Abstract
Isolated complex I deficiency is the most frequently observed oxidative phosphorylation defect in children with mitochondrial disease, leading to a diverse range of clinical presentations, including Leigh syndrome. For most patients the genetic cause of the biochemical defect remains unknown due to incomplete understanding of the complex I assembly process. Nonetheless, a plethora of pathogenic mutations have been described to date in the seven mitochondrial-encoded subunits of complex I as well as in 12 of the nuclear-encoded subunits and in six assembly factors. Whilst several mitochondrial DNA mutations are recurrent, the majority of these mutations are reported in single families. We have sequenced core structural and functional nuclear-encoded subunits of complex I in a cohort of 34 paediatric patients with isolated complex I deficiency, identifying pathogenic mutations in 6 patients. These included a novel homozygous NDUFS1 mutation in an Asian child with Leigh syndrome, a previously identified NDUFS8 mutation (c.236C>T, p.P79L) in a second Asian child with Leigh-like syndrome and six novel, compound heterozygous NDUFS2 mutations in four white Caucasian patients with Leigh or Leigh-like syndrome. Three of these children harboured an identical NDUFS2 mutation (c.875T>C, p.M292T), which was also identified in conjunction with a novel NDUFS2 splice site mutation (c.866+4A>G) in a fourth Caucasian child who presented to a different diagnostic centre, with microsatellite and single nucleotide polymorphism analyses indicating that this was due to an ancient common founder event. Our results confirm that NDUFS2 is a mutational hotspot in Caucasian children with isolated complex I deficiency and recommend the routine diagnostic investigation of this gene in patients with Leigh or Leigh-like phenotypes.
Keywords: mitochondrial disease, Leigh syndrome, complex I deficiency, NDUFS2, recurrent mutation
Introduction
Complex I [nicotinamide adenine dinucleotide (NADH)–ubiquinone oxidoreductase] is the first, largest (∼1 MDa) and most intricate multimeric component of the mitochondrial respiratory chain, responsible for the transfer of electrons from NADH to ubiquinone coupled with the translocation of protons across the inner mitochondrial membrane. The enzyme is L-shaped, with a hydrophilic peripheral arm that protrudes into the mitochondrial matrix and a hydrophobic arm nestled in the inner mitochondrial membrane. Human complex I consists of 45 subunits, of which 14 highly conserved subunits are thought to form the minimal functional core of the complex (ND1-6, ND4L, NDUFS1-3, NDUFS7-8, NDUFV1-2) (Walker, 1992; Carroll et al., 2006). Seven of the complex I subunits are encoded by the mitochondrial genome (ND1-6 and ND4L), whilst all other subunits and all accessory factors necessary for the correct assembly of complex I are encoded by nuclear genes.
Structural integrity is important for complex I functionality and, given the enzyme’s structural complexity, it is not surprising that isolated complex I deficiency is the most frequently observed oxidative phosphorylation disorder in children with mitochondrial disease (Kirby et al., 1999; Triepels et al., 2001). A wide variety of clinical presentations are associated with isolated complex I deficiency, including hepatopathy, cardiomyopathy, fatal congenital lactic acidosis and Leigh syndrome, where neurological features such as central hypopnoea, dystonia, hypotonia, dysphagia and myopathy are common (Distelmaier et al., 2009). Identifying the plethora of mutations responsible for complex I deficiency is challenging but essential for providing accurate and helpful diagnostic counselling. Mitochondrial DNA (mtDNA) mutations, found in all seven mtDNA genes (Kirby et al., 1999; Lebon et al., 2003; McFarland et al., 2004), are thought to account for ∼25% of cases (Lebon et al., 2003; Bugiani et al., 2004; McFarland et al., 2004), suggesting that nuclear gene defects must be responsible for the remainder of observed cases. To date, pathogenic nuclear DNA mutations have been reported in 12 complex I subunit genes, NDUFS1-4, NDUFS6-8, NDUFV1-2, NDUFA1-2 and NDUFA11 (Loeffen et al., 1998a, 2001; Schuelke et al., 1999; Triepels et al., 1999; Benit et al., 2001, 2003, 2004; Kirby et al., 2004; Fernandez-Moreira et al., 2007; Berger et al., 2008; Hoefs et al., 2008), and six complex I assembly factor genes, NDUFAF1, NDUFAF2, NDUFAF3, NDUFAF4, C8orf38 and C20orf7 (Ogilvie et al., 2005; Dunning et al., 2007; Pagliarini et al., 2008; Saada et al., 2008, 2009; Sugiana et al., 2008; Gerards et al., 2009).
Here, we describe the repeated finding of a heterozygous NDUFS2 nuclear complex I gene mutation (c.875T>C, p.M292T), which, in conjunction with secondary unique heterozygous mutations in the same gene, causes isolated complex I deficiency in four unrelated white Caucasian patients.
Patients
A cohort of 34 paediatric patients with identified isolated complex I deficiency in skeletal muscle was selected for this study (selected patients are presented in Table 1). The cohort was clinically heterogeneous, presenting with a variety of phenotypes, including Leigh syndrome, isolated hypertrophic cardiomyopathy and congenital lactic acidosis. In each case, sequencing of the entire mitochondrial genome had previously excluded the possibility of mtDNA point mutations as the cause of the biochemical defect. Written informed consent was obtained from all families in accordance with the Declaration of Helsinki and the study was approved by the local Ethics Committee.
Table 1.
Mitochondrial respiratory chain complex I activities in patient skeletal muscle homogenates
| Patient | Mutated gene |
Muscle homogenate |
|
|---|---|---|---|
| Complex Ia | Complex I/Complex IIb | ||
| 3 | NDUFS2 | 0.021 (20%) | 0.313 |
| 5 | NDUFS1 | 0.024 (23%) | 0.273 |
| 19 | NDUFS2 | 0.028 (27%) | 0.389 |
| 22 | NDUFS2 | 0.014 (13%) | 0.130 |
| 27 | NDUFS2 | 0.017 (16%) | 0.155 |
The activity of complex I in skeletal muscle from Patient 34 was assessed in a separate diagnostic centre. It was shown to be 30% of control values. Complex I is expressed as nanomoles NADH oxidized/min/unit citrate synthase. Complex II is calculated as nanomoles 2,6-dichlorophenolindophenol reduced/min/unit citrate synthase. Figures in brackets represent the percentage residual complex I activity expressed in muscle.
a Control range (n = 25): 0.104 ± 0.036.
b Control range (n = 25) : 0.52–0.95.
Case reports
Patient 3
This female was the second child of non-consanguineous Caucasian parents who have a healthy son. Her birth weight at 3.03 kg was on the 25th centile, but she fed poorly and vomited repeatedly, so that by 16 weeks her weight was on the 2nd centile. She presented at 8 months with failure to thrive, vomiting and developmental delay. Examination revealed fine sparse hair, generalized hypotonia and intermittent bilateral pendular nystagmus. Plasma and CSF lactate were elevated at 11.66 and 5.56 mmol/l, respectively, and cranial MRI scan demonstrated symmetrical abnormal signal in the cerebral peduncles, dorsal pons and upper medulla, with a lactate peak clearly visible on magnetic resonance spectroscopy. Nasogastric tube feeding was commenced, but episodes of vomiting continued regularly and were often accompanied by a severe acidosis, encephalopathy and developmental regression. At the age of 22 months, the patient developed central hypopnoea and died.
Patient 5
This full-term female infant presented at age 4 months with axial hypotonia, poor feeding, vomiting and failure to thrive. She subsequently developed intermittent nystagmus and would take up fixation only briefly. Fundoscopy showed no abnormalities, and tone in her limbs was normal, as were tendon reflexes. Head growth fell from 25th to 0.4th centile and cranial MRI (at 8 months) showed extensive symmetrical abnormalities in the cerebral peduncles, anterior pons, posterior limbs of the internal capsules and extensive T2 signal change in the white matter of the cerebellar hemispheres. She then developed spasticity in her limbs with brisk tendon reflexes. Echocardiography was normal. The patient’s feeding was further complicated by dysphagia, and nasogastric tube feeding was initiated. At 10.5 months a further sudden deterioration led to central hypopnoea, limb hypotonia, encephalopathy and death 24 h later. The blood and CSF lactate concentrations were raised at 3.3 and 4.7 mmol/l, respectively. Pyruvate dehydrogenase activity in cultured fibroblasts was normal.
The patient’s parents are first cousins and they have a healthy older son. Unfortunately, their second daughter was also affected (Fig. 1A). Again, she was born at term and appeared normal for the first 2 months. Thereafter, her feeding deteriorated and she developed truncal hypotonia and increasing irritability. Subsequently, she also showed poor head growth, and visual fixation ceased at ∼6 months. At 7 months, she developed limb hypertonia. She was managed with nasogastric tube feeding and baclofen but continued to deteriorate and died aged 10 months.
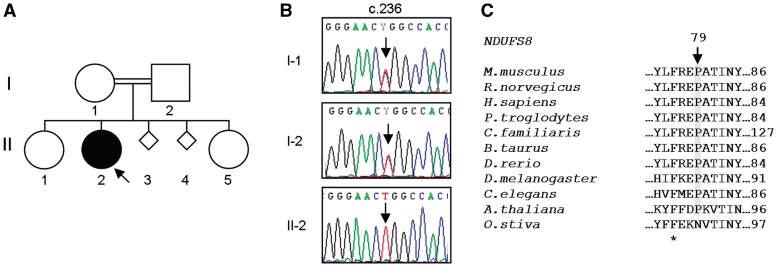
Figure 1.
NDUFS8 homozygous mutation, c.236C>T (p.P79L). (A) Family 34 pedigree, (B) sequence electropherograms of NDUFS8 gene with c.236 position highlighted in patient and family members and (C) amino acid alignment of NDUFS8 orthologues. Alignments were generated with ClustalW using GenBank sequences from Mus musculus (NP_659119.2), Rattus norvegicus (NP_001099792.1), Homo sapiens (NP_002487.1), Pan troglodytes (NP_001065248), Canis lupus familiaris (XP_852209.1), Bos taurus (NP_777243.2), Danio rerio (NP_998304.1), Drosophila melanogaster (NP_524719.1), Arabidopsis thaliana (NP_178022.1), Oryza sativa (NP_001051420.1) and Caenorhabditis elegans (NP_498595.1). Position p.79 (Homo sapiens) is indicated. Conserved sites are shown (asterisk).
Patient 19
This female is the youngest of six children and the only child of the current non-consanguineous relationship. With the exception of a half-brother with attention deficit disorder, the other siblings are healthy, as are both parents. She was born by breech delivery at full term, but did not require resuscitation. She was initially slow to feed, and gross motor skills were delayed due to four-limb dystonia. She did not walk independently until 3 years and remains unsteady at 9 years. Speech has also been delayed and further hampered by a severe dysarthria secondary to dystonia of her face and neck muscles, but communication is facilitated by the use of a computerized communication device. Since the age of 6 years she has had a number of seizures, although her EEG remains normal. Cranial MRI revealed bilateral symmetrical increased T2 signal in the basal ganglia. At the age of 9 years, she attends mainstream school and is independently mobile, albeit with frequent falls.
Patient 22
This female is the only child of healthy non-consanguineous parents. Psychomotor delay, learning difficulties and episodes of tonic upward eye deviation were noted from infancy. She developed progressive dystonia affecting all four limbs, optic nerve hypoplasia, dysarthria and dysphagia, requiring a gastrostomy at the age of 11 years. Cranial MRI at 18 months demonstrated bilateral low-density lesions in the cerebral peduncles with high T2 signal in the thalami and frontal lobes. These changes resolved by 5 years, but new symmetrical lesions in the heads of the caudate and lentiform nuclei were observed bilaterally and persisted when the scan was repeated at 7.5 years. Blood and CSF lactate concentrations were normal, as was plasma biotinidase activity, but activity of the pyruvate dehydrogenase complex in fibroblasts was slightly decreased. Immunostaining for the E1alpha subunit showed no evidence of mosaicism.
Patient 27
This female was the second child of healthy non-consanguineous parents; her brother is in good health. Born by elective Caesarean section at 36 weeks gestation, weighing 1.49 kg, she required no resuscitation at birth, but 2 h later was unwell with an elevated plasma lactate of 18 mmol/l. Plasma ammonia and glucose concentrations were normal, but urinary excretion of malate, fumarate and other tricarboxylic acid cycle intermediates was increased. A mild metabolic acidosis persisted (plasma lactate 6–9 mmol/l) despite treatment with sodium bicarbonate. Swallowing subsequently deteriorated and feeding was further compromised by gastro-oesophageal reflux and vomiting. Cranial MRI was normal. Echocardiography demonstrated mild left ventricular hypertrophy. At 3.5 months of age, she had a respiratory arrest and died. Pyruvate dehydrogenase complex activity in fibroblasts was decreased, but genetic investigations proved normal. Mitochondrial respiratory chain analysis of a skeletal muscle biopsy demonstrated an isolated deficiency of complex I.
Patient 34
This female was the second child of healthy consanguineous Pakistani parents who have two other healthy daughters. Born at 38 weeks gestation, she weighed 2.5 kg on the 9th centile and at 4 weeks she remained jaundiced with poor weight gain. She was admitted to hospital for artificial ventilation following an aspiration event that resulted in a respiratory arrest. Hyperkalaemia with persistent acidosis and raised serum (7.4 mmol/l) and CSF lactate (5.5 mmol/l) were noted. Swallowing remained unsafe and she failed to thrive despite nasogastric tube feeds. The development of seizures prompted an EEG, which demonstrated a diffusely slow record with occasional spikes. She was subsequently readmitted to hospital on several occasions, with hypoventilation and apnoea, and died following a respiratory arrest at the age of 3 months. Extensive investigations including biotinidase, acylcarnitine profile and amino acids were normal in blood. Urinary organic acids and activity of pyruvate dehydrogenase in fibroblasts were also normal, but mitochondrial respiratory chain analysis of skeletal muscle revealed an isolated complex I deficiency (30% of control values).
Patient 35
This male is described elsewhere (Patient 17 in Salemi R et al., submitted for publication). He presented at 34 months of age with developmental delay, ataxia, nystagmus, optic atrophy and a mild, persistent metabolic acidosis. Plasma lactate level was 3.2 mmol/l and CSF lactate was 2.7 mmol/l. Urinary organic acids plus plasma and urinary amino acids were all normal. CT brain scan without contrast was normal at 6 years of age.
Materials and methods
Respiratory chain biochemistry
The activities of the respiratory chain complexes I–IV and the matrix marker citrate synthase were determined in frozen skeletal muscle homogenates from all 34 patients as previously described (Kirby et al., 2007).
DNA extraction and whole genome amplification
Total genomic DNA was extracted from either muscle or blood samples of 34 paediatric patients using the Qiagen DNA mini kit (Qiagen, Crawley, UK) or Nucleon BACC3 Blood and Cell Culture DNA kit (Tepnel, Manchester, UK), respectively.
Whole genome amplification
Prior to polymerase chain reaction (PCR) amplification of nuclear targets, whole genomes were amplified by multiple displacement amplification using Φ29 polymerase (REPLI-g® Mini kit; Qiagen), as per the manufacturer’s instructions.
Polymerase chain reaction amplification
The entire mitochondrial genome was amplified using 36 sets of overlapping M13-tailed primers (Supplementary Tables 1 and 2). The coding regions of nine nuclear encoded complex I genes (NDUFS1-4, NDUFS7, NDUFS8, NDUFV1, NDUFV2 and NDUFAB1) were amplified using intronic M13-tailed primers, designed to flank the coding sequences of the genes (Hinttala et al., 2005) (Supplementary Table 3). All PCRs were carried out in 25 μl volumes containing 0.65 U AmpliTaq Gold polymerase (Applied Biosystems, Warrington, UK), 1× AmpliTaq Gold buffer, 0.8 μM of each primer, 100 μM deoxynucleoside triphosphates and 30-100 ng DNA (mtDNA PCR) or 1 μl of amplified DNA (nuclear gene PCR). Where appropriate, reactions were supplemented with 2% dimethyl sulphoxide (Supplementary Table 3).
Sequencing
All PCR products were purified (ExoSapIT, GE Healthcare, Buckinghamshire, UK) and sequenced with BigDye Terminator cycle sequencing chemistries (Applied Biosystems) on an Applied Biosystems ABI3100 Genetic Analyser. Sequence data were analysed using SeqScape software (v2.1.1, Applied Biosystems) and compared with the GenBank reference sequences: NC_012920 (revised Cambridge reference sequence for human mtDNA), NM_005006.5 (NDUFS1), NM_004550.3 (NDUFS2), NM_004551.1 (NDUFS3), NM_002495.1 (NDUFS4), NM_024407.3 (NDUFS7), NM_002496.1 (NDUFS8), NM_007103.2 (NDUFV1), NM_021074.2 (NDUFV2) and NM_004994.1 (NDUFAB1).
Nuclear haplotype analysis
Patients 3, 19, 22 and 35 and their parents were genotyped for the microsatellite markers D1S2635, D1S2707, D1S2771, D1S2705, D1S2675, D1S2844 and D1S1677 and the single nucleotide polymorphisms (SNPs) rs686015, rs10594, rs11582932, rs1041068, rs12402879, rs352685, rs512645, rs836, rs11576830, rs3924264, rs2501875, rs3935401, rs382627, rs2007773, rs6683580, rs12407444, rs2341481, rs4657136, rs1932933 and rs2341744. SNPs with a minor allele frequency of 0.15–0.48 and located at ∼100 kb intervals up- and downstream of NDUFS2 were chosen. Primer sequences for the microsatellite markers and map positions were obtained from the UniSTS database of the National Center for Biotechnology Information (Supplementary Table 4); primer sequences for the SNPs were designed using Primer3 software (v0.4.0.) (Rozen and Skaletsky, 2000) (Supplementary Table 5). Forward primers for the microsatellite markers were 5′-6-FAM-labelled; forward and reverse primers for the SNP analysis were M13-tailed. Reactions were carried out in 25 μl volumes containing 1 U GoTaq® polymerase (Promega, Southampton, UK), 1× GoTaq® buffer, 0.2 μM of each primer, 100 μM deoxynucleoside triphosphates and 20–100 ng DNA. PCR products (1 μl) were electrophoresed on an ABI3130xl Genetic Analyser (Applied Biosystems) with the GeneScan 500 ROX size standard (Applied Biosystems). Data were analysed using GeneMapper v4.0 software (Applied Biosystems).
Polymerase chain reaction/restriction fragment length polymorphism analysis
The NDUFS1 c.1222C>T mutation was confirmed by PCR/restriction fragment length polymorphism (RFLP) analysis. The assay was performed as previously described (Taylor et al., 2003) with modifications. A 218-base pair fragment encompassing the mutation site was amplified using the M13-tailed forward primer 5′-TGTAAAACGACGGCCAGTCATGGATTCTCTGTATGTCTTAAT-3′ and reverse primer 5′-ACCAACCTCTTTCGAATTCTAGC-3′. PCR amplifications were performed with an annealing temperature of 59°C and PCR products were digested with 10 U NlaIII (New England Biolabs, Hitchin, UK). Wild-type amplicons contain a single NlaIII recognition site and on digestion two fragments of 25 and 193 bp are generated. The c.1222C>T mutation creates an additional NlaIII site, and amplicons harbouring this mutation are digested into three fragments of 25, 45 and 148 bp.
Cell culture
Cultured skin fibroblasts from Patients 3, 5, 19, 22 and 27 and from two normal, paediatric controls were cultured as previously described (Taylor et al., 2003).
RNA extraction and reverse transcription
Total RNA was extracted from fibroblasts using Trizol (Invitrogen, Paisley, UK) according to the manufacturer’s instructions. Reverse transcription of the messenger RNA was performed using the Superscript® II reverse transcriptase kit (Invitrogen).
Measurement of reactive oxygen species levels and mitochondrial mass
Reactive oxygen species levels in cultured skin fibroblasts from Patients 3, 5, 19, 22 and 27 were measured as described previously (Saretzki et al., 2008). Briefly, cells (2 × 105) were stained at 37°C in serum-free Dulbecco’s modified Eagle’s medium (Gibco, Paisley, UK) with 5 μM MitoSOX (Molecular Probes, Eugene, OR, USA) for 15 min to measure mitochondrial superoxide levels or with 10 μM of nonyl acridine orange (Molecular Probes) for 10 min to ascertain mitochondrial mass. Cells were analysed in a flow cytometer (Partec, Münster, Germany) using blue excitation and the green (FL1) and red (FL3) emission channels. System alignment and gain settings were adjusted using fluorescent beads. Cells were gated in forward scatter/side scatter, and the median of the gated fluorescence peak was used. Data were corrected by subtraction of autofluorescence levels in unstained cells. Neonatal human skin fibroblasts (Control 1), MRC-5 lung fibroblasts (Control 3, both from the American Type Tissue Collection, Rockville, Maryland, USA) and paediatric skin fibroblasts from a healthy child (Control 2) were used as controls. To determine statistical significance, two-sample unpaired Student t-tests between each patient and control sample were performed. A Bonferroni corrected P-value ≤ 0.05 was considered statistically significant.
Blue-native polyacrylamide gel electrophoresis
Mitochondria-enriched fractions were prepared from 80% confluent 225 cm2 flasks of patient and control fibroblasts and processed as described elsewhere (Nijtmans et al., 2002). Sample protein concentrations were determined by Bradford assay. Protein samples (75 μg/lane) were separated on 4–13% blue-native gradient gels, cast and run as previously described (Nijtmans et al., 2002). To assess complex I assembly, electrophoresed native gels were incubated for 1 h with dissociating reagent (1% sodium dodecyl suphate, 1% β-mercaptoethanol) prior to protein transfer onto polyvinylidene fluoride membranes (Immobilon-P, Millipore Corporation, Watford, UK). Immunodetection was performed using primary monoclonal antibodies raised against the complex I NDUFA9 and complex II 70 kDa subunits (MitoSciences, Cambridge BioScience, Cambridge, UK). ECL Plus™ (GE Healthcare Life Sciences, Buckinghamshire, UK) chemiluminescence was detected on a Storm 480 PhosphorImager (Molecular Dynamics, Sunnyvale, California, USA). To determine in-gel complex I activity, assays of NADH dehydrogenase activity were performed in-gel with NADH and nitrotetrazolium blue (Sigma, Poole, UK) as substrates, as previously described (Calvaruso et al., 2008).
Sodium dodecyl sulphate polyacrylamide gel electrophoresis
Cell lysates were prepared from 80% confluent 75 cm2 flasks of patient and control fibroblasts in 200 μl lysis buffer (42.5 mM Tris–HCl pH 7.5, 127.5 mM NaCl, 1.7 mM MgCl2, 1% Nonidet P-40 and Roche ethylenediaminetetraacetic acid protease inhibitor cocktail). Samples were vortexed briefly, spun at 560 g for 2 min at 4°C and the supernatants retained. Protein concentrations were determined by Bradford assay. Equal amounts of protein (10–15 μg) were loaded and resolved on 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (PAGE) gels. Following wet transfer onto polyvinylidene fluoride membranes western blotting was performed using monoclonal antibodies against complex I NDUFS9, NDUFS3 and NDUFB8 and complex II 70 kDa subunits (MitoSciences). A monoclonal antibody raised against β-actin, clone AC-15, was also used (Sigma). ECL Plus (GE Healthcare Life Sciences) chemiluminescence was detected on a Storm 480 PhosphorImager (Molecular Dynamics).
Results
Sequence variations in nuclear-encoded complex I subunits
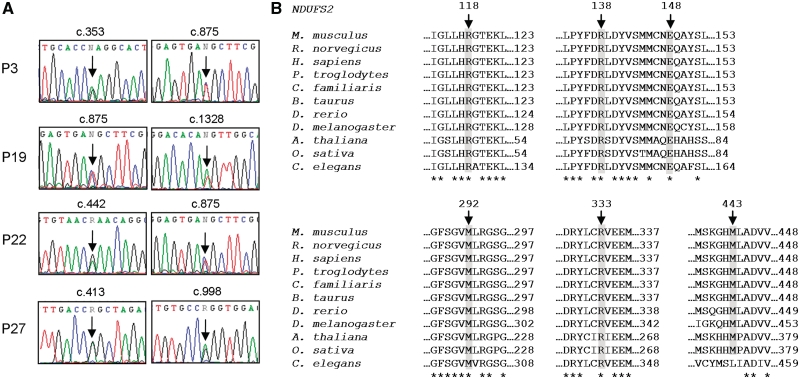
Nine nuclear-encoded complex I subunits (NDUFS1, NDUFS2, NDUFS3, NDUFS4, NDUFS7, NDUFS8, NDUFV1, NDUFV2 and NDUFAB1) were sequenced in a cohort of 34 paediatric patients with demonstrated isolated complex I deficiency in skeletal muscle. Twenty-four sequence variants were identified, almost half of which are synonymous substitutions (Table 2). Of the remaining 13 non-synonymous substitutions, 4 are described SNPs (NDUFS2 p.P20T and p.P352A, NDUFS7 p.P23L and NDUFV2 p.V29A), whilst the homozygous NDUFS8 sequence change in Patient 34 (p.P79L; Fig. 1) has been previously described by Loeffen et al. (1998a) as one of two heterozygous mutations implicated in the pathogenesis of a patient with Leigh-like syndrome. The final 8 sequence changes are, to our knowledge, reported here for the first time. One homozygous NDUFS1 sequence variant, c.1222C>T (predicting p.R408C), was identified in Patient 5 (Fig. 2). Two heterozygous NDUFS2 sequence variants were discovered in Patient 27, c.413G>A (p.R138Q) and c.998G>A (p.R333Q) (Fig. 3A), and a single heterozygous NDUFS2 substitution, c.422A>G (p.Y141C), was detected in Patient 16. Patients 3, 19 and 22 all had an identical NDUFS2 transition, c.875T>C (p.M292T), in addition to secondary unique heterozygous sequence variants in the same gene, namely c.353G>A (p.R118Q), c.1328T>A (p.M443K) and c.442G>A (p.E148K), respectively (Fig. 3A). Interestingly, sequencing of a separate cohort of patients with isolated complex I deficiency (n = 34) in a diagnostic centre in Melbourne (Australia) also identified the NDUFS2 c.875T>C transition in a white Caucasian patient (Patient 35), in conjunction with a second unique heterozygous substitution, c.866+4A>G, located within the 5′-splice site of intron 6. DNA from this patient was included in the haplotype studies described below.
Table 2.
Nuclear complex I gene sequence variants
| Gene | Sequence variant | refSNP ID | Amino acid change |
Patients |
|
|---|---|---|---|---|---|
| Homozygous | Heterozygous | ||||
| Synonymous sequence variants | |||||
| NDUFAB1 | c.102C>T | 31 | |||
| c.108C>G | rs466719 | 10 | |||
| NDUFS1 | c.414 T>C | rs11548670 | 1, 17 | ||
| c.966G>T | rs11548668 | 1, 4, 9, 10, 15, 17, 22, 24, 25, 30 | 3, 5, 7, 12, 13, 16, 23, 26, 31, 32, 33 | ||
| c.1251A>G | rs4147719 | 15, 22, 25, 30 | 1, 3, 4, 5, 9, 12, 13, 16, 17, 18, 23, 24, 26, 31, 32, 34 | ||
| c.1371G>A | 25 | ||||
| NDUFS2 | c.1289C>T | rs1136207 | 3, 13, 16, 19, 22, 27, 28, 29 | ||
| NDUFS4 | c.12G>C | rs2279516 | 6, 8, 9, 12, 14, 21, 23, 26, 27 | 1, 2, 3, 4, 5, 7, 10, 16, 17, 18, 20, 22, 24, 25, 28, 29, 31, 32, 33, 34 | |
| c.198A>C | rs31304 | 1, 2, 3, 4, 5, 6, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 | |||
| c.312A>G | rs31303 | 2, 6, 8, 9, 10, 12, 13, 14, 22, 23, 24, 26, 27, 28, 29, 31 | 1, 3, 4, 5, 7, 11, 15, 16, 17, 25, 32, 33, 34 | ||
| NDUFV2 | c.201A>T | rs41274300 | 3, 8, 15, 22 | 4, 7, 9, 12, 13, 24 | |
| Non-synonymous sequence variants | |||||
| NDUFS1 | c.1222C>T | p.R408C | 5 | ||
| NDUFS2 | c.58C>A | rs11538340 | p.P20 T | 1, 2, 29, 34 | |
| c.353G>A | p.R118Q | 3 | |||
| c.413G>A | p.R138Q | 27 | |||
| c.422A>G | p.Y141C | 16 | |||
| c.442G>A | p.E148 K | 22 | |||
| c.875T>C | p.M292T | 3, 19, 22 | |||
| c.998G>A | p.R333Q | 27 | |||
| c.1054C>G | rs11576415 | p.P352A | 20 | 6, 7, 11, 16, 27 | |
| c.1328 T>A | p.M443 K | 19 | |||
| NDUFS7 | c.68C>T | rs1142530 | p.P23 L | 3, 4, 5, 8, 9, 12, 16, 27, 29, 33 | 2, 6, 7, 13, 17, 23, 25, 34 |
| NDUFS8 | c.236C>T | rs28939679 | p.P79L | 34 | |
| NDUFV2 | c.86 T>C | rs906807 | p.V29A | 1, 3, 5, 6, 7, 9, 10, 12, 15, 16, 17, 18, 19, 20, 21, 22, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33 | 2, 4, 11, 13, 14, 23, 34 |
Sequence variants selected for further investigation and the previously reported pathogenic NDUFS8 mutation (p.P79L) are highlighted in bold.
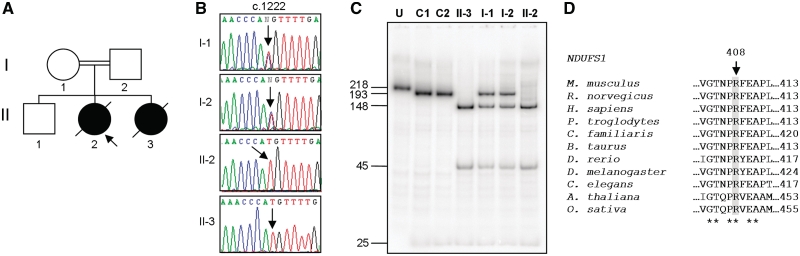
Figure 2.
NDUFS1 homozygous mutation, c.1222C>T (p.R408C). (A) Family 5 pedigree, (B) sequence electropherograms of NDUFS1 gene with c.1222 position highlighted in patient and family members, (C) confirmation of homozygous c.1222C>T mutation by PCR–RFLP. U = uncut; C1 = Control 1; C2 = Control 2. RFLP products were separated through a 12% non-denaturing polyacrylamide gel. The wild-type PCR product contains a single NlaIII site, which cuts the 218-bp amplicon into two fragments of 193 and 25 bp. In amplicons harbouring the c.1222C>T mutation, a second NlaIII site is created that cuts the 193-bp fragment into two smaller fragments of 148 and 45 bp. Fragment sizes (bp) are shown on the left; (D) amino acid alignment of NDUFS1 orthologues. Alignments were generated with ClustalW using GenBank sequences from Mus musculus (NP_663493.1), Rattus norvegicus (NP_001005550.1), Homo sapiens (NP_004997.4), Pan troglodytes (XP_516047.2), Canis lupus familiaris (XP_859697.1), Bos taurus (NP_777245.1), Danio rerio (NP_001007766.1), Drosophila melanogaster (NP_727255.1), Arabidopsis thaliana (NP_568550.1), Oryza sativa (NP_001051072.1) and Caenorhabditis elegans (NP_503733.1). Position p.408 (Homo sapiens) is indicated. Conserved sites are indicated (asterisk).
Figure 3.
NDUFS2 heterozygous mutations, c.353G>A (p.R118Q), c.413G>A (p.R138Q), c.442G>A (p.E148 K), c.875T>C (p.M292T), c.998G>A (p.R333Q) and c.1328 T>A (p.M443 K). (A) Sequence electropherograms of NDUFS2 gene with c.353, c.413, c.442, c.875, c.998 and c.1328 positions highlighted in Patients (P) 3, 19, 22 and 27; (B) amino acid alignments of NDUFS2 orthologues. Alignments were generated with ClustalW using GenBank sequences from Mus musculus (NP_694704.1), Rattus norvegicus (NP_001011907.1), Homo sapiens (NP_004541.1), Pan troglodytes (XP_001173728.1), Canis lupus familiaris (XP_536138.2), Bos taurus (NP_001068605.1), Danio rerio (NP_001018481.1), Drosophila melanogaster (NP_651926.1), Arabidopsis thaliana (NP_085511.1), Oryza sativa (YP_514643.1) and Caenorhabditis elegans (NP_498423.1). Positions p.118, p.138, p.148, p.292, p.333 and p.443 (Homo sapiens) are indicated. Conserved sites are indicated (asterisk).
Sequence variants NDUFS1 c.1222C>T and NDUFS2 c.353G>A, c.413G>A, c.442G>A, c.875T>C, c.998G>A and c.1328T>A were selected for further investigation. To exclude the possibility of ‘allele dropout’, the homozygous NDUFS1 c.1222C>T transition was verified by PCR/RFLP (Fig. 2C). All seven substitutions occur at highly evolutionarily conserved sites (Figs 2D and 3B) and neither the NDUFS1 sequence variant nor the NDUFS2 substitutions were identified in panels of 120 and 380, respectively, healthy, ethnically matched control chromosomes (data not shown).
Further characterization of the NDUFS2 c.422A>G mutation in Patient 16 was not performed due to a lack of observation of disease in either parent and the absence of an additional potentially pathogenic heterozygous change within NDUFS2 or any of the other investigated genes. Following confirmation of the recessive inheritance of the NDUFS8 mutation in Patient 34 and in view of the highly conserved nature of the affected residue (Fig. 1B and C), no further investigations were carried out on this patient, as the pathogenicity of this mutation has previously been proven (Loeffen et al., 1998a; Ahlers et al., 2000; Ugalde et al., 2004a).
Inheritance of NDUFS1 and NDUFS2 alleles
Parental genomic DNA samples were sequenced to confirm recessive inheritance of the NDUFS1 and NDUFS2 alleles. Both the mother and father of Patient 5 were revealed to be carriers of the NDUFS1 c.1222C>T transition (Fig. 2B and C). Unfortunately, DNA was unavailable from her unaffected brother; however, her clinically affected sister was similarly homozygous for this sequence variant. For all five NDUFS2 patients, each parent was shown to be heterozygous for only one of the two substitutions identified in respective patients (data not shown). During our investigations, the mother of Patient 3 conceived a third pregnancy and the parents requested prenatal genetic testing. Analysis of a chorionic villus biopsy failed to identify either of the familial NDUFS2 mutations (c.353G>A or c.875T>C).
Nuclear and mitochondrial DNA haplotype analysis
To determine whether the repeatedly observed NDUFS2 c.875T>C transition could be attributed to a common founder event, Patients 3, 19, 22 and 35 and their parents were initially genotyped for microsatellite markers located upstream (D1S2635, D1S2707, D1S2771 and D1S2705) and downstream (D1S2675, D1S2844 and D1S1677) of NDUFS2 (Supplementary Table 4). D1S2705 was the only common flanking marker in all four patients. Due to the shortfall of microsatellite markers within the genomic region surrounding the NDUFS2 gene, a more exhaustive investigation was subsequently carried out by SNP analysis, using SNPs spaced ∼100 kb apart upstream (rs686015, rs10594, rs11582932, rs1041068, rs12402879, rs352685, rs512645, rs836 and rs11576830) and downstream (rs2501875, rs3935401, rs382627, rs2007773, rs6683580, rs12407444, rs2341481, rs4657136, rs1932933 and rs2341744) of NDUFS2. The intragenic NDUFS2 SNPs, rs3924264 and rs1136207, were also assessed (Supplementary Table 5). Up to 12 SNPs, spanning a region ∼480-kb upstream and ∼520-kb downstream of NDUFS2, appeared to co-segregate with the c.875C>T sequence variant in all four patients, suggesting a mutation-specific haplotype [rs352685 (G), rs512645 (A), D1S2705 (156), rs836 (G), rs11576830 (C), rs3924264 (A), rs1136207 (T), rs2501875 (G), rs3935401 (T), rs382627 (C/T), rs2007773 (A), rs6683580 (A) and rs12407444 (A); Supplementary Fig. 1].
Mitochondrial DNA haplotypes were determined for each c.875T>C patient from mtDNA polymorphisms. All patients belonged to independent Caucasian mitochondrial haplogroups, namely V (Patient 3), K (Patient 19), J (Patient 22) and U (Patient 35).
Fibroblast complex I activity
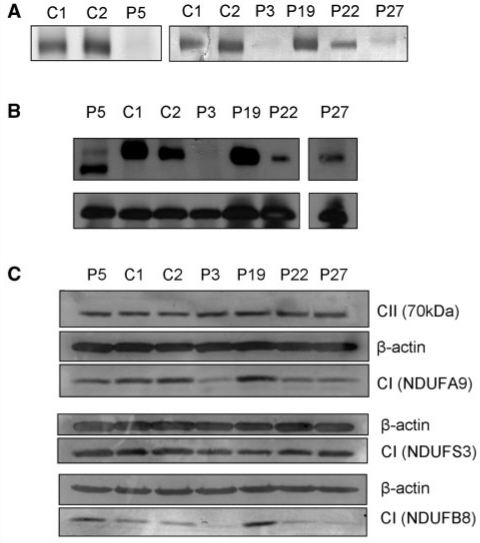
Investigation of complex I assembly and stability in muscle was not possible due to insufficient patient samples. However, cultured skin fibroblasts were available from Patients 3, 5, 19, 22 and 27. As cell lines do not consistently exhibit the complex I deficient phenotype (Mazzella et al., 1997; Benit et al., 2001; Antonicka et al., 2003), the activity of the complex was first assessed in these fibroblasts. In-gel complex I activity assays were performed on protein samples separated on 1D blue-native PAGE. Decreased complex I activity was identified in cell lines from Patients 3, 5 and 27, and to a lesser extent in cells from Patient 22 (Fig. 4A). Fibroblasts of Patient 19 demonstrated complex I activity comparable to controls.
Figure 4.
Complex I in-gel activity and assembly in patient fibroblasts. (A) Complex I in-gel activity. Mitochondria-enriched protein fractions (75 μg) extracted from patient (P) and control (C) fibroblasts were separated on 5–13% 1D blue-native PAGE gels. Gels were histochemically stained for 2 h with 2 mM Tris–HCl (pH 7.4), 0.1 mg/ml NADH and 2.5 mg/ml nitrotetrazolium blue to assess complex I activity. (B) Complex I assembly. Mitochondria-enriched protein fractions (75 μg) extracted from patient and control fibroblasts were separated on 5–13% 1D blue-native PAGE gels. Proteins were transferred to a polyvinylidine fluoride membrane and probed with antibodies raised against complex I NDUFS3 and complex II 70 kDa subunits. (C) Complex I subunit expression. Total cellular protein lysates (10 μg) of patient and control fibroblasts were separated on 10% sodium dodecyl sulphate–PAGE gels then transferred to a polyvinylidine fluoride membrane. Western blotting was performed with antibodies raised against complex I subunits NDUFS9, NDUFS3 and NDUFB8, complex II 70 kDa subunit and β-actin.
Transcription of mutant and wild-type NDUFS2 alleles
To ensure that all NDUFS2 alleles (both wild-type and mutant) were being transcribed, total RNA was extracted from fibroblasts of Patients 3, 19, 22 and 27. Following DNase1 treatment and reverse transcription of the RNA, regions of complementary DNA encompassing the putative pathogenic NDUFS2 mutations were sequenced. In all cases, both NDUFS2 alleles were found to be expressed (data not shown).
Assembly of complex I
Complex I assembly in patient and control fibroblasts was assessed by western analysis of a 1D blue-native PAGE gel (Fig. 4B). An antibody against NDUFS3 was used to visualize complex I. To confirm loading, an antibody against the 70 kDa subunit of complex II was used. No complex I was detected in fibroblasts of Patient 3. Fully assembled complex I was present in fibroblasts of Patient 19 at levels comparable to controls. Intact complex I was also observed in fibroblasts of Patients 22 and 27, albeit at noticeably lower levels than controls. Whilst a marked decrease in fully assembled complex I was evident in fibroblasts of Patient 5, high levels of a lower molecular weight sub-complex were also observed.
Complex I subunit protein expression
Protein expression levels of complex I subunits NDUFA9, NDUFS3 and NDUFB8 were assessed by sodium dodecyl sulphate–PAGE in patient and control fibroblasts (Fig. 4C). To check loading, the expression levels of the 70 kDa subunit of complex II and β-actin were also analysed. No difference in NDUFS3 protein levels was observed in patient cells relative to controls. NDUFA9 and NDUFB8 protein levels were reduced in fibroblasts of Patients 3, 22 and 27 when compared with controls. No marked changes in NDUFA9 protein levels were evident in fibroblasts of Patient 5, whereas NDUFB8 levels were slightly increased. In fibroblasts of Patient 19, the expression levels of both NDUFA9 and NDUFB8 subunits were strongly increased relative to controls.
Reactive oxygen species levels
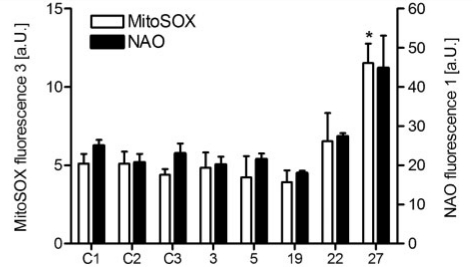
Three control fibroblasts [healthy neonatal (C1), paediatric (C2) or MRC-5 embryonic lung fibroblasts (C3)] demonstrated consistent values of MitoSOX and nonyl acridine orange staining, which were used as standards for cells without any mitochondrial defect. Cells from Patients 3, 5, 19 and 22 did not show any elevated levels of mitochondrial superoxides, while Patient 27 fibroblasts displayed an approximate 2.5-fold increase in MitoSOX staining (Bonferroni corrected P < 0.05; Fig. 5). Analysis of mitochondrial mass revealed no significant differences in patient fibroblasts when compared with controls, although nonyl acridine orange staining of fibroblasts of Patient 27 was slightly elevated.
Figure 5.
Reactive oxygen species levels and mitochondrial mass in patient fibroblasts. Mitochondrial superoxide levels were determined using MitoSOX (white bars) and mitochondrial mass was assessed with nonyl acridine orange (black bars) in fibroblasts of Patients 3, 5, 19, 22 and 27. Values are mean and standard error from at least three independent experiments. a.U. = arbitrary units; C1 = Control 1 (human neonatal skin fibroblasts); C2 = Control 2 (healthy paediatric skin fibroblasts); C3 = Control 3 (MRC-5 lung fibrobasts); NAO = nonyl acridine orange; *Bonferroni corrected P < 0.05.
Discussion
We report on the sequencing of functionally important nuclear-encoded complex I genes in a clinically heterogeneous cohort of 34 paediatric patients with isolated complex I deficiency in skeletal muscle. Our investigations identified the previously reported NDUFS8 mutation (c.236C>T, p.P79L) (Loeffen et al., 1998a) in a homozygous state in an Asian patient with Leigh-like syndrome as well as seven novel, potentially pathogenic mutations in five additional patients, namely one homozygous NDUFS1 change (c.1222C>T, p.R408C) in an Asian Leigh syndrome patient from a consanguineous family and six compound heterozygous NDUFS2 substitutions in four unrelated white Caucasian infants with Leigh or Leigh-like syndrome (c.353G>A and c.875T>C, c.875T>C and c.1328T>A, c.442G>A and c.875T>C, and c.413G>A and c.998G>A). Interestingly, the NDUFS2 c.875T>C (p.M292T) mutation was found in four unrelated patients identified at two different diagnostic centres.
Until now, NDUFS2 mutations have been specifically associated with encephalopathy and hypertrophic cardiomyopathy (Loeffen et al., 2001). We demonstrate in this report, however, that compound heterozygous NDUFS2 mutations can also occur in cases of Leigh and Leigh-like syndromes. Several lines of evidence support the pathogenicity of the novel NDUFS2 substitutions described: the conservation of the affected amino acids across different species, the absence of the sequence variants in 380 control alleles and the independent inheritance of each individual sequence variant all strongly indicate that these mutations are responsible for the isolated complex I deficiency observed. The complex I assembly data are also indicative of NDUFS2 disruptions in these patients. NDUFS2 is located in the hydrophilic arm of complex I, close to the membrane domain (Guenebaut et al., 1997), and forms part of an early peripheral arm subassembly with subunits NDUFS3, NDUFS7 and NDUFS8 (Ugalde et al., 2004b). Previous blue-native PAGE studies have demonstrated that mutations within these subunits result in markedly reduced levels of fully assembled complex I (Procaccio and Wallace, 2004; Ugalde et al., 2004a; Lebon et al., 2007). In keeping with these findings, we observed reductions in full-size complex I levels in patient fibroblasts that exhibited a complex I deficiency. Moreover, we demonstrated by western analysis that expression levels of subunits NDUFA9 and NDUFB8, known to integrate later in the complex I assembly pathway than NDUFS2 (Lazarou et al., 2007; Vogel et al., 2007), were decreased relative to controls in these cells. These data suggest NDUFA9 and NDUFB8 are more rapidly degraded in the complex I-deficient cell lines, possibly as a consequence of not being maintained within a stable, fully assembled enzyme complex. Invariant NDUFS3 protein levels indicate mutant NDUFS2 can thus support the assembly of only an early peripheral arm sub-complex. In the case of Patient 19, whose fibroblasts were not complex I deficient, the increased levels of NDUFA9 and NDUFB8 may reflect a compensatory mechanism the cells have acquired to overcome the NDUFS2 mutations.
Due to the large number of constitutive complex I subunits and associated assembly factors, the vast majority of mutations associated with isolated complex I deficiency that have been reported to date are unique. Several recurrent mtDNA mutations have been identified (Chol et al., 2003; Lebon et al., 2003; Sarzi et al., 2007) and recently, Berger et al. (2008) uncovered an NDUFA11 intronic splice site mutation in three unrelated consanguineous families. For the first time, we have identified multiple occurrences of nuclear coding sequence mutations in NDUFS8 and in NDUFS2. The NDUFS2 mutation (p.M292T) was associated with Leigh and Leigh-like syndromes in four unrelated Caucasian families. Microsatellite and SNP data suggest this transition is associated with a particular chromosome 1 haplotype (spanning ∼1 Mb around NDUFS2), indicative of an ancient common founder for this mutation within the European population, a notion that is further corroborated by the diverse mtDNA haplogroups of these patients. Prediction of the structural and functional effects of this common NDUFS2 sequence variant (c.875T>C) using PredictProtein (http://www.predictprotein.org) (Rost et al., 2004) identified that an amino acid change from methionine to threonine at residue 292 may result in the introduction of a protein kinase C phosphorylation site (Toll-like receptor) at position p.292. NDUFS2 already contains a conserved protein kinase C phosphorylation site (soluble leptin receptor) at the C-terminus of the protein (Loeffen et al., 1998b); the introduction of an additional site may affect the regulatory function of the protein, but this has yet to be evaluated.
NDUFS1, along with NDUFS4, NDUFS6 and NDUFV1, is located at the tip of the hydrophilic arm of complex I and, in contrast with NDUFS2, incorporates much later in the complex I assembly sequence. Consequently, mutations within these subunits are characterized by an accumulation of a ∼830 kDa enzymatically inactive sub-complex intermediate (Ugalde et al., 2004a; Iuso et al., 2006). Sub-complex accumulation in fibroblasts of Patient 5 is therefore indicative of a mutation in a late-incorporating complex I subunit such as NDUFS1, as are the protein expression data, which demonstrate no change in NDUFS3 or NDUFA9 levels, two subunits thought to assemble into complex I before and at the same time, respectively, as NDUFS1 (Lazarou et al., 2007; Vogel et al., 2007). Absence of the c.1222C>T transition in 60 healthy ethnically matched controls further corroborates the pathogenicity of the mutation, as does its presence in both alleles of an affected sibling, whilst both unaffected parents are heterozygous. In addition, the transition is likely to cause the substitution of a conserved positively charged arginine residue to an uncharged, sulphur-containing cysteine amino acid, the structural consequences of which could include aberrant disulphide bond formation, altered binding or disrupted iron–sulphur cluster coordination.
Reported effects of complex I deficiency on levels of reactive oxygen species production in patient fibroblasts are contradictory. Increases in reactive oxygen species production have been demonstrated in patient fibroblasts harbouring mutations in various complex I subunits including NDUFS1, NDUFS2, NDUFS4 and NDUFV1 (Iuso et al., 2006; Verkaart et al., 2007); however, these same reports and others also describe no changes in intracellular reactive oxygen species levels in cells with NDUFS4, NDUFA1 or NDUFV1 mutations (Iuso et al., 2006; Verkaart et al., 2007; Moran et al., 2010). In accordance with these conflicting data, we found an increased level of oxidative stress in only one of the patient cell lines (Patient 27; NDUFS2, p.R138Q and p.R333Q). While the elevated reactive oxygen species levels in these cells most likely reflect increased mitochondrial proliferation rather than increased reactive oxygen species production per mitochondrion (Fig. 5), our results and others suggest different mechanisms of disease pathogenesis, even for defects within the same structural subunit of complex I. The specific location of a mutation within a given subunit is seemingly important in influencing reactive oxygen species production; however, further investigations are still necessary to uncover the exact pathophysiological mechanisms of reactive oxygen species production in complex I deficient cells.
In our cohort of 34 patients with isolated complex I deficiency, we identified a nuclear genetic aetiology in 17.5% of cases, comparable to other nuclear gene sequencing studies in which the frequency of genetically diagnosed patients ranged from 7.7% to 25% (Bugiani et al., 2004; Ugalde et al., 2004a; Martin et al., 2005). Similarly, Benit et al. (2004) identified a nuclear genetic defect in 17% of their complex I deficient patients using denaturing high-performance liquid chromatography. Despite more widespread nuclear gene screening, it is estimated that underlying genetic defects (either mitochondrial or nuclear) are still identified in only 50% of all complex I-deficient cases (Thorburn et al., 2004). This low frequency is undoubtedly due to the fact that only a handful of the structural subunits and assembly factors of complex I are analysed in each study. In order to improve mutation ‘hit rates’, diagnostic strategies need to routinely include screening of all complex I-associated genes. It is therefore imperative that more high-throughput and rapid approaches be adopted, such as next generation sequencing, tiling arrays or Surveyor™ nuclease technology, which utilizes a celery endonuclease highly specific for DNA mismatches that may be created following hybridization of patient and wild-type DNA (Pagniez-Mammeri et al., 2009). Concerted efforts to further our understanding of complex I assembly and uncover all factors implicated in this process are also necessary. Of note is the recent approach taken by Pagliarini et al. (2008) to elucidate complex I function, which relied on the phylogenetic profiling of MitoCarta, a comprehensive compendium of the mitochondrial proteome. These evolutionary analyses unearthed several strong candidates for involvement in complex I activity, of which three have subsequently been defined as disease-causing genes, C8orf38 (Pagliarini et al., 2008), C3orf60 (Saada et al., 2009) and C20orf7 (Sugiana et al., 2008).
In summary, we report here on the genetic screening of a cohort of 34 paediatric patients with isolated complex I deficiency. A plethora of novel compound heterozygous NDUFS2 mutations were uncovered, including the c.875T>C (p.M292T) transition that was repeatedly identified in four unrelated white Caucasian infants with Leigh or Leigh-like syndrome. Homozygous substitutions in NDUFS1 (c.1222C>T, p.R408C) and NDUFS8 (c.236C>T, p.P79L) were also discovered in two patients from unrelated consanguineous families who presented with Leigh and Leigh-like syndrome, respectively. Based on our results, we propose that NDUFS2 is a mutation hotspot among white Caucasian patients with isolated complex I deficiency and Leigh or Leigh-like syndrome and recommend routine screening of the gene for patients with isolated complex I deficiency.
Funding
Wellcome Trust Programme Grant (074454/Z/04/Z; to Z.M.A.C-L. and R.W.T.); the Newcastle upon Tyne Hospitals NHS Foundation Trust Special Trustees; United Kingdom National Commissioning Group ‘Rare Mitochondrial Disorders of Adults and Children’ Diagnostic Service (http://www.mitochondrialncg.nhs.uk); Australian National Health and Medical Research Council Principal Research Fellowship (to D.R.T.).
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Acknowledgements
The authors wish to thank Professor Yanick Crow and Professor Patrick Chinnery for their help in providing appropriate ethnically matched control DNA samples, and Dr Gavin Hudson for advice regarding haplotyping.
Glossary
Abbreviations
- mtDNA
mitochondrial DNA
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- RFLP
restriction fragment length polymorphism
- SNP
single nucleotide polymorphism
References
- Ahlers PM, Garofano A, Kerscher SJ, Brandt U. Application of the obligate aerobic yeast Yarrowia lipolytica as a eucaryotic model to analyse Leigh syndrome mutations in the complex I core subunits PSST and TYKY. Biochim Biophys Acta. 2000;1459:258–65. doi: 10.1016/s0005-2728(00)00160-2. [DOI] [PubMed] [Google Scholar]
- Antonicka H, Ogilvie I, Taivassalo T, Anitori RP, Haller RG, Vissing J, et al. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J Biol Chem. 2003;278:43081–8. doi: 10.1074/jbc.M304998200. [DOI] [PubMed] [Google Scholar]
- Benit P, Chretien D, Kadhom N, de Lonlay-Debeney P, Cormier-Daire V, Cabral A, et al. Large-scale deletion and point mutations of the nuclear NDUFV1 and NDUFS1 genes in mitochondrial complex I deficiency. Am J Hum Genet. 2001;68:1344–52. doi: 10.1086/320603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benit P, Slama A, Cartault F, Giurgea I, Chretien D, Lebon S, et al. Mutant NDUFS3 subunit of mitochondrial complex I causes Leigh syndrome. J Med Genet. 2004;41:14–7. doi: 10.1136/jmg.2003.014316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benit P, Steffann J, Lebon S, Chretien D, Kadhom N, de Lonlay P, et al. Genotyping microsatellite DNA markers at putative disease loci in inbred/multiplex families with respiratory chain complex I deficiency allows rapid identification of a novel nonsense mutation (IVS1nt -1) in the NDUFS4 gene in Leigh syndrome. Hum Genet. 2003;112:563–6. doi: 10.1007/s00439-002-0884-2. [DOI] [PubMed] [Google Scholar]
- Berger I, Hershkovitz E, Shaag A, Edvardson S, Saada A, Elpeleg O. Mitochondrial complex I deficiency caused by a deleterious NDUFA11 mutation. Ann Neurol. 2008;63:405–8. doi: 10.1002/ana.21332. [DOI] [PubMed] [Google Scholar]
- Bugiani M, Invernizzi F, Alberio S, Briem E, Lamantea E, Carrara F, et al. Clinical and molecular findings in children with complex I deficiency. Biochim Biophys Acta. 2004;1659:136–47. doi: 10.1016/j.bbabio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Calvaruso MA, Smeitink J, Nijtmans L. Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods. 2008;46:281–7. doi: 10.1016/j.ymeth.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine complex I is a complex of 45 different subunits. J Biol Chem. 2006;281:32724–7. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- Chol M, Lebon S, Benit P, Chretien D, de Lonlay P, Goldenberg A, et al. The mitochondrial DNA G13513A MELAS mutation in the NADH dehydrogenase 5 gene is a frequent cause of Leigh-like syndrome with isolated complex I deficiency. J Med Genet. 2003;40:188–91. doi: 10.1136/jmg.40.3.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelmaier F, Koopman WJ, van den Heuvel LP, Rodenburg RJ, Mayatepek E, Willems PH, et al. Mitochondrial complex I deficiency: from organelle dysfunction to clinical disease. Brain. 2009;132 (Pt 4):833–42. doi: 10.1093/brain/awp058. [DOI] [PubMed] [Google Scholar]
- Dunning CJ, McKenzie M, Sugiana C, Lazarou M, Silke J, Connelly A, et al. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 2007;26:3227–37. doi: 10.1038/sj.emboj.7601748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moreira D, Ugalde C, Smeets R, Rodenburg RJ, Lopez-Laso E, Ruiz-Falco ML, et al. X-linked NDUFA1 gene mutations associated with mitochondrial encephalomyopathy. Ann Neurol. 2007;61:73–83. doi: 10.1002/ana.21036. [DOI] [PubMed] [Google Scholar]
- Gerards M, Sluiter W, van den Bosch BJ, de Wit E, Calis CM, Frentzen M, et al. Defective complex I assembly due to C20orf7 mutations as a new cause of Leigh syndrome. J Med Genet. 2010;47:507–12. doi: 10.1136/jmg.2009.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenebaut V, Vincentelli R, Mills D, Weiss H, Leonard KR. Three-dimensional structure of NADH-dehydrogenase from Neurospora crassa by electron microscopy and conical tilt reconstruction. J Mol Biol. 1997;265:409–18. doi: 10.1006/jmbi.1996.0753. [DOI] [PubMed] [Google Scholar]
- Hinttala R, Uusimaa J, Remes AM, Rantala H, Hassinen IE, Majamaa K. Sequence analysis of nuclear genes encoding functionally important complex I subunits in children with encephalomyopathy. J Mol Med. 2005;83:786–94. doi: 10.1007/s00109-005-0712-y. [DOI] [PubMed] [Google Scholar]
- Hoefs SJ, Dieteren CE, Distelmaier F, Janssen RJ, Epplen A, Swarts HG, et al. NDUFA2 complex I mutation leads to Leigh disease. Am J Hum Genet. 2008;82:1306–15. doi: 10.1016/j.ajhg.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuso A, Scacco S, Piccoli C, Bellomo F, Petruzzella V, Trentadue R, et al. Dysfunctions of cellular oxidative metabolism in patients with mutations in the NDUFS1 and NDUFS4 genes of complex I. J Biol Chem. 2006;281:10374–80. doi: 10.1074/jbc.M513387200. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, Thorburn DR. Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology. 1999;52:1255–64. doi: 10.1212/wnl.52.6.1255. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Salemi R, Sugiana C, Ohtake A, Parry L, Bell KM, et al. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J Clin Invest. 2004;114:837–45. doi: 10.1172/JCI20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby DM, Thorburn DR, Turnbull DM, Taylor RW. Biochemical assays of respiratory chain complex activity. Methods Cell Biol. 2007;80:93–119. doi: 10.1016/S0091-679X(06)80004-X. [DOI] [PubMed] [Google Scholar]
- Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol. 2007;27:4228–37. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebon S, Chol M, Benit P, Mugnier C, Chretien D, Giurgea I, et al. Recurrent de novo mitochondrial DNA mutations in respiratory chain deficiency. J Med Genet. 2003;40:896–9. doi: 10.1136/jmg.40.12.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebon S, Minai L, Chretien D, Corcos J, Serre V, Kadhom N, et al. A novel mutation of the NDUFS7 gene leads to activation of a cryptic exon and impaired assembly of mitochondrial complex I in a patient with Leigh syndrome. Mol Genet Metab. 2007;92:104–8. doi: 10.1016/j.ymgme.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Loeffen J, Elpeleg O, Smeitink J, Smeets R, Stockler-Ipsiroglu S, Mandel H, et al. Mutations in the complex I NDUFS2 gene of patients with cardiomyopathy and encephalomyopathy. Ann Neurol. 2001;49:195–201. doi: 10.1002/1531-8249(20010201)49:2<195::aid-ana39>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Loeffen J, Smeitink J, Triepels R, Smeets R, Schuelke M, Sengers R, et al. The first nuclear-encoded complex I mutation in a patient with Leigh syndrome. Am J Hum Genet. 1998a;63:1598–608. doi: 10.1086/302154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffen J, van den Heuvel L, Smeets R, Triepels R, Sengers R, Trijbels F, et al. cDNA sequence and chromosomal localization of the remaining three human nuclear encoded iron sulphur protein (IP) subunits of complex I: the human IP fraction is completed. Biochem Biophys Res Commun. 1998b;247:751–8. doi: 10.1006/bbrc.1998.8882. [DOI] [PubMed] [Google Scholar]
- Martin MA, Blazquez A, Gutierrez-Solana LG, Fernandez-Moreira D, Briones P, Andreu AL, et al. Leigh syndrome associated with mitochondrial complex I deficiency due to a novel mutation in the NDUFS1 gene. Arch Neurol. 2005;62:659–61. doi: 10.1001/archneur.62.4.659. [DOI] [PubMed] [Google Scholar]
- Mazzella M, Cerone R, Bonacci W, Caruso U, Munnich A, Rustin P, et al. Severe complex I deficiency in a case of neonatal-onset lactic acidosis and fatal liver failure. Acta Paediatr. 1997;86:326–9. doi: 10.1111/j.1651-2227.1997.tb08901.x. [DOI] [PubMed] [Google Scholar]
- McFarland R, Kirby DM, Fowler KJ, Ohtake A, Ryan MT, Amor DJ, et al. De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Ann Neurol. 2004;55:58–64. doi: 10.1002/ana.10787. [DOI] [PubMed] [Google Scholar]
- Moran M, Rivera H, Sanchez-Arago M, Blazquez A, Merinero B, Ugalde C, et al. Mitochondrial bioenergetics and dynamics interplay in complex I-deficient fibroblasts. Biochim Biophys Acta. 2010;1802:443–53. doi: 10.1016/j.bbadis.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Henderson NS, Holt IJ. Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods. 2002;26:327–34. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- Ogilvie I, Kennaway NG, Shoubridge EA. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest. 2005;115:2784–92. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–23. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagniez-Mammeri H, Lombes A, Brivet M, Ogier-de Baulny H, Landrieu P, Legrand A, et al. Rapid screening for nuclear genes mutations in isolated respiratory chain complex I defects. Mol Genet Metab. 2009;96:196–200. doi: 10.1016/j.ymgme.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Procaccio V, Wallace DC. Late-onset Leigh syndrome in a patient with mitochondrial complex I NDUFS8 mutations. Neurology. 2004;62:1899–901. doi: 10.1212/01.wnl.0000125251.56131.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32 (Web Server issue):W321–6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Saada A, Edvardson S, Rapoport M, Shaag A, Amry K, Miller C, et al. C6ORF66 is an assembly factor of mitochondrial complex I. Am J Hum Genet. 2008;82:32–8. doi: 10.1016/j.ajhg.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saada A, Vogel RO, Hoefs SJ, van den Brand MA, Wessels HJ, Willems PH, et al. Mutations in NDUFAF3 (C3ORF60), encoding an NDUFAF4 (C6ORF66)-interacting complex I assembly protein, cause fatal neonatal mitochondrial disease. Am J Hum Genet. 2009;84:718–27. doi: 10.1016/j.ajhg.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saretzki G, Walter T, Atkinson S, Passos JF, Bareth B, Keith WN, et al. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells. 2008;26:455–64. doi: 10.1634/stemcells.2007-0628. [DOI] [PubMed] [Google Scholar]
- Sarzi E, Brown MD, Lebon S, Chretien D, Munnich A, Rotig A, et al. A novel recurrent mitochondrial DNA mutation in ND3 gene is associated with isolated complex I deficiency causing Leigh syndrome and dystonia. Am J Med Genet A. 2007;143:33–41. doi: 10.1002/ajmg.a.31565. [DOI] [PubMed] [Google Scholar]
- Schuelke M, Smeitink J, Mariman E, Loeffen J, Plecko B, Trijbels F, et al. Mutant NDUFV1 subunit of mitochondrial complex I causes leukodystrophy and myoclonic epilepsy. Nat Genet. 1999;21:260–1. doi: 10.1038/6772. [DOI] [PubMed] [Google Scholar]
- Sugiana C, Pagliarini DJ, McKenzie M, Kirby DM, Salemi R, Abu-Amero KK, et al. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am J Hum Genet. 2008;83:468–78. doi: 10.1016/j.ajhg.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Giordano C, Davidson MM, d’Amati G, Bain H, Hayes CM, et al. A homoplasmic mitochondrial transfer ribonucleic acid mutation as a cause of maternally inherited hypertrophic cardiomyopathy. J Am Coll Cardiol. 2003;41:1786–96. doi: 10.1016/s0735-1097(03)00300-0. [DOI] [PubMed] [Google Scholar]
- Thorburn DR, Sugiana C, Salemi R, Kirby DM, Worgan L, Ohtake A, et al. Biochemical and molecular diagnosis of mitochondrial respiratory chain disorders. Biochim Biophys Acta. 2004;1659:121–8. doi: 10.1016/j.bbabio.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Triepels RH, van den Heuvel LP, Loeffen JL, Buskens CA, Smeets RJ, Rubio Gozalbo ME, et al. Leigh syndrome associated with a mutation in the NDUFS7 (PSST) nuclear encoded subunit of complex I. Ann Neurol. 1999;45:787–90. doi: 10.1002/1531-8249(199906)45:6<787::aid-ana13>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Triepels RH, Van Den Heuvel LP, Trijbels JM, Smeitink JA. Respiratory chain complex I deficiency. Am J Med Genet. 2001;106:37–45. doi: 10.1002/ajmg.1397. [DOI] [PubMed] [Google Scholar]
- Ugalde C, Janssen RJ, van den Heuvel LP, Smeitink JA, Nijtmans LG. Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum Mol Genet. 2004a;13:659–67. doi: 10.1093/hmg/ddh071. [DOI] [PubMed] [Google Scholar]
- Ugalde C, Vogel R, Huijbens R, Van Den Heuvel B, Smeitink J, Nijtmans L. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum Mol Genet. 2004b;13:2461–72. doi: 10.1093/hmg/ddh262. [DOI] [PubMed] [Google Scholar]
- Verkaart S, Koopman WJ, van Emst-de Vries SE, Nijtmans LG, van den Heuvel LW, Smeitink JA, et al. Superoxide production is inversely related to complex I activity in inherited complex I deficiency. Biochim Biophys Acta. 2007;1772:373–81. doi: 10.1016/j.bbadis.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Vogel RO, Dieteren CE, van den Heuvel LP, Willems PH, Smeitink JA, Koopman WJ, et al. Identification of mitochondrial complex I assembly intermediates by tracing tagged NDUFS3 demonstrates the entry point of mitochondrial subunits. J Biol Chem. 2007;282:7582–90. doi: 10.1074/jbc.M609410200. [DOI] [PubMed] [Google Scholar]
- Walker JE. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.