Abstract
Chlorpyrifos (CPF) is a widely used organophosphorus insecticide (OP) and putative developmental neurotoxicant in humans. The acute toxicity of CPF is elicited by acetylcholinesterase (AChE) inhibition. We characterized dose-related (0.1, 0.5, 1 and 2 mg/kg) gene expression profiles and changes in cell signaling pathways 24 hr following acute CPF exposure in seven day-old rats. Microarray experiments indicated that approximately 9% of the 44,000 genes were differentially expressed following either one of the four CPF dosages studied (546, 505, 522, and 3,066 genes with 0.1, 0.5, 1.0 and 2.0 mg/kg CPF). Genes were grouped according to dose-related expression patterns using K-means clustering while gene networks and canonical pathways were evaluated using Ingenuity Pathway Analysis®. Twenty clusters were identified and differential expression of selected genes was verified by RT-PCR. The four largest clusters (each containing from 276–905 genes) constituted over 50% of all differentially expressed genes and exhibited up-regulation following exposure to the highest dosage (2 mg/kg CPF). The total number of gene networks affected by CPF also rose sharply with the highest dosage of CPF (18, 16, 18 and 50 with 0.1, 0.5, 1 and 2 mg/kg CPF). Forebrain cholinesterase (ChE) activity was significantly reduced (26%) only in the highest dosage group. Based on magnitude of dose-related changes in differentially expressed genes, relative numbers of gene clusters and signaling networks affected, and forebrain ChE inhibition only at 2 mg/kg CPF, we focused subsequent analyses on this treatment group. Six canonical pathways were identified that were significantly affected by 2 mg/kg CPF (MAPK, oxidative stress, NFKB, mitochondrial dysfunction, arylhydrocarbon receptor and adrenergic receptor signaling). Evaluation of different cellular functions of the differentially expressed genes suggested changes related to olfactory receptors, cell adhesion/migration, synapse/synaptic transmission and transcription/translation. Nine genes were differentially affected in all four CPF dosing groups. We conclude that the most robust, consistent changes in differential gene expression in neonatal forebrain across a range of acute CPF dosages occurred at an exposure level associated with the classical marker of OP toxicity, AChE inhibition. Disruption of multiple cellular pathways, in particular cell adhesion, may contribute to the developmental neurotoxicity potential of this pesticide.
Introduction
Organophosphorus insecticides (OPs) are a major class of pesticides used in agricultural, industrial and household applications worldwide. In 2001, approximately 73 million pounds of organophosphorus insecticides (OPs) were used in the US alone (Kiely et al., 2004). Concerns for higher sensitivity to OPs in young children led to the withdrawal of many household applications for OPs (U.S. EPA. 2000, 2002). While restrictions on household applications have undoubtedly been effective in risk management, OPs remain a major insecticide in agricultural and other settings with the potential for widespread exposure.
OPs are primarily neurotoxicants. The classic mechanism of acute neurotoxicity of OPs is initiated by inhibition of the enzyme acetylcholinesterase (AChE), leading to accumulation of the neurotransmitter acetylcholine and resulting cholinergic signs of toxicity (reviewed in Mileson et al., 1998). A number of studies suggest that inhibition of AChE during nervous system development could disrupt neurodevelopmental outcome (Jones et al., 1995; Koenigsberger et al., 1997; Sternfeld et al., 1998; Das and Barone, 1999; Bigbee et al., 2000: Howard et al., 2005; Paranaou and Layer, 2008). Moreover, some OPs can interact with other non-acetylcholinesterase macromolecules to potentially influence nervous system function and/or neurodevelopment (Pope, 1999; Casida and Quistad, 2005; Pope et al., 2005). Developing individuals have a relatively low capacity for OP detoxification, potentially increasing sensitivity to many OPs (Benke and Murphy, 1975; Karanth et al., 2000, 2001; Pope et al., 2005; James et al., 2005). A number of experimental and epidemiological studies suggest that OPs including chlorpyrifos (CPF) can disrupt neurodevelopment (Colt et al., 2004; Slotkin et al., 2004; Weiss et al., 2004; Young et al., 2005; Whyatt et al. 2003, 2004, 2005; Jacobson and Jacobson 2006; Rauh et al., 2006). Epidemiological studies associating adverse neurological outcomes with markers of CPF exposure suggested that exposures far below those sufficient to inhibit acetylcholinesterase may lead to disruption of neurodevelopment (Berkowitz et al., 2004; Whyatt et al., 2004, 2005: Rauh et al. 2006). Moreover, the initial inhibition of acetylcholinesterase and consequent enhancement of cholinergic signaling could lead to subsequent changes in other downstream neurotransmitter signaling pathways. Thus, neurodevelopmental effects of CPF and OPs could be due to acetylcholinesterase and/or non-acetylcholinesterase related actions.
A powerful approach for identifying potential targets/pathways leading to developmental neurotoxicity following low-level OP exposure is through microarray technology, yielding an overall genome-wide expression profile in a single platform. OP-induced oxidative stress, altered expression of transcription factors, and disruption of transmitter, cytokine and hormone cell signaling cascades have been previously proposed to contribute to altered neurodevelopment following OP exposure in both animal and cell culture models (Mense et al., 2006; Slotkin et al., 2007; 2009a; 2009b). We proposed that changes in gene expression in the neonatal rat brain following one of a range of acute CPF dosages would exhibit a classical dose-response relationship, with the most consistent changes being associated with exposures sufficient to inhibit brain acetylcholinesterase. Microarray technology and cluster/pathway analysis was used to identify potential sensitive genes/pathways in the forebrain of neonatal rats following acute low-level CPF exposure, with subsequent verification of selected genes using real time PCR.
Methods
Chemicals
Chlorpyrifos was purchased from Chem Service (West Chester, PA) and kept dessicated under nitrogen at 4°C. Acetylcholine iodide (acetyl-3H, specific activity 76 Ci/mmol) was purchased from Perkin Elmer (Wellesley, MA). Whole-genome rat microarray 60-mer oligonucleotide (4 × 44k) slides were purchased from Agilent Technologies (Santa Clara, CA). Two-channel labeling and hybridization kits and the wash buffers were purchased from Agilent Technologies. Primers for real-time PCR were purchased from Integrated DNA Technologies (San Diego, CA) and a cDNA synthesis kit was purchased from Invitrogen (Carlsbad, CA). Realtime PCR kits were purchased from Qiagen (Valencia, CA). All other chemicals were purchased from Sigma Chemical Company (St. Louis, MO).
Animal Treatments
All experiments were carried out in accordance with protocols established in the NIH/NRC Guide for the Care and Use of Laboratory Animals and approved by the local Institutional Animal Care and Use Committee. Untimed-pregnant Sprague-Dawley rats were purchased from Harlan (Indianapolis, IN) and housed in a temperature-controlled room in breeding cages with a 12 h light-dark cycle (0700–1900 h). Animals had free access to food and water. The day after birth, all pups were randomized and redistributed to include 9–10 pups per dam. On postnatal day 7 (PND 7) pups were treated by oral gavage with either vehicle (peanut oil, 1 ml/kg) or CPF (0.1, 0.5, 1 or 2 mg/kg) in peanut oil. Twenty four hours later, forebrain was dissected by making a coronal cut at the anterior border of the cerebellum and then sagittally into two halves: one half was stored in RNAlater (Ambion, Austin, TX) at 4°C for subsequent RNA extraction while the other half was stored at −80°C for biochemical assays. Body weights were measured prior to dosing and at 24 hours after dosing.
Tissue preparations and biochemical assays
Forebrain cholinesterase activity was measured using the radiometric method of Johnson and Russell (1975) with [3H] acetylcholine as the substrate (1 mM final concentration, Won et al., 2001). Carboxylesterase activity was measured in the same tissue samples by the method of Clement and Erhardt (1990) using p-nitrophenylacetate as substrate (Karanth and Pope, 2000). Total protein content was estimated by the method of Lowry and coworkers (1951) using bovine serum albumin as standard.
RNA extraction and microarray analysis
Total RNA was isolated using Trizol solution (Invitrogen Life technology, Carlsbad, CA) and the quality of RNA was assessed by running a denaturing formaldehyde/agarose gel. RNA samples with an A260/A280 ratio between 1.9 and 2.0 (estimated using NanoDrop ND-1000 spectrophotometer) and having intact ribosomal RNA bands were selected for use in preparing labeled cRNAs for hybridization. We used a two-color system (cy5 for CPF treatment RNA: cy3 for control RNA) to determine relative expression ratios between control and treatment groups. There were five biological replicates (n=5 pups) performed for each CPF dosage. RNA amplification was done using the Low RNA Input-Fluorescence Linear Amplification Kit (Agilent) with amplification and labeling by fluorescent dyes being carried out according to the manufacturer’s instructions. Hybridization was done using In situ Hybridization Kit-Plus (Agilent) at 60°C for 17 h. The washes were performed using Agilent’s wash buffers according to the wash protocol and Agilent’s Stabilization and Drying solution. The arrays were scanned using a Scan Array Express from Perkin-Elmer (Gaithersburg, MA).
Data analysis from the scanned slides
The images were processed using GenePix Pro 4.0 (Axon Inc.). Pre-processing of data used GenePix Autoprocessor (GPAP; http://darwin.biochem.okstate.edu/gpap/). This analysis included: 1) removal of data where the fluorescence signal intensity in both channels was less than the background plus two standard deviations; 2) removal of data points where the signal was less than 200 Relative Fluorescence Units in both channels; 3) removal of bad quality spots flagged during processing of the image using GenePix Pro; 4) log2 transformation of the background subtracted cy5/cy3 median intensity ratios. Following pre-processing, the expression results across replicates for each treatment were averaged and normalized using print tip LOWESS normalization (Yang et al., 2002) using the GPAP web site.
The classical approach of choosing differentially expressed genes on the basis of fold change in log2 ratios can result in false-positives or false-negatives. A statistical calculation was therefore conducted for each gene based on the Empirical Bayes approach using the R-statistical language (http://www.r–project.org) and the Bioconductor software package (http://www.bioconductor.org) through the GPAP web site. Each treatment sample was analyzed for transcriptional differences relative to the pooled control sample using Bioconductor. A False Discovery Rate (FDR) of 5% (p ≤ 0.05) or less was applied for confident identification of differentially expressed genes with a change of 1.7-fold in expression levels.
This dataset containing 3,932 differentially expressed genes having 1.7-fold change (log2 ratio of 0.8) and a p value of ≤ 0.05 was subjected to K-means cluster analysis based on CPF dosing groups using Genesis software, which generated twenty clusters using 50 iterations (Sturn et al., 2002). The dataset of selected genes was further subjected to improved annotation by using biological annotation programs including DAVID (Database for Annotation, Visualization and Integrated Discovery) provided by the National Institute of Allergy and Infectious Diseases (http://david.abcc.ncifcrf.gov/).
Ingenuity Pathway Analysis (http://www.ingenuity.com) was used to identify biologically relevant networks and canonical pathways affected by early CPF exposure. Selected genes with unique gene identifiers (Agilent probe set ID) and their corresponding fold change values were uploaded as tab-delimited text file where the gene identifiers were mapped to its corresponding gene object in the Ingenuity database. These were then called “focus genes” and used as starting points to query the database to generate biological networks with a statistical score provided for each network, indicating the probability of the focus genes within a network being due to random chance (a score > 2 signified less than 1% chance that the focus genes were randomly linked). Similarly, canonical pathways were developed and evaluated using the Fisher’s test to calculate p-values.
Real-time PCR
Selected differentially expressed genes across different clusters and pathways were confirmed by real-time PCR analysis. Primers were designed using the full-length rat cDNAs available from GenBank. Genome sequences were compared to mRNA sequence using the SPIDEY program from NCBI to establish the exon-intron boundaries. Primers were chosen using the software from Integrated DNA Technologies. The same preparations of RNA extracted for microarray experiments were used for RT-PCR with three independent biological replicates, each biological replicate having two technical replicates. Two µg of RNA was used to synthesize cDNA in a 25-µl reaction volume using iScript™ cDNA synthesis kit (Bio-Rad), which included removal of genomic DNA contamination before cDNA synthesis.
RT-PCR reactions were performed with the ABI 7700 (Perkin-Elmer Applied Biosystems, USA) using SYBR green PCR kit (Qiagen) and gene specific primers in 96-well plates in a total reaction volume of 25 µl. PCR cycling conditions were as follows: 95°C for 2 min, 95°C for 8 min, 94°C for 30 s, 59–65°C for 30 s (depending on the primer set) and 72°C for 30 s. Melt-curve analysis was performed which confirmed the specific amplification and single gene product in each case. The average threshold cycle (Ct) values from all the replicates were used to calculate the fold-changes relative to the internal control (18S rRNA). The comparative ΔΔCt method was used to quantify the level of gene expressions; 2− ΔΔCt was used to calculate the relative fold change in gene expression.
Statistical Analyses
Analysis of ChE and RT-PCR data was done by one-way ANOVA followed by Dunnett’s test using the JMP statistical package (SAS, 1995), where a p value <0.05 was considered statistically significant.
Results
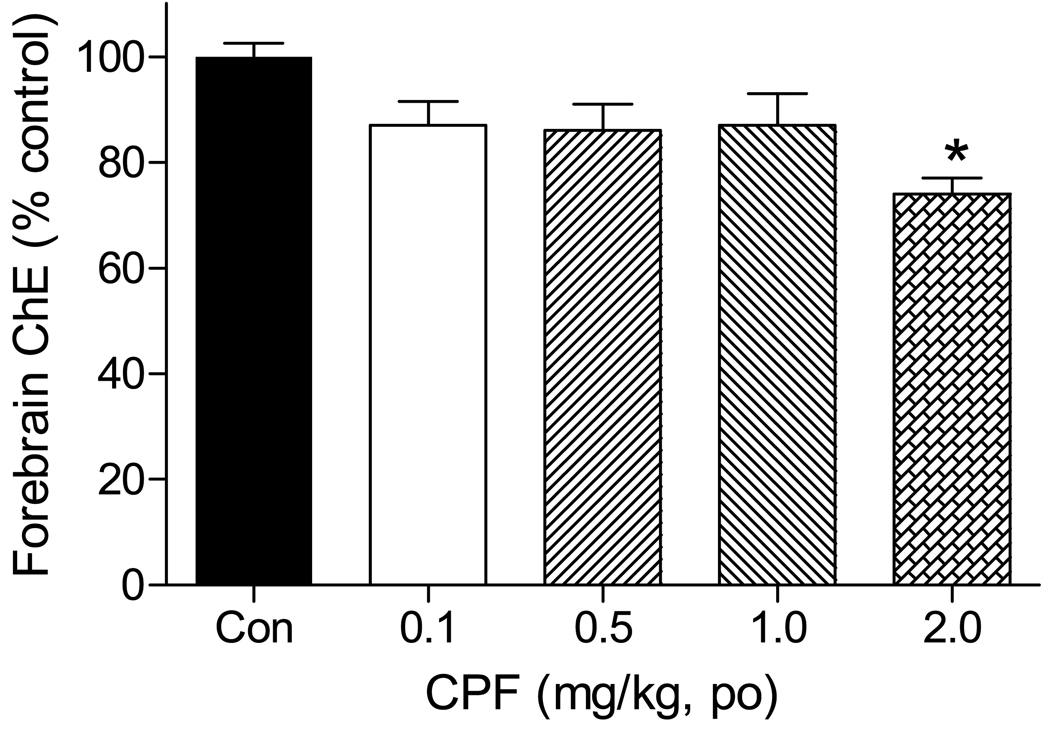
No overt signs of toxicity were noted in any pups following CPF dosing, and there were no significant changes in body weight in any CPF treatment groups (data not shown). No significant brain ChE inhibition was seen 24 hours following 0.1–1 mg/kg CPF, whereas the highest dosage (2 mg/kg CPF) elicited a slight but significant reduction in forebrain cholinesterase activity (26% inhibition; Figure 1). Carboxylesterases are important OP detoxifying enzymes, highly sensitive to inhibition by many OPs (Clement, 1984; Pond et al., 1995). No significant inhibition of brain carboxylesterase activity was noted in any of the CPF treatment groups (data not shown). Thus, functional and biochemical findings suggest that the dosages of CPF used in this study constituted low-level exposures, below or just at the level of inhibiting brain AChE activity.
Figure 1. Effects of CPF on forebrain cholinesterase activity.
Seven day-old rats were treated with 0, 0.1, 0.5, 1, 2 mg/kg CPF (oral gavage) as described in Materials and Methods and forebrain was collected 24 hours later. ChE activity was measured by the radiometric method and values expressed as percent of contemporaneous controls. Data represent mean ± SE (n=5/treatment group). An asterisk indicates a significant difference (p<0.05) between control and treatment groups. The activity of ChE in the controls was 56 ± 1.2 nmol acetylcholine hydrolyzed/min/mg of protein.
Out of 44,000 genes on the array, 3,932 were identified as differentially expressed in at least one treatment group, based on 1.7-fold change (log2 ratio of 0.8) in expression and a significant p value of ≤ 0.05 (see Table S1 for listing of all differentially expressed genes). The highest dosage (2 mg/kg CPF) was associated with the greatest number of differentially expressed genes (3,066 [2,553 up, 513 down]), compared to lesser but relatively similar numbers of differentially expressed genes in the lower dosage groups (0.1 mg/kg, 546 [380 up, 166 down]; 0.5 mg/kg, 505 [260 up, 245 down]; 1 mg/kg, 522 [283 up, 239 down]).
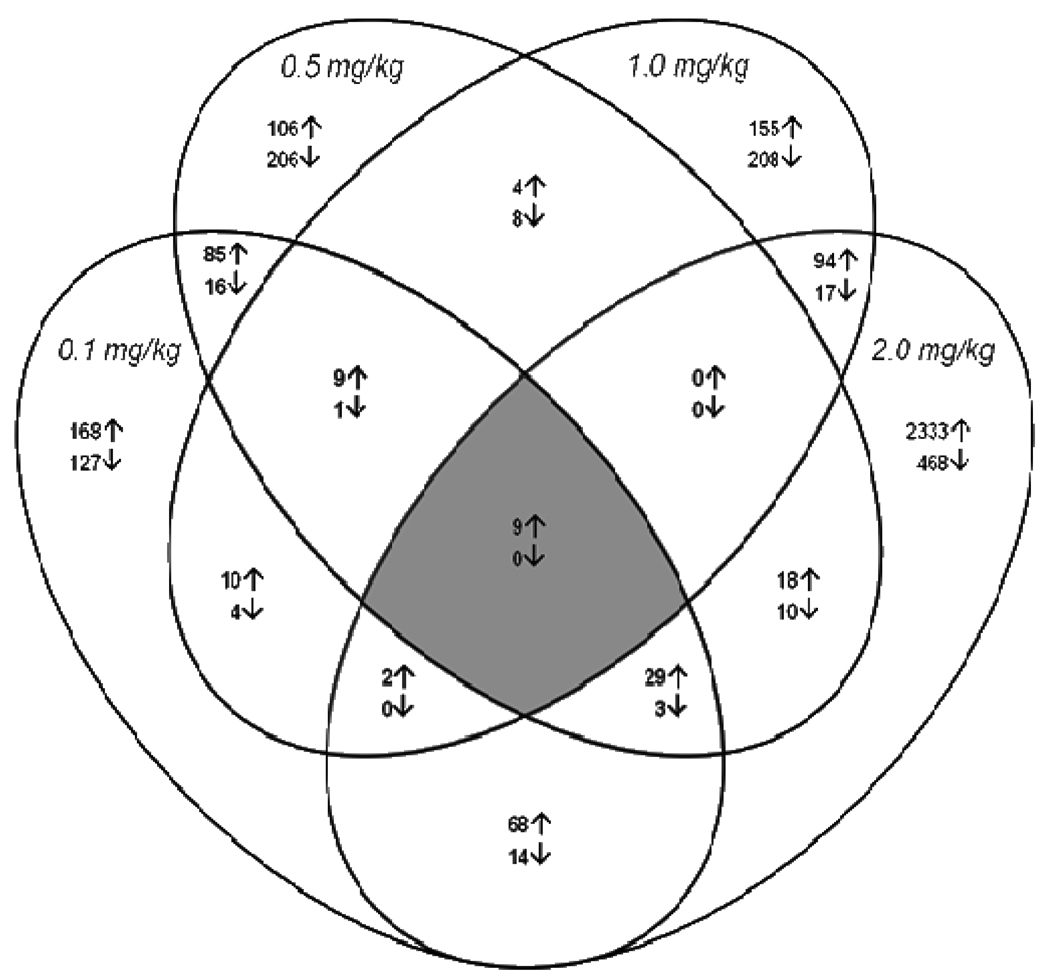
Figure 2 is a Venn diagram showing overlap of gene expression changes with the four CPF dosing conditions. As shown in this figure, there was relatively little overlap in differentially expressed genes across the 20-fold CPF dosage range. For example, across all four dosages (0.1, 0.5, 1 and 2 mg/kg), out of the 3,932 differentially expressed genes, only nine were commonly up-regulated. Comparing between consecutive dosages (similar to the approach used in Stapleton and Chan, 2009), there was some overlap between 0.1 and 0.5 mg/kg (85 up-regulated/16 down-regulated), between 0.5 and 1 mg/kg groups (4 up-regulated/8 down-regulated) and between 1 and 2 mg/kg (94 up-regulated/17 down-regulated) genes.
Figure 2. Venn diagram showing the differentially expressed genes across all the dosages.
This diagram illustrates the overlap of forebrain gene expression changes across dosages following CPF exposure. Upward arrows in the diagram indicate up-regulation while downward arrows indicate down-regulation.
K-means cluster analysis
Clustering using K-means analysis was conducted to evaluate groups of genes based on similar dose-related differences in expression. Genes are initially divided into a number (k) of user-defined groups containing equal numbers of genes. The centroid of each cluster is calculated as the average of the expression profiles. Genes are then reassigned to the cluster in which the centroid is the most similar to that gene. Group centroids are then recalculated and the process is reiterated until cluster compositions converge (Do and Choi, 2008; Zhu et al., 2008)
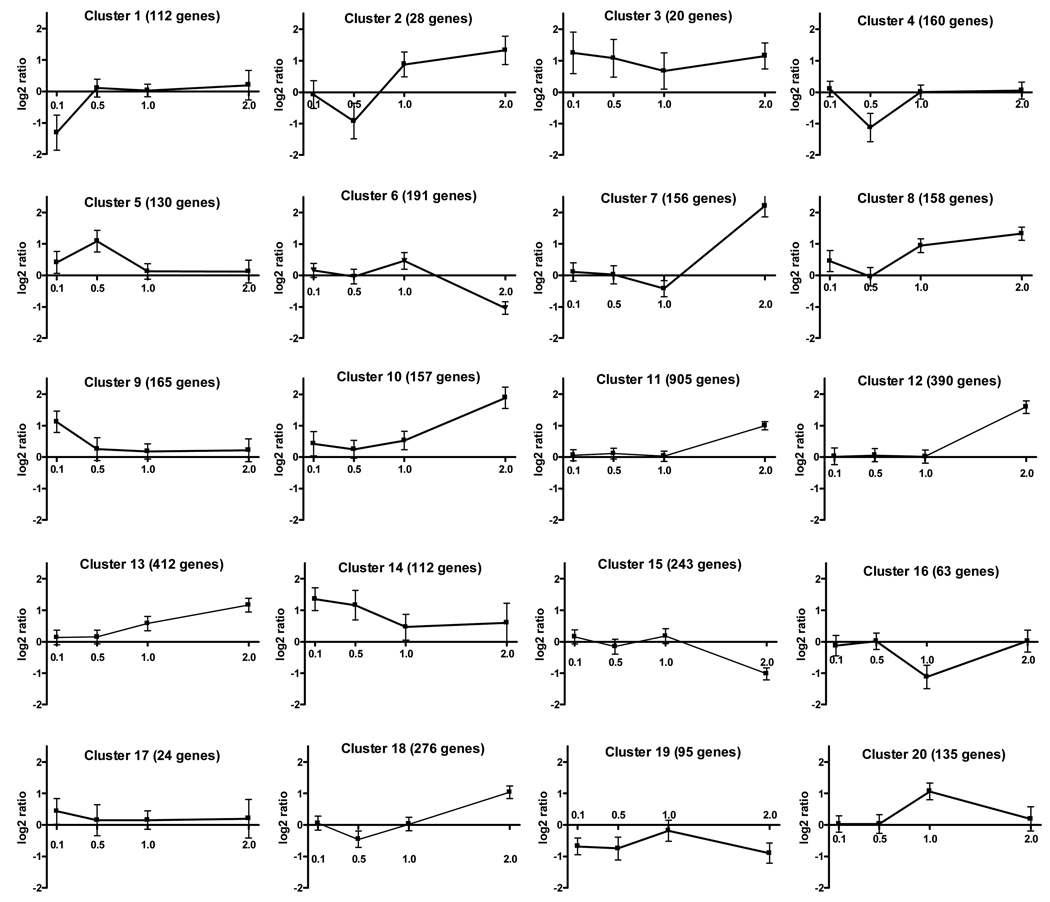
Figure 3 shows 20 gene clusters, with the total numbers of genes in each provided in the title of each pane. Of the 20 clusters, the top four (clusters 11, 12, 13 and 18; based on total numbers of genes within each) contained over 50% of all differentially expressed genes, with all of these clusters showing up-regulation in the 2 mg/kg CPF exposure group. Five others (clusters 6, 7, 8, 10 and 15) also showed high numbers of either up-regulated or down-regulated genes following 2 mg/kg CPF.
Figure 3. K-means cluster analysis of gene expression changes.
K-means cluster analysis was conducted on the 3,932 genes which were differentially expressed. On the X-axis different dosages are shown and on the Y-axis the log2 ratios of gene expression are provided. The Genesis program generated twenty clusters, with the greatest number of gene changes noted in the 2 mg/kg CPF dosage group in clusters 11>13>12>18 (ranked according to number of genes included in each, as shown in the title of each plate).
Cluster 11 was by far the largest, with 905 (871 unique) genes up-regulated at the highest CPF dosage (2 mg/kg). Examination of the differentially expressed genes in this cluster suggested three function-related groupings: 1) olfactory receptors, 2) synapse/synaptic transmission and 3) cell adhesion/migration. There were 34 different olfactory receptor genes in this cluster. In fact, of the seven top clusters showing up-regulation, all contained a number (3–34, 82 total) of different olfactory receptor genes, while the top two clusters showing down-regulation (clusters 6 and 15) contained no olfactory receptor genes. A number of synapse-relevant genes were noted in cluster 11 including adrenergic α1b (Adra1b), α2a (Adra2a) and β1 (Adrb1) receptors, the cholinergic muscarinic M5 (Chrm5) and nicotinic α7 (Chrna7), the GABA-B1 (Gabbr1) and GABA-C rho 1 (Gabrr1) receptor subunits, the glutamate NMDA2D receptor (Grin2d), the galanin 1 receptor (Galr1), adenylyl cyclase 5 (Adcy5), adenylyl cyclase associated protein 1 (Cap1) and SNAP25-interacting protein (Snip). Genes related to cell adhesion were also prominent, including integrin beta 1 binding protein 2 (Itgb1bp2), integrin alpha 5 (Itga5), ankyrin repeat and kinase domain containing 1 (Ankk1), cell adhesion molecule-related/down-regulated by oncogenes (Cdon), as well as similar to protocadherin gamma subfamily B6 (Pcdhgb6), similar to protocadherin beta 8 (Pcdhb8) and similar to catenin alpha 3 (Rgd1562230). Cluster 11 also contained three transcription/translation-related genes, the Sp1 transcription factor (Sp1), Sp7 transcription factor (Sp7), and runt-related transcription factor 3 (Runx3). Fibroblast growth factors (Fgf2 and Fgf23), mitogen-activated kinase 6 (Mapk6) and mitogen-activated protein kinase kinase 4 (Mkk4), heme oxygenase 1 (Ho1), and glutathione-S-transferase mu 5 (Gstm5) were also included in this cluster.
Cluster 13 contained the second most differentially expressed genes (412) showing up-regulation following 2 mg/kg CPF. Again, olfactory receptor genes were prominently represented, with 18 different genes in the cluster. Synapse-related genes were numerous including the GABA-A receptor theta subunit (Gabrq), dopamine D2 receptor (Drd2), angiotensin II type 1 receptor (Agtr1), and both calcium/calmodulin-dependent protein kinase II γ (CamkIIγ) and protein kinase Cγ (Prkcγ). Only one adhesion-related gene was noted in cluster 12, protocadherin gamma subfamily C3 (Pcdh2). Mitogen-activated protein kinase 1 (Mapk1), mitogen-activated protein kinase kinase kinase 4 (Mkkk4), and the neuron-specific growth suppressor necdin (Ndn) were also in this cluster.
Cluster 12 had the next highest number of up-regulated genes (390). There were nine olfactory receptor genes but few genes related to either cell adhesion or synapse/signal transduction, other than phospholipase C γ2 (Plcg2) and the neurexin binding protein, neurexophilin 4 (Nxph4). Artemin (Artn, a glial-derived neurotrophic factor), nerve growth factor beta (Ngfb), super oxide dismutase 2 (Sod2) and two important OP biotransformation-related genes, i.e., carboxylesterase 1 (Ces1) and paraoxonase 1 (Pon1) were also in this cluster.
Cluster 18 was the fourth largest cluster (276 genes). Again, olfactory receptor genes (nine) were prominent. Synapse-related genes included the glutamate NMDA2B receptor (Grin2b), somatostatin 4 receptor (Sstr4), neurexophilin 3 (Nxph3) and phospholipase A2 group IVB (Pla2g4b). Several cell adhesion-related genes were noted including integrin alpha 5 (Itga5), a disintegrin-like and metallopeptidase with thromobospondin type 1 motif 13 (Adamts13), ankyrin-2 (Ank2) and tight junction protein 1 (Tjp1). Other selected genes in this cluster included transforming growth factor beta (Tgfb), leptin (Lep), the pleiotroic protein RAS-related C3 botulinum substrate 2 (Rac2) and heat shock protein 4 (Hspa4).
Clusters 7, 8 and 10 had relatively similar numbers of differentially expressed genes (156, 158 and 157 respectively). Selected genes in cluster 7 included three olfactory receptor genes, protein kinase A anchor protein 3 (Akap3), protein tyrosine phosphatase receptor H (Ptprh), unc-5 homolog D (Unc5d), the cholinergic nicotinic receptor β1 subunit (Chrnb1), and the adhesion-related genes protocadherin gamma subfamily A5 (Pcdhga5) and paxillin (Pxn). Selected genes in cluster 8 included six olfactory receptor genes, phosphotriesterase (Pter, an enzyme involved in OP hydrolysis), synaptotagmin 8 (Syt8), protein tyrosine kinase 6 (Ptk6), guanylate cyclase activator 2a (Guca2a), neuregulin 2 (Nrg2), and olfactomedin-like 1 (Olfml1). Selected genes in cluster 10 included three olfactory receptor genes, heat shock 27kD protein member 7 (Hspb7), copper containing amine oxidase 3 (Aoc3), beta adrenergic receptor kinase 1 (Adrkb1) and aryl hydrocarbon receptor (Ahr).
As noted above, an obvious difference in differentially expressed genes between clusters showing down-regulation (Clusters 6 and 15) compared to those showing up-regulation following 2 mg/kg CPF was that no olfactory receptor genes were noted. Clusters 6 and 15 did, however, contain a number of synapse/synaptic transmission-related genes. Cluster 6 contained genes for synaptophysin-like protein (Sypl1), synaptotagmin binding cytoplasmic RNA interacting protein (Syncrip), and protein kinase A anchor protein 9 (Akap9). Cluster 15 included synaptotagmin II (Syt2), synaptotagmin XII (Syt12), syntaxin 6 (Stx6), unc-13 homolog C (Unc13c), G-protein coupled receptor kinase-interactor 2 (Git2), the glutamate NMDA2C receptor (Grin2c), striatin (calmodulin binding protein 3, Strn3), dynamin 1-like (Dnm1l), and seizure related 6 homolog (Sez6). Clusters 6 and 15 also contained a number of cell adhesion-related genes including ankyrin repeat and MYND domain containing 2 (Ankmy2), ankyrin repeat and BTB domain containing 2 (Abtb2), similar to ankyrin repeat domain 40 (Ankrd40), protocadherin 9 (Pcdh9), neurotrimin (Ntm), junctional adhesion molecule 3 (Jam3), and RAS-GTPase-activating protein SH3-domain binding protein (G3bp1). Cluster 6 showed prominent changes in transcription/translation-related genes including transcription factor 12 (Tcf12), transcription factor 19 (Tcf19), transcription elongation regulator 1 (Tcerg1), eukaryotic translation initiation factor 3 subunit 10 (Eif3a), eukaryotic translation initiation factor 4 subunit gamma 1 (Eif4g1) and a number of heat shock-related genes including heat shock protein 1 (Hspd1), heat shock protein 1 alpha (Hsp90aa1), heat shock 70kD protein 1A (Hspa1a), heat shock 70kD protein 4-like (Hspa4l), and similar to heat shock protein 1 (Loc501135). The cell cycle regulatory proteins cyclin B1 (Ccnb1) and cyclin B2 (Ccnb2) were also in cluster 6. Cluster 15 contained growth factor-related genes including nuclear factor of kappa light chain gene enhancer in B-cells p105 (Nfkb1), neurotrophic tyrosine kinase receptor type 2 (Ntrk2), and inhibitor of kappaB kinase gamma (Ikbkg).
Following cluster analysis, we evaluated gene networks and canonical signaling pathways across dosages to determine potentially sensitive targets and molecular interconnections using Ingenuity Pathway Analysis®. The numbers of gene networks affected across CPF dosages were 18, 16, 18 and 50 following 0.1, 0.5, 1 and 2 mg/kg CPF, respectively. Similar to both the relative numbers of differentially expressed genes across dosages, and the relative numbers of clusters as well as numbers of genes within the clusters showing changes following exposure to 2 mg/kg CPF, it appeared that a point of departure was exhibited with the highest exposure level, i.e., the number of networks affected was notably increased with the highest CPF dosage. As 2 mg/kg CPF a) elicited the greatest number of differentially expressed genes in the top clusters identified, b) was associated with markedly higher numbers of affected gene networks, and c) was the only dosage associated with significant inhibition of forebrain cholinesterase activity, we focused subsequent evaluations on changes following this exposure level.
Using Ingenuity Pathway Analysis®, six canonical pathways affected by 2 mg/kg CPF were identified with significant p values (in rank order of lower to higher p values, p < 0.000001 – p < 0.003): MAPK signaling < oxidative stress < NF-κB signaling < mitochondrial dysfunction < arylhydrocarbon receptor signaling < alpha adrenergic receptor signaling. Tables 1 and 2 show the top 30 up-regulated and down-regulated genes following 2 mg/kg CPF exposure (ranked by the magnitude of fold-change). A number of these highly up- or down-regulated genes were contained within the top clusters described above.
Table 1.
Top 30 upregulated genes following chlorpyrifos (2 mg/kg, po).
| Genebank Accn | Description | Fold Δ | Symbol |
|---|---|---|---|
| AI716801 | beta Adrenergic Receptor kinase 1 | 7.88 | Adrbk1 |
| AF387339 | HLA-B associated transcript 1A - | 7.71 | Bat1 |
| AW141787 | Bacuoloviral IAP repeat-containing 6 | 7.64 | Birc6 |
| BM388896 | Catenin (Cadherin associated protein), β 1 | 7.48 | Ctnnb1 |
| NM_001014771 | Envelope glycoprotein syncytin-A | 7.03 | Gm52 |
| AI231033 | Synaptojanin 1 | 6.66 | Synj1 |
| XM_342966 | Heat shock 27kD protein | 6.49 | Hspb7 |
| XM_233587 | Hypothetical protein FLJ32784 | 6.28 | Vwa5b1 |
| XM_221633 | Single-minded 2 | 6.28 | Sim2 |
| BC091428 | T-cell receptor beta chain | 6.09 | Tcbr |
| AW143700 | Helix-loop-helix ubiquitous kinase | 6.03 | Chuk |
| AW520784 | Zinc transporter 3 | 5.95 | Slc30a3 |
| NM_133396 | Testis-specific kinase 2 | 5.94 | Tesk2 |
| NM_001012147 | Paxillin | 5.87 | Pxn |
| AI045814 | Vanin 1 | 5.84 | Vnn1 |
| XM_573266 | Hypothetical protein FLJ20507 | 5.82 | Tmem127 |
| BM390441 | SRY-box containing gene 18 | 5.81 | Sox18 |
| NM_019208 | Multiple endocrine neoplasia 1 | 5.74 | Men1 |
| NM_053937 | Potassium voltage-gated channel, subfamily H, member 6 | 5.72 | Kcnh6 |
| NM_001025713 | Similar to melanoma antigen family A, 10 | 5.69 | Magea10 |
| AW141698 | Stearoyl-CoA Desaturase 2 | 5.69 | Scd2 |
| BI293306 | ATPase H+transporting, V1 subunit C, isoform 1 | 5.69 | Atp6v1c1 |
| BQ194049 | Tubulin Kinase 1 | 5.69 | Ttbk1 |
| AI411542 | Six transmembrane epithelial antigen of prostrate 2 | 5.51 | Steap2 |
| XM_577714 | Similar to SPBPJ4664.02 | 5.50 | SPBPJ4664.02 |
| NM_130414 | ATP-binding cassette, sub-family G, member 8 | 5.42 | Abcg8 |
| BM986233 | Calreticulin | 5.37 | Calr |
| XM_238575 | Hypothetical protein LOC301230 | 5.36 | LOC301230 |
| NM_013149 | Aryl hydrocarbon receptor | 5.35 | Ahr |
| BQ207094 | Serum response factor | 5.31 | Srf |
Table 2.
Top 30 downregulated genes following chlorpyrifos (2 mg/kg, po).
| Genebank Accn |
Description | Fold Δ |
Symbol |
|---|---|---|---|
| NM_019218 | Neurogenic differentiation 1 | −3.45 | Neurod1 |
| NM_017333 | Endothelin receptor type B | −3.35 | Ednrb |
| M58436 | Serine-threonine phosphatase catalytic subunit PP-1c | −3.29 | Pp1c |
| NM_020082 | RNase A family 4 | −3.20 | Rnase4 |
| NM_053296 | Glycine receptor, beta subunit | −3.15 | Glrb |
| NM_017009 | Glial fibrillary acidic protein | −3.13 | Gfap |
| NM_001009264 | Morf4 family associated protein 1 | −3.07 | Mrfap1 |
| NM_053655 | Dynamin 1 | −3.06 | Dnml1 |
| XM_238649 | Eukaryotic translation initiation factor 3, subunit 10 (theta) | −2.83 | Eif3s10 |
| XM_001063323 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) |
−2.80 | Elavl4 |
| XM_001074330 | Ethanolamine kinase 1 | −2.79 | Etnk1 |
| XM_214245 | MYC binding protein 2 | −2.78 | Mycbp2 |
| XM_001060756 | Eukaryotic translation initiation factor 4 gamma, 1 | −2.73 | Eif4g1 |
| XM_341663 | DnaJ (Hsp40) homolog, subfamily B, member 1 | −2.67 | Dnajb1 |
| XM_001060518 | SH3-binding domain glutamic acid-rich protein | −2.66 | Sh3bgrl |
| NM_019161 | Cadherin 22 | −2.65 | Cdh22 |
| NM_019149 | Matrin 3 | −2.61 | Matr3 |
| XM_001073523 | 3-oxoacid CoA transferase 1 | −2.54 | Oxct1 |
| XM_225735 | Ring finger and KH domain | −2.53 | Rkhd2 |
| XM_213849 | Nuclear factor I/X | −2.50 | Nfix |
| NM_031798 | Solute carrier family 12, member 2 | −2.50 | Slc12a2 |
| XM_221532 | TBC1 domain family, member 23 | −2.49 | Tbc1d23 |
| XM_001077084 | Trinucleotide repeat containing 6B | −2.47 | Tnrc6b |
| NM_022397 | Heterogeneous nuclear ribonucleoprotein F | −2.46 | Hnrnpf |
| NM_171991 | Cyclin B1 | −2.45 | Ccnb1 |
| XM_001061855 | MIR-interacting saposin-like protein (Transmembrane protein 4) | −2.45 | LOC68500 1 |
| NM_001013207 | RNA-binding region (RNP1, RRM) | −2.44 | Rnpc2 |
| XM_001054517 | Transmembrane emp24 protein transport domain | −2.44 | Tmed7 |
| XR_007953 | Heat shock protein 1 (chaperonin) | −2.43 | Hspd1 |
| XM_214535 | Aldehyde dehydrogenase family 7 | −2.39 | Aldh7a1 |
Table 3 shows the 9 genes that were differentially expressed among all four dosage groups: all were upregulated. Interestingly, a number of cell adhesion-related genes including focal adhesion kinase (Fak1), glycoprotein IX (Gcp9), glypican 3 (Gp3), leucine rich repeat and fibronectin III domain containing 1 (Lrfn1) and adhesion molecule with Ig like domain 3 (Amigo3) were upregulated across all four CPF dosages.
Table 3.
Genes differentially expressed in common among all four chlorpyrifos dosing groups (0.1, 0.5, 1 and 2 mg/kg, po).
| Genebank Accn |
Description | Fold Δ |
Symbol | |||
|---|---|---|---|---|---|---|
| 0.1 | 0.5 | 1 | 2 | |||
| XM_229333 | 60S ribosomal protein L19 | 2.6 8 |
4.0 0 |
2.38 | 5.4 2 |
Rpl19 |
| XM_576094 | Serine/threonine-protein phosphatase 4 regulatory subunit 4 |
2.4 4 |
2.4 3 |
2.00 | 2.8 8 |
Ppp4r4 |
| BF290004 | PTK2 protein tyrosine kinase 2, aka focal adhesion kinase 1 |
2.2 4 |
1.8 7 |
2.12 | 2.9 1 |
Fak1 |
| XM_222750 | Zinc finger, C2H2 type, similar to zinc finger protein 648 |
2.4 4 |
2.4 1 |
2.07 | 2.6 3 |
LOC1003648 22 |
| AW919536 | Methylcytosine dioxygenase TET1 | 2.5 6 |
2.4 5 |
1.98 | 2.1 4 |
Tet1 |
| NM_178144 | Adhesion molecule with Ig like domain 3 | 2.7 5 |
2.2 1 |
3.29 | 2.1 9 |
Amigo3 |
| XM_344874 | Synaptic adhesion-like molecule | 3.4 9 |
2.9 7 |
2.62 | 2.1 5 |
Lrfn1 |
| NM_00103182 5 |
Glycoprotein 9 | 2.0 4 |
1.8 8 |
1.86 | 2.0 0 |
Gp9 |
| AI02940 | Glypican 3 | 2.4 5 |
1.9 9 |
1.87 | 1.8 1 |
Gpc3 |
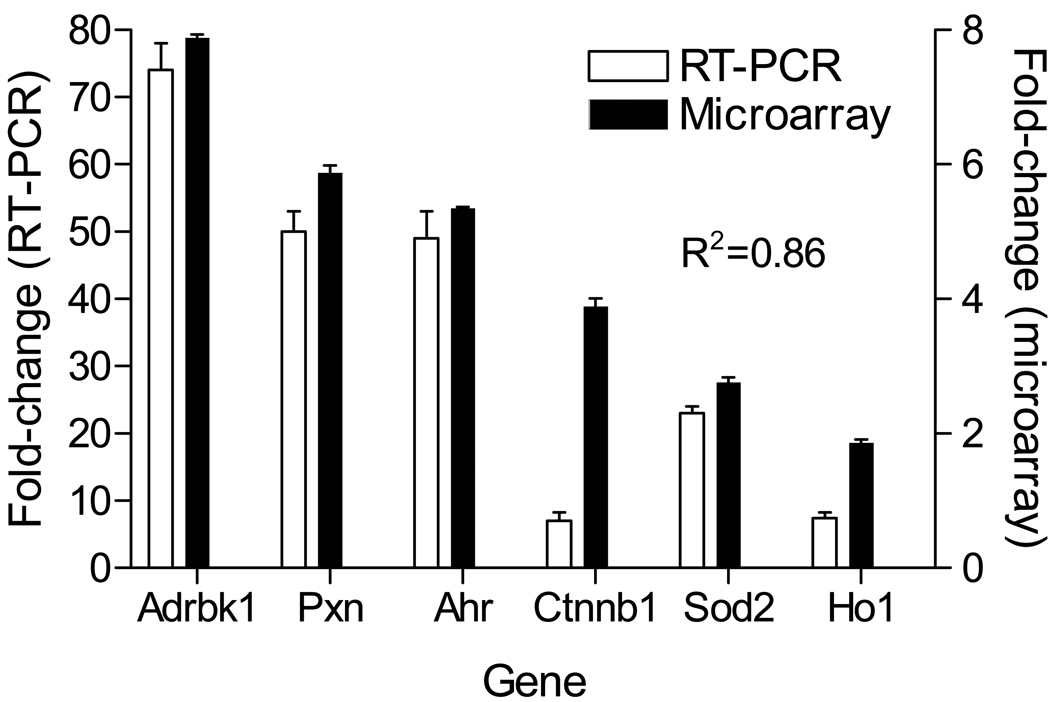
To confirm the differential expression of genes detected by microarray analysis, we sampled six genes (Adbrk1, Ahr, Pxn, Ctnnb1, Ho1 and Sod2) across the different clusters for verification using real-time PCR. Figure 4 shows comparative changes in these genes as determined by microarray and by RT-PCR. In all cases, results from RT-PCR generally agreed with changes in the microarray analysis although the absolute degree of change differed markedly between the methods.
Figure 4. Real time PCR of selected genes.
Real-time PCR was performed on Adrbk1, Ahr, Pxn, Ctnnb1, Ho1, and Sod2. The left Y-axis represents the fold-change with RT-PCR (mean ± SE) while the right Y-axis shows fold-change for microarray results (mean ± SE) in the same genes.
Discussion
Chlorpyrifos (CPF) is a common organophosphorus insecticide (OP), used worldwide in agricultural, industrial and household applications. Experimental and epidemiological studies suggest that early CPF exposures may lead to neurodevelopmental toxicity (Colt et al., 2004; Slotkin et al., 2004; Weiss et al., 2004; Young et al., 2005; Whyatt et al. 2003, 2004, 2005; Jacobson and Jacobson 2006; Rauh et al., 2006). Indeed, a number of epidemiological studies reported an association between umbilical cord/maternal plasma chlorpyrifos levels and adverse neurodevelopmental outcomes in children, with exposures apparently far below those necessary to inhibit acetylcholinesterase (Whyatt et al., 2003, 2004; Rauh et al., 2006). We hypothesized however that gene expression changes in neonatal rat brain following acute CPF exposure would exhibit a similar dose threshold as brain acetylcholinesterase inhibition. Neonatal rats were treated acutely (on postnatal day 7) with CPF (0.1, 0.5, 1 or 2 mg/kg) and tissues were sampled 24 hours later for biochemical and molecular assessments. Only the highest dosage (2 mg/kg) significantly reduced brain cholinesterase 24 hours after exposure (Figure 1). Carboxylesterase, a sensitive non-cholinesterase enzyme, was not significantly inhibited in these same tissues. Body weight was also not affected by CPF, and no overt signs of cholinergic toxicity were noted. Together, these data suggested that the CPF dosages used did not substantially influence cholinergic signaling through acetylcholinesterase inhibition or elicit any cholinergic toxicity.
In total, about 9% of all genes (3,932/44,000) were differentially expressed following CPF exposure. There were substantially more differentially expressed genes following 2 mg/kg CPF compared to lower, relatively similar numbers of differentially expressed genes in the lower exposure groups (0.1, 0.5 and 1 mg/kg CPF). Furthermore, there appeared to be relatively little overlap of differentially expressed genes among the four dosage groups (Figure 2). K-means analysis was conducted to further evaluate dosage-related changes in gene expression. Of the twenty clusters, we noted that the top 4 clusters represented about 50% of all genes differentially affected, all showing upregulation following 2 mg/kg CPF. Five other clusters also showed either up- or down-regulation of ≥156 genes in the 2 mg/kg CPF group. We then used Ingenuity Pathway Analysis® to evaluate gene networks affected by CPF. This analysis again suggested a break point across exposure levels, with markedly higher numbers of networks affected following 2 mg/kg CPF exposure compared to lesser, relatively similar changes in the lower dosing groups. Together, these dosage-related biochemical and molecular endpoints all suggested a departure in the dose-response curve with the highest dosage of CPF. Based on these findings, we further evaluated the differentially expressed genes in the top clusters as well as canonical signaling pathways affected by the highest dosage of CPF (2 mg/kg).
When examining genes within the different clusters, one of the most obvious changes was a notable up-regulation of olfactory receptor genes. All of the top gene clusters showing up-regulation following 2 mg/kg CPF (i.e., 7, 8, 10, 11, 12, 13, 18) contained a number of olfactory receptor genes, comprising almost 4% (34 of 905) of the differentially expressed genes in the largest cluster (cluster 11, Figure 3). In contrast, in the two clusters showing down-regulation (6 and 15), no olfactory genes were differentially affected. There are hundreds of different G protein-coupled olfactory receptors involved in odor perception (Buck and Axel, 1991; Su et al., 2009). The up-regulation of these different receptors by early CPF exposure could potentially lead to altered olfaction. Possible effects of early CPF exposure on development of olfactory systems are unknown, but should be investigated in future studies.
Across the top clusters showing up-regulation, genes related to synapse/signal transduction were highly represented. Genes for adrenergic (Adra1b, Adra2a, Adrb1), cholinergic (Chrm5, Chrna7, Chrnb1), glutamatergic (Grin2b, Grin2d), GABAergic (Gabrq, Gabbr1, Gabrr1), dopaminergic (Drd2), somatostatin (Sstr4), and galanin (Galr1) receptors were all up-regulated. The up-regulation of multiple transmitter receptor subtypes could potentially lead to altered synaptic signaling and interconnections during critical periods of neurodevelopment. It should be noted that changes in expression of neurotransmitter receptor genes following CPF exposure have been previously reported. In a focused gene expression study, daily administration of CPF (1 mg/kg on postnatal days 1–4) led to changes in gene expression for cholinergic, serotonergic, adrenergic and dopaminergic receptors (Slotkin and Seidler, 2007). These authors reported no change in expression of Adra1b, about a 15% increase in Adra2a, and about a 5% reduction in Adrb1. In contrast, we detected a 2.2-, 2.5- and 1.9-fold increase in these same genes. Interestingly, while we found that acute CPF (2 mg/kg) increased the expression of Chrm5 (muscarinic M5 receptor, > 2-fold), these authors reported about a 30% reduction in expression of Chrm5. Slotkin and Seidler (2007) also reported robust changes in expression of nicotinic receptor genes, with 11/14 showing changes (about 30–60% change from control) following CPF dosing. Of note, they reported about 30% reduction in Chrna7 (alpha 7 receptor), while we noted a 2.0-fold increase. We noted a 4.1-fold increase in expression of the beta 1 subunit (Chrnb1) but this receptor was not evaluated in the study by Slotkin and Seidler (2007). Slotkin and Seilder (2007) generally reported lesser effects on dopaminergic receptors, but detected a slight reduction (about 10%) in Drd2. In contrast, we noted a 2.8-fold increase in expression of Drd2. Generally, differential expression of genes for some of the same neurotransmitter receptors was noted in both studies, whether CPF was given acutely (on postnatal day 7 with evaluation on day 8, described herein) or repeatedly (on postnatal days 1–4 with evaluation on day 5, Slotkin and Seidler, 2007), but the results were typically in the opposite direction and the magnitude of changes was quite different.
Along with these cell surface receptors, a number of signal transduction-associated genes including those related to cAMP and cGMP signaling (Adcy5, Cap1, Akap3, Guca2), and phosphoprotein metabolism (Camk2γ, Prkc γ, Ptk6, Ptprh) were up-regulated. Synaptic vesicle exocytosis-related genes (Snip, Syt8) and the G-protein coupled receptor kinase 2 (GRK2, Adrkb1), involved in initial steps of desensitization of a number of G-protein coupled receptors, were also up-regulated. In fact, Adrkb1 was the most highly up-regulated gene (7.9-fold) noted out of all the clusters. Interestingly, Slotkin and Seidler (2007) reported about a 15% reduction in Adrkb1 expression in forebrain following repeated CPF exposures. We previously reported that chlorpyrifos oxon disrupted GRK2-mediated phosphorylation of muscarinic M2 receptors in vitro (Udarbe Zamora et al., 2008). Other highly up-regulated synapse-related genes (Table 1) included Synj1 (6.7-fold) and Slc30a3 (6-fold). The products of these genes are both associated with synaptic vesicle functions (Synj1, syanaptojanin, a phosphatase involved in synaptic vesicle endocytosis and Slc30a3, zinc transporter 3, involved in synaptic vesicle uptake of zinc, in particular in glutamatergic neurons; Mani et al., 2007; Linkous et al., 2008; Bitanihirwe and Cunningham MG, 2009). Obviously, marked changes in the expression of various neurotransmitter receptors as well as related cell signaling components could have the potential to affect neurodevelopment.
Genes involved in cell adhesion/migration also exhibited differential regulation following CPF exposure. These included Jam3, a number of integrin-related genes (Itgb1bp2, Itga5, Adamts13), ankyrin-related genes (Ankk1, Ank2), protocadherins (Pcdhb8, Pcdhga5, Pcdh8, Pcdhg2), Cdon, Tjp1, Olfml1 and Rgd1562230. The product of Jam3 may have a role in cell polarity during development via interactions with integrins such as integrin beta 1, and appears to specifically bind with other proteins during tight junction formation (Ebnet et al., 2001). Protein products of the ankyrin-associated genes interact with other proteins, forming complexes among integral membrane/cell adhesion proteins, signaling molecules, and the cytoskeleton, leading to organized protein networks (Cunha and Mohler, 2009).
The protocadherins represent the largest subfamily of the cadherin adhesion protein superfamily and are predominantly expressed in the brain (Morishita and Yagi, 2007; Yagi, 2008). During neurodevelopment, gamma protocadherins are up-regulated during neuronal differentiation, while the alpha protocadherins are markedly down-regulated during myelination. The alpha protocadherin subfamily is also extensively expressed in serotonergic neurons (Morishita and Yagi, 2007). In a mutant mouse model lacking the cytoplasmic region-encoding exons common to all alpha protocadherins, distribution of serotergic innervation was markedly affected in globus pallidus, hippocampus and dentate gyrus, suggesting a role for these adhesion proteins in appropriate targeting of serotonergic fibers during postnatal development (Katori et al., 2009). A number of papers (Raines et al., 2001; Aldridge et al. 2003, 2004, 2005; Slotkin and Seidler, 2005) have reported changes in serotonergic signaling systems following early postnatal exposure to CPF in rats. Protocadherins thus appear to participate in development of neuronal diversity and establishing circuitry. An inappropriate increase in expression of these genes could potentially disrupt the timing and extent of neuronal differentiation and innervation to projection areas.
Other adhesion-related genes up-regulated by CPF included Cdon (cell adhesion molecule-related/down-regulated by oncogenes, an Ig/fibronectin III adhesion molecule), tjp1 (tight junction protein 1, also known as zona occludens 1), olfml1 (olfactomedin-like 1, an extracellular matrix protein first identified in olfactory epithelium), Rgd1562230 (catenin alpha 3, an integrin binding protein), and Pxn (paxillin), a focal adhesion protein. Focal adhesions are multiprotein complexes that coordinate integrins, extracellular matrix proteins and the cytoskeleton (Harjanto and Zaman, 2010). Moreover, gender-selective differences in paxillin have been correlated with sexual differentiation in brain regions (e.g., hypothalamus) that develop perinatally in a sex-related manner (Speert et al., 2007). Some studies have reported gender-specific effects of early CPF exposures (Dam et al., 2000; Raines et al., 2001; Slotkin et al., 2001; Levin et al., 2001, 2002). As these protein-protein interactions play key roles in cell movement and division, up-regulation of adhesion-related components could potentially lead to inappropriate cell-cell connections, disrupted cytoskeletal networks and deficits in growth cone development, all potentially influencing neuronal growth and connectivity.
As noted before, in the two top clusters exhibiting down-regulation following 2 mg/kg CPF (clusters 6 and 15), there was a notable absence of olfactory receptor genes (in contrast to clusters showing up-regulation). Similar to the clusters showing up-regulation, however, a number of genes related to synapse/signal transduction were included in clusters 6 and 15. A number of genes for synaptic proteins including synaptotagmin II (Syt2), synaptotagmin XII (Syt12), syntaxin 6 (Stx6), synaptophysin-like protein (Sypl1), synaptotagmin binding cytoplasmic RNA interacting protein (Syncrip), protein kinase A anchor protein 9 (Akap9), NMDA2C receptor (Grin 2c), dynamin 1-like (Dnml1), striatin (Strn3), unc-13 homolog (Unc13c) and seizure related homolog 6 (Sez6) were all down-regulated. The synaptotagmin family of calcium binding proteins is well known to participate in evoked neurotransmitter release (Sudhof, 2004). Striatal inhibitory neurons in mice lacking synaptotagmin 2 showed a delayed time course of transmitter release (a reduced slow component of Ca2+-dependent release). In contrast, synaptotagmin 12 (Syt12) is different in that it does not bind calcium, but it has been shown to play a role in the regulation of spontaneous transmitter release (Maximov et al., 2007). Dong et al., (2005) reported that changes in Syt12 expression may contribute to neurodevelopmental effects of hypothyroidism. Kabayama and coworkers (2008) reported that syntaxin 6 played a role in neurite extension in PC12 cells in response to nerve growth factor exposure.
Adhesion/migration-related genes including ankyrin-related genes (Ankmy2, Abtb2, Ankrd40), protocadherin 9 (Pcdh9), neurotrimin (Ntm), RAS-GTPase-activating protein SH3-domain binding protein (G3bp1) and G-protein coupled receptor kinase-interactor 2 (Git2) were also down-regulated and contained in these two clusters. Directed cell migration requires coordination of both trophic factors and cell adhesion signaling (Yu et al., 2009). The G-protein receptor kinase-interactor 2 (GIT-2) is tyrosine phosphorylated in response to platelet-derived growth factor (PDGF) stimulation in mouse fibroblasts and shown necessary for directed cell migration in a wound repair model. Focal adhesion kinase (Fak) and Src family kinases-mediated, PDGF-dependent GIT-2 tyrosine phosphorylation was necessary for PDGF-stimulated GIT2 binding to paxillin, suggesting GIT-2 and paxillin both participate in growth factor/cell adhesion signaling, cell polarity and directional cell migration. Moreover, GIT-2-paxillin interactions mediated subsequent cell responses by inhibiting Rac1 GTPase activity (Kabayama et al., 2008). Rho GTPases including Rac1 and Rac2 (up-regulated in Cluster 16) are important in regulation of cadherin-based cell-cell adhesion (Takaishi et al. 1997, Yasuda et al. 2000). Our findings suggest that CPF dysregulates the expression of a number of genes associated with cell adhesion.
Numerous in vitro and in vivo studies indicate that axonal outgrowth of neurons during neurodevelopment depends on cell adhesion proteins (Matasunaga et al., 1988; Tomaselli et al., 1988; Inoue and Sanes, 1997; Weiner et al., 2005; Boscher and Mège, 2008; Barnes et al., 2010). A number of papers reported inhibition of axonal/neurite outgrowth by CPF (Song et al., 1998; Das and Barone, 1999; Crumpton et al., 2000). Interestingly, Howard et al. (2005) reported that in primary cultures of embryonic rat sympathetic neurons, CPF and CPF-oxon reduced axonal outgrowth but increased dendritic growth stimulated by bone morphogenetic protein. Recent global gene array studies with either acute CPF in adult rats (Stapleton and Chan, 2009) or repeated prenatal CPF exposures in developing mice (Moriera et al., 2010) also noted a number of changes in genes related to cell adhesion. Changes in adhesion protein interactions could lead to altered growth and targeting of axons during neurodevelopment and changes in neural circuitry.
Several genes important in transcription and translation were also within clusters 6 and 15, i.e., those top clusters showing down-regulation following 2 mg/kg CPF. Transcription elongation regulator 1 (Tcerg1), eukaryotic translation initiation factor 3 subunit 10 (Eif3s10) and eukaryotic translation initiation factor 4 subunit gamma 1 (Eif4sg1), transcription factor 19 (Tcf19), and the basic helix-loop-helix (bHLH) transcription factor 12 (Tcf12) were all markedly down-regulated following CPF exposure. Moreover, a number of transcription/translation-related genes were among the top 30 down-regulated genes (Table 2) including the bHLH transcription factor Neurod1 (the most highly down-regulated gene from all clusters), Ednrb, Eif3s10, Eif4g1, Elav4, Mycbp2, Matr3, Rkhd2, Nfix, Tnrc6b, Hnrnpf and Rnpc2. All of these genes encode proteins involved in RNA or protein expression/processing. It must be noted, however that several genes related to transcription/translation were also highly up-regulated (see Table 1), including the bHLH transcription factors Sim2 and Ahr, and others including Tmem127, Sox18, Men1, and Srf. Coordinated changes in transcriptional/translational activity necessary during neurodevelopment could potentially be disrupted by CPF via changes in the expression of any number of these genes. It is also noted that one of the most highly down-regulated genes (see Table 2) was Gfap (glial fibrillary acidic protein, an astrocytic intermediate protein and cell marker), possibly an indicator of decreased glial cell proliferation.
Pathway analysis detected six signaling pathways with the largest number of differentially expressed genes: MAPK signaling, oxidative stress, NF-κB signaling, mitochondrial dysfunction, arylhydrocarbon hydroxylase receptor signaling, and alpha adrenergic receptor signaling. MAPK signaling is a major signaling pathway for growth factor receptors including nerve growth factor, brain-derived neurotrophic factor and epidermal growth factor (Kaplan and Miller, 2000; Ferguson, 2003). The small GTP binding protein Ras and adaptor proteins bind to plasma membrane bound tyrosine receptor kinases, which activate the Ras protein to in turn recruit and activate Raf, a MAPK kinase kinase. Raf can then activate MEK (a MAPK kinase), which then activates MAPK, also known as ERK1/2. The end result of this sequence of serine/threonine/tyrosine phosphorylations is that phospho-ERK1/2 enters the nucleus to phosphorylate transcription factors to modulate gene expression. Several studies suggest growth factor/trophic factor signaling may be affected by early postnatal CPF exposures (Betancourt et al., 2006, 2007; Slotkin et al., 2007, 2008). As MAPK signaling was the top canonical pathway detected by Ingenuity Pathway Analysis and is coupled to growth factor/neurotrophic factor function, alterations in MAPK signaling could be important in the overall neurodevelopmental effects of early CPF exposure.
Oxidative stress was also detected as a signaling pathway affected by CPF exposure. A number of studies suggest that oxidative stress may participate in CPF-induced developmental neurotoxicity (Slotkin et al., 2007, 2009b). The developing brain is likely more vulnerable to oxidative stress due to relatively low concentrations of detoxifying enzymes and cellular antioxidants (Gupta, 2004). Interestingly, Moreiera and coworkers (2010) found no evidence of changes in gene categories related to oxidative stress in the prenatal mouse exposure model. Our findings suggest however that the highest dosage of CPF (2 mg/kg) affected gene expression in oxidative stress-related pathways.
Disruption of the other canonical signaling pathways could also contribute to expression of neurodevelopmental toxicity. The Ah receptor gene (Ahr) was highly up-regulated following 2 mg/kg CPF (see Table 1). The Ah receptor (AHR) has been associated with the toxicity of tetrachlorodibenzodioxin (TCDD, dioxin) and related xenobiotics (e.g., co-planar PCBs) for decades (reviewed in Beischlag et al., 2008). AHR is a ligand-activated bHLH transcription factor which, upon xenobiotic binding, translocates to the nucleus and regulates genes involved in xenobiotic metabolism, cellular proliferation and differentiation (Hankinson et al., 1995; Swanson et al., 1993). In a study evaluating over 200 pesticides as possible ligands of the AHR (Takeuchi et al., 2008), CPF exhibited AHR-mediated transcriptional activity. The AHR crosstalks with multiple signaling pathways, including MAPK (Henklova et al., 2008; Puga et al., 2009) and NF-kB (Tian et al., 1999; Tian, 2009). While the role of AHR in xenobiotic toxicity has been extensively studied, more recent studies indicate that AHR may have diverse physiological roles (Beishlag et al., 2008; Bock and Köhle, 2009; Fujii-Kuriyama and Kawajiri, 2010). AHR is present in mouse cerebellum during postnatal maturation, peaking from postnatal day 3–10, a period of active granule neuroblast maturation (Williamson et al., 2005) and TCDD reduced thymidine incorporation and granule neuroblast survival in a concentration-dependent manner, suggesting possible disruption of granule cell neurogenesis through an AHR-mediated pathway. Several studies suggest sex-dependent neurobehavioral differences in adults following postnatal CPF exposure (Garcia et al., 2003; Aldridge et al., 2005). Cross-talk between estrogen receptor and AHR at promoter sites appears important in the regulation of gene transcription of cypD19 (aromatase), an enzyme located in brain responsible for estrogen synthesis (Cheshenko et al., 2007). CPF-induced changes in AHR signaling and effects on transcription of steroidogenic enzymes could potentially lead to such functional alterations.
As with other microarray studies evaluating effects of CPF, our results suggest a great number of potential gene expression changes in the neonatal rat brain following acute CPF exposure. In the global gene array study by Stapleton and Chan (2009) evaluating gene changes in adult rats following acute chlorpyrifos exposure, only 277 differentially expressed genes were reported. It should be noted that these authors only recorded changes in differentially expressed genes if they occurred with consecutive dosages, i.e., a change in expression had to be shared with two consecutive dose levels before it was considered. As we noted above, there appeared to be relatively few commonly expressed genes among the different CPF dosing groups (Figure 2). Selecting differentially expressed genes based on similar changes with consecutive dosing groups would have substantially reduced the numbers of differentially expressed genes reported in our study. In the prenantal mouse following repeated CPF exposures (Moreiera et al., 2010), chlorpyrifos (10 mg/kg/day) led to over 2,600 differentially expressed genes, i.e., a number relatively similar to changes noted in our study. Thus, while the numbers of genes affected in our study seems high, they are in rough agreement with changes noted in two previous studies evaluating global gene changes in brain following chlorpyrifos exposure in rodents.
The multitude of differentially expressed genes among all four CPF treatment groups illustrates the sensitivity of this approach using a platform of >44,000 genes. A major advantage of using such a global approach in the evaluation of gene changes following xenobiotic exposure is that complex interconnections among gene signaling networks and pathways can be studied. This level of complexity also presents a major challenge, i.e., how to analyze and interpret a huge amout of data on gene changes across multiple levels. We concluded that the minimal degree of overlap among differentially expressed genes across all four CPF dosages (Figure 2), along with the “up-tick” in response with numbers of differentially expressed genes, gene clusters (Figure 3), gene networks and target enzyme inhibition with the highest dosage (2 mg/kg, Figure 1), all indicated that gene expression changes in the lower dosage groups were discontinuous across those dosing conditions and may not represent early and consistent treatment-related changes relevant to developmental neurotoxicity potential. With that assumption, we “narrowed” our subsequent focus to evaluation of only the high dose group (i.e., only about 3,000 differentially expressed genes).
Stapleton and Chan (2009) and Moreiera and coworkers (2010) both reported an inverted U-shaped dose-response relationship between CPF exposure and total numbers of differentially expressed genes in brain. The greatest number of changes in the prenatal mouse exposure model (gestational days 6–17 with analysis on day 18) was noted with the LOAEL for brain acetylcholinesterase inhibition (10 mg/kg/day), with higher exposure levels (i.e., 12 and 15 mg/kg/day) leading to less than 10% of the changes noted with 10 mg/kg/day exposures (Moreiera et al., 2010). In adult rat brain, Stapleton and Chan (2009) identified differentially expressed genes as those that were significantly affected over at least two consecutive CPF dosages (i.e., 0.5, 1, 5, 10, 30 and 50 mg/kg, with changes measured at four days after dosing). Using this criterion, they similarly reported an inverted dose-response relationship, with peak numbers of differentially expressed genes with 5–10 mg/kg CPF, and lesser changes with 30–50 mg/kg CPF. These investigators did not measure acetylcholinesterase inhibition. Using this same strain of rats and the same route of administration (oral), Timchalk and coworkers (2002) did however, report significant brain acetylcholinesterase inhibition following either 5 or 10 mg/kg CPF. Thus, both of these global gene array studies report peak changes in the numbers of genes affected with CPF dosages sufficient to inhibit brain acetylcholinesterase. It is unclear whether we would have detected a similar drop in the numbers of genes affected if we had included dosages higher than 2 mg/kg.
Examination of differentially expressed genes in the top gene clusters affected by 2 mg/kg CPF exposure identified functional areas including olfactory receptors, synapse/signal transduction, cell adhesion and transcription/translation. Olfactory perception may be a sensitive endpoint for early CPF exposure. Unfortunately, olfaction is not routinely evaluated in toxicity or epidemiology studies. A number of neurotransmitter receptors, both excitatory and inhibitory, and related synaptic proteins (e.g., SNARE proteins) were both up- and down-regulated by CPF. Obviously, such changes in neurotransmission-related functions could influence neurodevelopment. Dysregulation of genes involved in transcription and translation could lead to altered numbers of neurons or glial cells in the adult brain, altered distribution of cell types in different regions, and multiple changes based on alterations in translation of cell-specific proteins that may only have a limited window of time to function appropriately. Perhaps more prominently noted were genes associated with cell adhesion. Cell-cell interactions are important in numerous processes during neurodevelopment including cell migration, synapse formation, dendrite/axonal extension, tight junction formation, and others.
While we felt it was reasonable to focus on gene expression changes following 2 mg/kg CPF for the reasons stated above, genes that were commonly affected across all CPF dosages could be of high importance in the developmental neurotoxicity of CPF. There were nine genes that were differentially expressed in the 0.1, 0.5, 1 and 2 mg/kg CPF treatment groups (Table 3). Interestingly, cell-adhesion related genes again appeared prominent in this group of differentially expressed genes. Focal adhesion kinase (Fak1, also known as protein tyrosine kinase 2) is thought to be pivotally involved in cell migration (Hsia et al., 2009). Paxillin (Pxn), one of the most highly upregulated genes (Table 1), is a substrate for phosphorylation by focal adhesion kinase. Glypican 3 is a proteoglycan signaling co-receptor involved in cell adhesion and migration (Lander et al., 1996; Mythreye and Blobe, 2009). Three of the genes (Gp9, Amigo3, Lrnf1) upregulated by all four CPF dosages encode proteins with leucine-rich repeats, i.e., motifs with known adhesive functions (Hickey et al., 1989; Tumbarello et al., 2002; Chen et al., 2006). Thus, whether we focused on gene expression changes following the highest dosage, or on differentially expressed genes shared in common across all dosages of CPF, changes in the expression of genes associated with cell adhesion and related signaling suggest a target in the neurodevelopmental toxicity potential for this pesticide. Further studies on the effects of chlorpyrifos on cell adhesion-related processes are warranted.
There are a number of limitations to this study. While real-world exposures to CPF and other pesticides in the context of neurodevelopment would be repeated in nature, gene expression changes following acute exposure could represent initial early changes less potentially confounded by later responses. As with any study, timing of evaluations is important. Evaluating gene expression changes 24 hours after CPF exposure could lead to a different interpretation than that from studies examining gene expression changes at earlier or later time-points. Similarly, the estimate of cholinesterase inhibition is influenced by the time after dosing when enzyme activity is measured. In our case (cholinesterase was assayed 24 hours after dosing), we might expect greater inhibition following 2 mg/kg CPF, or significant inhibition with the lower dosages, if activity was evaluated earlier. Zheng et al., (2000) reported, however that a similar dosage of CPF (1.5 mg/kg, po) did not significantly inhibit cholinesterase activity in the cortex of 7-day old rats evaluated 4 hours after dosing. The route of exposure is another variable differing among studies. Many CPF toxicity studies have utilized subcutaneous dosing while others (including the present study) used oral dosing, and there can be toxicokinetic differences between the two. There are thus ample reasons to anticipate differences in gene expression changes noted among different studies based on differences in route of exposure, exposure level, acute vs. repeated dosing, timing of measurements, and other factors. Furthermore, the forebrain is not homogeneous but has subregions with distinct anatomical organizations, interconnections and functions, utilizing different signaling systems and containing different cell types. Gene expression changes in one subregion of the forebrain could be markedly different than in another subregion, and analysis of the forebrain as a whole could confound overall interpretations. Finally, gene expression changes may not necessarily translate directly or proportionately into ultimate changes in the products of those genes.
In summary, our findings along with results from other studies using different exposure models indicate that expression of a wide array of genes in the developing brain can be affected by early CPF exposure. While lower CPF dosages that did not elicit significant brain cholinesterase inhibition (0.1, 0.5 and 1 mg/kg) led to significant gene expression changes, there were similar numbers of genes affected across this 10-fold dosage range and relatively little overlap of shared genes. The present findings suggesting a point of departure for several endpoints with the highest dosage evaluated (2 mg/kg CPF), along with findings in the other global gene array studies cited above, suggest that consistent, extensive changes in gene expression following acute CPF exposure in the neonatal rat brain are likely to occur at exposure levels very near those that inhibit brain acetylcholinesterase, the primary target for cholinergic toxicity. Changes in the expression of numerous genes associated with cell adhesion may be particularly important in the neurodevelopmental toxicity of this pesticide.
Supplementary Material
Acknowledgements
This work was supported by a seed grant from the Oklahoma State University Center for Veterinary Health Sciences, the Oklahoma State University Board of Regents and by grant ES009119 (CNP) from the National Institute of Environmental Health Sciences (NIEHS). The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of NIEHS. We would like to thank Dr. Peter Hoyt, Director of the Microarray Core facility in the Dept. of Biochemistry and Molecular Biology for useful discussions regarding the microarray experiments and their analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldridge JE, Seidler FJ, Meyer A, Thillai I, Slotkin TA. Serotonergic systems targeted by developmental exposure to chlorpyrifos: effects during different critical periods. Environ. Health Perspect. 2003;111:1736–1743. doi: 10.1289/ehp.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge JE, Seidler FJ, Slotkin TA. Developmental exposure to chlorpyrifos elicits sex-selective alterations of serotonergic synaptic function in adulthood: critical periods and regional selectivity for effects on the serotonin transporter, receptor subtypes, and cell signaling. Environ. Health Perspect. 2004;112:148–155. doi: 10.1289/ehp.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge JE, Levin ED, Seidler FJ, Slotkin TA. Developmental exposure of rats to chlorpyrifos leads to behavioral alterations in adulthood, involving serotonergic mechanisms and resembling animal models of depression. Environ. Health Perspect. 2005;113:527–531. doi: 10.1289/ehp.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SH, Price SR, Wentzel C, Guthrie SC. Cadherin-7 and cadherin-6B differentially regulate the growth, branching and guidance of cranial motor axons. Development. 2010;137:805–814. doi: 10.1242/dev.042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag TV, Morales JL, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benke GM, Murphy SD. The influence of age on the toxicity and metabolism of methyl parathion and parathion in male and female rats. Toxicol. Appl. Pharmacol. 1975;31:254–269. doi: 10.1016/0041-008x(75)90161-1. [DOI] [PubMed] [Google Scholar]
- Berkowitz GS, Wetmur JG, Birman-Deych E, Obel J, Lapinski RH, Godbold JH, Holzman IR, Wolff MS. In utero pesticide exposure, maternal paraoxonase activity, and head circumference. Environ Health Perspect. 2004;112:388–391. doi: 10.1289/ehp.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AM, Burgess SC, Carr RL. Effect of developmental exposure to chlorpyrifos on the expression of neurotrophin growth factors and cell-specific markers in neonatal rat brain. Toxicol Sci. 2006;92:500–506. doi: 10.1093/toxsci/kfl004. [DOI] [PubMed] [Google Scholar]
- Betancourt AM, Filipov NM, Carr RL. Alteration of neurotrophins in the hippocampus and cerebral cortex of young rats exposed to chlorpyrifos and methyl parathion. Toxicol Sci. 2007;100:445–455. doi: 10.1093/toxsci/kfm248. [DOI] [PubMed] [Google Scholar]
- Bigbee JW, Sharma KV, Chan EL, Bögler O. Evidence for the direct role of acetylcholinesterase in neurite outgrowth in primary dorsal root ganglion neurons. Brain Res. 2000;861:354–362. doi: 10.1016/s0006-8993(00)02046-1. [DOI] [PubMed] [Google Scholar]
- Bitanihirwe BK, Cunningham MG. Zinc: the brain’s dark horse. Synapse. 2009;63:1029–1049. doi: 10.1002/syn.20683. [DOI] [PubMed] [Google Scholar]
- Bock KW, Köhle C. The mammalian aryl hydrocarbon (Ah) receptor: from mediator of dioxin toxicity toward physiological functions in skin and liver. Biol Chem. 2009;390:1225–1235. doi: 10.1515/BC.2009.138. [DOI] [PubMed] [Google Scholar]
- Boscher C, Mège RM. Cadherin-11 interacts with the FGF receptor and induces neurite outgrowth through associated downstream signalling. Cell Signal. 2008;20:1061–1072. doi: 10.1016/j.cellsig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Casida JE, Quistad GB. Serine hydrolase targets of organophosphorus toxicants. Chem Biol Interact. 2005;157–158:277–283. doi: 10.1016/j.cbi.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Chen Y, Aulia S, Li L, Tang BL. AMIGO and friends: an emerging family of brain-enriched, neuronal growth modulating, type I transmembrane proteins with leucine-rich repeats (LRR) and cell adhesion molecule motifs. Brain Res Rev. 2006;51:265–274. doi: 10.1016/j.brainresrev.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Cheshenko K, Brion F, Le Page Y, Hinfrey N, Pakdel F, Kah O, Segner H, Eggen RI. Expression of zebra fish aromatase cyp19a and cyp19b genes in response to the ligands of estrogen receptor and aryl hydrocarbon receptor. Toxicol. Sci. 2007;96:255–267. doi: 10.1093/toxsci/kfm003. [DOI] [PubMed] [Google Scholar]
- Clement JG. Role of aliesterase in organophosphate poisoning. Fundam. Appl. Toxicol. 1984;4:S96–S105. doi: 10.1016/0272-0590(84)90141-6. [DOI] [PubMed] [Google Scholar]
- Clement JG, Erhardt N. Serum carboxylesterase activity in various strains of rats: sensitivity to inhibition by CBDP (2-/o-cresyl/4H:1:3:2-benzodioxaphosphorin-2-oxide) Arch. Toxicol. 1990;64:414–416. doi: 10.1007/BF01973466. [DOI] [PubMed] [Google Scholar]
- Colt JS, Lubin J, Camann D, Davis S, Cerhan J, Severson RK, Cozen W, Hartge P. Comparison of pesticide levels in carpet dust and self-reported pest treatment practices in four US cities. J. Expo Anal. Environ Epidemiol. 2004;14:74–83. doi: 10.1038/sj.jea.7500307. [DOI] [PubMed] [Google Scholar]
- Crumpton TL, Seidler FJ, Slotkin TA. Is oxidative stress involved in the developmental neurotoxicity of chlorpyrifos? Brain Res. Dev Brain Res. 2000;121:189–195. doi: 10.1016/s0165-3806(00)00045-6. [DOI] [PubMed] [Google Scholar]
- Cunha SR, Mohler PJ. Ankyrin protein networks in membrane formation and stabilization. J. Cell Mol Med. 2009;13:4364–4376. doi: 10.1111/j.1582-4934.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam K, Seidler FJ, Slotkin TA. Chlorpyrifos exposure during a critical neonatal period elicits gender-selective deficits in the development of coordination skills and locomotor activity. Brain Res Dev Brain Res. 2000;121:179–187. doi: 10.1016/s0165-3806(00)00044-4. [DOI] [PubMed] [Google Scholar]
- Das KP, Barone S., Jr Neuronal differentiation in PC12 cells is inhibited by chlorpyrifos and its metabolites: is acetylcholinesterase inhibition the site of action? Toxicol. Appl. Pharmacol. 1999;160:217–230. doi: 10.1006/taap.1999.8767. [DOI] [PubMed] [Google Scholar]
- Do JH, Choi DK. Clustering approaches to identifying gene expression patterns from DNA microarray data. Mol Cells. 2008;25:279–288. [PubMed] [Google Scholar]
- Dong H, Wade M, Williams A, Lee A, Douglas GR, Yauk C. Molecular insight into the effects of hypothyroidism on the developing cerebellum. Biochem Biophys Res Commun. 2005;330:1182–1193. doi: 10.1016/j.bbrc.2005.03.099. [DOI] [PubMed] [Google Scholar]
- Ebnet K, Suzuki A, Horikoshi Y, Hirose T, Meyer Zu, Brickwedde MK, Ohno S, Vestweber D. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM) EMBO J. 2001;20:3738–3748. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SS. Receptor tyrosine kinase transactivation: fine-tuning synaptic transmission. Trends Neurosci. 2003;26:119–122. doi: 10.1016/S0166-2236(03)00022-5. [DOI] [PubMed] [Google Scholar]
- Fujii-Kuriyama Y, Kawajiri K. Molecular mechanisms of the physiological functions of the aryl hydrocarbon (dioxin) receptor, a multifunctional regulator that senses and responds to environmental stimuli. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:40–53. doi: 10.2183/pjab.86.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SJ, Seidler FJ, Slotkin TA. Developmental neurotoxicity elicited by prenatal or postnatal chlorpyrifos exposure; effects on neurospecific proteins indicate changing vulnerabilities. Environ. Health Perspect. 2003;111:297–303. doi: 10.1289/ehp.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RC. Brain regional heterogeneity and toxicological mechanisms of organophosphates and carbamates. Toxicol. Mech Methods. 2004;14:103–143. doi: 10.1080/15376520490429175. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Harjanto D, Zaman MH. Matrix mechanics and receptor-ligand interactions in cell adhesion. Org Biomol Chem. 2010;8:299–304. doi: 10.1039/b913064k. [DOI] [PubMed] [Google Scholar]
- Henklová P, Vrzal R, Ulrichová J, Dvorák Z. Role of mitogen-activated protein kinases in aryl hydrocarbon receptor signaling. Chem. Biol. Interact. 2008;172:93–104. doi: 10.1016/j.cbi.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Hickey MJ, Williams SA, Roth GJ. Human platelet glycoprotein IX: an adhesive prototype of leucine-rich glycoproteins with flank-center-flank structures. PNAS. 1989;86:6773–6777. doi: 10.1073/pnas.86.17.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard AS, Bucelli R, Jett DA, Bruun D, Yang D, Lein PJ. Chlorpyrifos exerts opposing effects on axonal and dendritic growth in primary neuronal cultures. Toxicol. Appl. Pharmacol. 2005;207:112–124. doi: 10.1016/j.taap.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Hsia DA, Mitra SK, Hauck CR, Streblow DN, Nelson JA, Ilic D, Huang S, Li E, Nemerow GR, Leng J, Spencer KS, Cheresh DA, Schlaepfer DD. Differential regulation of cell motility and invasion by FAK. J Cell Biol. 2003;160:753–767. doi: 10.1083/jcb.200212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPA. Ingenuity Pathway Analysis. ( http://www.ingenuity.com)
- Inoue A, Sanes JR. Lamina-specific connectivity in the brain: regulation by N-cadherin, neurotrophins, and glycoconjugates. Science. 1997;276:1428–1431. doi: 10.1126/science.276.5317.1428. [DOI] [PubMed] [Google Scholar]
- Jacobson SW, Jacobson JL. New evidence of effects of organophosphate pesticides on neurodevelopment in children: commentary on the article by Kofman et al. on page 88. Pediatr. Res. 2006;60:22–23. doi: 10.1203/01.pdr.0000220353.05515.d4. [DOI] [PubMed] [Google Scholar]
- James SJ, Slikker W, Melynk S, New E, Pogribna E, Jernigan S. Thimerosal neurotoxicity is associated with glutathione depletion: protection with glutathione precursors. Neurotoxicology. 2005;26:1–8. doi: 10.1016/j.neuro.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Russell RL. A rapid, simple radiometric assay for cholinesterase, suitable for multiple determinations. Anal. Biochem. 1975;64:229–238. doi: 10.1016/0003-2697(75)90423-6. [DOI] [PubMed] [Google Scholar]
- Jones SA, Holmes C, Budd TC, Greenfield SA. The effect of acetylcholinesterase on outgrowth of dopaminergic neurons in organotypic slice culture of rat mid-brain. Cell Tissue Res. 1995;279:323–330. doi: 10.1007/BF00318488. [DOI] [PubMed] [Google Scholar]
- Kabayama H, Tokushige N, Takeuchi M, Mikoshiba K. Syntaxin 6 regulates nerve growth factor-dependent neurite outgrowth. Neurosci Lett. 2008;436:340–344. doi: 10.1016/j.neulet.2008.03.061. [DOI] [PubMed] [Google Scholar]
- Kaplan DR, Miller FD. Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol. 2000;10:381–391. doi: 10.1016/s0959-4388(00)00092-1. [DOI] [PubMed] [Google Scholar]
- Karanth S, Pope CN. Carboxylesterase and A esterase activities during maturation and aging: relationship to the toxicity of chlorpyrifos and parathion in rats. Toxicol Sci. 2000;58:282–289. doi: 10.1093/toxsci/58.2.282. [DOI] [PubMed] [Google Scholar]
- Karanth S, Olivier K, Liu J, Pope C. In vivo interaction between chlorpyrifos and parathion in adult rats: sequence of administration can markedly influence toxic outcome. Toxicol. Appl. Pharmacol. 2001;177:247–255. doi: 10.1006/taap.2001.9312. [DOI] [PubMed] [Google Scholar]
- Katori S, Hamada S, Noguchi Y, Fukuda E, Yamamoto T, Yamamoto H, Hasegawa S, Yagi T. Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J. Neurosci. 2009;29:9137–9147. doi: 10.1523/JNEUROSCI.5478-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielly T, Donaldson D, Grube A. Office of prevention, Pesticides, and Toxic substances. Washington, DC: USEPA; 2004. Pesticides Industry sales and Usage: 2000 and 2001 market estimates. 20. [Google Scholar]
- Koenigsberger C, Chiappa S, Brimijoin S. Neurite differentiation is modulated in neuroblastoma cells engineered for altered acetylcholinesterase expression. J. Neurochem. 1997;69:1389–1397. doi: 10.1046/j.1471-4159.1997.69041389.x. [DOI] [PubMed] [Google Scholar]
- Lander AD, Stipp CS, Ivins JK. The glypican family of heparin sulfate proteoglycans: major cell-surface proteoglycans of the developing nervous system. Perspect Develop Neurobiol. 1996;3:347–358. [PubMed] [Google Scholar]
- Levin ED, Addy N, Nakajima A, Christopher NC, Seidler FJ, Slotkin TA. Persistent behavioral consequences of neonatal chlorpyrifos exposure in rats. Brain Res Dev Brain Res. 2001;130:83–89. doi: 10.1016/s0165-3806(01)00215-2. [DOI] [PubMed] [Google Scholar]
- Levin ED, Addy N, Baruah A, Elias A, Christopher NC, Seidler FJ, Slotkin TA. Prenatal chlorpyrifos exposure in rats causes persistent behavioral alterations. Neurotoxicol. Teratol. 2002;24:733–741. doi: 10.1016/s0892-0362(02)00272-6. [DOI] [PubMed] [Google Scholar]
- Linkous DH, Flinn JM, Koh JY, Lanzirotti A, Bertsch PM, Jones BF, Giblin LJ, Frederickson CJ. Evidence that the ZNT3 protein controls the total amount of elemental zinc in synaptic vesicles. J Histochem Cytochem. 2008;56:3–6. doi: 10.1369/jhc.6A7035.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough N, Farr Al, Randall RJ. Protein measurement with Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mani M, Lee SY, Lucast L, Cremona O, Di Paolo G, De Camilli P, Ryan TA. The dual phosphatase activity of synaptojanin1 is required for both efficient synaptic vesicle endocytosis and reavailability at nerve terminals. Neuron. 2007;56:1004–1018. doi: 10.1016/j.neuron.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga M, Hatta K, Nagafuchi A, Takeichi M. Guidance of optic nerve fibres by N-cadherin adhesion molecules. Nature. 1988;334:62–64. doi: 10.1038/334062a0. [DOI] [PubMed] [Google Scholar]
- Maximov A, Shin OH, Liu X, Südhof TC. Synaptotagmin-12, a synaptic vesicle phosphoprotein that modulates spontaneous neurotransmitter release. J. Cell Biol. 2007;176:113–124. doi: 10.1083/jcb.200607021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mense S, Sengupta A, Lan C, Zhou M, Bentsman G, Volsky DJ, Whyatt RM, Perera FP, Zhang Li. The common insecticides cyfluthrin and chlorpyrifos alter the expression of a subset of genes with diverse functions in primary human astrocytes. Toxicol. Sci. 2006;93:125–135. doi: 10.1093/toxsci/kfl046. [DOI] [PubMed] [Google Scholar]
- Mileson BE, Chambers JE, Chen WL, Dettbarn W, Ehrich M, Eldefrawi AT, Gaylor DW, Hamernik K, Hodgson E, Karczmar AG, Padilla S, Pope CN, Richardson RJ, Saunders DR, Sheets LP, Sultatos LG, Wallace KB. Common mechanism of toxicity: a case study of organophosphorus pesticides. Toxicol. Sci. 1998;41:8–20. doi: 10.1006/toxs.1997.2431. [DOI] [PubMed] [Google Scholar]
- Moreira EG, Yu X, Robinson JF, Griffith W, Hong SW, Beyer RP, Bammler TK, Faustman EM. Toxicogenomic profiling in maternal and fetal rodent brains following gestational exposure to chlorpyrifos. Toxicol. Appl. Pharmacol. 2010 doi: 10.1016/j.taap.2010.03.015. Mar 27. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita H, Yagi T. Protocadherin family: diversity, structure, and function. Curr Opin Cell Biol. 2007;19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Mythreye K, Blobe GC. Proteoglycan signaling co-receptors: roles in cell adhesion, migration and invasion. Cell Signal. 2009;21:1548–1558. doi: 10.1016/j.cellsig.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraoanu LE, Layer PG. Acetylcholinesterase in cell adhesion, neurite growth and network formation. FEBS J. 2008;275:618–624. doi: 10.1111/j.1742-4658.2007.06237.x. [DOI] [PubMed] [Google Scholar]
- Pond AL, Chambers HW, Chambers JE. Organophosphate detoxication potential of various rat tissues via A-esterase and aliesterase activities. Toxicol. Lett. 1995;78:245–252. doi: 10.1016/0378-4274(95)03327-h. [DOI] [PubMed] [Google Scholar]
- Pope CN. Organophosphorus pesticides: do they all have the same mechanism of toxicity. J. Toxicol Environ Health. 1999;2:161–181. doi: 10.1080/109374099281205. [DOI] [PubMed] [Google Scholar]
- Pope C, Karanth S, Liu J. Pharmacology and toxicology of cholinesterase inhibitors: uses and misuses of a common mechanism of action. Environ. Toxicol. Pharmacol. 2005;19:433–446. doi: 10.1016/j.etap.2004.12.048. [DOI] [PubMed] [Google Scholar]
- Puga A, Ma C, Marlowe JL. The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem Pharmacol. 2009;77:713–722. doi: 10.1016/j.bcp.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines KW, Seidler FJ, Slotkin TA. Alterations in serotonin transporter expression in brain regions of rats exposed neonatally to chlorpyrifos. Brain Res Dev Brain Res. 2001;130:65–72. doi: 10.1016/s0165-3806(01)00211-5. [DOI] [PubMed] [Google Scholar]
- Rauh VA, Garfinkel R, Perera FP, Andrews HF, Hoepner L, Barr DB, Whitehead R, Tang D, Whyatt RW. Impact of prenatal chlorpyrifos exposure on neurodevelopment in the first 3 years of life among inner-city children. Pediatrics Dec. 2006;118(6):e1845–e1859. doi: 10.1542/peds.2006-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS. Statistics and Graphics Guide, Version 3. Cary, NC: SAS Institute; 1995. SAS. [Google Scholar]
- Slotkin TA, Cousins MM, Tate CA, Seidler FJ. Persistent cholinergic presynaptic deficits after neonatal chlorpyrifos exposure. Brain Res. 2001;902:229–243. doi: 10.1016/s0006-8993(01)02387-3. [DOI] [PubMed] [Google Scholar]
- Slotkin TA. Cholinergic systems in brain development and disruption by neurotoxicants: nicotine, environmental tobacco smoke, organophosphates. Toxicol. Appl. Pharmacol. 2004;198:132–151. doi: 10.1016/j.taap.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. The alterations in CNS serotonergic mechanisms caused by neonatal chlorpyrifos exposure are permanent. Brain Res Dev Brain Res. 2005;158:115–119. doi: 10.1016/j.devbrainres.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. Comparative developmental neurotoxicity of organophosphates in vivo: Transcriptional responses of pathways for brain cell development, cell signaling, cytotoxicity and neurotransmitter systems. Brain Research Bulletin. 2007;72:232–274. doi: 10.1016/j.brainresbull.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ, Fumagalli F. Targeting of neurotrophic factors, their receptors, and signaling pathways in the developmental neurotoxicity of organophosphates in vivo and in vitro. Brain Res Bull. 2008;76:424–438. doi: 10.1016/j.brainresbull.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. Protein kinase C is a target for diverse developmental neurotoxicants: Transcriptional responses to chlorpyrifos, diazinon, dieldrin and divalent nickel in PC12 cells. Brain Research. 2009a;1263:23–32. doi: 10.1016/j.brainres.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. Oxidative and excitatory mechanisms of developmental neurotoxicity: transcriptional profiles for chlorpyrifos, diazinon, dieldrin, and divalent nickel in PC12 cells. 2009b;117:587–595. doi: 10.1289/ehp.0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Violin JD, Seidler FJ, Slotkin TA. Modeling the developmental neurotoxicity of chlorpyrifos in vitro: macromolecule synthesis in PC12 cells. Toxicol. Appl. Pharmacol. 1998;151:182–191. doi: 10.1006/taap.1998.8424. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Speert DB, Konkle AT, Zup SL, Schwarz JM, Shiroor C, Taylor ME, McCarthy MM. Focal adhesion kinase and paxillin: novel regulators of brain sexual differentiation? Endocrinology. 2007;148:3391–3401. doi: 10.1210/en.2006-0845. [DOI] [PubMed] [Google Scholar]
- Stapleton AR, Chan VT. Subtoxic chlorpyrifos treatment resulted in differential expression of genes implicated in neurological functions and development. Arch Toxicol. 2009;83:319–333. doi: 10.1007/s00204-008-0346-2. [DOI] [PubMed] [Google Scholar]
- Sternfeld M, Ming G, Song H, Sela K, Timberg R, Poo M, Soreq H. Acetylcholinesterase enhances neurite growth and synapse development through alternative contributions of its hydrolytic capacity, core protein, and variable C termini. J Neurosci. 1998;18:1240–1249. doi: 10.1523/JNEUROSCI.18-04-01240.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. Cluster analysis of microarray data. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Su Z, Yuan Y, Chen J, Cao L, Zhu Y, Gao L, Qiu Y, He C. Reactive astrocytes in glial scar attract olfactory ensheathing cells migration by secreted TNF-alpha in spinal cord lesion of rat. PLoS One. 2009;4(12):e8141. doi: 10.1371/journal.pone.0008141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson HI, Tullis K, Denison MS. Binding of transformed Ah receptor complex to a dioxin responsive transcriptional enhancer: evidence for two distinct heteromeric DNA-binding forms. Biochemistry. 1993;47:12841–12849. doi: 10.1021/bi00210a037. [DOI] [PubMed] [Google Scholar]
- Takaishi K, Sasaki T, Kotani H, Nishioka H, Takai Y. Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J. Cell Biol. 1997;139:1047–1059. doi: 10.1083/jcb.139.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi S, Iida M, Yabushita H, Matsuda T, Kojima H. In vitro screening for aryl hydrocarbon receptor agonistic activity in 200 pesticides using a highly sensitive reporter cell line, DR-EcoScreen cells, and in vivo mouse liver cytochrome P450-1A induction by propanil, diuron and linuron. Chemosphere. 2008;74:155–165. doi: 10.1016/j.chemosphere.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. Ah Receptor and NF-kB Interactions, a Potential Mechanism for Dioxin Toxicity. J. of Biol. Chemistry. 1999;274:510–515. doi: 10.1074/jbc.274.1.510. [DOI] [PubMed] [Google Scholar]
- Tian Y. Ah receptor and NF-kB interplay on the stage of epigenome. Biochem. Pharmacol. 2009;77:670–680. doi: 10.1016/j.bcp.2008.10.023. [DOI] [PubMed] [Google Scholar]
- Timchalk C, Nolan RJ, Mendrala AL, Dittenber DA, Brzak KA, Mattsson JL. A Physiologically based pharmacokinetic and pharmacodynamic (PBPK/PD) model for the organophosphate insecticide chlorpyrifos in rats and humans. Toxicol Sci. 2002;66:34–53. doi: 10.1093/toxsci/66.1.34. [DOI] [PubMed] [Google Scholar]
- Tomaselli KJ, Neugebauer KM, Bixby JL, Lilien J, Reichardt LF. N-cadherin and integrins: two receptor systems that mediate neuronal process outgrowth on astrocyte surfaces. Neuron. 1988;1:33–43. doi: 10.1016/0896-6273(88)90207-3. [DOI] [PubMed] [Google Scholar]
- Tumbarello DA, Brown MC, Turner CE. The paxillin LD motifs. FEBS Lett. 2002;513:114–118. doi: 10.1016/s0014-5793(01)03244-6. [DOI] [PubMed] [Google Scholar]
- Udarbe Zamora EM, Liu J, Pope CN. Effects of chlorpyrifos oxon on M2 muscarinic receptor internalization in different cell types. J Toxicol Environ Health A. 2008;71:1440–1447. doi: 10.1080/15287390802328887. [DOI] [PubMed] [Google Scholar]
- U.S. EPA. Chlorpyrifos: Re-evaluation Report of the FQPA Safety Factor Committee. HED Doc. No. 014077. Washington, DC: U.S. Environmental Protection Agency; 2000. [Google Scholar]
- U.S. EPA. Chlorpyrifos: End-Use Products Cancellation Order. Fed Reg. 2002;67:3698–3700. Available: http://www.epa.gov/fedrgstr/EPA-PEST/2002/January/Day-25/p1764.htm.
- Weiner JA, Wang X, Tapia JC, Sanes JR. Gamma protocadherins are required for synaptic development in the spinal cord. Proc. Natl. Acad. Sci. 2005;102:8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B, Amler S, Amler RW. Pesticides. Pediatrics. 2004;113:1030–1036. [PubMed] [Google Scholar]
- Whyatt RM, Barr DB, Camann DE, Kinney PL, Barr JR, Andrews HF, Hoepner LA, Garfinkel R, Hazi Y, Reyes A, Ramirez J, Cosme Y, Perera FP. Contemporary-use pesticides in personal air samples during pregnancy and blood samples at delivery among urban minority mothers and newborns. Environ Health Perspect. 2003;111:749–756. doi: 10.1289/ehp.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt RM, Rauh V, Barr DB, Camann DE, Andrews HF, Garfinkel R, Hoepner LA, Diaz D, Dietrich J, Reyes A, Tang D, Kinney PL, Perera FP. Prenatal insecticide exposures and birth weight and length among an urban minority cohort. Environ Health Perspect. 2004;112:1125–1132. doi: 10.1289/ehp.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt RM, Camann DE, Perera FP, Rauh VA, Tang D, Kinney PL, Garfinkel R, Andrews H, Hoepner L, Barr DB. Biomarkers in assessing residential insecticide exposures during pregnancy and effects on fetal growth. Toxicol. Appl. Pharmacol. 2005;206:246–254. doi: 10.1016/j.taap.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Williamson MA, Gasiewicz TA, Opanashuk LA. Aryl hydrocarbon receptor expression and activity in cerebellar granule neuroblasts: implications for development and dioxin neurotoxicity. Toxicol Sci. 2005;83:340–348. doi: 10.1093/toxsci/kfi031. [DOI] [PubMed] [Google Scholar]
- Won YK, Liu J, Olivier K, Jr, Zheng Q, Pope CN. Age-related effects of chlorpyrifos on acetylcholine release in rat brain. Neurotoxicology. 2001;22:39–48. doi: 10.1016/s0161-813x(00)00009-7. [DOI] [PubMed] [Google Scholar]
- Yagi T. Clustered protocadherin family. Dev Growth Differ. 2008;50 Suppl. 1:S131–S140. doi: 10.1111/j.1440-169X.2008.00991.x. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Ohtsuka T, Inoue E, Yokoyama S, Sakisaka T, Kodama A, Takaishi K, Takai Y. Importance of spatial activation of Cdc42 and rac small G proteins by frabin for microspike formation in MDCK cells. Genes Cells. 2000;5:583–591. doi: 10.1046/j.1365-2443.2000.00349.x. [DOI] [PubMed] [Google Scholar]
- Young JG, Eskenazi B, Gladstone EA, Bradman A, Pedersen L, Johnson C, Barr DB, Furlong CE, Holland NT. Association between in utero organophosphate pesticide exposure and abnormal reflexes inneonates. Neurotoxicology. 2005;26:199–209. doi: 10.1016/j.neuro.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Yu JA, Deakin NO, Turner CE. Paxillin-kinase-linker tyrosine phosphorylation regulates directional cell migration. Mol Biol Cell. 2009;20:4706–4719. doi: 10.1091/mbc.E09-07-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Olivier K, Won YK, Pope CN. Comparative cholinergic neurotoxicity of oral chlorpyrifos exposures in preweanling and adult rats. Toxicol Sci. 2000;55:124–132. doi: 10.1093/toxsci/55.1.124. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wang Z, Miller DJ, Clarke R, Xuan J, Hoffman EP, Wang Y. A ground truth based comparative study on clustering of gene expression data. Front Biosci. 2008;13:3839–3849. doi: 10.2741/2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.