Abstract
A new member of the phlebovirus genus, tentatively named Granada virus, was detected in sandflies collected in Spain. By showing the presence of specific neutralizing antibodies in human serum collected in Granada, we show that Granada virus infects humans. The analysis of the complete genome of Granada virus revealed that this agent is likely to be a natural reassortant of the recently described Massilia virus (donor of the long and short segments) with a yet unidentified phlebovirus (donor of the medium segment).
Introduction
The genus Phlebovirus of the family Bunyaviridae comprises 37 virus species in 9 serocomplexes.1 Fifteen additional viruses are registered as tentative species.
Rift Valley Fever virus (RVFV), an emerging deadly pathogen for cattle, sheep, and humans, is the type species and the most studied member of the genus.2 Another phlebovirus, Toscana virus (TOSV), can cause meningitis, encephalitis, or meningoencephalitis in humans.3–7 Human TOSV infection is reported in France, Portugal, Spain, and Italy.8–11 TOSV belongs to the Naples serocomplex that also includes Sandfly Fever Naples virus (SFNV), the cause of a self-limited febrile condition known as phlebotomus, papatacci, or sandfly fever.4 Charrel and others12 described a new member of this serocomplex: Massilia virus (MASV). MASV was isolated from Phlebotomus spp. sandflies and shown to circulate in southeastern France.
Recently, we identified the presence of MASV-like sequences in sandflies in the northeast part of Spain, near France.13 To determine the geographical distribution of this agent, ascertain if it was MASV or a different virus, and assess its capacity to infect humans, we surveyed sandflies for phleboviruses and performed human seroprevalence studies. Through these efforts, we detected and isolated a new agent, Granada virus (GRV), obtained the complete coding sequence of one strain (GRV 25), and found serological evidence of human GRV infection. Although no relation with human disease is yet established, the presence of antibodies in human sera raises questions about its possible pathogenicity and indicates a need for further investigation.
Materials and Methods
Sandfly collection, nucleic acids extraction, and isolation of viruses in cell culture.
Phlebotomine sandflies were captured using Centers for Disease Control and Prevention (CDC) light traps from June to October of 2003 and 2004 in Granada province (southeast Spain). All traps were placed in the vicinity of animals (horses, pigs, dogs, chicken, turkeys, sheep, rabbits, or goats) in human (residential or rural) or animal dwellings. Sandflies were captured from dusk to dawn. Traps were immediately transported to the laboratory to pool the individuals (50–100 individuals per pool) by sex and trapping area. A total of 103 pools were used for phleboviruses investigation by reverse transcription polymerase chain reaction (RT-PCR) and viral culture.14 Twenty-two pools obtained in 2003 consisted only of female sandflies; in 2004, we collected 42 male-only and 39 female-only pools. Briefly, phlebotomines were introduced in vials with sterile crystal beads and 0.5 mL minimal essential media (Sigma-Aldrich, Madrid, Spain), 20% bovine fetal serum (Reactiva SA; Biological Industries, Spain), and antibiotic mix (0.4 mg/mL gentamicin, 0.5 mg/mL vancomicin, and 2.5 µg/mL amphotericin B or 10% penicillin and streptomycin; BioWhittaker, Barcelona, Spain). Vials were vortexed and centrifuged at 13,000 rpm for 5 minutes. The pellet with the phlebotomines was used for nucleic acids extraction (QIAmp Viral RNA system; QIAGEN) and subsequent generic RT-PCR.15 A 200-μL aliquot of the supernatant was inoculated in tubes with African green monkey kidney cells (Vero cells; see below). Tube cultures were incubated at 37°C and examined daily to observe the appearance of cythopathic effect (CPE). Tubes were tested for phleboviruses by generic RT-PCR.15 Five pools (containing 50–100 individuals per pool) of female sandflies were also obtained in Ibiza (Balearic Islands) during 2004 (June and July). Ibiza samples were only used for nucleic acids extraction (QIAmp Viral RNA system; QIAGEN).
For cell-culture experiments involving TOSV, the strain ISS.Phl.3 was used. For the molecular studies, other phleboviruses were also tested: Anhanga virus (ANHV), Bujaru virus (BUJV), Candiru virus (CDUV), Icoaraci virus (ICOV), Itaporanga virus (ITPV), Chagres virus (CHGV), Salehabad virus (SALV), and Arumawot virus (AMTV). All of these viruses were obtained from the ATCC (LGC, Spain). GRV25 was isolated from a pool of female sandflies captured in Alfacar, Granada in the month of June in 2004 as previously described. All viruses were grown in Vero cells.
Molecular identification of phleboviruses.
A generic RT-nested PCR for phleboviruses was used for detection.15 Subsequent sequencing allowed preliminary identification of the virus species. Virus sequences were obtained using generic primers binding to the long (L) segment. Sequences obtained were compared with available phleboviral sequences by pairwise sequence comparison. The simple P distance model of substitution was used to calculate distances (MEGA4 software).16 Genbank accession numbers are D10759 (Uukuniemi virus, UUKUV), DQ363408 (Punta Toro virus, PTV), X56464 (RVFV), GU065633–GU065645 (different Spanish isolates of TOSV), GU65646, FJI53280, and 1284836 (Spanish isolates of TOSV obtained from sandflies), X68414 (Italian TOSV), FJ153281 and DQ195277 (TOSV from France), EF095551 and AY293623 (Sandfly fever Sicilian virus, SFSV), VEU725771 (MASV), EU266620 (Arbia virus, ARBV), DQ656070 (SFNV), and GU143710–GU143718 (corresponding to BUJV, ANHV, CHGV, ITPV, SALV, ICOV, and CDUV). GU143719–GU143723 correspond to sequences of B105-05, B68-03, B43-02, B151-04, and B79-02 that are phleboviruses similar to MASV and GRV previously found in Barcelona, Catalonia.13 GU135606 (GRV25) and 1284882, 1284889, 1284913, 1284919, 1284920, 1284927, and 1284930 (corresponding to sequences of GR49-04, GR44-04, GR36-04, GR65-04, GR52-04, GR29-04, and GR98-04 that correspond to the sequences of other positive pools found in Granada) have been also included.
Genomic sequencing.
RNA was extracted from the supernatant of cells infected by selected viruses. Briefly, after digestion with DNase I to eliminate human chromosomal DNA, RNA preparations were amplified by means of RT-PCR with the use of random primers. Amplification products were pooled and sequenced with the use of the GSL FLX platform (454 Life Sciences), but DNA fragmentation was omitted. After trimming to remove sequences derived from the amplification primer and after filtration to eliminate highly repetitive sequences, the dataset was analyzed by subtracting fragments that matched human sequences, clustering non-redundant sequences, and assembling them into contiguous sequences. Genbank accession numbers for GRV25 are GU135606–GU135608.
Phylogenetic analysis of complete sequences.
Original sequence data were first analyzed by the CHROMAS software (version 1.3, McCarthy 1996; School of Biomolecular and Biomedical Science, Faculty of Science and Technology, Griffith University, Brisbane, Queensland, Australia); forward and reverse sequence data of each sample were aligned using the program SEQMAN (DNASTAR Inc. Software, Madison, WI). These sequences were compared with all published phleboviral sequences recovered from GenBank. They were aligned using CLUSTAL X, version 1.83.17 Phylogenetic analyses were performed using the Tamura Nei model of nucleotide substitution.18 Programs from the MEGA package (version 4) were used to produce phylogenetic trees, reconstructed through the neighbor-joining (NJ) method. The statistical significance of a particular tree topology was evaluated by bootstrap resampling of the sequences 1,000 times.
Serological assays.
Indirect immunofluorescence assays.
Vero E6 cells infected with GRV or TOSV were used for detecting immunoglobulin G (IgG) antibodies by indirect immunofluorescence assay (IFA). Sera were tested at 1:10 dilution, and rabbit anti-human IgG conjugated with fluorescein isothiocyanate (Dako, Denmark) was used. All positive samples at 1:10 dilution were titrated to endpoint using 2-fold dilutions. The reading was done at a magnification of 200×.
Enzyme-linked immunosorbent assay.
Anti-TOSV IgM or IgG were detected by commercial enzymatic immunoassays (Enzywell Toscana Virus IgG/IgM, Diesse, Italy). The IgG test was based in an indirect method, and the IgM was based in a capture approach. Both assays used a recombinant nucleoprotein of TOSV.19 Samples were tested diluted 1:100, and the results were calculated by using cutoff controls.
Microneutralization assay.
A collection of 248 human sera previously described in Sanbonmatsu-Gamez and others14 was assayed. The serum collection corresponds to healthy individuals from the metropolitan area of Granada collected in 2003. A serum microneutralization test was done using 2-fold serial dilutions of serum in minimal essential medium (MEM) and a constant virus dose. Sera were heat-inactivated by incubation at 55°C for 30 minutes, serially diluted 2-fold in MEM, and tested for neutralization capacity using 100 TCID50 of TOSV ISS.Phl.3 or GRV25 in Vero cells.
Results
Identification of GRV in sandflies.
A total of 103 pools from Granada and 5 pools from Balearic Islands (Ibiza) were analyzed by consensus PCR. Sequences from TOSV were detected in three pools captured in 2004 from Granada (two female-only pools and one male-only pool; accession numbers GU65646, FJI53280, and 1284836). Additionally, another group of sequences was also obtained in 2004 from eight pools in Granada (five female-only pools and three male-only pools; GU135606, 1284882, 1284889, 1284913, 1284919, 1284920, 1284927, and 1284930 corresponding to sequences of GRV25, GR44-04, GR36-04, GR65-04, GR52-04, GR29-04, GR49-04, and GR98-04) and in one female-only pool obtained in Ibiza during 2004 (BAL13-04). Because the sequences available for other members of this genus in this gene are scant, a collection of yet uncharacterized phleboviruses (BUJV, ANHV, CHGV, ITPV, SALV, ICOV, and CDUV; Genbank accession numbers GU143710–GU143718) was also analyzed. These new sequences and those in GenBank were used in the initial pairwise test to calculate the phylogenetic distances. By this approach, we determined that GRV and the other group of sequences found in Granada and Ibiza were from members of the SFNV serocomplex, related to MASV and sequences of viruses previously found in Barcelona (Catalonia) (Table 1). The distance within this group of sequences formed by MASV, GRV25, and related sequences is 0.033, and the distance within the group of TOSV sequences is quite similar (0.036). The distances between the different viruses range from 0.293 (between ARBV and SALV) to 0.586 (between UUKUV and sequences of BUJV, CHGV, and ARBV). Distances from GRV/MASV to TOSV and to SFNV are 0.351 and 0.364, respectively, whereas distances between TOSV and SFNV are 0.297 (Table 1).
Table 1.
Genetic distances between different phleboviruses calculated in a fragment of L gene
| TOSV | SFSV | SFNV | PTV | RVFV | UUKUV | BUJV | ANHV | CHGV | AMTV | ITPV | SALV | ICOV | CDV | ARBV | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SFSV | 0.420 | ||||||||||||||
| SFNV | 0.297 | 0.480 | |||||||||||||
| PTV | 0.430 | 0.470 | 0.419 | ||||||||||||
| RVFV | 0.411 | 0.384 | 0.434 | 0.414 | |||||||||||
| UUKUV | 0.536 | 0.520 | 0.530 | 0.530 | 0.545 | ||||||||||
| BUJV | 0.418 | 0.439 | 0.429 | 0.465 | 0.424 | 0.586 | |||||||||
| ANHV | 0.378 | 0.399 | 0.419 | 0.389 | 0.444 | 0.520 | 0.439 | ||||||||
| CHGV | 0.443 | 0.424 | 0.480 | 0.505 | 0.439 | 0.586 | 0.404 | 0.444 | |||||||
| AMTV | 0.448 | 0.429 | 0.465 | 0.444 | 0.419 | 0.545 | 0.434 | 0.449 | 0.500 | ||||||
| ITPV | 0.432 | 0.432 | 0.409 | 0.374 | 0.409 | 0.571 | 0.429 | 0.379 | 0.444 | 0.414 | |||||
| SALV | 0.456 | 0.455 | 0.460 | 0.419 | 0.449 | 0.540 | 0.419 | 0.414 | 0.475 | 0.394 | 0.419 | ||||
| ICOV | 0.412 | 0.359 | 0.434 | 0.500 | 0.444 | 0.545 | 0.449 | 0.429 | 0.470 | 0.460 | 0.439 | 0.444 | |||
| CDV | 0.407 | 0.424 | 0.439 | 0.389 | 0.394 | 0.530 | 0.414 | 0.424 | 0.449 | 0.424 | 0.414 | 0.409 | 0.460 | ||
| ARBV | 0.456 | 0.439 | 0.495 | 0.455 | 0.455 | 0.586 | 0.444 | 0.455 | 0.475 | 0.394 | 0.429 | 0.293 | 0.455 | 0.374 | |
| GR/MAS | 0.351 | 0.394 | 0.364 | 0.457 | 0.355 | 0.544 | 0.428 | 0.429 | 0.468 | 0.447 | 0.474 | 0.498 | 0.433 | 0.417 | 0.491 |
Sequences used in the analysis are described in Materials and Methods. SFSV corresponds to sequences of Sabin and Chios strains; GR/MAS includes GRV, MASV, and sequences of B105-05, B68-03, B43-02, B151-04, B79-02, BAL13-04, GR49-04, GR44-04, GR36-04, GR65-04, GR52-04, and GR29-04. In the TOSV group, Spanish, Italian, and French isolates are included.
Phylogenetic analysis of the complete sequence of Granada virus.
Complete genome-sequencing analysis was used for full characterization of the strain GRV25. All gene segments cluster within the SFNV complex. The L, nucleoprotein (N), and nonstructural (NSs) genes have high homology to MASV (Table 2). The medium (M) segment does not have high homology. Whereas the P distances at the nucleotide level between GRV and MASV for the L, N, and NSs were 4.3%, 3.1%, and 4.9%, respectively, the calculated P distance in M segment between MASV and GRV was 37.6% at the nucleotide level and 39.7% at the amino acid level. The average M-segment P distance at the nucleotide level between MASV and TOSV is 42.5% and between TOSV and GRV is 42.9%. The differences in genetic distances are also shown in Figure 1. This difference is compatible with a reassortment event.
Table 2.
Sequence differences observed among phleboviruses
| Comparison | L segment | M segment | Ns gene | N gene | ||||
|---|---|---|---|---|---|---|---|---|
| Mean (%) | SD (%) | Mean (%) | SD (%) | Mean (%) | SD (%) | Mean (%) | SD (%) | |
| Within RVFV | 8.3 | 0.2 | 6.8 | 0.3 | 2.6 | 0.3 | 2.2 | 0.3 |
| Amino acid difference | 1.6 | 0.2 | 3.9 | 0.4 | 1.8 | 0.4 | 0.4 | 0.2 |
| Within TOSV | 4.7 | 0.1 | 2.9 | 0.1 | 8.5 | 0.6 | 3.2 | 0.4 |
| Amino acid difference | 2.6 | 0.1 | 0.8 | 0.1 | 5.3 | 0.8 | 0.0 | 0.0 |
| Between GRV and MASV | 4.3 | 0.2 | 37.6 | 0.8 | 4.9 | 0.7 | 3.1 | 0.6 |
| Amino acid difference | 1.1 | 0.2 | 39.7 | 1.3 | 3.2 | 0.3 | 0.4 | 0.4 |
| Between GRV and TOSV | 26.8 | 0.5 | 42.9 | 0.9 | 48.6 | 1.7 | 25.6 | 1.6 |
| Amino acid difference | 17.1 | 0.8 | 44.1 | 1.7 | 54.9 | 2.6 | 16.2 | 2.5 |
| Between GRV and RVFV | 41.8 | 0.6 | 54.5 | 0.7 | 69.4 | 1.6 | 44.1 | 1.7 |
| Amino acid difference | 45.4 | 1.1 | 65.5 | 0.3 | 87.0 | 2.0 | 49.8 | 0.3 |
| Between MASV and TOSV | 26.5 | 0.5 | 42.5 | 0.9 | 48.4 | 1.6 | 24.3 | 1.6 |
| Amino acid difference | 17.1 | 0.8 | 44.0 | 1.7 | 53.1 | 2.6 | 15.7 | 2.5 |
| Between MASV and RVFV | 41.7 | 0.6 | 53.9 | 0.7 | 68.6 | 1.7 | 44.1 | 1.7 |
| Amino acid difference | 45.6 | 1.1 | 64.7 | 1.3 | 86.6 | 2.0 | 49.4 | 0.3 |
| Between TOSV and RVFV | 42.6 | 0.6 | 55.5 | 0.9 | 67.6 | 1.6 | 45.6 | 1.9 |
| Amino acid difference | 45.6 | 1.1 | 64.1 | 1.6 | 83.4 | 2.1 | 49.1 | 3.3 |
Percent similarity of the sequences is shown. To estimate the evolutionary divergence over sequence pairs between groups, the average number of nucleotide differences per site over all sequence pairs between groups and among groups was calculated. Standard error estimates are shown in the second column and were obtained by a bootstrap procedure (500 replicates). For each pairwise sequence comparison, all positions containing alignment gaps and missing data were eliminated. SD = standard deviation.
Figure 1.
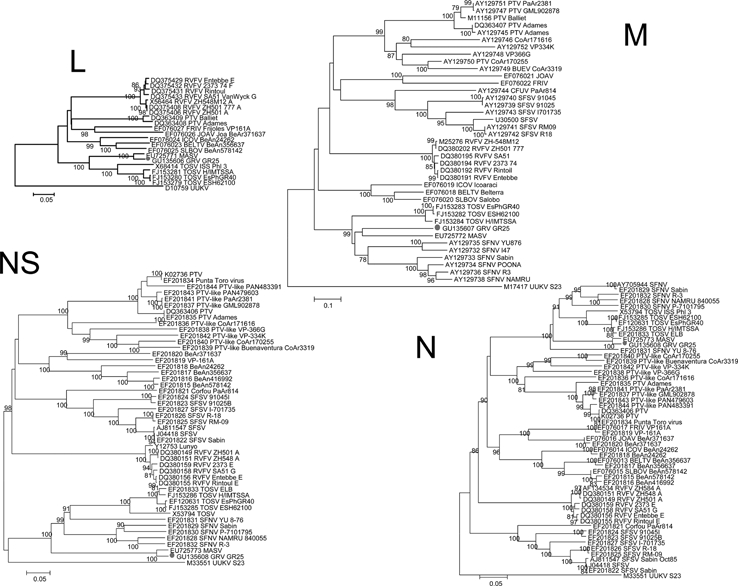
Phylogenetic analysis of phlebovirus. Coding regions for nucleoprotein (N) gene, nonstructural (NSs) gene, large (L) segment, and medium (M) segment were studied by using distance MEGA (www.megasoftware.net); relationships between different strains are shown. Each sequence used shows the GenBank accession number followed by the name of the virus according to the International Committee on Taxonomy of Viruses and the corresponding strain. RVFV = Rift Valley Fever virus; PTV = Punta Toro virus; SFSV = Sandfly Fever Sicilian virus; UUKV = Uukuniemi virus; SFNV = Sandfly Fever Naples virus; FRIV = Frijoles virus; ICOV = Icoaraci virus; JOAV = Joa virus; BELTV = Belterra virus; SLBOV = Salobo virus; BUEV = Buenaventura virus; TOSV = Toscana virus; GRV = Granada virus; MASV = Massilia virus. Scale bars indicate nucleotide substitutions per site.
Detection of antibodies against GRV.
Because of its similarity with other SFNV human pathogens, we used an IFA to search for antibodies against GRV in a bank of sera from healthy individuals collected in Granada in 2003; 37 of 248 (14.9%) sera were positive. To test whether these results reflected cross-reactivity with TOSV, the other member of the same serocomplex circulating in southern Spain, we assayed the positive sera for neutralizing antibodies against GRV. Five samples were positive. The presence of total (IFA) or neutralizing (NT) antibodies against TOSV was also determined in these samples (Table 3). Three of five (samples 484, 956, and 908) were immunoreactive with TOSV in IFA and enzyme-linked immunosorbent assay (ELISA) but negative in NT. One (sample 487) was negative in IFA, ELISA, and NT for TOSV. One (sample 905) was positive in IFA, ELISA, and NT. The neutralization titer was 1/128 for GRV and 1/256 for TOSV.
Table 3.
Results of serological tests
| Sample | IFA GRV | NT GRV | ELISA TOSV | IFA TOSV | NT TOSV |
|---|---|---|---|---|---|
| 487 | 1/80 | 1/128 | − | − | − |
| 905 | 1/160 | 1/128 | + | 1/80 | 1/256 |
| 484 | 1/20 | 1/64 | + | 1/20 | − |
| 956 | 1/40 | 1/16 | + | 1/10 | − |
| 908 | 1/80 | 1/8 | + | 1/20 | − |
The results obtained by ELISA, IFA, and neutralization assays are shown.
Discussion
Here, we report isolation of a new phlebovirus, GRV, from sandflies collected in southeastern Spain. Sequence analysis indicates that GRV is a member of the SFNV serocomplex and represents the first example of phleboviral reassortment in nature between different species. Serosurveillance showed the presence of specific neutralizing antibodies against GRV in healthy human subjects in the same geographical location.
Description of new species of phleboviruses should be approached with caution, because the genetic and serologic information is scarce. The International Committee for Taxonomy of Viruses distinguishes phlebovirus species based on a 4-fold difference in neutralization tests. Because reagents and isolates needed to perform these tests were limited, alternative methods for virus speciation based on genetic distance had been used for other viruses and have been proposed for phleboviruses.12,20–22 By that proposal, a PCR product representing NT 2095–2296 of the L segment of TOSV (X68414) would be sufficient to suggest species delineation. In that model, P distance values of more than 15% would be used to suggest a candidate new species. However, in the case of phleboviruses and other segmented viruses and given the possibilities of reassortment, we believe that full genome sequencing and serological characterization should be requested for confirmation of taxonomic status. In fact, our data strongly support the need for caution in using this approach. If we would have based our analysis only in the L data, we would have concluded that GRV was MASV.
When L-gene analyses of a MASV-like virus isolated from sandflies suggested the presence of a novel virus, we pursued full genomic characterization. Although GRV is nearly identical to MASV in the L and short (S) segments, there is a profound difference in the M segments of MASV and GRV. This difference is compatible with a reassortment between different species. RVFV reassortment between different strains of this RVFV has been described in nature, culture, and dually infected mosquitoes.23–25 Interspecies reassortants are also described in Orthobunyaviruses.26,27 If we accept the premise that the distant M segment parental donor is a different species of phlebovirus, this would be the first report of a natural interspecies reassortant in the phlebovirus genus.
The M segment of phleboviruses encodes two glycoproteins, G1 and G2, and a non-structural protein, NSm. As with other members of the Bunyaviridae, the phlebovirus envelope glycoproteins are important for viral infection, pathogenesis, and immunity; they serve as neutralizing and hemagglutinin-inhibiting antibody targets and are exposed to selective pressure.28–31 Neutralizing epitopes typically depend on the tertiary structure of proteins, and even a single amino acid substitution may be important for biological activity.32,33 Although neutralization assays are well-established methods for differentiating phleboviruses, our results also illustrate the potential risk associated with relying on serology alone. If we would have relied completely on virus neutralization as a way to delineate species, we would not have identified GRV as a virus reassortant. Thus, we suggest the need of a method for classification of bunyaviruses (and other segmented viruses) that embraces both serological and genetic data.
It is likely that many new phleboviruses are yet to be discovered. The putative donor of the segment M of MASV or GRV is presumably one of them. Similar sequences to GRV and MASV were detected in sandflies of Granada and two additional geographical locations in Spain (Barcelona in Catalonia and Ibiza in the Balearic Islands).13 There is no available data on the M gene sequence of these viruses. Thus, they could be strains of MASV or GRV. Additional phleboviral survey of sandflies in the area will be needed to understand their origin. We should also point out that because of the scarce amount of genetic information among members of the genus, it is possible that our sequences might be related to other uncharacterized members of the Naples serocomplex. Karimabad or Tehran viruses fall into this category. They are not available to us, and therefore, neither genetic nor antigenic comparison was carried out. However, given that phleboviruses are normally geographically restricted, the lack of detection of these viruses within the Mediterranean basin makes this hypothesis improbable. Full genomic characterization of all the members of the genus will solve these issues.
The sequences of viruses found in Granada similar to MASV or GRV have been detected in males and females, suggesting vertical and/or venereal transmission. Identification of the species of sandflies could not be done in any of these pools, but in Spain in a previous study, Phlebotomus perniciosus represents the 68% of sandflies captured in this region.14 Therefore, the most probable vector for this new virus is P. perniciosus, although this should be tested.
In the original report of MASV, Charrel and others12 explored the possibility of MASV causing disease in humans. Four hundred seventy-seven cerebrospinal fluids from individuals with infectious central nervous system diseases were assayed; zero were positive. We did not examine samples from individuals with disease but instead, chose to focus on evidence of infection in asymptomatic human volunteers. In IFA, 14.9% (37 of 248 samples) were positive for GRV. Five had specific GRV-neutralizing antibodies. Four were also TOSV-positive by ELISA or IFA, but only one of four had TOSV-neutralizing antibodies. These findings confirm that GRV infects humans but provides no insight into whether it can be implicated in human disease. These findings also indicate the pitfalls associated with relying on N proteins-based ELISA. N is one of the most conserved proteins in phleboviruses; thus, serology based on N may be unsuited to discriminate infection below the genus level.34 GRV pathogenicity remains unknown, but we believe these results warrant the need of future research in this regard.
Acknowledgments
The authors thank all members of the Red de Enfermedades Víricas Transmitidas por Artrópodos y Roedores, a multidisciplinary group founded by the Fondo de Investigaciones Sanitarias (FIS), Spanish Ministry of Health (G03/059). This work was founded also by Instituto de Salud “Carlos III” and is part of the work carried out within the Red de Investigación Cooperativa en Enfermedades Tropicales (RICET) network (FIS C03/04 and RD06/0021). Work in the Center for Infection and Immunity in the Mailman School of Public Health of Columbia University is supported by National Institutes of Health Grants AI051292 and AI57158 (Northeast Biodefense Center–Lipkin), the US Department of Defense, and Google.org. Finally, we also thank Alla Tashmukhamedova for her help in sequencing.
Disclaimer: Samples used were made anonymous to follow bioethics guides, and no conflict of interest related to this article was declared.
Footnotes
Authors' addresses: Ximena Collao, Antonio Tenorio, and María Paz Sánchez-Seco, Laboratory of Arbovirus and Imported Viral Diseases, National Center of Microbiology, Institute of Health “Carlos III,” Madrid, Spain, E-mails: ximecf1@hotmail.com, atenorio@isciii.es, and paz.sanchez@isciii.es. Gustavo Palacios and W. Ian Lipkin, Center for Infection and Immunity, Mailman School of Public Health, Columbia University, New York, New York, E-mails: gp2050@columbia.edu and wil2001@columbia.edu. Fernando de Ory, Serology Section, National Center of Microbiology, Institute of Health “Carlos III,” Madrid, Spain, E-mail: fory@isciii.es. Sara Sanbonmatsu, Mercedes Pérez-Ruiz, and José María Navarro, Service of Microbiology, Universitary Hospital “Virgen de las Nieve,” Granada, Spain, E-mails: sbmsara@yahoo.es, mercedes.perez.ruiz.sspa@juntadeandalucia.es, and josem.navarro.sspa@juntadeandalucia.es. Ricardo Molina, Service of Parasitology, National Center of Microbiology, Institute of Health “Carlos III,” Madrid, Spain, E-mail: rmolina@isciii.es. Stephen K. Hutchison, 454 Life Sciences, Branford, CT, E-mail: stephen.hutchison@roche.com. Ximena Collao actual address: Virology, Medicine School, Universidad de Valparaíso, Valparaiso, Chile.
References
- 1.Nichol ST, Beaty BJ, Elliott RM, Goldbach R, Plyusin A, Schmaljohn C, Tesh R. In: Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. San Diego, CA: Elsevier Academic Press; 2005. pp. 695–723. (Family Bunyaviridae). [Google Scholar]
- 2.Gould EA, Higgs S. Impact of climate change and other factors on emerging arbovirus diseases. Trans R Soc Trop Med Hyg. 2009;103:109–121. doi: 10.1016/j.trstmh.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valassina M, Meacci F, Valensin PE, Cusi MG. Detection of neurotropic viruses circulating in Tuscany: the incisive role of Toscana virus. J Med Virol. 2000;60:86–90. doi: 10.1002/(sici)1096-9071(200001)60:1<86::aid-jmv14>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 4.Nicoletti L, Ciufolini MG, Verani P. Sandfly fever viruses in Italy. Arch Virol Suppl. 1996;11:41–47. doi: 10.1007/978-3-7091-7482-1_5. [DOI] [PubMed] [Google Scholar]
- 5.Dionisio D, Valassina M, Ciufolini MG, Vivarelli A, Esperti F, Cusi MG, Marchi A, Mazzoli F, Luppi C. Encephalitis without meningitis due to sandfly fever virus serotype toscana. Clin Infect Dis. 2001;32:1241–1243. doi: 10.1086/319759. [DOI] [PubMed] [Google Scholar]
- 6.Braito A, Ciufolini MG, Pippi L, Corbisiero R, Fiorentini C, Gistri A, Toscano L. Phlebotomus-transmitted toscana virus infections of the central nervous system: a seven-year experience in Tuscany. Scand J Infect Dis. 1998;30:505–508. doi: 10.1080/00365549850161539. [DOI] [PubMed] [Google Scholar]
- 7.Braito A, Corbisiero R, Corradini S, Fiorentini C, Ciufolini MG. Toscana virus infections of the central nervous system in children: a report of 14 cases. J Pediatr. 1998;132:144–148. doi: 10.1016/s0022-3476(98)70500-1. [DOI] [PubMed] [Google Scholar]
- 8.Hemmersbach-Miller M, Parola P, Charrel RN, Paul Durand J, Brouqui P. Sandfly fever due to toscana virus: an emerging infection in southern France. Eur J Intern Med. 2004;15:316–317. doi: 10.1016/j.ejim.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Ehrnst A, Peters CJ, Niklasson B, Svedmyr A, Holmgren B. Neurovirulent toscana virus (a sandfly fever virus) in Swedish man after visit to Portugal. Lancet. 1985;1:1212–1213. doi: 10.1016/s0140-6736(85)92886-7. [DOI] [PubMed] [Google Scholar]
- 10.Mendoza-Montero J, Gamez-Rueda MI, Navarro-Mari JM, de la Rosa-Fraile M, Oyonarte-Gomez S. Infections due to sandfly fever virus serotype toscana in Spain. Clin Infect Dis. 1998;27:434–436. doi: 10.1086/514684. [DOI] [PubMed] [Google Scholar]
- 11.Nicoletti L, Verani P, Caciolli S, Ciufolini MG, Renzi A, Bartolozzi D, Paci P, Leoncini F, Padovani P, Traini E, Baldereschi M, Balducci M. Central nervous system involvement during infection by phlebovirus toscana of residents in natural foci in central Italy (1977–1988) Am J Trop Med Hyg. 1991;45:429–434. doi: 10.4269/ajtmh.1991.45.429. [DOI] [PubMed] [Google Scholar]
- 12.Charrel RN, Moureau G, Temmam S, Izri A, Marty P, Parola P, da Rosa AT, Tesh RB, Lamballerie X. Massilia virus, a novel phlebovirus (Bunyaviridae) isolated from sandflies in the Mediterranean. Vector Borne Zoonotic Dis. 2009;9:519–530. doi: 10.1089/vbz.2008.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Seco MP, Vazquez A, Collao X, Hernandez L, Aranda C, Ruiz S, Escosa R, Marques E, Bustillo MA, Molero F, Tenorio A. Surveillance of arboviruses in Spanish wetlands: detection of new flavi- and phleboviruses. Vector Borne Zoonotic Dis. 2010;10:203–206. doi: 10.1089/vbz.2008.0188. [DOI] [PubMed] [Google Scholar]
- 14.Sanbonmatsu-Gamez S, Perez-Ruiz M, Collao X, Sanchez-Seco MP, Morillas-Marquez F, de la Rosa-Fraile M, Navarro-Mari JM, Tenorio A. Toscana virus in Spain. Emerg Infect Dis. 2005;11:1701–1707. doi: 10.3201/eid1111.050851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez-Seco MP, Echevarria JM, Hernandez L, Estevez D, Navarro-Mari JM, Tenorio A. Detection and identification of toscana and other phleboviruses by RT-nested-PCR assays with degenerated primers. J Med Virol. 2003;71:140–149. doi: 10.1002/jmv.10465. [DOI] [PubMed] [Google Scholar]
- 16.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 19.Valassina M, Soldateschi D, Dal Maso GM, Santini L, Bianchi S, Valensin PE, Cusi MG. Diagnostic potential of toscana virus N protein expressed in Escherichia coli. J Clin Microbiol. 1998;36:3170–3172. doi: 10.1128/jcm.36.11.3170-3172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domingo C, Palacios G, Jabado O, Reyes N, Niedrig M, Gascon J, Cabrerizo M, Lipkin I, Tenorio A. Use of a short fragment of the C-terminal E gene for detection and characterization of two new lineages of dengue virus 1 in India. J Clin Microbiol. 2006;44:1519–1529. doi: 10.1128/JCM.44.4.1519-1529.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casas I, Avellon A, Mosquera M, Jabado O, Echevarria JE, Campos RH, Rewers M, Perez-Breña P, Lipkin I, Palacios G. Molecular identification of adenoviruses in clinical samples by analyzing a partial hexon genomic region. J Clin Microbiol. 2005;43:6176–6182. doi: 10.1128/JCM.43.12.6176-6182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palacios G, Jabado O, Cisterna D, de Ory F, Renwick N, Echevarria JE, Castellanos A, Mosquera M, Freire MC, Campos RH, Lipkin I. Molecular identification of mumps virus genotypes from clinical samples: standardized method of analysis. J Clin Microbiol. 2005;43:1869–1878. doi: 10.1128/JCM.43.4.1869-1878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sall AA, Zanotto PM, Sene OK, Zeller HG, Digoutte JP, Thiongane Y, Bouloy M. Genetic reassortment of Rift Valley fever virus in nature. J Virol. 1999;73:8196–8200. doi: 10.1128/jvi.73.10.8196-8200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saluzzo JF, Smith JF. Use of reassortant viruses to map attenuating and temperature-sensitive mutations of the Rift Valley fever virus MP-12 vaccine. Vaccine. 1990;8:369–375. doi: 10.1016/0264-410x(90)90096-5. [DOI] [PubMed] [Google Scholar]
- 25.Turell MJ, Saluzzo JF, Tammariello RF, Smith JF. Generation and transmission of Rift Valley fever viral reassortants by the mosquito Culex pipiens. J Gen Virol. 1990;71:2307–2312. doi: 10.1099/0022-1317-71-10-2307. [DOI] [PubMed] [Google Scholar]
- 26.Briese T, Kapoor V, Lipkin WI. Natural M-segment reassortment in Potosi and Main Drain viruses: implications for the evolution of orthobunyaviruses. Arch Virol. 2007;152:2237–2247. doi: 10.1007/s00705-007-1069-z. [DOI] [PubMed] [Google Scholar]
- 27.Briese T, Bird B, Kapoor V, Nichol ST, Lipkin WI. Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa. J Virol. 2006;80:5627–5630. doi: 10.1128/JVI.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keegan K, Collett MS. Use of bacterial expression cloning to define the amino acid sequences of antigenic determinants on the G2 glycoprotein of Rift Valley fever virus. J Virol. 1986;58:263–270. doi: 10.1128/jvi.58.2.263-270.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Battles JK, Dalrymple JM. Genetic variation among geographic isolates of Rift Valley fever virus. Am J Trop Med Hyg. 1988;39:617–631. doi: 10.4269/ajtmh.1988.39.617. [DOI] [PubMed] [Google Scholar]
- 30.Pifat DY, Osterling MC, Smith JF. Antigenic analysis of Punta Toro virus and identification of protective determinants with monoclonal antibodies. Virology. 1988;167:442–450. [PubMed] [Google Scholar]
- 31.Besselaar TG, Blackburn NK. Topological mapping of antigenic sites on the Rift Valley fever virus envelope glycoproteins using monoclonal antibodies. Arch Virol. 1991;121:111–124. doi: 10.1007/BF01316748. [DOI] [PubMed] [Google Scholar]
- 32.Schmaljohn CS, Chu YK, Schmaljohn AL, Dalrymple JM. Antigenic subunits of Hantaan virus expressed by baculovirus and vaccinia virus recombinants. J Virol. 1990;64:3162–3170. doi: 10.1128/jvi.64.7.3162-3170.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horling J, Lundkvist A. Single amino acid substitutions in Puumala virus envelope glycoproteins G1 and G2 eliminate important neutralization epitopes. Virus Res. 1997;48:89–100. doi: 10.1016/s0168-1702(97)01436-6. [DOI] [PubMed] [Google Scholar]
- 34.Collao X, Palacios G, Sanbonmatsu-Gamez S, Perez-Ruiz M, Negredo AI, Navarro-Mari JM, Grandadam M, Aransay A, Lipkin I, Tenorio A, Sanchez-Seco MP. Genetic diversity of toscana virus. Emerg Infect Dis. 2009;15:574–577. doi: 10.3201/eid1504.081111. [DOI] [PMC free article] [PubMed] [Google Scholar]


