Summary
The overproduction, purification and in vitro characterization of the polyene glycosyltransferases (GTs) AmphDI and NysDI are reported. Enabled by a novel nucleotidyltransferase mutant (RmlA Q83D) for the chemoenzymatic synthesis of unnatural GDP-sugar donors, in conjunction with polyene GT-catalyzed sugar exchange/reverse reactions, the donor and acceptor specificity of these novel enzymes were subsequently probed. The evaluation of polyene GT aglycon and GDP-sugar donor specificity revealed some tolerance to aglycon structural diversity but stringent sugar specificity, culminating in new polyene analogs in which L-gulose or D-mannose replace the native sugar D-mycosamine.
Keywords: Antifungal, carbohydrate, sugar, glycosylation, drug, anti-infective
INTRODUCTION
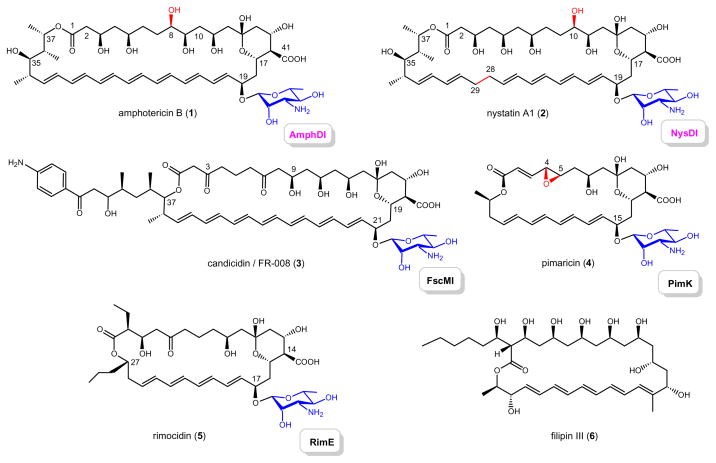
The polyene macrolide antibiotics are a family of diverse natural products primarily produced by Streptomyces and closely related bacteria.[1–3] As exemplified by amphotericin B (AmB, 1), nystatin A1 (2), candicidin or FR008 (3), pimaricin (4), rimocidin (5) and filipin III (6) (Scheme 1), a polyhydroxylated, polyunsaturated macrolactone ring core is the structural signature of family members, and most are decorated with the unique deoxyaminosugar mycosamine.[2] While polyene macrolide antibiotics are most noted for their antifungal properties, these metabolites display several other biological activities including antivirus, antiprotozoal, and even antiprion activity.[4–8] Their primary mechanism of action derives from unique interactions between polyene molecules and specific sterol-containing membranes to generate lethal transmembrane channels wherein selectivity derives from a preference for ergosterol-containing membranes.[3, 4, 7, 8] Remarkably, even after a half century of clinical use of 1,[4] the development of resistance to polyenes has been sparse.[9] However, the clinical utility of polyenes remains severely restricted by compound solubility and dose-dependent side-effects, most notably nephrotoxicity.[4, 5] Thus, the development of formulations and/or analogs to reduce unwanted side-effects and/or improve selectivity remains an active area of research.[4, 6]
Scheme 1. Naturally-occurring polyene macrolides.
Amphotericin B (1), nystatin A1 (2), candicidin/FR-008 (3), pimaricin (4), rimocidin (5) and filipin III (6) are all produced by Streptomyces strains and genetic loci for 1–5 have been characterized. The corresponding mycosaminyltransferases (AmphDI, NysDI, FscMI, PimK and RimE, respectively) responsible for glycoside formation are highlighted.
Toward this goal, the most common synthetic strategy for polyene derivatization has relied upon semisynthetic derivatization of the natural product core scaffold carboxyl (e.g. C41 of 1 and/or the C3′ amine of the appended aminosugar (e.g. mycosamine in 1).[10–20] In addition to providing analogs with altered antifungal properties, the outcome of such studies has also challenged the dogma pertaining to the intramolecular C3′ ammonium-C41 carboxylate interaction in channel assemblage.[16, 17] As an alternative to synthesis, the genetic loci encoding for pimaricin (4),[21, 22] nystatin (2),[23] AmB (1)[24] and candicidin/FR-008 (3),[25–27] have been partially or fully characterized,[3, 28] enabling both the elucidation of key post-PKS modification steps in polyene biosyntheses and the directed engineering of unique polyene analogs.[27–39] The cumulative SAR based upon this diverse array of semisynthetic and engineered polyene derivatives has also clearly illuminated the critical role of the amino-sugar moiety for antifungal activity.[13, 15, 32]
The growing appreciation of the importance of natural product sugar moieties has spurred the development of methods for natural product glycosylation and glycodiversification – ranging from new synthetic methodologies to enzyme-intensive approaches.[40–42] While there exists a C35-mycarosyl-substituted nystatin analog with improved antifungal potency,[31] few reported examples exist in which the natural polyene mycosamine has been successfully replaced by an alternative sugar.[27, 32] In addition, although the functions of enzymes that catalyze the attachment to polyenes (glycosyltransferases or GTs, Scheme 1) have been inferred via in vivo genetics, they have evaded in vitro biochemical characterization in part, due to the lack of sugar nucleotide substrate availability.[27, 32, 35, 38] Unlike most natural product GTs, which utilize pyrimidine-base sugar nucleotides, bioinformatics and biochemical characterization of the early steps in mycosamine biosynthesis implicate polyene GTs to utilize GDP-based sugar nucleotides.[23, 38] Herein we report the first in vitro characterization of two polyene GTs, AmphDI and NysDI. The aglycon and sugar nucleotide substrate specificity of these model polyene GTs were probed with a set of unique GDP-D- and L-sugars to reveal some tolerance to aglycon structural diversity but stringent GDP-sugar specificity. This study notably highlights the utility of a recently engineered nucleotidyltransferase variant to synthesize novel GDP-sugars and the application of these reagents,[43, 44] in conjunction with the reversibility of GT-catalyzed reactions,[45–49] to study purine sugar nucleotide-dependent GTs.
RESULTS
Over-production and purification of polyene GTs
The polyene GTs (AmphDI, NysDI, FscMI, PimK and RimE for 1–5, repectively, Scheme 1) share very high sequence (over 65% identity, Figure S1) and functional (mycosaminyl transfer, based upon in vivo gene inactivation)[27, 38] similarities. In an effort to study these novel catalysts in vitro, the genes of two polyene GTs, amphDI and nysDI,[23, 24] were PCR amplified from genomic DNA of the amphotericin producer Streptomyces nodosus ATCC 14899 and the nystatin producer Streptomyces noursei ATCC 11455, respectively. Heterologous expression N-His6-tagged AmphDI or NysDI in E. coli using a pET28a-based system provided only small amounts of the desired recombinant GTs (< 0.5 mg per liter of culture under optimized conditions) after affinity chromatography. The alignment of AmphDI and NysDI with 3 other polyene GTs (PimK,[22] RimE[50] and FscMI[27]) surprisingly revealed an extended N-terminus sequence lacking predicted structure [http://bioinf.cs.ucl.ac.uk/psipred/] (Figure S1). Expression of the two truncated GTs, designated as AmphDI-T2 and NysDI-T2 (starting from the ‘common’ methionine residue M21 for AmphDI and M44 for NysDI, Figure S1) under identical conditions led to 10–12 mg of the desired N-His6-tagged AmphDI-T2 and NysDI-T2 per liter E. coli culture (Figure S2).
Reversibility of polyene GT-catalyzed reactions
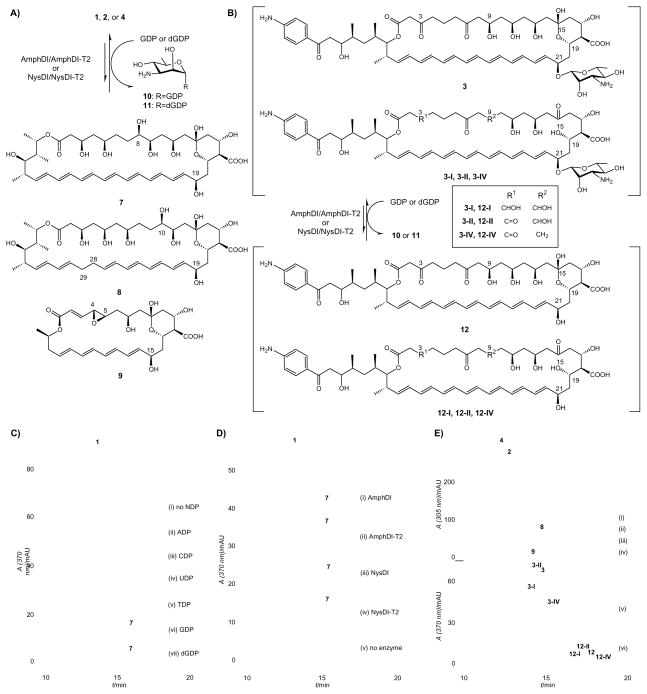
Given the difficulty to access polyene GT substrates (both the polyene aglycon acceptor and putative sugar nucleotide donor),[17, 51] we first investigated the reversibility of polyene GT-catalyzed reactions as recently described for other natural product GTs.[45–49] Specifically, polyene natural products were incubated with GTs in the presence of NDPs and the loss of mycosamine was assessed via HPLC (Figures 1A & 1B). For example, incubation of 20 μM AmB (1) with 5 μM AmphDI-T2 revealed a new product only in the presence of 2 mM GDP or dGDP at 30 °C for 6 h (Figure 1C, vi & vii), while no reaction was observed in the absence of NDPs (Figure 1C, i) or enzyme, or in the presence of alternative NDPs (ADP, CDP, UDP, and TDP, Figure 1C, ii–v). LC-MS of the new species was consistent with deglycosylated 1 aglycon, amphoteronolide B (7, Figure 1A, calculated 778.4; found [M-H]− 777.5, [M+Na]+ 801.5). Also consistent with a GT-catalyzed reverse reaction, the parallel formation of GDP-D-mycosamine (10) was verified via anion exchange HPLC chromatography and ESI-MS/MS analysis (10, calculated 588.1, found [M-H]− 587.0) (Figure S3). In a similar manner, reaction reversibility assessed in the presence of AmphDI, NysDI and NysDI-T2 revealed an absolute dependence upon GDP/dGDP (Figure 1D, i–iv), and enzyme (Figure 1D, v).
Figure 1. Polyene GT-catalyzed reverse reactions.
(A) Schematic of polyene GT-catalyzed conversion of 1, 2 or 4 to deglycosylated products 7, 8 or 9, respectively; (B) Schematic of polyene GT-catalyzed conversion of candicidin complex (3-I, 3-II, 3 and 3-IV) to deglycosylated complex (12-I, 12-II, 12 and 12-IV); (C) HPLC analyses of AmphDI-T2 NDP-specificity in GT-catalyzed reverse reactions. In this example, 20 μM AmB (1) was incubated with 5 μM of AmphDI-T2 without NDP (i) or in the presence of 1 mM of ADP (ii), CDP (iii), UDP (iv), TDP (v), GDP (vi) or dGDP (vii), at 30 °C overnight; (D) HPLC analyses of polyene GT-catalyzed reverse reactions with AmB (1) and different polyene GTs. For this study, 20 μM AmB (1) was incubated with 1 mM of GDP in the presence of 5 μM AmphDI (i), AmphDI-T2 (ii), NysDI (iii), NysDI-T2 (iv) or without GT (v), at 30 °C overnight; (E) HPLC analyses of AmphDI-T2 aglycon specificity in GT-catalyzed reverse reactions. In this study, 20 μM nystatin (2), 50 μM pimaricin (4) or 20 μM of candicidin complex (3-I, 3-II, 3 and 3-IV) were incubated with 1 mM GDP in the absence or presence of 5 μM AmphDI-T2: (i) 2, no enzyme (control), (ii) 2, AmphDI-T2, (iii) 4, no enzyme (control), (iv) 4, AmphDI-T2, (v) candicidin complex (3-I, 3-II, 3 and 3-IV), no enzyme (control), (iii) candicidin complex (3-I, 3-II, 3 and 3-IV), AmphDI-T2.
To probe aglycon tolerance, several other polyene macrolides were subjected to the same AmphDI, AmphDI-T2, NysDI and NysDI-T2 assay conditions. In this study, reaction reversibility was observed with both nystatin (2) and, to a lesser extent, pimaricin (4), by all four GTs tested (AmphDI, AmphDI-T2, NysDI and NysDI-T2) only in the presence of GDP or dGDP to provide nystatinolide (8, Figure 1A and 1E, calculated 780.4; found [M-H]− 779.4, [M+Na]+ 803.5) or pimaricin aglycon (9, Figures 1A and 1E, iv, calculated 520.2; found [M-H]− 519.2, [M+Na]+ 543.3), respectively. In similar manner, reaction reversibility was also established in the presence of GDP using the commercially available candicidin (a complex consisting of at least four major compounds - 3-I, 3-II, 3 and 3-IV, Figures 1B and 1E, v)[27] to provide the corresponding aglycons (12-I, 12-II, 12 and 12-IV, Figures 1B and 1E, vi), the mass ions of which were detectable via LC-MS (12-I, calculated 965.5, found [M-H]− 964.5; 12-II, calculated 963.5, found [M-H]− 962.4; 12, calculated 963.5, found [M-H]− 962.4; 12-IV, calculated 947.5, found [M-H]− 946.3). Finally, the activity of AmphDI, AmphDI-T2, NysDI and NysDI-T2 toward different polyene macrolides were compared under pre-steady-state conditions (20 μM polyene glycoside, 0.5 μM GT, 2 mM GDP, 30 °C, up to 1 h). As summarized in Figure S4, the truncated GTs outperformed their original ‘extended’ counterparts, the magnitude of which varied depending upon the polyene substrate employed.
Synthesis of GDP-sugars
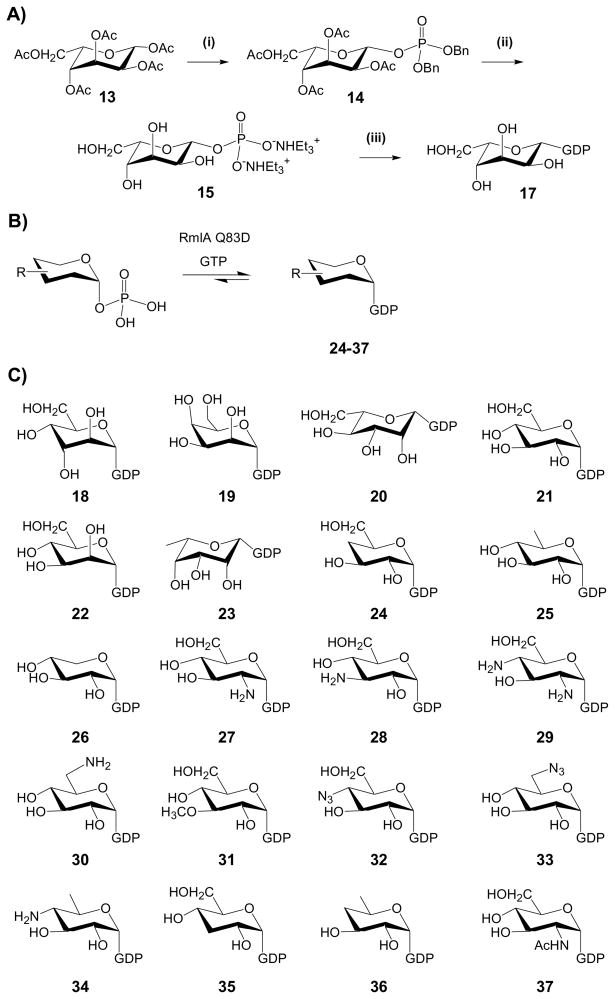
Consistent with previous postulations,[3, 38] the specific requirement of GDP (or dGDP) for the reversibility of AmphDI/NysDI-catalyzed reactions is consistent with GDP-mycosamine as the requisite sugar donor. To further probe the sugar nucleotide donor substrate flexibility of polyene GTs, a small set of GDP-sugars was subsequently generated using both chemical and enzymatic methods (Scheme 2). Together with 3 commercially available GDP-sugars (GDP-α-D-glucose, 21; GDP-α-D-mannose, 22; and GDP-β-L-fucose, 23), the combination of chemical and enzymatic strategies summarized in the next two paragraphs provided a set of 21 putative GDP-sugar donors (17–37, Scheme 2C) for this study.
Scheme 2. Chemical and chemoenzymatic preparation of GDP-sugars.
(A) The chemical synthesis of GDP-L-β-gulose (17). (i) Ac2O/pyridine; HBr/AcOH; HPO2(OBn)2, CF3SO3Ag, Me3C5H2N/CH2Cl2; (ii) H2/Pd-C; AG 50W-X8 (Et3NH+); (iii) GDP-morpholidate (16) and 1H-tetrazole/pyridine. (B) The chemoenzymatic synthesis of GDP-sugars. Generally, 6 mM of chemically synthesized sugar-1 phosphate was incubated with 8 mM of GTP in the presence of 20 μM RmlA mutant Q83D. (C) GDP-sugars employed in this work. GDP-D-mycosamine (10) was generated via reverse GT-catalysis, GDP-D-glucose (18), GDP-D-mannose (19) and GDP-L-fucose (22) were commercially available; GDP-L-gulose (17), GDP-D-altrose (20), GDP-D-talose (21) and GDP-L-mannose (23) were chemically synthesized (scheme A) and GDP-sugars 24–35 were enzymatically synthesized (scheme B).
A conventional morpholidate-dependent coupling strategy was applied for the chemical synthesis of several GDP-sugars (Scheme 2A).[52, 53] The syntheses for the required α-D-altrose-1-phosphate, α-D-talose-1-phosphate and β-L-mannose-1-phosphate precursors have been previously reported.[53–57] Following an identical strategy, peracylated β-L-gulose (13) was converted to the protected sugar phosphate (14) in two steps (55% yield), the deprotection of which led to a triethylammonium sugar phosphate (15) (85% yield). Coupling the target sugar-1-phosphates with the guanosine 5′-monophosphomorpholidate (1.6 eq) provided the desired GDP-sugars (17–20) in 45–65% yield. Following this route, 4 sugar nucleotides, GDP-β-L-gulose (17), GDP-α-D-altrose (18), GDP-α-D-talose (19) and GDP-β-L-mannose (20) were generated for this study.
A recently described nucleotidyltransferase mutant (RmlA Q83D)[43] with enhanced activity toward GTP was also employed in the synthesis of GDP-sugars for this study (Scheme 2B). Following the established protocol,[43] sugar 1-phosphates incubated with GTP and purified RmlA Q83D at 37 °C overnight provided 14 additional unique GDP-sugars (Scheme 2C, Figure S5, 24–37). Among them, 10 sugar nucleotides (24–32) were produced in good yields (ranging from 23–96%, Table S1 and Figure S5), while less than 10% conversion was observed for remaining 4 sugar-1-phosphates (33–37, Table S1 and Figure S5). All GDP-sugar nucleotide products were confirmed by LC-MS and ESI MS/MS spectrometry also confirmed typical secondary fragment ions for [GDP-H]− (442) and [GMP-H]− (362) for all reaction products (Table S1).
Polyene GT-catalyzed sugar exchange and glycoside formation
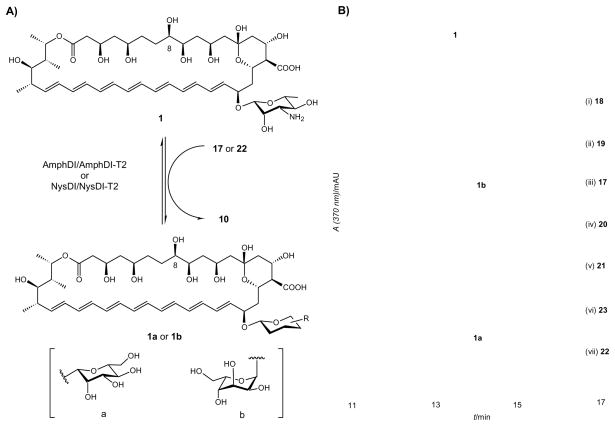
GT-mediated sugar exchange enables the exchange of the native sugar within a native natural product glycoside with exogenous carbohydrates supplied as NDP-sugars.[45, 49] Following this same protocol, the donor substrate flexibility of polyene GTs was probed via a sugar exchange reaction (Figure 2A) using the putative GDP-sugar donors described in the previous section. Specifically, 20 μM AmB (1) and 5 μM AmphDI-T2 were incubated individually with GDP-sugar donors (2 mM for 17–23, 30 – 300 μM for 24–33, < 30 μM for 34–37, Scheme 2C) at 30 °C overnight. Analysis of the reactions by RP-HPLC revealed new products only in the presence of GDP-α-D-mannose (22, Figure 2B, vii) or GDP-β-L-gulose (17, Figure 2B, iii), the identities of which were confirmed by LC-MS to be 1a (calcd. 940.5, found 939.4 [M-H]− and 963.4 [M+Na]+) and 1b (calcd. 940.5, found 939.5 [M-H]− and 963.5 [M+Na]+) (Figure 2A), respectively.
Figure 2. Polyene GT-catalyzed Sugar Exchange Reactions.
(A) Schematic of polyene GT-catalyzed sugar exchange reaction. (B) HPLC analyses of AmphDI-T2 catalyzed sugar exchange reactions. In this study, 20 μM AmB (1) was co-incubated with 5 μM of AmphDI-T2 in the presence of 1 mM of (i) GDP-α-D-altrose (18), (ii) GDP-α-D-talose (19), (iii) GDP-β-L-gulose (17), (iv) GDP-β-L-mannose (20), (v) GDP-α-D-glucose (21), (vi) GDP-β-L-glucose (23), (vii) GDP-α-D-mannose (22).
To assess the activity in a more conventional GT-catalyzed assay, a small amount of the acceptor amphoteronolide (7, 0.1 mg, 0.128 μmol, 12.8% overall yield), was partially purified from a 10 ml preparative AmphDI-T2 catalyzed reverse reaction (with 20 μM of 1). This is an advance over chemical routes for 7 preparation – for example, a recent chemical approach provided a 10.8% overall yield of 7 in 8 steps from 1.[17] Subsequently, 4 μM of the isolated aglycon (7) was incubated with 5 μM AmphDI-T2 and various GDP-sugar donors (2 mM for 17–23, 30 – 300 μM for 24–33, < 30 μM for 34–37). Under these conditions, substantial amounts of 1a and 1b (almost 100% conversion, Figure S6) were produced from sugars 17 and 22 while, consistent with ‘sugar exchange’ assays, all other donors (18–21, 23–37) failed to provide glycoside variants. Interestingly, a prior study in which disruption of mycosamine biosynthesis in S. nodosus led to a minor shunt metabolite with a mass consistent with a hexosyl-amphoteronolide A which was proposed to be mannosyl- or glucosyl-amphoteronolide.[30] The ability of AmphDI to accept GDP-mannose, but not GDP-Glc, supports the potential in vivo of mannosyl-amphoteronolide A but refutes the possibility of glucoside formation. When GDP-D-mannose was replaced by TDP-D-mannose (chemoenzymatically prepared from a RmlA reaction)[45, 46] in the assay with 7 and AmphDI-T2, no products were detectable, consistent with GDP-sugar specificity. In a similar manner, only 17 and 23 were identified as NysDI-T2 donor substrates to afford 1a and 1b.
DISCUSSION
Unlike the two-component GTs associated with the biosynthesis of many glycosaminyl-modified polyketides first described by Liu and coworkers,[58–61] AmphD1 and NysD1 do not require an auxiliary protein for in vitro activity. Consistent with prior bioinformatics[3, 21–28] and the recent biochemical characterization of a GDP-D-mannose 4,6-dehydratase (NysDIII) encoded by the nystatin biosynthetic gene cluster,[38] this study unequivocally confirms polyene GTs to be (d)GDP-sugar specific. Enabled by a recently reported RmlA mutant engineered to provide the ability to generate a repertoire of unnatural GDP-sugar donors,[43] the highlighted polyene GT-catalyzed sugar exchange and glycoside formation reactions required GDP-sugar donors. While it is typical for the forward and reverse reactions to utilize the same nucleotide,[45–48] there now exists one exception - a recent study with the calicheamicin GT CalG3 revealed nucleotide specificity of the reverse reaction to differ from the forward reaction.[49] Attempts toward differentially-glycosylated variants via pathway engineering have led to only a few polyene sugar variations to date – specifically, the replacement of D-mycosamine with 6-deoxy-D-Man, 3-keto-6-deoxy-D-Man polyenes and an uncharacterized hexose (putatively Man or Glc).[27, 30, 32] While the current study led to two new polyene sugar appendages (L-gulose and D-Man), it also suggests the stringent sugar nucleotide specificity of the polyene GTs may, in part, restrict the generation of variant glycosides via in vivo engineering or in vitro chemoenzymatic methods. However, the recent success of expanding the substrate promiscuity of a natural product GTs by directed evolution/engineering may present exciting new avenues to circumvent the sugar nucleotide stringency of polyene GTs and thereby further enhance their synthetic utility.[62, 63]
In contrast to their sugar nucleotide stringency, the demonstrated ability of Amph/NysDI to utilize AmB (1), nystatin A1 (2), candicidin members (Figure 1B) and pimaricin (4, Scheme 1), positions these polyene GTs among a growing list of natural product GTs with promiscuity toward aglycon acceptors, exemplified by GTs such as OleD[62–64] or VinC.[47, 65, 66] Related to this, previous polyene in vivo and in vitro biosynthetic studies established oxidative tailoring (e.g., 8-hydroxylation by AmphL in 1 and 10-hydroxylation by NysL in 2) to occur after mycosaminylation,[30, 37, 38] While the present study revealed hydroxylated aglycons (e.g. 7 and 8) to be substrates of AmphDI and NysDI in vitro. Thus, the ultimate order of the biosynthetic events (hydroxylation and mycosaminylation) in vivo may be dictated by the substrate specificity of the oxidases AmphL and NysL.
SIGNIFICANCE
This study extends the fundamental understanding of polyene biosynthesis and the potential for chemoenzymatic diversification of polyene-based antifungal drugs. In the context of biosynthesis, the first in vitro characterization of representative polyene GTs unequivocally confirmed, for the first time, these enzymes to be GDP-sugar dependent and also revealed the correct start codons for the previously identified amphDI and nysDI genes. In addition, the demonstrated aglycon flexibility of polyene GTs in vitro suggests the order of final tailoring steps implicated via in vivo studies (glycosylation followed by oxidation) must be dictated by oxidase, not GT, specificity. With respect to polyene diversification, this study highlights the utility of a recently engineered nucleotidyltransferase (RmlA) variant to synthesize novel GDP-sugars, and the application of these reagents in conjunction with the reversibility of GT-catalyzed reactions, to study purine sugar nucleotide-dependent GTs. Using these reagents, the evaluation of polyene GT aglycon and sugar nucleotide substrate specificity revealed some tolerance to aglycon structural diversity but stringent sugar specificity, culminating in new polyene analogs in which L-gulose or D-mannose replace the native sugar D-mycosamine.
EXPERIMENTAL SECTION
Materials and general methods
E. coli DH5α and BL21(DE3) competent cells were purchased from Invitrogen (Carlsbad, CA). Streptomyces nodosus ATCC 14899 and Streptomyces noursei ATCC 11455 were purchased from the American Type Culture Collection (ATCC) (Manassas, VA). The pET-28a Escherichia coli expression vector was purchased from Novagen (Madison, WI). Primers were ordered from Integrated DNA Technology (Coralville, IA). Pfu DNA polymerase was purchased from Stratagene (La Jolla, CA). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). Candicidin was purchased from U. S. Pharmacopeia (Rockville, MD). Other polyene macrolide antibiotics such as amphotericin B (1), nystatin A1 (2), pimaricin (4), filipin III (6) and sugar nucleotides GDP-D-glucose (21), GDP-D-mannose (22) and GDP-L-fucose (23) were purchased from Sigma (St. Louis, MO).
For chemical syntheses, all moisture sensitive reactions were performed in flame-dried glassware under an atmosphere of Ar. Reactions were generally concentrated under reduced pressure using a Büchi rotary evaporator at water aspirator pressure (< 20 torr) followed by removal of residual volatile materials under high vacuum (via a standard belt-drive oil pump, < 1 torr). Analytical thin layer chromatography (TLC) was performed on E. Merck TLC plates pre-coated with silica gel 60 F254 (250 μm thickness) and column chromatography (FCC) was performed on Silicycle silica gel (40–60 μm, 60 Å pore size). All reagents were purchased from Aldrich (Milwaukee, WI), Sigma (St. Louis, MO), or Fisher Scientific (Pittsburg, PA) and used without further purification.
Analytical HPLC was run on a Varian Prostar 210/216 system connected to a Prostar 330 photodiode array detector (Varian, Walnut Creek, CA). Mass spectra (MS) were obtained by using electrospray ionization on Agilent 1100 HPLC-MSD SL quadrupole mass spectometer (Agilent Technologies, Palo Alto, CA) connected with a UV/Vis diode array detector. Proton nuclear magnetic resonance (1H NMR) and carbon NMR (13C NMR) spectra were recorded on Varian UNITYINOVA 400 MHz and 500 MHz spectrometers in deuterated sovents. Chemical shifts are reported in parts per million (ppm, δ) relative to residual solvent peaks (CHCl3: 1H: δ 7.26, 13C: δ 77.0; H2O: 1H: δ 4.78).
Cloning, expression and purification of polyene GTs
The genomic DNA was isolated from the amphotericin producer S. nodosus ATCC 14899 and the nystatin producer S. noursei ATCC 11455 strains, respectively, following a literature procedure.[67] The amphDI and nysDI genes were amplified from genomic DNA of the corresponding producers with Pfu DNA polymerase, by using the following primer pairs: 5′-cgacttcatatgggtggacgcgaggcg - 3′ (amphDI_F, forward, NdeI) and 5′-ggacatcctagatctcctcggtcagtcgtttgc -3′ (amphDI_R, reverse, BglII) for amphDI (1.45 kb); 5′-gtgccggcatatgacccttccttccgg - 3′ (nysDI_F, forward, NdeI) and 5′-gggttttggatcctcctcggtcagtcggtt -3′ (nysDI_R, reverse, BamHI) for nysDI (1.52 kb); respectively. PCR products were digested with NdeI/BglII (for amphDI) or NdeI/BamHI (for nysDI) and ligated into the pET28a expression vector (NdeI/BamHI) to give plasmids pCST551 (NysDI) and pCST571 (AmphDI), respectively. For the truncated NysDI, a 1.39 kb nysDI-T2 DNA fragment was PCR amplified from pCST551 using primer pairs: 5′-gtgttgcatatgggcgcgaatcggcg - 3′ (nysDI-T2_F, forward, NdeI) and 5′-gggttttggatcctcctcggtcagtcggtt -3′ (nysDI-T2_R, reverse, BamHI). Similarly, a truncated 1.39 kb amphDI-T2 DNA fragment was PCR amplified from pCST571 using primer pairs: 5′-gtgttgcatatgggcgcgcacagg - 3′ (amphDI-T2_F, forward, NdeI) and 5′-5′-ggacatcctagatctcctcggtcagtcgtttgc -3′ (amphDI-T2_R, reverse, BglII). Subsequently, PCR products were digested with NdeI/BamHI (for nysDI-T2) or NdeI/BglII (for amphDI-T2) and ligated into the pET28a expression vector (NdeI/BamHI) to give plasmids pCST556 (NysDI-T2), pCST576 (AmphDI-T2), respectively.
For AmphDI production, a single transformant of E. coli BL21(DE3)/pCST571 was inoculated into LB medium (4 ml) supplemented with kanamycin (50 μg/ml) and grown with shaking at 37 °C overnight. The precultures were used to inoculate LB medium (1 L) containing kanamycin (50 μg/ml) which was grown with shaking at 18 °C to an OD600 of 0.5 – 0.7. Protein expression was induced with the addition of isopropyl-β-D-thiogalactopyranoside (IPTG, 0.3 mM) followed by growth for an additional 20 h. The cells obtained from 1 L of culture were washed twice with buffer A (20 mM NaH2PO4, pH 7.5, 500 mM NaCl, 10 mM imidazole) and resuspended in buffer A (30 ml) supplemented with lysozyme (1 mg/ml). After 10 min incubation on ice, the cells were lysed via 3 rounds of French-press (1,200 psi, Thermo IEC) and the insoluble material was removed by centrifugation at 30,000 g for 1 hr (4°C). The supernatants were loaded onto the HisTrap HT column (1 ml, GE Healthcare) and the N-(His)10-tagged AmphDI was eluted with a linear gradient of imidazole (10 – 500 mM) in buffer A by a FPLC system (GE Healthcare). The purified protein was desalted through PD-10 column (GE Healthcare) and stored in the buffer containing 50 mM Tris-HCl (pH 8.0) and 10% glycerol until use. Protein concentration was measured by Bradford assay. N-(His)6-tagged NysDI, NysDI-T2 and AmphDI-T2 were produced and purified following the same protocol from E. coli BL21(DE3) strains harboring pCST551, pCST556 and pCST576, respectively.
Chemical synthesis of sugar phosphates (Scheme 2A)
The syntheses for the required α-D-altrose-1-phosphate, α-D-talose-1-phosphate and β-L-mannose-1-phosphate precursors have been previously reported.[54–57]
Dibenzyl-(2,3,4,6-tetra-O-acetyl-β-L-gulopyranosyl) phosphate (14)
Peracylated β-L-gulose (13, 351 mg, 0.9 mmol) was dissolved in acetic acid (2 mL) to which 33% HBr in acetic acid (1 mL) was added dropwise at 0 °C. The reaction was allowed to warm to room temperature with stirring for 2 hr, diluted with cold CHCl3 (100 mL), and washed successively with cold saturated NaHCO3 solution (3 × 20 mL), H2O (20 mL) and brine (20 mL). The organics were dried over anhydrous Na2SO4 and concentrated and the crude gulopyranosyl bromide was used directly. A mixture of dibenzyl phosphate (300 mg, 1.08 mmol), silver trifilate (300 mg, 1.17 mmol), 2,4,6-collidine (0.23 mL, 1.74 mmol) and activated 4 Å molecular sieves (400 mg) in dry CH2Cl2 (10 mL) was stirred at room temperature under argon in the absence of light for 1 hr. The mixture was then cooled to −40 °C to which a solution of crude protected pyranosyl bromide in dry CH2Cl2 (5 mL) was added dropwise. The reaction mixture was kept at −40 °C for 2 hr and then allowed to warm to room temperature with stirring overnight. The filtrate was diluted CH2Cl2 (with 100 mL) and washed with saturated CuSO4 (2 × 30 mL), H2O (20 mL), and brine (20 mL). The organics were dried over Na2SO4, concentrated and purified by silica gel chromatography (hexane/EtOAc 1:1~1:1.5) to give 300 mg 14 (55% for two steps). [α]D = 1.5 (c = 1, CHCl3); 1H NMR (CDCl3) 7.30~7.22 (m, 10H), 5.59 (dd, J = 7.2, 8.3 Hz, 1H), 5.35 (m, 1H), 5.06 (m, 3H), 4.97(d, J = 7.2 Hz, 2H), 4.92 (m, 1H), 4.29 (t, J = 5.7 Hz, 1H), 4.12 (dd, J = 11.5, 5.6 Hz, 1H), 4.03 (dd, J = 7.3, 11.5 Hz, 1H), 2.11 (s, 3H), 2.08 (s, 3H), 1.90 (s, 3H), 1.81 (s, 3H); 31P NMR (CDCl3) 2.17; MS: calcd. C28H33O13PNa 631.2, found m/z 631.3 (M+Na).
Triethylammonium-(β-L-gulopyranosyl)phosphate (15)
Compound 14 (260 mg, 0.43 mmol) was dissolved in MeOH (5 mL), 1N NaHCO3 solution (1.2 mL) to which 10% Pd/C was added (90 mg). The mixture was stirred overnight at room temperature under hydrogen atmosphere after which the catalyst was removed by filtration and the filtrate concentrated to approximately 3 mL volume. The solution was cooled to 0 °C and 1 N NaOH (2.5 mL) was added dropwise with stirring. The mixture was stirred for an additional 2 hr and neutralized with 1 N HOAc. The resulting solution was then submitted to anion exchange chromatography (Dowex 1 × 8, 1.2 × 12 cm) eluted with H2O (100 mL), 0.1 M NH4HCO3 (100 mL), 0.2 M NH4HCO3 (100 mL), and 0.3 M NH4HCO3 (100mL). The product containing fractions (which eluted with 0.2 M NH4HCO3) were pooled and co-evaporated with EtOH several times to remove residual NH4HCO3. The obtained sugar phosphate sodium salt was transformed into a triethylamine salt by passing through a AG 50W-X8 cation-exchange column (Et3NH+ type, 1.5 × 10cm) eluted with ddH2O. The product containing fractions (5 × 20 mL) were pooled and lyophilized to give 135 mg product (87% yield). 1H NMR (D2O) 5.16 (t, J = 7.8 Hz, 1H), 4.04 (m, 2H), 3.79 (m, 2H), 3.68 (m, 2H), 3.17 (q, J = 7.2 Hz, 10H), 1.25 (t, J = 7.2 Hz, 15H); 13C NMR (D2O) 95.72, 74.80, 71.10, 69.50 (x 2), 61.64, 46.93, 8.54; 31P NMR (D2O) 2.64; MS: calcd for C6H12O9P 259.0, found m/z 258.7 (M+H+).
Chemical synthesis of GDP-sugars (Scheme 2A)
A mixture of triethylammonium sugar phosphate (e.g. 15) and 4-morpholine-N,N′-dicyclohexylcarboxamidinium guanosine 5′-monophosphomorpholidate (1.6 eq) was coevaporated with dry pyridine (3 mL) three times after which 1H-tetrazole (3 eq) and dry pyridine (3 mL) were added and the solution stirred at room temperature. After three days, the mixture was diluted with water, evaporated and purified on by Bio-Gel P-2 column chromatography (1.5 × 150 cm, 1 mL min−1) eluted with 0.05 M NH4HCO3. The product containing fractions (which eluted between 160–180 mL) were collected and lyophilized to afford the desired product. The typical yield of this procedure ranged from 46–65%.
Guanosine 5′-β-L-gulopyranosyl diphosphate (17)
Using the general procedure, triethylammonium-β-L-gulopyranosyl phosphate 15 (50 mg, 0.14 mmol) gave 40 mg of the desired product 17 (46 %). 1H NMR (D2O) 8.10 (s, 1H), 5.93 (dd, 1H, J = 1.6, 6.2 Hz), 5.28 (dt, 1H, J = 1.7, 8.1 Hz), 4.53 (m, 1H), 4.23 (m, 3H), 4.07 (m, 2H), 3.82 (m, 2H), 3.73 (m, 2H); 31P NMR (D2O) −12.1, −13.5; 13C NMR (D2O) 160.0, 155.0, 153.0, 136.7, 115.3, 95.7, 87.9, 84.9, 75.7, 74.8, 71.9, 71.5, 70.1, 69.7, 66.4, 62.3; HRMS: calcd for C16H24N5O16P2 604.0693, found 604.0708 (M+H).
Guanosine 5′-α-D-altropyranosyl diphosphate (Scheme 2C, 18)
Using the general procedure, triethylammonium-α-D-altropyranosyl phosphate (25 mg, 0.07 mmol) gave 26.5 mg of the desired product 18 (61.5 %). 1H NMR (D2O) 8.09 (s, 1H), 5.91 (d, 1H, J = 6.2 Hz), 5.40 (d, 1H, J = 8.2 Hz), 4.51 (dd, 1H, J = 3.3, 5.1 Hz), 4.33 (m, 1H), 4.20 (dd, 2H, J = 3.3, 5.2 Hz), 3.99 (dd, 2H, J = 2.0, 4.0 Hz), 3.94 (t, 1H, J = 3.7 Hz), 3.83 (m, 2H), 3.74 (dd, 1H, J = 5.5, 12.4 Hz); 31P NMR (D2O) −10.5, −12.9; 13C NMR (D2O) 159.6, 154.6, 152.4, 138.2, 116.9, 96.9, 87.6, 84.4, 74.5, 71.1, 70.8, 70.6, 66.0, 64.8, 61.6; HRMS: calcd for C16H24N5O16P2 604.0693, found 604.0710 (M+H).
Guanosine 5′-α-D-talopyranosyl diphosphate (Scheme 2C, 19)
Using the general procedure, triethylammonium-α-D-talopyranosyl phosphate (50 mg, 0.14 mmol) gave 56 mg of the desired product 19 (65 %). 1H NMR (D2O) 8.09 (s, 1H), 5.92 (d, 1H, J = 6.0 Hz), 5.60 (d, 1H, J = 7.9 Hz), 4.50 (m, 1H), 4.34 (dd, 1H, J = 2.0, 3.1 Hz), 4.20 (m, 2H), 4.10 (m, 2H), 3.95 (m, 2H), 3.89 (s, 2H), 3.80 (dd, 1H, J = 7.7, 12.0 Hz), 3.72 (dd, 1H, J = 4.7, 12.0 Hz); 31P NMR(D2O) −10.6, 13.0; 13C NMR (D2O) 158.0, 153.0, 150.9, 136.7, 115.3, 96.1, 86.0, 82.9, 72.9, 71.7, 69.5, 69.5, 68.5, 64.5, 63.8, 60.5; HRMS: calcd for C16H24N5O16P2 604.0693, found 604.0700 (M+H+).
Chemoenzymatic synthesis of GDP-sugars (Scheme 2B)
A set of GDP-sugars were generated following protocols previously described for dTDP/UDP-sugars.[54–56, 68–70] The reaction was carried out in Tris-HCl buffer (50 mM, pH 8.0) containing 5 mM MgCl2, 1 U inorganic pyrophosphatase, 10 μM of purified RmlA Q83D,[43] 8 mM sugar-1-phosphate and 6 mM GTP, and incubated at 37 °C for 2 hrs. The formation of sugar nucleotides (24 – 37, Scheme 2C) was analyzed by HPLC using an anion exchange column (SphereClone SAX, 5 μm, 250 × 4.60 mm, H2O with 0% – 100% 600 mM ammonium formate gradient over 25 min, 1 mL/min, A254).
Polyene GT assays
Reverse or sugar exchange assays with polyene GTs (AmphDI, NysDI, AmphDI-T2 and NysDI-T2) were performed in a total volume of 100 μl containing 20–50 μM of polyene glycosides (1–4) and 2 mM of NDPs or various GDP-sugar donors (2 mM for 17–23, and 30 – 300 μM for 24–33, less than 30 μM for 34–37, Scheme 2C) with incubation at 30 °C overnight in the presence of 5 μM of polyene GTs (AmphDI, NysDI, AmphDI-T2 and NysDI-T2), in Tris-HCl buffer (50 mM, pH 8.0) containing 1 mM of MgCl2. For forward assays, partially purified acceptor amphoteronolide (7, 4 μM) was incubated with 5 μM AmphDI-T2 and various GDP-sugar donors (2 mM for 17–23, 30 – 300 μM for 24–33, < 30 μM for 34–37). For all assays, the assay mixtures without addition of polyene GTs served as controls. The reactions were subsequently quenched by the addition of MeOH (100 μl) and were centrifuged to remove proteins. The formation of new products was monitored by reverse phase HPLC (Phenomenex Luna C18, 5μm, 250 × 4,6 mm, 0.1% TFA (A) and 10 % –100 % CH3CN (B) over 30 min, 1 mL/min, 370 or 305 nm). The conversion rate was calculated by dividing the integrated area of glycosylated product with the sum of integrated area of product and the remaining substrate. The newly-formed products were analyzed by LC-MS (ESI) in negative (−) mode.
Supplementary Material
Acknowledgments
We thank the University of Wisconsin–Madison School of Pharmacy Analytical Facility for analytical support and Drs. Byron R. Griffith and Gavin J. Williams for many helpful discussions. This research was supported in part by National Institutes of Health Grants AI52218 and GM70637 (to J.S.T.) and a National Cooperative Drug Discovery Group Grant (U19 CA113297) from the National Cancer Institute. RM was a National Institutes of Health Molecular Bioscience Training Grant trainee (GM07215). J.S.T is a UW HI Romnes Fellow.
References
- 1.Hamilton-Miller JM. Bacteriol Rev. 1973;37:166. [Google Scholar]
- 2.Gil JA, Martin JF. Polyene antibiotics. In: Strohl WR, editor. Biotechnology of Antibiotics. 2. Marcel Dekker; New York: 1997. pp. 551–576. [Google Scholar]
- 3.Aparicio JF, Mendes MV, Anton N, Recio E, Martin JF. Curr Med Chem. 2004;11:1645. doi: 10.2174/0929867043365044. [DOI] [PubMed] [Google Scholar]
- 4.Lemke A, Kiderlen AF, Kayser O. Appl Microbiol Biotechnol. 2005;68:151. doi: 10.1007/s00253-005-1955-9. [DOI] [PubMed] [Google Scholar]
- 5.Cereghetti DM, Carreira EM. Synthesis. 2006:914. [Google Scholar]
- 6.Torrado JJ, Espada R, Ballesteros MP, Torrado-Santiago S. J Pharm Sci. 2007:96. doi: 10.1002/jps.21179. in press. [DOI] [PubMed] [Google Scholar]
- 7.Zotchev SB. Curr Med Chem. 2003;10:211. doi: 10.2174/0929867033368448. [DOI] [PubMed] [Google Scholar]
- 8.Baginski M, Czub J, Sternal K. Chem Rec. 2006;6:320. doi: 10.1002/tcr.20096. [DOI] [PubMed] [Google Scholar]
- 9.Ellis D. J Antimicrob Chemother. 2002;49(Suppl 1):7. doi: 10.1093/jac/49.suppl_1.1. [DOI] [PubMed] [Google Scholar]
- 10.Cheron M, Cybulska B, Mazerski J, Grzybowska J, Czerwinski A, Borowski E. Biochem Pharmacol. 1988;37:827. doi: 10.1016/0006-2952(88)90168-2. [DOI] [PubMed] [Google Scholar]
- 11.Cybulska B, Bolard J, Seksek O, Czerwinski A, Borowski E. Biochim Biophys Acta. 1995;1240:167. doi: 10.1016/0005-2736(95)00170-0. [DOI] [PubMed] [Google Scholar]
- 12.Falk R, Domb AJ, Polacheck I. Antimicrob Agents Chemother. 1999;43:1975. doi: 10.1128/aac.43.8.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szlinder-Richert J, Mazerski J, Cybulska B, Grzybowska J, Borowski E. Biochim Biophys Acta. 2001;1528:15. doi: 10.1016/s0304-4165(01)00166-0. [DOI] [PubMed] [Google Scholar]
- 14.Mazerski J, Bolard J, Borowski E. Biochim Biophys Acta. 1995;1236:170. doi: 10.1016/0005-2736(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 15.Paquet V, Carreira EM. Org Lett. 2006;8:1807. doi: 10.1021/ol060353o. [DOI] [PubMed] [Google Scholar]
- 16.Matsumori N, Sawada Y, Murata M. J Am Chem Soc. 2005;127:10667. doi: 10.1021/ja051597r. [DOI] [PubMed] [Google Scholar]
- 17.Palacios DS, Anderson TM, Burke MD. J Am Chem Soc. 2007;129:13804. doi: 10.1021/ja075739o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paquet V, Volmer AA, Carreira EM. Chemistry. 2008;14:2465. doi: 10.1002/chem.200701237. [DOI] [PubMed] [Google Scholar]
- 19.Szpilman AM, Cereghetti DM, Wurtz NR, Manthorpe JM, Carreira EM. Angew Chem. 2008;120:4407. doi: 10.1002/anie.200800589. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed Engl. 2008;47:4335. [Google Scholar]
- 20.Szpilman AM, Manthorpe JM, Carreira EM. Angew Chem. 2008;120:4411. doi: 10.1002/anie.200800590. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed Engl. 2008;47:4339. [Google Scholar]
- 21.Aparicio JF, Colina AJ, Ceballos E, Martin JF. J Biol Chem. 1999;274:10133. doi: 10.1074/jbc.274.15.10133. [DOI] [PubMed] [Google Scholar]
- 22.Aparicio JF, Fouces R, Mendes MV, Olivera N, Martin JF. Chem Biol. 2000;7:895. doi: 10.1016/s1074-5521(00)00038-7. [DOI] [PubMed] [Google Scholar]
- 23.Brautaset T, Sekurova ON, Sletta H, Ellingsen TE, StrLm AR, Valla S, Zotchev SB. Chem Biol. 2000;7:395. doi: 10.1016/s1074-5521(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 24.Caffrey P, Lynch S, Flood E, Finnan S, Oliynyk M. Chem Biol. 2001;8:713. doi: 10.1016/s1074-5521(01)00046-1. [DOI] [PubMed] [Google Scholar]
- 25.Hu Z, Bao K, Zhou X, Zhou Q, Hopwood DA, Kieser T, Deng Z. Mol Microbiol. 1994;14:163. doi: 10.1111/j.1365-2958.1994.tb01276.x. [DOI] [PubMed] [Google Scholar]
- 26.Campelo AB, Gil JA. Microbiology. 2002;148:51. doi: 10.1099/00221287-148-1-51. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Huang X, Zhou X, Bai L, He J, Jeong KJ, Lee SY, Deng Z. Chem Biol. 2003;10:1065. doi: 10.1016/j.chembiol.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Aparicio JF, Caffrey P, Gil JA, Zotchev SB. Appl Microbiol Biotechnol. 2003;61:179. doi: 10.1007/s00253-002-1183-5. [DOI] [PubMed] [Google Scholar]
- 29.Mendes MV, Recio E, Fouces R, Luiten R, Martin JF, Aparicio JF. Chem Biol. 2001;8:635. doi: 10.1016/s1074-5521(01)00033-3. [DOI] [PubMed] [Google Scholar]
- 30.Byrne B, Carmody M, Gibson E, Rawlings B, Caffrey P. Chem Biol. 2003;10:1215. doi: 10.1016/j.chembiol.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Bruheim P, Borgos SE, Tsan P, Sletta H, Ellingsen TE, Lancelin JM, Zotchev SB. Antimicrob Agents Chemother. 2004;48:4120. doi: 10.1128/AAC.48.11.4120-4129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmody M, Murphy B, Byrne B, Power P, Rai D, Rawlings B, Caffrey P. J Biol Chem. 2005;280:34420. doi: 10.1074/jbc.M506689200. [DOI] [PubMed] [Google Scholar]
- 33.Mendes MV, Anton N, Martin JF, Aparicio JF. Biochem J. 2005;386:57. doi: 10.1042/BJ20040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seco EM, Cuesta T, Fotso S, Laatsch H, Malpartida F. Chem Biol. 2005;12:535. doi: 10.1016/j.chembiol.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Seco EM, Fotso S, Laatsch H, Malpartida F. Chem Biol. 2005;12:1093. doi: 10.1016/j.chembiol.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Borgos SE, Tsan P, Sletta H, Ellingsen TE, Lancelin JM, Zotchev SB. J Med Chem. 2006;49:2431. doi: 10.1021/jm050895w. [DOI] [PubMed] [Google Scholar]
- 37.Volokhan O, Sletta H, Ellingsen TE, Zotchev SB. Appl Environ Microbiol. 2006;72:2514. doi: 10.1128/AEM.72.4.2514-2519.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nedal A, Sletta H, Brautaset T, Borgos SE, Sekurova ON, Ellingsen TE, Zotchev SB. Appl Environ Microbiol. 2007;73:7400. doi: 10.1128/AEM.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Power P, Dunne T, Murphy B, Lochlainn LN, Rai D, Borissow C, Rawlings B, Caffrey P. Chem Biol. 2008;15:78. doi: 10.1016/j.chembiol.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Griffith BR, Langenhan JM, Thorson JS. Curr Opin Biotechnol. 2005;16:622. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Salas JA, Mendez C. Trends Microbiol. 2007;15:219. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Thibodeaux CJ, Melancon CE, Liu HW. Nature. 2007;446:1008. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 43.Moretti R, Thorson JS. J Biol Chem. 2007;282:16942. doi: 10.1074/jbc.M701951200. [DOI] [PubMed] [Google Scholar]
- 44.Moretti R, Thorson JS. Anal Biochem. 2008;377:251. doi: 10.1016/j.ab.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS. Science. 2006;313:1291. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- 46.Zhang C, Albermann C, Fu X, Thorson JS. J Am Chem Soc. 2006;128:16420. doi: 10.1021/ja065950k. [DOI] [PubMed] [Google Scholar]
- 47.Minami A, Kakinuma K, Eguchi T. Tetrahedron Lett. 2005;46:6187. [Google Scholar]
- 48.Zhang C, Fu Q, Albermann C, Li L, Thorson JS. ChemBioChem. 2007;8:385. doi: 10.1002/cbic.200600509. [DOI] [PubMed] [Google Scholar]
- 49.Zhang C, Goff R, Griffith BR, Albermann C, Thorson JS. Chem Biol. 2008 doi: 10.1016/j.chembiol.2008.06.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seco EM, Perez-Zuniga FJ, Rolon MS, Malpartida F. Chem Biol. 2004;11:357. doi: 10.1016/j.chembiol.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 51.Nicolaou KC, Chakraborty TK, Ogawa Y, Daines RA, Simpkins NS, Furst GT. J Am Chem Soc. 1988;110:4660. [Google Scholar]
- 52.Khaled A, Ivannikova T, Auge C. Carbohydr Res. 2004;339:2641. doi: 10.1016/j.carres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Y, Thorson JS. J Org Chem. 1998;63:7568. doi: 10.1021/jo981265n. [DOI] [PubMed] [Google Scholar]
- 54.Jiang J, Biggins JB, Thorson JS. J Am Chem Soc. 2000;122:6803. [Google Scholar]
- 55.Jiang J, Biggins JB, Thorson JS. Angew Chem. 2001;113:1550. [PubMed] [Google Scholar]; Angew Chem Intl Ed. 2001;40:1502. [Google Scholar]
- 56.Albermann C, Jiang J, Thorson JS. ChemBioChem. 2003;4:443. doi: 10.1002/cbic.200200566. [DOI] [PubMed] [Google Scholar]
- 57.Hoffmeister D, Yang J, Liu L, Thorson JS. Proc Natl Acad Sci USA. 2003;100:13184. doi: 10.1073/pnas.2235011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borisova SA, Zhao LS, Melançon CE, Kao CL, Liu H-w. J Am Chem Soc. 2004;126:6534. doi: 10.1021/ja049967j. [DOI] [PubMed] [Google Scholar]
- 59.Melançon CE, Takahashi H, Liu H-w. J Am Chem Soc. 2004;126:16726. doi: 10.1021/ja043900e. [DOI] [PubMed] [Google Scholar]
- 60.Yuan YQ, Chung HS, Leimkuhler C, Walsh CT, Kahne D, Walker S. J Am Chem Soc. 2005;127:14128. doi: 10.1021/ja053704n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu W, Leimkuhler C, Gatto GJ, Oberthür M, Kahne D, Walsh CT. Chem Biol. 2005;12:527–534. doi: 10.1016/j.chembiol.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 62.Williams GJ, Zhang C, Thorson JS. Nat Chem Biol. 2007;3:657. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- 63.Williams GJ, Goff RD, Zhang C, Thorson JS. Chem Biol. 2008;15:393. doi: 10.1016/j.chembiol.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang M, Proctor MR, Bolam DN, Errey JC, Field RA, Gilbert HJ, Davis BG. J Am Chem Soc. 2005;127:9336. doi: 10.1021/ja051482n. [DOI] [PubMed] [Google Scholar]
- 65.Minami A, Uchida R, Eguchi T, Kakinuma K. J Am Chem Soc. 2005;127:6148. doi: 10.1021/ja042848j. [DOI] [PubMed] [Google Scholar]
- 66.Minami A, Eguchi T. J Am Chem Soc. 2007;129:5102. doi: 10.1021/ja0685250. [DOI] [PubMed] [Google Scholar]
- 67.Pospiech A, Neumann B. Trends Genet. 1995;11:217. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 68.Barton WA, Lesniak J, Biggins JB, Jeffrey PD, Jiang J, Rajashankar KR, Thorson JS, Nikolov DB. Nat Struct Biol. 2001;8:545. doi: 10.1038/88618. [DOI] [PubMed] [Google Scholar]
- 69.Barton WA, Biggins JB, Jiang J, Thorson JS, Nikolov DB. Proc Natl Acad Sci USA. 2002;99:13397. doi: 10.1073/pnas.192468299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS. Nat Biotechnol. 2003;21:1467. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.