Abstract
The presence of 5-methylcytosine (5-mC) in DNA is a vital epigenetic mark in vertebrates. While the enzymes responsible for methylating DNA in vertebrates have been identified, the means by which this mark can be removed are still unclear. Recently, it has been shown that activation-induced cytidine deaminase (AID) contributes to the demethylation of DNA in certain systems. This enzyme has been intensely studied in its role as a key driver of antibody diversification in B cells, but recent observations from early development in zebrafish and mice as well as heterokaryons point to a role beyond immunology. This review takes stock of the reports linking AID and related deaminases to DNA demethylation, and describes the many important questions left to be answered in this field.
Keywords: Cytidine deaminase, AICDA, AID, active demethylation, epigenetic reprogramming
DNA cytosine methylation is a key epigenetic mark in vertebrates. It occurs predominantly in the sequence context CpG, allowing for symmetrical strand modification and thus heritable transmission of epigenetic information through cell divisions. The means by which cytosine methylation in DNA is regulated is an issue of great interest because of its importance in differentiation (Reik 2007), cell reprogramming (Simonsson and Gurdon 2004; Mikkelsen et al. 2008), retroelement suppression (Walsh et al. 1998), parental imprinting (Edwards and Ferguson-Smith 2007), and X-chromosome inactivation (Sado et al. 2000).
The majority (70%–80%) of cytosines in the sequence context CpG are methylated in differentiated cells in mammals (Ehrlich et al. 1982). Methylation levels are much lower in regions with high CpG density, termed “CpG islands” (Illingworth and Bird 2009). Methylation of such islands in the promoters of genes results in silencing (Bird 2002) and can regulate alternative promoter usage when found within gene bodies (Maunakea et al. 2010). Additionally, cytosine methylation in non-CpG contexts has been found at significant levels in human embryonic stem (ES) cells (Ramsahoye et al. 2000) and at detectable levels in differentiated cells (Lister et al. 2009).
Significant progress has been made in elucidating the mechanisms of establishing and maintaining DNA methylation in vertebrates (Law and Jacobsen 2010). The effectors of DNA cytosine methylation are the DNA methyltransferase (DNMT) family of enzymes (Goll and Bestor 2005). DNMT3A and DNMT3B act as de novo methyltransferases, establishing the basic somatic methylation pattern early in development. In contrast, DNMT1 serves as a “maintenance” methyltransferase, acting on hemimethylated DNA and thereby preserving methylation state down through multiple cell divisions.
DNA cytosine methylation increases rapidly around the time of implantation but thereafter remains grossly stable (Hemberger et al. 2009). At the blastocyst stage, coinciding with lineage fixation, the inner cell mass has gross methylation levels equivalent to those in fully differentiated cells (Geiman and Muegge 2010). Whether changes in DNA methylation at specific loci are a general mechanism of gene regulation in differentiated cells or one restricted to silencing of pluripotency-associated genes after early development is a controversial issue (Illingworth and Bird 2009).
The removal of DNA cytosine methylation in vertebrates is a far murkier subject than its addition (Ooi and Bestor 2008). It is clear that “passive,” or DNA replication-dependent, demethylation can occur if DNMTs are absent or prevented from accessing newly synthesized DNA. “Active,” or replication-independent, demethylation has been reported to occur in a number of contexts in somatic cells (Zhu 2009). However, it is unclear in these cases if decreased promoter methylation is a driver of increased transcription or a downstream consequence. While active demethylation in differentiated cells remains controversial (Ooi and Bestor 2008), there are several apparent examples in normal physiology: the paternal genome immediately after fertilization (Mayer et al. 2000; Oswald et al. 2000), the formation of primordial germ cells (PGCs) in the embryo (Hajkova et al. 2002), and the differentiation of peripheral blood monocytes to dendritic cells (Klug et al. 2010).
DNA demethylation has also been described in other important contexts. Aberrant hypomethylation of oncogenes, as well as hypermethylation of tumor suppressor genes, have both been implicated in oncogenesis (Ehrlich 2009). Additionally, it is clear that demethylation of the promoters of certain pluripotency-associated genes, such as Oct4 and Nanog, is a requisite for ex vivo cellular reprogramming (Mikkelsen et al. 2008). In the relatively slow process of reprogramming by transcription factor overexpression, this demethylation is thought to occur passively. However, an active mechanism is likely involved in the fast reprogramming processes of nuclear transfer and heterokaryon formation (Yamanaka and Blau 2010).
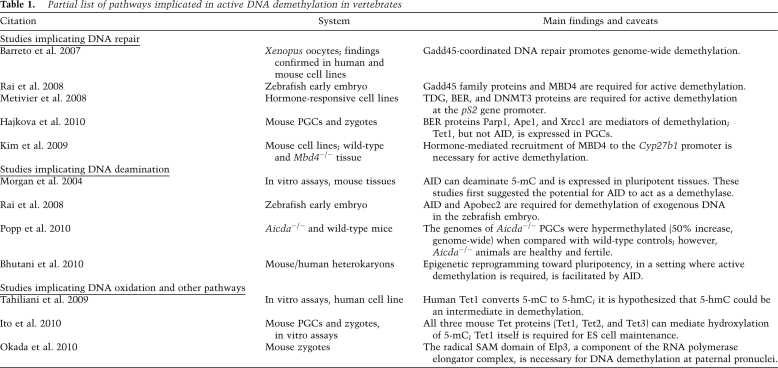
A number of potential mechanisms have been proposed for active demethylation in mammals (Table 1). The most direct posits that the methyl group could simply be enzymatically removed. The Alkb family of enzymes have been shown to demethylate 1-methyladenosine and 3-methylcytosine via oxidized intermediates, releasing the methyl carbon as formaldehyde and regenerating the original base (Falnes et al. 2002; Trewick et al. 2002). However, no comparable Alkb family enzyme that accepts 5-methylcytosine (5-mC) as a substrate has been observed in vertebrates. The methyl-CpG-binding protein MBD2 has been proposed as such a direct DNA demethylase (Bhattacharya et al. 1999), but these findings could not be replicated (Bird 2002). Furthermore, a number of studies indicate that DNA is broken and subsequently repaired during the course of demethylation (Kress et al. 2006; Barreto et al. 2007; Hajkova et al. 2010), suggesting that the entire base is removed. A hypothesis consistent with these reports is that DNA demethylation in mammals is achieved by targeted removal of 5-mC by a glycosylase without previous modification of the base. This is thought to be the dominant mechanism of active DNA demethylation in plants, with the DME/ROS1 family of glycosylases serving this function in Arabidopsis thaliana (Zhu 2009). MBD4 has been proposed as such a glycosylase in mammals (Kim et al. 2009), although previous in vitro work found this enzyme to be far more active on thymidine than 5-mC (Zhu et al. 2000).
Table 1.
Partial list of pathways implicated in active DNA demethylation in vertebrates
Yet another proposed class of mechanisms for DNA cytosine demethylation involves alteration of 5-mC followed by removal of the altered base and subsequent replacement with unmethylated cytosine. There is evidence that this may proceed by conversion of 5-mC to a 5-mC radical by the elongator complex member Elp3 (Okada et al. 2010), or to thymidine by DNMT3 family enzymes under conditions of low S-adenosylmethionine (Kangaspeska et al. 2008; Metivier et al. 2008). It has also been shown that 5-mC can be converted to 5-hydroxymethylcytosine (5-hmC) by the enzyme TET1 (Kriaucionis and Heintz 2009; Tahiliani et al. 2009), raising the possibility that demethylation could proceed through this intermediate. The requirement of TET1 for proper maintenance of methylation state at the Nanog promoter in ES cells lends credence to this model (Ito et al. 2010). Finally, a series of recent studies have suggested that the cytidine deaminase AID (activation-induced cytidine deaminase) and its relatives are also potential candidates for agents of active DNA demethylation in vertebrates.
The classical view of AID
AID was initially identified as a factor required for class switch recombination (CSR) and immunoglobulin (Ig) somatic hypermutation (SHM) (Muramatsu et al. 2000; Revy et al. 2000). The 24-kDa protein, encoded by the Aicda gene, is conserved among jawed vertebrates and is closely related to the APOBEC family of zinc-coordinating polynucleotide cytidine deaminases (Conticello 2008). It is well established that AID is able to convert cytosine to uracil in ssDNA, as demonstrated in Escherichia coli (Petersen-Mahrt et al. 2002) and in vitro (Bransteitter et al. 2003; Chaudhuri et al. 2003; Dickerson et al. 2003).
In the decade since AID's discovery, a broadly accepted model for its roles in antibody diversification has emerged (Delker et al. 2009). AID initiates CSR and SHM by conversion of cytosine to uracil in different regions of the Ig loci (Fig. 1A). CSR occurs as a result of the double-stranded breaks frequently produced in the course of repair of such lesions in the S regions of the IgH locus. Joining of breaks in different S regions results in a different constant region immediately downstream from the transcribed V(D)J and, consequently, to antibodies of a different isotype (Stavnezer et al. 2008). SHM is initiated by AID cytosine deamination within the V(D)J region of Ig loci. Repair of these lesions proceeds with an unusually high error rate, leading to mutations that can alter the affinity of the encoded antibody (Peled et al. 2008). Selection of cells bearing these mutated Igs accomplishes affinity maturation. AID is required for both of these processes: Aicda−/− mice exhibit a complete lack of secondary Ig isotypes and no somatic mutations in Ig variable regions during an immune response (Muramatsu et al. 2000). In humans, mutations in the AICDA gene result in a similar condition known as hyper-IgM syndrome type 2 (Revy et al. 2000).
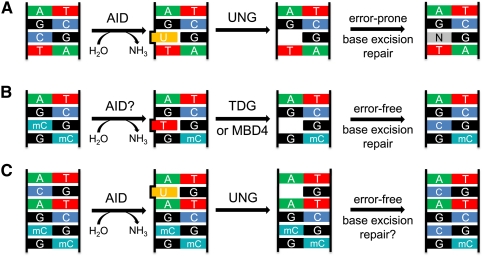
Figure 1.
AID can initiate distinct processes through a single enzymatic activity. (A) Deamination of cytosine yields uracil. Excision of uracil by UNG and repair of the subsequent abasic site leads to error-prone repair and SHM when it occurs in the V(D)J regions of the Ig loci. Although error-prone repair also gives rise to mutations at nearby bases, only the outcome of repair on C is shown. (B) Deamination of 5-mC by AID yields thymine. Excision of thymine by either of the mammalian T-G-specific glycosylases (TDG or MBD4) and subsequent error-free replacement with cytosine would yield loss of methylation at that base on one strand. (C) Deamination of cytosine followed by excision of uracil and error-free long-patch BER could lead to replacement and net demethylation of neighboring 5-mCs.
AID beyond the immune system
Despite the lack of an obvious nonimmune phenotype in AID-deficient mice, there are signs that AID has additional functions outside of antibody diversification. Although it is expressed at highest levels in germinal center B cells, it is also found in many other cell types; namely, oocytes, PGCs, ES cells (Morgan et al. 2004), breast tissue (Pauklin et al. 2009), and prostate epithelial cells (Lin et al. 2009), although its expression in PGCs has been challenged recently (Hajkova et al. 2010). As AID is a DNA mutator, its expression outside of B cells cannot be dismissed as inconsequential. Indeed, AID has been shown to act beyond the Ig loci, causing point mutations (Liu et al. 2008), double-stranded breaks (Hasham et al. 2010), and translocations (Pasqualucci et al. 2008; Lin et al. 2009; Robbiani et al. 2009) throughout the genome. Through these processes, AID has been shown to contribute to tumorigenesis (Okazaki et al. 2007). As such, expression of AID in non-antibody-producing cells would likely be selected against in evolution unless it had some function in these tissues. Further suggestions of functions for AID beyond the immune system come from studies of lower vertebrates. As in mice, AID is expressed during early development in Danio rerio (Rai et al. 2008), Xenopus laevis (Marr et al. 2007), and the newt Pleurodeles waltl (Bascove and Frippiat 2010). The broad conservation of this pattern of AID expression strongly suggests a function in early development.
The first direct evidence that AID might have functions beyond the standard model of antibody diversification came in 2004, when it was shown by Petersen-Mahrt and colleagues (Morgan et al. 2004) that AID, along with the related cytidine deaminase APOBEC1, can convert 5-mC in ssDNA to thymidine in vitro. This observation led to the proposal that these enzymes could be the elusive vertebrate DNA cytosine demethylase.
AID could initiate demethylation by a damage and repair mechanism similar to that used in SHM (Fig. 1B). The deamination of 5-mC by AID yields thymidine. This T would then be removed by a T-G mismatch-specific glycosylase, of which two, TDG and MBD4, are known in mammals (Millar et al. 2002; Hardeland et al. 2003). The resulting abasic site would then be replaced by an unmethylated cytidine via base excision repair (BER) processes, yielding the net removal of methylation without alteration of sequence. BER could also proceed through either short or long patch repair (Fortini and Dogliotti 2007), potentially yielding demethylation of multiple neighboring cytosines in the latter case (Fig. 1C). This mechanism would allow for demethylation after deamination of cytosine or 5-mC, and could give rise to the appearance of processive demethylation, despite originating from a single deamination event.
It may seem counterintuitive to invoke the mechanism of SHM to explain that of DNA demethylation, which proceeds with fidelity. However, the fact that AID-induced lesions are apparently resolved through multiple pathways makes this more plausible. At a small number of loci—most dramatically, Ig—repair of AID-induced lesions is particularly error-prone, while, at other genomic locations, it results in almost undetectable levels of mutation (Liu et al. 2008). How these different pathways are selected is still unclear, but, given that high-fidelity repair predominates at most sites, it is likely the norm, while frequent mutation as observed at the Ig loci is the special case.
AID-dependent demethylation in lower vertebrates
The AID model of demethylation received a measure of in vivo validation from work in D. rerio early embryos (Rai et al. 2008). Introduction of a methylated DNA fragment into single-cell embryos induces expression of AID along with related putative cytidine deaminases Apobec2a and Apobec2b. Additionally, overexpression of AID along with the T-G glycosylase MBD4 leads to efficient demethylation of a methylated DNA fragment, with observable conversion of 5-mC to T. Morpholino knockdown of AID or MBD4 in D. rerio single-cell stage embryos results in hypermethylation of the promoter of the neurogenesis-related transcription factor neurod2 at 80% epiboly. While the large number of cell divisions between morpholino injection and the observed difference in methylation at neurod2 seems to point toward a passive mechanism, both AID and MBD4 were also detectable at the locus by chromatin immunoprecipitation (ChIP), suggesting that the promoter's methylation state was dynamic through development and not just the result of perturbations at the single-cell stage. Consequently, AID morphants displayed severe defects in neurogenesis consistent with decreased neurod2 expression. The tissue- and locus-specific phenotype suggests that active demethylation may serve as a mechanism for regulating specific lineage decisions, as opposed to only resetting methylation state grossly in the zygote and germline.
Intriguingly, Rai et al. (2008) also found that the Gadd45 family of proteins, previously implicated in active demethylation in X. laevis (Barreto et al. 2007), were involved in AID-dependent demethylation. Combined knockdown of four of the six Gadd45 family members found in zebrafish sharply reduced demethylation of a reporter and increased methylation of the genome as a whole. It was thus suggested that Gadd45 serves as a scaffold to couple AID and MBD4, supported by the facts that Gadd45α promotes their association with a methylated reporter plasmid and that the three proteins also coimmunoprecipitate. The apparent physical interaction of AID and MBD4, enforced by Gadd45, provides a potential mechanism for the tight coupling of deamination to repair that would be necessary for demethylation without attendant widespread mutation, as has been noted (Law and Jacobsen 2010).
AID-dependent demethylation in mammalian PGCs
Although there is no direct evidence for deaminase-dependent effects on cytosine methylation in mammalian somatic tissue, a recent study has implicated AID in establishment of the hypomethylated state of mouse PGCs (Popp et al. 2010). In this study, genomic DNA from sperm, total fetus, placenta, and male and female embryonic day 13.5 (E13.5) PGCs from wild-type and Aicda−/− mice was bisulfite-converted and sequenced using the Illumina ultrahigh-throughput platform, a method termed BS-Seq or MethylC-Seq (Cokus et al. 2008; Lister et al. 2008). The resulting data provide a genome-wide map of cytosine methylation at single-nucleotide resolution. While a lack of sequencing depth precluded quantitative measures of methylation for every genomic cytosine, the coverage was sufficient to detect a significant increase in methylation in Aicda−/− PGCs. This difference was more pronounced for female than male PGCs, and was roughly homogeneous throughout the genome. The broad nature of the methylation increase implies that AID functions without regard to specific loci, and thus is a plausible component of germline methylation erasure.
While the evidence that AID has a role in demethylation is suggestive, there are still considerable caveats. Most significantly, a recent report detected no AID expression in mouse PGCs at E11.5, although a small level of AID expression was observed at E12.5 (Hajkova et al. 2010). As the epigenetic reprogramming of PGCs begins at E11.5, AID cannot be the sole agent of demethylation. This idea is consistent with the occurrence of significant, albeit reduced, demethylation in the PGCs of Aicda−/− mice compared with somatic cells. Whether AID-independent demethylation in these cells is a result of 5-mC deamination by another member of the AID/APOBEC family or is due to any of the mechanisms mentioned above is an open question. It is also possible that the AID/APOBEC family of deaminases have differing target gene preferences for demethylation, and thus play complementary roles in the genome-wide removal of cytosine methylation.
Another issue raised by these results is the functional importance of the observed PGC hypermethylation, as Aicda−/− mice are fully viable. Even if AID were the sole agent of active demethylation, it is not clear that loss of this mechanism would result in a drastic phenotype. This view is supported by evidence from A. thaliana, in which loss of all three 5-mC-removing glycosylases in vegetative tissue leads to viable plants displaying increased methylation only at certain loci, and no genome-wide increase (Penterman et al. 2007). As DNA methylation is essential for parental imprinting, Popp et al. (2010) suggested that the consequences of PGC AID deficiency may lie in retention of a parental-like epigenetic state. Data supporting this view lies in a small but significant difference that exists between wild-type and Aicda−/− mice in the relationship between litter size and birth weight. In normal mice, pups that are part of large litters tend to have lower birth weights, while in Aicda−/− mice this compensation is absent (Popp et al. 2010). Hypermethylated elements responsible for this phenotype have not been identified. New technologies for methylation-sensitive high-throughput sequencing (Flusberg et al. 2010) and the decreasing cost of “traditional” deep sequencing should make the quantitative, single-nucleotide resolution studies of PGC methylation feasible in the near future to address this question.
AID-dependent demethylation in heterokaryons
Corroboration of AID's capacity to demethylate DNA in mammalian cells came in a recent study of reprogramming during interspecies heterokaryon formation (Bhutani et al. 2010). This system uses polyethylene glycol fusion of mouse ES cells with human fibroblasts to induce reprogramming of the human nucleus to an ES-like state with high frequency. Because reprogramming is known to involve demethylation, heterokaryon formation allows for study of demethylation in mammalian cells, albeit in a nonphysiological context. Knockdown of AID was found to significantly inhibit reprogramming, as measured by transcript levels of the pluripotency markers Oct4 and Nanog. Methylation of the promoters of these genes was also significantly increased as a result of reduced levels of AID. As heterokaryons are nondividing, AID-dependent demethylation in this system is necessarily active.
AID is known to act only on ssDNA, which presumably occurs transiently in vivo during transcription (Shen et al. 2009). Yet AID-dependent demethylation clearly occurs at the promoters of silenced genes in heterokaryons in the absence of replication. Whether these promoters share distinguishing structural features that would give rise to transient ssDNA, such as Z/B DNA junctions (de Rosa et al. 2010) or other non-B structures (Raghavan et al. 2004), is not known. However, at least for the limited number of genes studied, there seems to be no clear relation between AID occupancy and transcription: While Oct4 and Nanog are both transcribed and occupied, Tp53 is transcribed but not occupied, and the constant (C) region of IgH is occupied but presumably not transcribed (although this was not investigated). Additionally, AID could be found by ChIP at the human fibroblast Oct4 and Nanog loci, but not at the same genes in the mouse ES nuclei. This provides evidence for AID's (or an AID-containing complex's) affinity for silenced (or at least methylated) genes. Whether this affinity is caused by inherent tendency of AID to bind 5-mC or a more intricately targeted system of reprogramming toward pluripotency also active in PGCs remains to be determined. Whatever the case may be, the behavior of AID in heterokaryons likely has implications for the biology of PGCs, and likely B cells as well.
Future directions
In certain lower vertebrates, AID appears to be involved in a process other than antibody diversification. Although the early embryos of X. laevis and D. rerio provide a more accessible in vivo system for studying active demethylation, the discrepancies between findings in these lower vertebrates and mammals calls into question the existence of a single demethylation mechanism shared between these groups. In particular, there is no evidence that any member of the Gadd45 family is involved in mediating demethylation in mammals. While Gadd45α overexpression alone has been shown to promote demethylation in X. laevis (Barreto et al. 2007), and is required for AID- and MBD4-dependent demethylation in zebrafish, it also appears to be dispensable for normal regulation of cytosine methylation in mice (Engel et al. 2009). Furthermore, the gene is not expressed in the mouse oocyte (Jin et al. 2008), precluding its involvement in post-fertilization paternal genome demethylation. One hypothesis that would account for the differences between observations in mice and zebrafish is that AID is redundant with other APOBEC family deaminases in demethylation (Morgan et al. 2004). Phylogenetics lends some support to this idea, as there are several members of the APOBEC gene family that first appear in eutherians (Conticello et al. 2005).
Even restricting the discussion to mammalian systems, there are significant questions regarding AID's role in demethylation. If AID is acting as a demethylase in PGCs and heterokaryons, the question arises as to whether it does the same in B cells, given that there is enough AID in activated B cells to mutate a number of non-Ig loci (Liu et al. 2008) and cause double-stranded breaks throughout the genome in the absence of proper DNA repair (Hasham et al. 2010). One possible explanation is that there are yet unidentified cell type-specific cofactors of AID that either promote demethylation in pluripotent cells or suppress this activity in B cells. It is also conceivable that demethylation might occur in B cells at some rate, with the effect being less dramatic due to more frequent DNA methylation, resulting in a dynamic equilibrium.
Whatever the case, AID's likely role in demethylation is pertinent to those interested in epigenetics, development, and immunology. The identification of a promising candidate for vertebrate DNA cytidine demethylase has raised more questions than it has answered; elucidation of the particulars of AID's relationship to DNA demethylation will surely be the subject of much inquiry in the coming years.
Acknowledgments
We thank Jayanta Chaudhuri, Claire Hamilton, and Brad Rosenberg for thoughtful comments on the manuscript. Work relating to AID and its roles in Ig mutation and cancer in F.N.P.'s laboratory is supported by NCI grant no. CA098495 and the Starr Cancer Consortium. E.L.F. is supported by a predoctoral training grant from the Cancer Research Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1963010.
References
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. 2007. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445: 671–675 [DOI] [PubMed] [Google Scholar]
- Bascove M, Frippiat JP 2010. Molecular characterization of Pleurodeles waltl activation-induced cytidine deaminase. Mol Immunol 47: 1640–1649 [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M 1999. A mammalian protein with specific demethylase activity for mCpG DNA. Nature 397: 579–583 [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM 2010. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463: 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci 100: 4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422: 726–730 [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG 2008. The AID/APOBEC family of nucleic acid mutators. Genome Biol 9: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol 22: 367–377 [DOI] [PubMed] [Google Scholar]
- Delker RK, Fugmann SD, Papavasiliou FN 2009. A coming-of-age story: Activation-induced cytidine deaminase turns 10. Nat Immunol 10: 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosa M, de Sanctis D, Rosario AL, Archer M, Rich A, Athanasiadis A, Carrondo MA 2010. Crystal structure of a junction between two Z-DNA helices. Proc Natl Acad Sci 107: 9088–9092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN 2003. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med 197: 1291–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC 2007. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol 19: 281–289 [DOI] [PubMed] [Google Scholar]
- Ehrlich M 2009. DNA hypomethylation in cancer cells. Epigenomics 1: 239–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C 1982. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res 10: 2709–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, Tront JS, Erinle T, Nguyen N, Latham KE, Sapienza C, Hoffman B, Liebermann DA 2009. Conserved DNA methylation in Gadd45a−/− mice. Epigenetics 4: 98–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PØ, Johansen RF, Seeberg E 2002. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419: 178–182 [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7: 461–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini P, Dogliotti E 2007. Base damage and single-strand break repair: Mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 6: 398–409 [DOI] [PubMed] [Google Scholar]
- Geiman TM, Muegge K 2010. DNA methylation in early development. Mol Reprod Dev 77: 105–113 [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117: 15–23 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA 2010. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329: 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland U, Bentele M, Jiricny J, Schar P 2003. The versatile thymine DNA-glycosylase: A comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res 31: 2261–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasham MG, Donghia NM, Coffey E, Maynard J, Snow KJ, Ames J, Wilpan RY, He Y, King BL, Mills KD 2010. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat Immunol 11: 820–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W 2009. Epigenetic dynamics of stem cells and cell lineage commitment: Digging Waddington's canal. Nat Rev Mol Cell Biol 10: 526–537 [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP 2009. CpG islands–‘a rough guide.’ FEBS Lett 583: 1713–1720 [DOI] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Guo C, Pfeifer GP 2008. GADD45A does not promote DNA demethylation. PLoS Genet 4: e1000013 doi: 10.1371/journal.pgen.1000013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G 2008. Transient cyclical methylation of promoter DNA. Nature 452: 112–115 [DOI] [PubMed] [Google Scholar]
- Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, et al. 2009. DNA demethylation in hormone-induced transcriptional derepression. Nature 461: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Klug M, Heinz S, Gebhard C, Schwarzfischer L, Krause SW, Andreesen R, Rehli M 2010. Active DNA demethylation in human postmitotic cells correlates with activating histone modifications, but not transcription levels. Genome Biol 11: R63 doi: 10.1186/gb-2010-11-6-r63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress C, Thomassin H, Grange T 2006. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci 103: 11112–11117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. 2009. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139: 1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature 451: 841–845 [DOI] [PubMed] [Google Scholar]
- Marr S, Morales H, Bottaro A, Cooper M, Flajnik M, Robert J 2007. Localization and differential expression of activation-induced cytidine deaminase in the amphibian Xenopus upon antigen stimulation and during early development. J Immunol 179: 6783–6789 [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T 2000. Demethylation of the zygotic paternal genome. Nature 403: 501–502 [DOI] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. 2008. Cyclical DNA methylation of a transcriptionally active promoter. Nature 452: 45–50 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A 2002. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science 297: 403–405 [DOI] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK 2004. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: Implications for epigenetic reprogramming. J Biol Chem 279: 52353–52360 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563 [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y 2010. A role for the elongator complex in zygotic paternal genome demethylation. Nature 463: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki IM, Kotani A, Honjo T 2007. Role of AID in tumorigenesis. Adv Immunol 94: 245–273 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH 2008. The colorful history of active DNA demethylation. Cell 133: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J 2000. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10: 475–478 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC III, Nussenzweig MC, Dalla-Favera R 2008. AID is required for germinal center-derived lymphomagenesis. Nat Genet 40: 108–112 [DOI] [PubMed] [Google Scholar]
- Pauklin S, Sernandez IV, Bachmann G, Ramiro AR, Petersen-Mahrt SK 2009. Estrogen directly activates AID transcription and function. J Exp Med 206: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD 2008. The biochemistry of somatic hypermutation. Annu Rev Immunol 26: 481–511 [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL 2007. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci 104: 6752–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418: 99–103 [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463: 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR 2004. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature 428: 88–93 [DOI] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR 2008. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135: 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R 2000. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci 97: 5237–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102: 565–575 [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, et al. 2009. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell 36: 631–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E 2000. X inactivation in the mouse embryo deficient for Dnmt1: Distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol 225: 294–303 [DOI] [PubMed] [Google Scholar]
- Shen HM, Poirier MG, Allen MJ, North J, Lal R, Widom J, Storb U 2009. The activation-induced cytidine deaminase (AID) efficiently targets DNA in nucleosomes but only during transcription. J Exp Med 206: 1057–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsson S, Gurdon J 2004. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol 6: 984–990 [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE 2008. Mechanism and regulation of class switch recombination. Annu Rev Immunol 26: 261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B 2002. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419: 174–178 [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117 [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM 2010. Nuclear reprogramming to a pluripotent state by three approaches. Nature 465: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK 2009. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet 43: 143–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Angliker H, Schwarz S, Thiry S, Siegmann M, Jost JP 2000. 5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence. Nucleic Acids Res 28: 4157–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]