Abstract
Ribonucleic acid (RNA) editing is a mechanism that generates RNA and protein diversity, which is not directly encoded in the genome. The most common type of RNA editing in vertebrates is the conversion of adenosine to inosine in double-stranded RNA which occurs in the higher eukaryotes. This editing is carried out by the family of adenosine deaminase acting on RNA (ADAR) proteins. The most-studied substrates of ADAR proteins undergo editing which is very consistent, highly conserved, and functionally important. However, editing causes changes in protein-coding regions only at a small proportion of all editing sites. The vast majority of editing sites are in noncoding sequences. This includes microRNAs, as well as the introns and 3′ untranslated regions of messenger RNAs, which play important roles in the RNA-mediated regulation of gene expression.
Introduction
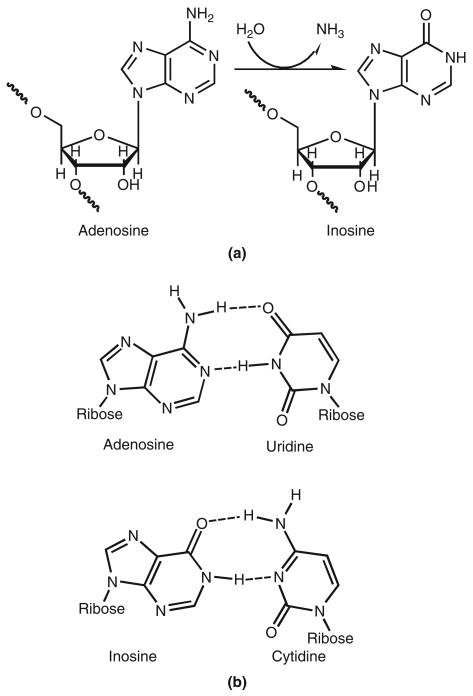
After transcription of a eukaryotic ribonucleic acid (RNA) molecule from deoxyribonucleic acid (DNA), the newly formed transcript undergoes a number of modifications that impact the final RNA or protein product. RNA splicing, similar to the cut-and-paste operation of a word processor, creates large-scale rearrangements of the original RNA message. Single-nucleotide editing processes, similar to a search-and-replace operation, selectively insert and delete single nucleotides, or convert one nucleotide to another. These processes are mechanisms for generating a diverse set of RNA and protein products from a limited number of DNA genes. With the realization that the human genome contains not more than 25,000 genes,1 it is clear that the complexity of the higher eukaryotes must come from such diversification phenomena, and not from differences in gene number. The first RNA editing process discovered in mammals was the deamination of cytidine (C) by APOBEC proteins to form uracil (U). However, the most prevalent type of RNA editing in the higher eukaryotes is the deamination of adenosine (A) to form inosine (I) (Figure 1(a)) in double-stranded (ds) RNA. This reaction is catalyzed by the family of adenosine deaminases acting on RNA (ADARs).2–4
FIGURE 1.
Deamination of adenosine by adenosine deaminases acting on RNAs (ADARs) alters base-pairing preferences. (a) Hydrolytic deamination of adenosine yields inosine. (b) While adenosine base pairs with uridine, inosine forms Watson–Crick base pairing with cytidine, analogous to that of guanosine.
The effects of this modification are twofold. A-to-I editing changes the informational content of the RNA molecule, as inosine preferentially base pairs with cytidine (Figure 1(b)), and is therefore interpreted as guanosine(G) by the translational and splicing machinery. The three-dimensional structure of the dsRNA, which determines its interactions with RNA-binding proteins, is also altered by the addition or removal of bulges formed by mismatched base pairs. Editing efficiency (the extent of conversion from A to I) varies depending on substrate, developmental timing, and location, allowing mixed populations of products to exist, and for these populations to change in response to changing conditions. Editing can affect anywhere from 0% to 100% of an RNA population, whereas a variation in the genome fixes the heterogeneity at exactly 50%, or none at all. It was initially thought that the only function of ADARs was to modify protein-coding genes. Recent discoveries have overthrown this view, by revealing a large number of editing sites in noncoding sequences5–7 and numerous regulatory interactions with the RNA interference (RNAi) and microRNA (miRNA) biogenesis pathways.8
The RNA Editing Reaction
Adenosine deaminases acting on RNA
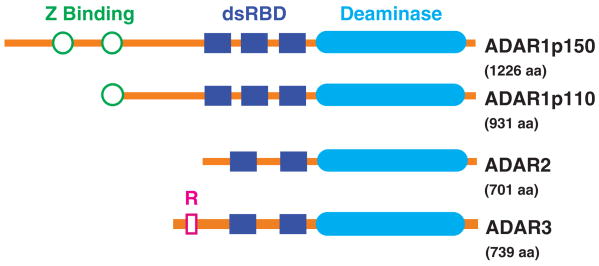
The A-to-I editing reaction is catalyzed by the family of ADAR enzymes. These proteins, which are conserved across many eukaryotes, contain a C-terminal catalytic domain, as well as several dsRNA-binding domains (Figure 2). There are three vertebrate ADAR genes, which give rise to several ADAR proteins. The ADAR1 protein has long (p150) and short (p110) isoforms, which result from the use of alternative promoters and start codons. ADAR2 and ADAR1p110 are mainly present in the nucleus, whereas ADAR1p150, driven by an interferon-inducible promoter, is present in both nucleus and cytoplasm, and is upregulated upon cellular stress or viral infection.9 ADAR1 and ADAR2 are expressed in most tissues, whereas ADAR3 is only found in the central nervous system. While ADAR1 and ADAR2 must form homodimers for activity, ADAR3 remains in monomeric form.10 All known editing sites have been attributed to ADAR1 or ADAR2 activity, and ADAR3 shows no deaminase activity in vitro, possibly due to its inability to dimerize, leaving its function unknown.11
FIGURE 2.
Human ADAR proteins. All three proteins share the deaminase domain (filled ovals), which catalyzes the editing reaction, and two or more double-stranded (ds) RNA-binding domains (filled rectangles), which mediate binding to the substrate RNA. The Z-DNA-binding domains (unfilled circles) of ADAR1 may localize it to newly synthesized transcripts,12 or assist in binding of short RNAs.13 The two isoforms of this protein are the result of alternative splicing that causes translation from different start codons. The R domain (unfilled rectangle) of ADAR3 has been reported to bind to single-stranded RNA,11 but the protein's function remains unknown.
Substrate specificity
ADARs interact with their dsRNA substrates through dsRNA-binding domains (dsRBDs). The structural features of the RNA helix bury sequence-specific functional groups deep inside the molecule, making it highly unlikely that ADAR proteins bind to their substrates in a sequence-specific manner.14 Instead, editing activity appears to be determined by the stability of the substrate dsRNA. This is supported by the highly efficient and random editing of adenosines in perfectly complementary long dsRNA molecules15,16 (Figure 4(f)). The editing process tends to destabilize the substrate RNA,5 and likely proceeds until the enzyme can no longer recognize the substrate as dsRNA. DsRNA structures that contain bulges and loops are edited at specific adenosines17 because it takes only a few deaminations to reduce their stability below the dsRNA threshold recognized by ADARs. Substrate specificity differs between the functional forms of ADAR,18–20 possibly due to the different number and spacing of dsRBDs, which may allow discrimination between different dsRNA structures and stabilities (see Figure 2).
FIGURE 4.
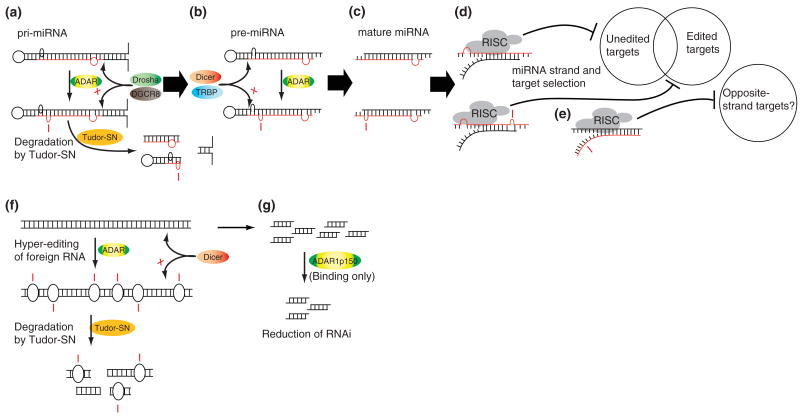
Effects of A-to-I RNA editing on noncoding transcripts. (a) primary microRNA (pri-miRNAs) are subject to editing by ADARs, which may block the drosha cleavage step and subject the transcript to degradation by Tudor-SN (tudor staphylococcal nuclease), as in the case of miR142.35 (b) If drosha cutting is unaffected by editing, or if the pre-miRNA itself is edited, processing by dicer may be blocked as in the case of miR151.36 (c) If mature edited miRNAs are produced, (d) the targets of the miRNA can be changed, as in the case of miR376a-5p.37 (e) Alterations in miRNA stability could hypothetically affect active-strand selection.8 (f) Long dsRNAs, such as viral RNAs, are very efficiently and nonspecifically edited by ADARs, resulting in inhibition of RNAi,33 and degradation by Tudor-SN.38,39 (g) ADAR1p150 can tightly bind small interfering RNAs (siRNAs) without editing them, reducing their concentration and thereby decreasing the effectiveness of RNA interference (RNAi).
The results of in vitro studies, and the sequences of known in vivo substrates have demonstrated preferences for certain flanking nucleotides at editing sites.15,21 As mentioned earlier, such sequence-specific interactions are unlikely to be mediated by the dsRNA-binding domains. These preferences might be explained by the proposed catalytic model for the deaminase domain, in which the target adenosine is flipped from the inside of the helix into the enzyme's active site,15 perhaps allowing for sequence-specific interactions with neighboring nucleotides. This suggests that the deaminase domain confers some additional specificity to editing site selection based on the immediate sequence context of the site.
Regulation of Protein Function by ADARs
Recoding of protein sequence
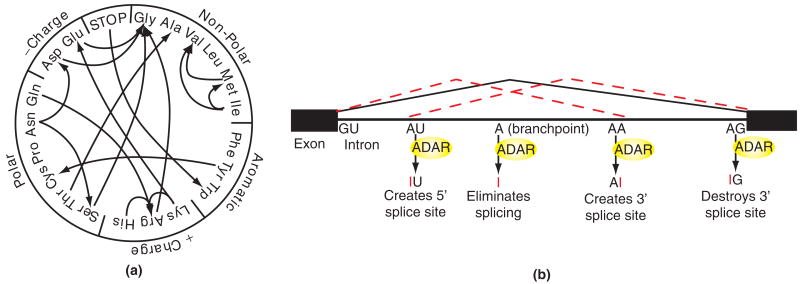
Before splicing of eukaryotic transcripts, stem-loop structures are often formed between intronic and exonic regions of the pre-mRNA (messenger RNA). These structures can be edited by ADARs and the resulting inosine is interpreted as guanosine by the ribosome, because of its Watson–Crick base pairing with cytidine (see Figure 1). Editing in coding regions may result in amino acid substitutions (see Figure 3(a)). The canonical examples of this phenomenon are the mRNAs for glutamate receptor (GluR) and serotonin receptor subtype 2C (5-HT2CR), which contain several specific editing sites, resulting in many protein isoforms with varying functionality.22,23 The importance of editing at these sites is underscored by the phenotype of ADAR2 knockout mice. These mice have reduced editing levels at the Q/R site of GluR-B, which is normally 100% edited in vivo. The mice are seizure-prone, and die within 3 weeks of birth.24 In humans, defects in editing have been linked to a skin pigmentation disorder (dyschromatosis symmetrica hereditaria) and a number of neurological diseases including epilepsy, schizophrenia, and amyotrophic lateral sclerosis.25
FIGURE 3.
Effects of A-to-I RNA editing on protein-coding transcripts. (a) Codon changes that can result from A-to-I RNA editing, based on interpretation of inosine as guanosine by translational machinery. Amino acids are grouped according to charge and hydrophobic properties. (b) Generation of alternative splice isoforms by RNA editing. The solid bent line indicates the intron which is removed in the absence of editing. Dotted lines indicate alternative splice isoforms resulting from editing. Editing can create new 5′ donor or 3′ acceptor sites, or eliminate splicing altogether by editing the internal branch point adenosine; 3′ acceptor sites can also be abolished.3 (Reprinted with permission from Ref 3. Copyright 2005 Elsevier).
Alteration of Splice Sites
Intronic regions of coding sequence are also subject to editing, which can have profound effects on splicing. Editing has the potential to modify 5′ and 3′ splice sites, or to eliminate splicing altogether via modification of splice site donor or acceptor sequences3,26 (see Figure 3(b)). The most clear-cut case is the self-regulation of ADAR2, which edits its own mRNA, leading to generation of an alternative splice acceptor site. Translation of this alternatively spliced mRNA results in production of truncated protein lacking dsRBDs and the deaminase domain, due to a frameshift.26
Editing may also have a subtle effect on splicing kinetics, by destabilizing intronic RNA duplexes and thereby providing the splicing machinery with better access to the RNA. This model is supported by the preferential splicing of edited transcripts in the brains of ADAR2 knockout mice.24 In accordance with this, it has been proposed that the Z-DNA-binding domains of ADAR1p150 (see Figure 2) serve to localize it to sites of active transcription, where the Z-conformation of DNA is stabilized by the supercoiling behind the active polymerase.27 Thus ADAR1 has a chance to act on the pre-mRNA before splicing occurs.12
Effects on Protein Evolution
Unlike a mutation in the DNA, A-to-I editing usually leaves some proportion of the original transcript, allowing for the creation of new functions without destroying older functions that may be necessary for survival. After a genomic G-to-A mutation, the A nucleotide may become an editing site, thereby lessening the effect of the mutation.2 The reverse could also occur, where selective pressure increases the editing frequency at a site over time, easing the transition to a genomically encoded G.28 Editing of the GluR-B Q/R site supports both scenarios. This is seen in the case of ADAR2 knockout mice, whose phenotype can be rescued by creating a genomic A-to-G mutation at the GluR-B Q/R site.24 This site is completely edited in normal mice, and may have evolved as a mutation corrected by editing. This shows that, at least in some cases, the function of RNA editing can be performed equally well by a change in the genomic sequence of the substrate. A-to-I and C-to-U editing are the most common forms of RNA editing in animals, which may explain the prevalence of A/G and C/T variants in related genomes.28
Interactions with Noncoding RNA
Despite their important functions, the ∼30 known protein-coding targets of A-to-I editing do not account for the large amount of inosine detected in mammalian RNAs.29 A combination of biochemical and bioinformatics studies have identified more than 12,000 new editing sites, most of which are located in noncoding regions such as introns, 5′ and 3′ untranslated regions (UTRs), and repetitive sequences such as human Alu elements.5–7 The wide variety of functions performed by noncoding RNAs, many of which are double-stranded, suggest that ADARs may play regulatory roles in many processes.
Effects on RNAi
RNAi is the process by which ∼22 bp dsRNAs called small interfering RNAs (siRNAs) direct the degradation of homologous mRNA transcripts.30,31 In addition to being an important viral defense and gene regulatory mechanism in the cell, RNAi has become an immensely powerful and widely used experimental tool. RNAi can be induced by long dsRNA, which is cleaved by the ribonuclease Dicer into siRNAs (see Figure 4(f) and (g)), which direct destruction of the original transcript and any other identical sequences.32 Long dsRNA is also a preferred substrate for ADARs, which can nonspecifically edit more than 50% of adenosines in such molecules15,16 (see Figure 4(f)). Such extensive editing has been shown in vitro to suppress dicing of these RNAs, and thus to suppress RNAi induced by them.33 In addition to competing with Dicer for long dsRNA substrates, ADAR1p150 has been shown to bind tightly to siRNAs, decreasing their effective concentration in the cytoplasm, thereby reducing the efficiency of RNAi13 (see Figure 4(g)). Strains of Caenorhabditis elegans in which active ADAR genes have been deleted to show defects in chemotaxis (the ability to seek out or avoid certain substances). These defects are eliminated in strains which also have defects in RNAi machinery.34 This suggests that the chemotaxis phenotype is a result of hyperactivity in an RNAi pathway which is normally inhibited by ADARs.
Effects on miRNA Processing and Target Selection
MiRNAs are small dsRNAs encoded by eukaryotic genomes, which regulate gene expression via an RNAi-like pathway. They need not be perfectly complementary to their targets, so a single miRNA can affect a large number of mRNAs. The miRNAs are transcribed into long primary transcripts called primary miRNAs (pri-miRNAs), which form a stem-loop secondary structure. The pri-miRNAs are cleaved by the nuclear Drosha-DGCR8 complex to yield ∼60–70nt miRNA precursors (pre-miRNAs), which are transported into the cytoplasm by Exportin5-Ran-GTP. The pre-miRNA is then cleaved by the cytoplasmic Dicer-TRBP complex into mature miRNAs: dsRNAs ∼22bp in length, with 2-nucleotide 3′ overhangs. The miRNA is then loaded into the RNA-induced silencing complex (RISC) which selects the active miRNA strand. The miRNA guides RISC to its target site, usually in the 3′ UTR of an mRNA, resulting in translational repression or mRNA degradation.30,40
The dsRNA regions of pri- and pre-miRNAs allow ADARs to interact with the miRNA biogenesis pathway. A-to-I editing has been detected in numerous endogenous pri-miRNAs,41,42 and there is in vitro evidence for editing at the pre-miRNA stage.36 These editing events can affect miRNA processing by inhibiting the Drosha35 or Dicer36 cleavage steps (see Figure 4(a) and (b)), thereby reducing levels of the mature miRNA. Since the transcription of miRNAs is often controlled by the promoters of other genes,40 modulation of processing is thought to be the main method of regulating miRNA levels. If processing is not affected by these editing events, then mature miRNAs with A-to-I substitutions are expressed37,43 (see Figure 4(c)). These miRNAs can silence a set of genes different from that of their unedited counterpart (see Figure 4(d)). In the case of miRNA 376a, the edited form of the mature miRNA gains the ability to downregulate the mRNA of the PRPS1 gene.37 PRPS1 is involved in purine metabolism, whose end product is uric acid. Its overexpression in humans is associated with buildup of purines and uric acid, resulting in gout and in some cases neurodevelopmental impairment.44 Mice lacking ADAR2, the responsible editing enzyme, have increased levels of PRPS1 protein and uric acid in their brains compared to wild-type mice. The effect of uric acid on the brain is unclear, but some studies have shown reduced levels in patients with multiple sclerosis, suggesting that it may have a neuroprotective effect.45
In most cases, only one strand of the miRNA is thought to be active, while the other is degraded. Selection of the active miRNA strand is based on the thermodynamic stability of the 5′ ends of the miRNA,40 thus the structural changes caused by editing could change the effective strand,8 which is likely to drastically change the genes silenced by the miRNA (see Figure 4(e)). The 3′ UTRs of mRNAs are known to be frequently edited5–7 and genomic A-to-G changes in 3′ UTRs have been shown to create new miRNA target sites,46 so it is possible that editing of 3′ UTRs might enhance or suppress silencing of specific mRNAs.47 Similarly, editing may destabilize secondary structure in 3′ UTRs, allowing RISC to access previously inaccessible target sites, and possibly preventing Drosha and Dicer from cleaving dsRNA regions of mRNAs. Through all of these mechanisms, A-to-I RNA editing has the potential to rapidly change gene expression levels in response to stimuli.
Role of RNA Editing in Innate Immune Response
Many viruses contain RNA genomes, or replicate their genomes through an RNA intermediate, which is often double-stranded in form. Such long dsRNAs are rarely produced endogenously by the cell,48 and are quickly recognized by many cellular systems. ADARs can carry out hyperediting of such transcripts, potentially triggering their degradation by the inosine-specific nuclease Tudor-SN (tudor staphylococcal nuclease)38,39,49 (see Figure 4(f)). Viral RNAs extracted from the brains of measles patients show a large number of U-to-C and A-to-G conversions,50 consistent with hyperediting of both the sense and antisense viral genomes.51,52 Since the virus is cytoplasmic, this editing is likely to be carried out by interferon-inducible ADAR1p150, which may be expressed in response to viral infection. An interesting exception is the hepatitis delta virus, which takes advantage of the editing machinery to edit a stop codon (UAG) to a tryptophan codon (UGG), resulting in synthesis of a longer viral antigen. In other words, this virus hijacks the host's editing machinery to carry out an essential step in its life cycle.53 The recent discovery of interferon-inducible miRNAs that directly target viral RNAs54 raises the possibility of a cooperative interaction between miRNAs and RNA editing in the immune response. On the other hand, a liver miRNA (miR122) has been discovered that facilitates replication of hepatitis C virus.55 Although that miRNA is not known to be edited, an increase in cytoplasmic ADAR1 could result in editing of other such pre-miRNAs, thereby slowing viral replication.
Conclusion
Even though editing sites have been discovered in a large number of RNA transcripts, the effects of A-to-I RNA editing remain undetermined in most cases. Editing sites in some coding targets show clear phenotypes when defective in model organisms, but they make up only a tiny fraction of all editing sites. ADAR1 knockout mice die as embryos because of widespread apoptosis.56,57 The cause of this phenotype is unknown. The newly discovered interactions between ADAR1 and noncoding RNA might be responsible, or the real cause may lie in an undiscovered function of ADAR1 or RNA editing in general.
Because of the high background of edited repetitive sequence in the transcriptome, the extent of editing in substrates such as coding regions and miRNAs is unclear, and editing may affect other classes of RNAs. It is possible that some of these interactions are accidental, due to the nonspecific nature of the dsRNA-binding domains of ADARs. In the case of the widespread editing of repetitive sequences, A-to-I editing may protect the genome against transposable elements7 by destabilizing RNA with repetitive sequences. However, it is quite possible that such editing events are a side effect of the huge amounts of repetitive sequence in the human genome. In the past decade, our knowledge of the crucial regulatory roles and potential clinical applications of RNA-dependent pathways has exploded. Determining the extent and physiological significance of RNA editing in such pathways is therefore of critical importance to those who study or use these technologies.
References
- 1.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- 4.Gerber AP, Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- 5.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Halleger M. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 6.Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci U S A. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maas S, Rich A, Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J Biol Chem. 2003;278:1391–1394. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- 8.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho DSC, Yang W, Lee JT, Shiekhattar R, Murray JM, Nishikura K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem. 2003;278:17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 11.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc Natl Acad Sci U S A. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Wang Q, Howell KL, Lee JT, Cho DS. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J Biol Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polson AG, Bass BL. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikura K, Yoo C, Kim U, Murray JM, Estes PA. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991;10:3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann KA, Bass BL. The importance of internal loops within RNA substrates of ADAR1. J Mol Biol. 1999;291:1–13. doi: 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- 18.Lai F, Chen CX, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melcher T, Maas S, Herb A, Sprengal R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 20.Gerber A, O'Connell MA, Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 21.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 22.Reenan RA. The RNA world meets behavior: A–> I pre-mRNA editing in animals. Trends Genet. 2001;17:53–56. doi: 10.1016/s0168-9525(00)02169-7. [DOI] [PubMed] [Google Scholar]
- 23.Seeburg PH, Hartner J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr Opin Neurobiol. 2003;13:279–283. doi: 10.1016/s0959-4388(03)00062-x. [DOI] [PubMed] [Google Scholar]
- 24.Higuchi M, Maas S, Single FN, Hartner J, Roznor A. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 25.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 27.Rahmouni AR, Wells RD. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989;246:358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- 28.Tian N, Wu X, Zhang Y, Jin Y. A-to-I editing sites are a genomically encoded G: implications for the evolutionary significance and identification of novel editing sites. RNA. 2008;14:211–216. doi: 10.1261/rna.797108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul MS, Bass BL. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 31.Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 32.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 33.Scadden AD, Smith CW. RNAi is antagonized by A–> I hyper-editing. EMBO Rep. 2001;2:1107–1111. doi: 10.1093/embo-reports/kve244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawahara Y, Zinshteyn B, Sethupathy P, Iizaza H, Hatzigeorgiou AG, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 39.Scadden AD, O'Connell MA. Cleavage of dsRNAs hyper-edited by ADARs occurs at preferred editing sites. Nucleic Acids Res. 2005;33:5954–5964. doi: 10.1093/nar/gki909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 41.Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 44.Ahmed M, Taylor W, Smith PR, Becker MA. Accelerated transcription of PRPS1 in X-linked overactivity of normal human phosphoribosylpyrophosphate synthetase. J Biol Chem. 1999;274:7482–7488. doi: 10.1074/jbc.274.11.7482. [DOI] [PubMed] [Google Scholar]
- 45.Spitsin S, Koprowski H. Role of uric acid in multiple sclerosis. Curr Top Microbiol Immunol. 2008;318:325–342. doi: 10.1007/978-3-540-73677-6_13. [DOI] [PubMed] [Google Scholar]
- 46.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 47.Liang H, Landweber LF. Hypothesis: RNA editing of microRNA target sites in humans? RNA. 2007;13:463–467. doi: 10.1261/rna.296407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawahara Y, Nishikura K. Extensive adenosine-to-inosine editing detected in Alu repeats of antisense RNAs reveals scarcity of sense-antisense duplex formation. FEBS Lett. 2006;580:2301–2305. doi: 10.1016/j.febslet.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scadden AD, Smith CW. Specific cleavage of hyper-edited dsRNAs. EMBO J. 2001;20:4243–4252. doi: 10.1093/emboj/20.15.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter MA. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bass BL, Weintraub H, Cattaneo R, Billeter MA. Biased hypermutation of viral RNA genomes could be due to unwinding/modification of double-stranded RNA. Cell. 1989;56:331. doi: 10.1016/0092-8674(89)90234-1. [DOI] [PubMed] [Google Scholar]
- 52.Cattaneo R, Schmid A, Spielhofer P, Kaelin K, Baczko K. Mutated and hypermutated genes of persistent measles viruses which caused lethal human brain diseases. Virology. 1989;173:415–425. doi: 10.1016/0042-6822(89)90554-0. [DOI] [PubMed] [Google Scholar]
- 53.Polson AG, Bass BL, Casey JL. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 54.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 56.Wang Q, Miyakoda M, Yang W, Khillian J, Stachura DL. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 57.Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]