Abstract
The metastasis suppressor NM23-H1 possesses three enzymatic activities in vitro, a nucleoside diphosphate kinase (NDPK), a protein histidine kinase, and a more recently characterized 3’–5’ exonuclease. While the histidine kinase has been implicated in suppression of motility in breast carcinoma cell lines, potential relevance of the NDPK and 3’-5 exonuclease to metastasis suppressor function has not been addressed in detail. To this end, site-directed mutagenesis and biochemical analyses of bacterially-expressed mutant NM23-H1 proteins have identified mutations that disrupt the 3’–5’ exonuclease alone (Glu5 to Ala, or E5A), the NDPK and histidine kinase activities tandemly (Y52A, H118F), or all three activities simultaneously (K12Q). While forced expression of NM23-H1 potently suppressed spontaneous lung metastasis of subcutaneous tumor explants derived from the human melanoma cell line 1205LU, no significant metastasis suppressor activity was obtained with the exonuclease-deficient variants E5A and K12Q. The H118F mutant which lacked both the NDPK and histidine kinase while retaining the 3’-5 exonuclease, also exhibited compromised suppressor activity. In contrast, each mutant retained the ability to suppress motility and invasive characteristics of 1205LU cells in culture, indicating the NM23-H1 molecule possesses an additional activity(s) mediating these suppressor functions. These studies provide the first demonstration that the 3’-5 exonuclease activity of NM23-H1is necessary for metastasis suppressor function, and further indicate cooperativity of the three enzymatic activities of the molecule on suppression of the metastatic process.
Keywords: metastasis, metastasis suppressor, NM23, melanoma, exonuclease, nucleoside diphosphate kinase
Introduction
Metastasis suppressors are a class of genes defined by their ability to selectively inhibit the metastatic process with little or no impact on primary tumor growth.1 The first metastasis suppressor gene to be identified was nm23-M1, which was discovered by virtue of its low expression in K-1735-derived melanoma cell lines with elevated metastatic potential.2 Subsequent studies have confirmed the human homologue nm23-H1 exhibits potent metastasis suppressor activity in breast carcinoma and melanoma cell lines, and is underexpressed in multiple forms of metastatic cancer.3 While the mechanisms underlying metastasis suppressor activity of NM23-H1 are not fully understood, the protein exhibits three different enzymatic activities in vitro that represent potential antimetastatic functions. First to be described was its nucleoside diphosphate kinase (NDPK) activity, which maintains balance in intracellular nucleotide pools by catalyzing transfer of γ-phosphate between nucleoside triphosphates and diphosphates.4 While a role for the NDPK in metastasis suppression has been challenged,5,6,7 the concept has yet to be addressed directly with in vivo models of metastatic growth. NM23-H1 is also reported to exhibit a protein histidine kinase activity that mediates its antimotility function8, possibly via serine phosphorylation of the kinase suppressor of ras (KSR) and suppression of ras-initiated signaling mechanisms.9 More recently, a 3’–5’ exonuclease activity has also been described for NM23-H1, which shows a preference for DNA substrates containing overhanging 3’-termini.10 3’–5’ exonucleases are associated with proofreading processes during DNA repair and replication,11 and consistent with such a function we have shown that the yeast NM23 homologue, ynk1, indeed possesses antimutator activity.12 Although this potential role of the 3’–5’ exonuclease in maintenance of genomic integrity suggests a novel mechanism of metastasis suppression, such has yet to be demonstrated experimentally. This DNase activity has has also been implicated in the granzyme A-initiated, caspase-independent pathway of apoptosis.13 While this apoptotic pathway has been shown to be operative in the cellular response to attack by cytotoxic T cells, its role in cancer and metastasis remains to be addressed.
Also of potential importance to metastasis suppressor activity are a host of physical interactions of NM23-H1 with other proteins. In this regard, NM23-H1 has been shown to bind with the Ras GTPase protein Rad14, the Rac-specific nucleotide exchange factor Tiam115, the Epstein-Barr virus protein EBNA-3C16, the Kinase Suppressor of Ras protein (KSR)9, and the multifunctional enzyme Prune17. Interestingly, three of these (Rad, Tiam1, Ksr) are involved in Ras-MAPK signaling , suggesting a modulatory role for NM23-H1 in that cascade. The physical interaction between Prune and NM23-H1 has been documented in the nucleus, suggesting the potential for functional interactions between them in the putative genomic surveillance functions of NM23-H1. Moreover, Prune is a metastasis promoter, suggesting the equilibrium between these proteins may regulate metastatic potential18.
To date, site-directed mutagenesis studies of NM23-H1 have identified multiple amino acid residues obligatory for the three known enzymatic activities. NDPK activity of both NM23-H1 and NM23-H2 isoforms requires a catalytic H118 residue19, as well as K12, Y52, Arg88, and Asn115,20,21,10 the latter being implicated in nucleotide binding by crystal structure analysis.22 The histidine kinase requires the catalytic H118, as well as S120 and P96,23 the latter of which is targeted by the killer of prune mutation (Kpn) in Drosophila melanogaster, P96S.24 We have shown the 3’–5’ exonuclease activity of NM23-H1 requires K12,10 implicated previously as the catalytic residue for nuclease activity of the NM23-H2 isoform20, while another report25 demonstrated contributions from additional residues E5, F33, D54, D121 and E129. However, no NM23-H1 mutants have been characterized fully in terms of all three enzymatic activities. Herein, we report the results of a complete biochemical analysis for a comprehensive panel of NM23-H1 mutants. Moreover, we show that mutations which disrupt the 3’–5’ exonuclease and, probably the NDPK activity, greatly diminish metastasis suppressor activity. Finally, our study reveals that none of the enzymatic activities are sufficient individually for metastasis suppressor function in vivo, strongly suggesting functional cooperativity between them.
Material and Methods
DNA and Site-directed Mutagenesis
cDNAs encoding NM23-H1 variants Q17N, Y52A, P96S, R88A and R105A were generated by the overlap extension modification of polymerase chain reaction as described 26, and were inserted in frame between the NdeI and BamHI sites of the E. coli expression plasmid pET3c (New England Biolabs, Ipswich, MA). E5A and D54A mutants were constructed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). cDNA inserts were sequenced to verify accurate construction of mutations. pET3c plasmids containing wild-type NM23-H1 and H118F were kindly provided by E. Postel (University of Medicine and Dentistry of New Jersey, New Brunswick, NJ), while construction of K12Q was described previously.10 NM23-H1 and mutant proteins were expressed in E. coli and purified by automated chromatography on DEAE-Sephacel and hydroxylapatite columns. This procedure provides preparations of essentially homogeneous NM23-H1 protein, as assessed by Coomassie blue staining and as described previously by our laboratory27,10 and others.28,29 For stable transfection, cDNAs encoding NM23-H1 variants were cloned into pCI-EGFP (provided by S. Kraner, Univ. of KY), which contains an internal ribosome entry sequence (IRES) for coexpression with enhanced green fluorescence protein (EGFP). pSV2neo (Clontech, Mountain View, CA) was employed for selection of stable transfectants with the neomycin analogue, G418.
Structural Analysis of NM23-H1 Variants: Gel Filtration HPLC and Circular Dichroism Analyses
Purified wild-type or mutant forms of NM23-H1 were analyzed by gel filtration chromatography as described10 using a Shodex gel filtration KW-800 HPLC column (Showa Denko, New York, NY), preequilibrated in 50 mM Tris, pH 7.5, 0.1 M KCl. Molecular weights were estimated relative to a standard curve generated with molecular weight standards (12.5– 200 kDa; Sigma, St. Louis, MO). Circular dichroism analyses were conducted as described10 using a Jasco J-810 spectrometer, with each individual spectrum representing the average of thirty replicate measurements.
Nucleoside Diphosphate Kinase and Histidine-Dependent Protein Kinase Assays
NDPK activity of NM23-H1 and mutant variants was measured as described,4 and adapted for use of 96-well plates and an automated microplate reader10. Histidine-dependent protein kinase activity was measured as described23 with minor modifications. [γ-32P]ATP (3000 Ci/mmol) was diluted to a specific activity of 50 Ci/mmol using unlabeled 10 mM ATP lithium salt (Roche; > 95% ATP, < 3.5% ADP, < 1.5% AMP). Autophosphorylation of NM23-H1 was achieved by incubation of 20 µg NM23-H1 with 500 µCi [γ-32P]ATP for 15 minutes at room temperature in 100 µl of 20 mM Tris-HCl pH 8.0, 5 mM MgCl2, 1 mM DTT. Assays were conducted with 2 × 105 cpm of phosphorylated NM23-H1 (40–140 pmol) and a 5-fold molar excess of NM23-H2 in a 30 µl volume of buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT). Reactions were stopped with SDS sample buffer without boiling, followed by electrophoresis of 20 µl aliquots in 13% SDS polyacrylamide gels.
3’–5’ Exonuclease Assays
Assays were performed as described27 with a single-stranded, 33-base oligodeoxyribonucleotide substrate corresponding to the noncoding strand of the 5’SHS silencer element from the platelet-derived growth factor-A gene.30 5’-termini were radiolabeled with [γ-32P]ATP and T4 polynucleotide kinase. Reactions were conducted at ambient room temperature (21–23°C) in a 15 µl volume containing 20 mM Hepes buffer (pH 7.9), 10–20 fmol of 5’-[32P]-labeled oligonucleotide, 2 mM MgCl2 and 100 mM KCl, and were initiated by addition of 0.5 µg NM23-H1 (320 nM). Reaction products were resolved on 20% sequencing gels and visualized by phosphoimaging.
Cell culture and stable transfection
A panel of human melanoma cell lines was provided generously by M. Herlyn (Wistar Inst.), which included those derived from melanomas of radial growth phase (RGP: WM35, WM3211 and sbcl2), vertical growth phase (VGP: WM278, WM793, WM1366 and WM3248), and metastatic (WM9, WM164, WM239, WM1158, 451LU, and 1205LU) origins. Melanoma cell lines were cultured in growth medium composed of MCDB153/L15 (v/v: 4/1; Sigma, St. Louis, MO) supplemented with 2 mM CaCl2, insulin (5 µg/ml) and 2% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA). For stable transfections, pCI-EGFP-based NM23-H1 expression vectors were mixed with pSV2neo (1: 0.3 µg ratio) and the liposomal reagent Fugene 6 (Roche, Nutley, NJ), and incubated with 1205LU and WM793 melanoma cells for 48 hours at 37°C. Colonies surviving G418 (250 µg/ml) were pooled and subjected to fluorescence-activated cell sorting (FACS; FACSCalibur Flow Cytometer, Becton-Dickinson, Mountain View, CA) to obtain EGFP-positive, mixed populations of stable transfectants. A consistent window of fluorescence intensity was employed for sorting to assure similar levels of NM23-H1 expression in the stably transfected cell lines.
SDS-PAGE and immunoblot analysis
Whole cell lysates were prepared from cultures (75–80% confluence) using M-PER lysis buffer (Pierce, Rockford, IL). Protein concentrations were determined by Bradford microassay (Bio-Rad, Richmond, CA) and measuring absorbance at 620 nm (Anthos Reader 2001, Anthos Labtech). Fifty µg of extract protein was subjected to SDS-PAGE (15% gels) under reducing conditions, followed by semi-dry blotting to nitrocellulose filters (Bio-Rad). NM23-H1 variants were detected by incubation with a 1:500 dilution of polyclonal rabbit anti-NM23 antibody (Ab-1; Labvision, Fremont, CA) for 1 hour at room temperature, followed by washing and incubation with a 1:500 dilution of goat anti-rabbit IgG-horseradish peroxidase conjugate (Santa Cruz, Santa Cruz, CA) for 1 hour, and use of the Supersignal West Pico chemiluminscence kit (Pierce). For two-dimensional SDS-PAGE, 50 µg of cell extract or purified recombinant NM23-H1 proteins were loaded in 350 µl of rehydration buffer containing 8 M urea, 4% CHAPS, 100 mM dithiothreitol (DTT), 0.2 % Bio-lytes, and 0.001% bromphenol blue. ReadyStrip IPG strips (17 cm, pH 4–7) were placed into rehydration buffer in a tray channel with 3 ml of mineral oil applied to the top, followed by rehydration for 12 hours at 50 volts (all reagents from Bio-Rad). Electrophoresis in the first dimension was conducted in an isolectric focusing apparatus (Protean, Bio-Rad). Isoelectric focusing strips were soaked for 10 minutes in solubilization buffer (375 mM Tris-HCl, pH 8.8, 6 M urea, 2% SDS, 2% DTT, 20% glycerol) followed by 10 minutes with alkylation buffer (same as alkylation buffer, except for substitution of DTT with 2% iodoacetamide). Second-dimension SDS-PAGE and anti-NM23-H1 immunoblot analyses were performed as described above.
Cell proliferation, motility and invasion assays
Cell proliferation was assessed by MTS assay using the CellTiter96 AQueous One reagent (Promega, Madison, WI). Cell motility was assessed for 104 melanoma cells in the upper chambers of transwell 24-well plates (6.5 mm diameter, 8.0 µm pore size, Corning, Lowell, MA), in growth medium supplemented with 0.1% FBS. Lower chambers were loaded with 0.6 ml of growth medium plus 2% FBS and 5 µg/ml insulin as chemoattractants. After 24 hours, cells were removed from the upper membrane surface with a cotton swab, followed by fixation of cells on the lower surface with 70% methanol for 1 hour and staining with hematoxylin for 1 hour. Stained cells were counted by light microscopy, with 5 random fields (10 × 10) counted per well. Cell invasiveness was determined over 24 h using BioCoat invasion chambers in a 24-well format (Becton-Dickinson, San Jose, CA). Plates were incubated at 37°C for 18 hours prior to addition of 104 cells in 0.1 ml of growth medium to the upper compartment. Lower compartments were loaded with 0.6 ml of growth medium containing 10% FBS, with all subsequent steps conducted as in the motility assays. Results of proliferation, motility and invasion assays represent the mean of three or more experiments, with triplicate wells per treatment within experiments.
Tumor and metastasis assays
Mouse care, surgery and injection protocols were approved by the Institutional Animal Care and Use Committee at the University of Kentucky (protocols 00319M2001 and 00801M2004). To produce tumor explants, 2 × 106 cells were suspended in 100 µl of phosphate-buffered saline, loaded into a 1 ml tuberculin syringe with a 26.5-gauge needle, and injected subcutaneously into the right flank of female athymic nude mice at 7–8 weeks of age (nu/nu, Harlan, Indianapolis, IN).31,32 For spontaneous metastasis formation, tumor explants were excised surgically upon reaching 800 mm3 in volume, usually within 28–30 days of initial cell injection. After 3 months, mice were killed by sodium pentobarbital injection, lungs were excised, and were stained in Bouin’s fixative for enhanced visualization of surface metastases. Visible metastatic lesions (> 1 mm diameter) were counted using a dissecting microscope.
Measurement of recombinant NM23-H1 mRNAs by quantitative PCR (Q-PCR)
Total cellular RNA was prepared from primary tumor tissue (100–200 mg) or metastatic nodules (<50 mg) using the RNeasy Midi kit (Qiagen, Valencia, CA). Reverse transcription PCR of RNA was conducted with random hexamer primers (Applied Biosystems, Foster City, CA). Q-PCR was performed with a TaqMan probeset spanning the small t intron, with a 5' sense primer (AGTCTCGAACTTAAGCTGCAGAAG) located within the cytomegalovirus promoter region, an antisense primer (TCTAGCCTTAAGAGCTGTAATTGAACTG) located within the 5' untranslated sequence of NM23-H1, and an MGB probe (ACTGGGCAGGGTGTCCA) located four nucleotides upstream of the antisense primer. NM23-H1 mRNA levels were normalized to endogenous beta-2-microglobulin mRNA (B2M TaqMan reagents; Applied Biosystems).
Results
Expression and purification of NM23-H1 variants harboring point mutations
To identify variants of NM23-H1 with selective lesions in each of its three enzymatic activities, mutant cDNAs were first generated by site-directed mutagenesis, expressed in E. coli, and prepared in purified form for functional analysis. A representation of targeted residues is provided in the context of the three-dimensional structure of the NM23-H1 molecule (Fig. 1a and b). Mutants glutamate5-to-alanine (E5A) and K12Q were generated for this analysis, for which profound deficiencies in 3’–5’ exonuclease activity and normal NDPK activity had been reported, but for which assessments of histidine protein kinase activity were lacking. Also mutated were residues required for nuclease activity of the NM23-H2 homolog,33,21 including a Q17N variant, which displays dysfunctional nuclease but normal NDPK activities, and R88A and R105A, which exhibit both nuclease and NDPK deficits. Residues Y52 and D54 were targeted due to their presence within the αA helix domain (Fig. 1b), for which we noted considerable identity with the consensus EXOIII domain of 3’–5’ exonucleases, D/EIRGY/FIDL/I.11 The Y52A mutation had been shown previously to disrupt NDPK activity of NM23-H2.33
FIGURE 1.
Location of amino acid residues in NM23-H1 targeted by site-directed mutagenesis. (a) Shown is a three-dimensional surface model of the NM23-H1 hexamer, with one subunit represented in a ribbon format. Amino acid residues targeted by mutagenesis in this study are highlighted with a ball-and-stick format. Negative charges are identified in red and positive charges in blue . (b) The amino acid sequence of NM23-H1 is displayed in a linear alignment with secondary structure elements. Residues targeted by site-directed mutagenesis are underlined and highlighted in bold font.
Prior to analysis of metastasis suppressor function for the NM23-H1 variants, it was essential to verify that the mutations did not introduce alterations in secondary or oligomeric structure. Circular dichroism (CD) spectrometry revealed that secondary structure estimates for four mutants (E5A, Q17N, Y52A and P96S) were not significantly different from that of the wild-type NM23-H1 protein (Fig. 2 and Table I). Likewise, the apparent molecular weights (i.e. oligomeric structures) of E5A, Q17N and P96S (85–88 kDa) were not significantly different from wild-type NM23-H1 as determined by gel filtration high performance liquid chromatography (HPLC), although a slightly lower apparent molecular weight was observed for the Y52A mutant (78 kDa). A prior study from our laboratory has shown that secondary and oligomeric structures of the K12Q and H118F mutants are not significantly different from wild-type NM23-H1.10 Thus, any observed effects of these mutations on protein function are most likely attributable to specific roles of the residues targeted and not to nonspecific effects on overall protein structure.
FIGURE 2.
Analysis of secondary structure for NM23-H1 mutant variants by circular dichroism spectrometry. Wild-type (WT) and mutant variants E5A, Q17N, Y52A and P96S were expressed in E. coli, purified to near homogeneity, and subjected to circular dichroism spectrometry, as described previously.10
TABLE I.
Estimates of molecular weight and protein secondary structure for NM23-H1 variants
| Protein | MW1 | CD spectrometry2 | |||
|---|---|---|---|---|---|
| a Helix | β Sheet | Turns | Random | ||
| WT | 87.7 ± 2.1 | 14.2 ± 2.8 | 32.6 ± 5.3 | 20.0 ± 0.1 | 32.6 ± 3.8 |
| E5A | 85.1 ± 7.1 | 10.7 ± 2.2 | 35.0 ± 2.5 | 19.4 ± 0.6 | 32.9 ± 1.8 |
| Q17N | 86.3 ± 1.5 | 8.8 ± 2.6 | 40.2 ± 4.3 | 18.7 ± 2.0 | 32.3 ± 0.7 |
| Y52A | 78.4 ± 3.0* | 11.4 ± 4.1 | 35.9 ± 1.9 | 19.4 ± 1.4 | 30.7 ± 1.1 |
| P96S | 84.7 ± 0.4 | 13.7 ± 1.7 | 33.0 ± 1.2 | 19.8 ± 1.3 | 32.7 ± 2.0 |
Results are expressed in kDa as the mean (± SD) of three replicate determinations by gel filtration HPLC.
Secondary structure content is expressed as a percent of the total structure (mean ± SD).
Mean within a column is significantly different (p ≤ 0.05), as determined by one-way ANOVA.
Identification of amino acid residues required for NDPK, 3’–5’ exonuclease, and histidine kinase activities of NM23-H1
Direct enzymatic assay of NDPK activity for mutants K12Q , H118F, and Y52A revealed nearly total losses in function, as seen previously,34,10,25 while the P96S mutant exhibited a lesser decrease to 20% of wild-type (Table II). With respect to 3’–5’ exonuclease activity, the E5A and K12Q mutations were strongly inhibitory, with reductions to 12% and 18% of wild-type, respectively (Fig. 3a–c and Table II), as previously reported.10,25 In contrast, the Q17N (Fig.3a and c, Table II) and the R88A and R105A mutations (data not shown) had little effect on 3’–5’ exonuclease activity, in contrast with their previously reported disruptive effects on the nuclease activity of NM23-H2. The Y52A and P96S mutations also had no significant impact on 3’–5’ exonuclease activity, the former excluding functional relevance of the EXOIII-like motif. The hierarchy of histidine protein kinase activity across the panel of NM23-H1 variants was remarkably similar to that of NDPK activities, with K12Q, Y52A and H118F displaying the strongest inhibition, and P96S to a lesser extent (Fig. 3d and Table 1). In summary, these biochemical analyses identified four classes of mutants with: 1) selective deficiency in 3’–5’ exonuclease activity (E5A), 2) complete deficiencies in both NDPK and histidine kinase (Y52A, H118F), 3) more moderate deficiencies in NDPK and histidine kinase (P96S), and 4) complete deficiencies in all three activities (K12Q).
TABLE II.
Ndpk, histidine kinase and exonuclease activities of Nm23-H1 variants
| Protein | NDPK1 | Histidine kinase2 | Exonuclease3 |
|---|---|---|---|
| WT | 627 ± 36d (100) | 9.9 (100) | 28.4 ± 4.0 (100) |
| E5A | 438 ± 31c (70) | 8.2 (83) | 3.4 ± 1.5* (12) |
| K12Q | 14 ± 0.9a (2) | 0.9 (9) | 5.2 ± 3.5* (18) |
| Q17N | 516 ± 10c (82) | 6.5 (66) | 31.4 ± 6.0 (110) |
| Y52A | B.D.a | 0.4 (4) | 25.3 ± 3.9 (89) |
| P96S | 127 ± 5b (20) | 1.1 (11) | 24.9 ± 5.1 (88) |
| H118F | B.D.4 | B.D. | 33.0 ± 9.0 (116) |
NDPK activity is expressed in units/mg (mean ± S.E.), derived from at least three replicate measurements and three independent protein preparations. Activity expressed relative to wild-type is displayed in parentheses.
Histidine kinase activity is expressed as the percent of 32P radiolabeled NM23-H2 substrate converted to a phosphorylated per minute.
Exonuclease activity is expressed in fmol of 32P liberated from radiolabeled nucleotide substrate per 5 min (mean ± SE) as determined with three replicate measurements for at least three independent protein preparations.
B.D., below detection.
Means not bearing a common superscript are significantly different (p ≤ 0.01), as determined by one-way ANOVA and mean separation by Student’s t-Test.
Means bearing an asterisk within a column are significantly different (p ≤ 0.05), as determined by one-way ANOVA and mean separation by Student’s t-Test.
FIGURE 3.
Determination of 3’–5’ exonuclease and protein histidine kinase activity for selected NM23-H1 mutant variants. (a) Purified preparations of wild-type or mutant NM23-H1 proteins (500 ng) were incubated with 10 fmol of a single-stranded, 5’-radiolabeled DNA substrate for the indicated times at room temperature, as described in Materials and Methods. Extent of DNA cleavage was analyzed by electrophoresis through denaturing 10% polyacrylamide gels and visualization by phosphorimaging. P, radiolabeled probe. (b) 3’–5’ exonuclease assay of E5A mutant is shown. C, Quantitation of results in panels A and B are shown, expressed as total nucleotides (nt) cleaved. (c) Protein histidine kinase activity was analyzed for wild-type and mutant variants of NM23-H1. NM23-H1 proteins were auto-radiolabeled with [32P-γ] ATP, followed by incubation with unlabeled NM23-H2 substrate for the indicated times.
Suppression of cell motility and invasiveness by NM23-H1 does not require residues E5, K12, P96 or H118
To determine the contributions of the three enzymatic activities of NM23-H1 to metastasis suppressor activity, mutants E5A, K12Q, P96S and H118F were introduced by stable transfection into the human melanoma cell line, 1205LU. This line was chosen by virtue of its deficient expression of both the NM23-H1 and NM23-H2 isoforms (Supporting Information Fig. 1a), and its previously described metastatic growth properties in athymic nude mice.35 Low expression of NM23-H1 and NM23-H2 was observed in 1205LU and two other metastatic human melanoma cell lines (WM239 and WM1158), as compared with melanoma cell lines derived from radial (RGP) or vertical growth phase (VGP) tumors (Supporting Information Fig. 1a). Mixed populations of 1205LU transfectants were obtained following transfection with an expression vector containing the desired NM23-H1 variant cDNAs in linkage with the EGFP reporter gene, followed by enrichment for EGFP-positive cells by fluorescence-activated cell sorting (FACS; see Material and Methods for details). Each line exhibited approximately 3-fold increases in expression over endogenous NM23-H1 protein, as verified by one- and two-dimensional SDS-polyacrylamide gel electrophoresis (Supporting Information Fig. 1b,c).
Proliferation rates of the 1205LU cell lines were unaltered by expression of NM23-H1 or mutant variants (Fig. 4a) as shown previously,3 as were their growth rates as tumor explants in athymic nude mice (Fig. 4b) However, wild-type NM23-H1 was a potent suppressor of both motility (Fig. 5a) and invasiveness (Fig. 5b) of 1205LU cells in culture. Interestingly, each of the mutants analyzed (E5A, K12Q, P96S and H118F) also exhibited full motility and invasion suppressor activity. We have more recently observed identical results with the same panel of NM23-H1 variants in a scratch/wound-healing model of cell motility in another melanoma cell line WM793 (J. McCorkle, unpublished observations). These findings indicate that the NDPK, histidine kinase, and 3’–5’ exonuclease activities of NM23-H1 do not contribute to suppression of motility and invasion in the specific setting of 1205LU melanoma cells, and strongly suggest a novel mechanism in this cell line which remains to be elucidated.
FIGURE 4.
Forced expression of NM23-H1 variants has minimal effects on the transformed phenotype of 1205LU melanoma cells in culture and as tumor explants in athymic nude mice. (a) Cell proliferation assays were conducted on 1205LU cells using the MTS procedure, as described in Materials and Methods. (b) Data represents the mean tumor volume obtained over the indicated time courses following subcutaneous injection into athymic nude mice of the 1205LU parent and transfected derivatives expressing wild-type NM23-H1 (H1-WT), P96S and H118F variants (left panel), and in a separately conducted series of experiments, H1-WT, E5A, and K12Q mutants (right panel). No significant differences between growth curves were detected between any of the cell lines in panels a and b, as determined by two-way ANOVA.
FIGURE 5.
NM23-H1 inhibits motility and invasive capacities of 1205LU melanoma cells in a manner independent of its 3’–5’ exonuclease, NDPK and histidine kinase activities. (a) The 1205LU panel of cell lines were evaluated for motility and (b) invasive characteristics in Boyden chamber assays. Asterisks denote means that are significantly different (p < 0.05) from all other treatment means within a panel, as determined by one-way ANOVA and Student’s t-Test.
Amino acid residues E5, K12 and H118 are required for suppression of spontaneous metastasis of 1205LU cells in vivo
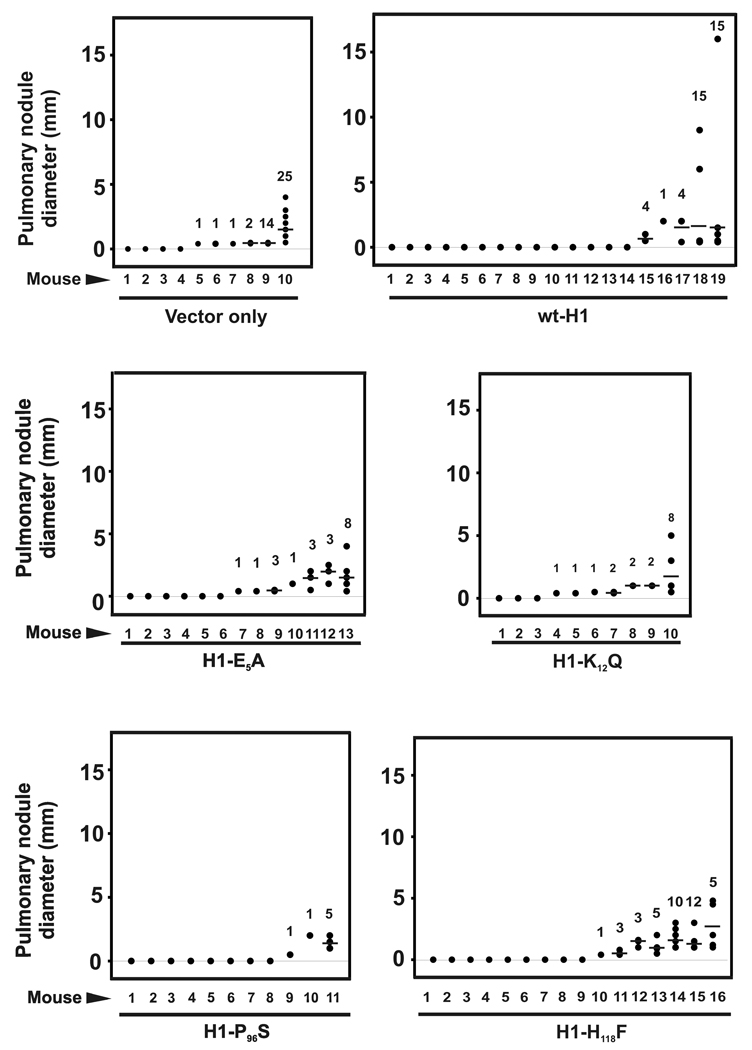
To assess the metastatic potential of the 1205LU panel of cell lines, a spontaneous metastasis (i.e. via subcutaneous tumor explants) approach were undertaken in athymic nude mice. Forced overexpression of wild-type NM23-H1 strongly inhibited spontaneous metastasis (25% incidence, Table III) compared to the relatively high penetrance seen in parent and empty vector-transfected 1205LU cell lines (65 and 60%, respectively). Two of the mutants (E5A and K12Q) failed to exert measurable metastasis suppressor activity, while the NDPK-and histidine kinase-null H118F mutant exhibited only weak suppressor activity at best (WT vs. H118F, p < 0.07). The P96S mutant, which retained residual NDPK and histidine kinase activities and full exonuclease activity, possessed full metastasis suppressor activity.The loss of spontaneous metastasis suppressor activity for the E5A mutant, which harbors a selective lesion in 3’–5’ exonuclease activity but normal NDPK and histidine kinase activities, strongly suggests a critical role for the 3’–5’ exonuclease. Consistent with that interpretation was the loss in suppressor activity for the K12Q mutant, which is also deficient in 3’–5’ exonuclease, as well as the NDPK and histidine kinase functions. Although incidence of spontaneous metastasis was suppressed strongly by wild-type and P96S forms of NM23-H1, lesions that did arise in these groups were not different in number per lung or size from those seen in the vector-transfected control group (Fig. 6).
TABLE III.
3’–5’ exonuclease activity of NM23-H1 is necessary for suppression of spontaneous metastasis of 1205lu melanoma cells.
| 1205Lu cell lines | Exogenous H1- mediated activities |
Spontaneous metastases† |
|||
|---|---|---|---|---|---|
| NDPK | HisK | EXO | % | (incidence) | |
| parent | none | 65a | (17/26) | ||
| vector alone | none | 60a,b | (6/10) | ||
| Wild-type H1 | + | + | + | 25c | (12/48) |
| E5A | + | + | − | 50a,b | (13/26) |
| K12Q | − | − | − | 58a,b | (14/24) |
| P96S | +/− | +/− | + | 25c | (6/24) |
| H118F | − | − | + | 46a–c | (14/24) |
Spontaneous metastasis denotes lung metastases obtained from seeding by primary tumors derived from subcutaneous explantation of tumor cells. The number and size of macrometastases was not statistically different (p ≥ 0.05) between the cell lines as determined by the Mann-Whitney rank sum test. The apparent difference between H118F and wild-type NM23-H1 nearly achieved statistical significance (p ≥ 0.07).
Means not showing a common superscript are significantly different (p ≤ 0.01) as determined by Fisher’s exact test.
n.d., not determined
FIGURE 6.
Number and diameter of pulmonary metastatic lesions in spontaneous metastasis assay. Closed circles represent diameters for individual nodules, horizontal bars represent the average number of visible metastases per lung, and values listed above each column of data indicate the total number of nodules observed within a given lung. No significant differences between the cell lines were found in either the number of lesions per lung or the average lesion size, as determined by Kruskal-Wallis one-way ANOVA (p > 0.05).
Similar levels of transgene expression were verified across the cell line panel in primary tumor explants (p > 0.05) as measured by quantitative polymerase chain reaction using primers specific for the recombinant transcripts (Supporting Information Fig. 2a), indicating the transfected cells maintained transgene expression during the lung seeding period. A trend toward lower expression of the K12Q mutant was observed, suggesting expression of this mutant might be less stable. Persistence of recombinant NM23-H1 mRNA expression was further verified in a small subset of metastatic nodules from the wild-type- and E5A-expressing cell lines (Supporting Information Fig. 2b), strongly suggesting that the increased metastatic incidence seen with the E5A mutant was not caused by loss of its expression.
Discussion
To elucidate mechanisms underlying metastasis suppressor activity of NM23-H1, a panel of mutant variants was constructed and characterized biochemically in terms of the three primary enzymatic activities of the protein (NDPK, histidine kinase and 3’–5’ exonuclease). The most selective lesion was obtained with E5A, which was deficient in 3’–5’ exonuclease activity with little attenuation of the NDPK or protein histidine kinase activities. The juxtaposition of two residues critical for 3’–5’ exonuclease activity, E5 and K12, at opposite ends of the β1 strand (Supplemental Figure 1B) indicates this region is a critical component of the enzymatic active site. Previous studies of the NM23-H2 isoform have demonstrated formation of a covalent complex between K12 and DNA during initial nucleolytic attack,28 consistent with conservation of the β1 strand as a critical nuclease domain for both the H1 and H2 isoforms.
Surprisingly, the NDPK and histidine kinase activities could not be dissociated by mutation, with a nearly identical profile for these two activities across all mutants studied. Our study revealed a requirement of K12 and Y52 for histidine kinase activity, and the lack of involvement of P96 and Q17 in the 3’–5’ exonuclease activity of NM23-H1. The H118F mutant was particularly informative in light of its full 3’–5’ exonuclease activity in the context of inactive NDP and histidine kinase functions. Metastasis suppressor activity of H118F appeared to be impaired (p < 0.07), suggesting that the NDPK activity, or least the catalytic H118 residue, is required for full metastasis suppression. Importantly, this indicates that the 3’–5’ exonuclease activity alone, which is intact in the H118F mutant, is not sufficient for full suppressor activity. Conversely, suppressor activity of the E5A mutant (exonuclease-deficient, NDPK/histidine kinase intact) was significantly impaired, indicating that the NDPK and histidine kinase activities were also insufficient as suppression mediators. Together, these findings strongly suggest cooperation between the 3’–5’ exonuclease, NDPK and/or the histidine kinase, in the molecular mechanism of suppression. Interestingly, none of the NM23-H1 point mutations studied (E5A, K12Q, P96S, H118F) inhibited cell motility or invasiveness of the 1205LU cell line in transwell assays, suggesting an underlying mechanism independent of the NDPK, histidine kinase or 3’–5’ exonuclease functions. Consistent with our results, the H118F mutation fails to disrupt NM23-H1-mediated suppression of motility in the prostate carcinoma cell line, DU145, indicating nonessentiality of the NDPK and histidine kinase functions in that setting as well.7 These results are distinct from reports of reduced motility suppressor activity for the P96S variant in human MDA-MB-435 (breast carcinoma or melanoma origin) and L9981 (large cell lung carcinoma) cells,36,37 suggesting differences between the different cell types studied. A key implication of our findings is that motility- and invasion-suppressing activities of NM23-H1 in cell culture are not equivalent with metastasis suppressor activity in vivo. For example, the E5A and K12Q were fully active as suppressors of motility and invasion of 1205LU cells in culture, yet they both failed to suppress spontaneous metastasis in athymic nude mice. While our results reinforce the inadequacy of motility and invasion in as surrogate cell culture indices for the full metastatic process, it should be reinforced they do not exclude potential important contributions of antimotility or anti-invasion functions of NM23-H1 in vivo.
The biochemical data of this study provide a strong argument in support of important roles of the 3’–5’ exonuclease and possibly the NDPK activity for metastasis suppressor activity of NM23-H1. However, the possibility cannot be excluded that the mutations employed herein may also impact on other functions of the molecule. One possibility is that these mutations could impair important physical interactions with other modulators of metastasis. Such physical interactions could take the form of NM23-H1 as a factor which physically binds and sequesters a metastasis-promoting protein or complex (e.g. Prune; ref. 17,18). A detailed mapping of contact residues on NM23-H1 and on such known, as well as yet to be discovered binding partners of NM23-H1, should be informative in this regard. Mutants generated for this purpose could be exploited by forced expression and analysis of metastatic competence in an approach similar to that employed in the current study.
3’–5’ exonucleases are critical for maintenance of genomic stability via proofreading or other related functions in DNA repair, replication and recombination. Accordingly, loss of NM23-H1 expression and its cognate 3’–5’ exonuclease activity might be envisioned to promote genomic instability and malignant progression. Indeed, we have observed recently that loss of NM23 expression is required for repair of DNA damage induced by ultraviolet radiation and etoposide in the yeast Saccharomyces cerevisiae,12 as well as in the human melanoma cell line WM793 (Jarrett et al., unpublished observations). One possibility is that 1205LU cells may be poised for progression to metastasis, but may require further genetic alterations in vivo for competence. Alternatively, the possibility remains open that the 3’–5’ exonuclease activity may exert a metastasis suppressor-relevant function(s) that does not involve DNA repair processes. Further investigation into the cellular and biochemical function(s) of the 3’–5’ exonuclease should be informative in resolving these possibilities.
Supplementary Material
Acknowledgments
We thank Kristen Ormerod for technical assistance, Ioan Lascu for providing the three-dimensional representation of NM23-H1 displayed in this paper, and the late Dr. Stephen Zimmer for his helpful advice in the early stages of this work. The work was supported by NIH grant R01 CA83237 from the National Cancer Institute (D. Kaetzel). This work is submitted in partial fulfillment of the requirements of the Department of Molecular and Biomedical Pharmacology Graduate Program (J.R.M. and M.N.). Author contributions: Q.Z., J.R.M., M.N. and M.Y. performed research; Q.Z., J.R.M., M.N., M.Y. and D.M.K. designed research, analyzed data and wrote the article.
Grant sponsor: U.S. Public Health Service Grant, National Cancer Institute; Grant number: R01 CA83237; Grant sponsor: Kentucky Lung Cancer Research Program.
Abbreviations
- CD
circular dichroism
- FACS
fluorescence-activated cell sorting
- FBS
fetal bovine serum
- HPLC
high performance liquid chromatography
- kDa
kilodalton
- NDPK
nucleoside diphosphate kinase
- PBS
phosphate-buffered saline
- Q-PCR
quantitative polymerase chain reaction
- RGP
radial growth phase
- VGP
vertical growth phase
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
References
- 1.Steeg PS, Ouatas T, Halverson D, Palmieri D, Salerno M. Metastasis suppressor genes: basic biology and potential clinical use. Clin Breast Cancer. 2003;4:51–62. doi: 10.3816/cbc.2003.n.012. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS, Bevilacqua G, Kopper L, Thorgerisson UR, Talmadge JE, Liotta LA, Sobel M. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–205. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 3.Hartsough MT, Steeg PS. Nm23/nucleoside diphosphate kinase in human cancers. J Bioenerg Biomembr. 2000;32:301–308. doi: 10.1023/a:1005597231776. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal RP, Robinson B, Parks RE. Nucleoside diphosphokinase from erythrocytes. Methods Enzymol. 1978;51:376–386. doi: 10.1016/s0076-6879(78)51051-3. [DOI] [PubMed] [Google Scholar]
- 5.Sastre-Garau X, Lacombe ML, Jouve M, Veron M, Magdelenat H. Nucleoside diphosphate kinase/NM23 expression in breast cancer: lack of correlation with lymph-node metastasis. Int J Cancer. 1992;50:533–538. doi: 10.1002/ijc.2910500406. [DOI] [PubMed] [Google Scholar]
- 6.Leone A, Flatow U, VanHoutte K, Steeg PS. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: effects on tumor metastatic potential, colonization and enzymatic activity. Oncogene. 1993;8:2325–2333. [PubMed] [Google Scholar]
- 7.Lee HY, Lee H. Inhibitory activity of nm23-H1 on invasion and colonization of human prostate carcinoma cells is not mediated by its NDP kinase activity. Cancer Lett. 1999;145:93–99. doi: 10.1016/s0304-3835(99)00236-0. [DOI] [PubMed] [Google Scholar]
- 8.Wagner PD, Vu ND. Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J Biol Chem. 1995;270:21758–21764. doi: 10.1074/jbc.270.37.21758. [DOI] [PubMed] [Google Scholar]
- 9.Hartsough MT, Morrison DK, Salerno M, Palmieri D, Ouatas T, Mair M, Patrick J, Steeg PS. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277:32389–32399. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- 10.Ma D, McCorkle JR, Kaetzel DM. The metastasis suppressor NM23-H1 possesses 3'-5' exonuclease activity. J Biol Chem. 2004;279:18073–18084. doi: 10.1074/jbc.M400185200. [DOI] [PubMed] [Google Scholar]
- 11.Shevelev IV, Hübscher U. The 3'-5' exonucleases. Nat Rev Mol Cell Biol. 2002;3:1–12. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- 12.Yang M, Jarrett SG, Craven R, Kaetzel DM. YNK1, the yeast homolog of human metastasis suppressor NM23, is required for repair of UV radiation- and etoposide-induced DNA damage. Mutat Res. 2009;660:74–78. doi: 10.1016/j.mrfmmm.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 14.Tseng YH, Vicent D, Zhu J, Niu Y, Adeyinka A, Moyers JS, Watson PH, Kahn CR. Regulation of growth and tumorigenicity of breast cancer cells by the low molecular weight GTPase Rad and nm23. Cancer Res. 2001;61:2071–2079. [PubMed] [Google Scholar]
- 15.Otsuki Y, Tanaka M, Yoshii S, Kawazoe N, Nakaya K, Sugimura H. Tumor metastasis suppressor nm23H1 regulates Rac1 GTPase by interaction with Tiam1. Proc Natl Acad Sci U S A. 2001;98:4385–4390. doi: 10.1073/pnas.071411598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian C, Cotter MA, Robertson ES. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat Med. 2001;7:350–355. doi: 10.1038/85499. [DOI] [PubMed] [Google Scholar]
- 17.D'Angelo A, Garzia L, Andre A, Carotenuto P, Aglio V, Guardiola O, Arrigoni G, Cossu A, Palmieri G, Aravind L, Zollo M. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell. 2004;5:137–149. doi: 10.1016/s1535-6108(04)00021-2. [DOI] [PubMed] [Google Scholar]
- 18.Galasso A, Zollo M. The Nm23-H1-h-Prune complex in cellular physiology: a 'tip of the iceberg' protein network perspective. Mol Cell Biochem. 2009;329:149–159. doi: 10.1007/s11010-009-0115-4. [DOI] [PubMed] [Google Scholar]
- 19.Gilles AM, Presecan E, Vonica A, Lascu I. Nucleoside diphosphate kinase from human erythrocytes. Structural characterizatioin of the two polypeptide chains responsible for heterogeneity of the hexameric enzyme. J Biol Chem. 1991;266:8784–8789. [PubMed] [Google Scholar]
- 20.Postel EH, Abramczyk BM, Levit MN, Kyin S. Catalysis of DNA cleavage and nucleoside triphosphate synthesis by NM23-H2/NDP kinase share an active site that implies a DNA repair function. Proc Natl Acad Sci U S A. 2000;97:14194–14199. doi: 10.1073/pnas.97.26.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Postel EH, Abramczyk BA, Gursky SK, Xu Y. Structure-based mutational and functional analysis identify human NM23-H2 as a multifunctional enzyme. Biochemistry. 2002;41:6330–6337. doi: 10.1021/bi025606+. [DOI] [PubMed] [Google Scholar]
- 22.Morera S, Lacombe ML, Xu Y, LeBras G, Janin J. X-ray structure of human nucleoside diphosphate kinase B complexed with GDP at 2 A resolution. Structure. 1995;3:1307–1314. doi: 10.1016/s0969-2126(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 23.Freije JM, Blay P, MacDonald NJ, Manrow RE, Steeg PS. Site-directed mutation of Nm23-H1. Mutations lacking motility suppressive capacity upon transfection are deficient in histidine-dependent protein phosphotransferase pathways in vitro. J Biol Chem. 1997;272:5525–5532. doi: 10.1074/jbc.272.9.5525. [DOI] [PubMed] [Google Scholar]
- 24.Rosengard AM, Krutzch HC, Shearn A, Biggs JR, Barker E, Margulies IMK, King CR, Liotta LA, Steeg PS. Reduced Nm23/Awd protein in tumor metastases and aberrant Drosophila development. Nature. 1989;342:177–180. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- 25.Yoon JH, Singh P, Lee DH, Qiu J, Cai S, O'Connor TR, Chen Y, Shen B, Pfeifer GP. Characterization of the 3' --> 5' exonuclease activity found in human nucleoside diphosphate kinase 1 (NDK1) and several of its homologues. Biochemistry. 2005;44:15774–15786. doi: 10.1021/bi0515974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the PCR. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 27.Ma D, Xing Z, Liu B, Pedigo N, Zimmer S, Bai Z, Postel E, Kaetzel DM. NM23-H1 cleaves and represses transcriptional activity of nuclease-hypersensitive elements in the PDGF-A promoter. J Biol Chem. 2002;277:1560–1567. doi: 10.1074/jbc.M108359200. [DOI] [PubMed] [Google Scholar]
- 28.Postel EH. Cleavage of DNA by human NM23-H2/nucleoside diphosphate kinase involves formation of a covalent protein-DNA complex. J Biol Chem JID - 2985121R. 1999;274:22821–22829. doi: 10.1074/jbc.274.32.22821. [DOI] [PubMed] [Google Scholar]
- 29.Postel EH. Cleavage of DNA by human NM23-H2/nucleoside diphosphate kinase involves formation of a covalent protein-DNA complex. J Biol Chem. 1999;274:22821–22829. doi: 10.1074/jbc.274.32.22821. [DOI] [PubMed] [Google Scholar]
- 30.Liu B, Maul RS, Kaetzel DM., Jr Repression of platelet-derived growth factor A-chain gene transcription by an upstream silencer element. J Biol Chem. 1996;271:26281–26290. doi: 10.1074/jbc.271.42.26281. [DOI] [PubMed] [Google Scholar]
- 31.Kath R, Jambrosic JA, Holland L, Rodeck U, Herlyn M. Development of invasive and growth factor-independent cell variants from primary human melanomas. Cancer Res. 1991;51:2205–2211. [PubMed] [Google Scholar]
- 32.Schilling D, Reid JDI, Hujer A, Morgan D, DeMoll E, Bummer P, Fenstermaker RA, Kaetzel DM. Loop III region of platelet-derived growth factor (PDGF) B-chain mediates binding to PDGF receptors and heparin. Biochem J. 1998;333:637–644. doi: 10.1042/bj3330637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Postel EH, Weiss VH, Beneken J, Kirtane A. Mutational analysis of NM23-H2/NDP kinase identifies the structural domains critical to recognition of a c-myc regulatory element. Proc Natl Acad Sci USA. 1996;93:6892–6897. doi: 10.1073/pnas.93.14.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacDonald NJ, De La Rosa A, Benedict MA, Freije JM, Krutsch H, Steeg PS. A serine phosphorylation of Nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J Biol Chem. 1993;268:25780–25789. [PubMed] [Google Scholar]
- 35.Herlyn D, Iliopoulos D, Jensen PJ, Parmiter A, Baird J, Hotta H, Adachi K, Ross AH, Jambrosic J, Koprowski H. In vitro properties of human melanoma cells metastatic in nude mice. Cancer Res. 1990;50:2296–2302. [PubMed] [Google Scholar]
- 36.MacDonald NJ, Freije JM, Stracke ML, Manrow RE, Steeg PS. Site-directed mutagenesis of nm23-H1. Mutation of proline 96 or serine 120 abrogates its motility inhibitory activity upon transfection into human breast carcinoma cells. J Biol Chem. 1996;271:25107–25116. doi: 10.1074/jbc.271.41.25107. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Q, Yang X, Zhu D, Ma L, Zhu W, Sun Z, Yang Q. Double mutant P96S/S120G of Nm23-H1 abrogates its NDPK activity and motility-suppressive ability. Biochem Biophys Res Commun. 2007;356:348–353. doi: 10.1016/j.bbrc.2007.02.066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.