Abstract
The nuclear p68 RNA helicase is a prototypical member of the DEAD box family of RNA helicases. P68 RNA helicase has been implicated in cell proliferation and early organ development and maturation. However, the functional role of p68 RNA helicase in these biological processes at the molecular level is not well understood. We previously reported that tyrosine phosphorylation of p68 RNA helicase mediates the effects of PDGF in induction of EMT by promoting β-catenin nuclear translocation (Yang et.al. Cell 127:139-155 2006). Here we report that phosphorylation of p68 RNA helicase at Y593 up-regulates transcription of the Snail1 gene. The phosphorylated p68 activates transcription of the Snail1 gene by promoting HDAC1 dissociation from the Snail1 promoter. Our results showed that p68 interacted with the nuclear remodeling and deacetylation complex MBD3:Mi-2/NuRD. Thus, our data suggested that a DEAD box RNA unwindase can potentially regulate gene expression by functioning as a protein ‘displacer’ to modulate protein-protein interactions at the chromatin remodeling complex.
Keywords: P68 RNA helicase, E-cadherin, Snail1, transcription activation, DEAD-box, HDAC1, Mi-2/NuRD
Introduction
E-cadherin, a prototypical member of the cadherin family, is the key component of the epithelial cell-cell adhesion junction. During embryonic development and tissue remodeling, the expression of E-cadherin is repressed. As a consequence, the strong adhesions of the epithelial cells are weakened. The cells adopt a fibroblast-like morphology. This is the so-called Epithelial-Mesenchymal-Transition (EMT) process (Kang & Massague, 2004; Radisky, 2005). Loss of E-cadherin expression or function constitutes one main reason for epithelial carcinoma progression to an invasive metastatic status (Kang & Massague, 2004; Rodrigo et al., 1999). Both expression and function of E-cadherin are regulated at multiple levels (Bryant & Stow, 2004; Davis et al., 2003). A zinc-finger transcription factor, Snail1, and its closely related family have been demonstrated to play a key role in downregulation of E-cadherin gene transcription ( Peinado et al., 2004; De Craene et al., 2005). It was revealed that Snail1 mediates E-cadherin repression by recruiting histone deacetylase (HDAC) to the E-cadherin promoter (Peinado et al., 2004). Repression of E-cadherin by Snail1 leads to Epithelial-Mesenchymal Transition (EMT). As a master regulator for EMT, expression of Snail1 is stimulated by signaling pathways of a number of growth factors (De Craene et al., 2005), such as, EGF, FGF and TGF-β (Ciruna & Rossant, 2001; Lu et al., 2003; Zavadil & Bottinger, 2005). Cellular levels of Snail1 are regulated via a number of different mechanisms, including gene transcription and protein turn-over in cells (Barbera et al., 2004; Zhou et al., 2004). Most recently, Fujita and co-workers demonstrated that MTA3, a member of the metastasis associated gene family, regulates Snail1 expression by targeting the nuclear remodeling and deacetylation complex MBD3:Mi-2/NuRD-HDAC1 to the Snail1 promoter in breast cancer cells (Fujita et al., 2003).
The nuclear p68 RNA helicase (ref to as p68) is a prototypical member of the DEAD box family of RNA helicases (Crawford et al., 1982; Lane & Hoeffler, 1980). As an early example of a cellular RNA helicase, the ATPase and the RNA unwinding activities of p68 RNA helicase were documented (Ford et al., 1988; Hirling et al., 1989; Iggo & Lane, 1989). Expression of p68 correlates with cell proliferation and early organ maturation (Stevenson et al., 1998). The protein was also shown to potentially play a critical role in the tumorigenesis process (Causevic et al., 2001; Dubey et al., 1997; Wei & Hu, 2001). It has been demonstrated by numerous laboratories that p68 has a functional role in transcriptional regulation of a number of genes, including Estrogen Receptor alpha (ERα) (Endoh et al., 1999) and several p53-dependent genes (Bates et al., 2005). The protein was also shown to interact with p300/CBP and the RNA polymerase II holoenzyme (Rossow & Janknecht, 2003). The molecular mechanism by which p68 is involved in transcriptional regulation is not clear. Interestingly, p68 was detected to interact with histone deacetylase 1 (HDAC), indicating that the protein may have a functional role in regulation of gene expression by chromatin remodeling (Wilson et al., 2004). We previously reported that p68 is phosphorylated at multiple amino acid residues, including serine/threonine and tyrosine (Yang & Liu, 2004; Yang et al., 2005b). Tyrosine phosphorylation of p68 correlates with tumor progression (Yang et al., 2005a). In the present study, we present evidence to show that the phosphor-p68 represses E-cadherin expression by regulating transcription of the Snail1 gene. Phosphorylation of p68 at Y593 promoted dissociation of HDAC1 from Snail1 promoter. P68 RNA helicase interacted with the nuclear remodeling and deacetylation MBD3:Mi-2/NuRD complex, suggesting a potential role of phosphor-p68 in dissociating HDAC1 from the MBD3:Mi-2/NuRD-HDAC1 complex at the Snail1 promoter. Our study revealed a close correlation between the Snail1 expression levels and the phosphorylation levels of p68, both correlated closely with cancer metastasis.
Results
The phosphor-p68 repressed E-cadherin by upregulating transcription of Snail1
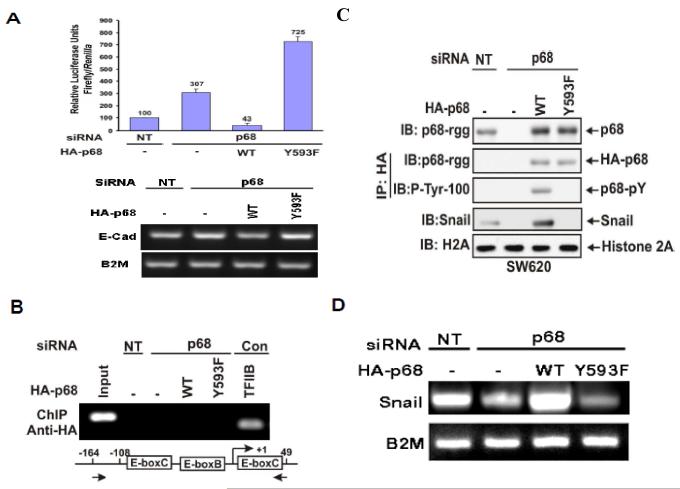
We previously reported that phosphorylation of p68 at Y593 mediates growth factor PDGF stimulated EMT by promoting β-catenin nuclear translocation (Yang et al., 2006). The phosphor-p68 represses expression of epithelial marker, E-cadherin. We sought to understand the molecular mechanism by which the phosphor-p68 regulates E-cadherin expression. We first tested whether the effects of p68 phosphorylation on the changes in E-cadherin expression was due to transcription of the E-cadherin gene. The transcription activity of the E-cadherin promoter in a metastatic colon cancer cell line SW620 was examined. We used a luciferase reporter fused to the E-boxes of the E-cadherin promoter (Fearon, 2003) in cells in which p68 was knocked down and HA-p68s wild type or Y593F mutant was exogenously expressed in p68 knockdown cells (SW620−p68/+wt and SW620−p68/+Y593F). E-cadherin transcription activity was not dramatically affected by p68 knockdown but was substantially down-regulated in SW620−p68/+wt cells (Fig. 1A). On the other hand, E-cadherin transcription was significantly upregulated in SW620−p68/+Y593F cells (Fig. 1A). We tested whether phosphor-p68 regulated E-cadherin transcription directly or if the regulatory effects were mediated through other cellular factor(s). To this end, we examined whether phosphor-p68 interacted with the promoter of the E-cadherin gene by chromatin immunoprecipitation (ChIP). It was clear that neither p68 wild-type nor the Y593F mutant interacted with the E-cadherin promoter (Fig. 1B). Thus, it was likely that phosphor-p68 regulated E-cadherin expression indirectly.
Figure 1.
The phosphor-p68 down-regulated E-cadherin by upregulation of transcription of Snail1.
(A upper panel) Luciferase reporter of E-boxes (E-cadherin promoter) was cotransfected into p68 knockdown SW620 cells along with HA-p68s, wt or mutant (indicated). The luciferase activity was expressed as relative luciferase activity (numbers on top of bars) by comparing the luciferase activity of SW620 cells without p68 knockdown (NT) and without HA-p68s expression (define as 100). The lower panel shows the mRNA expression of E-cadherin under the same conditions. (B) Chromatin immunoprecipitations (ChIP) of the E-cadherin promoter by anti-HA antibody in SW620 cells with/without (p68/NT) p68 knockdown. The HA-p68s (WT or Y593F mutant) were exogenously expressed. The primers positions for PCRs were indicated by arrows. ChIP by mouse IgG and antibody against TFIIB, which binds to the GAPDH promoter, were used as controls. Inputs were PCR products from DNA extracts without ChIP. (C) & (D) Expression of Snail1 was examined by immunoblot of cell lysate (C) and RT-PCR of total RNAs (D) prepared from SW620 cells with/without (p68/NT) p68 knockdown. The HA-p68s (WT or Y593F mutant) was exogenously expressed. The expression and tyrosine phosphorylation of HA-p68s were examined by IB of immunoprecipitated HA-p68s (IP:HA). Total p68 level was detected by IB using monoclonal antibody p68-rgg (IB:p68). IB of histone 2A (H2A) was a loading control. RT-PCR detection of B2M mRNA in the RNA samples was a control for PCR reaction and loading.
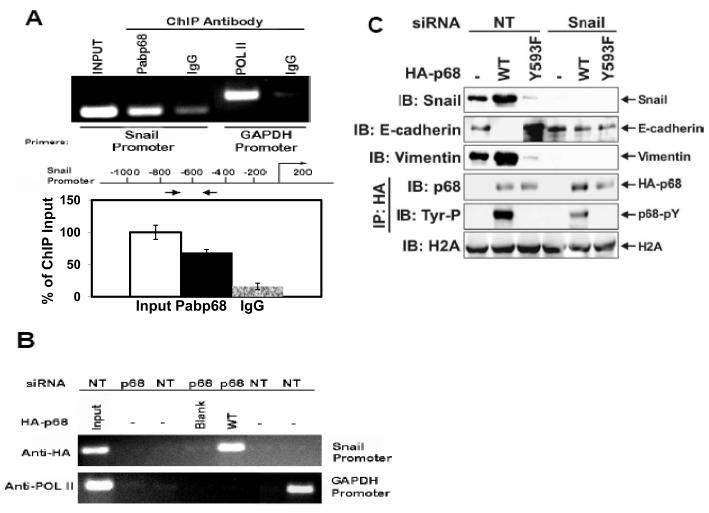
Snail1/Slug and SIP1 are the master regulators that regulate E-cadherin transcription ( Thiery & Chopin, 1999; Bolos et al., 2003; Fearon, 2003). In a ChIP-on-chip assay, we detected that p68 interacted with the Snail1 promoter (Data not shown). Thus, we reasoned whether the phosphor-p68 regulated E-cadherin through regulation of Snail1 expression. We examined the effects of p68 phosphorylation on the expression of Snail1. Immunoblotting showed that cellular levels of Snail1 were increased in the SW620−p68/+wt cells and decreased in the SW620−p68/+Y593F cells (Fig. 1C). The Snail1 upregulation correlated with the p68 expression and phosphorylation at Y593 (Fig. 1C). Our results indicated that phosphorylation of p68 at Y593 may regulate the transcriptional activity of the Snail1 gene. Regulation of transcription of the Snail1 gene by phosphor-p68 was also confirmed by RT-PCR detection of the Snail1 mRNA in SW620, SW620−p68/+wt, or SW620−p68/+Y593F cells (Fig. 1D). To test whether phosphor-p68 regulated Snail1 transcription directly, we performed ChIP experiments with the Snail1 promoter using the antibody Pabp68. Clearly, p68 precipitated with the Snail1 promoter (Fig. 2A). The similar experiments were also carried out with exogenously expressed HA-p68 in SW620 cells using anti-HA antibody. P68 precipitated with the Snail1 promoter (Fig. 2B), indicating that p68 was directly involved in the transcriptional regulation of the Snail1 gene.
Figure 2.
P68 interacted with Snail1 promoter.
(A) & (B) Chromatin immunoprecipitations (ChIP) of the Snail1 promoter by antibody Pabp68 (A, Anti-p68) and by anti-HA antibody (B, Anti-HA) in SW620 cells with/without (p68/NT) p68 knockdown. The HA-p68s (WT or Y593F mutant) were exogenously expressed. The primer positions for Snail1 PCR were indicated by arrows. ChIP by mouse IgG and antibody against RNA polymerase II (POL II), which binds to the GAPDH promoter, were used as controls. Inputs were PCR products from DNA extracts without ChIP (use 10% of sample). The lower panel shows the quantization ChIP results by real-time PCR. The ChIP quantization is expressed as a percentage of the input by defining the input as 100. The error bars represent standard deviation of results from three independent measurements. (C) Cellular levels of E-cadherin (second panel from top) and vimentin (third panel from top) were detected by IBs of cellular extracts made from SW620 cells with/without (Snail1/NT) Snail1 siRNA knockdown and exogenous expression of HA-p68s (WT or Y593F mutant). Tyrosine phosphorylation of HA-p68s was analyzed by IB of anti-HA IPs via antibody p-Tyr-100 (fifth panel from top). IB of histone 2A (H2A) was loading control.
If the phosphor-p68 repressed E-cadherin through transcriptional regulation of the Snail1 gene, we reasoned that Snail1 expression must be required for the effects of the Y593 phosphorylation on the E-cadherin repression. To test this conjecture, we examined the effects of the p68 phosphorylation on the cellular level of E-cadherin in SW620 cells with/without Snail1 knockdown by RNAi. We found that Snail1 was upregulated, E-cadherin was repressed, and vimentin was upregulated by expression of wild-type p68 in SW620 cells. In contrast, expression of p68 Y593F mutant in SW620 cells led to Snail1 repression, E-cadherin upregulation, and vimentin repression (Fig. 2C). However, knockdown of Snail1 by RNAi abolished the effects of exogenous expression of HA-p68s (wild-type and Y593F mutant) on cellular levels of E-cadherin and vimentin (Fig. 2C). The results supported the conclusion that regulating the transcription of Snail1 mediated the regulatory effects of the phosphorylated p68 in repressing E-cadherin expression. Interestingly, we repeatedly observed that there was a very low level of expression of E-cadherin in cells where Snail1 was knocked down (Fig. 2C), suggesting that there is an alternative mechanism which represses E-cadherin expression in Snail1 knockdown cells. This alternative mechanism is not regulated by phosphor-p68.
P68 associates with the MBD3:Mi2/NuRD complex
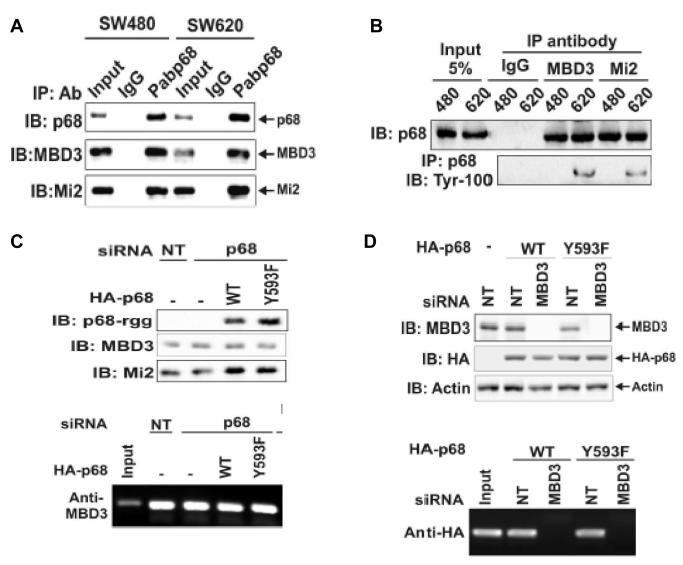
The preceding experiments suggested the role of phosphor-p68 in repression of E-cadherin through the regulation of transcription of the Snail1 gene. To further understand the molecular mechanism by which phosphor-p68 regulated transcription of the Snail1 gene, we attempted to probe the protein or protein complex that interacted with phosphor-p68 at the Snail1 promoter. Recently, Fujita and co-workers demonstrated that MTA3 targeted the nuclear remodeling and deacetylation complex Mi-2/NuRD to the Snail1 promoter, and directly regulated Snail1 gene transcription (Fujita et al., 2003). Thus, we asked whether phosphor-p68 interacted with the MBD3:Mi-2/NuRD complex in SW620 cells. To this end, we carried out co-immunoprecipitation with the nuclear extracts made from SW620 cells using PAbp68. As a comparison, the co-immunoprecipitation experiments were also performed with nuclear extracts made from SW480, a cell line derived from the tissue of the same patient from whom SW620 was derived. SW480, however, was derived from tissue of non-metastatic adenocarcinoma. The tyrosine phosphorylation of p68 was almost undetectable in SW480 cells (data not shown, also see Fig. 5A). It was clear that the antibody against p68 precipitated MBD3 and Mi-2 in the extracts made from both SW480 and SW620 cells (Fig. 3A). To confirm the co-immunoprecipitation results, we performed co-immunoprecipitation using antibodies against MBD3 and Mi-2. The co-precipitation of p68 with MBD3 and Mi-2 in the extracts made from SW480 and SW620 cells was clearly evident (Fig. 3B). These coimuunoprecipitation experiments suggested that p68 interacted with Mi-2/NuRD complex.
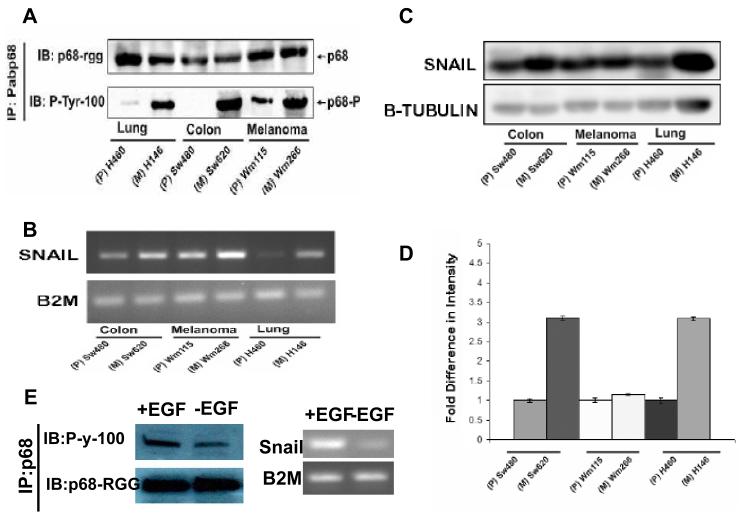
Figure 5.
P68 phosphorylation correlates with Snail1 expression in metastatic and non-metastatic cancer cells.
(A) P68 tyrosine phosphorylation in three pair of cancer cell lines (indicated). The tyrosine phosphorylation of p68 was detected by immunoblot (via the anti-phosphor-tyrosine antibody; IB:P-tyr-100) of p68 that was immunoprecipitated from nuclear extracts of the cells using the antibody Pabp68 (IP:Pabp68). Immunoblot of p68 in the IPs using the antibody p68-rgg (IB:p68- rgg) was the loading control. (B) The expression levels of Snail1 mRNA in the three pair of cancer cells (indicated) were detected by RT-PCR of total RNAs isolated from the cells. The RT-PCR detections of mRNA of B2M (B2M) gene in the cells were the controls. (C) The cellular Snail1 protein levels in the three pair of cancer cells (indicated) were examined by immunoblot of the cell lysate prepared from the cells using antibody against Snail1 (IB:Snail1). IB of β-tubulin was used as the loading control. (D) is the quantification (average) of the immunoblot signals of Snail1 after normalizing to the loading control β-tubulin blots. The error bars represent standard deviation of results from three independent immunoblots. (E) (Left) Tyrosine phosphorylation of p68 in SW480 cells that were treated (+EGF) or untreated (−EGF) with EGF (25 ng/ml) was analyzed by Immunoprecipitation of p68 (IP:p68) from the cell lysates followed by immunoblot using the antibody P-y-1oo (IB:P-y-100). Immunoblot of p68 (IB:p68-RGG) indicated the precipitated p68. (Right) Expression of Snail1(Snail) in SW480 cells that were treated (+EGF) or untreated (−EGF) with EGF (25 ng/ml) was analyzed by RT-PCR of the total RNA isolated from cell lysates. RT-PCR analyses of B2M mRNA in the EGF treated or untreated cells is a control.
Figure 3.
P68 interacted with the MBD3:Mi-2/NuRD complex.
(A) Co-IPs of MBD3 and Mi-2 with p68 in SW480 and SW620 cells were detected by IB of p68 co-immunoprecipitates using appropriate antibodies (indicated). P68 was precipitated by polyclonal antibody Pab-p68. Rabbit IgG was used as a negative control IP antibody. Inputs were the IBs of extracts without IP. (B) Co-IPs of p68 with MBD3 and Mi-2 in SW620 (620) and SW480 (480) cells were detected by IBs of co-immunoprecipitates of antibodies (anti-MBD3 and anti-Mi-2) using monoclonal antibody p68-rgg. Mouse IgG was used as control IP antibody. The inputs were the IBs of extracts without IP. The tyrosine phosphorylation of p68 was detected by IB of PAbp68 immunoprecipitated p68 using antibody Tyr-100. (C) Upper panel, cellular levels of MBD3, HDAC1, and exogenously expressed HA-p68s (WT or Y593F mutant) were analyzed via IBs using appropriate antibodies (indicated). The IBs were performed with cellular extracts made from SW620 cells that were treated with p68 siRNA (p68) or non-targeting siRNA (NT). Lower panel, Interactions of MBD3 with Snail1 promoter in the cells treated as described in upper panels were detected by ChIP assays using anti-MBD3 antibody (Anti-MBD3). (D) Upper panel, cellular levels of MBD3 and exogenously expressed HA-p68s (WT or Y593F mutant) (without endogenous p68 knockdown) were analyzed via IBs using appropriate antibodies (indicated). The IBs were performed with cellular extracts made from SW620 cells that were treated with MBD3 siRNA (MBD3) or non-targeting siRNA (NT). IB of Actin was a loading control. Lower panel, Interactions of HA-p68 with Snail1 promoter in the cells treated as described in upper panels were detected by ChIP assays using anti-HA antibody (Anti-HA).
We next asked whether the interaction between p68 and MBD3:Mi-2/NuRD is required for the association of p68 and/or MBD3:Mi-2/NuRD with the Snail1 promoter. We performed the ChIP experiments in SW620 cells in which either p68 or MBD3 was knocked down by RNAi. Immunoblotting demonstrated an efficient knock down of these two proteins (over 90%) (Fig 3D and Fig 1C). We then exogenously expressed HA-p68s (wt or Y593F mutant) in MBD3 and p68 knockdown cells. Anti-HA antibody did not precipitate the Snail1 promoter in MBD3 knockdown cells (Fig. 3D, Upper panel). However, antibody against MBD3 did precipitate the Snail1 promoter in the p68 knockdown cells (Fig. 3D). Expression of p68 (wt or Y593F mutant) did not affect the MBD3-Snail1 promoter precipitation (Fig. 3D). The data suggested that association of p68 with the MBD3:Mi-2/NuRD is not required for the recruitment of MBD3:Mi-2/NuRD complex to the Snail1 promoter. In contrast, MBD3 was required for association of p68 with Mi-3/NuRD complex and the Snail1 promoter.
HDAC1 dissociated from the Snail1 promoter in the presence of phosphor-p68
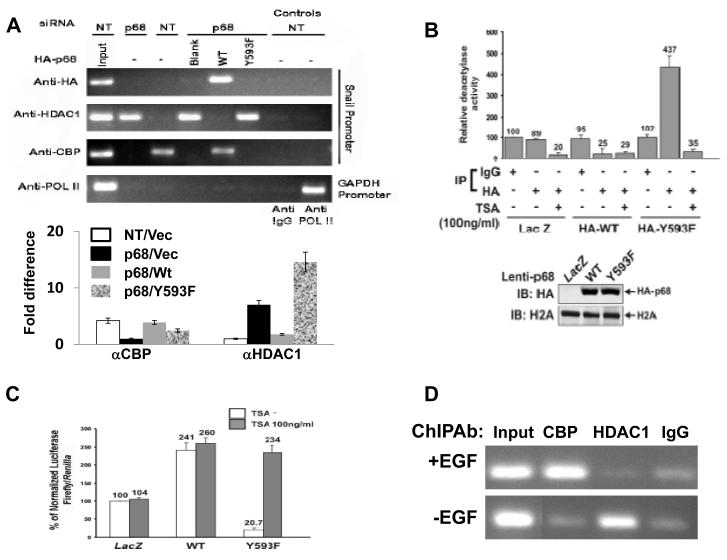
An important cellular activity of Mi-2/NuRD complex is histone deacetylation via its associated HDAC1 and HDAC2 (Bowen et al., 2004; Xue et al., 1998). To determine whether phospho-p68 plays a role in regulating the function of the Mi-2/NuRD complex at the Snail1 gene promoter, we first probed the association of HDAC1 with the Snail1 promoter in SW620−p68, SW620−p68/+wt, and SW620−p68/+Y593F cells by ChIP experiments using antibody against HDAC1. It was clear that p68 knockdown dramatically enhanced the association HDAC1 with the Snail1 promoter. However, expression of HA-p68 wt in the p68 kockdown cells diminished the association of HDAC1 with the Snail1 promoter. The HDAC1 associated with the Snail1 promoter in the cells expressing Y593F mutant after endogenous p68 knockdown but not the p68 wild-type (Fig. 4A). The experiments suggested that the phosphor-p68 promoted HDAC1 dissociation from the Snail1 promoter. Interestingly, the Y593F mutant did not interact with the Snail1 promoter in the ChIP assay (Fig. 4A), suggesting that phosphorylation of p68 is required for p68 association with the Snail1 promoter. Examination of the association of CREB binding protein (CBP) with the Snail1 promoter in SW620−p68, SW620−p68/+wt, and SW620−p68/+Y593F cells revealed that dissociation of HDAC1 from Snail1 promoter facilitated by the p68 phosphorylation increased the binding of CBP to the Snail1 promoter (Fig. 4A), consistent with active transcription of the Snail1 gene. We previously demonstrated that EGF treatment led to the increase in the p68 phosphorylation (Yang et al., 2006). Thus, we probed the effects of EGF treatment on interaction between HDAC1 and the Snail1 promoter. ChIP assays indeed showed that EGF treatment led to a substantial decrease in HDAC1 and the Snail1 promoter interaction, while a strong increase in CBP and Snail1 promoter interaction (Fig. 4D).
Figure 4.
Phosphor-p68 dissociated HDAC1 from the Snail1 promoter.
(A) ChIP of the Snail1 promoter using Anti-HA (HA), anti-HDAC1 (HDAC1), and anti-CBP (CBP) antibodies in SW620 cells. SW620 cells were treated with p68 siRNA (p68) or non-targeting siRNA (NT). HA-p68s (WT or Y593F mutant) or empty vector (Blank) was exogenously expressed in p68 knockdown cells. Inputs were PCR products from SW620 DNA extracts without ChIP. ChIP assays with IgG were the negative controls. ChIP of GAPDH promoter in the cells using anti-POL II (POL II) antibody was a control ChIP experiment. (A, Lower panel) Quantization ChIP results by real-time PCR. The ChIP quantization is expressed as fold difference by defining the lowest ChIP value in each antibody group (αCBP or αHDAC1) as 1 after normalizing ChIP value in each antibody group to the inputs within the group. The error bars represent standard deviation of results from three independent measurements. NT/p68 means the cells were treated with non-target or p68 target RNAi. Vec, wt, or Y593F represents p68 wt, Y593F mutant, or vector was expressed in the cells. (B) Deacetylase activities of co-immunoprecipitates by mouse IgG (IgG, as control IP antibody) and anti-HA antibody (HA) from cellular extracts made from SW620 cells were analyzed. HA-p68s (WT or Y593F mutant) were stably expressed using Lenti-viral system. LacZ is a control expression protein. The immunoprecipitates were treated/untreated (+/−) with 100 ng/ml of TSA. The deacetylase activity (numbers on top of bars) was expressed as relative deacetylase activity by define the activity of co-IP by mouse IgG without TSA treatment without HA-p68 expression as 100. The upper panel is IB analyses of stable expression of HA-p68 in SW620 cells. IB of H2A is a loading control. (C) Luciferase reporter of the Snail1 promoter was transfected into SW620 cells in which HA-p68, wt or mutant (indicated) was stably expressed. Twenty four hours post transfection, cells were treated/untreated (filled bars/open bars) with TSA (100 ng/ml) overnight. Luciferase activities were then analyzed. The luciferase activity was expressed as relative luciferase activity (numbers on top of bars) by compared to the luciferase activity of SW620 cells without HA-p68s expression and TSA treatment (define as 100). (D) ChIP of the Snail1 promoter using anti-HDAC1 (HDAC1) and anti-CBP (CBP) antibodies in SW480 cells that were treated (+EGF) or untreated (−EGF) (EGF 15 ng/ml). ChIP using rabbit IgG was a control ChIP experiment. In (B) and (C), the error bars represent standard deviation of results from three independent measurements.
We questioned whether the p68 phosphorylation at Y593 affected the HDAC1 activity at the Snail1 promoter. To this end, the HA-p68s (wt or Y593F mutant) were stably expressed in SW620 cells using a commercially available lentiviral system. The expression level of p68 wt/mutant was high as revealed by the immunoblot with the anti-HA antibody (Fig. 4B). The expressed p68s were immunoprecipitated from nuclear extracts by anti-HA. The deacetylase activity of the immunoprecipitates was analyzed by HDAC Activity Colorimetric Assay kit. It was evident that overexpression of wt p68 suppressed the deacetylase activities by over 3 folds (Fig. 4B, comparing LacZ/IP:HA to HA-WT/IP:HA). In contrast, overexpression of Y593F mutant led to the increase of deacetylase activities by over 5 fold (Fig. 4B, comparing LacZ/IP:HA to HA-Y593F/IP:HA) or by 17 folds (Fig. 4B, comparing HA-WT/IP:HA to HA-Y593F/IP:HA). The increase in the deacetylase activities was sensitive to TSA (an HDAC inhibitor) treatment (Fig. 4B), supporting that an HDAC activity was co-precipitated with HA-Y593F but not with HA-WT. To further investigate whether the p68 phosphorylation affected the HDAC activity at the Snail1 promoter, using the Snail1 promoter/luciferase reporter construct, we measured the Snail1 promoter activity in the presence and absence of the HDAC inhibitor TSA in SW620 cells in which p68 wt/mutant was overexpressed. The data indicated that the phosphor-p68 indeed affected the deacetylase activities at the Snail1 promoter (Fig. 4C).
P68 phosphorylation correlates with Snail1 expression in metastatic cancer cells
Snail1 expression has been demonstrated to be associated with cancer metastasis in various different cancer types (Barrallo-Gimeno & Nieto, 2005; Miyoshi et al., 2005; Nyormoi & Bar-Eli, 2003). We previously reported that p68 phosphorylation at tyrosine residues correlates with cancer progression. Thus, we sought to determine whether there is a correlation between phosphorylation of p68 at tyrosine 593 and Snail1 expression levels, and whether both will correlate with cancer metastasis. To this end, we utilized three pair of cell lines that were derived from different cancer types. SW480 and SW620 are colon cancer cell lines derived from the same cancer patient. WM115 and WM266 are melanoma cell lines from the same patient. H146 and H460 are lung cancer cell lines derived from the same patient. Among these three pair of cell lines, SW620, WM266, and H460 were derived from metastatic cancer tissue, while the other three were derived from cancer before metastasis. Protein extracts and total RNA samples were made from these cells. Immunoprecipitation of p68 followed by immunoblot of the IPs using antibody P-Tyr-100 demonstrated that there were substantially higher p68 tyrosyl phosphorylation levels in the three cell lines that were derived from metastasis cancer than that in cell lines derived from corresponding non-metastatic cancer (Fig. 5A). Examination of Snail1 mRNA levels in these three pairs of cell lines by RT-PCR indicated that the Snail1 mRNA levels were substantially higher in SW620, WM266 and H460 cells than in corresponding non-metastatic cells (Fig. 5B), correlating with the tyrosine phosphorylation of p68 in these cells. We further analyzed the cellular Snail1 levels in the extracts made from the three pairs of cells by immunoblot analyses using antibody against Snail1. It was clear that the cellular Snail1 levels in the metastatic cancer cells (SW620 and H460) were much higher than that of matched non-metastastic cells (SW480, H146) (Fig. 5C). In the pair of melanoma cells, the Snail1 levels are higher in metastatic cells than in non-metastatic cells. However, the difference was less dramatic (Fig. 5C). Thus, our experiments demonstrated a close correlation between phosphorylation of p68 at tyrosine residues and Snail1 expression, and both the Snail1 expression and p68 tyrosyl phosphorylation correlated with cancer metastasis. To further correlation of Snail1 expression and p68 phosphorylation, SW480 cells were stimulated by EGF treatment. Phosphorylation of p68 and Snail1 expression were examined. Consistent with our previous observation (Yang et al., 2006), the p68 tyrosine phosphorylation responsed well to EGF stimulations in the cells. Snail expression was also upregulated upon the growth factor treatment. The results further support a correlation between p68 tyrosine phosphorylation and Snail1 expression (Fig. 5E).
Discussion
In this report, we demonstrated that phosphorylation of p68 RNA helicase repressed E-cadherin expression by up-regulating transcription of the Snail1 gene. The phosphor-p68 activates transcription of the Snail1 gene by dissociating HDAC1 from the Snail1 promoter. P68 RNA helicase has been implicated in transcriptional regulation of many genes (Bates et al., 2005; Endoh et al., 1999; Metivier et al., 2003). However, it is not known how a DEAD box RNA helicase functions in transcriptional regulation. A recent observation made by Allen C. Spradling’s laboratory reveals that drosophila p68 may have a role in unwinding the RNA transcripts from its DNA template, facilitating a quick reset of the nucleosome structure, which is transcriptionally inactive (Buszczak & Spradling, 2006). Our observations may reveal another model for the functional role of p68 in transcriptional regulation. P68 RNA helicase may have a role in remodeling or re-arranging the protein complex that assembles at the gene promoter.
There were two possible explanations for the observed dissociation of HDAC1 from the Snail1 promoter in SW620 cells. (1) The unphosphorylated p68 recruited HDAC1 to the promoter. The phosphor-p68 could not function as a recruiter. (2) The phosphor-p68 may ‘displace’ HDAC1 from the Snail1 promoter. The unphosphorylated p68 could not function as a protein ‘displacer’. Although we observed that the phosphor-p68 acquired β-catenin binding dependent ATPase activity (data not shown), indicating a possibility that phosphor-p68 can use protein-binding as substrate to stimulate its ATPase activity for the protein ‘displacement’, additional experiments are required to prove that the phosphor-p68 can indeed displace HDAC1 from Mi-2/NuRD complex. Histone deacetylases are enzymes that modify chromatin structure and subsequently regulate gene expression. HDACs are usually recruited to a particular regulatory site with their associated multi-protein complexes, such as NuRD or Sin3 complex (Knoepfler & Eisenman, 1999; Narlikar et al., 2002). While most studies concentrated on the mechanism by which the HDAC activity and its associated complex are recruited to a specific gene promoter (Forsberg & Bresnick, 2001; Kurdistani & Grunstein, 2003; Neely & Workman, 2002), our studies suggested a possible mechanism of action by which HDACs can be displaced from their associated complex by a DEAD box helicase. Given that tyrosine phosphorylations of p68 were closely associated with cancer development (Yang et al., 2005a), it is tempting to speculate that displacement of HDACs by the phosphor-p68 is a dysregulated route for tumor progression through activation of specific genes. Whether p68 is a constitutive member of the NuRD complex is an open question. P68 was not identified in the originally isolated NuRD complex (Bowen et al., 2004; Tong et al., 1998; Xue et al., 1998; Zhang et al., 1999). Our experiments demonstrated co-immunoprecipitation of p68 with the Mi-2 and MBD3, the components of NuRD complex. This seems to argue that p68 is a part of protein components of the NuRD complex. In contrast, knockdown of p68 did not affect the association of the MBD3:Mi-2/NuRD complex with Snail1 promoter, suggesting that p68 is not required for the NuRD complex to be recruited to the Snail1 promoter. This observation argues that p68 may not be a constitutive component of the NuRD complex. The best explanation is that p68 only associates with a subset of NuRD complexes. Most recently, p68 was detected to interact with HDAC1 in a promoter-specific manner (Wilson et al., 2004), which also seems to support the notion. It was not known whether the detected interaction of p68 with HDAC1 was direct or mediated by other protein factors. Knockdown of MBD3 abolished the association of p68 at the Snail1 promoter, suggesting that p68 was recruited to the Snail1 promoter by a component of the NuRD complex. Taken together, we speculated that p68 RNA helicase is recruited to the MBD3:Mi-2/NuRD complex in a promoter specific manner. An alternative explanation is that phosphorylated p68 RNA helicase may compete with HDAC1 to assemble to the Mi-2/NuRD complex. The unphosphorylated p68 and Y593F mutant are unable to compete. Thus, there may be two populations of Mi-2/NuRD complexes in SW620 cells. One contains phosphor-p68 and the other contains HDAC1.
Methods
Reagents and antibodies
TSA was purchased from PeproTech. Both polyclonal antibody PAbp68 and monoclonal antibody p68-rgg against human p68 were raised against bacterially expressed His-tagged Cterminal domain of p68 (Invitrogen, Auburn University Hybridoma Facility). Commercial antibodies used in this study were purchased from Santa Cruz (Actin, against human β-actin, HDAC1, GAPDH against human GAPDH, ChIP grade monoclonal against HDAC1, Mi-2, SNAI 1 against Snail1), Cell Signaling Technology (p-Tyr-100, against phosphor-tyrosine, HDAC1, polyclonal against HDAC1, H2A, against human histone 2A), Imgenex (MBD3, ChIP grade), BD Bioscience (E-cadherin and Vimentin), Roche (12CA5, against HA-tag ChIP grade), Abcam (KAT3A/CBP, against CBP) and Upstate (HA, against HA-tag).
Cell culture and RNA interference
SW620 and SW480 cells were purchased from ATCC and grown by following the vendor’s instructions. All DNAs or RNAs transfections were performed using Lipofectamine 2000 by following the manufacturer’s instructions (Invitrogen). For the siRNA experiments, cells were grown to 50% confluence and transfected with siRNA (100 pM). The duplex RNA oligonucleotides for RNAi were purchased from Dharmacon siGENOME™ and SMARTpool®. A duplex RNA oligonucleotides with random sequence (non-targeting, NT) provided by the vendor was included in all siRNA knockdown experiments as negative controls. For transient expression of wild-type p68 or mutants in p68 knockdown cells, the cells were transfected with the indicated plasmid DNA 24 hours after the cells were transfected with siRNA and harvested after an additional 48 hours incubation.
Subcellular extracts, Immunoprecipitation, Immunoblot
All subcellular extracts or whole cell extracts were made freshly after appropriate treatments (indicated in figures). Subcellular extracts were prepared using commercially available cell extracting kit and by following the vendor’s instructions (Active motif). The protein concentration of the extracts was determined using the Bradford assay (Bio-Rad). Immunoprecipitation experiments and immunoblot analyses were performed as described in previous studies (Liu et al., 1998). The blotting signals were detected using SuperSignal West Dura Extended Duration Substrate (Pierce).
Luciferase reporter assays
Before cells were appropriately treated (indicated in figures), cells were transfected with 1 μg of the indicated reporter plasmid and 0.01 μg of pRL null, which expresses Renilla luciferase from Renilla reniformis as an internal control. The total amount of plasmid DNA was adjusted with pcDNA3-β-Galactosidase. Firefly and Renilla luciferase activities present in cellular lysates were assayed using the Dual-Luciferase Reporter System (Promega). Data were represented as Firefly luciferase activity normalized by Renilla luciferase activity.
Expression of p68s by lentiviral system
Stable overexpression of HA-tagged p68 wild-type or Y593F mutant was carried out using the ViralPower lentiviral expression system (Invitrogen) by following the manufacturer’s instructions. The ORFs of p68 wild type or Y593F mutant with N-terminal HA-tag were cloned into pLenti6/TOPO (Invitrogen). The infections of cells with the lentiviruses that carry pLenti6-p68 were carried out in the presence of 6 μg/mL of polybrene and 10 mM HEPES. Following transduction, the cells were selected by 8 μg/mL of Blasticidin (Invitrogen).
RT-PCR
Cells were appropriately treated, then total RNA was extracted using the total RNA extraction kit (Qiagen). The RNA was quantified and then converted to cDNA using the Improm II reverse transcription system (Promega) following the manufacturer’s protocol. The cDNA was then used in the final PCR reaction. The cycles were an initial denaturing of 94 °C for 2 minutes followed by 25 cycles of 94 °C for 15s, 55 °C for 30s, and 72 °C for 1m with an additional extension time of 5m added after the last cycle. Densitometry was performed using the ImageJ program. Primers used were: Snail1 (sense 5′-TCTAGGCCCTGGCTGCTAC-3′ antisense 5′-GCCTGGCACTGGTACTTCTT-3′); B2M (sense 5′- TGCTGTCTCCATGTTTGATGTATCT-3′) antisense 5′-TCTCTGCTCCCCACCTCTAAGT-3′). Primers for B2M were used as PCR and loading controls. Controls for the RT reaction (not shown) contained no template or no reverse transcriptase.
Chromatin Immunoprecipitation (ChIP)
The ChIP experiments were performed using ChIP-IT™ Kit (Activemotif). The precipitation of Snail1 or E-cadherin promoters was determined by PCR using primers spanning nt −680 – −541 of the Snail1 promoter (sense 5′-GGGTGCTCTTGGCTAGCTG-3′ antisense 5′- CTGGAGAGCGTGGCATTG-3′) or nt −164 – +49 of the E-cadherin promoter (sense 5′- GTCACCGCGTCTATGCGAGGCCG-3′, Antisense 5′- GGACACTCGAACGCCTTCAGTCAAGT-3′). TFIIB/Pol II antibodies and mouse IgG were used as control antibodies (included in the kit). ChIP-IT’s negative control primers flank a region of genomic DNA between the GAPDH gene and CNAP1 gene. The primers were provided by the vendor (sense 5′-ATGGTTGCCACTGGGGATCT-3′ antisense 5′- TGCCAAAGCCTAGGGGAAGA-3′).
HDAC activity assay
SW620 cells were lysed in RIPA buffer (Upstate) 48 hr after transfection as described above. The lysate was then diluted in RIPA buffer and HA-tagged proteins immunoprecipitated with anti-HA polyclonal antibody (Upstate). HDAC activities were determined by HDAC Activity Colorimetric Assay Kit (BioVision) according to manufacturer’s instructions. Antibodybound beads were washed in HDAC assay buffer prior to being added to the 96-well plate, to remove immunoprecipitation buffer. Reactions were incubated for 30 min at 37°C with or without the addition of 1 μM TSA. Samples were read in a VICTOR3™ plate reader (PerkinElmer) at 405 nm. Typically each assay was performed 3 times.
Acknowledgments
We thank Roger Bridgeman for antibody p68-rgg production. We also thank Birgit Neuhaus for assistance in confocal imaging. This manuscript is greatly improved by critical comments from Jenny Yang, Mike Kirberger, Julian A. Johnson, and Heena Dey. This work is supported in part by research grants from National Institute of Health (GM063874), (CA118113), and Georgia Cancer Coalition to ZR Liu.
References
- Barbera MJ, Puig I, Dominguez D, Julien-Grille S, Guaita-Esteruelas S, Peiro S, Baulida J, Franci C, Dedhar S, Larue L, de Herreros A Garcia. Oncogene. 2004;23:7345–54. doi: 10.1038/sj.onc.1207990. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. Development. 2005;132:3151–61. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, Gregory DJ, Lane DP, Perkins ND, Fuller-Pace FV. Embo J. 2005;24:543–53. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA. Biochim Biophys Acta. 2004;1677:52–7. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Stow JL. Trends Cell Biol. 2004;14:427–34. doi: 10.1016/j.tcb.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Spradling AC. Genes Dev. 2006;20:977–89. doi: 10.1101/gad.1396306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causevic M, Hislop RG, Kernohan NM, Carey FA, Kay RA, Steele RJ, Fuller-Pace FV. Oncogene. 2001;20:7734–43. doi: 10.1038/sj.onc.1204976. [DOI] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Crawford L, Leppard K, Lane D, Harlow E. J Virol. 1982;42:612–20. doi: 10.1128/jvi.42.2.612-620.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. J Cell Biol. 2003;163:525–34. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B, van Roy F, Berx G. Cell Signal. 2005;17:535–47. doi: 10.1016/j.cellsig.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Dubey P, Hendrickson RC, Meredith SC, Siegel CT, Shabanowitz J, Skipper JC, Engelhard VH, Hunt DF, Schreiber H. J Exp Med. 1997;185:695–705. doi: 10.1084/jem.185.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, Yanagisawa J, Metzger D, Hashimoto S, Kato S. Mol Cell Biol. 1999;19:5363–72. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fearon ER. Cancer Cell. 2003;3:307–10. doi: 10.1016/s1535-6108(03)00087-4. [DOI] [PubMed] [Google Scholar]
- Ford MJ, Anton IA, Lane DP. Nature. 1988;332:736–8. doi: 10.1038/332736a0. [DOI] [PubMed] [Google Scholar]
- Forsberg EC, Bresnick EH. Bioessays. 2001;23:820–30. doi: 10.1002/bies.1117. [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. Cell. 2003;113:207–19. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- Hirling H, Scheffner M, Restle T, Stahl H. Nature. 1989;339:562–4. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- Iggo RD, Lane DP. Embo J. 1989;8:1827–31. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Massague J. Cell. 2004;118:277–9. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Eisenman RN. Cell. 1999;99:447–50. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Grunstein M. Nat Rev Mol Cell Biol. 2003;4:276–84. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- Lane DP, Hoeffler WK. Nature. 1980;288:167–70. doi: 10.1038/288167a0. [DOI] [PubMed] [Google Scholar]
- Liu ZR, Sargueil B, Smith CW. Mol Cell Biol. 1998;18:6910–20. doi: 10.1128/mcb.18.12.6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Ghosh S, Wang Z, Hunter T. Cancer Cell. 2003;4:499–515. doi: 10.1016/s1535-6108(03)00304-0. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Miyoshi A, Kitajima Y, Kido S, Shimonishi T, Matsuyama S, Kitahara K, Miyazaki K. Br J Cancer. 2005 doi: 10.1038/sj.bjc.6602266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cell. 2002;108:475–87. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Neely KE, Workman JL. Biochim Biophys Acta. 2002;1603:19–29. doi: 10.1016/s0304-419x(02)00067-7. [DOI] [PubMed] [Google Scholar]
- Nyormoi O, Bar-Eli M. Clin Exp Metastasis. 2003;20:251–63. doi: 10.1023/a:1022991302172. [DOI] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Mol Cell Biol. 2004;24:306–19. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky D. J Cell Sci. 2005;118:4325–26. doi: 10.1242/jcs.02552. [DOI] [PubMed] [Google Scholar]
- Rodrigo I, Cato AC, Cano A. Exp Cell Res. 1999;248:358–71. doi: 10.1006/excr.1999.4438. [DOI] [PubMed] [Google Scholar]
- Rossow KL, Janknecht R. Oncogene. 2003;22:151–6. doi: 10.1038/sj.onc.1206067. [DOI] [PubMed] [Google Scholar]
- Stevenson RJ, Hamilton SJ, MacCallum DE, Hall PA, Fuller-Pace FV. J Pathol. 1998;184:351–9. doi: 10.1002/(SICI)1096-9896(199804)184:4<351::AID-PATH1235>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Chopin D. Cancer Metastasis Rev. 1999;18:31–42. doi: 10.1023/a:1006256219004. [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Nature. 1998;395:917–21. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Wei Y, Hu MH. Yi Chuan Xue Bao. 2001;28:991–6. [PubMed] [Google Scholar]
- Wilson BJ, Bates GJ, Nicol SM, Gregory DJ, Perkins ND, Fuller-Pace FV. BMC Mol Biol. 2004;5:11. doi: 10.1186/1471-2199-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. Mol Cell. 1998;2:851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu ZR. Mol Cancer Res. 2005a;3:355–63. doi: 10.1158/1541-7786.MCR-05-0022. [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu ZR. Cell Signal. 2005b doi: 10.1016/j.cellsig.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu ZR. Cell. 2006;127:139–55. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- Yang L, Liu ZR. Protein Expr Purif. 2004;35:327–33. doi: 10.1016/j.pep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Bottinger EP. Oncogene. 2005;24:5764–74. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Genes Dev. 1999;13:1924–35. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Nat Cell Biol. 2004;6:931–40. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]