ma X, Dong Y, Matzuk MM, Kumar TR 2004 Targeted disruption of luteinizing hormone β-subunit leads to hypogonadism, defects in gonadal steroidogenesis, and infertility. Proc Natl Acad Sci USA 101:17294–17299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combelles CM, Carabatsos MJ, Kumar TR, Matzuk MM, Albertini DF 2004 Hormonal control of somatic cell oocyte interactions during ovarian follicle development. Mol Reprod Dev 69:347–355 [DOI] [PubMed] [Google Scholar]
- Wiater E, Vale W 2008 Activins and inhibins. In: Derynck R, Miyazono K, eds. The TGFB Family. 1st ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 79–120 [Google Scholar]
- Matzuk MM, Kumar TR, Bradley A 1995 Different phenotypes for mice deficient in either activins or activin receptor type II. Nature 374:356–360 [DOI] [PubMed] [Google Scholar]
- Pangas SA, Jorgez CJ, Tran M, Agno J, Li X, Brown CW, Kumar TR, Matzuk MM 2007 Intraovarian activins are required for female fertility. Mol Endocrinol 21:2458–2471 [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A 1992 α-Inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature 360:313–319 [DOI] [PubMed] [Google Scholar]
- Kumar TR, Wang Y, Matzuk MM 1996 Gonadotropins are essential modifier factors for gonadal tumor development in inhibin-deficient mice. Endocrinology 137:4210–4216 [DOI] [PubMed] [Google Scholar]
- Nagaraja AK, Agno JE, Kumar TR, Matzuk MM 2008 Luteinizing hormone promotes gonadal tumorigenesis in inhibin-deficient mice. Mol Cell Endocrinol 294:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M, Middlebrook BS, Matzuk MM, Pangas SA 2009 Loss of inhibin α uncouples oocyte-granulosa cell dynamics and disrupts postnatal folliculogenesis. Dev Biol 334:458–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, Kumar TR, Shou W, Coerver KA, Lau AL, Behringer RR, Finegold MJ 1996 Transgenic models to study the roles of inhibins and activins in reproduction, oncogenesis, and development. Recent Prog Horm Res 51:123–154; discussion 155–157 [PubMed] [Google Scholar]
- Pierson TM, Wang Y, DeMayo FJ, Matzuk MM, Tsai SY, Omalley BW 2000 Regulable expression of inhibin A in wild-type and inhibin alpha null mice. Mol Endocrinol 14:1075–1085 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2[-ΔΔC(T)] method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Shindo T, Kurihara H, Kuno K, Yokoyama H, Wada T, Kurihara Y, Imai T, Wang Y, Ogata M, Nishimatsu H, Moriyama N, Oh-hashi Y, Morita H, Ishikawa T, Nagai R, Yazaki Y, Matsushima K 2000 ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J Clin Invest 105:1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittaz L, Russell DL, Wilson T, Brasted M, Tkalcevic J, Salamonsen LA, Hertzog PJ, Pritchard MA 2004 Adamts-1 is essential for the development and function of the urogenital system. Biol Reprod 70:1096–1105 [DOI] [PubMed] [Google Scholar]
- Brown HM, Dunning KR, Robker RL, Pritchard M, Russell DL 2006 Requirement for ADAMTS-1 in extracellular matrix remodeling during ovarian folliculogenesis and lymphangiogenesis. Dev Biol 300:699–709 [DOI] [PubMed] [Google Scholar]
- Mayer J, Hamel MG, Gottschall PE 2005 Evidence for proteolytic cleavage of brevican by the ADAMTSs in the dentate gyrus after excitotoxic lesion of the mouse entorhinal cortex. BMC Neurosci 6:52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey KL, Hecht JT 2008 The role of cartilage oligomeric matrix protein (COMP) in skeletal disease. Curr Drug Targets 9:869–877 [DOI] [PubMed] [Google Scholar]
- Leask A, Abraham DJ 2006 All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci 119:4803–4810 [DOI] [PubMed] [Google Scholar]
- Harlow CR, Davidson L, Burns KH, Yan C, Matzuk MM, Hillier SG 2002 FSH and TGF-β superfamily members regulate granulosa cell connective tissue growth factor gene expression in vitro and in vivo. Endocrinology 143:3316–3325 [DOI] [PubMed] [Google Scholar]
- Slee RB, Hillier SG, Largue P, Harlow CR, Miele G, Clinton M 2001 Differentiation-dependent expression of connective tissue growth factor and lysyl oxidase messenger ribonucleic acids in rat granulosa cells. Endocrinology 142:1082–1089 [DOI] [PubMed] [Google Scholar]
- Diaz FJ, Wigglesworth K, Eppig JJ 2007 Oocytes determine cumulus cell lineage in mouse ovarian follicles. J Cell Sci 120:1330–1340 [DOI] [PubMed] [Google Scholar]
- Burns KH, Agno JE, Sicinski P, Matzuk MM 2003 Cyclin D2 and p27 are tissue-specific regulators of tumorigenesis in inhibin α knockout mice. Mol Endocrinol 17:2053–2069 [DOI] [PubMed] [Google Scholar]
- Sriraman V, Rudd MD, Lohmann SM, Mulders SM, Richards JS 2006 Cyclic guanosine 5′-monophosphate-dependent protein kinase II is induced by luteinizing hormone and progesterone receptor-dependent mechanisms in granulosa cells and cumulus oocyte complexes of ovulating follicles. Mol Endocrinol 20:348–361 [DOI] [PubMed] [Google Scholar]
- Shimada M, Yanai Y, Okazaki T, Yamashita Y, Sriraman V, Wilson MC, Richards JS 2007 Synaptosomal-associated protein 25 gene expression is hormonally regulated during ovulation and is involved in cytokine/chemokine exocytosis from granulosa cells. Mol Endocrinol 21:2487–2502 [DOI] [PubMed] [Google Scholar]
- Meunier H, Cajander SB, Roberts VJ, Rivier C, Sawchenko PE, Hsueh AJ, Vale W 1988 Rapid changes in the expression of inhibin α-, βA-, and βB-subunits in ovarian cell types during the rat estrous cycle. Mol Endocrinol 2:1352–1363 [DOI] [PubMed] [Google Scholar]
- Woodruff TK, D'Agostino J, Schwartz NB, Mayo KE 1989 Decreased inhibin gene expression in preovulatory follicles requires primary gonadotropin surges. Endocrinology 124:2193–2199 [DOI] [PubMed] [Google Scholar]
- Meunier H, Roberts VJ, Sawchenko PE, Cajander SB, Hsueh AJ, Vale W 1989 Periovulatory changes in the expression of inhibin α-, βA-, and βB-subunits in hormonally induced immature female rats. Mol Endocrinol 3:2062–2069 [DOI] [PubMed] [Google Scholar]
- Li Q, Kumar R, Underwood K, O'Connor AE, Loveland KL, Seehra JS, Matzuk MM 2007 Prevention of cachexia-like syndrome development and reduction of tumor progression in inhibin-deficient mice following administration of a chimeric activin receptor type II-murine Fc protein. Mol Hum Reprod 13:675–683 [DOI] [PubMed] [Google Scholar]
- Coerver KA, Woodruff TK, Finegold MJ, Mather J, Bradley A, Matzuk MM 1996 Activin signaling through activin receptor type II causes the cachexia-like symptoms in inhibin-deficient mice. Mol Endocrinol 10:534–543 [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Finegold MJ, Mather JP, Krummen L, Lu H, Bradley A 1994 Development of cancer cachexia-like syndrome and adrenal tumors in inhibin-deficient mice. Proc Natl Acad Sci USA 91:8817–8821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey GJ, Krasnow JS, Beattie WG, Richards JS 1990 Aromatase cytochrome P450 in rat ovarian granulosa cells before and after luteinization: adenosine 3′,5′-monophosphate-dependent and independent regulation. Cloning and sequencing of rat aromatase cDNA and 5′ genomic DNA. Mol Endocrinol 4:3–12 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick SL, Richards JS 1991 Regulation of cytochrome P450 aromatase messenger ribonucleic acid and activity by steroids and gonadotropins in rat granulosa cells. Endocrinology 129:1452– 1462 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Robayna IJ, Alliston TN, Buse P, Firestone GL, Richards JS 1999 Functional and subcellular changes in the A-kinase-signaling pathway: relation to aromatase and Sgk expression during the transition of granulosa cells to luteal cells. Mol Endocrinol 13:1318–1337 [DOI] [PubMed] [Google Scholar]
- El-Hefnawy T, Zeleznik AJ 2001 Synergism between FSH and activin in the regulation of proliferating cell nuclear antigen (PCNA) and cyclin D2 expression in rat granulosa cells. Endocrinology 142:4357–4362 [DOI] [PubMed] [Google Scholar]
- Park Y, Maizels ET, Feiger ZJ, Alam H, Peters CA, Woodruff TK, Unterman TG, Lee EJ, Jameson JL, Hunzicker-Dunn M 2005 Induction of cyclin D2 in rat granulosa cells requires FSH-dependent relief from FOXO1 repression coupled with positive signals from Smad. J Biol Chem 280:9135–9148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TR, Palapattu G, Wang P, Woodruff TK, Boime I, Byrne MC, Matzuk MM 1999 Transgenic models to study gonadotropin function: the role of follicle-stimulating hormone in gonadal growth and tumorigenesis. Mol Endocrinol 13:851–865 [DOI] [PubMed] [Google Scholar]
- Dierich A, Sairam MR, Monaco L, Fimia GM, Gansmuller A, LeMeur M, Sassone-Corsi P 1998 Impairing follicle-stimulating hormone (FSH) signaling in vivo: targeted disruption of the FSH receptor leads to aberrant gametogenesis and hormonal imbalance. Proc Natl Acad Sci USA 95:13612–13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel MH, Wootton AN, Wilkins V, Huhtaniemi I, Knight PG, Charlton HM 2000 The effect of a null mutation in the follicle-stimulating hormone receptor gene on mouse reproduction. Endocrinology 141:1795–1803 [DOI] [PubMed] [Google Scholar]
- Grieshaber NA, Boitano S, Ji I, Mather JP, Ji TH 2000 Differentiation of granulosa cell line: follicle-stimulating hormone induces formation of lamellipodia and filopodia via the adenylyl cyclase/cyclic adenosine monophosphate signal. Endocrinology 141:3461–3470 [DOI] [PubMed] [Google Scholar]
- Grieshaber NA, Ko C, Grieshaber SS, Ji I, Ji TH 2003 Follicle-stimulating hormone-responsive cytoskeletal genes in rat granulosa cells: class I β-tubulin, tropomyosin-4, and kinesin heavy chain. Endocrinology 144:29–39 [DOI] [PubMed] [Google Scholar]
- Shiota M, Tanihiro T, Nakagawa Y, Aoki N, Ishida N, Miyazaki K, Ullrich A, Miyazaki H 2003 Protein tyrosine phosphatase PTP20 induces actin cytoskeleton reorganization by dephosphorylating p190 RhoGAP in rat ovarian granulosa cells stimulated with follicle-stimulating hormone. Mol Endocrinol 17:534–549 [DOI] [PubMed] [Google Scholar]
- Albertini DF, Combelles CM, Benecchi E, Carabatsos MJ 2001 Cellular basis for paracrine regulation of ovarian follicle development. Reproduction 121:647–653 [DOI] [PubMed] [Google Scholar]
- Shikone T, Matzuk MM, Perlas E, Finegold MJ, Lewis KA, Vale W, Bradley A, Hsueh AJ 1994 Characterization of gonadal sex cord-stromal tumor cell lines from inhibin-α and p53-deficient mice: the role of activin as an autocrine growth factor. Mol Endocrinol 8:983–995 [DOI] [PubMed] [Google Scholar]
- Pangas SA, Li X, Umans L, Zwijsen A, Huylebroeck D, Gutierrez C, Wang D, Martin JF, Jamin SP, Behringer RR, Robertson EJ, Matzuk MM 2008 Conditional deletion of Smad1 and Smad5 in somatic cells of male and female gonads leads to metastatic tumor development in mice. Mol Cell Biol 28:248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M, Pangas SA 2009 New insights into the regulatory roles of TGFβ family members in folliculogenesis. WIREs Syst Biol Med 2:117–125 [DOI] [PubMed] [Google Scholar]
- Edson MA, Nalam RL, Clementi C, Franco HL, Demayo FJ, Lyons KM, Pangas SA, Matzuk MM 2010 Granulosa cell-expressed BMPR1A and BMPR1B have unique functions in regulating fertility but act redundantly to suppress ovarian tumor development. Mol Endocrinol 24:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Nakamura M, Igarashi S, Miyamoto K, Eto Y, Ibuki Y, Minegishi T 1994 Effect of activin on luteinizing hormone-human chorionic gonadotropin receptor messenger ribonucleic acid in granulosa cells. Endocrinology 134:2329–2335 [DOI] [PubMed] [Google Scholar]
- Tamura K, Matsushita M, Endo A, Kutsukake M, Kogo H 2007 Effect of insulin-like growth factor-binding protein 7 on steroidogenesis in granulosa cells derived from equine chorionic gonadotropin-primed immature rat ovaries. Biol Reprod 77:485–491 [DOI] [PubMed] [Google Scholar]
- Burns KH, Owens GE, Ogbonna SC, Nilson JH, Matzuk MM 2003 Expression profiling analyses of gonadotropin responses and tumor development in the absence of inhibins. Endocrinology 144:4492–4507 [DOI] [PubMed] [Google Scholar]
- Chu S, Rushdi S, Zumpe ET, Mamers P, Healy DL, Jobling T, Burger HG, Fuller PJ 2002 FSH-regulated gene expression profiles in ovarian tumours and normal ovaries. Mol Hum Reprod 8:426–433 [DOI] [PubMed] [Google Scholar]
- Reinholz MM, Zschunke MA, Roche PC 2000 Loss of alternately spliced messenger RNA of the luteinizing hormone receptor and stability of the follicle-stimulating hormone receptor messenger RNA in granulosa cell tumors of the human ovary. Gynecol Oncol 79:264–271 [DOI] [PubMed] [Google Scholar]
- Richards JS 2007 Genetics of ovulation. Semin Reprod Med 25:235–242 [DOI] [PubMed] [Google Scholar]
- Richards JS, Hernandez-Gonzalez I, Gonzalez-Robayna I, Teuling E, Lo Y, Boerboom D, Falender AE, Doyle KH, LeBaron RG, Thompson V, Sandy JD 2005 Regulated expression of ADAMTS family members in follicles and cumulus oocyte complexes: evidence for specific and redundant patterns during ovulation. Biol Reprod 72:1241–1255 [DOI] [PubMed] [Google Scholar]
- Doyle KM, Russell DL, Sriraman V, Richards JS 2004 Coordinate transcription of the ADAMTS-1 gene by luteinizing hormone and progesterone receptor. Mol Endocrinol 18:2463–2478 [DOI] [PubMed] [Google Scholar]
- Irving-Rodgers HF, Hummitzsch K, Murdiyarso LS, Bonner WM, Sado Y, Ninomiya Y, Couchman JR, Sorokin LM, Rodgers RJ 2010 Dynamics of extracellular matrix in ovarian follicles and corpora lutea of mice. Cell Tissue Res 339:613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Naito I, Momota R, Sado Y, Hasegawa H, Ninomiya Y, Ohtsuka A 2007 The distribution of type IV collagen α chains in the mouse ovary and its correlation with follicular development. Arch Histol Cytol 70:243–253 [DOI] [PubMed] [Google Scholar]
- Welt CK, Taylor AE, Fox J, Messerlian GM, Adams JM, Schneyer AL 2005 Follicular arrest in polycystic ovary syndrome is associated with deficient inhibin A and B biosynthesis. J Clin Endocrinol Metab 90:5582–5587 [DOI] [PubMed] [Google Scholar]
- Papachroni KK, Piperi C, Levidou G, Korkolopoulou P, Pawelczyk L, Diamanti-Kandarakis E, Papavassiliou AG 6 July 2009 Lysyl oxidase interacts with AGEs signaling to modulate collagen synthesis in polycystic ovarian tissue. J Cell Mol Med 10.1111/j. 1582–4934.2009.00841.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerboom D, Paquet M, Hsieh M, Liu J, Jamin SP, Behringer RR, Sirois J, Taketo MM, Richards JS 2005 Misregulated Wnt/β-catenin signaling leads to ovarian granulosa cell tumor development. Cancer Res 65:9206–9215 [DOI] [PubMed] [Google Scholar]
- Tian C, Zhou ZG, Meng WJ, Sun XF, Yu YY, Li L, Luo HZ, Yang L, Zhou B, Gu J 2007 Overexpression of connective tissue growth factor WISP-1 in Chinese primary rectal cancer patients. World J Gastroenterol 13:3878–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennica D, Swanson TA, Welsh JW, Roy MA, Lawrence DA, Lee J, Brush J, Taneyhill LA, Deuel B, Lew M, Watanabe C, Cohen RL, Melhem MF, Finley GG, Quirke P, Goddard AD, Hillan KJ, Gurney AL, Botstein D, Levine AJ 1998 WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci USA 95:14717–14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Nakachi K, Wang H, Elashoff R, Koeffler HP 2001 Elevated levels of connective tissue growth factor, WISP-1, and CYR61 in primary breast cancers associated with more advanced features. Cancer Res 61:8917–8923 [PubMed] [Google Scholar]
- Mason HD, Willis DS, Beard RW, Winston RM, Margara R, Franks S 1994 Estradiol production by granulosa cells of normal and polycystic ovaries: relationship to menstrual cycle history and concentrations of gonadotropins and sex steroids in follicular fluid. J Clin Endocrinol Metab 79:1355–1360 [DOI] [PubMed] [Google Scholar]
- Wachs DS, Coffler MS, Malcom PJ, Shimasaki S, Chang RJ 2008 Increased androgen response to follicle-stimulating hormone administration in women with polycystic ovary syndrome. J Clin Endocrinol Metab 93:1827–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.