Abstract
Africa is the source of all modern humans, but characterization of genetic variation and of relationships among populations across the continent has been enigmatic. We studied 121 African populations, four African American populations, and 60 non-African populations for patterns of variation at 1327 nuclear microsatellite and insertion/deletion markers. We identified 14 ancestral population clusters in Africa that correlate with self-described ethnicity and shared cultural and/or linguistic properties. We observed high levels of mixed ancestry in most populations, reflecting historical migration events across the continent. Our data also provide evidence for shared ancestry among geographically diverse hunter-gatherer populations (Khoesan speakers and Pygmies). The ancestry of African Americans is predominantly from Niger-Kordofanian (~71%), European (~13%), and other African (~8%) populations, although admixture levels varied considerably among individuals. This study helps tease apart the complex evolutionary history of Africans and African Americans, aiding both anthropological and genetic epidemiologic studies.
Modern humans originated in Africa ~200,000 years ago and then spread across the rest of the globe within the past ~100,000 years (1). Thus, modern humans have existed continuously in Africa longer than in any other geographic region and have maintained relatively large effective population sizes, resulting in high levels of within-population genetic diversity (1, 2). Africa contains more than 2000 distinct ethnolinguistic groups representing nearly one-third of the world's languages (3). Except for a few isolates that show no clear relationship with other languages, these languages have been classified into four major macrofamilies: Niger-Kordofanian (spoken across a broad region of Africa), Afroasiatic (spoken predominantly in Saharan, northeastern, and eastern Africa), Nilo-Saharan (spoken predominantly in Sudanic, Saharan, and eastern Africa), and Khoesan (languages containing click-consonants, spoken by San in southern Africa and by Hadza and Sandawe in eastern Africa) (fig. S1) (4).
Despite the importance of African population genetics, the pattern of genome-wide nuclear genetic diversity across geographically and ethnically diverse African populations is largely uncharacterized (1, 2, 5). Because of considerable environmental diversity, African populations show a range of linguistic, cultural, and phenotypic variation (1, 2, 4). Characterizing the pattern of genetic variation among ethnically diverse African populations is critical for reconstructing human evolutionary history, clarifying the population history of Africans and African Americans, and determining the proper design and interpretation of genetic disease association studies (1, 6), because substructure can cause spurious results (7). Furthermore, variants associated with disease could be geographically restricted as a result of new mutations, genetic drift, or region-specific selection pressures (1). Thus, our in-depth characterization of genetic structure in Africa benefits research of biomedical relevance in both African and African-diaspora populations.
We genotyped a panel of 1327 polymorphic markers, consisting of 848 microsatellites, 476 indels (insertions/deletions), and three SNPs (single-nucleotide polymorphisms), in 2432 Africans from 113 geographically diverse populations (fig. S1), 98 African Americans, and 21 Yemenites (table S1). To incorporate preexisting African data and to place African genetic variability into a worldwide context, we integrated these data with data from the panel of markers genotyped in 952 worldwide individuals from the CEPH-HGDP (Centre d'Étude du Polymorphisme Humain–Human Genome Diversity Panel) (8-10) in 432 individuals of Indian descent (11) and in 10 Native Australians (tables S1 and S2).
African variation in a worldwide context
African and African American populations, with the exception of the Dogon of Mali, show the highest levels of within-population genetic diversity (θ = 4Neμ, where is the level of genetic diversity based on variance of microsatellite allele length, Ne is the effective population size, and μ is the microsatellite mutation rate) (figs. S2 and S3). In addition, genetic diversity declines with distance from Africa (fig. S2, A to C), consistent with proposed serial founder effects resulting from the migration of modern humans out of Africa and across the globe (9, 11-13). Within Africa, genetic diversity estimated from expected heterozygosity significantly correlates with estimates from microsatellite variance (fig. S4) (4) and varies by linguistic, geographic, and subsistence classifications (fig. S5). Three hunter-gatherer populations (Baka Pygmies, Bakola Pygmies, and San) were among the five populations with the highest levels of genetic diversity based on variance estimates (fig. S2A) (4). In addition, more private alleles exist in Africa than in other regions (fig. S6A). Consistent with bidirectional gene flow (14), African and Middle Eastern populations shared the greatest number of alleles absent from all other populations (fig. S6B). Within Africa, the most private alleles were in southern Africa, reflecting those in southern African Khoesan (SAK) San and !Xun/Khwe populations (fig. S6C) (12). Eastern and Saharan Africans shared the most alleles absent from other African populations examined (fig. S6D).
The proportion of genetic variation among all studied African populations was 1.71% (table S3). In comparison, Native American and Oceanic populations showed the greatest proportion of genetic variation among populations (8.36% and 4.59%, respectively), most likely due to genetic drift (9, 15, 16). Distinct patterns of the distribution of variation among African populations classified by geography, language, and subsistence were also observed (4).
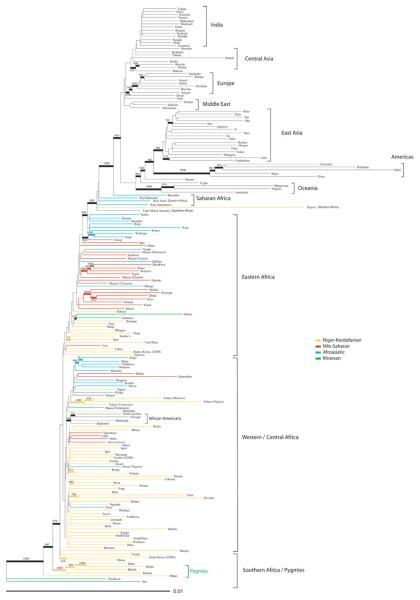
Phylogenetic trees constructed from genetic distances between populations generally showed clustering by major geographic region, both on a global scale and within Africa (Fig. 1 and figs. S7 and S8). Within Africa, the two SAK populations cluster together and are the most distant from other populations, consistent with mitochondrial DNA (mtDNA), Y chromosome, and autosomal chromosome diversity studies, indicating that SAK populations have the most diverged genetic lineages (12, 17-21). The Pygmy populations cluster near the SAK populations in the tree constructed from D2 genetic distances (Fig. 1), whereas the Hadza and Sandawe cluster near the SAK populations in the tree constructed from RST genetic distances (fig. S8) (4). Note that population clustering in the tree may reflect common ancestry and/or admixture. African populations with high levels of non-African admixture [e.g., the Cape Mixed Ancestry (CMA) population, commonly referred to as “Cape Coloured” in South Africa] cluster in positions that are intermediate between Africans and non-Africans, whereas the African American populations, which are relatively less admixed with non-Africans, cluster more closely with West Africans. Additionally, populations with high levels of genetic drift (i.e., the Americas, Oceania, and Pygmy, Hadza, and SAK hunter-gatherers) have longer branch lengths.
Figure 1.
Neighbor-joining tree from pairwise D2 genetic distances between populations (65). African population branches are color-coded according to language family classification. Population clusters by major geographic region are noted; bootstrap values above 700 out of 1000 are indicated by thicker lines and bootstrap number.
Geographic distances (great circle routes) and genetic distances (δμ)2 between population pairs were significantly correlated, consistent with an isolation-by-distance model (figs. S9 to S11 and table S4) (13). A heterogeneous pattern of correlations across global regions was observed, consistent with a previous study (16); the strongest correlations were in Europe and the Middle East (Spearman's ρ = 0.88 and 0.83, respectively; P ≤0.0001 for both), followed by Africa (Spearman's ρ = 0.40; P < 0.0001). Correlations were not significant for central Asia or India. Within Africa, the strongest correlations between genetic and geographic distances were in Saharan Africa and central Africa (Spearman's ρ = 0.76 and 0.55, respectively; P < 0.0001 for both) (fig. S11 and table S4). The smallest correlation was observed in eastern Africa (ρ = 0.19; P < 0.0001).
Genetic structure on a global level
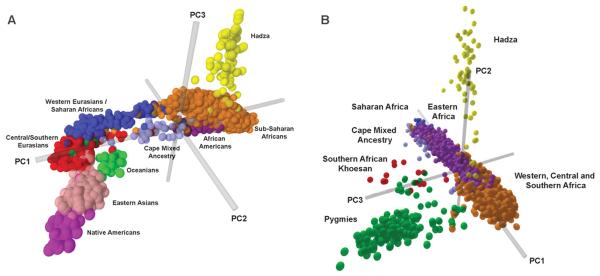
Global patterns of genetic structure and individual ancestry were inferred by principal components analysis (PCA) (22) (Fig. 2A) and a Bayesian model-based clustering approach with STRUCTURE (23) (Figs. 3 and 4 and figs. S12 to S14). Worldwide, 72 significant principal components (PCs) were identified by PCA (P < 0.05) (22). PC1 (accounting for 19.5% of the extracted variation) distinguishes Africans from non-Africans. The CMA and African American individuals cluster between Africans and non-Africans, reflecting both African and non-African ancestry. PC2 (5.01%) distinguishes Oceanians, East Asians, and Native Americans from others. PC3 (3.5%) distinguishes the Hadza hunter-gatherers from others. The remaining PCs each extract less than 3% of the variation, and the 22nd to 72nd PCs extract less than 1% combined, with some minor PCs corresponding to regional and/or ethnically defined populations, consistent with STRUCTURE results below.
Figure 2.
Principal components analysis (22) created on the basis of individual genotypes. (A) Global data set and (B) African data set.
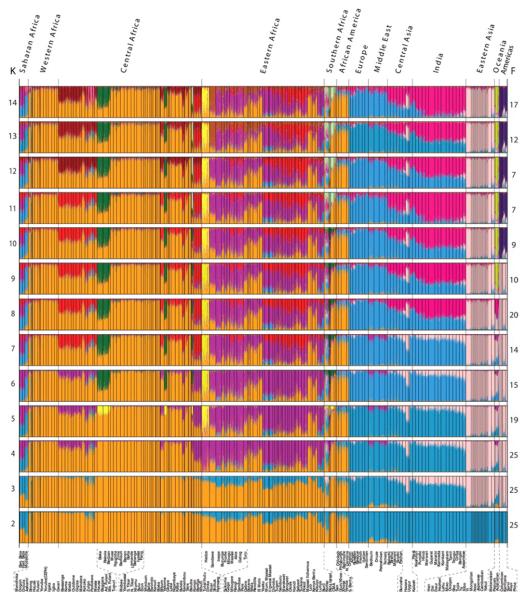
Figure 3.
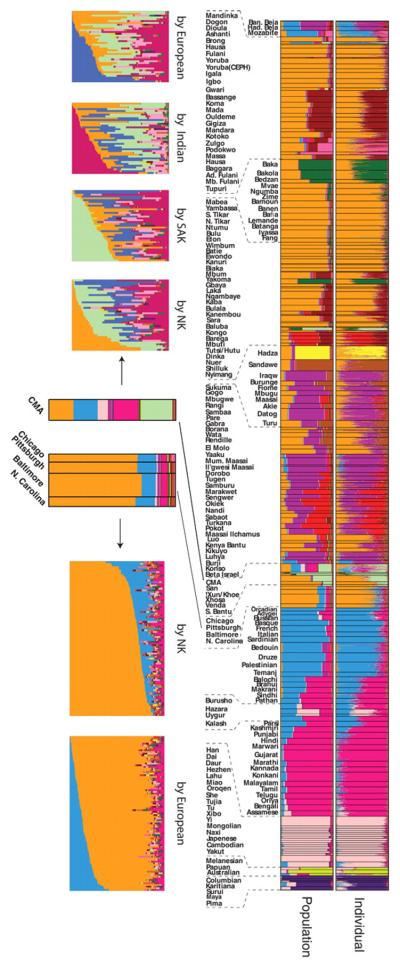
STRUCTURE analysis of the global data set with 1327 markers genotyped in 3945 individuals. Each vertical line represents an individual. Individuals were grouped by self-identified ethnic group (at bottom) and ethnic groups are clustered by major geographic region (at top). Colors represent the inferred ancestry from K ancestral populations. STRUCTURE results for K = 2 to 14 (left) are shown with the number of similar runs (F) for the primary mode of 25 STRUCTURE runs at each K value (right).
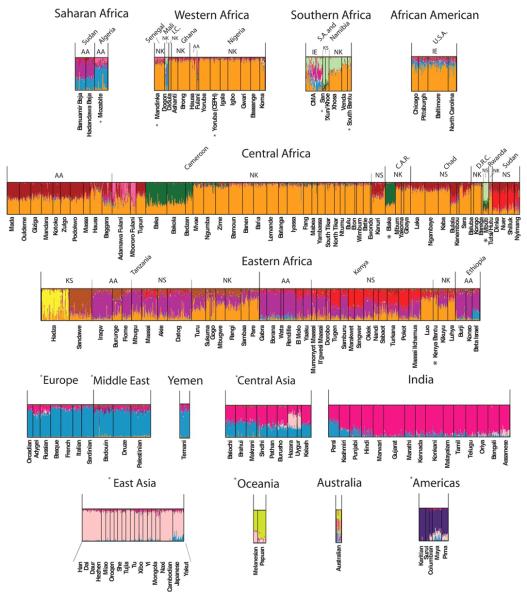
Figure 4.
Expanded view of STRUCTURE results at K = 14. Populations from the CEPH diversity panel are identified by asterisks. Languages spoken by populations are classified as Niger-Kordofanian (NK), Nilo-Saharan (NS), Afroasiatic (AA), Khoesan (KS), or Indo-European (IE).
STRUCTURE analysis revealed 14 ancestral population clusters (K = 14) on a global level (Figs. 3 and 4) (4). Middle Eastern and Oceanic populations exhibit low levels of East African ancestry up to K = 8, consistent with possible gene flow into these regions and with studies suggesting early migration of modern humans into southern Asia and Oceania (16, 24). The Hadza, and to a lesser extent the Pygmy, SAK, and Sandawe hunter-gatherers, are distinguished at K = 5. The 11th cluster (K = 11) distinguishes Mbuti Pygmy and SAK individuals, indicating common ancestry of these geographically distant hunter-gatherers. A number of Africans (predominantly CMA, Fulani, and eastern Afroasiatic speakers) exhibit low to moderate levels of European–Middle Eastern ancestry, consistent with possible gene flow from those regions. We found more African substructure on a global level (nine clusters) than previously observed (9-12, 20). A phylogenetic tree of genetic distances from inferred ancestral clusters (fig. S14) indicates that within Africa, the Pygmy and SAK associated ancestral clusters (AACs) form a clade, as do the Hadza and Sandawe AACs and the Nilo-Saharan and Chadic AACs, reflecting their ancient common ancestries.
Genetic structure within Africa
PCA of genetic variation within Africa indicated the presence of 43 significant PCs (P < 0.05 with a Tracy-Widom distribution). PC1 (10.8% of the extracted variation) distinguishes eastern and Saharan Africa from western, central, and southern Africa (Fig. 2B). The second PC (6.1%) distinguishes the Hadza; the third PC (4.9%) distinguishes Pygmy and SAK individuals from other Africans. The fourth PC (3.7%) is associated with the Mozabites, some Dogon, and the CMA individuals, who show ancestry from the European–Middle Eastern cluster. The fifth PC (3.1%) is associated with SAK speakers. The 10th PC was of particular interest (2.2%) because it associates with the SAK, Sandawe, and some Dogon individuals, suggesting shared ancestry.
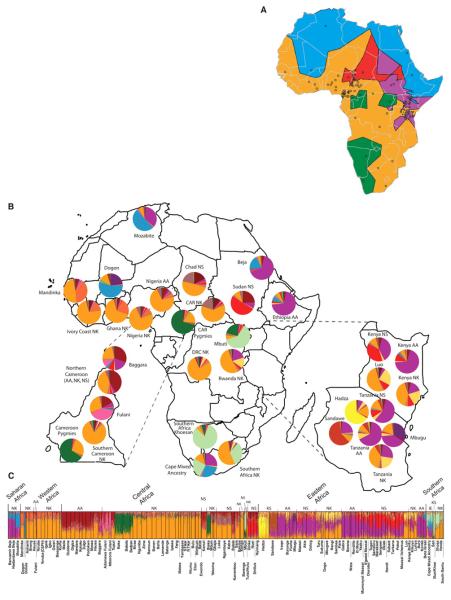
We incorporated geographic data into a Bayesian clustering analysis, assuming no admixture (TESS software) (25) and distinguished six clusters within continental Africa (Fig. 5A). The most geographically widespread cluster (orange) extends from far Western Africa (the Mandinka) through central Africa to the Bantu speakers of South Africa (the Venda and Xhosa) and corresponds to the distribution of the Niger-Kordofanian language family, possibly reflecting the spread of Bantu-speaking populations from near the Nigerian/Cameroon highlands across eastern and southern Africa within the past 5000 to 3000 years (26, 27). Another inferred cluster includes the Pygmy and SAK populations (green), with a noncontiguous geographic distribution in central and southeastern Africa, consistent with the STRUCTURE (Fig. 3) and phylogenetic analyses (Fig. 1). Another geographically contiguous cluster extends across northern Africa (blue) into Mali (the Dogon), Ethiopia, and northern Kenya. With the exception of the Dogon, these populations speak an Afroasiatic language. Chadic-speaking and Nilo-Saharan–speaking populations from Nigeria, Cameroon, and central Chad, as well as several Nilo-Saharan–speaking populations from southern Sudan, constitute another cluster (red). Nilo-Saharan and Cushitic speakers from the Sudan, Kenya, and Tanzania, as well as some of the Bantu speakers from Kenya, Tanzania, and Rwanda (Hutu/Tutsi), constitute another cluster (purple), reflecting linguistic evidence for gene flow among these populations over the past ~5000 years (28, 29). Finally, the Hadza are the sole constituents of a sixth cluster (yellow), consistent with their distinctive genetic structure identified by PCA and STRUCTURE.
Figure 5.
Geographic and genetic structure of populations within Africa. (A) Geographic discontinuities among African populations using TESS, assuming a model of no population admixture (25). Circles indicate location of populations included in the study. (B) Inferred proportions of ancestral clusters from STRUCTURE analysis at K = 14 for individuals grouped by geographic region and language classification. Classifications of languages spoken by self-identified ethnic affiliation in the Africans are as in Fig. 1. (C) Inferred proportion of ancestral clusters in individuals from STRUCTURE analysis at K = 14.
STRUCTURE analysis of the Africa data set indicated 14 ancestral clusters (Fig. 5, B and C, and figs. S15 to S18). Analyses of subregions within Africa indicated additional substructure (figs. S19 to S29). At low K values, the Africa-wide STRUCTURE results (fig. S15) recapitulated the PCA and worldwide STRUCTURE results. However, as K increased, additional population clusters were distinguished (4): the Mbugu [who speak a mixed Bantu and Cushitic language (30), shown in dark purple]; Cushitic-speaking individuals of southern Ethiopian origin (light purple); Nilotic Nilo-Saharan–speaking individuals (red); central Sudanic Nilo-Saharan–speaking individuals (tan); and Chadic-speaking and Baggara individuals (maroon). At K = 14, subtle substructure between East African Bantu speakers (light orange) and West Central African Bantu speakers (medium orange), and individuals from Nigeria and farther west, who speak various non-Bantu Niger-Kordofanian languages (dark orange), was also apparent (Fig. 5, B and C). Bantu speakers of South Africa (Xhosa, Venda) showed substantial levels of the SAK and western African Bantu AACs and low levels of the East African Bantu AAC (the latter is also present in Bantu speakers from Democratic Republic of Congo and Rwanda). Our results indicate distinct East African Bantu migration into southern Africa and are consistent with linguistic and archeological evidence of East African Bantu migration from an area west of Lake Victoria (28) and the incorporation of Khoekhoe ancestry into several of the Southeast Bantu populations ~1500 to 1000 years ago (31).
High levels of heterogeneous ancestry (i.e., multiple cluster assignments) were observed in nearly all African individuals, with the exception of western and central African Niger-Kordofanian speakers (medium orange), who are relatively homogeneous at large K values (Fig. 5C and fig. S15). Considerable Niger-Kordofanian ancestry (shades of orange) was observed in nearly all populations, reflecting the recent spread of Bantu speakers across equatorial, eastern, and southern Africa (27) and subsequent admixture with local populations (28). Many Nilo-Saharan–speaking populations in East Africa, such as the Maasai, show multiple cluster assignments from the Nilo-Saharan (red) and Cushitic (dark purple) AACs, in accord with linguistic evidence of repeated Nilotic assimilation of Cushites over the past 3000 years (32) and with the high frequency of a shared East African–specific mutation associated with lactose tolerance (33).
Our data support the hypothesis that the Sahel has been a corridor for bidirectional migration between eastern and western Africa (34-36). The highest proportion of the Nilo-Saharan AAC was observed in the southern and central Sudanese populations (Nuer, Dinka, Shilluk, and Nyimang), with decreasing frequency from northern Kenya (e.g., Pokot) to northern Tanzania (Datog, Maasai) (Fig. 5, B and C, and fig. S15). Additionally, all Nilo-Saharan–speaking populations from Kenya, Tanzania, southern Sudan, and Chad clustered with west central Afroasiatic Chadic–speaking populations in the global analysis at K ≤11 (Fig. 3), which is consistent with linguistic and archeological data suggesting bidirectional migration of Nilo-Saharans from source populations in Sudan within the past ~10,500 to 3000 years (4, 29). The proposed migration of proto-Chadic Afroasiatic speakers ~7000 years ago from the central Sahara into the Lake Chad Basin may have resulted in a Nilo-Saharan to Afroasiatic language shift among Chadic speakers (37). However, our data suggest that this shift was not accompanied by large amounts of Afroasiatic gene flow. Other populations of interest, including the Fulani (Nigeria and Cameroon), the Baggara Arabs (Cameroon), the Koma (Nigeria), and Beja (Sudan), are discussed in (4).
Genetic structure in East Africa
East Africa, the hypothesized origin of the migration of modern humans out of Africa, has a remarkable degree of ethnic and linguistic diversity, as reflected by the greatest level of regional substructure in Africa (figs. S15, S16, and S19 to S21). The diversity among populations from this region reflects the proposed long-term presence of click-speaking Hadza and Sandawe hunter-gatherers and successive waves of immigration of Cushitic, Nilotic, and Bantu populations within the past 5000 years (4, 29, 32, 38, 39). Within eastern Africa, including southern and central Sudan, clustering is primarily associated with language families, including Niger-Kordofanian, Afroasiatic, Nilo-Saharan, and two click-speaking hunter-gatherer groups: the Sandawe and Hadza (figs. S19 to S21). However, individuals from the Afroasiatic Cushitic Iraqw and Gorowa (Fiome) and the Nilo-Saharan Datog, who are in close geographic proximity, also cluster. Additionally, several hunter-gatherer populations were distinct, including the Okiek, Akie, and Yaaku and El Molo. Of particular interest is the common ancestry of the Akie (who have remnants of a Cushitic language) and the Eastern Cushitic El Molo and Yaaku at K = 9, consistent with linguistic data suggesting that these populations originated from southern Ethiopia and migrated into Kenya and Tanzania within the past ~4000 years (4, 29, 32, 39).
Origins of hunter-gatherer populations in Africa
Our analyses demonstrate potential shared ancestry of a number of populations who practice (or until recently practiced) a traditional hunting and gathering lifestyle. For example, we observed a Hadza AAC (yellow) at K = 5 and K = 3 in the global and African STRUCTURE analyses, respectively (Fig. 3 and fig. S15), which is at moderate levels (0.18 to 0.32) in the SAK and Pygmy populations and at low levels (0.03 to 0.04) in the Sandawe and neighboring Burunge with whom the Sandawe have admixed (tables S8 and S9). The SAK and Pygmies continue to cluster at higher K values (Fig. 3 and fig. S15) and in the TESS (Fig. 5A) and phylogenetic (Fig. 1) analyses, consistent with an exclusively shared Y chromosome lineage (B2b4) (40). Additionally, we observed clustering of the SAK, Sandawe, and Hadza in the RST phylogenetic tree (fig. S8) and of the SAK, Sandawe, and Mbuti Pygmies at low K values in the secondary modes of Africa STRUCTURE analyses (fig. S16), consistent with observed low frequency of the Khoesan-specific mitochondrial haplotype (L0d) in the Sandawe (18, 19), the presence of Khoesan-related rock art near the Sandawe homeland (41), and similarities between the Sandawe and SAK languages (42). These results suggest the possibility that the SAK, Hadza, Sandawe, and Pygmy populations are remnants of a historically more widespread proto–Khoesan-Pygmy population of hunter-gatherers. Analyses of mtDNA and Y chromosome lineages in the Khoesan-speaking populations suggest that divergence may be >35,000 years ago (4, 17-19). The shared ancestry, identified here, of Khoesan-speaking populations with the Pygmies of central Africa suggests the possibility that Pygmies, who lost their indigenous language, may have originally spoken a Khoesan-related language, consistent with shared music styles between the SAK and Pygmies (4, 43).
Shared ancestry of western and eastern Pygmies, who do not become differentiated until larger K values in STRUCTURE analyses (Fig. 3 and fig. S15), was also supported by the phylogenetic trees (Fig. 1 and figs. S7 and S8), consistent with mtDNA and autosomal studies indicating that the western and eastern Pygmies diverged >18,000 years ago (44-47). Western Pygmy populations usually clustered (Fig. 3 and fig. S15), consistent with a proposed recent common ancestry within the past ~3000 years (48). However, subtle substructure within the western Pygmies was apparent in the analysis of central Africa (fig. S24), probably due to recent geographic isolation and genetic drift. Asymmetric Bantu gene flow into Pygmy populations was also observed, with Bantu ancestry ranging from 0.13 in Mbuti to 0.54 in the Bedzan (table S8), consistent with prior studies (40, 44, 49, 50).
The Hadza, with a census size of ~1000, were genetically distinct on a global level with STRUCTURE, PCA, and TESS (Figs. 2 to 5), consistent with linguistic data indicating that the Hadza language is divergent from or unrelated to other Khoesan languages (42, 51, 52). The Hadza, who have maintained a traditional hunter-gatherer lifestyle, show low levels of asymmetric gene flow from neighboring populations, whereas the Sandawe, with a census size of >30,000 (39), show evidence of bidirectional gene flow with neighboring populations, from whom they may have adopted mixed farming technologies (Figs. 3 to 5 and fig. S15). In fact, we observed high levels of the Sandawe AAC in northern Tanzania and low levels in northern Kenya and southern Ethiopia (Fig. 3 and fig. S15) (K = 8 to 13), consistent with linguistic and genetic data suggesting that Khoesan populations may once have extended from Somalia through eastern Africa and into southern Africa (28, 38, 53-55). Although the Hadza and Sandawe show evidence of common ancestry (Fig. 1 and figs. S7, S8, S14, S18, and S21), we observe no evidence of recent gene flow between them despite their geographic proximity, consistent with mtDNA and Y chromosome studies indicating divergence >15,000 years ago (19). The origins of other African hunter-gatherer populations (Dorobo, Okiek, Yaaku, Akie, El Molo, and Wata) are discussed in (4).
Origins of human migration within and out of Africa
The geographic origin for the expansion of modern humans was inferred, as in (13), from the correlation between genetic diversity and geographic position of populations (r) (figs. S30 and S31). Both the point of origin of human migration and waypoint for the out-of-Africa migration were optimized to fit a linear relationship between genetic diversity and geographic distance (4). This analysis indicates that modern human migration originated in southwestern Africa, at 12.5°E and 17.5°S, near the coastal border of Namibia and Angola, corresponding to the current San homeland, with the waypoint in northeast Africa at 37.5°E, 22.5°N near the midpoint of the Red Sea (figs. S2C, S30, and S31). However, the geographic distribution of genetic diversity in modern populations may not reflect the distribution of those populations in the past, although our waypoint analysis is consistent with other studies suggesting a northeast African origin of migration of modern humans out of Africa (1, 56).
Correlation between genetic and linguistic diversity in Africa
Genetic clustering of populations was generally consistent with language classification, with some exceptions (Fig. 1 and fig. S32). For example, the click-speaking Hadza and Sandawe, classified as Khoesan, were separated from the SAK populations in the D2 and (δμ)2 phylogenetic trees (Fig. 1) and fig. S7). However, this observation is consistent with linguistic studies indicating that these Khoesan languages are highly divergent (42, 51) and may reflect gene flow between the Hadza and Sandawe with neighboring populations in East Africa subsequent to divergence from the SAK. Additionally, the Afroasiatic Chadic–speaking populations from northern Cameroon cluster close to the Nilo-Saharan–speaking populations from Chad, rather than with East African Afroasiatic speakers (Fig. 1), consistent with a language replacement among the Chadic populations.
Other divergences between genetic and linguistic classifications include the Pygmies, who lost their indigenous language and adopted the neighboring Niger-Kordofanian language (27), and the Fulani, who speak a West African Niger-Kordofanian language but cluster near the Chadic- and Central Sudanic–speaking populations in the phylogenies (Fig. 1 and figs. S7 and S8), consistent with Y chromosome studies (34). Additionally, the Nilo-Saharan–speaking Luo of Kenya show predominantly Niger-Kordofanian ancestry in the STRUCTURE analyses (orange) (Figs. 3 and 4, Fig. 5, B and C, and fig. S15) and cluster together with eastern African Niger-Kordofanian–speaking populations in the phylogenetic trees (Fig. 1 and figs. S7 and S8).
Both language and geography explained a significant proportion of the genetic variance, but differences exist between and within the language families (table S5 and fig. S33, A to C) (4). For example, among the Niger-Kordofanian speakers, with or without the Pygmies, more of the genetic variation is explained by linguistic variation (r2 = 0.16 versus 0.11, respectively; P < 0.0001 for both) than by geographic variation (r2 = 0.02 for both; P < 0.0001 for both), consistent with recent long-range Bantu migration events. The reverse was true for Nilo-Saharan speakers (r2 = 0.06 for linguistic distance versus 0.21 for geographic distance; P < 0.0001 for both), possibly due to admixture among Nilo-Saharan–, Cushitic-, and Bantu-speaking populations in eastern Africa, which might reduce the variation explained by language. The Afroasiatic family had the highest r2 for both linguistic and geographic distances (0.20 and 0.34, respectively). However, when subfamilies were analyzed independently, the Chadic-speaking populations showed a strong association between geography and genetic variation (0.39), but not between linguistic and genetic variation (0.0012), as expected on the basis of a possible language replacement, whereas the Cushitic-speaking populations were significant for both (0.29 and 0.27, respectively) (4).
Genetic ancestry of African Americans and CMA populations
In contrast to prior studies of African Americans (57-61), we inferred African American ancestry with the use of genome-wide nuclear markers from a large and diverse set of African populations. African American populations from Chicago, Baltimore, Pittsburgh, and North Carolina showed substantial ancestry from the African Niger-Kordofanian AAC, most common in western Africa (means 0.69 to 0.74), and from the European–Middle Eastern AAC (means 0.11 to 0.15) (Fig. 6 and tables S6 and S8), consistent with prior genetic studies and the history of the slave trade (4, 57-62). European and African ancestry levels varied considerably among individuals (Fig. 6). We also detected low levels of ancestry from the Fulani AAC (means 0.0 to 0.03, individual range 0.00 to 0.14), Cushitic AAC (means 0.02, individual range 0.00 to 0.10), Sandawe AAC (means 0.01 to 0.03, individual range 0.0 to 0.12), East Asian AAC (means 0.01 to 0.02, individual range 0.0 to 0.08), and Indian AAC (means 0.04 to 0.06, individual range 0.01 to 0.17) (table S6) (4). We observed very low levels of Native American ancestry, although other U.S. regions may reveal Native American ancestry (57).
Figure 6.
Analyses of Cape Mixed Ancestry (CMA) and African American populations. Frequencies of inferred ancestral clusters are shown for K = 14 with the global data set for individuals (top row) and proportion of AACs in self-identified populations (bottom row). The proportions of AACs in the CMA and African American populations are highlighted in the center bottom row; proportions of AACs in individuals, sorted by Niger-Kordofanian, European, SAK, and/or Indian ancestry, are shown to the left and right, bottom row.
Supervised STRUCTURE analysis (fig. S34) (4) was used to infer African American ancestry from global training populations, including both Bantu (Lemande) and non-Bantu (Mandinka) Niger-Kordofanian–speaking populations (fig. S34 and table S7). These results were generally consistent with the unsupervised STRUCTURE analysis (table S6) and demonstrate that most African Americans have high proportions of both Bantu (~0.45 mean) and non-Bantu (~0.22 mean) Niger-Kordofanian ancestry, concordant with diasporas originating as far west as Senegambia and as far south as Angola and South Africa (62). Thus, most African Americans are likely to have mixed ancestry from different regions of western Africa. This observation, together with the subtle substructure observed among Niger-Kordofanian speakers, will make it a challenge to trace the ancestry of African Americans to specific ethnic groups in Africa, unless considerably more markers are used.
The CMA population shows the highest levels of intercontinental admixture of any global population, with nearly equal high levels of SAK ancestry (mean 0.25, individual range 0.01 to 0.48), Niger-Kordofanian ancestry (mean 0.19, individual range 0.01 to 0.71), Indian ancestry (mean 0.20, individual range 0.0 to 0.69), and European ancestry (mean 0.19, individual range 0.0 to 0.86) (Fig. 6 and tables S6 and S8). The CMA population also has low levels of East Asian ancestry (mean 0.08, individual range 0.0 to 0.21) and Cushitic ancestry (mean 0.03, individual range 0.0 to 0.40). These results are consistent with the supervised STRUCTURE analyses (fig. S34 and table S7) and with the history of the CMA population (4, 63).
The genetic, linguistic, and geographic landscape of Africa
The differentiation observed among African populations is likely due to ethnicity, language, and geography, as well as technological, ecological, and climatic shifts (including periods of glaciation and warming) that contributed to population size fluctuations, fragmentations, and dispersals in Africa (1, 4, 34, 64). We observed significant associations between genetic and geographic distance in all regions of Africa, although their strengths varied. We also observed significant associations between genetic and linguistic diversity, reflecting the concomitant spread of languages, genes, and often culture [e.g., the spread of farming during the Bantu expansion (28)]. Of interest for future anthropological studies are the cases in which populations have maintained their culture in the face of extensive genetic introgression (e.g., Maasai and Pygmies) and populations that have maintained both cultural and genetic distinction (e.g., Hadza).
Given the extensive amount of ethnic diversity in Africa, additional sampling—particularly from underrepresented regions such as North and Central Africa—is important. Because of the extensive levels of substructure in Africa, ethnically and geographically diverse African populations need to be included in resequencing, genome-wide association, and pharmacogenetic studies to identify population- or region-specific functional variants associated with disease or drug response (1). The high levels of mixed ancestry from genetically divergent ancestral population clusters in African populations could also be useful for mapping by admixture disequilibrium. Future large-scale resequencing and genotyping of Africans will be informative for reconstructing human evolutionary history, for understanding human adaptations, and for identifying genetic risk factors (and potential treatments) for disease in Africa.
Supplementary Material
Acknowledgments
We thank the thousands of people who donated DNA samples used in this study. We thank D. Bygott, S. J. Deo, D. Guracha, J. Hanby, D. Kariuki, P. Lufungulo, A. Mabulla, A. A. Mohamed, W. Ntandu, L. A. Nyindodo, C. Plowe, and A. Tibwitta for assisting with sample collection; K. Panchapakesan and L. Pfeiffer for assistance in sample preparation; S. Dobrin for assistance with genotyping; J. Giles, J. Bartlett, N. Kodaman, and J. Jarvis for assistance with analyses; and N. Rosenberg, J. Pritchard, A. Brooks, J. S. Friedlaender, J. Jarvis, C. Lambert, B. Payseur, N. Patterson, and J. Plotkin for helpful suggestions and discussions. Conducted in part using the ACCRE computing facility at Vanderbilt University, Nashville, TN. Supported by L. S. B. Leakey and Wenner Gren Foundation grants, NSF grants BCS-0196183, BSC-0552486, and BCS-0827436, NIH grants R01GM076637 and 1R01GM083606-01, and David and Lucile Packard and Burroughs Wellcome Foundation Career Awards (S.A.T.); NIH grant F32HG03801 (F.A.R.); and NIH grant R01 HL65234 (S.M.W. and J.H.M.). Genotyping was supported by the NHLBI Mammalian Genotyping Service. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. The project included in this manuscript has been funded in part with federal funds from the National Cancer Institute under contract N01-CO-12400. Original genotype data are available at http://research.marshfieldclinic.org/genetics/genotypingData_Statistics/humanDiversityPanel.asp. Data used for analyses in the current manuscript are available at www.med.upenn.edu/tishkoff/Supplemental/files.html and at http://chgr.mc.vanderbilt.edu/page/supplementary-data.
References and Notes
- 1.Campbell MC, Tishkoff SA. Annu. Rev. Genomics Hum. Genet. 2008;9:403. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed FA, Tishkoff SA. Curr. Opin. Genet. Dev. 2006;16:597. doi: 10.1016/j.gde.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Ethnologue ( www.ethnologue.com)
- 4.See supporting material.
- 5.Tishkoff SA, Williams SM. Nat. Rev. Genet. 2002;3:611. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- 6.Sirugo G, et al. Hum. Genet. 2008;123:557. doi: 10.1007/s00439-008-0511-y. [DOI] [PubMed] [Google Scholar]
- 7.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Am. J. Hum. Genet. 2000;67:170. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cann HM, et al. Science. 2002;296:261. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg NA, et al. Science. 2002;298:2381. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 10.Rosenberg NA, et al. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg NA, et al. PLoS Genet. 2006;2:e215. doi: 10.1371/journal.pgen.0020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakobsson M, et al. Nature. 2008;451:998. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 13.Ramachandran S, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15942. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster P, Romano V. Science. 2007;316:50. doi: 10.1126/science.316.5821.50. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, et al. PLoS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedlaender JS, et al. PLoS Genet. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behar DM, et al. Am. J. Hum. Genet. 2008;82:1130. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. Mol. Biol. Evol. 2007;24:757. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- 19.Tishkoff SA, et al. Mol. Biol. Evol. 2007;24:2180. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 20.Li JZ, et al. Science. 2008;319:1100. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 21.Chen YS, et al. Am. J. Hum. Genet. 2000;66:1362. doi: 10.1086/302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson N, Price AL, Reich D. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard JK, Stephens M, Donnelly P. Genetics. 2000;155:945. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forster P, Matsumura S. Science. 2005;308:965. doi: 10.1126/science.1113261. [DOI] [PubMed] [Google Scholar]
- 25.Francois O, Ancelet S, Guillot G. Genetics. 2006;174:805. doi: 10.1534/genetics.106.059923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehret C. Int. J. Afr. Hist. Stud. 2001;34:5. [Google Scholar]
- 27.Klieman KA. “The Pygmies Were Our Compass”: Bantu and Batwa in the History of West Central Africa, Early Times to c. 1900 C.E. Heinemann; Portsmouth, NH: 2003. [Google Scholar]
- 28.Ehret C. An African Classical Age: Eastern and Southern Africa in World History, 1000 B.C. to A.D. 400. Univ. Press of Virginia; Charlottesville, VA: 1998. [Google Scholar]
- 29.Ehret C. In: Culture History in the Southern Sudan. Mack J, Robertshaw P, editors. British Institute in Eastern Africa; Nairobi: 1983. pp. 19–48. [Google Scholar]
- 30.Ehret C. The Historical Reconstruction of Southern Cushitic Phonology and Vocabulary. Reimer; Berlin: 1980. [Google Scholar]
- 31.Ehret C. Southern African Humanities. 2008;30:7. [Google Scholar]
- 32.Ehret C. Ethiopians and East Africans: The Problem of Contacts. East African Publishing House; Nairobi: 1974. [Google Scholar]
- 33.Tishkoff SA, et al. Nat. Genet. 2007;39:31. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hassan HY, Underhill PA, Cavalli-Sforza LL, Ibrahim ME. Am. J. Phys. Anthropol. 2008;137:316. doi: 10.1002/ajpa.20876. [DOI] [PubMed] [Google Scholar]
- 35.Bereir RE, et al. Eur. J. Hum. Genet. 2007;15:1183. doi: 10.1038/sj.ejhg.5201892. [DOI] [PubMed] [Google Scholar]
- 36.Cerny V, Salas A, Hajek M, Zaloudkova M, Brdicka R. Ann. Hum. Genet. 2007;71:433. doi: 10.1111/j.1469-1809.2006.00339.x. [DOI] [PubMed] [Google Scholar]
- 37.Ehret C. In: West African Linguistics: Studies in Honor of Russell. Schuh G, Newman P, Hyman L, editors. Ohio State Univ.; Columbus, OH: 2006. pp. 56–66. [Google Scholar]
- 38.Ambrose SH. In: The Archaeological and Linguistic Reconstruction of African History. Ehret C, Posnansky M, editors. Univ. of California Press; Berkeley, CA: 1982. pp. 104–157. [Google Scholar]
- 39.Newman JL. The Peopling of Africa. Yale Univ. Press; New Haven, CT: 1997. [Google Scholar]
- 40.Wood ET, et al. Eur. J. Hum. Genet. 2005;13:867. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- 41.Lim L. Brown University; 1992. thesis. [Google Scholar]
- 42.Ehret C. Sprache Gesch. Afrika. 1986;7:105. [Google Scholar]
- 43.Lomax A, et al. Folk Song Style and Culture. National Association for the Advancement of Science; Washington, DC: 1968. pp. 16–18. 26, 91–92. [Google Scholar]
- 44.Quintana-Murci L, et al. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1596. [Google Scholar]
- 45.Batini C, et al. Mol. Phylogenet. Evol. 2007;43:635. doi: 10.1016/j.ympev.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 46.Destro-Bisol G, et al. Am. Nat. 2004;163:212. doi: 10.1086/381405. [DOI] [PubMed] [Google Scholar]
- 47.Patin E, et al. PLoS Genet. 2009;5:e1000448. doi: 10.1371/journal.pgen.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verdu P, et al. Curr. Biol. 2009;19:312. doi: 10.1016/j.cub.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 49.Destro-Bisol G, et al. Mol. Biol. Evol. 2004;21:1673. doi: 10.1093/molbev/msh186. [DOI] [PubMed] [Google Scholar]
- 50.Coia V, et al. Am. J. Hum. Biol. 2004;16:57. doi: 10.1002/ajhb.10236. [DOI] [PubMed] [Google Scholar]
- 51.Sands B. In: Language, Identity and Conceptualization Among the Khoisan. Schladt M, editor. Vol. 15. Rudiger Kupper; Köln, Germany: 1998. pp. 266–283. [Google Scholar]
- 52.Elderkin ED. Sprache Gesch. Afrika. 1982;4:67. [Google Scholar]
- 53.Scozzari R, et al. Am. J. Hum. Genet. 1999;65:829. doi: 10.1086/302538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA. Am. J. Hum. Genet. 2002;70:265. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavalli-Sforza LL, Piazza A, Menozzi P. History and Geography of Human Genes. Princeton Univ. Press; Princeton, NJ: 1994. [Google Scholar]
- 56.Walter RC, et al. Nature. 2000;405:65. doi: 10.1038/35011048. [DOI] [PubMed] [Google Scholar]
- 57.Parra EJ, et al. Am. J. Phys. Anthropol. 2001;114:18. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 58.Salas A, et al. Am. J. Phys. Anthropol. 2005;128:855. doi: 10.1002/ajpa.20117. [DOI] [PubMed] [Google Scholar]
- 59.Lind JM, et al. Hum. Genet. 2007;120:713. doi: 10.1007/s00439-006-0261-7. [DOI] [PubMed] [Google Scholar]
- 60.Smith MW, et al. Am. J. Hum. Genet. 2004;74:1001. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parra EJ, et al. Am. J. Hum. Genet. 1998;63:1839. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trans-Atlantic Slave Trade Database ( www.slavevoyages.org/tast/index.faces)
- 63.Nurse GT, Weiner JS, Jenkins T. The Peoples of Southern Africa and Their Affinities. Oxford Univ. Press; New York: 1985. [Google Scholar]
- 64.Mellars P. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9381. doi: 10.1073/pnas.0510792103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reynolds JB, et al. Genetics. 1983;105:767. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.