Abstract
Signal transduction networks can be perturbed biochemically, genetically, and pharmacologically to unravel their functions. But at the systems level, it is not clear how such perturbations are best implemented to extract molecular mechanisms that underlie network function. Here, we combined pairwise perturbations with multiparameter phosphorylation measurements to reveal causal mechanisms within the signaling network response of cardiomyocytes to coxsackievirus B3 (CVB3) infection. Using all possible pairs of six kinase inhibitors, we assembled a dynamic nine-protein phosphorylation signature of perturbed CVB3 infectivity. Cluster analysis of the resulting dataset showed repeatedly that paired inhibitor data were required for accurate data-driven predictions of kinase substrate links in the host network. With pairwise data, we also derived a high-confidence network based on partial correlations, which identified phospho-IκBα as a central “hub” in the measured phosphorylation signature. The reconstructed network helped to connect phospho-IκBα with an autocrine feedback circuit in host cells involving the proinflammatory cytokines, TNF and IL-1. Autocrine blockade substantially inhibited CVB3 progeny release and improved host cell viability, implicating TNF and IL-1 as cell autonomous components of CVB3-induced myocardial damage. We conclude that pairwise perturbations, when combined with network-level intracellular measurements, enrich for mechanisms that would be overlooked by single perturbants.
Keywords: pairwise perturbation, signaling network, systems biology, viral myocarditis, picornaviridae
Intracellular signal transduction is achieved through binary protein–protein interactions that connect together to form networks (1). Signaling pathways often have multiple layers of feedback control, and cross-talk between pathways is usually extensive (2). This complexity enables diverse stimuli to be integrated when key cell decisions must be made (3, 4). However, conceptually, a “tangled” signaling circuit makes it difficult to assign unambiguous roles to individual proteins in a straightforward way.
Viruses and other pathogens have evolved effective strategies to subvert host signaling networks for their own purposes (5, 6). Infection is therefore equivalent to a systems-level perturbation, which can be useful for understanding network function when combined with targeted interventions. The question is how best to deploy these perturbations in a way that takes network complexity into consideration but also quickly identifies mechanistic connections between signals and cell outcomes.
One observation that has emerged from recent studies of networks is the surprising richness of information that is captured when pairs of inputs or perturbations are considered (4, 7–14). Two-factor screens typically reveal phenotypic synergies in only ∼10–25% of all possible nC2 combinations (7, 15). However, pairwise stimulation and observation appears to enrich for network states that are especially relevant to predicting function (9, 10, 13). Dual perturbation approaches can directly assign functional synergies in cell phenotype (7, 14), but explaining the basis of such observations is difficult without follow-up experiments at the molecular level. It has not been determined whether a more comprehensive mechanistic understanding could be gained by collecting intracellular measurements for all pairwise conditions from the start.
Here, we pursued this question through an in vitro model of cardiomyocyte infection with the pathogen coxsackievirus B3 (CVB3). CVB3 is among the most common causes of viral myocarditis in infants and young children, often leading to acute heart failure and sudden death (16). To examine the direct mechanisms whereby CVB3 disrupts the host cell signaling network and causes tissue damage, we developed a pairwise pharmacological approach to perturb cardiomyocyte signaling during CVB3 infection. Using a panel of small-molecule kinase inhibitors, our goal was to identify which protein kinases activated by CVB3 were responsible for the phosphorylation signatures and toxicity observed in host cells.
We found multiple instances in which data from inhibitor pairs were required for accurate clustering-based assignments of kinase substrate interactions. Moreover, using graphical Gaussian modeling (GGM) to reconstruct pairwise interactions based on partial correlations within the inhibitor dataset, we were able to reveal an extracellular positive feedback circuit for CVB3 cardiotoxicity. CVB3 drives release of the proinflammatory cytokines, TNF, and IL-1, which act as autocrine effectors to augment virus-induced cell death. Our results suggest that pairwise perturbations may be more effective at uncovering molecular mechanisms within signaling networks than an equivalent numbers of single agents.
Results
Pairwise Pharmacological Perturbation of Cardiomyocytes Infected with CVB3.
To investigate the host network response to CVB3 in a uniform cell population, we used HL-1 murine cardiomyocytes (17). These cells can be infected with CVB3, support viral replication, and undergo virus-induced cell death (18).
We sought targeted perturbations that could be introduced rapidly into cells and combined easily. Therefore, we avoided slow overexpression or knockdown approaches that might yield adaptations in the underlying network. Focusing on small-molecule inhibitors, we targeted six kinase signaling pathways previously implicated in different facets of CVB3 infectivity: Akt–GSK3 (19), IKK–NF-κB (20), Src-family kinases (21, 22), p38–MK2 (23, 24), JNK (24), and MEK–ERK (25). Small-molecule inhibitors are selective, but rarely specific (26, 27). In fact, some of the inhibitors we used have unknown mechanisms of action (28) or are known to perturb multiple kinases aside from the primary target (26, 27, 29). Consequently, the purpose of the small molecules was not to assign roles directly to the presumed target kinase. Instead, inhibitor combinations were used as a modular way to dampen the capacity of cells to signal through different branches of the overall network.
We pretreated cells with single inhibitors or a paired combination and monitored the dynamics of nine phosphorylation events over a 24-h time course of CVB3 infection. Time points were selected to capture the major stages of the viral life cycle: viral docking to the host cell [∼10 min postinfection (p.i.)], host cell endocytosis (∼1 h p.i.), synthesis of viral RNA (∼8 h p.i.), synthesis of viral proteins such as VP1 capsid (∼16 h p.i.), and viral progeny release (VPR) (∼24 h p.i.). We directly tracked the phosphorylation of five kinases—Akt, GSK3β, p38, JNK, and ERK—whose activity we perturbed pharmacologically. We also measured the phosphorylation status of two proteins, Hsp-27 and IκBα, which are reliable indicators of p38–MK2 and IKK activity, respectively (30–32). Lastly, we quantified key phosphorylation sites on the transcription factors, CREB and ATF-2. These two proteins lie downstream of multiple kinase signaling pathways (see below) and are important for integrating the host response to CVB3 infection (33, 34).
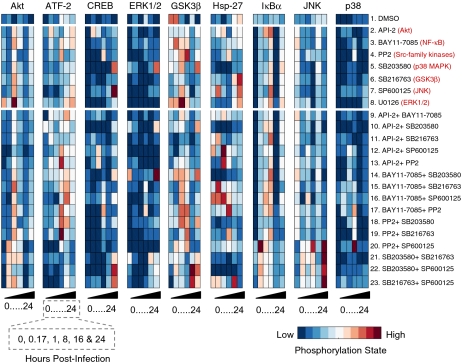
We found that inhibitor pairs showed remarkably different patterns of phosphoprotein dynamics in response to CVB3 infection compared with the corresponding single inhibitors (Fig. 1). For example, pretreatment of cells with any single inhibitor in the panel caused an increase in Akt phosphorylation at 1 h p.i. (conditions 2–8) compared with control (condition 1). However, this spike in phospho-Akt was dampened when single inhibitors were combined with the Akt inhibitor API-2 (28) (conditions 9–13), even though API-2 by itself led to increased Akt phosphorylation at 1 h p.i. (condition 2). Similarly, the nonspecific JNK inhibitor SP600125 (26) and GSK3 inhibitor SB216763 (27) each increased Hsp-27 phosphorylation (conditions 6–7) at 24 h p.i. compared with control. However, combined inhibition with SP600125 and SB216763 appeared to accelerate Hsp-27 phosphorylation (condition 23), such that phospho-Hsp-27 was decreased relative to control at 24 h p.i. Overall, we found that dual inhibitor treatments caused changes in CVB3-induced signaling that were not quantitatively predictable from single inhibitors (Fig. S1). This suggested that inhibitor pairs had revealed network-level behaviors that otherwise would have been missed.
Fig. 1.
Single and pairwise small-molecule perturbation of a dynamic nine-protein phosphorylation signature induced by CVB3 infection. HL1 cells were pretreated with one of seven small-molecule kinase inhibitors or a paired combination of these inhibitors for 0.5 h, infected with CVB3 (M.O.I. = 9), and then assessed for the indicated phosphoproteins by phospho-ELISA. Data are shown as mean of two or four independent experiments and were standardized by the z-score function in MATLAB as described in Methods.
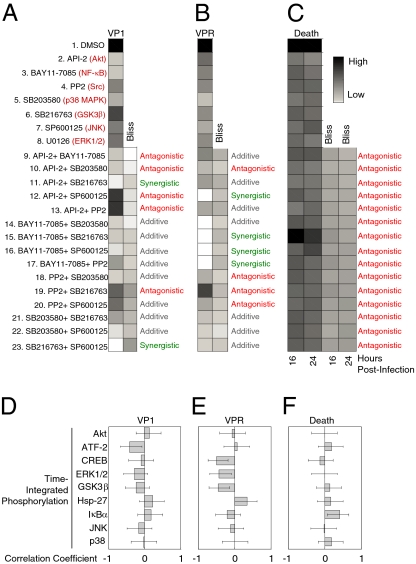
Next, we complemented the intracellular signaling dataset with three functional readouts of productive CVB3 infection: VP1 capsid protein expression, viral progeny release (VPR), and cytotoxicity determined by loss of MTS positivity (Fig. 2 and Fig. S2). We found that all single inhibitor treatments substantially reduced VP1 expression, VPR, and toxicity caused by CVB3 (P < 0.05). When these data were used to predict dual inhibitor responses under the assumption of Bliss independence (35), we found many instances of significant synergy or antagonism (P < 0.05). Interestingly, the pattern of nonadditivity across inhibitor pairs depended on the readout. For instance, SP600125 plus SB216763 was strongly synergistic for reducing VP1 expression (Fig. 2A). However, the same combination showed an additive VPR response and antagonistic cytotoxicity (Fig. 2 B and C). Our measurements of CVB3 infectivity further support the conclusion that paired signaling inhibitors establish network states that cannot be achieved or predicted if the same inhibitors are used individually (15).
Fig. 2.
Paired inhibitor combinations cause synergistic or antagonistic inhibition of viral protein expression, progeny release, and cardiotoxicity. HL1 cells were pretreated and infected as described in Fig. 1. (A) VP1 capsid protein expression, (B) VPR, and (C) cell death were measured by Western blotting, plaque assay, and MTS assay, respectively (Methods). Data are presented as the mean of three independent replicates. Both VP1 expression and VPR were normalized to the DMSO-treated control. Cell death was normalized to the CVB3-infected DMSO-treated control. Bliss predictions of independence were performed as described (35), and significant differences between measured responses and Bliss predictions were calculated by two-tailed t test with unequal variance. Measured responses significantly below Bliss prediction were defined as synergistic inhibition (green), whereas those significantly above were antagonistic (red), and those not significantly different were additive (gray). Time-integrated phosphorylation profiles of proteins measured in Fig. 1 were correlated with (D) VP1 capsid protein expression, (E) VPR, and (F) cell death at 16 h. Pearson correlation coefficients are shown ±90% Fisher z-transformed confidence intervals.
To examine simple signal–response relationships between measured phosphoprotein dynamics and cell phenotype, we correlated each time-integrated phosphorylation profile with VP1, VPR, and cell death across all 23 conditions (Fig. 2 D–F). We observed weak covariation between individual phosphoprofiles and CVB3-induced cell outputs with |R| < 0.5 for all correlations examined. This indicates that the host response is distributed across multiple signaling circuits with no individual pathway dominating in the network as a universal control point (3). We conclude that systematic intracellular perturbations are not an effective means for observing strong links between individual signals and cell responses.
Hierarchical Deconstruction of the CVB3-Induced Signaling Network.
We next sought to determine whether dual inhibitor treatments could enrich for correlations that were causal within the signaling dataset. Our predictions focused on identifying the context-dependent kinases responsible for the activating phosphorylation of ATF-2 on Thr69/Thr71 and of CREB on Ser133 (Fig. 1 and Table S1). Active ATF-2– and CREB-mediated gene expression is an important component of the antiviral and pathogenic responses to CVB3 infection (33, 34).
ATF-2 and CREB phosphorylation sites act as points of convergence for multiple kinase signaling pathways. ATF-2 can be phosphorylated by the proline-directed JNK (36), p38 (37), and ERK (38) MAPK pathways. Conversely, CREB is phosphorylated by basophilic kinases, such as PKA (39), RSK2 downstream of ERK (40), and MSK1 downstream of ERK and p38 (41). The pathways mediating ATF-2 and CREB phosphorylation in a given context depend on which kinases are activated and accessible during a biological event. Many of the candidate ATF-2 and CREB kinases (or their upstream activators) were measured in our dataset (Fig. 1 and Table S1). We therefore could test whether data from the dual inhibitor conditions were particularly important for correctly identifying the dominant ATF-2 and CREB kinases during CVB3 infection.
We used hierarchical clustering as a simple pairwise analysis tool for assembling groups of phosphoproteins with similar measurement patterns (42). Because different kinase pathways could be important for ATF-2 and CREB phosphorylation at different stages of CVB3 infection, we clustered the phosphoprotein data for each individual time point (Fig. S3). We built clusters from four different variations of the inhibitor dataset. First, we analyzed the complete dataset comprised of 23 conditions (seven single inhibitor treatments, 15 dual inhibitor treatments, and one control). Then, we subsampled the complete dataset three different ways, using only treatments with single inhibitors (plus control), only treatments with double inhibitors, or with mixtures of treatments involving single and double inhibitors. We controlled for differences in sample size by repeatedly taking random subsets of double and single + double inhibitor treatments that matched the size of the single inhibitor treatments (eight conditions). We averaged the clustering results of many random samplings to achieve a size-adjusted clustering dendrogram for double and single + double treatments (Methods). Lastly, we inspected the position of ATF-2 and CREB in the resulting dendrograms and predicted that the nearest measured kinase (or activation readout) would be the most important biochemically. These predictions were tested by adding inhibitors, which were chemically distinct from those used in the original dataset (Fig. 1), shortly before the selected time point of CVB3 infection.
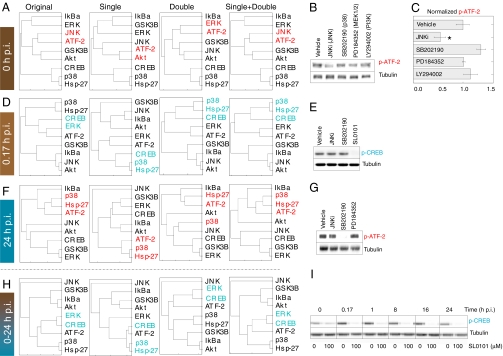
We found that data from paired inhibitors improved the accuracy of clustering-based predictions in several distinct ways. For example, at 0 h p.i., double inhibitor treatments appeared to synergize with single treatments independent of the overall sample size to link ATF-2 with JNK (Fig. 3A). Upon treatment of cells with a JNK peptide inhibitor (JNKi), we reproducibly detected a 2-fold decrease in phospho-ATF-2 (P < 0.01, Fig. 3 B and C). The decrease was specific to JNK inhibition because ATF-2 phosphorylation was not significantly perturbed by inhibitors of the p38, MEK–ERK, or PI3K–Akt pathways (Fig. 3B). This indicates that JNK signaling partly controls basal ATF-2 phosphorylation that precedes CVB3 infection.
Fig. 3.
Pairwise inhibitor data enable accurate, context-dependent predictions of ATF-2 and CREB kinases. (A) Combined single and double inhibitor data uniquely predicts JNK as an ATF-2 kinase at 0 h p.i. (B and C) JNK inhibition reduces basal ATF-2 phosphorylation in HL1 cardiomyocytes. Densitometry of phospho-ATF-2 (p-ATF-2) was quantified and is shown as the mean ± SEM of three independent samples. (D) Double inhibitor treatments increase the measured data space to predict that CREB phosphorylation lies downstream of ERK rather than p38 at 0.17 h p.i. (E) Inhibition of the ERK substrate RSK with SL0101 abolishes CREB phosphorylation at 0.17 h p.i. (F) p38 is predicted to be an ATF-2 kinase at 24 h p.i. irrespective of the inhibitor data used. (G) ATF-2 phosphorylation is specifically reduced by the p38 inhibitor SB202190. (H) Double inhibitor treatments predict that signaling downstream of ERK globally regulates CREB phosphorylation in response to CVB3 infection. (I) Inhibition of the ERK substrate RSK with SL0101 blocks CREB phosphorylation throughout CVB3 infection. For A, D, F, and H, phosphoproteins were clustered at the indicated time points using a Euclidean distance metric and average linkage. The dendrograms were built from the complete original dataset of single and double inhibitors (original) or subsampled as single-, double-, or single + double inhibitor subsets as described in Methods. The nearest kinases or activation reporters to ATF-2 or CREB on the resulting dendrograms are highlighted in red and blue, respectively. For B, C, E, G, and I, HL1 cells were pretreated and infected as described in Fig. 1, except that inhibitors were added 1 h before the indicated time point p.i. Inhibitors were used at the following concentrations: JNKi, 1 μM; SB202190, 10 μM; SL0101, 100 μM; PD184352, 10 μM; and LY294002, 10 μM. Both ATF-2 and CREB phosphorylation (p-) were measured by Western blotting with tubulin used as a loading control.
A second, more practical advantage of paired inhibitors related to increasing the data “space” with which to define protein clusters (Fig. 1) (42). By using pairwise combinations, we could augment the breadth of experimental conditions without adding more inhibitors to the panel. Importantly, we found that the information contained in these added conditions was meaningful. For instance, at 0.17 h p.i., phospho-CREB clustered with p38–MK2 activation for all size-adjusted, eight-sample clusters, irrespective of which inhibitor treatments were included (Fig. 3D). By contrast, when the complete dataset was used, we found that phospho-CREB clustered with MEK–ERK activation, suggesting that RSK was the relevant CREB kinase (40). Treatment of cells with inhibitors of p38 (43) or RSK (44) revealed that RSK inhibition abolished CREB phosphorylation, whereas p38 inhibition had no effect (Fig. 3E). These results show that RSK is a key CREB kinase for the early response to CVB3 infection, which was predicted when the inhibitor training set was expanded to include paired combinations.
Accurate kinase substrate predictions did not universally require pairwise data. For example, at 24 h p.i., all of the clustering variants grouped ATF-2 phosphorylation with p38–MK2 signaling (Fig. 3F). Accordingly, p38 inhibition substantially decreased phospho-ATF-2 during the late stages of CVB3 infection, whereas JNK or MEK–ERK inhibition did not (Fig. 3G). We note that the same p38 inhibitor had no effect on ATF-2 phosphorylation before CVB3 infection (Fig. 3B), illustrating the context-dependent role of individual kinases on shared substrates.
To test the accuracy toward more global predictions of mechanism, we combined the time course measurements into a single dataset and subsampled the inhibitor conditions as before (Fig. 3H). With time-aggregated data, we found that ERK specifically clustered with CREB phosphorylation because of the information contained in double inhibitor treatments. When RSK was inhibited shortly before each time point of CVB3 infection, we found that CREB phosphorylation was uniformly abolished (Fig. 3I), as predicted by the double inhibitor data. Thus, in contrast to the context-specific kinases of ATF-2 (Fig. 3 B and G), our results support RSK as the dominant CREB kinase throughout CVB3 infection.
Host Signaling Network Reconstruction Using Paired Inhibitor Data.
The CVB3 clustering results suggested that causal connections became enriched when paired inhibitors were included. However, clustered dendrograms focus on the most dominant positive covariations in a dataset and do not account for shared correlations among measured variables (42). This is problematic for network reconstruction (45) because anticorrelations (as in Fig. 2 D–F) are important for defining inhibitory edges between nodes. Furthermore, ignoring shared correlations makes it impossible to distinguish whether two nodes are directly connected or whether a third correlated variable intervenes between them.
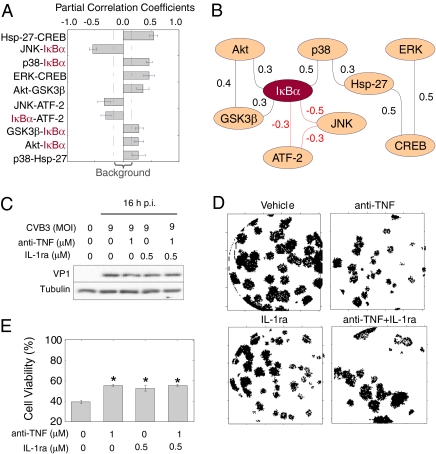
To accommodate these scenarios, we used GGM (46) to derive a candidate host cell signaling network from the phosphoprotein dataset. GGM assigns edges between nodes based on their partial correlation—the pairwise correlation that remains after considering the correlations that two variables share with other variables in the dataset. Of 36 possible pairwise edges between the nine phosphoproteins, we identified 10 whose partial correlations were significantly above background (P < 0.05) (Fig. 4A). Eight of 10 partial correlations were consistent with biochemical mechanisms that have been reported in the literature (Table S2) although only one had been specifically implicated in CVB3 pathogenesis (20). Visual inspection of the resulting GGM network revealed phospho-IκBα as a network “hub” that was densely connected with other proteins in the dataset (Fig. 4B). Notably, the assigned edges between IκBα and p38, JNK, and ATF-2 were not indicated by cluster-based analysis of the same data (Fig. 3H), demonstrating the value of the GGM approach for network reconstruction.
Fig. 4.
Data-driven interactions between phosphoprotein pairs reveal a convergent autocrine circuit through TNF, IL-1, and phospho-IκBα that promotes CVB3 progeny release and cardiotoxcity. (A) Significant partial correlation coefficients in the phosphoprotein network based on single and paired inhibitor data. Partial correlation coefficients are shown as the mean ± SD as determined by bootstrapping. The range of background partial correlations (dashed line) was calculated by shuffling the starting dataset before performing the analysis. (B) Phospho-IκBα is a hub in the measured CVB3-induced network. An undirected graph of the partial correlations from A is shown, where the black and red lines indicate positive and negative partial correlations, respectively. (C) Autocrine TNF and IL-1 blockade does not affect VP1 expression. HL1 cells were pretreated with TNF-neutralizing antibody (anti-TNF, 1 μM), IL-1ra (0.5 μM), or anti-TNF + IL-1ra for 1 h and then infected with CVB3 (M.O.I. = 9) as described in Fig. 1. VP1 expression at 16 h p.i. was quantified by Western blotting with tubulin used as a loading control. (D) Autocrine TNF and IL-1 blockade inhibits CVB3 progeny release. A digitized, representative CVB3 plaque assay is shown from HL1 cells treated with the indicated inhibitors and infected with CVB3 for 24 h. (E) Autocrine TNF and IL-1 blockade significantly improves cell viability in CVB3-infected cells. Data are presented as the mean ± SD of three independent replicates. Asterisks indicate P < 0.05 by paired t test. Treatment with anti-TNF or IL-1ra without CVB3 infection did not affect measured viability (not shown).
IκBα phosphorylation is a key intermediate step toward activation of NF-κB (47), a transcription factor that is critical for inflammatory gene expression (48). CVB3 infection itself did not strongly induce phospho-IκBα (Fig. 1), in agreement with previous work showing that host cell IκBα levels are reduced by a viral protease encoded by the CVB3 genome (49). However, upon pretreatment of cells with various signaling inhibitors that disrupt CVB3 infection, we observed a spike of IκBα phosphorylation at ∼8 h p.i. IκBα phosphorylation could conceivably stem from expression of viral proteins, but many inhibitor conditions that strongly induced phospho-IκBα also blocked expression of viral proteins (e.g., conditions 2, 11, and 14) (Figs. 1 and 2A). This raised the possibility that phospho-IκBα arose from stimuli that were endogenous to host cells, thereby contributing to its centrality in the GGM network (Fig. 4B).
Two major inducers of IκBα phosphorylation are the proinflammatory cytokines TNF and IL-1 (48). Moreover, in response to pathogenic and inflammatory stimuli, both TNF and IL-1 can be released to signal in an autocrine manner (50, 51). We tested whether these cytokines could be involved in CVB3 pathogenesis by blocking autocrine signaling with a neutralizing antibody to TNF and a naturally occurring IL-1 receptor antagonist (IL-1ra). Although neither perturbation affected CVB3-induced VP1 expression (Fig. 4C), we observed potent inhibition of VPR by plaque forming assay when either cytokine receptor was blocked (Fig. 4D). The decrease in VPR further coincided with significant increases in cell viability upon CVB3 infection (P < 0.05) (Fig. 4E). Thus, autocrine TNF and IL-1 signaling is critical for establishing a host cell signaling network (Fig. 4B) that enables CVB3 propagation, release, and cardiotoxicity. More generally, our work provides evidence that coupling intracellular measurements with pairwise pharmacological perturbations may be particularly efficient at revealing molecular mechanisms between signaling proteins.
Discussion
Interrogating signal transduction by small-molecule pairs is a convenient method for rapidly exploring network states and their associated cell outcomes. Our results illustrate that inhibitor pairs reveal information about a signaling network that improves the prediction of biochemical mechanisms. If signaling profiles of dual inhibitors were simply linear superpositions of the individual agents, then there would be no gain in useful information. A recent report has suggested such additivity in the intracellular response to drugs (52). However, this work focused on protein expression levels rather than posttranslational modifications and examined only three targeted signaling inhibitors (one of which showed clear nonadditivity). Although gradual changes in protein expression may be roughly approximated by linear superposition, our data show that direct perturbations of kinase activity are frequently nonadditive.
There are advantages to exploiting the nonlinearity of signaling inhibitors when designing experiments. For example, with 10 inhibitors, there are (102 − 10)/2 = 45 additional pairwise opportunities to further characterize the systems-level properties of a network. Using this number of inhibitor pairs is easier, more cost-effective, and (for some analyses) more valuable than adding 45 single inhibitors to study network function. The mechanisms revealed by such studies will depend heavily on the choice of inhibitors and their effects on the measured intracellular pathways, emphasizing the importance of experimental design when using a pairwise approach. The key is to view these interventions as general perturbations to the network rather than as experiments that will directly assign molecular mechanisms.
Still higher combinations of inhibitors could readily be tested as we did for pairs here, but it is uncertain whether these conditions would reveal additional molecular insights. Chatterjee et al. (8) recently showed that higher-order combinations of input stimuli can be quantitatively predicted from pairwise data, suggesting that nonlinear information processing may normally stop at pairs (11). However, small-molecule inhibitors differ from extracellular stimuli in that inhibitors intentionally push the signaling network into states not normally occupied physiologically. The benefit of using more than two small molecules will ultimately depend on the overall connectivity of the network and the ability of the inhibitor panel to disrupt redundant or compensatory pathways within cells.
Our pairwise analysis of the host signaling response to CVB3 infection highlighted a central role for phospho-IκBα downstream of autocrine TNF and IL-1 signaling. For nearly two decades, it has been known that TNF and IL-1 promote myocarditis caused by CVB3 (53). Circulating TNF and IL-1 levels rise substantially upon CVB3 infection (54), but the major source of these cytokines in the heart has been thought to be infiltrating monocytes (55). Our results with cultured cells suggest that cardiomyocytes may “prime” monocyte infiltration and their own toxicity by autocrine stimulation with inflammatory cytokines. In the future, pairwise inhibitor strategies could be applied to identify the relevant CVB3-dependent pathways that drive autocrine TNF and IL-1 signaling in the heart.
Methods
Cells and Viruses.
HL1 cells were obtained from Dr. William C. Claycomb. (Louisiana State University Medical Center, New Orleans, LA) and were maintained as described previously (17). CVB3 (Kandolf strain) was propagated in HeLa cells and virus titers were determined by the plaque assay.
Viral Infection and Perturbations.
HL1 cells were sham-infected with PBS or infected with CVB3 (multiplicity of infection [M.O.I.] = 9) after pretreatment with one of 23 different experimental conditions (one control, seven single signaling inhibitors, or 15 combinations of two signaling inhibitors). The chemical inhibitors included API-2 (reported Akt inhibitor, 1 μM), BAY11-7085 (IκB, 10 μM), SB203580 (p38, 50 μM), SB216763 (GSK3β, 10 μM), SP600125 (JNK, 50 μM) PP2 (Src-family kinases, 10 μM), and U0126 (MEK1/2, 20 μM) were obtained from Tocris Biosciences. SL0101 (RSK, 100 μM), LY 294002 (PI3K, 10 μM), and SB202190 (p38, 10 μM) were purchased from Calbiochem. PD184352 (MEK1/2, 10 μM) was obtained from Santa Cruz Biotechnology. Both anti–TNF-α (1 μM) and IL-1ra (0.5 μM) were obtained from R&D Systems.
Phospho-ELISA.
Cell lysates of infected HL1 cells were collected at 0, 0.17, 1, 8, 16, and 24 h p.i. After normalizing protein concentration, samples were analyzed by phospho-ELISA (Biosource) for the phosphorylation levels of Akt (Ser473), ATF2 (Thr69/Thr71), CREB (Ser133), ERK1/2 (Thr185/Tyr187), GSK3β (Ser9), Hsp27 (Ser82), IκBα (Ser32), JNK1/2 (Thr183/Tyr185), and p38 MAPK (Thr180/Tyr182) according to the manufacturer's instructions.
Western Blot Analysis.
Cell lysates were prepared as described previously (24). Equal amounts of protein were subjected to SDS-polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (GE Healthcare). Membranes were blocked for 1 h with nonfat dry milk solution (3% in PBS) containing 0.1% Tween-20. Blots were then incubated with one of the following primary antibodies: anti-VP1 (1:1,000; Dako), anti–β-actin (1:5,000; Sigma), anti–phospho-ATF2 (1:1,000, Thr69/Thr71; Cell Signaling), anti–phospho-CREB (1:1,000. Ser133; Cell Signaling), or anti-tubulin (1:5,000; Cell Signaling) for 1 h, followed by incubation for 1 h with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz). Immunoreactive bands were visualized by enhanced chemiluminescence (Pierce) on a ChemiGenius2 CCD camera-based detection system. Where indicated, band intensities were quantified by densitometry with ImageJ.
Plaque Assays.
CVB3 titers in cell supernatants were determined on monolayers of HeLa cells by an agar overlay plaque assay in triplicate (24). Briefly, samples were serially diluted 10-fold and overlaid on 90–95% confluent monolayers of HeLa cells in six-well plates and incubated for 1 h. Medium was aspirated, HeLa cells were washed with PBS twice, and 2 mL of complete DMEM containing 0.75% agar was overlaid onto each well. Cells were incubated at 37 °C for 72 h, fixed with Carnoy's fixative (75% ethanol, 25% acetic acid) for 30 min, and stained with 1% crystal violet. Plaques were counted, and viral concentrations were calculated as plaque-forming unit per milliliter.
Cell Viability Assays.
HL-1 cells were grown in 12-well plates and infected with CVB3 (M.O.I = 9) for 16 and 24 h after pretreatment with inhibitors. The MTS solutions (1:5) were added to wells for 2.5 h and then transferred to 96-well plates. Cell viabilities of infected and noninfected cells were assessed by MTS assay (CellTiter 96; Promega, Inc.).
Hierarchical Clustering.
Phosphoprotein data were standardized as z-scores before clustering. Hierarchical clustering was performed with the clustergram function in MATLAB using a Euclidean distance metric and average linkage. For subsampled dendrograms (Fig. 3), 10,000 different submatrices were randomly generated from original data by the function, and the mean Euclidean distances were used as the basis for the subsampled clustering with average linkage.
GGM.
The GeneNet package (56) was used in R to calculate significant partial correlation coefficients for signaling molecules. Background partial correlation coefficients were determined by random sampling of 100 different submatrices of the original dataset.
Supplementary Material
Acknowledgments
We thank Drs. Ynuk Bosse, Jeff Saucerman, and Scott Tebbutt for reading the manuscript; Dr. Farhad B. Hashemi for intellectual support; and Cheryl Lane for technical assistance. This work was supported by grants from the Heart and Stroke Foundation of British Columbia and Yukon (to B.M.M.), the Canadian Institutes of Health Research (to B.M.M.), National Institutes of Health Director's New Innovator Award Program Grant 1-DP2-OD006464-01 (to K.A.J.), the Pew Scholars Program in the Biomedical Sciences (K.A.J.), and the David and Lucile Packard Foundation (K.A.J.). F.S.G. is supported by a Doctoral Award from Tehran University of Medical Science-Iran. D.M. is supported by Canadian Institutes of Health Research, Integrated and Mentored Pulmonary and Cardiovascular Training (IMPACT), and Heart and Stroke Foundation Postdoctoral Fellowships. H.L. is a Michael Smith Foundation for Health Research Scholar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006478107/-/DCSupplemental.
References
- 1.Pawson T. Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 2.Saez-Rodriguez J, et al. Discrete logic modelling as a means to link protein signalling networks with functional analysis of mammalian signal transduction. Mol Syst Biol. 2009;5:331. doi: 10.1038/msb.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janes KA, et al. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 4.Natarajan M, Lin KM, Hsueh RC, Sternweis PC, Ranganathan R. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8:571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 5.Mattoo S, Lee YM, Dixon JE. Interactions of bacterial effector proteins with host proteins. Curr Opin Immunol. 2007;19:392–401. doi: 10.1016/j.coi.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Singh A, Weinberger LS. Stochastic gene expression as a molecular switch for viral latency. Curr Opin Microbiol. 2009;12:460–466. doi: 10.1016/j.mib.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borisy AA, et al. Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci USA. 2003;100:7977–7982. doi: 10.1073/pnas.1337088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee MS, Purvis JE, Brass LF, Diamond SL. Pairwise agonist scanning predicts cellular signaling responses to combinatorial stimuli. Nat Biotechnol. 2010;28:727–732. doi: 10.1038/nbt.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaudet S, et al. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol Cell Proteomics. 2005;4:1569–1590. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: Evolutionary units of three-dimensional structure. Cell. 2009;138:774–786. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janes KA. Paring down signaling complexity. Nat Biotechnol. 2010;28:681–682. doi: 10.1038/nbt0710-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janes KA, Reinhardt HC, Yaffe MB. Cytokine-induced signaling networks prioritize dynamic range over signal strength. Cell. 2008;135:343–354. doi: 10.1016/j.cell.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneidman E, Berry MJ, 2nd, Segev R, Bialek W. Weak pairwise correlations imply strongly correlated network states in a neural population. Nature. 2006;440:1007–1012. doi: 10.1038/nature04701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St Onge RP, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh P, Tschumi AI, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat Genet. 2006;38:489–494. doi: 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- 16.Esfandiarei M, McManus BM. Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol. 2008;3:127–155. doi: 10.1146/annurev.pathmechdis.3.121806.151534. [DOI] [PubMed] [Google Scholar]
- 17.Claycomb WC, et al. HL-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan J, et al. A phosphorothioate antisense oligodeoxynucleotide specifically inhibits coxsackievirus B3 replication in cardiomyocytes and mouse hearts. Lab Invest. 2004;84:703–714. doi: 10.1038/labinvest.3700083. [DOI] [PubMed] [Google Scholar]
- 19.Esfandiarei M, et al. Protein kinase B/Akt regulates coxsackievirus B3 replication through a mechanism which is not caspase dependent. J Virol. 2004;78:4289–4298. doi: 10.1128/JVI.78.8.4289-4298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esfandiarei M, et al. Coxsackievirus B3 activates nuclear factor kappa B transcription factor via a phosphatidylinositol-3 kinase/protein kinase B-dependent pathway to improve host cell viability. Cell Microbiol. 2007;9:2358–2371. doi: 10.1111/j.1462-5822.2007.00964.x. [DOI] [PubMed] [Google Scholar]
- 21.Opavsky MA, et al. Enhanced ERK-1/2 activation in mice susceptible to coxsackievirus-induced myocarditis. J Clin Invest. 2002;109:1561–1569. doi: 10.1172/JCI13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu P, et al. The tyrosine kinase p56lck is essential in coxsackievirus B3-mediated heart disease. Nat Med. 2000;6:429–434. doi: 10.1038/74689. [DOI] [PubMed] [Google Scholar]
- 23.Marchant D, et al. Bosentan enhances viral load via endothelin-1 receptor type-A-mediated p38 mitogen-activated protein kinase activation while improving cardiac function during coxsackievirus-induced myocarditis. Circ Res. 2009;104:813–821. doi: 10.1161/CIRCRESAHA.108.191171. [DOI] [PubMed] [Google Scholar]
- 24.Si X, et al. Stress-activated protein kinases are involved in coxsackievirus B3 viral progeny release. J Virol. 2005;79:13875–13881. doi: 10.1128/JVI.79.22.13875-13881.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo H, et al. Coxsackievirus B3 replication is reduced by inhibition of the extracellular signal-regulated kinase (ERK) signaling pathway. J Virol. 2002;76:3365–3373. doi: 10.1128/JVI.76.7.3365-3373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: An update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bain J, et al. The selectivity of protein kinase inhibitors: A further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, et al. Akt/protein kinase B signaling inhibitor-2, a selective small molecule inhibitor of Akt signaling with antitumor activity in cancer cells overexpressing Akt. Cancer Res. 2004;64:4394–4399. doi: 10.1158/0008-5472.CAN-04-0343. [DOI] [PubMed] [Google Scholar]
- 29.Pierce JW, et al. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 30.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 31.Mercurio F, et al. IKK-1 and IKK-2: Cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 32.Stokoe D, Engel K, Campbell DG, Cohen P, Gaestel M. Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett. 1992;313:307–313. doi: 10.1016/0014-5793(92)81216-9. [DOI] [PubMed] [Google Scholar]
- 33.Reimold AM, Kim J, Finberg R, Glimcher LH. Decreased immediate inflammatory gene induction in activating transcription factor-2 mutant mice. Int Immunol. 2001;13:241–248. doi: 10.1093/intimm/13.2.241. [DOI] [PubMed] [Google Scholar]
- 34.Yang D, et al. Viral myocarditis: Identification of five differentially expressed genes in coxsackievirus B3-infected mouse heart. Circ Res. 1999;84:704–712. doi: 10.1161/01.res.84.6.704. [DOI] [PubMed] [Google Scholar]
- 35.Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;26:585–615. [Google Scholar]
- 36.Gupta S, Campbell D, Dérijard B, Davis RJ. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 37.Raingeaud J, et al. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 38.Ouwens DM, et al. Growth factors can activate ATF2 via a two-step mechanism: Phosphorylation of Thr71 through the Ras-MEK-ERK pathway and of Thr69 through RalGDS-Src-p38. EMBO J. 2002;21:3782–3793. doi: 10.1093/emboj/cdf361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 40.Xing J, Ginty DD, Greenberg ME. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 41.Deak M, Clifton AD, Lucocq LM, Alessi DR. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- 43.Lee JC, et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 44.Smith JA, et al. Identification of the first specific inhibitor of p90 ribosomal S6 kinase (RSK) reveals an unexpected role for RSK in cancer cell proliferation. Cancer Res. 2005;65:1027–1034. [PubMed] [Google Scholar]
- 45.Papin JA, Hunter T, Palsson BO, Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- 46.Ma S, Gong Q, Bohnert HJ. An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 2007;17:1614–1625. doi: 10.1101/gr.6911207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 48.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: The control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 49.Zaragoza C, et al. Viral protease cleavage of inhibitor of kappaBalpha triggers host cell apoptosis. Proc Natl Acad Sci USA. 2006;103:19051–19056. doi: 10.1073/pnas.0606019103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 51.Janes KA, et al. The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell. 2006;124:1225–1239. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 52.Geva-Zatorsky N, et al. Protein dynamics in drug combinations: A linear superposition of individual-drug responses. Cell. 2010;140:643–651. doi: 10.1016/j.cell.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 53.Lane JR, Neumann DA, Lafond-Walker A, Herskowitz A, Rose NR. Interleukin 1 or tumor necrosis factor can promote Coxsackie B3-induced myocarditis in resistant B10.A mice. J Exp Med. 1992;175:1123–1129. doi: 10.1084/jem.175.4.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lundgren M, Darnerud PO, Blomberg J, Friman G, Ilbäck NG. Sequential changes in serum cytokines reflect viral RNA kinetics in target organs of a coxsackievirus B infection in mice. J Clin Immunol. 2009;29:611–619. doi: 10.1007/s10875-009-9294-8. [DOI] [PubMed] [Google Scholar]
- 55.Henke A, et al. Coxsackievirus B3-induced production of tumor necrosis factor-alpha, IL-1 beta, and IL-6 in human monocytes. J Immunol. 1992;148:2270–2277. [PubMed] [Google Scholar]
- 56.Opgen-Rhein R, Strimmer K. From correlation to causation networks: A simple approximate learning algorithm and its application to high-dimensional plant gene expression data. BMC Syst Biol. 2007;1:37. doi: 10.1186/1752-0509-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.