Abstract
We report a simple, low-cost, rapid, and mask-free method to fabricate two-dimensional (2D) and three-dimensional (3D) microfluidic chip for biological analysis researches. In this fabrication process, a laser system is used to cut through paper to form intricate patterns and differently configured channels for specific purposes. Bonded with cyanoacrylate-based resin, the prepared paper sheet is sandwiched between glass slides (hydrophilic) or polymer-based plates (hydrophobic) to obtain a multilayer structure. In order to examine the chip’s biocompatibility and applicability, protein concentration was measured while DNA capillary electrophoresis was carried out, and both of them show positive results. With the utilization of direct laser cutting and one-step gas-sacrificing techniques, the whole fabrication processes for complicated 2D and 3D microfluidic devices are shorten into several minutes which make it a good alternative of poly(dimethylsiloxane) microfluidic chips used in biological analysis researches.
INTRODUCTION
During the recent years, the development in microfluidics has stimulated growing enthusiasm in low-cost, straightforward, and rapid prototyping of microfluidic devices.1 Among various kinds of methods, soft-lithography, which uses a soft polymer such as poly(dimethylsiloxane) (PDMS) to imprint and transfer the structure on a well patterned mold, is the most popular and successful one.2, 3, 4, 5 It is so widely employed into the microfluidic researches owning to its high reliability and advantageous properties of PDMS microfluidic devices;6, 7, 8 however, there remain shortcomings. For example, it is difficult to construct complex three-dimensional (3D) structures by multilayer bonding; accompanying this usually requires a silane coupling agent to treat the PDMS surface, as well as laborious step-by-step bonding of the structures themselves,9, 10 the whole fabrication process is not very efficient, for the patterned mold requires several hours or even days to deal with mask making and photolithography. This time consuming problem could become severe especially in the case when, with the research progresses, chip design needs continuous optimization due to unknown problems.
To complement the new functionalities and applications being developed in the microfluidic researches, especially the converted all-in-one system that is envisaged, many rapid and inexpensive chip making methods such as mechanical and direct laser cutting technology11, 12 are introduced into this field. One kind of these methods is “print-to-cast,”13, 14, 15, 16, 17, 18, 19, 20, 21, 22 with which microfluidic devices are usually made with paper, transparent overhead-projector film, polymethyl methacrylate (PMMA) plate, and other inexpensive, easily machinable and accessible materials.23, 24, 25, 26, 27, 28 Taking the thermal printer-based chip fabrication process, for example,16 it utilizes a wax printer to directly pattern hydrophobic walls of wax in the hydrophilic paper. The fluid transports by means of capillary action in the porous cellulose. It is less time consuming, less expensive, and largely simplifies the fluid injection facilities comparing to soft-lithography PDMS microfluidic devices. In spite of this, limitations exist. For example, the fluid cannot form continuous flow in the paper, which means that droplet cannot be formed and the liquid transport velocity depends on the passive capillary action and could not be changed, the cellulose fiber has absorption on many molecules, the chip is usually an open system, and easy to be affected by outside environment. Therefore, this kind of method is mostly used to develop simple, inexpensive point-of-care chips for fast diagnostic application. An alternative fabricating method is realized with double-side pressure sensitive adhesive (PSA) tape.18, 29 This method inherits the advantages of print-to-cast as low-cost, straightforward, and rapid. Continuous flow could be formed as it was in PDMS devices. Nonetheless, bridge bonding between different materials (e.g., between glass and PMMA) is difficult, the bonding strength is weak and sensitive to environmental humidity and temperature, and the chemical composition of the adhesive is usually too complex for biological samples used in biological analysis researches.
In the present research, we proposed a one-step gas-sacrificing glue bonding method to fabricate microfluidic systems for academic researches. With this method, not only intricate hydrophilic or hydrophobic channels can be easily fabricated, 3D structures are also realized by one-step bonding of multiple layers together. The whole process could be completed within 1 h and cost less than 1 US Dollar. The bonding material is cyanoacrylate-based resin (Aron Alpha Co.,West Jefferson, OH), a kind of rapid-polymerizing liquid glue. This kind of glue cannot only adhere to various kinds of material surface and harden within several minutes without incident under exposure both to aqueous and solvent systems but also offer excellent bonding strength. In the medical field, cyanoacrylate-based resin is wildly exploited for surgery where it bonds human tissues together without wire suture. In the present study, we applied the cyanoacrylate-based resin bonding technology to microfluidic chip fabrication. We first evaluated the biocompatibility of materials employed in the chip fabrication, including A4 print paper, filter paper, and cyanoacrylate-based resin. To do it, we used polymerase chain reaction (PCR) as a model in the investigation of the PCR-inhibitory effect. Protein detection and DNA capillary electrophoresis are carried out to show the realistic bioapplicability for academic researches. A prototype of three-dimensional structure was also fabricated in a single step which offers the opportunity to realize even more complex functions in order for further researches.
EXPERIMENTAL METHODS AND RESULTS
Laser cutting
We used a CO2 laser (Versa Laser System, model VR3.50, Universal Laser Systems, Ltd., Scottsdale, AZ) to cut through film (paper sheet and overhead-projector film) in continuous mode. Laser cutting technique, comparing the photolithography or soft-lithography techniques, is very efficient and convenient to make hollow microfluidic channels. For example, a size of 2 cm×2 cm microfluidic chip will take less than 3 min for laser cutting to make all the channels and patterns on it without any other chemical or physical pretreatment. The focal spot size of our laser beam was about 30 μm in width. The laser power we used could vary from 5 to 20 W along with the cutting velocity from 0.25 to 25 mm∕s. To cut through print paper, 5 W power with velocity of 0.76 mm∕s could easily burn and produce uniform grooves exactly along the entire length of the working surface. By adjusting cutting powers and velocities, intricate designs could be formed without any stress or deformation. The channel dimensions are a function not only of the laser power and the cutting speed but also of the composition and thickness of the film. For example, with 100 μm thick A4 print paper sheet (FUJI XEROX, Japan), the smallest size obtainable is around 50 μm in width, whereas for 250 μm thick filter paper, it is around 100 μm.30, 31
One-step gas-sacrificing glue bonding
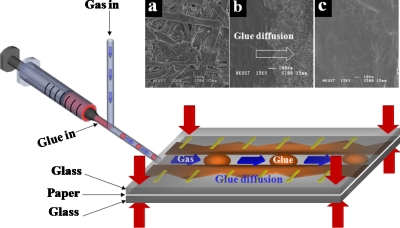
In the present study, laser-cut paper was sandwiched between two glass slides in which inlet and outlet holes had been drilled, forming a simple two-dimensional (2D) three-layered structure (Fig. 1). These three layers were pressed together at the four corners by four clamps. These clamps would offer a pressure of more than 200 kPa on the three layers. The cyanoacrylate-based resin was loaded with a syringe connected to one inlet of a “T” channel. Compressed air was infused into the other one. A syringe pump restricted the flow rate of acrylic resin while the gas flow rate was controlled by means of a gas valve (V290-4EHQ, Shinyeong Mechatronics Co., Ltd., Siheung, South Korea). By balancing the flow rates of the glue and gas into the two inlets, a segmented flow of glue in the paper channels could be achieved.32 This can be seen in Fig. 1, which shows that the fast-adhesive glue was sheared into plugs and pushed by the gas flow along the channel, the glue simultaneously diffusing into the paper laterally along the channel, due to the capillary effect. Normally, the gas pressure was set 20 kPa at the beginning to prevent the blockage of the channel. After most of the area was bonded, the gas pressure was decreased to about 2 kPa. Then the glue diffused to the whole piece of paper and formed a smooth channel. The diffusion process was proved by the scanning electron microscope (SEM) images in Figs. 1a, 1b, 1c. The gas-sacrificing means that the compressed gas serves as a sacrificing medium to prevent the cyanoacrylate-based resin from solidifying inside the microchannels. Furthermore, if leakage happens, the gas will carry the segmented flow of glue to the leakage and block it automatically. Extra glue will be pushed out from the outlets. After 1 or 2 min, the glue eventually solidifies and adheres tightly to the upper and lower layers.
Figure 1.
Chip bonding method for simple 2D chip. The T shape junction was used to form segment glue. The two glass layers and the paper layer were pressed together. (a) SEM picture of paper surface. (b) SEM picture of paper with glue diffusion. (c) SEM picture of paper after full diffusion of glue. After solidification, the pores inside the paper fibers were filled, forming a smooth surface.
The bonding strength of the resin was tested using a single straight channel with an inlet and an outlet located at each end, respectively. Air was injected through the inlet and the pressure was monitored using the “Druck DPI 104” digital pressure sensor (GE Druck, Leicester, U.K.). When adequate pressure reached a critical value, the layer bonding was broken, resulting in gas leakage. It was found that the resin bonding strength between paper and PMMA was much greater than that between paper and glass slides, as can be seen in reference to the data in Table 1.
Table 1.
Comparisons of bonding strength of microfluidic chips carried out with different materials.
| Chip materials | Silicon-silicona | PDMS | Paper-glass | Paper-PMMA | |
|---|---|---|---|---|---|
| Method | Low temperature (400 °C) | Half-cure bonding | Plasma bonding | Acrylic resin | Acrylic resin |
| Bonding strength | 25 MPa | 50 kPa | 200 kPa | 5 MPa | 8 MPa |
The silicon-silicon bonding strength data were cited in Ref. 33.
Fabrication of microfluidic chips with 2D and 3D structures
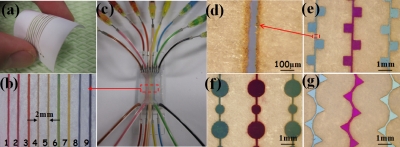
To fabricate a 2D microfluidic chip [Figs. 2a, 2b, 2c], we first used a laser to cut nine parallel channels through 3×5 cm square paper. The widths of channels were set at 100 μm, and the distance between two channels was about 2 mm. The well-bonded microfluidic 2D chip with nine channels sandwiched with two PMMA slides is shown in Fig. 2c. We found that channel blockage usually occurred during the bonding process, when multiple channels existed. Therefore, a pure gas flow was continuously applied. For example, when bonding channel 2 shown in Fig. 2b, pure gas flow was applied to channels 1 and 3 in order to blow away excess glue solution. Alternative channel configurations can easily be achieved via the same process [see Figs. 2d, 2e, 2f, 2g].
Figure 2.
(a) Parallel channels cut through paper using laser system. (b) Magnified pictures corresponding to selected area in (c). The distance between the two channels was 2 mm. (c) Prototypes of two-dimensional microfluidic chip. (d) Magnified pictures corresponding to selected area in (e). The channel size was 80 μm. (e), (f), and (g) are magnified pictures of paper chips with square, circular, and triangle channels, respectively.
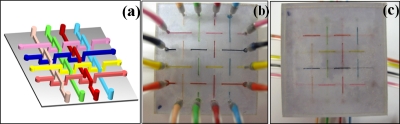
Fabrication of a 3D microfluidic chip [Fig. 3a] is not more than complex multiple channel chips in the 2D case. In the present study, three pieces of laser-cut paper were precisely overlapped. The middle layer paper, with several holes of 150 μm diameter, conducted and constrained the fluid flow between the upper and lower channels. To avoid channel blockage and increase the bonding efficiency, the glue plugs, carried by compressed air, were input into all of the channels simultaneously. Upon solidification, a 3D paper chip with multicrossed channels was realized [Figs. 3b, 3c].
Figure 3.
(a) Schematic picture of three-dimensional chip. Differently colored channels intersected and overlapped with each other without connecting. (b) Top-view of three-dimensional acrylic resin microfluidic chip. Each channel was filled with one color of fluid, corresponding to (a). (c) Bottom view of three-dimensional acrylic resin microfluidic chip.
PCR enzymatic biocompatibility of chip material
Biocompatibility of materials can be accessed by the interaction with DNA, proteins (enzymes), or cells as usually reported. The contact of materials with biomolecular reaction components may result in adsorption or inhibition of biomolecules.34, 35 The materials used in the chip fabrication, which included acrylic resin, various papers, and acrylic-resin-impregnated papers, were tested to find any PCR-inhibitory effects. The first PCR mixture was prepared without bovine serum albumin (BSA); the second contained BSA at a final concentration of 2000 μg∕ml. The PCR mixtures were added to the material fragments. After incubation at room temperature for 30 min, the materials were removed and PCR was performed on a bench thermocycler (MyGenie 96 Gradient Thermal Block, Bioneer Corporation, Daejeon, South Korea). pEYFP-C1 vector (total size: 4.731 kbp) having been purchased from Clontech Laboratories, Inc. (Mountain View, CA), PCR amplification experiments were carried out with pEYFP-C1 plasmid containing a 589 bp cytomegalovirus (CMV) fragment. The oligonucleotides used in the amplification were CMV368F and CMV409R. The PCR consisted of the following components in their final concentrations: 1 μM primers (Life Technologies Co., Carlsbad, CA), 3.5 mM MgCl2(Kapa Biosystems, Cambridge, MA), 0.2 mMdNTP (Takara Bio Inc., Otsu, Japan), 1× fast buffer I reaction buffer, up to 0.2 mM (0.008%) cresol red (Sigma-Aldrich, Co., St. Louis, MO), 1.2Mbetaine (Sigma), 2×106 pEYFP-C1 template molecules, and 0.05 U∕μlSpeedStar HS DNA polymerase (Takara Bio Inc.). After the PCR, the amplification products were loaded directly onto the gel for visualization.
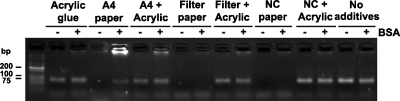
The biocompatibility testing results for the different materials can be seen in reference to Fig. 4. It is well-known that most biofriendly materials exhibit similar signals regardless of the inclusion or not of BSA in the PCR mixture: these are cyanoacrylate-based resin and paper impregnated with the resin. That signal is comparable to the one of no-additive control. Cyanoacrylate-based resin is an inert material,36 and as such, is not expected to interact with PCR components. Without BSA, no-signal was obtained in the PCR mix for all of the three kinds of papers investigated. Interestingly, a signal was obtained in the PCR carried out with BSA for the A4 printing paper. BSA is thought to compete with DNA polymerase for adsorption at chip walls and, thus, to improve PCR yields. BSA also acts as a polymerase competitor in inhibitor chelation. Additionally, BSA thickens the PCR mix, facilitates primer annealing, stabilizes both DNA and DNA polymerase, and, in so doing, acts as an osmoprotectant. Overall, the results showed that resin-impregnated paper behaves differently from pure paper, specifically in avoiding the PCR-inhibitory effect, and thus is biocompatible with the enzymes.
Figure 4.
PCR biocompatibility of materials used in the study. PCR product detection was achieved by running the samples in 4% agarose gel containing SYBR Safe DNA stain (Life Technologies) and by subsequent gel imaging. A low-molecular-weight DNA ladder New England Biolabs (NEB) was applied as a reference in estimating the sizes of the DNA fragments. The reaction outcome can be seen in the PCRs with (+) and without (−) BSA. A4 print paper: cellulose-based paper; Filter paper: alpha-cotton-cellulose-based paper; NC paper: nitrocellulose-based paper.
BSA assay and protein detection
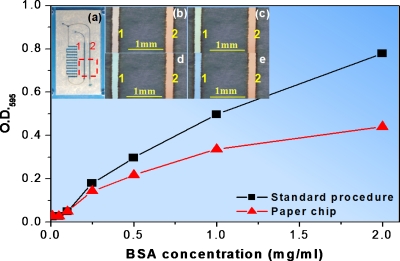
To demonstrate the realistic bioapplicability of materials used, the Bradford protein assay was used. The Bradford protein assay is a well-known method of biological analysis by cause of convenience and simplicity. It is based on the color change of the Coomassie Brilliant Blue G-250 dye (from 450 to 595 nm) in response to various concentrations of BSA.37 In our experiment, the Bradford protein assay dye reagent concentrate solution (Bio-Rad, Hercules, CA) contained 25% Coomassie Brilliant Blue G-250 dye, 50% phosphoric acid, and 25% methanol. The BSA dilution series (2000, 1000, 500, 250, 100, 50, and 10 μg∕ml) and the blank were prepared in synthetic urine solution. The synthetic urine solution contained 14.1 g NaCl, 2.8 g KCl, 17.3 g urea, 1.9 ml of 25% v∕v ammonia solution, 0.6 g CaCl2, and 0.43 g MgSO4, filled to 1 l with the addition of 0.02M HCl solution, for a final pH of 7.0. 10 μl of each solution was mixed with 500 μl of diluted dye in tubes and our chips, respectively. The measurements were carried out using a spectrophotometer (Spectronic GENESYS 10 Bio UV-Visible Spectrophotometer, Thermo Electron Corporation, Marietta, OH). Optical density signals (absorption at 595 nm) were directly taking from tubes and our chips. The results are shown in Fig. 5. It can be seen that the signal intensity taken from the chip was a little smaller compared to that from tubes as a consequence of small volume reagents. This did not affect the accuracy of the analysis using our chips. For an unknown BSA dilution, the measurement result error is only 0.82%, compared to the standard method (72.8 μg∕ml in chip and 73.4 μg∕ml in tube). Furthermore, the color changes in the chip were clearly visible with naked eyes [Figs. 5b, 5c, 5d, 5e]. It served as a good qualitative measurement method to tell the existence and approximate concentration of bovine serum albumin.
Figure 5.
Signal intensity dependency from protein concentration in solution. Absorbance intensity of BSA solution in microfluidic channel 2 in (a) (red) and tubes (black). The measurements were performed using a spectrophotometer at 595 nm. (b)–(e) are magnified pictures corresponding to the selected area in (a) to show the color changes in the chip as a qualitative measurement of protein. Coomassie Brilliant Blue G-250 dye solution was first infused into the channel 2 as a reference, mixed with BSA dilutions in the left area of the chip and then flew into channel 1(a). (b) Background (no protein), color of dye diluted by synthetic urine. (c) Synthetic urine solution with BSA concentration of 500 μg∕ml. (d) Synthetic urine solution with BSA concentration of 1000 μg∕ml (e) Synthetic urine solution with BSA concentration of 2000 μg∕ml.
DNA capillary electrophoresis
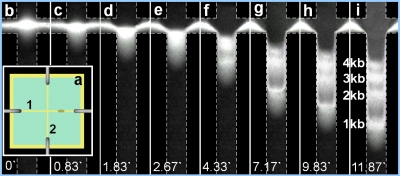
Capillary electrophoresis is a highly versatile analytical technique to study a wide range of biological analytes (proteins, peptides, DNA, cell, etc.). As one of the most important modern biotechnologies, DNA∕protein electrophoresis is widely used for biochemical reactions and biomolecules analysis. Many adaptations of microchip-based capillary electrophoresis in biology analysis applications including genotyping, pathogen detection∕identification, and DNA sequencing have been reported.38, 39 In our study, the double strand DNA fragments, EGFP (4.7 kb) and Mcl-1 (1 kb), were generated by restriction enzymes EcoRI and HindIII (Roche) digestion of plasmid DNA pEGFP-Mcl-1.40 Briefly, the mixture of 2 μg of pEGFP-Mcl-1, 2 μl of 10× buffer B (Roche), and 1 μl of EcoRI and HindIII, respectively, were supplemented with MilliQ water to achieve a total volume of 20 μl and then incubated at 37 °C overnight. After reaction, the mixture was then subjected to PCR product purification kit (Qiagen, Santa Clarita, CA) and stored in freezer for subsequent analysis. The other two fragments (2 and 3 kb) were generated by similar approach. These DNA fragments were labeled with YOYO-1 intercalating dye (Invitrogen Corp., Carlsbad, CA), which made the DNA visible under the excitation of UV irradiation at the wavelength of 260 nm. The gel was prepared with 1% 2-hydroxyethyl cellulose (average Mv ∼1 300 000, Sigma-Aldrich, Co., St. Louis, MO) in 1×TAE running buffer and loaded into a cross-channel filter-paper chip instead of print paper to avoid fluorescence [Fig. 6a]. Electric field was applied by four needle electrodes connected to the beginning and end of each channel. DNA sample was first injected into the loading channel [channel 1 in Fig. 6a] through the tiny window at the right side of the cross while a low voltage (∼20 V) was applied along the channel for 1–2 min to concentrate the DNA sample at the crossing area. Then a potential of 50 V was then applied along the separation channel [channel 2 in Fig. 6a]. The DNA fragments were separated and four distinct bands could be seen upon the application of the 50 V potential for around 12 s. The time-lapse images were shown in Figs. 6b, 6c, 6d, 6e, 6f, 6g, 6h, 6i.
Figure 6.
DNA electrophoresis results in microfluidic chip. (a) Top-view of cross-channel filter-paper chip. Channel 1 for DNA fragment injection and concentration and channel 2 for separation. (b)–(i) are time-lapse images of DNA separation in channel 2 after electric field applied.
CONCLUSIONS
In the present research, we proposed a one-step gas-sacrificing glue bonding method to fabricate microfluidic systems for academic researches. The whole process of fabrication only took a few minutes by means of laser cutting and one-step gas-sacrificing glue bonding technique. 2D and 3D microfluidic chips with varying configurations could easily be fabricated within several minutes. The biocompatibility of the chips was tested by PCR experiments. The expected, confirmatory results were obtained in microfluidic chip for detection of protein concentrations. DNA electrophoresis was also carried out well. The results show that the microfluidic devices, fabricated by this method, are applicable as a good alternative for PDMS microfluidic devices in biological analysis researches.
ACKNOWLEDGMENTS
This publication is based on work partially supported by Award No. SA-C0040∕U.K.-C0016 made by King Abdullah University of Science and Technology (KAUST) and the Hong Kong RGC under Grant No. HKUST 603608. This work was also partially supported by the Nanoscience and Nanotechnology Program at HKUST.
References
- Yager P., Edwards T., Fu E., Helton K., Nelson K., Tam M. R., and Weigl B. H., Nature (London) 442, 412 (2006). 10.1038/nature05064 [DOI] [PubMed] [Google Scholar]
- McDonald J. C., Duffy D. C., Anderson J. R., Chiu D. T., Wu H., Schueller O. J. A., and Whitesides G. M., Electrophoresis 21, 27 (2000). [DOI] [PubMed] [Google Scholar]
- Xia Y. and Whitesides G. M., Angew. Chem., Int. Ed. 37, 550 (1998). [DOI] [PubMed] [Google Scholar]
- Duffy D., McDonald J., Schueller O., and Whitesides G. M., Anal. Chem. 70, 4974 (1998). 10.1021/ac980656z [DOI] [PubMed] [Google Scholar]
- Xia Y. and Whitesides G. M., Annu. Rev. Mater. Sci. 28, 153 (1998). 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- Chung B. G., Flanagan L. A., Rhee S. W., Schwarts P. H., Lee A. P., Monuki E. S., and Jeon N. L., Lab Chip 5, 401 (2005). 10.1039/b417651k [DOI] [PubMed] [Google Scholar]
- Rhee S. W., Taylor A. M., Tu C. H., Cribbs D. H., Cotman C. W., and Jeon N. L., Lab Chip 5, 102 (2005). 10.1039/b403091e [DOI] [PubMed] [Google Scholar]
- Larry J. M., Matthew E. S., Jonathan V. S., Ralph G. N., and Martha U. G., Lab Chip 8, 987 (2007). [Google Scholar]
- Anderson J. R., Chiu D. T., Jackman R. J., Cherniavskaya O., McDonald J. C., Wu H., Whitesides S. H., and Whitesides G. M., Anal. Chem. 72, 3158 (2000). 10.1021/ac9912294 [DOI] [PubMed] [Google Scholar]
- Luo Y. and Zare R. N., Lab Chip 8, 1688 (2008). 10.1039/b807751g [DOI] [PubMed] [Google Scholar]
- Klank H., Kutter J. P., and Geschke O., Lab Chip 2, 242 (2002). 10.1039/b206409j [DOI] [PubMed] [Google Scholar]
- Rötting O., Röpke W., Becker H., and Gärtner C., Microsyst. Technol. 8, 32 (2002). 10.1007/s00542-002-0106-9 [DOI] [Google Scholar]
- Lu Y., Shi W., Qin J., and Lin B., Anal. Chem. 82, 329 (2010). 10.1021/ac9020193 [DOI] [PubMed] [Google Scholar]
- Martinez A. W., Phillips S. T., and Whitesides G. M., Proc. Natl. Acad. Sci. U.S.A. 105, 19606 (2008). 10.1073/pnas.0810903105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrilho E., Martinez A. W., and Whitesides G. M., Anal. Chem. 81, 7091 (2009). 10.1021/ac901071p [DOI] [PubMed] [Google Scholar]
- Lu Y., Shi W., Jiang L., Qin J., and Lin B., Electrophoresis 30, 1497 (2009). 10.1002/elps.200800563 [DOI] [PubMed] [Google Scholar]
- Sia S. K., Linder V., Parviz B. A., Siegel A., and Whitesides G. M., Angew. Chem., Int. Ed. 43, 498 (2004). 10.1002/anie.200353016 [DOI] [PubMed] [Google Scholar]
- Yuen P. K. and Goral V. N., Lab Chip 10, 384 (2010). 10.1039/b918089c [DOI] [PubMed] [Google Scholar]
- Martinez A. W., Phillips S. T., Carrilho E., Thomas S. W., Sindi H., and Whitesides G. M., Anal. Chem. 80, 3699 (2008). 10.1021/ac800112r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Tian J., Nguyen T., and Shen W., Anal. Chem. 80, 9131 (2008). 10.1021/ac801729t [DOI] [PubMed] [Google Scholar]
- Martinez A. W., Phillips S. T., Butte M. J., and Whitesides G. M., Angew. Chem., Int. Ed. 46, 1318 (2007). 10.1002/anie.200603817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. W., Phillips S. T., Wiley B. J., Gupta M., and Whitesides G. M., Lab Chip 8, 2146 (2008). 10.1039/b811135a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vullev V. I., Wan J., Heinrich V., Landsman P., Bower P. E., Xia B., Millare B., and Jones G., J. Am. Chem. Soc. 128, 16062 (2006). 10.1021/ja061776o [DOI] [PubMed] [Google Scholar]
- Bartholomeusz D. A., Boutté R. W., and Andrade J. D., J. Microelectromech. Syst. 14, 1364 (2005). 10.1109/JMEMS.2005.859087 [DOI] [Google Scholar]
- Witek M. A., Hupert M. L., Park D. S., Fears K., Murphy M. C., and Soper S. A., Anal. Chem. 80, 3483 (2008). 10.1021/ac8002352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch J. S., Rosenberger F., Highsmith W. E., Kimball C., DeVoe D. L., and Lee C. S., Lab Chip 5, 392 (2005). 10.1039/b416682e [DOI] [PubMed] [Google Scholar]
- Metz S., Holzer R., and Renaud P., Lab Chip 1, 29 (2001). 10.1039/b103896f [DOI] [PubMed] [Google Scholar]
- Rolland J. P., Dam R. M. V., Schorzman D. A., Quake S. R., and DeSimone J. M., J. Am. Chem. Soc. 126, 2322 (2004). 10.1021/ja031657y [DOI] [PubMed] [Google Scholar]
- Weigl B. H., Bardell R. L., Schulte T. H., Battrell C. F., and Hayenga J., Biomed. Microdevices 3, 267 (2001). 10.1023/A:1012448412811 [DOI] [Google Scholar]
- Wada H., Sasaki H., and Kamijoh T., Solid-State Electron. 43, 1655 (1999). 10.1016/S0038-1101(99)00115-X [DOI] [Google Scholar]
- Choudhury I. A. and Shirley S., Opt. Laser Technol. 42, 503 (2010). 10.1016/j.optlastec.2009.09.006 [DOI] [Google Scholar]
- Garstecki P., Fuerstman M. J., Stone H. A., and Whitesides G. M., Lab Chip 6, 437 (2006). 10.1039/b510841a [DOI] [PubMed] [Google Scholar]
- Wei J., Den S. S., Tan C. M., and Wong C. K., 2004 Electronics Packaging Technology Conference, 2004, pp. 189–192.
- Shoffner M. A., Cheng J., Hvichia G. E., Kricka L. J., and Wilding P., Nucleic Acids Res. 24, 375 (1996). 10.1093/nar/24.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding P., Shoffner M. A., and Kricka L. J., Clin. Chem. 40, 1815 (1994). [PubMed] [Google Scholar]
- Joseph C., Jefferson A. D., and Cantoni M. B., Proceedings of the First International Conference on Self Healing Materials, 18–20 April 2007.
- Bradford M., Anal. Biochem. 72, 248 (1976). 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Dolnik V. and Liu S. R., J. Sep. Sci. 28, 1994 (2005). 10.1002/jssc.200500243 [DOI] [PubMed] [Google Scholar]
- Handbook of Capillary and Microchip Electrophoresis and Associated Microtechniques, edited by Landers J. P. (Taylor & Francis, Boca Raton, FL, 2008). [Google Scholar]
- Zhou L., Chan W. K., Xu N., Xiao K., Luo H., Luo K. Q., and Chang D. C., Life Sci. 83, 394 (2008). 10.1016/j.lfs.2008.07.011 [DOI] [PubMed] [Google Scholar]