Abstract
The 7-methylguanosine cap structure at the 5′-end of eukaryotic mRNAs is a critical determinant of their stability and translational efficiency1–3. It is generally believed that 5’-end capping is a constitutive process that occurs during mRNA maturation and lacks the need for a quality control mechanism to ensure its fidelity. We recently reported that the yeast Rai1 protein has pyrophosphohydrolase activity towards mRNAs lacking a 5’-end cap4. Here we show that, in vitro as well as in yeast cells, Rai1 possess a novel decapping endonuclease activity that can also remove the entire cap structure dinucleotide from an mRNA. Interestingly this activity is targeted preferentially towards mRNAs with unmethylated caps in contrast to the canonical decapping enzyme, Dcp2, that targets mRNAs with a methylated cap. Capped but unmethylated mRNAs generated in yeast cells with a defect in the methyltransferase gene are more stable in a rai1 gene disrupted background. Moreover, rai1Δ yeast cells with wild-type capping enzymes show significant accumulation of mRNAs with 5’-end capping defects under nutritional stress conditions of glucose or amino acid starvation. These findings provide evidence that 5’-end capping is not a constitutive process that necessarily always proceeds to completion and demonstrates that Rai1 plays an essential role in clearing mRNAs with aberrant 5’-end caps. We propose Rai1 is involved in a hitherto-uncharacterized quality control process that ensures mRNA 5’-end integrity by an aberrant-cap mediated mRNA decay mechanism.
Keywords: mRNA decapping, mRNA decay, mRNA quality control, decapping endonuclease
The stability and translational efficiency of eukaryotic mRNAs are significantly influenced by the 5’-end cap1–3. The cap is cotranscriptionally added and consists of a guanine nucleoside methylated at the N-7 position attached to the terminal nucleoside of the RNA by an unusual 5′-5′ pyrophosphate linkage5–6. Capping is carried out by the combination of three enzymatic activities5,7 consisting of a triphosphatase, guanylyltransferase and methyltransferase8. Three different proteins carry out the distinct activities in yeast while the triphosphatase and guanylyltransferase activities are carried out by a single bifunctional protein in mammals9. The presence of the methyl group on the cap is essential for recognition by the cap binding proteins, CBP and eIF4E as well as the scavenger decapping enzyme DcpS10–11. Removal of the cap is a regulated process catalyzed by the Dcp2 decapping enzyme to release m7GDP and monophosphate RNA12–14. The exposed 5′ monophosphate RNA is subsequently subjected to degradation by the cytoplasmic 5′ to 3′ exoribonuclease Xrn1 to clear the mRNA body15–16. Interestingly, Dcp2 functions on a cap substrate with an N7 methyl moiety and does not function on an unmethylated cap13. This latter point raises an interesting question regarding the fate of mRNAs aberrantly lacking cap methylation.
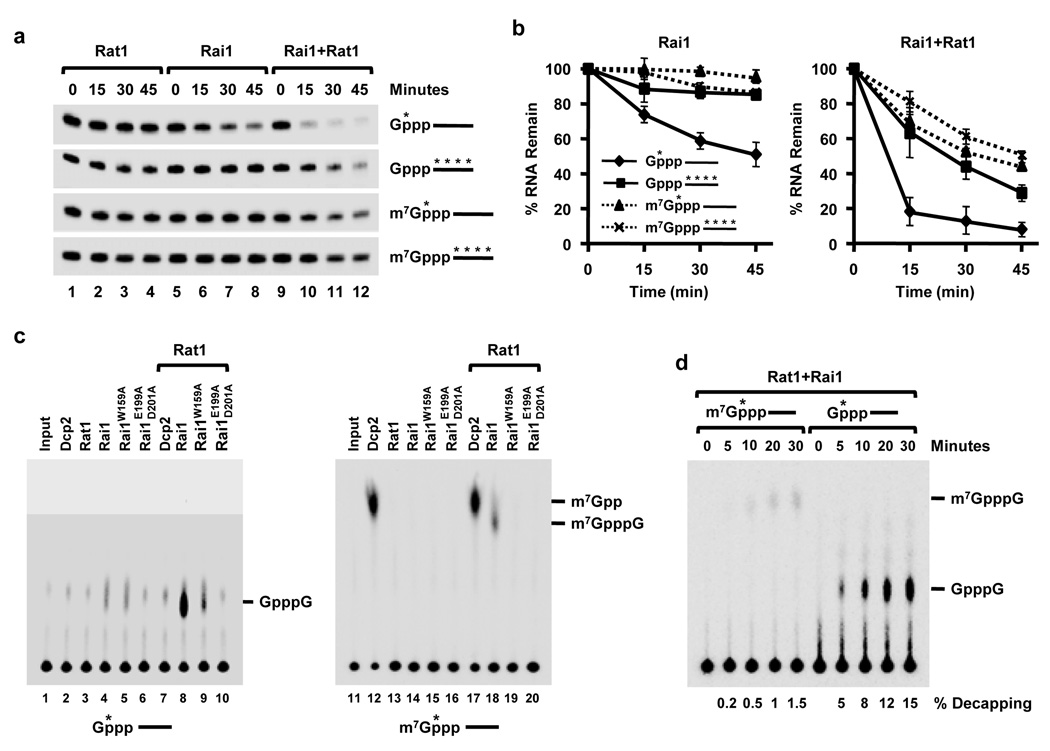
Rai1 is a pyrophosphohydrolase that hydrolyzes the 5’-end triphosphate of an uncapped RNA to release diphosphate and a monophosphorylated 5’-end RNA which could be degraded by the Rai1-interacting 5’ to 3’ nuclear exoribonuclease Rat14, suggesting the possible role of Rai1 in a quality control mechanism for 5’-end capping. To address this possibility, the activity of Rai1 on unmethylated capped mRNAs was tested. In vitro generated capped RNAs containing or lacking the N7 methyl moiety (m7GpppRNA and GpppRNA, respectively) were labeled either within the 5’ cap at the γ phosphate relative to the mRNA, or uniformly labeled throughout the RNA body, and incubated with recombinant Rat1, Rai1 or both proteins simultaneously. As expected, Rat1 did not appreciably decrease the level of either full-length RNA (Fig. 1a, lanes 1–4) since they lack the required 5’-end monophosphate. Incubation of the RNAs with Rai1, however, revealed a striking decrease in the level of 5’-end labeled unmethylated RNA (Fig. 1a lanes 5–8 and Fig. 1b), which was further stimulated by Rat1 (Fig. 1a lanes 9–12 and Fig. 1b). The preferential decay of unmethylated capped RNA was a function of Rai1; modest levels of reaction products were detected when this RNA was incubated with Rai1 protein (Fig. 1c, lanes 4) or a single point mutant compromised in its ability to interact with Rat1 (Rai1W159A; lane 5). As expected, background levels of activity were detected from the catalytically inactive Rai1 mutant protein containing substitutions in the cation binding residues4 (Rai1E199A/D201A, lane 6). Significantly, Rai1 hydrolysis activity was stimulated by Rat1 (lane 8) and the stimulation was attenuated with a mutant Rai1 compromised in its ability to interact with Rat1 (lane 9). Consistent with the decay results above, undetectable levels of reaction products were observed on 5’ end methyl-capped RNAs (lane 14) with a relatively modest increase following addition of Rat1 (lane 18). A direct comparison of methylated and unmethylated capped RNA decapping revealed at least a 10-fold greater activity on the cap lacking a methyl moiety (Fig. 1d). Moreover, the activity is restricted to capped RNA and does not function on cap structure, as hydrolysis of cap structures lacking the linked RNA was not detected (Fig. S1). Collectively, these data demonstrate that Rai1 can preferentially remove the cap from an unmethylated 5′-end capped RNA and that this activity is enhanced by Rat1.
Figure 1. Rai1 preferentially hydrolyzes unmethylated capped RNA.
(a) In vitro transcribed 32P-cap-labeled or uniform-labeled pcP RNAs with a methylated or unmethylated cap were subjected to Rat1 or Rai1 proteins and the decay of the RNAs followed at the indicated times. The RNAs used are denoted on the right and the asterisk represents the position of the 32P labeling. Quantitation of the amount of RNA remaining in the assays in (a) following Rai1 or Rai1 + Rat1 treatment are graphed in (b). Data are from three independent experiments normalized to a 32P-labeled DNA oligonucleotide loading control included in the stop buffer and presented relative to time zero. Error bars represent +/− standard deviation (SD). (c) In vitro decapping assays were carried out at 37°C for 15 min as in (a) with the indicated proteins and decapping products were resolved by PEI-TLC developed in 0.45 M (NH4)2SO4. Migration of the cap analog markers are shown on the right. Human Dcp2 was used as a methyl capped RNA decapping positive control (panels 2, 7 and panels 12, 17). (d) In vitro decapping assay using 32P-cap-labeled methylated or unmethylated 5’ capped pcP RNAs were carried out with 50nM Rat1 and Rai1 recombinant proteins for the indicated times and resolved as in (c) above. Percent decapping of three independent experiments are presented on the bottom.
Surprisingly, enzymatic tests confirmed the Rai1 decapping products corresponded to cap analogue, GpppG or m7GpppG (Fig. S2). Therefore, unlike the pyrophosphohydrolase activity of Dcp2 decapping, which releases m7Gpp12–13, or of Rai1 itself on a 5’ triphosphate RNA, which releases PPi diphosphate4, the presence of a cap guanosine on the triphosphorylated 5′-end of an RNA converts the pyrophosphohydrolase activity of Rai1 into a phosphodiesterase decapping endonuclease that releases the entire cap structure (GpppN) from an unmethylated capped RNA with comparable efficiencies (Figure S3). Nevertheless, similar to canonical decapping, the decapping endonuclease activity of Rai1 also generates a 5’ monophosphorylated RNA that is a substrate for Rat1 exoribonuclease-direct mRNA decay.
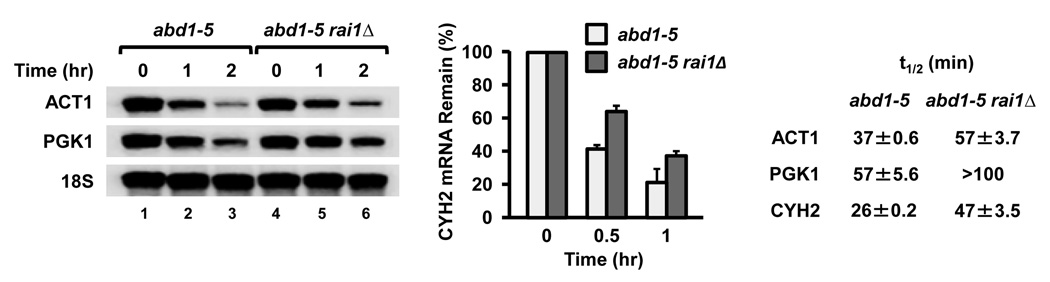
The Rai1-mediated selective hydrolysis of mRNA caps lacking the methyl residue was also observed in cells using a yeast abd1-5 strain harboring a temperature-sensitive cap methyl transferase. The abd1–5 strain is unable to methylate the 5’ cap at the nonpermissive temperature and results in the reduction of PGK1 and ACT1 mRNA levels17. We reasoned stability of mRNAs in an abd1–5 rai1Δ double mutant background should increase if Rai1 was involved in hydrolyzing aberrant unmethylated capped cellular mRNAs that would be produced at the nonpermissive temperature. Following a 45-min shift to the nonpermissive temperature to enable accumulation of unmethylated capped mRNAs, thiolutin was added to block transcription and mRNA levels were determined. Consistent with a role of Rai1 in modulating the stability of unmethylated capped mRNAs in cells, a stabilization of the PGK1, ACT1 and CYH2 mRNAs was observed in the abd1–5 rai1Δ background (Fig. 2). These data demonstrate that the methylation state of the mRNA cap can determine its susceptibility to Rai1 and that Rai1 functions to normally clear aberrant mRNAs with an unmethylated 5’ cap.
Figure 2. Rai1 preferentially hydrolyzes unmethylated 5’ end capped mRNA in yeast cells.
Yeast strains harboring the temperature-sensitive ABD1 methyltransferase mutant allele, abd1-5, or the abd1–5 rai1Δ double mutation were grown at the 37°C nonpermissive temperature for 45 min prior to transcriptional block with 5 µg/ml thiolutin. RNA was isolated from cells at the indicated time points following thiolutin addition and levels of RNA remaining determined by Northern blot analysis (PGK1 and ACT1) or RT-qPCR (CYH2). Half-lives (t1/2) of the mRNAs were determined relative to the 18S rRNA and were derived from three independent experiments. The range of half-lives obtained are consistent with previously reported thiolutin-directed transcriptional arrest measurements30.
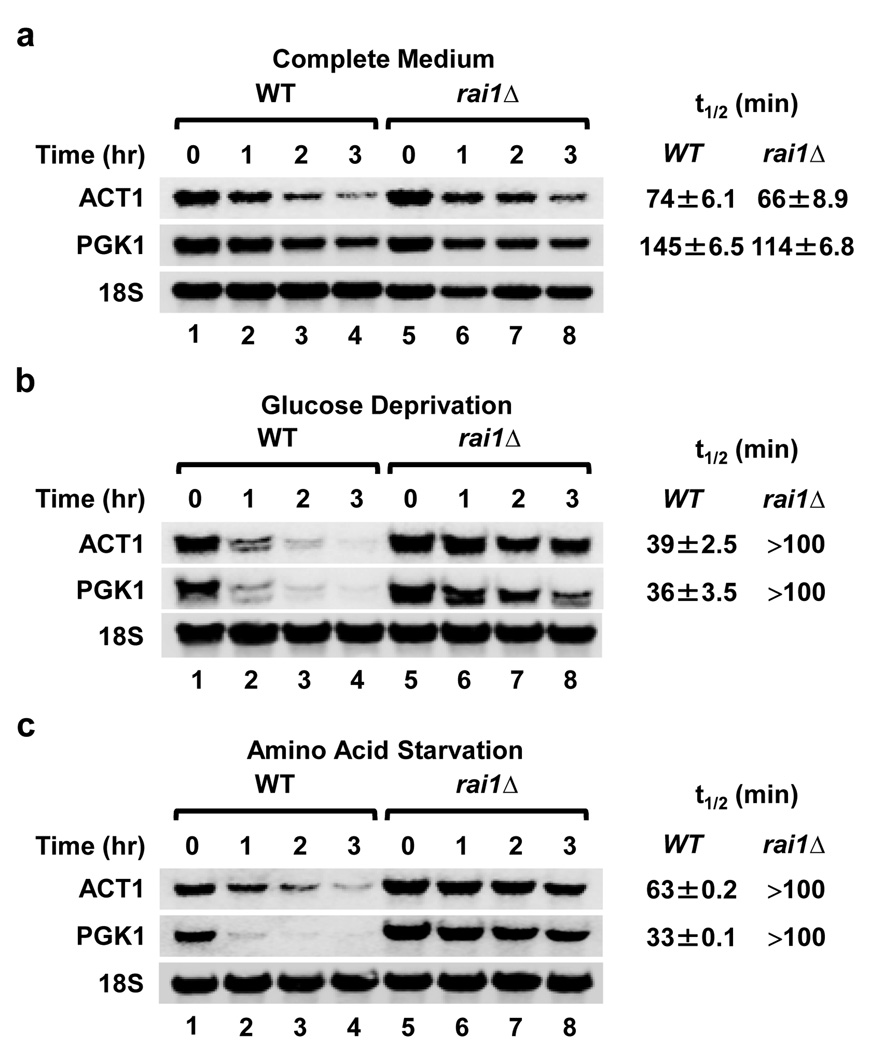
We next addressed the physiological significance of Rai1 decapping in mRNA stability. The stability of the PGK1 and ACT1 mRNAs was comparable regardless of whether Rai1 was present when cells were grown in complete media (Fig. 3a). However, significant stabilization of the mRNAs was observed when cells were exposed to nutritional stress. Both mRNAs were substantially more stable in the rai1Δ strain following glucose starvation (Fig. 3b) or amino acid starvation (Fig. 3c) relative to the wild-type strain grown in the same media. These results demonstrate the significance of Rai1 in the modulation of mRNA stability following exposure of yeast cells to stress. Although previous studies have indicated that mRNAs appear stable following nutrient stress18, the discrepancy between our results and that of others is based on the timing of the analysis where we set time zero at 45 min following removal of nutrient source rather than at the time of depletion. Incorporation of the lag period provided an opportunity for a majority of capped and methylated mRNAs that were transcribed prior to the onset of nutrient depletion to be cleared.
Figure 3. Rai1 functions to clear mRNAs in cells subjected to glucose or amino acid starvation.
Wild-type (WT) or rai1Δ yeast strains were grown in complete medium at 30°C to an OD600 of 0.6 and subsequently cultured in complete medium (a), glucose minus medium (b) or amino acid minus medium (c) for 30 min followed by addition of 5 µg/ml thiolutin. Total RNAs were isolated and detected at the indicated times following thiolutin addition by Northern Blot analysis and RNA half-lives (t1/2) quantified from three independent experiments normalized relative to the 18S rRNA are presented.
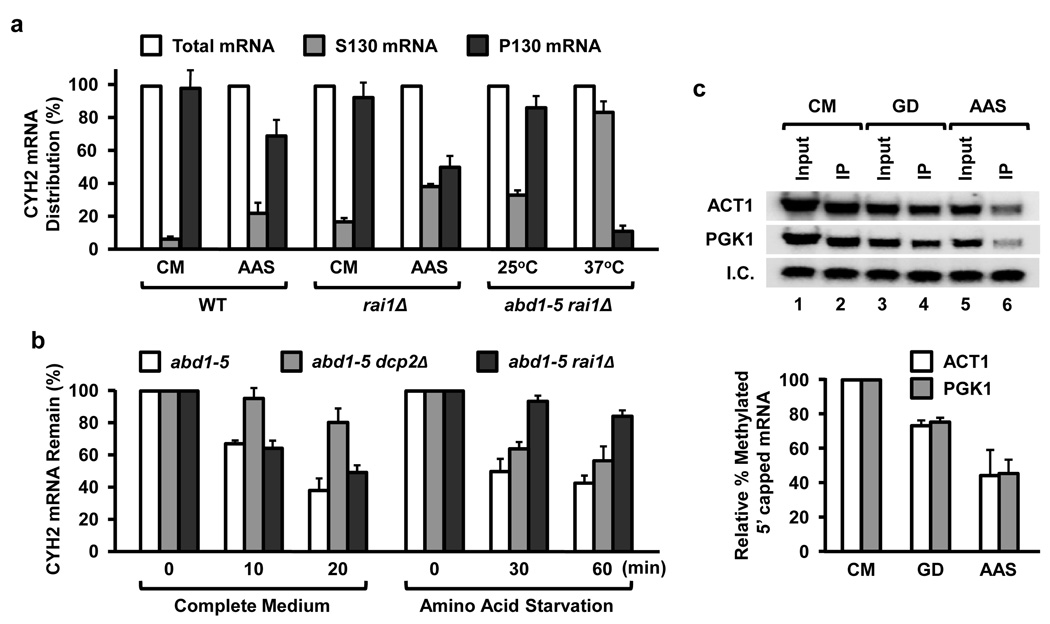
A surprising implication of these results is that mRNAs with an aberrant 5’ end are generated in yeast cells following nutrient starvation. To begin testing this hypothesis, steady-state RNA was isolated from rai1Δ cells grown for 45 min in either complete medium or nutrient-deprived medium. Cells were fractionated into polysome-containing P130 pellet and soluble S130 mRNP fraction. The relative ratio of the CYH2 mRNA distribution shifted from the polysome-containing fraction to the mRNP fraction in amino acid starved cells, and this difference was further evident in the rai1Δ strain (Fig. 4a) suggesting nutrient starvation promotes the generation of aberrantly capped mRNA. The presence of an aberrant 5’ cap following nutrient starvation was further substantiated in a strain disrupted for the methyl-cap specific decapping enzyme, Dcp2. The increased mRNA stability following nutrient starvation in the rai1Δ background was not observed in a dcp2Δ background (Fig. 4b) indicating the presence of an aberrant 5’ cap resistant to Dcp2 upon nutrient starvation. Furthermore, determination of the steady-state levels of methylated capped PGK1 and ACT1 mRNAs by immunopurification with an anti-cap column, which specifically retains methylated but not unmethylated capped RNA (Fig. S4), showed a 25% and 55% reduction in methylated species when rai1Δ cells were grown in glucose- or amino acid-deficient medium relative to cells grown in complete medium (Fig. 4c). Therefore, despite the increase in ACT1 and PGK1 mRNA stabilities in the rai1Δ cells upon nutrient starvation (Fig. 3), a decrease in the extent of methylated capped mRNA is observed under these conditions (Fig. 4c), indicating that nutrient starvation leads to the accumulation of mRNAs with aberrant 5’ ends. In addition, absence of the methyl moiety does not appear to be a consequence of demethylation following proper capping, since the ratio of spliced and unspliced CYH2 mRNAs containing an aberrant cap remained constant under both normal and nutrient starvation (Fig. S5). Therefore, the aberrant cap does not appear to be generated from a methylated cap precursor.
Figure 4. Aberrantly capped mRNA levels increase in cells exposed to nutrient starvation.
(a) Amino acid starvation shifts mRNAs into the soluble mRNP fraction. The mid-log phase yeast strains were shifted to the indicated medium and grown for 45 min prior to fractionation. RNA was isolated from polysome-containing fractions sedimenting at 130,000 × g (P130) and the supernatant (S130) fraction, which contained the soluble mRNP. Distribution of the CYH2 mRNA from each fraction was determined by quantitative RT-PCR. The abd1–5 rai1Δ double mutant strain grown at the permissive 25°C (methylated capped mRNA) or non-permissive 37°C (unmethylated capped mRNA) were used as a positive control. Results of three independent experiments are presented with error bars denoting +/− SD. (b) Aberrantly capped mRNAs are minimally affected by the Dcp2 decapping enzyme. The indicated strains were grown in complete medium at 22°C to an OD600 of 0.6 and subsequently cultured in the same medium or amino acid minus medium for 45 min followed by the addition of thiolutin. The levels of CYH2 mRNA were determined by quantitative RT-PCR as in (a) above. (c) Methylated capped RNA was immunopurified utilizing monoclonal anti-trimethylguanosine antibody column from cells grown at the denoted culture conditions for 45 min and were detected by Northern Blot analysis. Quantitations for the mRNA cap methylation were normalized to total input RNA and 32P-labeled methylated capped pcP RNA internal control (I.C.) and derived from three independent experiments. The error bars represent +/− SD.
Overall, we have shown that mRNA cap methylation is a regulated process that can be diminished upon exposure of yeast cells to glucose or amino acid starvation. Our data, combined with reports demonstrating a stimulation in general cap methylation by Impα19 or specific methylation by c-Myc20 and transcriptionally-linked uncapped RNAs21 indicates that addition of the cap and methyl moiety are regulated processes that impact overall gene expression. Moreover, the accumulation of aberrantly capped mRNAs upon nutrient starvation and the significance of Rai1 in clearing these mRNAs define a novel regulatory mechanism to potentially control the translation of mRNA transcribed during stress. Furthermore, Rai1 defines a novel class of decapping endonuclease that specifically removes the cap structure and promotes the decay of mRNAs possessing unmethylated capped 5’-end. The monophosphorylated 5’-end containing mRNA products generated by Rai1 are likely degraded by Rat1, although cytoplasmic recapping22 and recycling into the translationally competent mRNA pool cannot be ruled out. Collectively, our data suggest Rai1 initiates a quality control mechanism that clears aberrant 5’ end-containing mRNAs, whether they lack a cap4 or contain an unmethylated cap, and defines a new aberrant-cap mediated mRNA decay surveillance mechanism that functions under normal and stress conditions to ensure the integrity of mRNA 5’-ends.
METHODS SUMMARY
RNA in vitro Decay Assay
The indicated recombinant proteins (50 nM) were incubated with 32P-cap-labeled or 32P-uniform-labeled pcDNA2 polylinker (pcP) generic RNA containing either m7Gppp or Gppp at the 5’ end in decapping buffer as previously described23. The decay products were resolved by 8% urea denaturing polyacrylamide gel electrophoresis to visualize the RNA or by polyethyleneimine-cellulose TLC (PEI-TLC) plates developed in 0.45 M (NH4)2SO4 at room temperature to detect decapping products as noted. Quantifications were carried out using a Molecular Dynamics PhosphorImager (Storm860) with ImageQuant-5 software.
Yeast growth conditions and RNA isolation
Yeast strains were grown in complete medium at 30°C to mid-log and switched to medium lacking glucose or amino acids as described24–25. Following 30-min growth in the new medium conditions, transcription was terminated with the addition of thiolutin (5 µg/ml) and cells were harvested at 0, 1, 2 or 3 hr time points. In Fig. 2, abd1–5 temperature sensitive strains were grown in complete medium lacking tryptophan until mid-log phase at 25°C and shifted to the nonpermissive 37°C for 45 min to abolish methyl transferase activity. Cells are viable for several hours at 37°C17. In Fig. 4b, strains were cultured at 22°C to further minimize the ts phenotype. The cells were harvested by centrifugation and stored as pellets at −80°C. Total RNAs were isolated with the acidic hot phenol method26.
Cap antibody immunoprecipitations and Northern blotting
Methylated capped mRNA was immunoprecipitated (IP) from 20µg yeast total RNA with an agarose-conjugated 2,2,7-trimethylguanosine antibody column (Calbiochem, San Diego, CA) which immunpurifies monomethyl capped mRNA as previously described27–28. RNAs were detected by Northern blotting carried out with [α-32P]dCTP-labeled gene specific DNA probes as described29.
Methods
Yeast Strains
The genotypes of all the Saccharomyces cerevisiae strains used in this study are listed in Supplemental Table 1. The abd1–5 strain has been reported31. The rai1 gene disruption construct was isolated from the chromosomal DNA −708 to +600 of yeast knock-out strain YGL246C using PCR amplification by forward primer 5’-CCATTTCTAACAAAGTGTACCAACGAGAAACG-3’ and reverse primer 5’-CCGCAAGATGCTAGATTAGCCCGAC-3’. The dcp2 gene disruption construct was isolated from the chromosomal DNA of a heterozygous diploid dcp2 knockout strain YNL118C by PCR amplification with a forward primer 5’-AGCTCATAGATAATCGTCGTAAGGCTGACAC-3’ and a reverse primer 5’-TCAAGTATGGCTAAGCCGTCACAATGTC-3’. The amplified fragments were inserted into pSTBlue-1 vector (EMD, Germany) to create the plasmids pSTB-raiΔ::kanMAX4 and pSTB-dcp2Δ::kanMAX4. The raiΔ mutant strain and abd1–5 raiΔ or abd1–5 dcp2Δ double mutant strains were generated by transformation of ABD1 wild-type and abd1–5 strains respectively with either the rai1 or dcp2 disruption cassette DNA fragments obtained by digesting pSTB-raiΔ::kanMAX4 with HpaI or digesting pSTB-dcp2Δ::kanMAX4 with KpnI/XhoI.
Protein Expression and Purification
Recombinant His-tagged Rat1, Rai1, and Rai1 mutant (Rai1W156A and Rai1E199A/D201A) proteins were expressed in E. coli BL21 (DE3) Rosetta cells with pET26b vector (EMD, Germany) and purified by Ni-NTA (Qiagen, Valencia, CA) and gel filtration (Sephacryl S-300, GE Healthcare) chromatography as described previously32.
RNA Generation
The pcDNA3 polylinker was amplified by PCR with SP6 promoter primer and T7 promoter primer containing 16 cytosines at the 5′ end and was used as template to reverse transcript RNA with SP6 RNA polymerase as previously described to generate the pcP RNA33. The resulting RNA following transcription with SP6 polymerase will yield an RNA containing 16 guanosines at the 3’ end to minimize 3’ end endonucleolytic decay34. 5’-end N7-methylated or unmethylated 32P-cap-labeled pcP RNAs were generated with the vaccinia virus capping enzyme by the inclusion or omission of the methyl donor S-adenosylmethionine (SAM) in the presence of [α-32P]GTP35. 5’-end capped N7-methylated or unmethylated 32P-uniform-labeled pcP RNAs were generated by transcription with SP6 RNA polymerase under standard conditions that included [α-32P]UTP and either m7GpppG or GpppG cap analog in the respective reactions.
RNA in vitro Decay Assay
His-tagged Rat1, Rai1 or Rai1 mutant recombinant proteins (50 nM) were incubated with 32P-cap-labeled or 32P-uniform-labeled pcP RNAs in decapping buffer as previously described36. Following incubation at 37°C for the indicated times, the decay reactions were terminated with phenol:chloroform extractions and resolved by 8% denaturing polyacrylamide gel electrophoresis35 or polyethyleneimine-cellulose TLC (PEI-TLC) plates developed in 0.45 M (NH4)2SO4 at room temperature37. The dried gels or TLC plates detected by a Molecular Dynamics PhosphorImager (Storm860) and quantified using ImageQuant-5 software.
Yeast growth conditions and RNA isolation
Yeast strains were grown in standard media lacking tryptophan until mid-log phase at 25°C. Following pelleting and washing, the cells were resuspended into pre-warmed standard media lacking tryptophan, or glucose deprivation medium, or Yeast Nitrogen Base (YNB) amino acid deprivation medium (Invitrogen, Carlsbad, CA) for nutrient stress conditions as noted. Following a 45 min incubation at 30°C, transcription was stopped by the addition of transcriptional inhibitor thiolutin (Sigma-Aldrich) to a final concentration of 5 µg/ml. The cells were harvested 0, 1, 2, 3 hr post-thiolutin addition and RNAs isolated. abd1–5 and abd1–5 raiΔ strains were grown at 25°C in standard media lacking tryptophan until mid-log phase and shifted to 37°C for 2 hr to abolish methyl transferase activity; thiolutin was added and RNA isolated at the indicated times. Yeast total RNAs were isolated with the acidic hot phenol method38 with modifications as described39.
Northern Blotting
Twenty micrograms of total yeast RNA isolated from the indicated strains and time points were resolved on a 1% formaldehyde denaturing agarose gel and transferred to a Hybond-N membrane (GE Healthcare Life Science, NJ, USA) for Northern Blot analysis as described previously40. Specific mRNAs were detected by [α-32P]dCTP-labeled gene specific DNA probes. Quantifications were carried out using a Molecular Dynamics PhosphorImager (Storm860) using ImageQuant-5 software.
Real-Time Quantitative Reverse Transcription and PCR
Yeast total RNA isolated with acidic hot phenol method or immunoprecipiation-isolated RNAs were reverse transcribed into cDNA with M-MLV reverse transcriptase (Promega) according to the manufacturer's instructions. Real-time PCR was performed with iTaq Supermix (Biorad) according to the manufacturer's instructions. Data were computed by the comparative Ct method. The CYH2 mRNA was amplified with forward primer 5’-TAGAGGTATGGCCGGTGGTC-3’ and reverse primer 5’-ACCCAAGATCTTACCGTAACC-3’. The CYH2 intron containing pre-mRNA was amplified with forward primer 5’-TCAAATGGTTGTAGAGAGCGC-3’ and reverse primer 5’-AGTTCAAGACTGGCTTCCAG-3’. Forward primer 5’-TTCTGGCTAACCTTGAGTCC-3’ and reverse primer 5’-AAAACGTCCTTGGCAAATGC-3’ were used for amplify the internal control 18S rRNA.
P130 and S130 fraction
Yeast cultures grown to mid-log, were transferred to either complete medium or nutrient-deprived medium and grown an additional 45 min prior to harvesting. The cells were harvested and total extracts were generated with the glass bead method41 in the presence of 0.1 mg/ml cycloheximide, 1× protease inhibitor (Roche, Germany) and 0.8 U/µl RNase inhibitor (Promega). The resulting extract was fractionated into a polysome-containing P130 pellet and soluble S130 mRNP fraction by centrifugation at 130,000 × g at 4°C for 30 min. RNAs were isolated as described above.
Cap analogue generation, phosphatase and NDPK treatments
32P-labeled cap analogs m7GpppG, GpppG and GpppGp were generated by hydrolyzing 32P-cap labeled methylated or unmethylayed pcP RNA (m7G*ppp RNA or G*ppp RNA) with nuclease P1 (Roche) or RNase T1 (Roche) as previously described37. Phosphatase and NDPK treatments were carried out with 1 U calf intestinal alkaline phosphatase (NEB) or 1 U nucleoside diphosphate kinase (Sigma) in the presence of 1 mM ATP at 37°C for 30 min as previously described34.
Supplementary Material
Acknowledgements
We thank B. Schwer for the abd1–5 yeast strain and A. Shatkin for helpful discussions. This research was supported by grants from the NIH to LT (GM077175) and MK (GM67005).
Footnotes
Full Methods and any associated references are available in the online version of the paper at …‥
Supplementary Information is linked to the online version of the paper at …….
Author Contributions. X.J. and M.K. conceived the project, analyzed the data and wrote the manuscript. X.J. carried out the experiments. S.X. and L.T. provided the recombinant proteins. C.O. and C.E.M. generated the yeast mutant strains. All authors discussed the results and commented on the manuscript.
The authors declare no competing financial interests.
References
- 1.Liu H, Kiledjian M. Decapping the message: a beginning or an end. Biochem Soc Trans. 2006;34:35–38. doi: 10.1042/BST20060035. [DOI] [PubMed] [Google Scholar]
- 2.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 3.Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 4.Xiang S, et al. Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 7.Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 8.Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat Rev Mol Cell Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- 9.Yue Z, et al. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci U S A. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodfellow IG, Roberts LO. Eukaryotic initiation factor 4E. Int J Biochem Cell Biol. 2008;40:2675–2680. doi: 10.1016/j.biocel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer PM. Cap in hand: targeting eIF4E. Cell Cycle. 2009;8:2535–2541. doi: 10.4161/cc.8.16.9301. [DOI] [PubMed] [Google Scholar]
- 12.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. Embo J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 16.Hsu CL, Stevens A. Yeast cells lacking 5'-->3' exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5' cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwer B, Saha N, Mao X, Chen HW, Shuman S. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics. 2000;155:1561–1576. doi: 10.1093/genetics/155.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilgers V, Teixeira D, Parker R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen Y, Shatkin AJ. Cap methyltransferase selective binding and methylation of GpppG-RNA are stimulated by importin-alpha. Genes Dev. 2000;14:2944–2949. doi: 10.1101/gad.848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol. 2007;27:2059–2073. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jimeno-Gonzalez S, Haaning LL, Malagon F, Jensen TH. The yeast 5'-3' exonuclease Rat1p functions during transcription elongation by RNA polymerase II. Mol Cell. 2010;37:580–587. doi: 10.1016/j.molcel.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Otsuka Y, Kedersha NL, Schoenberg DR. Identification of a cytoplasmic complex that adds a cap onto 5'-monophosphate RNA. Mol Cell Biol. 2009;29:2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piccirillo C, Khanna R, Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. Rna. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natarajan K, et al. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 27.He F, Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol. 2001;21:1515–1530. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Kiledjian M. Scavenger decapping activity facilitates 5' to 3' mRNA decay. Mol Cell Biol. 2005;25:9764–9772. doi: 10.1128/MCB.25.22.9764-9772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiao X, et al. Modulation of neuritogenesis by a protein implicated in X-linked mental retardation. J Neurosci. 2009;29:12419–12427. doi: 10.1523/JNEUROSCI.5954-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwer B, Saha N, Mao X, Chen HW, Shuman S. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics. 2000;155:1561–1576. doi: 10.1093/genetics/155.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang S, et al. Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl Acad. Sci. USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piccirillo C, Khanna R, Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2c. RNA. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Meth. Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 39.Amin-ul Mannan M, Sharma S, Ganesan K. Total RNA isolation from recalcitrant yeast cells. Anal. Biochem. 2009;389:77–79. doi: 10.1016/j.ab.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Jiao X, et al. Modulation of neuritogenesis by a protein implicated in X-linked mental retardation. J. Neurosci. 2009;29:12419–12427. doi: 10.1523/JNEUROSCI.5954-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S, Williams CJ, Wormington M, Stevens A, Peltz SW. Monitoring mRNA decapping activity. Methods. 1999;17:46–51. doi: 10.1006/meth.1998.0706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.