DELLA degradation, a key event in gibberellin (GA) signaling, is triggered by the GA-dependent interaction between the GA receptor GID1 and the DELLA/TVHYNP motif of DELLA. The interaction between GID1 and the DELLA/TVHYNP motif is found to enable the C-terminal GRAS domain of DELLA to interact with GID1, which leads to the degradation of the DELLA protein SLR1 via the SCFGID2 E3 ligase.
Abstract
The DELLA protein SLENDER RICE1 (SLR1) is a repressor of gibberellin (GA) signaling in rice (Oryza sativa), and most of the GA-associated responses are induced upon SLR1 degradation. It is assumed that interaction between GIBBERELLIN INSENSITIVE DWARF1 (GID1) and the N-terminal DELLA/TVHYNP motif of SLR1 triggers F-box protein GID2-mediated SLR1 degradation. We identified a semidominant dwarf mutant, Slr1-d4, which contains a mutation in the region encoding the C-terminal GRAS domain of SLR1 (SLR1G576V). The GA-dependent degradation of SLR1G576V was reduced in Slr1-d4, and compared with SLR1, SLR1G576V showed reduced interaction with GID1 and almost none with GID2 when tested in yeast cells. Surface plasmon resonance of GID1-SLR1 and GID1-SLR1G576V interactions revealed that the GRAS domain of SLR1 functions to stabilize the GID1-SLR1 interaction by reducing its dissociation rate and that the G576V substitution in SLR1 diminishes this stability. These results suggest that the stable interaction of GID1-SLR1 through the GRAS domain is essential for the recognition of SLR1 by GID2. We propose that when the DELLA/TVHYNP motif of SLR1 binds with GID1, it enables the GRAS domain of SLR1 to interact with GID1 and that the stable GID1-SLR1 complex is efficiently recognized by GID2.
INTRODUCTION
Gibberellins (GAs) are a large family of tetracyclic diterpenoid plant hormones that induce a wide range of plant growth responses, including seed germination, stem elongation, leaf expansion, flowering, pollen maturation, and transition from vegetative growth to flowering (Richards et al., 2001; Thomas et al., 2005). Through molecular genetic studies on GA-insensitive mutants of rice (Oryza sativa) and Arabidopsis thaliana, factors important for GA signaling have been identified, which seem to be conserved among vascular plants (Hirano et al., 2007; Yasumura et al., 2007). The key regulator of the GA-signaling pathway is the negative regulator of GA signaling, DELLA, which consists of an N-terminal DELLA/TVHYNP motif and a C-terminal GRAS domain, defining DELLA proteins as members of the GRAS family of putative transcriptional regulators (Peng et al., 1997; Ikeda et al., 2001; Chandler et al., 2002; reviewed in Hirano et al., 2008). DELLA protein is rapidly degraded when plants are treated with GA and as a consequence, GA-associated responses are induced (Dill et al., 2001; Silverstone et al., 2001; Gubler et al., 2002; Itoh et al., 2002). Other factors, represented by rice GIBBERELLIN INSENSITIVE DWARF2 (GID2) and Arabidopsis SLEEPY1 (SLY1), are F-box components of Skp1-Cullin-F box protein (SCF) E3 ubiquitin ligases responsible for targeting DELLA proteins to the proteasome for degradation (McGinnis et al., 2003; Sasaki et al., 2003). Recently, a soluble GA receptor, GID1, was identified in rice and in several other plant species (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Nakajima et al., 2006; Hirano et al., 2007; Aleman et al., 2008; Chandler et al., 2008). The observation that GID1 and the DELLA/TVHYNP motif of DELLA physically interact in a GA-dependent manner was first made in a yeast two-hybrid (Y2H) assay (Ueguchi-Tanaka et al., 2005) and then by x-ray crystallography of GID1 forming a complex with GA and the N-terminal portion of the DELLA protein (Murase et al., 2008). This interaction seems to be important for degradation of DELLA because various in frame or amino acid changes in the DELLA/TVHYNP motif lead to reduced DELLA protein degradation (Peng et al., 1997; Dill et al., 2001; Gubler et al., 2002; Asano et al., 2009). On the other hand, the C-terminal GRAS domain of DELLA is known to be necessary for GA repression activity, since mutations in the GRAS domain often lead to a loss-of-function slender phenotype (Silverstone et al., 1998; Ikeda et al., 2001; Bassel et al., 2008; Weston et al., 2008).
Based on these observations, this model of GA signaling is proposed as follows. In the absence of GA, DELLA represses GA action. When GA is present, the GA-bound GID1 receptor interacts with the DELLA/TVHYNP motif of DELLA protein. This interaction triggers DELLA protein degradation through the SCFGID2/SLY1 proteasome pathway, and GA action occurs. A recent addition to this model is that interaction of DELLA with GID1 per se (i.e., in the absence of GID2/SLY1 activity) seems to reverse the repressive activity of DELLA protein without requiring DELLA protein degradation (Ariizumi et al., 2008; Ueguchi-Tanaka et al., 2008). Taking into consideration that DELLA is more abundant than GID1 in plant cells (see Supplemental Figure 1 online), degradation of DELLA is likely to have two important functions in GA signaling: to completely diminish the repression activity of DELLA and to produce free GID1 by specifically degrading the DELLA within the DELLA-GID1 complex. In either case, DELLA degradation by GA in association with GID1 and F-box protein is considered to be the key event in GA signaling.
Many biological processes are regulated by the SCF proteasome system, in which F-box proteins provide the specificity for which target to degrade (reviewed in Ravid and Hochstrasser, 2008). Since degradation of target proteins is strictly regulated both temporally and spatially, the target proteins must be recognized by F-box protein only under favorable conditions. Such selective recognition is accomplished mainly by posttranslational modification of target proteins, such as phosphorylation, oxidation, and glycosylation (Ravid and Hochstrasser, 2008). In GA signaling, it is not yet clear how GID2/SLY1 recognizes DELLA prior to degrading it (Itoh et al., 2005). However, as stated above, the GA-dependent interaction of GID1 with the DELLA/TVHYNP motif of DELLA seems to be necessary for DELLA degradation: SLENDER RICE1 (SLR1) containing a deletion of this motif is no longer degraded upon GA treatment. In this article, we identified a gain-of-function dwarf mutant in rice with a mutation in the region encoding the GRAS domain of SLR1. Analyses of this mutant suggest that binding of the DELLA/TVHYNP motif to GID1 is not sufficient for SLR1 to be efficiently recognized by GID2 and that a stable interaction between GID1 and SLR1 through the GRAS domain of SLR1 is also essential. Our results suggest that the interaction between the GRAS domain and GID1 serves as the recognition signal for targeting of SLR1 by GID2.
RESULTS
Isolation and Characterization of a Novel Rice GA-Insensitive Mutant
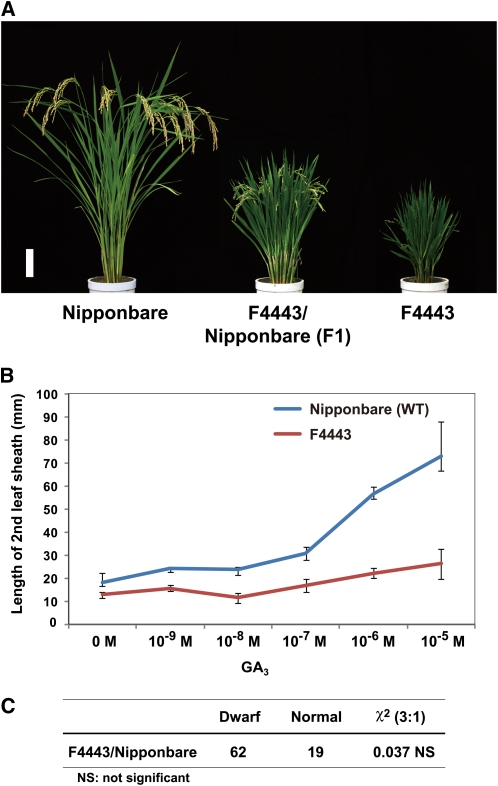
A rice semidominant dwarf mutant, F4443, was identified from >2000 Nipponbare M2 lines mutagenized with N-methyl-N-nitrosourea (MNU). Mutant plants were about one-quarter the height of the original strain, with wide, dark-green leaf blades similar to other GA-related mutants previously characterized (Figure 1A). The response of F4443 to exogenously applied GA3 was much lower than that of the wild-type plant (Figure 1B), indicating that this mutant has a defect in GA responsiveness. When F4443 was crossed with the original strain (Nipponbare), the F1 plants showed intermediate plant height relative to the parents (Figure 1A). In the F2 progeny, the segregation ratio of dwarf (including the semidwarf phenotype of F1 plants) to normal was 62:19, which corresponded to the expected 3:1 segregation ratio for a single dominant gene (χ2 = 0.037; Figure 1C). These genetic results demonstrate that this mutation behaves in an incomplete dominant manner.
Figure 1.
Gross Morphology of Rice Semidominant Dwarf Mutant F4443, Its Response to GA3 Treatment, and Segregation Ratio of F2 Plants.
(A) Gross morphology of a wild-type Nipponbare (left), F1 plant derived from a cross between F4443 and Nipponbare (center), and F4443 homozygous plant (right). Bar = 10 cm.
(B) Elongation of second leaf sheath in response to GA3 treatment. Nipponbare was used as a control. Data are means ± sd; n = 10.
(C) Segregation of F2 progeny of a self-pollinated F1 plant (F4443 × Nipponbare).
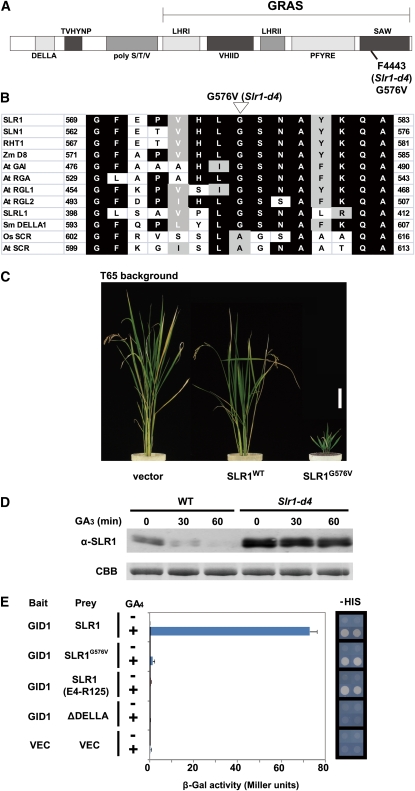
Because rice and Arabidopsis GA-insensitive mutants that behave in a dominant manner often have mutations in the genes encoding DELLA proteins (Peng et al., 1997, 1999; Asano et al., 2009), we directly sequenced the rice DELLA protein gene, SLR1, of this mutant. We identified one nucleotide substitution, which exchanges the Gly at position 576 with Val (G576V) in the C-terminal SAW subdomain of GRAS (Figure 2A). This Gly is conserved among all the known vascular plant DELLA proteins (Figure 2B), suggesting that the G576V mutation might be the cause of the reduced responsiveness to GA. When tested in a transgenic rice experiment, the overexpression of the SLR1G576V gene in wild-type T65 plants caused a severe GA-insensitive dwarf phenotype, whereas SLR1 overexpressed in the wild type showed only a moderate dwarf phenotype compared with vector control plants (Figure 2C). Based on these results, we renamed F4443 as Slr1-d4. Although many dominant mutants of DELLA proteins have been isolated, all of the mutants identified previously (including rice Slr1-d1, -d2, and -d3) contain in-frame deletions or amino acid substitutions located in the DELLA/TVHYNP motif of the N-terminal portion of the protein (Peng, et al., 1997; Dill et al., 2001; Gubler et al., 2002; Asano et al., 2009); the only exception is a Brassica mutant located in the VHIID domain (Muangprom et al., 2005). Indeed, although we have identified several mutations in the SAW domain of SLR1, so far, all of them have been loss-of-function mutants that produce a slender phenotype, and a gain-of-function dwarf mutant had not been identified until this study (Ikeda et al., 2001).
Figure 2.
The Dwarf Phenotype of F4443 Is Caused by a Mutation in the SLR1 SAW Domain, Which Leads to a Reduced Interaction with GID1.
(A) Schematic structure of SLR1. The protein in F4443 (Slr1-d4) contains a G576V substitution (SLR1G576V).
(B) Comparison of the region around G576 of DELLA proteins from various plant species. Rice (Kamiya et al., 2003) and Arabidopsis (Di Laurenzio et al., 1996) SCR, which belong to another GRAS protein family, are also shown. Zm, Zea mays; At, Arabidopsis thaliana; Sm, Selaginella moellendorffii; Os, Oryza sativa.
(C) Gross morphology of transgenic plants at harvest. FLAG-tagged SLRWT and FLAG-tagged SLR1G576V were each overproduced in wild-type T65 rice. Vector, T65 transformed with proAct-FLAG/pCAMBIA control vector. Bar = 10 cm.
(D) Degradation of SLR1 and SLR1G576V protein upon GA3 treatment in rice callus. Nipponbare and Slr1-d4 calli were incubated with 10−5 M GA3 for the indicated times, and the crude protein extracts were subject to immunoblot analysis using an anti-SLR1 antibody. The loading control of Coomassie blue (CBB) staining is shown in the bottom panel.
(E) Interaction between GID1 and SLR1G576V with (+) or without (−) 10−4 M GA4. Left, β-Gal activity detected in a liquid assay with yeast strain Y187 transformants (means ± sd; n = 3). Right, Growth of yeast strain AH109 transformants on –HIS plates. GID1 was used as bait, and SLR1 and its mutants were used as prey. SLR1 (E4-R125), DELLA/TVHYNP domain; ΔDELLA, SLR1 containing a deletion in the DELLA domain (from D39 to A55).
Because of the unusual properties of Slr1-d4, we examined the molecular mechanism of GA insensitivity conferred by this gain-of-function mutation using calli produced from wild-type and Slr1-d4 plants. Immunoblot analysis using polyclonal anti-SLR1 antibodies revealed that only slight degradation of the Slr1-d4 protein (SLR1G576V) had occurred even after 60 min, while the wild-type protein (SLR1WT) was almost completely degraded within 60 min of GA3 treatment (Figure 2D). We considered the failure of SLR1G576V degradation to be due to failure in its interaction with either GID1 or GID2. We first examined the interaction with GID1 by Y2H using two different detection systems. When we used a liquid assay (based on β-galactosidase [β-Gal] activity) to detect the interaction between GID1 and SLR1, interaction was observed only for the combination of GID1 and SLR1WT in the presence of GA; mutated SLR1s such as SLR1G576V, the N-terminal portion of SLR1 containing the DELLA/TVHYNP motif [from E4 to R125, designated SLR1 (E4-R125)], and SLR1 truncated in the DELLA domain (ΔDELLA) did not interact with GID1 (Figure 2E, left panel). On the other hand, when we performed Y2H using a plate assay, SLR1WT, SLR1 (E4-R125), and SLR1G576V all interacted with GID1, whereas ΔDELLA did not (Figure 2E, right panel). According to the current GA perception model, the interaction between GID1 and DELLA protein occurs through the DELLA/TVHYNP motif of DELLA in the presence of GA (Ueguchi-Tanaka et al., 2007; Murase et al., 2008). The results of the plate assay are consistent with this model and indicate that the N-terminal part of SLR1 containing the DELLA/TVHYNP motif is both necessary and sufficient for interaction with GID1. On the other hand, the results of the liquid assay suggest that the interaction of SLR1G576V with GID1 is different from that of SLR1WT and similar to that of the DELLA/TVHYNP motif alone. This led us to speculate that there are two types of GID1-SLR1 interaction, stable and unstable, and that the C-terminal part of SLR1 might be necessary for stable interaction with GID1 (see below).
Analysis of the GID1-SLR1 Interaction by Surface Plasmon Resonance
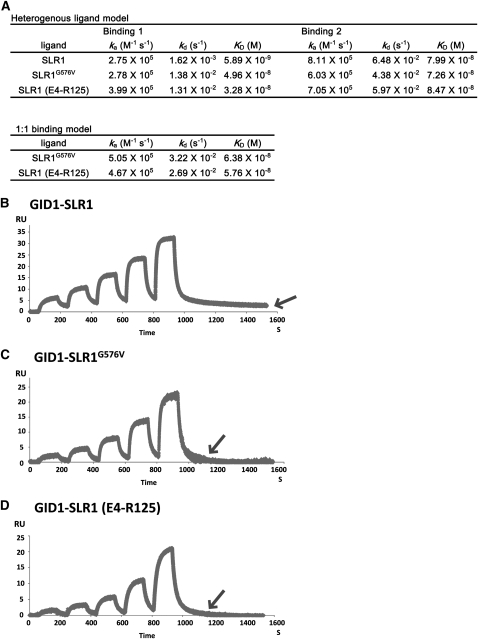
The Y2H liquid assay indicated that the GID1-SLR1G576V interaction is less stable than the GID1-SLR1WT interaction in the presence of GA (Figure 2E). To investigate the biochemical difference between these interactions, we used surface plasmon resonance (SPR). Glutathione S-transferase (GST)-fused SLR1 and its mutants were used as ligands, GID1 was used as an analyte, and the interactions were measured using the single-cycle kinetic method (Karlsson et al., 2006). For the GID1-SLR1 interaction, we applied the heterogenous ligand model and calculated the three kinetic parameters, ka (association rate), kd (dissociation rate), and KD (kd/ka, binding affinity) of the two binding sites (Figure 3A). The binding showing higher binding affinity (KD) was designated as Binding 1; the binding showing lower KD was designated as Binding 2. For GID1-SLR1G576V and GID1-SLR1 (E4-R125) interactions, kinetic parameters were calculated based on the heterogenous ligand model and the 1:1 binding model. For both interactions, the parameters of the 1:1 binding model did not differ greatly from the parameters of Binding 1 or Binding 2 of the heterogenous ligand model. Therefore, we decided to apply the heterogenous ligand model to both interactions, for ease of comparing the kinetic parameters for the GID1-SLR1 interaction.
Figure 3.
Physicochemical Analysis of the Interaction between GID1 and SLR1WT, SLR1G576V, and SLR1 (E4-R125).
(A) Various kinetic constants of GID1-SLR1, GID1-SLR1G576V, and GID1-SLR1 (E4-R125) interactions in the presence of GA4. Parameters were calculated by the heterogenous ligand model for three interactions or by the 1:1 binding model for GID1-SLR1G576V and GID1-SLR1 (E4-R125) interactions. GST-fused SLR1 and SLR1 mutant proteins were used for the analysis.
(B) to (D) Interactions between GID1 and SLR1 proteins in the presence of GA4. Note that the response unit (RU) value of the GID1-SLR1WT interaction does not return to the baseline level at the end of the reaction due to a delayed dissociation rate (B) compared with GID1-SLR1G576V (C) and GID1-SLR1 (E4-R125) (D), as shown by arrows.
(B) GID1-SLR1WT interaction.
(C) GID1-SLR1G576V interaction.
(D) GID1-SLR1 (E4-R125) interaction.
Whereas there was relatively little variation among the three parameters for Binding 2 for GID1-SLR1WT, GID1-SLR1G576V, and GID1-SLR1 (E4-R125), the parameters for Binding 1 showed large variation, indicating that the parameters for Binding 1 predominantly characterize the GID1-SLR1 interaction. When we compared the kinetic parameters of Binding 1 for the three interactions (Figure 3A), the KD value of GID1-SLR1G576V (4.96 × 10−8) was >8 times higher than that of GID1-SLR1WT (5.89 × 10−9) and similar to that of GID1-SLR1 (E4-R125) (3.28 × 10−8). This corresponds well to the results observed in the Y2H assays (i.e., that the GID1-SLR1G576V complex is less stable than GID1-SLR1WT). The unstable GID1-SLR1G576V complex (higher KD value) depends mainly on having a higher kd (1.38 × 10−2) than GID1-SLR1WT (1.62 × 10−3), indicating that rapid dissociation of the GID1-SLR1G576V complex is a major reason for the lower affinity parameter of GID1-SLR1G576V. This is also supported by comparison of the GID1-SLR1WT sensorgram (Figure 3B) to that of GID1-SLR1G576V (Figure 3C). The response unit value of GID1-SLR1G576V rapidly returned to the baseline level after washing away the GID1 analyte with buffer, while that of GID1-SLR1WT did not return to baseline even after prolonged washing (indicated by an arrow in Figure 3B). The sensorgram and the parameters of GID1-SLR1G576V are similar to those of GID1-SLR1 (E4-R125) (Figure 3D), suggesting that the interaction between GID1 and SLR1G576V is essentially the same as that of GID1-SLR1 (E4-R125). These results indicate that the GID1-SLR1G576V interaction depends almost entirely on the DELLA/TVHYNP motif but hardly at all on the GRAS domain.
Development of a System to Detect SLR1-GID2 Interactions in Yeast
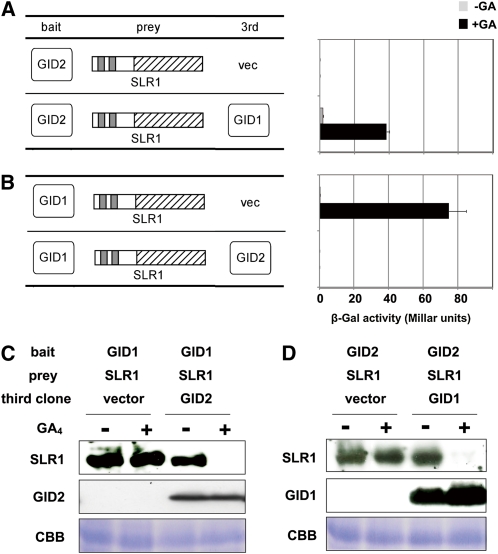
We next examined the SLR1G576V-GID2 interaction using a yeast three-hybrid assay (Y3H). When GID2 was used as bait and SLR1 as prey, no interaction was observed regardless of GA application (Figure 4A). When GID1 was expressed as a third clone, very low interaction was observed in the absence of GA, and it was drastically enhanced by GA, demonstrating that the interaction between GID2 and SLR1 occurs in a GA- and GID1-dependent manner in this assay system. Curiously, however, when we used GID1 as bait, SLR1 as prey, and GID2 as a third clone, the GID1-SLR1 interaction was not observed regardless of GA application, even although high activity was observed in the presence of GA when GID2 was replaced with an empty vector (Figure 4B). These conflicting results led us to suspect a problem with the Y3H assay itself. Because SLR1 degradation is mediated by the SCFGID2 complex in rice cells, we suspected that degradation of SLR1 might also occur in yeast cells when GID2 is present. To test this hypothesis, we analyzed the accumulation of SLR1 in yeast cells. Crude proteins were extracted from yeast cultured with or without 10−4 M GA4, and accumulation of SLR1 was detected immunologically (Figures 4C and 4D). As suspected, accumulation of SLR1 expressed as prey was drastically reduced by GA4 application in the presence of GID2, whereas in the absence of GID2, no GA-dependent disappearance was observed (Figure 4C). The same phenomenon was also observed in yeast cells expressing GID2 as bait, SLR1 as prey, and GID1 as a third protein (Figure 4D). Although the expression of bait protein was too low to be detected in either experiment, the amount of GID1 or GID2 expressed as a third clone did not change greatly upon GA application. These results demonstrate that SLR1 degradation occurs in yeast cells in a GA-dependent manner, as in plant cells, when GID1 and GID2 are coexpressed. Additionally, GID2 expressed as a third clone seems to degrade SLR1 more efficiently than when expressed as bait since interaction between GID2 and SLR1 were still observed even in the presence of GID1 and GA.
Figure 4.
GID1, GID2, and GA-Dependent Degradation of SLR1 in Yeast Cells.
(A) and (B) Interaction of SLR1-GID2 (A) and GID1-SLR1 (B) in yeast cells. Interaction of BD and AD fusion proteins in yeast cells with or without 10−4 M GA4 were scored using β-Gal activity (means ± sd; n = 3). Either GID2 or GID1 was used as bait, SLR1 was used as prey, and either GID1 or GID2 was expressed in yeast as a third clone.
(C) and (D) Accumulation of AD-HA-SLR1, HA-GID2, and HA-GID1 protein in yeast cells. Crude protein extracts from yeast grown in the absence or presence of 10−4 M GA4 were subject to immunoblot analysis and detected using the HA antibody for HA-SLR1 and HA-GID2, and anti-GID1 antibody for HA-GID1. The loading control of Coomassie blue (CBB) staining is shown in the bottom panels.
[See online article for color version of this figure.]
Identification of Essential Domains of GID2 for SLR1 Interaction
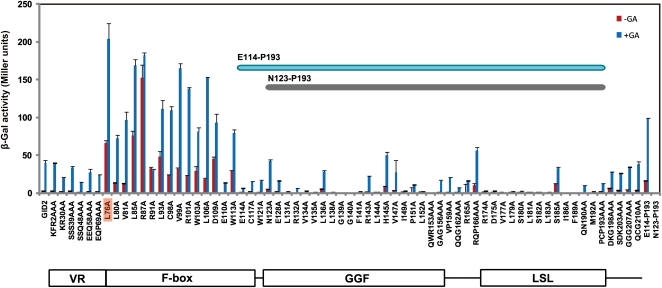
To accurately assay the SLR1-GID2 interaction in yeast, it was necessary to identify and mutate amino acids of GID2 important for SLR1 degradation, but which do not affect the interaction of GID2 with SLR1. We did this by performing Ala scanning of GID2. For this purpose, we selected amino acid residues relatively conserved among vascular plant GID2 proteins and exchanged each one with Ala (see Supplemental Figure 2 online). For residues not strictly conserved, we picked two or three contiguous amino acids as a single block, and the amino acids in each block were exchanged with the same number of Ala residues. Consequently, 68 mutagenized GID2 (mGID2) constructs were produced and used as bait proteins in the Y3H assay. As expected, most of the amino acid exchanges in the F-box domain increased β-Gal activity compared with intact GID2 (Figure 5; activity with intact GID2 is shown at the far left side), indicating that SLR1 degradation occurs in a GID2-dependent manner in yeast cells. GID2 carrying an L76A substitution (GID2L76A) showed the highest β-Gal activity, which was >5 times higher than for intact GID2. When accumulation of SLR1 was investigated in yeast expressing GID2L76A and GID1, no difference in SLR1 protein level was observed either before or after GA application (see Supplemental Figure 3 online). This demonstrates that GID2L76A can be used as a bait protein and that it eliminates SLR1 degradation without reducing its interaction with SLR1. For subsequent experiments, we used GID2L76A as bait to obtain accurate estimates of the SLR1-GID2 interaction.
Figure 5.
Ala Scanning Analysis of GID2 for SLR1-Interacting Activities.
SLR1-interacting activity of 68 mutated GID2s and the wild-type GID2 was assessed using a Y3H assay with GID2s as baits, the full-length SLR1 as prey, and GID1 expressed as a third clone (means ± sd; n = 3). Interactions were analyzed in the absence (red bars) or presence (blue bars) of 10−4 M GA4. Activity of wild-type GID2 is shown at the far left of the graph. The minimum region necessary for SLR1 interaction (E114-P193) is indicated within the graph. Activities of regions E114-P193 and N123-P193 are shown at the far right side of the graph. A schematic structure of GID2 is shown at the bottom.
The GID2 Ala scanning experiment also demonstrated that most of the conserved amino acids in the GGF and LSL domains are important for the interaction with SLR1 (Figure 5). Furthermore, the region ranging from E114-P193 containing the GGF and LSL domain was sufficient to interact with SLR1, whereas a more truncated peptide containing only amino acids N123-P193 completely lost the ability to interact with SLR1 (E114-P193 and N123-P193; bars at the far right side of Figure 5).
SLR1G576V Does Not Interact with GID2 Even in the Presence of GA and GID1
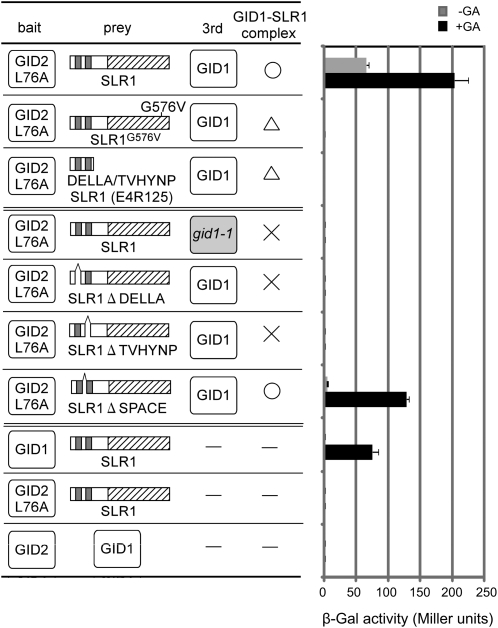
Using the improved conditions of Y3H (i.e., with GID2L76A as bait), we reexamined the interaction between GID2 and SLR1G576V. The interaction activity between GID2L76A and SLR1 was about 5 times higher than that of GID2WT-SLR1 (cf. Figure 4A and the top row of Figure 6), demonstrating that this Y3H system worked well. Even using this improved system, no interaction between GID2L76A and SLR1G576V was detected in the presence of GA and GID1, the same result seen using a truncated protein containing only the DELLA/TVHYNP domain (Figure 6, third row). This suggests that G576, located within the SAW subdomain of SLR1, is essential for binding of GID2 to SLR1. However, this alone does not demonstrate that G576 interacts directly with GID2. SLR1G576V was unable to form a stable complex with GID1 (Figures 2E and 3), which might have caused the failure of SLR1G576V to interact with GID2.
Figure 6.
Interaction of GID2L76A, SLR1, and GID1 in Yeast Cells.
Interactions of BD and AD fusion proteins with or without 10−4 M GA4 were scored by β-Gal activity (means ± sd; n = 3). For the Y3H assay, GID2L76A was used as bait, SLR1 (wild-type or mutant) was used as prey, and either GID1 or gid1-1 mutant protein was expressed as a third clone. The GA-dependent interaction of SLR1 and GID1 proteins is indicated with a circle (strong interaction) or triangle (weak interaction), and “×” indicates no interaction.
We further analyzed how interaction between SLR1 and GID1 affects the subsequent interaction with GID2. Under conditions of no interaction between GID1 and SLR1, as seen with GID1-ΔDELLA, GID1-ΔTVHYNP, and gid1-1 (mutant without GA binding activity)-SLR1, no SLR1-GID2L76A interaction was observed (Figure 6). On the other hand, GID2L76A was able to interact with SLR1-ΔSPACE, which can form a stable complex with GID1 (Ueguchi-Tanaka et al., 2007). These results support the commonly accepted model that interaction between GID1 and SLR1 via the DELLA/TVHYNP motif is a prerequisite for SLR1-GID2 interaction.
Role of GID1 in the SLR1-GID2 Interaction
Because the requirement for GID1-SLR1 formation might suggest a direct commitment of GID1 to the SLR1-GID2 interaction, we examined this possibility using a series of mutated GID1 (mGID1s) in which amino acid residues conserved among the rice GID1 protein and three Arabidopsis GID1 proteins were substituted with Ala (Ueguchi-Tanaka et al., 2007). In this experiment, we only used mGID1s that retained stable interaction activity with SLR1 because stable GID1-SLR1 interaction is necessary for SLR1-GID2 formation. Of the 21 mGID1s showing detectable interaction activity with SLR1 in a liquid Y2H assay (Ueguchi-Tanaka et al., 2007), all were able to prompt the interaction of GID2 and SLR1 in the Y3H assay (see Supplemental Figure 4 online), indicating that GID1 regions other than those involved in the SLR1 interaction are not necessary for SLR1-GID2 interaction. This result, together with the fact that GID1 did not directly interact with GID2 in the Y2H assay (Figure 6, bottom row), suggests that the primary function of GID1 in the SLR1-GID2 interaction is to act on SLR1 and make it recognizable to GID2.
Identification of SLR1 Regions Important for Interaction with GID1 and GID2
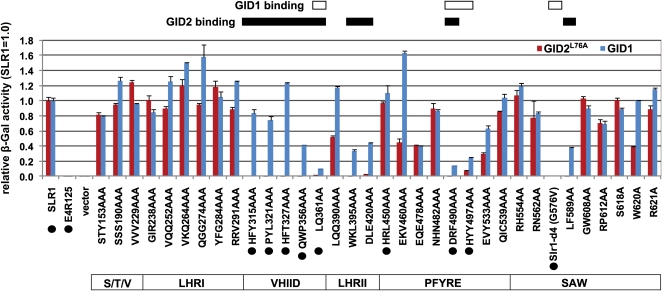
Since analysis of Slr1-d4 demonstrated the importance of the SLR1 GRAS domain for stable interaction with GID1 and also for interaction with GID2, we performed an Ala scanning experiment to learn whether there are other regions in the GRAS domain important for interaction with GID1 and GID2. According to comparative analysis of various GRAS family proteins, the GRAS domain has been divided into several subdomains: LHRI, VHIID, LHRII, PFYRE, and SAW (Bolle, 2004; see Figure 2A for a schematic of SLR1). However, the functional relevance of each of these subdomains remains to be discovered. In DELLA proteins, the DELLA/TVHYNP motif and the GRAS domain are connected by the poly S/T/V domain, a domain that shows large sequence variation among DELLA proteins. For Ala scanning, we mutagenized amino acids within the poly S/T/V domain and the GRAS domain that were conserved across the vascular plant DELLA proteins and that provided even coverage of all the subdomains (see Supplemental Figure 5 online). Based on these criteria, 41 mutagenized SLR1 proteins (mSLR1s) were produced and subjected to Y2H or Y3H analyses to investigate the interaction of mSLRs with GID1 or GID2, respectively (Figure 7). Out of 41 mSLR1s tested, seven constructs did not produce SLR1 proteins in yeast cells and were therefore not tested further (shown by red boxes in Supplemental Figure 5 online).
Figure 7.
Ala Scanning Analysis of SLR1 for GA-Dependent GID2- and GID1-Interacting Activities.
Interacting activities between 34 mutated SLR1s and GID1 or GID2L76A are shown by blue and red bars, respectively (means ± sd; n = 3). SLR1WT and SLR1 (E4-R125) were used as controls (far left). Activities are shown as a relative rate, with activities of wild-type SLR1 set as 1. To measure interactions between SLR1s and GID1, a Y2H assay was performed using GID1 as bait and the mutated SLR1 as prey in the presence of 10−4 M GA4. For interactions between SLR1s and GID2, a Y3H assay was performed using GID2L76A as bait and the mutated SLR1 as prey in the presence of GID1 and 10−4 M GA4. The structure of the SLR1 GRAS domain is shown at the bottom of the figure, and regions important for SLR1 to interact with GID1 or GID2 are shown in rectangles at the top. SLR1 mutants, which were analyzed with SPR (Table 1), are marked with black circles.
Ala exchanges of residues in the poly S/T/V and LHRI regions did not affect the interaction of SLR1 with either GID1 or GID2 (Figure 7), indicating that poly S/T/V and LHRI are not important for this interaction. On the other hand, four out of five exchanges of residues in the VHIID subdomain specifically reduced the interaction with GID2 (with only minor effects on the interaction with GID1), suggesting that this subdomain is important for the interaction with GID2. The only exception was LQ361-2, the most C-terminal region of the VHIID subdomain analyzed, which also showed reduced interaction with GID1 when substituted with Ala. Reduced interaction with GID2 was also observed for substitutions in the LHRII region, with partial reduction of GID1-SLR1 interaction also observed. This indicates that LHRII is also important for interaction with GID2 and may be partially involved in the stable interaction with GID1. In the case of the PFYRE and SAW subdomains, the overall trend of the mutant proteins is apparently different from that of the previous four subdomains, and there are two valleys of interaction with GID1 in each one. Exchanges of DRF490-2AAA or HYY497-9AAA in the PFYRE subdomain dramatically reduced the interaction with GID1 and nearly eliminated GID2 interaction. In the SAW subdomain, G576V abolished interaction with both GID1 and GID2, as previously observed. LF589-590AA eliminated the interaction with GID2, but the interaction with GID1 remained at about half the wild-type level.
We also analyzed the parameters of some of the mSLR1-GID1 interactions by SPR (Table 1). The parameters for Binding 2 of the GID1-SLR1 interaction did not differ much among these mSLR1s. On the other hand, the parameters for Binding 1 differed depending on the mutant protein. Overall, the mutant SLR1 proteins with higher GID1 interaction activity in the Y2H assay (Figure 7) had lower kd and KD values, whereas mutant proteins with lower GID1 interaction activities in the Y2H assay showed higher kd and KD. This confirms that Y2H:mSLR1s showing higher β-Gal activity in Y2H (e.g., HFY315-7AAA, PYL321-3AAA, HFT327-9AAA, QWP356-8AAA, and HRL450-2AAA) can form stable complexes with GID1, while mSLR1s showing lower activity (e.g., LQ361-2AA, DRF490-2AAA, and HYY497-9AAA) form unstable complexes.
Table 1.
Various Kinetic Constants of the Interaction between GID1 and mSLR1s Using SPR
| Binding 1 |
Binding 2 |
||||||
| Analyte | Ligand | ka (M−1 s−1) | kd (s−1) | KD (M) (kd/ka) | ka (M−1 s−1) | kd (s−1) | KD (M) (kd/ka) |
| GID1 | SLR1WT | 2.75 × 105 | 1.62 × 10−3 | 5.89 × 10−9 | 8.11 × 105 | 6.48 × 10−2 | 7.99 × 10−8 |
| GID1 | HFY315-7AAA | 2.43 × 105 | 1.30 × 10−3 | 5.36 × 10−9 | 5.19 × 105 | 4.67 × 10−2 | 9.00 × 10−8 |
| GID1 | PLY321-3AAA | 2.43 × 105 | 1.13 × 10−3 | 4.65 × 10−9 | 5.22 × 105 | 5.15 × 10−2 | 9.87 × 10−8 |
| GID1 | HFT327-9AAA | 2.35 × 105 | 1.95 × 10−3 | 8.29 × 10−9 | 5.71 × 105 | 5.39 × 10−2 | 9.44 × 10−8 |
| GID1 | QWP356-8AAA | 1.79 × 105 | 9.64 × 10−4 | 5.39 × 10−9 | 6.90 × 105 | 6.23 × 10−2 | 9.03 × 10−8 |
| GID1 | HRL450-2AAA | 1.31 × 105 | 1.06 × 10−3 | 8.09 × 10−9 | 6.50 × 105 | 6.09 × 10−2 | 9.37 × 10−8 |
| GID1 | SLR1 (E4-R125) | 3.99 × 105 | 1.31 × 10−2 | 3.28 × 10−8 | 7.05 × 105 | 5.97 × 10−2 | 8.47 × 10−8 |
| GID1 | SLR1G576V | 2.78 × 105 | 1.38 × 10−2 | 4.96 × 10−8 | 6.03 × 105 | 4.38 × 10−2 | 7.26 × 10−8 |
| GID1 | LQ361-2AA | 1.65 × 105 | 1.48 × 10−2 | 8.97 × 10−8 | 7.29 × 105 | 6.19 × 10−2 | 8.49 × 10−8 |
| GID1 | DRF490-2AAA | 4.35 × 105 | 1.01 × 10−2 | 2.32 × 10−8 | 5.75 × 105 | 5.26 × 10−2 | 9.15 × 10−8 |
| GID1 | HYY497-9AAA | 5.19 × 105 | 7.01 × 10−3 | 1.35 × 10−8 | 5.89 × 105 | 5.44 × 10−2 | 9.24 × 10−8 |
Transgenic Plants Expressing Mutated SLR1 Proteins (mSLR1s)
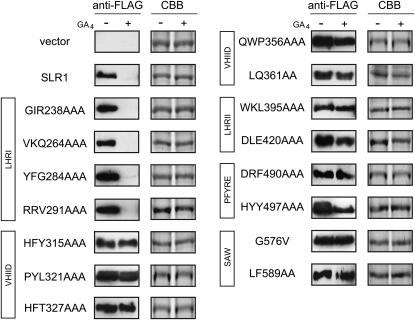
To confirm that the interaction pattern of mSLR1s with GID1 and GID2 in yeast and in vitro mimics their degradation in planta, we selected 15 mSLR1s that interacted with GID1 and GID2 in different manners and expressed each of them in a wild-type rice background. We analyzed the degradation rate of these mSLR1s in rice callus, which does not contain endogenous GA and is therefore a good system to evaluate the effect of exogenously treated GA (Itoh et al., 2005). Degradation of mSLR1 proteins with mutations in the LHRI domain (GIR238-240AAA, VKQ264-6AAA, YFG284-6AAA, and RRV291-3AAA) was similar to SLR1: treatment with 10−5 M GA4 for 12 h led to almost complete disappearance (Figure 8). Other mSLR1s that showed reduced interaction with GID1 and/or GID2 showed reduced degradation compared with SLR1, demonstrating that a defect in interaction with either GID1 or GID2 leads to reduced degradation of SLR1 in planta.
Figure 8.
Degradation of Ala-Mutated SLR1 Proteins in Rice Calli Treated with GA4.
Wild-type T65 rice calli overproducing FLAG-tagged SLR1 mutants with changes in the domains indicated in boxes were incubated with or without 10−5 M GA4 for 12 h, and the crude protein extracts were subject to immunoblot analysis using anti-FLAG-tag antibody. The loading control of Coomassie blue (CBB) staining is shown in the right-hand panels.
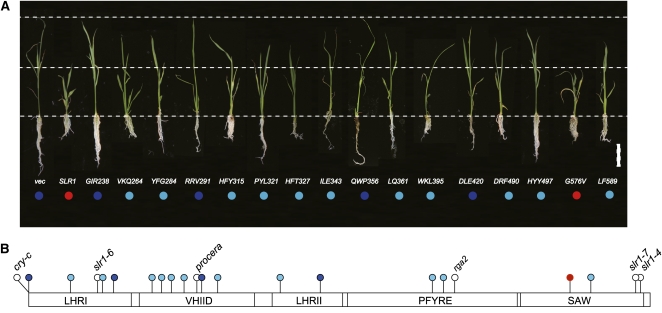
We also examined the suppressive activity of mSLR1s by examining the height of transformants expressing each type of mSLR1 compared with wild-type SLR1. Transformants were grown in the presence of uniconazole, a GA synthesis inhibitor, to control for the differences in the degradation rate of each mSLR1 in the presence of endogenous GA. We produced multiple independent transformants (>10 plants) for each mSLR1. Transformants expressing any of the mSLR1s showed milder dwarfism than the plants expressing SLR1WT (Figure 9A, second from left; see Supplemental Figure 6 online), with the exception of SLR1G576V (Figure 9A, red circles). Some of the mSLR1 transformant types almost completely lost suppressive activity against GA action (Figure 9A, dark-blue circles) and others partially lost this activity (Figure 9A, light-blue circles). This indicates that all of the GRAS subdomains are important for the suppressive function of SLR1 (Figure 9B). This corresponds well with the previous observations that in-frame or amino acid substitution mutations in various regions of the GRAS domain lead to loss-of-function mutants in various DELLA proteins (Silverstone et al., 1998; Ikeda et al., 2001; Bassel et al., 2008; Weston et al., 2008; Figure 9B).
Figure 9.
Regions Necessary for Repression Activity of SLR1 Are Scattered within the GRAS Domain.
(A) Gross morphology of transgenic seedlings grown under GA-deficient conditions. Seedlings were grown in the presence of 10−6 M uniconazole (an inhibitor of GA synthesis). Wild-type and mSLR1s fused with FLAG tag were overproduced in wild-type T65 rice. vec, T65 transformed with proAct-FLAG/pCAMBIA control vector. Repression activity of SLR1 and mSLR1 proteins was assessed by comparing the height of each transgenic plant to wild-type SLR1 transformants. Three-week-old seedlings, which exhibit the typical phenotype obtained for each protein tested, are shown. Red circle, level of repression activity similar to SLR1WT; light blue, repression activity decreased but still retained; dark blue, repression activity is almost eliminated. Bar = 5 cm.
(B) Results of repression activity in (A) were plotted onto the GRAS domain of SLR1. Mutation sites of loss-of-function mutants reported in DELLA proteins are also plotted. For mutants that are not derived from rice, the corresponding site in rice is plotted. Definition of colored circles is the same as in (A), except mutation sites of mutants from other studies are shown in white. cry-c, Pisum sativa DELLA protein CRY mutant (Weston et al., 2008); procera, Solanum lycopersicum DELLA protein PROCERA mutant (Bassel et al., 2008); rga2, Arabidopsis RGA mutant (Silverstone et al., 1998).
GA- and GID1-Dependent SLR1-GID2 Interaction in Vitro and in Vivo
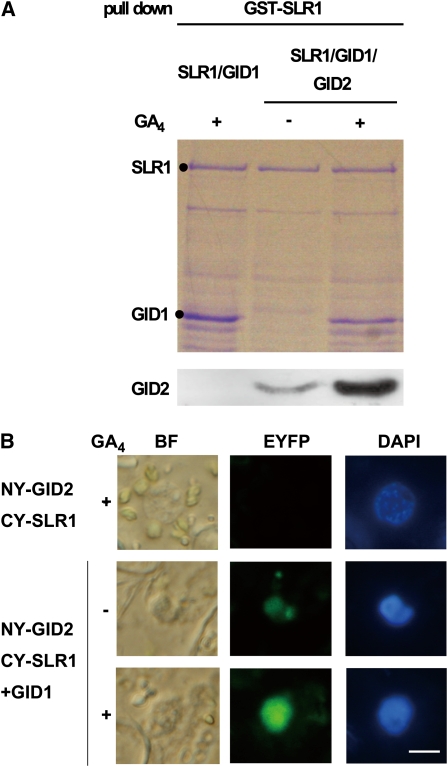
We next performed an in vivo pull-down assay to confirm the GA- and GID1-dependent SLR1-GID2 interaction in planta. Prior to the in vivo experiment, we conducted an in vitro pull-down assay using GID2, SLR1, and GID1 recombinant proteins produced in Escherichia coli. These proteins were produced together in E. coli with or without 10−4 M GA4, and subsequently the GST-SLR1 complex was purified. The expression of hemagglutinin epitope (HA)-tagged GID2 was low, so an HA antibody was used for detection. GID1 copurified with GST-SLR1 in a GA-dependent manner (Figure 10A) and the amount of GID2 copurified with GST-SLR1 was much greater in the presence than in the absence of GA. Interaction in the absence of GA has also been observed for Arabidopsis F-box protein SLY1 and DELLA protein GAI (Dill et al., 2004; Fu et al., 2004), suggesting that low levels of GA-independent interaction may be a common characteristic of F-box protein-DELLA protein interactions in vitro.
Figure 10.
GA-Dependent Interaction between GID2 and SLR1 in Vitro and in Vivo.
(A) In vitro pull-down assay showing GA-dependent interaction between GST-SLR1 and T7-tagged GID1 and between GST-SLR1 and HA tagged-GID2. GST-SLR1, T7 tagged-GID1, and HA tagged-GID2 were expressed together in E. coli and purified with glutathione beads. E. coli expressing GST-SLR1 and T7-tagged GID1 was used as a control. HA-tagged GID2 was detected with anti-HA antibody.
(B) BiFC analysis of in vivo interaction between GID2 and SLR1 in N. benthamiana leaf epidermis (Abe et al., 2005). BF, bright-field image; EYFP, EYFP fluorescence; DAPI, 4',6-diamidino-2-phenylindole; NY-GID2 CY-SLR1, expression of N•EYFP-GID2 and C•EYFP-SLR1 without GID1; NY-GID2 CY-SLR1+GID1, expression of N•EYFP-GID2 and C•EYFP-SLR1 with nontagged GID1. Leaves were sprayed with 10−4 M GA4 dissolved in ethanol (+) or with ethanol alone (–) 10 min before observation of the signals. Bar = 10 μm.
To examine the GID1-SLR1-GID2 interaction in vivo, we observed the GA-dependent interaction between GID2 and SLR1 using bimolecular fluorescence complementation (BiFC) (Abe et al., 2005). Cell suspensions of Agrobacterium tumefaciens carrying N-terminal enhanced yellow fluorescent protein (N•EYFP)-GID2 and C-terminal enhanced yellow fluorescent protein (C•EYFP)-SLR1 constructs were infiltrated into Nicotiana benthamiana leaf epidermal cells with or without Agrobacterium carrying the untagged GID1 construct. The YFP signal, caused by an interaction between N•EYFP-GID2 and C•EYFP-SLR1, was only detected in the leaf when Agrobacterium carrying GID1 was coinfiltrated (Figure 10B). Consistent with the result in yeast cells, application of GA4 enhanced the YFP signal. These results demonstrate that GID2 interacts with SLR1 in a GID1- and GA-dependent manner in planta, as predicted by the events that were observed in yeast (Figure 6).
DISCUSSION
In this study, we aimed to clarify the molecular mechanism whereby SLR1 is degraded following the GID1-DELLA/TVHYNP motif interaction (i.e., how SLR1 is targeted by GID2).
The GRAS Domain of SLR1 Stabilizes the Interaction between GID1 and SLR1
Using the mutant protein SLR1G576V, we demonstrated that interaction between GID1 and the DELLA/TVHYNP motif alone is insufficient to allow SLR1 to be recognized by GID2. The SPR experiment further showed that the GRAS domain of SLR1 functions to stabilize the interaction between SLR1 and GID1 by decreasing SLR1’s dissociation rate (kd). This suggests that SLR1 interacts with GID1 not only through the DELLA/TVHYNP motif but also through the GRAS domain. Since the GRAS domain alone cannot interact with GID1, the interaction of the DELLA/TVHYNP motif with GID1 may cause a conformational change of SLR1, making some of the GRAS domain region available to interact with GID1 to form a stable complex. In this study, we identified several possible GID1-interacting sites in the SLR1 GRAS domain (e.g., LQ361-2, DRF490-2, and HYY497-9), implying that there are several GID1-interacting sites within SLR1. It is interesting, however, that all three kinetic parameters of SLR1G576V are similar to those of the DELLA/TVHYNP motif alone, even although SLR1G576V is expected to contain all of the GID1-interacting sites except for G576. This leads us to speculate that the G576V substitution may inhibit the conformational change of SLR1 after the DELLA/TVHYNP motif–GID1 interaction. The G576V mutation may be unique in terms of conferring almost complete loss of interaction with GID1 and subsequent loss of interaction with GID2 but having no defect in SLR1 suppressive activity. By contrast, all of the other SLR1 mutations produced in this study that affected its interaction with GID1 or GID2 also caused some defects in SLR1 suppressive activity.
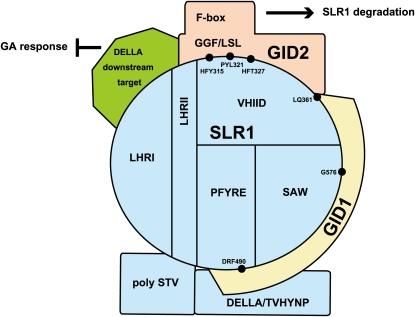
Domain Analysis of GA Signaling Components and a Model of GID1-SLR1-GID2 Complex Formation
We analyzed the regions of each protein important for the formation of the GID1-SLR1-GID2 complex. In GID2, the GGF and LSL domains are necessary for the interaction with SLR1, whereas the F-box is not very important for SLR1 interaction, and the VR domain is not necessarily required for GID2 function (Figure 5). These results are consistent with previous complementation tests of the gid2 mutant, in which the authors produced a GID2 deletion series (ΔVR-, ΔF-box-, ΔGGF-, and ΔLSL-GID2; Gomi et al., 2004).
For GID1, all of the mutated GID1 proteins that retained the ability to stably interact with SLR1 were able to prompt the SLR1-GID2 interaction when expressed as a third clone in Y3H (see Supplemental Figure 4 online). These results indicate that GID1 regions other than those involved in the GID1-SLR1 interaction are not necessary for the SLR1-GID2 interaction. This suggests that GID2 does not directly interact with GID1, or if it does, GID2 recognizes the GID1-SLR1 interacting sites.
In SLR1, the poly S/T/V and LHRI subdomains might not be involved in the interaction with either GID1 or GID2 (Figure 11). However, previous observations in transgenic plants showed that deletion of the whole LHRI domain results in GA insensitivity (Itoh et al., 2002; Wang et al., 2009). This conflicting result might be due to deletion of the LHRI domain affecting the overall structure of SLR1; another possibility is that LHRI regions other than those analyzed in this study are important for degradation.
Figure 11.
Molecular Model for the Formation of the GID1-SLR1-GID2 Complex.
The blue circle at the center of the diagram represents the GRAS domain of SLR1. See text for details.
On the other hand, VHIID and LHRII seem to be preferentially involved in the interaction with GID2 and might be the direct GID2 binding site (Figure 11). The C-terminal region of the VHIID subdomain (LQ361-2) is also involved in the interaction with GID1. Previously, Muangprom et al. (2005) identified a mutant in Brassica rapa that shows reduced degradation of DELLA protein. This mutant contains a Q-to-R mutation in the VHIID domain at the position corresponding to the Q362 position of rice SLR1, supporting that the C-terminal region of the VHIID domain (including LQ361-2) is involved in the stable interaction with GID1, although another possibility is that this region of the Brassica DELLA protein is involved in the interaction with the F-box protein. Our data also suggest the involvement of the PFYRE (DRF490-2) and SAW (G576) domains in the stable SLR1-GID1 interaction. In particular, G576, the mutation found in Slr1-d4, is expected to make a large contribution to the stable binding between GID1 and SLR1 compared with other identified sites, since G576V diminishes the interaction of SLR1 with GID2 in yeast cells.
In contrast with the pattern of GID1 and GID2 interactions, regions important for the suppressive activity of SLR1 were scattered all over the GRAS domain and were difficult to localize to specific regions. This suggests that the overall structure of SLR1 is important for the expression of its suppressive function.
Recently, PHYTOCHROME INTERACTING FACTOR3 (PIF3) and PIF4 of potato (Solanum tuberosum) and Arabidopsis were shown to interact with their own DELLA proteins (St RGA and At RGA, respectively), and further analysis in Arabidopsis reported that this interaction inhibited hypocotyl elongation (de Lucas et al., 2008; Feng et al., 2008). Interaction between potato RGA and PIF4 requires the LHRI domain of RGA (de Lucas et al., 2008), suggesting that one of the roles of LHRI is to interact with the direct downstream target of DELLA protein. Even though all of the exchanges of conserved amino acids with Ala caused decreases in SLR1 suppressive activity (Figure 9), the results of Ala scanning of SLR1 also indicated that the LHRI subdomain has a unique function. That is, LHRI is only involved in SLR1 suppressive activity, whereas other subdomains of GRAS are involved both in its suppressive function and in the interaction of SLR1 with GID1 and/or GID2. This suggests that the LHRI subdomain may be directly and specifically involved in SLR1 suppressive function by interacting with other protein(s), such as PIFs, to form the suppressive complex for GA action. By contrast, mutations in other subdomains may indirectly affect SLR1 suppressive function through a change in its overall structure, leading to deactivation of the repressive activity (Figure 9) but some still able to interact with GID1 and/or GID2 (Figure 7). It is interesting to speculate that conformational change of SLR1 after GID1-DELLA/TVHYNP motif interaction leads to loss of suppression function of SLR1 but in turn allows the GRAS domain to interact with GID1 and GID2 for degradation. This model is partly supported by the recent finding that binding of GID1 to SLR1 per se (i.e., in the absence of GID2 activity) results in the derepression of the repressive activity.
Target recognition of F-box protein must be strictly regulated to ensure that unwanted degradation of target protein does not occur. In contrast with recognizing posttranslational modifications of target proteins such as phosphorylation, oxidation, and glycosylation, our results suggest that GID2 recognizes SLR1 following GA-dependent GID1-SLR1 complex formation and that the GRAS domain bound to GID1 might serve as a recognition signal for GID2. The recognition of such a protein–protein interaction by an F-box protein is rare, and the molecular mechanism for this type of recognition is less understood than for recognition based on posttranslational modifications. One known example of such regulation is the MAT transcription factors of Saccharomyces cerevisiae (Johnson et al., 1998). MATα2 masks its degrading signal (degron) by heterodimerization with MATa1. The degron is exposed when MATα2 does not form a complex with MATa1, leading to degradation through the proteasome. Intriguingly, the output of this system and that of GA signaling are opposite: heterodimerization inhibits MATα2 recognition, whereas GID1-SLR1 complex formation is suggested to lead to SLR1 recognition by GID2. To our knowledge, such complex formation–dependent recognition of F-box protein has not been reported for any SCFs. Furthermore, compared with the MATα2 degradation system, recognition of SLR1 by GID2 requires an additional step: GID1 must first bind with GA to allow GID1-SLR1 formation. Although auxin and jasmonate signaling also use the ubiquitin-proteasome system, these phytohormones are considered to promote the direct connection between the F-box protein (receptor) and their targets (Chini et al., 2007; Tan et al., 2007; Thines et al., 2007) and thus are simpler than the GA perception system. Clarifying the molecular mechanism underlying the recognition of SLR1 by GID2 may explain why plants have evolved such a complex system to perceive GA signal.
METHODS
Plant Materials
The Slr1-d4 mutant was identified in a mutant library of Oryza sativa cv Nipponbare mutagenized by MNU. MNU treatment was conducted according to the method of Suzuki et al. (2008). Panicles were dipped in 1 mM MNU solution for 45 min at 25°C at ~6, 8, 10, 12, 14, 16, 18, and 20 h after flowering. Subsequently, panicles were washed with tap water, and the flowers grown to seed maturation were harvested.
Phylogenetic Analysis
The amino acid sequences of GID2 and its homologs, and the DELLA proteins, were aligned with ClustalW version 1.81 using the default parameters (Thompson et al., 1994; http://align.genome.jp/), followed by manual alignment. Alignments were drawn with BoxShade (http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html) using the default parameters.
GA Induction in Shoot Elongation
Seeds of the wild-type cv Nipponbare and Slr1-d4 were sterilized with 2.5% NaClO for 30 min, washed five times in sterile distilled water, and incubated at 4°C for 1 d. The seeds were then placed on 1% agar plates containing various concentrations of GA3 and grown under continuous fluorescent light at 30°C. After 10 d, the length of the second leaf sheath of each plant was measured.
Immunoblot Analysis of SLR1 Protein
Calli of cv Nipponbare and Slr1-d4 were used to detect GA-dependent degradation of SLR1. The calli were transferred to fresh N6D solid medium. After 3 d, the calli were treated with 10−5 M GA3 solution containing 0.02% Tween 20 at room temperature. The calli were then immediately frozen at −80°C until used for protein gel blot analysis.
Crude protein extracts of rice calli were prepared by grinding with liquid nitrogen using a mortar and a pestle in the presence of sea sand (particle size 425 to 850 μm; Wako Pure Chemical). An equal volume of 2× sample buffer (135 mM Tris-HCl, pH 6.8, 200 mM DTT, 4% SDS, 0.2% bromophenol blue, and 20% glycerol) was added, and samples were boiled for 5 min. After incubation for 5 min on ice, samples were centrifuged and the supernatants were collected. Protein samples were separated by 7.5% SDS-PAGE and transferred to Hybond enhanced chemiluminescence nitrocellulose membrane (GE Healthcare). To detect SLR1 and SLRG576V, the blots were treated with 5% skim milk in TBST (0.1% Tween 20 in 2 mM Tris-HCl, pH 7.6, and 13.7 mM NaCl) for 1 h and subsequently incubated with anti-Os SLR1 antiserum (1:5000 dilution) raised in rabbit (Itoh et al., 2002) for 1 h. Blots were washed three times with TBST for 15 min each. The membrane was incubated with goat anti-rabbit IgG horseradish peroxidase–conjugated secondary antibody (1:10,000 dilution) for 45 min, and blots were washed following the same procedure described above. All reactions were conducted at room temperature. Peroxidase activity was detected according to the instruction manual of SuperSignal West Dura Extended Duration Substrate (Pierce).
Plasmid Construction
Sequences of primers used in this study are listed in Supplemental Table 1 online. All PCR fragments were sequenced to confirm that no mutations were introduced.
Plasmids Used in Yeast Experiments
For the Y2H assay, pGADT7 (Clontech) and pGBKT7 (Clontech) were used as expression vectors, and for the Y3H assay, pGADT7 (Clontech) and pBRIDGE (pBr: Clontech) were used. SLR1G576V and SLR1 (E4-R125R) were PCR amplified using cDNA synthesized from Slr1-d4 and wild-type rice mRNA, respectively, and cloned into the NdeI-EcoRI and EcoRI-SmaI sites of pGADT7, respectively. Construction of pGBKT7 GID1, pGADT7-SLR1, -ΔDELLA, -ΔTVHYNP, and -ΔSPACE were described previously (Ueguchi-Tanaka et al., 2007). pGADT7 GID1 was constructed by PCR amplifying GID1 cDNA with EcoRI sites at both ends and cloning the resulting fragment into the EcoRI site of pGADT7. For the construction of pBr BD-GID2 and pBr BD-GID1, GID2 and GID1 cDNAs with an EcoRI and BamHI site at each end were cloned into the pBr vector to produce binding domain (BD)-fused constructs. pBr BD-GID2/3rd-GID1 and pBr BD-GID1/3rd-GID2 were constructed by PCR amplifying GID1 and GID2 cDNAs with BglII sites at both ends and cloning the resulting fragments into the BglII sites of pBr BD-GID2 and pBr BD-GID1, respectively. For the construction of the Ala-mutated SLR1s (mSLR1s) in the pGADT7 vector and the Ala-mutated GID2s (mGID2s) in the pBRIDGE vector carrying GID1, full-length wild-type SLR1 or GID2 was amplified by PCR and cloned into the pBluescript II SK+ vector (Stratagene). Full-length SLR1 or GID2 cDNA was then PCR amplified using one set of mutagenized primers corresponding to each mutation. The parental methylated and hemimethylated DNA in the PCR reaction mixture were digested with DpnI, and the mutated SLR1 or GID2 cDNA that could not be digested with DpnI was transformed into Escherichia coli XL10-Gold (Stratagene). After sequencing, each mutated SLR1 or GID2 cDNA was digested with EcoRI-SmaI and cloned into pGADT7 or pBRIDGE containing full-length GID1 cDNA downstream of the MET25 promoter (pBr BD/ 3rd-GID1, construction that can express GID1 protein as a third clone), respectively, to produce pGADT7-mSLR1s and pBr BD-mGID2s/3rd-GID1, respectively. pBr BD-GID2L76A was constructed by PCR amplifying GID2L76A with EcoRI and SmaI sites at 5′ and 3′ ends, respectively, and cloning the resulting fragment into the EcoRI-SmaI site of pBr vector. For the construction of mutated GID1s (including gid1-1:mGID1s) in pBr BD-GID2L76A, previously constructed mutated GID1s (Ueguchi-Tanaka et al., 2007) were used as DNA templates to be amplified by PCR and cloned downstream of the MET25 promoter of pBr BD-GID2L76A to produce pBr BD-GID2L76A/3rd-mGID1s. In detail, mGID1s carrying a SmaI site at each end were digested with SmaI and ligated into pBr BD-GID2L76A plasmid digested with BglII and blunted with T4 DNA polymerase (Takara).
Plasmids Used in SPR Analyses
Construction of pET32a GID1 was as previously described (Ueguchi-Tanaka et al., 2007). Intact and mutant SLR1 used in the SPR were expressed in the wheat germ extract system using the pEU101 vector (CellFree Sciences). GST-SLR1, GST-mSLR1s, and GST-SLR1 (E4-R125) in pEU101 were constructed by the following procedure. SLR1, mSLR1s, or SLR1 (E4-R125) were PCR amplified and cloned into the EcoRI-SmaI site of pGEX 6P-1 (GE Healthcare). These plasmids were used as templates to amplify GST-SLR1, GST-mSLR1s, and GST-SLR1 (E4-R125) by PCR. Each PCR product was digested with the appropriate restriction enzymes and cloned into pEU101.
Plasmids for Transgenic Plants
For rice transformation, proAct-FLAG/pCAMBIA was constructed and used as a binary vector. In detail, proAct1 of the Hm2 vector was PCR amplified and cloned into the EcoRI-XbaI site of pCAMBIA1380 to produce proAct/pCAMBIA. 3XFLAG was synthesized containing XbaI and SmaI sites at the 5′ and 3′ ends, respectively, and cloned into proAct/pCAMBIA to produce proAct-FLAG/pCAMBIA. SLR1, mSLR1, and SLR1 (E4-R125) were PCR amplified and cloned into the SmaI-SpeI site of proAct-FLAG/pCAMBIA.
Plasmids Used for in Vitro and in Vivo SLR1-GID2 Interaction
pGEX GST-SLR1+rbs-cMyc-GID1-10xHIS, a plasmid that polycistronically expresses SLR1 and GID1, was constructed by the following procedure. SLR1 cDNA was inserted into the pGEX 4T-1 vector (GE Healthcare) at the EcoRI site to produce pGEX GST-SLR1.The cMyc-GID1 fragment was amplified from pGBKT7 GID1 and cloned into the KpnI and BamHI sites of pET52b (Novagen) to produce pET52b cMyc-GID1. The rbs-cMyc-GID1-10xHIS region of pET52b cMyc-GID1 was PCR amplified and ligated into the SmaI site of pGEX GST-SLR1 to produce pGEX GST-SLR1+rbs-cMyc-GID1-10xHIS. pACYC184 T7-3xHA-GID2+rbs-SKp15, a plasmid that polycistronically expresses GID2 and Skp15, was constructed by the following procedure. The Os Skp15 fragment was PCR amplified using pGADT7 Os Skp15 as a template (Gomi et al., 2004) and cloned into the BamHI site of pET3d (Novagen) to produce pET3d-Skp15. rbs-Skp was PCR amplified using pET3d-Skp15 as template and cloned into the XhoI site of pGEX4T-1 to produce pGEX rbs-Skp15. 3xHA-GID2 was PCR amplified using 3xHA-GID2 in Hm2 vector as template (Gomi et al., 2004) and cloned into the EcoRI-SmaI sites of pGEX rbs-Skp15 to produce pGEX 3xHA-GID2+rbs-Skp15. The 3xHA-GID2+rbs-Skp15 fragment was PCR amplified with EcoRV sites at both ends and cloned into the EcoRV site of pET15b (Novagen) to produce pET15b T7-3xHA-GID2+rbs-Skp15. This plasmid was digested with SphI and HindIII and subcloned into the SphI-HindIII site of pACYC (Nippongene) to finally produce pACYC T7-3xHA-GID2+rbs-Skp15.
For constructs used in the BiFC experiment, the N-terminal half of the EYFP clone without a stop codon and the C-terminal half of the EYFP clone without a stop codon were kindly provided by T. Araki (Abe et al., 2005). To construct N_EYFP-GID2 (NY-GID2), the GID1 fragment of NY-GID1/pBI121 (Ueguchi-Tanaka et al., 2007) was replaced with the GID2 cDNA fragment. In detail, NY-GID1/pBI121 was digested with XbaI and SacI to cut out the GID1 fragment, and the GID2 cDNA fragment containing XbaI and SacI sites at its 5′ and 3′ ends was cloned into the digested plasmid. Construction of C_EYFP-SLR1 (CY-SLR1) containing full-length SLR1 was previously described (Ueguchi-Tanaka et al., 2007). The GID1 cDNA fragment was cloned into the SmaI-SacI site of pBI121 to produce GID1/pBI121.
Yeast Two-Hybrid and Three-Hybrid Assays
The Y2H assay was performed as described previously (Ueguchi-Tanaka et al., 2005) using BD Matchmaker Two-Hybrid System 3 (Clontech). Vector cassettes for DNA-BD and -AD (activation domain) were used as negative controls, and Saccharomyces cerevisiae strains AH109 and Y187 were used as the hosts for the plate assay and the liquid assay, respectively. 10−1 M GA4 dissolved in ethanol (GA4+ treatment) or ethanol alone (GA4− treatment) was added to the culture medium at a dilution rate of 1/1000. Expression of AD fusion proteins was confirmed by immunoblot analysis using anti-HA (Sigma-Aldrich) antibodies. Experiments were independently repeated at least three times. For the Y3H assay, the Matchmaker Yeast Three-Hybrid System (Clontech) was used. S. cerevisiae strain SFY576 was used as the host, and GA4 was added to the culture medium as described for the Y2H assay. Details of the methods used for the yeast assays can be found in the manufacturer’s instructions (Yeast Protocols Handbook PT3024-1; Clontech). Accumulation of HA-tagged AD fusion SLR1 protein, or HA-tagged GID1 and GID2 proteins expressed as a third clone, was detected by immunoblot analysis using anti-HA (Sigma-Aldrich) antibodies followed by a mouse secondary antibody (1:10,000; Pierce).
In Vitro Translation of GST-SLR1 and GST-mSLR1 mRNA
GST-SLR1 and GST-mSLR1s were transcribed in vitro using the reagents and enzymes supplied in the ENDEXT Wheat Germ Expression Premium Kit (CellFree Sciences). The transcribed mRNAs were then translated using the same kit. All the procedures were conducted according to the manufacturers’ instructions (http://www.cfsciences.com/jp/pdf/ENDEXT_PreExpKit_ver1-7.pdf). After translation, GST-SLR1 and GST mutant SLR1s were purified using glutathione Sepharose 4B beads as previously described (Ueguchi-Tanaka et al., 2007). The purified proteins were applied to a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with HBS-EP buffer (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, and 0.005% [v/v] Tween 20] and eluted with the same buffer at a flow rate of 0.5 mL per minute. The peak fraction containing SLR1 or SLR1 mutant protein was analyzed using SPR.
Affinity and Kinetic Studies
The interaction of immobilized GST-SLR1, GST-SLR1 (E4-R125), and GST-mSLR1 proteins with GID1 was assayed by a method based on SPR (Karlsson et al., 2006) using a biosensor instrument (Biacore T100; GE Healthcare). Binding was measured using the single-cycle kinetic method. Anti-GST antibody was immobilized to the CM5 sensor chip using a GST fusion capture kit (GE Healthcare), followed by immobilization of purified GST-SLR1, GST-SLR1 (E4-R125), or GST-mSLR1 proteins. Trx-HIS-GID1 expressed in E. coli was used as an analyte (Ueguchi-Tanaka et al., 2007). Association and dissociation profiles were obtained using a continuous flow of 30 μL per min. GID1 as analyte was applied at concentrations ranging from 0.0625 to 1 μg per mL in the presence of 10−4 M GA4. Kinetic parameters were obtained using Biacore T100 Evaluation Software version 2.
Overexpression of SLR1 and Mutated SLR1 Genes in Rice
proAct-FLAG, proAct-FLAG-SLR1, and proAct-FLAG-mSLR1s were introduced into wild-type T65 rice by Agrobacterium tumefaciens–mediated transformation (Hiei et al., 1994). For the detection of FLAG-tagged SLR1 proteins in the transgenic calli, the procedure described above was used with the following modifications: Wild-type T65 callus expressing the empty vector was used as a control, 10−5 M GA4 was treated for 12 h, anti-FLAG antibody raised in mouse (1:4000 dilution) (F1804; Sigma-Aldrich) was used as a primary antibody, and anti-mouse IgG horseradish peroxidase–conjugated antibody was used as a secondary antibody (1:10,000 dilution). To examine the repressive activity of mSLR1 proteins, transgenic plants were treated with 10−5 M uniconazole soon after plant regeneration, and plant height was measured after 3 weeks.
In Vitro Pull-Down Assay and BiFc Analysis
To produce recombinant GST-SLR1, cMyc-GID1-10xHIS, HA-GID2, and Skp15 together in E. coli, strain BL21 (DE3; Novagen) harboring pACYC184 T7-3xHA-GID2-rbs-SKp15 vector and/or pGEX GST-SLR-rbs-cMyc-GID1-10xHIS vector was incubated at 37°C in the presence of 10−4 M GA4 until reaching an OD600 of 0.4 to 0.6, shifted to 16°C, and incubated for an additional 12 h after 0.5 mM isopropyl-β-d-thiogalactopyranoside induction.
For the in vitro pull-down assays, E. coli cells were extracted and purified using the same method for purification of GST-SLR1 protein as previously described (Ueguchi-Tanaka et al., 2007), except that 10−4 M GA4 was kept in the buffer throughout the experiment. The purified samples were resolved using 12% SDS-PAGE and stained with Coomassie Brilliant Blue to detect SLR1 and GID1 protein or analyzed by immunoblot analysis using anti-HA antibody (Sigma-Aldrich) followed by a mouse secondary antibody (1:10,000; Pierce) to detect GID2 protein.
Infiltration of Nicotiana benthamiana leaf epidermal cells for the BiFC analysis was conducted as previously described (Ueguchi-Tanaka et al., 2007), except that an A. tumefaciens line containing a third plasmid, GID1/pBI121, was infiltrated along with A. tumefaciens containing NY-GID2 and CY-SLR1. Leaves were sprayed with 10−4 M GA4 dissolved in ethanol (+) or with ethanol alone (–) 10 min before observation of the signals.
Accession Numbers
GenBank/EMBL accession numbers and Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: SLR1 (BAE96289), GID1 (Q6L545), GID2 (Q7XAK4), Os Skp15 (AAT09201), SLRL1 (AAR31213), Os SCR (ABA91267), SLN1 (Q8W127), RHT1(Q9ST59), Zm D8 (Q9ST48), At GAI (AT1G14920), At RGA (AT2G01570), At RGL1 (AT1G66350), At RGL2 (AT3G03450), Sm DELLA1 (ABX10758), At SCR (AT3G54220), At SLY1 (AT4G24210), At SNZ (AT5G48170), and Sm GID2a (ABX10767). Sequence data obtained from The Computational Biology and Functional Genomics Laboratory (http://compbio.dfci.harvard.edu/tgi/plant.html) can be found under the following accession number: Medicago truncatula GID2 homolog (TC136403).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amount of SLR1 and GID1 Protein in Rice Cells.
Supplemental Figure 2. Comparison of Amino Acid Sequences of Vascular Plant GID2 Proteins.
Supplemental Figure 3. Mutation in the F-Box Domain of GID2 Abolishes the GA-, GID1-, and GID2-Dependent Degradation of SLR1.
Supplemental Figure 4. Effect of GID1 Amino Acid Substitution on SLR1-GID2 Interaction in Yeast.
Supplemental Figure 5. Comparison of Amino Acid Sequences of Vascular Plant DELLA Proteins.
Supplemental Figure 6. Comparison of Second Leaf Sheath Length of Mutated SLR1 Overproduced Transgenic Seedlings Grown under GA-Deficient Conditions.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Kozue Ohmae, Hiroko Ohmiya, and Hitomi Kihara, for their technical assistance and Koichiro Aya for the proAct-FLAG/pCAMBIA. This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Project 18107001 for M.M.) and the Target Protein Research Program (M.M.).
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Aleman L., Kitamura J., Abdel-mageed H., Lee J., Sun Y., Nakajima M., Ueguchi-Tanaka M., Matsuoka M., Allen R.D. (2008). Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Mol. Biol. 68: 1–16 [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Murase K., Sun T.-p., Steber C.M. (2008). Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20: 2447–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Hirano K., Ueguchi-Tanaka M., Angeles-Shim R.B., Komura T., Satoh H., Kitano H., Matsuoka M., Ashikari M. (2009). Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol. Genet. Genomics 281: 223–231 [DOI] [PubMed] [Google Scholar]
- Bassel G.W., Mullen R.T., Bewley J.D. (2008). procera is a putative DELLA mutant in tomato (Solanum lycopersicum): Effects on the seed and vegetative plant. J. Exp. Bot. 59: 585–593 [DOI] [PubMed] [Google Scholar]
- Bolle C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218: 683–692 [DOI] [PubMed] [Google Scholar]
- Chandler P.M., Harding C.A., Ashton A.R., Mulcair M.D., Dixon N.E., Mander L.N. (2008). Characterization of gibberellin receptor mutants of barley (Hordeum vulgare L.). Mol. Plant 1: 285–294 [DOI] [PubMed] [Google Scholar]
- Chandler P.M., Marion-Poll A., Ellis M., Gubler F. (2002). Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 129: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davierre J.M., Rodriguez-Falcon M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blazquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Dill A., Jung H.S., Sun T.P. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.-p. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Richards D.E., Fleck B., Xie D., Burton N., Harberd N.P. (2004). The Arabidopsis mutant sly1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K., Sasaki A., Itoh H., Ueguchi-Tanaka M., Ashikari M., Kitano H., Matsuoka M. (2004). GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37: 626–634 [DOI] [PubMed] [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.-L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.-P., Thomas S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Chandler P.M., White R.G., Llewellyn D.J., Jacobsen J.V. (2002). Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol. 129: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hirano K., et al. (2007). The GID1-mediated gibberellin perception mechanism is conserved in the lycophyte Selaginella moellendorfii but not in the bryophyte Physcomitrella patens. Plant Cell 19: 3058–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Ueguchi-Tanaka M., Matsuoka M. (2008). GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 13: 192–199 [DOI] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., Matsuoka M., Yamaguchi J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Sasaki A., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Hasegawa Y., Minami E., Ashikari M., Matsuoka M. (2005). Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 46: 1392–1399 [DOI] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P.R., Swanson R., Rakhilina L., Hochstrasser M. (1998). Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 94: 217–227 [DOI] [PubMed] [Google Scholar]
- Kamiya N., Itoh J., Morikami A., Nagato Y., Matsuoka M. (2003). The SCARECROW gene's role in asymmetric cell divisions in rice plants. Plant J. 36: 45–54 [DOI] [PubMed] [Google Scholar]
- Karlsson R., Katsamba P.S., Nordin H., Pol E., Myszka D.G. (2006). Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 349: 136–147 [DOI] [PubMed] [Google Scholar]
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T., Steber C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muangprom A., Thomas S.G., Sun T.P., Osborn T.C. (2005). A novel dwarfing mutation in a green revolution gene from Brassica rapa. Plant Physiol. 137: 931–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.-p., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Nakajima M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., et al. (1999). 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Ravid T., Hochstrasser M. (2008). Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards D.E., King K.E., Ait-ali T., Harberd N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 67–88 [DOI] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D.H., An G., Kitano H., Ashikari M., Matsuoka M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Silverstone A.L., Ciampaglio C.N., Sun T.-p. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Jung H.S., Dill A., Kawaide H., Kamiya Y., Sun T.P. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Eiguchi M., Kumamaru T., Satoh H., Matsusaka H., Moriguchi K., Nagato Y., Kurata N. (2008). MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics 279: 213–223 [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Thomas S.G., Rieu I., Steber C.M. (2005). Gibberellin metabolism and signaling. Vitam. Horm. 72: 289–338 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.I., Kitano H., Yamaguchi I., Matsuoka M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Hirano K., Hasegawa Y., Kitano H., Matsuoka M. (2008). Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Katoh E., Ohmiya H., Asano K., Saji S., Hongyu X., Ashikari M., Kitano H., Yamaguchi I., Matsuoka M. (2007). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1 and gibberellin. Plant Cell 19: 2140–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zhu D., Huang X., Li S., Gong Y., Yao Q., Fu X., Fan L.M., Deng X.W. (2009). Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston D.E., Elliot R.C., Lester D.R., Rameau C., Reid J.B., Murfet I.C., Ross J.J. (2008). The pea DELLA proteins LA and CRY are important regulators of gibberellin synthesis and root growth. Plant Physiol. 147: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura Y., Crumpton-Taylor M., Fuentes S., Harberd N.P. (2007). Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr. Biol. 17: 1225–1230 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.