Abstract
Leucine-rich repeat containing proteins are involved in immune response in many capacities. In insects, these include Toll-like receptors and the Anopheles gambiae proteins APL1 and LRIM1. Here we describe the identification and characterization of leureptin, a novel extracellular protein with 13 leucine-rich repeats from hemolymph of the insect Manduca sexta. After injection of bacteria, leureptin mRNA level increased in fat body, but protein levels in plasma decreased, an indication that leureptin is consumed during the immune response. Leureptin bound to bacterial lipopolysaccharide (LPS). Microscopy using leureptin antiserum showed that leureptin associates with hemocytes after injection of bacteria, an indication that leureptin is involved in hemocyte responses to bacterial infection. Sequence database searches suggest similar proteins are present in other Lepidopteran species.
Keywords: Leucine-rich repeat, Lipopolysaccharide, Hemolymph, Hemocyte, Innate immunity, Insect, Manduca sexta
Introduction
Immune responses in Manduca sexta include melanization, antimicrobial peptide production, and clotting as well as the hemocyte responses: phagocytosis, nodulation, and encapsulation. Prior to initiating an immune response, the insect must recognize the pathogen using proteins that bind conserved microbial surface molecules. Known pattern recognition proteins in M. sexta include hemolin, which contains four immunoglobulin domains; four c-type lectins, immunlectin 1–4; two beta glucan recognition proteins (βGRPs); and soluble peptidoglycan recognition proteins (Ragan et al., 2009). Hemolin binds to lipid A and the O-specific antigen portions of lipopolysaccharide as well as to lipotechoic acid (Schmidt et al., 1993, Daffre and Faye, 1997, Yu and Kanost, 2002). The initiating protease for one branch of phenoloxidase activation, HP14, can autoactivate in the presence of βGRP and microbial β-1,3-glucan (Wang and Jiang, 2010). The pathways for immune response to Lipopolysaccharide (LPS), a molecule on the surface of gram-negative bacteria, are still unclear in M. sexta and other insects. Also unclear are details surrounding the initiation of hemocyte responses. One family of proteins that may serve these functions is known as leucine-rich repeat (LRR) proteins.
LRR proteins are present in animals, plants, fungi, and some bacteria (Kobe et al., 2001) and well represented within organisms, accounting for approximately one percent of all genes in the insects Anopheles gambiae and Drosophila melanogaster (Zdobnov et al., 2002). The range of functions of LRR proteins is enormous, including protein-protein interactions, signal transduction, and cell adhesion (Buchanan and Gay, 1996). This functional versatility derives from a conserved three dimensional structure, a curved coil composed of repeating units of ~24 amino acid residues. The repeating unit contains both conserved and variable regions. The conserved portion corresponds to the LRR motif, LxxLxLxxNxL, where L is Leu, Ile, Phe, or Val and X is any amino acid (Kobe et al., 2001, Bella et al., 2008). The 2nd and 3rd conserved leucines are involved in forming beta strands, which all assemble to form a beta sheet on a concave face. Each beta strand is connected to the next by a loop formed from the less conserved region of the repeat. This loop can take on a variety of secondary structures (Bella et al., 2008, Kajava and Kobe, 2002, Hindle et al., 2009). The LRR region altogether resembles a curved solenoid and is well suited to protein-protein or protein-ligand interactions on the concave face as well as on other surfaces (Bella et al., 2008).
Two LRR proteins are involved in LPS binding and signaling in mammals, Toll-like receptor-4 (TLR-4) and the pattern recognition protein CD-14, which has soluble and GPI-anchored forms (Ferrero et al., 1990, Pugin et al., 1994). CD-14 forms a dimer, connected at the C-termini, and can bind LPS at each N-terminus (Kim et al., 2005). Membrane-bound CD-14 receives LPS from LPS binding protein then transfers LPS to MD-2, which binds TLR-4 and induces TLR-4 dimerization and rapid signaling by activation of transcription factors like nuclear factor-kappa B (NF-κB) (Tsukamoto et al., 2010). Slower signaling can occur in membrane CD14-negative cells and in the absence of LPS binding protein as long as soluble CD-14 is present; this slower activation does not require dimerization of TLR-4 (Tsukamoto et al., 2010).
All Toll-like receptors contain extracellular LRRs and intracellular TIR-domains (Leulier and Lemaitre, 2008). In humans, all 10 TLRs are involved in innate immunity through binding of microbial patterns or other danger signals, while in Drosophila melanogaster Toll1, one of 9 Toll-like proteins, is activated during immune response by binding of the cytokine spätzle (Leulier and Lemaitre, 2008, Pal and Wu, 2009). Active spätzle is generated by a microbe triggered serine protease cascade; intracellular Toll signaling occurs through activation of NF-κB, which leads to production of antimicrobial peptides like drosomycin (Leulier and Lemaitre, 2008). In Bombyx mori, 14 TLRs are present in the genome, six of which are in a cluster with TLRs known to be involved in immunity (Tanaka et al., 2008).
In addition to TLRs, many other proteins involved in immune responses contain both a leucine-rich repeat domain and a signaling domain. Vertebrate intracellular defense is mediated by NOD-like receptors, which contain LRRs (Istomin and Godzik, 2009). Plant intracellular defense involves large numbers of LRRs proteins with nucleotide binding domains (NB-LRRs), while cell-surface responses in plants are mediated by extracellular LRRs on pattern recognition receptors (Padmanabhan et al., 2009).
Other immune related LRR proteins include secreted, nonmembrane-bound extracellular proteins. LRIM1 and APL1C in Anopheles gambiae are involved directing deposition of thioester containing protein 1 (TEP1) on the surface of Plasmodium, the malaria parasite (Riehle et al., 2008, Fraiture et al., 2009, Povelones et al., 2009). LRIM1 and APL1C contain LRRs at the N-terminus, a coiled-coil domain at the C-terminus, and circulate together in hemolymph as a ~260 kDa complex, which is held together by disulfide binding; orthologs have not been detected outside of mosquito species (Povelones et al., 2009). Here we report the characterization of a novel extracellular protein from the insect, Manduca sexta, which contains 13 LRRs, is upregulated upon immune challenge, and binds to bacterial lipopolysaccharide.
Materials and Methods
Insects and Collection of hemolymph, hemocytes and fat body from M. sexta larvae
M. sexta eggs were originally obtained from Carolina Biological Supply and reared using established methods (Dunn and Drake, 1983). Hemolymph, hemocytes, and fat body were collected from day 2 fifth instar larvae as described previously (Zhu et al., 2003b).
cDNA library screening and sequence analysis
A cDNA clone (accession BI262751) in vector pGEM-T with a partial sequence of leureptin was isolated from a subtracted cDNA library designed to represent genes expressed in fat body in response to bacterial challenge (Zhu et al., 2003a). This clone was digested with RsaI to release a 439 bp leureptin fragment, which was labeled with [α-32P]dCTP and used to probe a bacteria-induced M. sexta larval fat body cDNA library in Uni-Zap XR (Stratagene). A total of 8×104 lambda phage plaques were screened following a standard protocol (Sambrook and Russell, 2001). A positive clone (Leu 10-4) with the longest cDNA insert was sequenced from both strands at the Iowa State University DNA sequencing facility. Sequence analysis was performed as described previously (Zhu et al., 2003b).
Expression of recombinant leureptin in E. coli
Leureptin cDNA sequence encoding the mature protein, residue 18 to 407, was amplified by PCR using a forward primer (5'-TCA GCC ATG GGA AGT CCA ACA TG-3') with an NcoI site and a reverse primer (5'-AAA CTG CAG CTA GTG ATT GTC CAC-3') with a PstI site and used for protein expression in E. coli using a previously described protocol (Zhu et al., 2003b). Purified recombinant leureptin (1 mg) was further resolved by SDS-PAGE, the leureptin protein band excised, and used for the production of polyclonal rabbit antiserum (Cocalico Biologicals, Inc).
Northern blotting analysis and RT-PCR
The expression pattern of leureptin was examined by Northern blot and RT-PCR as described previously (Zhu et al., 2003b). Briefly, total RNA (20 μg) was isolated from fat body or hemocytes 24 h after injection of fifth instar larvae with either 10 μl filter-sterilized saline (0.85% NaCl) or 100 μg M. luteus in 10 μl sterile saline. Total RNA was fractionated by electrophoresis on an 1% agarose gel containing formaldehyde, then transferred to a nylon membrane and probed with a 32P-labeled leureptin cDNA fragment. A duplicate membrane was probed with 32P-labeled ribosomal protein S3 (rpS3) cDNA to confirm equal mRNA loading. The autoradiography exposure time was 16 h for the fat body blot and 14 days for the hemocyte blot. For RT-PCR, day 2 fifth instar larvae were injected with 100 μg M. luteus, and fat body from four larvae was dissected at different times after injection (0.5 h–24 h). Total RNA from fat body dissected at 24 h after injection with saline was used as a control. PCR amplification with 20 cycles of 94°C for 30 s, 44°C for 30 s, 72°C for 90 s using leureptin gene-specific primers (forward: 5'-GAG GGG TCT AGT GTC AGA C-3'; reverse: 5'-CCA TTT CTG CTC AAA TCT AAT-3').
SDS-PAGE and immunoblotting
SDS-PAGE was performed using 10% gels and sample buffer containing β-mercaptoethanol. For immunoblotting, proteins were electrotransferred to a nitrocellulose membrane using the MilliBlot SDE Transfer System (Millipore) and developed using rabbit antiserum to leureptin and an AP conjugate substrate kit (Bio-Rad).
In vitro fat body culture and immunprecipitation
Fat body (0.1 g) from individual insects 24 h after injection with saline or M. luteus was rinsed in sterile PBS and placed in 2 ml of Grace's insect cell culture medium (GibcoBRL) containing 5 μg/ml L-methionine (Sigma), 100 μCi L-[35S]-methionine (1000 Ci/mmol; Amersham Pharmacia Biotech) and 100 μg/ml ampicillin. Fat bodies from three insects for each treatment were cultured separately in six-well tissue culture plate wells. The incubations were performed at 28°C with shaking at 100 rpm. After 24 h, the medium was collected and filtered. To remove proteins that bind non-specifically, 500 μl of the filtrate was mixed with 25 μl of rabbit preimmune serum at 4°C for 1 hr and then incubated with 300 μl of a protein A-Sepharose bead suspension in PBS (25 mg/ml; Sigma) at 4°C for 30 min. The beads and nonspecifically bound proteins were removed by centrifuging at 10, 000×g, 5 min, 4°C. Leureptin was immunoprecipitated from the supernatant by incubating with 48 μl leureptin antiserum at 4°C overnight, then 600 μl of protein A bead suspension were added and incubated for 30 min at 4°C. The antigen-antibody-protein A complex was pelleted by centrifuging at 10,000g for 5 min and washed three times with PBS. The complex was treated with 1 × SDS sample buffer, and one fifth of the complex was subjected to SDS/PAGE followed by autoradiography. After exposure, the bands were excised from the gel, and radioactivity was measured by liquid scintillation counting.
Purification of native leureptin form naive hemolymph
125 ml of diluted cell-free hemolymph (1:1 in PBS) collected from naive fifth instar larvae was dialyzed against 4 L of 50 mM sodium phosphate, pH 6.3, 10 mM NaCl at 4°C for 24 h, with four changes of dialysis buffer. The dialyzed plasma was applied to a 53 ml CM-Sepharose cation exchange chromatography column. The column was subsequently washed with five bed-volumes of 50 mM sodium phosphate, pH 6.3, 10 mM NaCl, and leureptin was eluted at 2 ml/min with a NaCl gradient of 10 mM-500 mM. The 4 ml fractions were collected and analyzed by SDS-PAGE and identified by immunoblotting using leureptin antiserum.
Fractions containing leureptin from the cation exchange chromatography were pooled. A total volume of 62 ml was dialyzed against 2 l of 10 mM sodium phosphate, pH 6.45, 20 mM NaCl at 4°C overnight, with two changes of dialysis buffer. The dialyzed sample was applied to a 20 ml hydroxylapatite chromatography column, followed by washing with three column-volumes of 10 mM sodium phosphate, pH 6.45, 20 mM NaCl. Leureptin was eluted at 0.45 ml/min with two sequential sodium phosphate gradients: 10 mM – 100 mM with total volume of 50 ml and 100 mM–400 mM with total volume of 50 ml. The fractions (2 ml) were analyzed by immunoblotting.
Four isoforms of leureptin were separated after hydroxylapatite chromatography. To further purify isoform 2, the fractions containing isoform 2 were pooled and concentrated to 230 μl. A 40 μl sample was applied each time to Bio-Sil SEC-250 HPLC gel filtration (300×7.8 mm; Biorad), which was previously equilibrated with 100 mM sodium phosphate, pH 6.4, 150 mM NaCl. Leureptin isoform 2 was eluted with the same buffer at 1 ml/min. Fractions of 0.25 ml were collected and analyzed by SDS-PAGE and identified by immunoblotting. Standards (Bio-Rad), including thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa) and vitamin B-12 (1.35 kDa), were separated under the same conditions.
Determination of amino-terminal sequences of purified leureptin isoforms
4.5 μg of each purified leureptin isoform was treated with SDS sample buffer containing 2-mercaptoethanol and resolved by SDS-PAGE on a 10% gel. The protein was then transferred onto polyvinylidene difluoride (PVDF) membrane (Bio-rad) and stained with 0.025% Coomassie blue R-250 in 40% methanol. The band was excised and subjected to automated Edman degradation on an Applied Biosystems Model 473 pulse-liquid sequencer.
Deglycosylation of purified leureptin isoforms
1.5 μg of each of the purified leureptin isoforms was denatured at 95°C, 5 min in the presence of 1% SDS and then cooled to room temperature. The denatured protein was subsequently incubated with 2 units of N-glycosidase F (Boehringer Mannheim) in a total volume of 100 μl of 0.1 M sodium phosphate buffer, pH 6.4, containing 20 mM NaCl, 20 mM EDTA, 1% β-mercaptoethanol, and 1% CHAPS. The reaction was carried out at 37°C for 24 h. The enzyme-treated isoforms and untreated isoforms were analyzed using SDS-PAGE.
Circular dichroism (CD)
CD spectra were obtained for leureptin isoform 1 at a concentration of 0.11 mg/ml in 0.1 M sodium phosphate, 0.15 M NaCl, pH 6.4. Spectra from 280 to 190 nm were collected in a Jasco J720 spectrometer using a quartz cuvette with a 1 mm light path by scanning at least 16 times at 50 nm/min. The instrument was calibrated using D-10-camphorsulfonic acid as a standard.
LPS binding assay
The LPS binding ability of leureptin was analyzed as previously described (Yu and Kanost, 2000). Two μg of LPS suspension in 50 μl water was added to wells of a flat bottom 96-well plate (Coaster, Fisher). After the water was completely evaporated at room temperature, the plate was incubated at 60°C for 30 min to immobilize LPS to the well. 200 μl of 1 mg/ml bovine serum albumin (BSA) in Tris buffer (TB; 50 mM Tris-Cl, 50 mM NaCl, pH 8.0) was added to each well and incubated at 37°C for 2 hr to block the nonspecific binding sites. The plate was then washed twice with 200 μl/well of TB. Purified leureptin isoform 1 was added at serial dilutions of 0.5 – 32 μg/ml in a total volume of 50 μl/well and incubated with immobilized LPS at room temperature for 4 hr, followed by washing the plate four times with 200 μl TB/well. In the control well, 50 μl of TB without leureptin was added. Leureptin antiserum (1:1000 diluted in TB containing 0.1 mg/ml BSA) and alkaline phosphatase-conjugated goat anti-rabbit IgG (1:3000 diluted in TB containing 0.1 mg/ml BSA; Biorad) were sequentially added at 100 μl/well and incubated at 37°C for 2 h respectively. After each antibody incubation the wells were washed four times with 200 μl of TB. The bound leureptin was detected by washing the plate with 10 mM diethanolamine, 0.5 mM MgCl2 twice at 200 μl/well before adding 50 μl of 1 mg/ml of p-nitro-phenylphosphate into each well. Absorbance at 405 nm was determined after every 5 minutes for 30 minutes using a microtiter plate reader (Bio-Tek Instrument, Inc.).
To examine the competition of free LPS for leureptin binding, 50 μl of free LPS suspension at concentrations from 0.1 μg/ml – 1 mg/ml was added to the LPS-coated well simultaneously with 32 μg/ml leureptin. The total amount of free LPS added ranged from 0.005 – 50 μg per well. The incubation, subsequent antibody reaction and color development were performed as described above. The percentage of leureptin bound to immobilized LPS was calculated by comparing to the control which contained only leureptin without free LPS.
Association of leureptin with hemocytes
Hemocytes were collected from fifth instar larvae at 24 h after injection with saline or M. luteus. The cells from two insects were resuspended in 800 μl Manduca saline solution (4 mM NaCl, 40 mM KCl, 18 mM MgCl2, 3 mM CaCl2, 1.7 mM PIPES, 0.1% polyvinylpyrrolidone, 5% sucrose), applied to glass slides at 8 μl/well, and allowed to settle for 20 min. An equal volume of 4% paraformaldehyde in Manduca saline without CaCl2 or MgCl2 was added and incubated for 20 min to fix the cells. After removing the solution and washing the hemocytes with 20 μl of Manduca saline, slides were blocked for 1 h at room temperature with 3% BSA in Manduca saline (30 μl/well). Slides were rinsed once with TBS (40 μl/well) and then put into a negative 70°C freezer for 30 min. Anti-leureptin IgG purified from rabbit antiserum using an ImmunoPure IgG (Protein A) Purification Kit (PIERCE) was diluted at 1:100 in TBS containing 1% BSA and applied to slides at 30 μl/well. The binding of antileureptin IgG to leureptin was carried out at 4° overnight. Slides were then washed with 40 μl TBS/well three times for 5 min each at room temperature, followed by adding 30 μl of the secondary antibody, fluorescein isothiocyanate (FITC) labeled goat-anti-rabbit- IgG (Sigma), diluted at 1:400 in TBS containing 1% BSA. The incubation was performed in the dark at room temperature for 4 h. The slide was then washed three times with 40 μl TBS/well. Slides were covered with coverslips and examined by phase contrast and fluorescent microscopy.
Results
cDNA cloning and sequence of leureptin
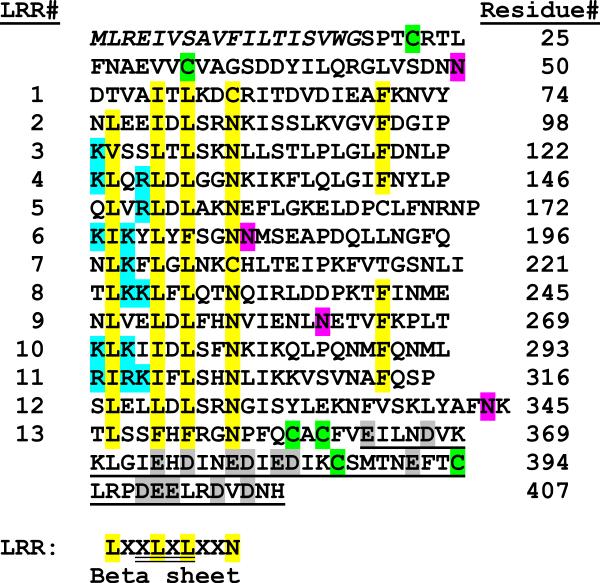
From our subtracted cDNA library of M. sexta fifth instar larval fat body we selected a cDNA clone with a 439 bp insert (BI262751) that was similar in sequence to a group proteins belonging to the LRR superfamily. The 439 bp fragment was then used as probe to screen a lambda phage cDNA library of E. coli-induced fat body to obtain full length clones. The longest clone, Leu 10-4, (AAO21503.1) contained a 1224-nucleotide open reading frame encoding a 407 amino acid residue polypeptide (Fig. 1) with a predicted amino-terminal secretion signal peptide of 18 residues, confirmed through N-terminal sequencing of purified protein (see below). A highly charged segment is found at the carboxyl-terminus, where 19 out of 45 residues (42%) are charged amino acids, and 14 of these are acidic (Fig. 1). A concentrated region of basic amino acids occurs in the N-terminal portion of repeats 3–9 and 11–12. Four potential N-glycosylation sites are present at Asn32, Asn165, Asn243 and Asn325. Without accounting for glycosylation, the calculated molecular mass of the mature protein is 44,520 Da and the isoelectric point is 7.3.
Fig. 1. M. sexta leureptin amino acid sequence and leucine rich repeats.
The complete amino acid sequence of M. sexta leureptin is shown with the 18-residue signal peptide in italics. The numbering of the amino acid residues is indicated on the right. The thirteen LRRs (LRR) have conserved hydrophobic residues, which are highlighted in yellow, and cysteines involved in N or C terminal caps are in green. The consensus for a typical LRR subfamily repeat is shown below with a region commonly found in a beta sheet double underlined. A carboxyl-terminal segment, enriched in negatively charged residues, is single underlined and acidic residues in that region are highlighted in gray. A region at the start of many of the LRRs has a concentrated number of basic residues highlighted in cyan. The four putative N-linked glycosylation sites are shown in magenta.
LRR motifs in leureptin
Leureptin amino acid residues 60–285 match the NCBI conserved domain database LRRs (LRRs), ribonuclease inhibitor (RI)-like subfamily (cd00116). Blastp results show some of the highest hits to LRR super family members derived from phylogenetically distant species, including many hits to hypothetical proteins from Branchiostoma floridae (top hit XP 002588853.1, e value 7e-29), a putative toll precursor from Pediculus humanus corporis (XP 002431445.1, e value 1e-25), carboxypeptidase N regulatory subunit from Rattus norvegicus (NP 001100555.1, e value 2e-24), LRR-containing protein 15 isoform b from Homo sapiens (NP 570843.2, e value 6e-24), and tartan/capricious-like protein from Tribolium castaneum (EFA04688.1, e value 5e-23).
Like other LRR containing proteins, leureptin has a high content (15.7%) of leucine residues (61 out of 389 residues in the mature protein). These leucine residues are distributed in a pattern of LxxLxLxxN. Thirteen repeats containing this pattern are arranged in tandem, eight of which have 24 residues, while the other internal repeats range from 23 to 28 residues long. Sequence alignment of all thirteen repeats gives rise to a consensus sequence that is very similar to the Typical LRR subfamily consensus, LxxLxLxxNxLxxLxxxxFxxLxx, which is also the subfamily most frequently found in Toll-like receptors (Bella et al., 2008, Kajava, 1998, Matsushima et al., 2007). We observed cysteine residues at the N and C termini, which are similar to clusters found in Toll-like receptors. The amino-terminal cysteine cluster, in the orientation Cx9C, is predicted to form a disulfide bond. The C-terminal LRR in both leureptin and Toll-like receptors matches LxxLxLxxNP(F/L)xCxCxxxx(F/L)xxxx, while the leureptin C-terminal cysteine pattern, CxCx25Cx7C, is quite similar to those found in Toll-like receptors (CxCx22–25Cx15–20C), with the exception of fewer amino acid residues between the last two cysteines (Matsushima et al., 2007, Ao et al., 2008b). Based on similarity to the LRR region of vertebrate Toll-like receptors, we predict disulfide bonds form between the first and third C-terminal cysteine residues and between the second and fourth (Matsushima et al., 2007).
Leureptin mRNA and protein expression
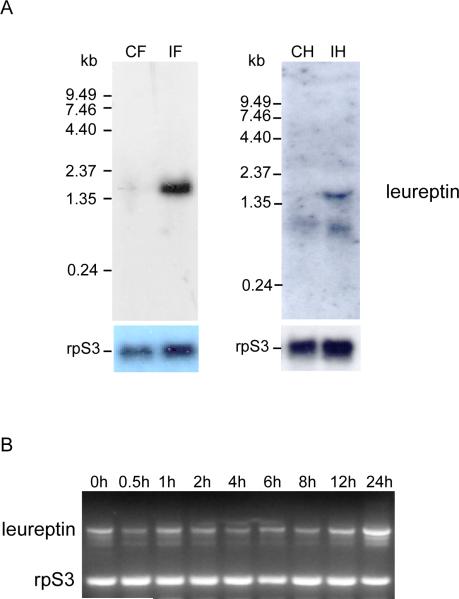
Northern blot analysis showed that a leureptin 1.5 kb mRNA was expressed in fat body after bacterial challenge, but was very faint in saline-injected insects (Fig. 2A). A longer autoradiography exposure time was required to detect a weak signal in RNA from bacteria-induced hemocytes, indicating that hemocytes express leureptin at a low level. When leureptin mRNA level in fat body was examined by RT-PCR (Fig. 2B), we observed that leureptin was constitutively expressed at a low level. The amount of leureptin mRNA was significantly elevated at 12 hr after injection of M. luteus and continued to increase up to 24 hr.
Fig. 2. Expression of leureptin mRNA.
(A) Northern blot analysis of leureptin mRNA expression was performed as described in Materials and Methods. CF: control fat body; IF: bacteria-induced fat body; CH: control hemocytes; IF: bacteria-induced hemocytes. (B) Detection of leureptin mRNA induction by RT-PCR. Day 2, fifth instar larvae were injected with M. luteus. Fat body was dissected at different times after the injection and used for RNA isolation and analysis by RT-PCR using gene-specific primers. Primers specific for rpS3 were used as an internal control.
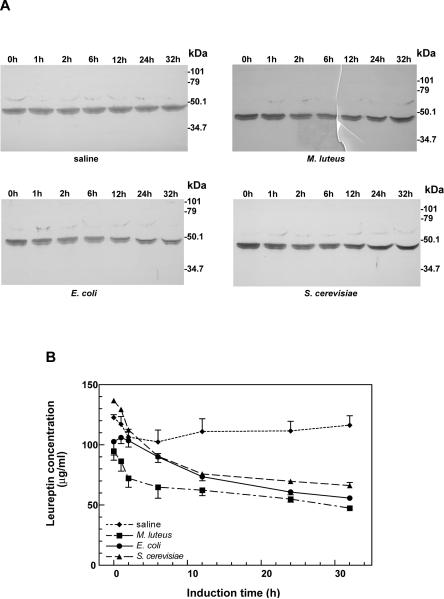
The expression of leureptin protein in M. sexta fifth instar larval plasma was examined by immunoblotting (Fig. 3). Leureptin was present in plasma of naive insects, and its concentration did not appear to change after injection of saline. However, unlike other immune-inducible genes identified so far in M. sexta, whose plasma protein levels increase during immune challenge, leureptin concentration began to decrease at 2 h and continued to decline up to 32 hr after injection of Gram-positive bacteria (M. luteus), Gram-negative bacteria (E. coli) or yeast (S. cerevisiae). Several bands at 48–50 kDa cross-reacted with antiserum to recombinant leureptin which suggests several isoforms exist in hemolymph (see below).
Fig. 3. The induction pattern of leureptin protein in plasma.
Day 2, fifth instar larvae were injected with saline, M. luteus, E. coli, or S. cerevisiae. Hemolymph was collected at different times after the injection, and the hemocytes were removed by centrifugation. The plasma was then separated by SDS-PAGE and leureptin protein was detected by immunoblot. (A) One typical blot from each group is shown. Numbers above the lanes indicate hours after injection. (B) Leureptin concentration was measured by immunoblot analysis, using known amounts of purified recombinant leureptin as standard. The error bars represent the standard error of the mean (n=4).
Secretion of leureptin by fat body in vitro
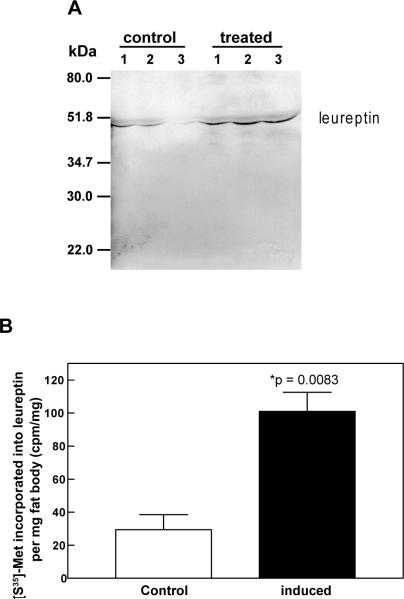
The decrease in leureptin concentration after microbial challenge may indicate that the constitutively expressed leureptin was consumed during a response to infection. To examine whether increased leureptin protein is synthesized and secreted following the induced expression of its mRNA, fat body from saline-injected (control) or M. luteus-injected (induced) fifth instar larvae were incubated for 24 h in culture medium containing [35S]-methionine. Leureptin immunoprecipitated from tissue culture medium was resolved by SDS-PAGE followed by autoradiography (Fig. 4A). A radioactive band at approximately 48 kDa was observed in each lane, indicating that leureptin was synthesized and secreted into medium by fat body from both control and M. luteus treated insects. More intense bands from culture medium of bacteria-challenged fat body samples demonstrated that leureptin was synthesized and secreted at a greater rate from the bacteria-challenged fat body than from fat body of control insects. Scintillation counting of the leureptin bands demonstrated that de novo synthesis and secretion of leureptin by fat body from bacteria-injected larvae was 3.4 fold greater than that by fat body from saline-injected controls (Fig. 4B).
Fig. 4. Synthesis and secretion of leureptin by cultured fat body.
Fat body was dissected 24 h after injection of day 2, fifth instar larvae with saline (control) or M. luteus (induced). Fat body was cultured at 28°C for 24 h in [35S]-methionine. Leureptin in the culture medium was then immunoprecipitated as described in materials and methods. (A) Immunoprecipitates corresponding to the protein secreted by 5 mg fat body (wet weight) were subjected to SDS/polyacrylamide gel and detected by autoradiography. The radioactive bands corresponding to leureptin are indicated by an arrow head. Each lane represents the immunoprecipitate from one insect fat body culture. (B) Quantification of leureptin synthesis and secretion. The radioactive leureptin bands were excised from polyacrylamide gel and subjected to liquid scintillation counting. The radioactivity incorporated into leureptin per mg fat body was calculated for each treatment. Error bars indicate standard error of the mean (n=3). The significance of difference between the two samples was determined by t test.
Purification of leureptin from larval plasma
To further characterize and study functions of leureptin, we purified the protein from naïve larvae. The plasma was dialyzed against 50 mM sodium phosphate buffer, 10 mM NaCl, pH 6.3, and then was separated by cation exchange chromatography on CM-Sepharose. All isoforms of leureptin bound to the column and eluted together in the sodium chloride gradient between 320–430 mM NaCl (Supplemental Fig. 1A). The fractions containing leureptin were then pooled, dialyzed against 10 mM sodium phosphate buffer, 20 mM NaCl, pH 6.45, and applied to a hydroxylapatite column, which separated four isoforms of leureptin into different fractions (Supplemental Fig. 1B). Isoform 1 eluted closely with isoform 2 between 230 mM-250 mM and 265 mM-290 mM sodium phosphate respectively. Isoform 3 was eluted followed by isoform 4 at 400 mM sodium phosphate. The four isoforms had slightly different mobility on SDS-PAGE. Isoforms 1, 3 and 4 were purified to near homogeneity after hydroxylapatite chromatography (Supplemental Fig. 1B), but some major contaminating proteins of ~20 kDa co-eluted together with isoform 2. These smaller protein contaminants were separated from isoform 2 by gel filtration HPLC (Supplemental Fig. 2) and comparison of the elution volume of leureptin isoform 2 with a set of protein standards indicated that the native molecular mass of leureptin isoform 2 is ~59 kDa, which suggests that it is a monomer in solution.
The amino-terminal sequences of all four purified isoforms were determined by automated Edman degradation. The ten amino acid residues at the amino-terminus, SPTXRTLFNA were the same for all of the isoforms and match the sequence for the mature protein predicted from the cDNA (Fig. 1). Cys at residue 4 was not detected, which suggests that it is part of a disulfide bond and forms an N-terminal cap (Matsushima 2007). Further sequencing of isoform 1 to residue 26 showed an amino acid sequence identical to that deduced from the cDNA.
The mass of the leureptin isoforms estimated by SDS/PAGE, 48–50 kDa, is larger than the calculated molecular mass of 44.5 kDa. This could be due to post-translational modification such as glycosylation. Considering the four potential N-glycosylation sites in the leureptin sequence, we tested whether this protein was N-glycosylated. Treatment of each purified isoform with N-glycosidase F resulted in a disappearance of the 50 kDa band and appearance of a band with an apparent mass of 47 kDa (Supplemental Fig. 3). This result indicates that all four isoforms of leureptin are N-glycosylated.
Secondary structure of leureptin
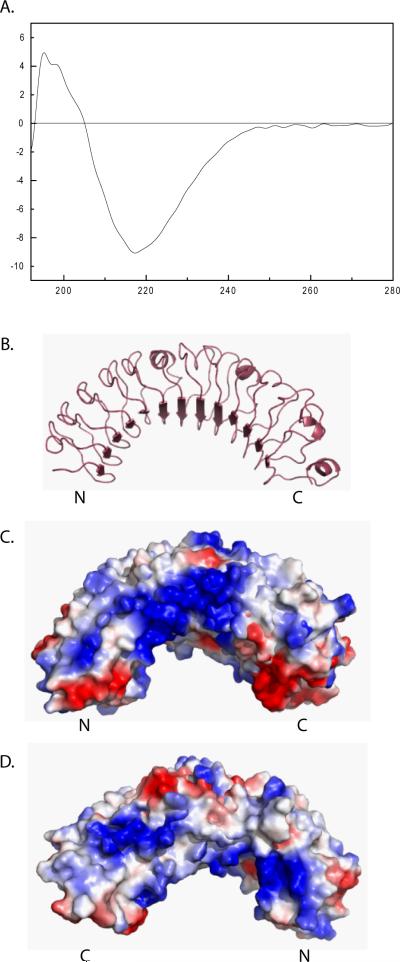
LRR proteins form a curved coil shape with beta sheets on the concave face. We investigated the secondary structure of leureptin using circular dichroism (CD) spectroscopy (Fig. 5A). The CD spectrum showed a strong negative peak at 217 nm and one strong positive peak between 195 nm and 200 nm, the characteristic features of a spectrum generated by β-sheet (Sreerama and Woody, 2004). Besides the major peaks, we also detected a negative shoulder around 221 nm, a positive shoulder between 200 and 210 nm and a negative band between 190–195 nm, which are the characters of β-turn Class B spectra (Woody, 1974). The features of spectra for α-helices, two strong minima at 208 nm and 222 nm and a maximum at 192 nm (Sreerama and Woody, 2004), were not observed in the leureptin CD spectrum. From the CD spectrum, it is very likely that leureptin adopts a structure formed predominantly from β-structure.
Fig 5. Leureptin structure.
(A) The CD spectrum of leureptin was obtained using 0.11 mg/ml of leureptin isoform 2 in 0.1 M sodium phosphate, 0.15 M NaCl, pH 6.4. Using Pymol we displayed a homology model of leureptin (Q86RS5) from N to C terminus in cartoon (B) and spacefill with electrostatics (C). Positive charge is shown in blue and negative charge is shown in red. Leureptin from C to N terminus is shown in spacefill with electrostatics in (D).
Four homology models for leureptin (Q86RS5) are present in the Swiss Model repository (Kiefer et al., 2009, Kopp and Schwede, 2004). The best fitting model, e-value 4.3E-43, uses a portion of the lingo receptor (PDB 2ID5) as the template (Mosyak et al., 2006). The match between leureptin and the template is based on HHSearch of predicted secondary structure (Kiefer et al., 2009). This model covers leureptin residues 50–366, which includes the 13 LRRs. The two sequences share 24% identity in this region. As expected for LRR structure, the 13 repeat sequences in this model form parallel beta strands containing the second conserved leucine in the consensus sequence (Fig. 5B).
Consistent with our CD data, the homology models show a large beta sheet forming a concave surface and little alpha helical content. Electrostatic analysis shows a distinctive positively charged region on the N-terminal side loop due to an abundance of Lys and Arg residues at positions 0, 2, or 3 in repeats 3–8, 10, and 11 (Fig. 5C). The opposite side of the protein, from C-terminus to N terminus, is less dramatically charged but also contains a positive patch near the N-terminus (Fig. 5D). The acidic region at the very C-terminus of the protein, from residues 363 to 407, is not present in this model.
LPS binding by leureptin
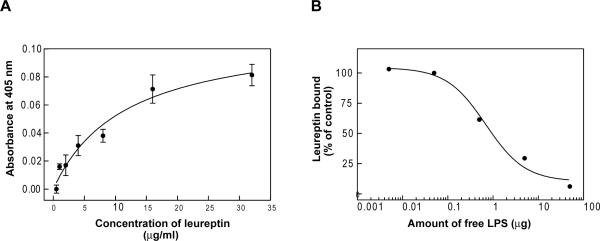
Two important proteins in LPS signaling in mammals are CD14 and TLR4, both LRR family members (Ferrero et al., 1993, Gioannini and Weiss, 2007). This prompted us to test whether leureptin can bind to LPS, using an ELISA-like plate assay in which LPS was immobilized in wells of a 96-well plate. After washing, leureptin antiserum was used to detect bound leureptin. Leureptin bound LPS in a concentration-dependent and saturable manner (Fig. 6A). To test the possibility that leureptin could bind nonspecifically to the well, we performed a competitive binding assay, in which different concentration of free LPS were added to the binding reactions. Free LPS competed effectively for leureptin binding to immobilized LPS (Fig.6B). Leureptin binding was inhibited by 50% at ~1μg of free LPS per well, approximately the same amount of LPS (2 μg) immobilized in the wells. Free peptidoglycan did not compete with leureptin's binding to LPS (data not shown).
Fig. 6. LPS binding ability of leureptin.
(A) LPS binding assay. Purified leureptin isoform 1 from plasma was added in serial dilutions into an LPS-coated microtiter plate, and the binding assay was carried out as described in materials and methods. Each point represents the mean of three individual measurements ± SEM. The line represents the nonlinear regression calculation of one-site binding curve (R2 = 0.87). (B) Competition of free LPS for leureptin binding. Free LPS (0.005 – 50 μg) was added to each well of an LPS-coated microtiter plate simultaneously with 35μg/ml leureptin. The amount of the bound leureptin was measured as in materials and methods. The percentage of leureptin bound to immobilized LPS was calculated by comparison to the control, which contained no free LPS. The line represents the calculation of nonlinear regression of one-site competition curve (R2 = 0.99).
Association by leureptin with hemocytes
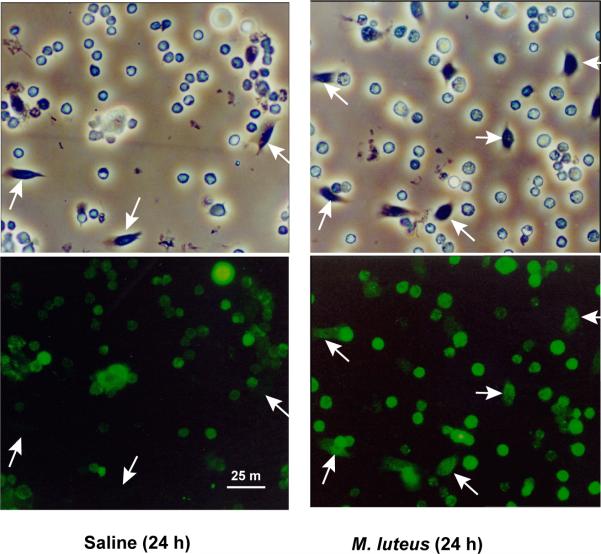
Leureptin protein level in hemolymph decreased after infection despite the induction of leureptin mRNA by bacterial challenge and accelerated synthesis of leureptin in fat body from bacteria-challenged insects. This suggests that leureptin is consumed, which may include being digested by proteases in the hemolymph or cleared from circulation. Immunoblots of plasma from insects challenged with bacteria have not shown any bands migrating at lower molecular mass, suggesting leureptin is not being degraded by proteases. One plausible explanation for the decreased circulating leureptin concentration is that after binding to the surface of microorganisms, leureptin associates with hemocytes. We utilized immunofluorescence microscopy to visualize the association of leureptin with hemocytes. Hemocytes collected from saline-injected (control) or Micrococcus-injected (challenged) fifth instar larvae at 24 h after injection were fixed on microscope slides and reacted with purified anti-leureptin IgG, followed by subsequent incubation with FITC-labeled goat-anti-rabbit antibody. The hemocytes from bacteria challenged larvae had an overall stronger fluorescent signal than the control group (Fig. 7). The fluorescence of granular hemocytes and plasmatocytes was much more intense in the challenged group than in the control group. The plasmatocytes were barely labeled from saline-injected larvae but clearly fluorescent from Micrococcus-injected larvae. Oenocytoids were labeled to similar degree in both control and bacteria-challenged samples. This result showed that leureptin binds to hemocytes, especially granulocytes and plasmatocytes, after bacterial challenge.
Fig. 7. Association of leureptin with hemocyte membranes.
Hemocytes collected 24 h after injection of saline or M. luteus were fixed on glass slides. Anti-leureptin IgG and FITC-labeled goat-anti-rabbit antibody were used to detect leureptin as described in materials and methods. The upper panels show hemocytes viewed by phase contrast, and the lower panels show hemocytes viewed by fluorescence microscopy. Both plasmatocytes (indicated by arrows) and granulocytes stained more intensely in bacteria-challenged insects. Oenocytoids had similar labeling intensity in control and treated samples.
Discussion
Our characterization of a novel LRR protein in Manduca sexta hemolymph reveals that leureptin mRNA and protein increase after immune challenge but that leureptin protein does not accumulate in plasma. We have also shown that leureptin is a soluble pattern recognition protein that can recognize LPS. After injection of bacteria, leureptin also increases association with certain populations of hemocytes, the granular cells and plasmatocytes, which are both implicated in encapsulation and phagocytosis. M. sexta hemocytes also contain at least one Toll-like receptor which is upregulated upon immune challenge (Ao et al., 2008b).
We propose that leureptin may bind bacteria and target them for immune effector responses. The binding of leureptin to hemocytes might trigger phagocytosis or encapsulation by hemocytes. Hemocyte binding may be a trigger for phagocytosis, nodule formation, or other signal transduction. Other LRR proteins have a documented role in phagocytosis. Listeria monocytogenes triggers its own uptake by phagocytosis to facilitate invasion of mammalian host cells through binding of the membrane-bound LRR protein internalin to E-cadherin on host cells (Kedzierski et al., 2004). Leureptin, however, does not appear to be membrane bound as it lacks a transmembrane region or region for a GPI anchor.
The large, positively charged region observed on the N-terminal side of leureptin differs from the surfaces in predicted structures of human NLRs and TLRs (Istomin and Godzik, 2009). A positive patch due to four basic residues is seen in polygalacturonase-inhibiting protein, an extracellular LRR protein from the plant Phaseolus vulgaris, where the positive cluster is predicted to ionically anchor the protein to pectin in the plant cell wall (Di Matteo et al., 2006). Mutation of the Arg and Lys residues decreases binding to polygalacturonic acid (Spadoni et al., 2006). It is possible that the positively charged patch of leureptin electrostatically associates with negatively charged bacterial surfaces such as the negative phosphate groups in lipopolysaccharide.
Soluble, secreted LRR proteins participate in A. gambiae immune response against Plasmodium species. LRIM1 and APL1C are LRR proteins with C-terminal coiled coil domains that together form a high molecular weight (~260 kDa) disulfide bond linked complex in plasma (Povelones et al., 2009). The LRIM1-APL1C complex appears to bind active thioester containing protein 1 (TEP1) and direct its deposition onto Plasmodium berghei tissues (Povelones et al., 2009). In the absence of the LRIM1-APL1 complex, active TEP1 binds to mosquito tissues (Fraiture et al., 2009). Interestingly, APL1C is also implicated in protection against Plasmodium yoelii while APL1A specifically protects against Plasmodium falciparum but not P. berghei or P. yoelii (Mitri et al., 2009). Orthologs of LRIM1 and APL1 have not been found outside of mosquito species (Povelones et al., 2009)
The domain structure of leureptin is simple: a secretion signal, the 13 LRRs, and a hydrophilic C-terminus enriched in negatively charged amino acids with no indication of covalent membrane linkage. We assume that proteins filling a similar function to leureptin in other species would have the same general structure. A total of 66 extracellular LRR proteins have been catalogued in D. melanogaster, only 15 of which appeared to be soluble rather than membrane bound (Dolan et al., 2007) and none of which showed particular similarity to leureptin. Blast searches of the NCBI EST database show putative homologs in other Lepidopteran insects, including Choristoneura fumiferana (FC942037.1), Heliconius erato (DT665984.3), Bicyclus anynana (GE671847.1), Spodoptera frugiperda (DY777187.1), and Heliothis virescens (GT207433.1), but we could not identify a homolog in Bombyx mori.
Recently a 16kDa protein, ML-1, was purified and characterized from M. sexta plasma (Ao et al., 2008a). This protein contains an ML (MD-2-related lipid-recognition) domain. It co-purified with leureptin through three chromatography steps, and the two proteins were finally separated by hydroxyapatite chromatography (Ao et al., 2008a). ML-1 is similar in sequence to MD-2 in mammals, which accepts LPS from CD-14 and binds TLR-4. Recombinant M. sexta ML-1 bound to immobilized LPS and immobilized lipid A (Ao et al., 2008a). Further research is required to investigate any direct interaction between leureptin and ML-1 and their role in LPS recognition.
In conclusion, we purified and characterized leureptin, a LRR containing protein in Manduca sexta that has putative orthologs in other Lepidopteran insects. Leureptin is upregulated upon bacterial challenge yet leureptin protein in the plasma decreases, indicating leureptin leaves circulation in the immune response. Leureptin binds to bacterial LPS and hemocytes, leading us to propose that leureptin may target bacteria for subsequent immune responses like phagocytosis and encapsulation.
Research Highlights.
Leureptin, a hemolymph plasma protein from Manduca sexta, contains 13 leucine-rich repeats.
Leureptin mRNA levels increase in fat body after bacterial injection but protein levels in plasma decrease, suggesting leureptin is consumed during the immune response.
Leureptin binds to bacterial lipopolysaccharide and associates with hemocytes after injection of bacteria.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ao JQ, Ling E, Rao XJ, Yu XQ. A novel ML protein from Manduca sexta may function as a key accessory protein for lipopolysaccharide signaling. Mol. Immunol. 2008a;10:2772–2781. doi: 10.1016/j.molimm.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao JQ, Ling E, Yu XQ. A Toll receptor from Manduca sexta is in response to Escherichia coli infection. Mol. Immunol. 2008b;2:543–552. doi: 10.1016/j.molimm.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell Mol. Life Sci. 2008;15:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan SG, Gay NJ. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog. Biophys. Mol. Biol. 1996;1–2:1–44. doi: 10.1016/s0079-6107(96)00003-x. [DOI] [PubMed] [Google Scholar]
- Daffre S, Faye I. Lipopolysaccharide interaction with hemolin, an insect member of the Ig-superfamily. FEBS Letters. 1997;408:127–130. doi: 10.1016/s0014-5793(97)00397-9. [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Bonivento D, Tsernoglou D, Federici L, Cervone F. Polygalacturonase-inhibiting protein (PGIP) in plant defence: a structural view. Phytochemistry. 2006;6:528–533. doi: 10.1016/j.phytochem.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Dolan J, Walshe K, Alsbury S, Hokamp K, O'Keeffe S, Okafuji T, Miller SF, Tear G, Mitchell KJ. The extracellular leucine-rich repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genomics. 2007:320. doi: 10.1186/1471-2164-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn PE, Drake D. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm, Manduca sexta. J. Invertebr. Pathol. 1983:77–85. [Google Scholar]
- Ferrero E, Hsieh CL, Francke U, Goyert SM. CD14 is a member of the family of leucine-rich proteins and is encoded by a gene syntenic with multiple receptor genes. J. Immunol. 1990;1:331–336. [PubMed] [Google Scholar]
- Ferrero E, Jiao D, Tsuberi BZ, Tesio L, Rong GW, Haziot A, Goyert SM. Transgenic mice expressing human CD14 are hypersensitive to lipopolysaccharide. Proc. Natl. Acad. Sci. U. S. A. 1993;6:2380–2384. doi: 10.1073/pnas.90.6.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraiture M, Baxter RH, Steinert S, Chelliah Y, Frolet C, Quispe-Tintaya W, Hoffmann JA, Blandin SA, Levashina EA. Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell. Host Microbe. 2009;3:273–284. doi: 10.1016/j.chom.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Gioannini TL, Weiss JP. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol. Res. 2007;1–3:249–260. doi: 10.1007/s12026-007-0069-0. [DOI] [PubMed] [Google Scholar]
- Hindle KL, Bella J, Lovell SC. Quantitative analysis and prediction of curvature in leucine-rich repeat proteins. Proteins. 2009;2:342–358. doi: 10.1002/prot.22440. [DOI] [PubMed] [Google Scholar]
- Istomin AY, Godzik A. Understanding diversity of human innate immunity receptors: analysis of surface features of leucine-rich repeat domains in NLRs and TLRs. BMC Immunol. 2009:48. doi: 10.1186/1471-2172-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 1998;3:519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Kobe B. Assessment of the ability to model proteins with leucine-rich repeats in light of the latest structural information. Protein Sci. 2002;5:1082–1090. doi: 10.1110/ps.4010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierski L, Montgomery J, Curtis J, Handman E. Leucine-rich repeats in host-pathogen interactions. Arch. Immunol. Ther. Exp. (Warsz) 2004;2:104–112. [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;(Database issue):D387–92. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Lee CJ, Jin MS, Lee CH, Paik SG, Lee H, Lee JO. Crystal structure of CD14 and its implications for lipopolysaccharide signaling. J. Biol. Chem. 2005;12:11347–11351. doi: 10.1074/jbc.M414607200. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001;6:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- Kopp J, Schwede T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 2004;(Database issue):D230–4. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Lemaitre B. Toll-like receptors--taking an evolutionary approach. Nat. Rev. Genet. 2008;3:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- Matsushima N, Tanaka T, Enkhbayar P, Mikami T, Taga M, Yamada K, Kuroki Y. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genomics. 2007:124. doi: 10.1186/1471-2164-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri C, Jacques JC, Thiery I, Riehle MM, Xu J, Bischoff E, Morlais I, Nsango SE, Vernick KD, Bourgouin C. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 2009;9:e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosyak L, Wood A, Dwyer B, Buddha M, Johnson M, Aulabaugh A, Zhong X, Presman E, Benard S, Kelleher K, Wilhelm J, Stahl ML, Kriz R, Gao Y, Cao Z, Ling HP, Pangalos MN, Walsh FS, Somers WS. The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition. J. Biol. Chem. 2006;47:36378–36390. doi: 10.1074/jbc.M607314200. [DOI] [PubMed] [Google Scholar]
- Padmanabhan M, Cournoyer P, Dinesh-Kumar SP. The leucine-rich repeat domain in plant innate immunity: a wealth of possibilities. Cell. Microbiol. 2009;2:191–198. doi: 10.1111/j.1462-5822.2008.01260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Wu LP. Pattern recognition receptors in the fly: lessons we can learn from the Drosophila melanogaster immune system. Fly (Austin) 2009;2:121–129. doi: 10.4161/fly.8827. [DOI] [PubMed] [Google Scholar]
- Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science. 2009;5924:258–261. doi: 10.1126/science.1171400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugin J, Heumann ID, Tomasz A, Kravchenko VV, Akamatsu Y, Nishijima M, Glauser MP, Tobias PS, Ulevitch RJ. CD14 is a pattern recognition receptor. Immunity. 1994;6:509–516. doi: 10.1016/1074-7613(94)90093-0. [DOI] [PubMed] [Google Scholar]
- Ragan EJ, An C, Jiang H, Kanost MR. Roles of haemolymph proteins in antimicrobial defences of Manduca sexta. In: Rolff J, Reynolds SE, editors. Insect Infection and Immunity. Oxford University Press; 2009. pp. 34–48. [Google Scholar]
- Riehle MM, Xu J, Lazzaro BP, Rottschaefer SM, Coulibaly B, Sacko M, Niare O, Morlais I, Traore SF, Vernick KD. Anopheles gambiae APL1 is a family of variable LRR proteins required for Rel1-mediated protection from the malaria parasite, Plasmodium berghei. PLoS One. 2008;11:e3672. doi: 10.1371/journal.pone.0003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: A laboratory manual. 3rd Edition Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Schmidt O, Faye I, Lindstrom Dinnetz I, Sun SC. Specific immune recognition of insect hemolin. Dev Comp Immunol. 1993;17:195–200. doi: 10.1016/0145-305x(93)90038-r. [DOI] [PubMed] [Google Scholar]
- Spadoni S, Zabotina O, Di Matteo A, Mikkelsen JD, Cervone F, De Lorenzo G, Mattei B, Bellincampi D. Polygalacturonase-inhibiting protein interacts with pectin through a binding site formed by four clustered residues of arginine and lysine. Plant Physiol. 2006;2:557–564. doi: 10.1104/pp.106.076950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004:318–351. doi: 10.1016/S0076-6879(04)83013-1. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ishibashi J, Fujita K, Nakajima Y, Sagisaka A, Tomimoto K, Suzuki N, Yoshiyama M, Kaneko Y, Iwasaki T, Sunagawa T, Yamaji K, Asaoka A, Mita K, Yamakawa M. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol. 2008;12:1087–1110. doi: 10.1016/j.ibmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, Fukudome K, Takao S, Tsuneyoshi N, Kimoto M. Lipopolysaccharide-binding protein-mediated Toll-like receptor 4 dimerization enables rapid signal transduction against lipopolysaccharide stimulation on membrane-associated CD14-expressing cells. Int. Immunol. 2010;4:271–280. doi: 10.1093/intimm/dxq005. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiang H. Binding properties of the regulatory domains in Manduca sexta hemolymph proteinase-14, an initiation enzyme of the prophenoloxidase activation system. Dev. Comp. Immunol. 2010;3:316–322. doi: 10.1016/j.dci.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XQ, Kanost MR. Binding of hemolin to bacterial lipopolysaccharide and lipoteichoic acid. Eur. J. Biochem. 2002;269:1827–1834. doi: 10.1046/j.1432-1033.2002.02830.x. [DOI] [PubMed] [Google Scholar]
- Yu XQ, Kanost MR. Immulectin-2, a lipopolysaccharide-specific lectin from an insect, Manduca sexta, is induced in response to gram-negative bacteria. J. Biol. Chem. 2000;48:37373–37381. doi: 10.1074/jbc.M003021200. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM, Mueller HM, Dimopoulos G, Law JH, Wells MA, Birney E, Charlab R, Halpern AL, Kokoza E, Kraft CL, Lai Z, Lewis S, Louis C, Barillas-Mury C, Nusskern D, Rubin GM, Salzberg SL, Sutton GG, Topalis P, Wides R, Wincker P, Yandell M, Collins FH, Ribeiro J, Gelbart WM, Kafatos FC, Bork P. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;5591:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Johnson TJ, Myers AA, Kanost MR. Identification by subtractive suppression hybridization of bacteria-induced genes expressed in Manduca sexta fat body. Insect Biochem. Mol. Biol. 2003a;5:541–559. doi: 10.1016/s0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wang Y, Gorman MJ, Jiang H, Kanost MR. Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase-activating proteinases. J. Biol. Chem. 2003b;47:46556–46564. doi: 10.1074/jbc.M309682200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.