Abstract
The serine protease inhibitor plasminogen activator inhibitor-1 (PAI-1) is increased in several cancers, including breast, where it is associated with a poor outcome. Metastatic breast cancer has a dismal prognosis, as evidenced by treatment goals that are no longer curative but are largely palliative in nature. PAI-1 competes with integrins and the urokinase plasminogen activator receptor on the surface of breast cancer cells for binding to vitronectin. This results in the detachment of tumor cells from the extracellular matrix, which is critical to the metastatic process. For this reason, we sought to isolate RNA aptamers that disrupt the interaction between PAI-1 and vitronectin. Through utilization of combinatorial chemistry techniques, aptamers have been selected that bind to PAI-1 with high affinity and specificity. We identified two aptamers, WT-15 and SM-20, that disrupt the interactions between PAI-1 and heparin, as well as PAI-1 and vitronectin, without affecting the antiprotease activity of PAI-1. Furthermore, SM-20 prevented the detachment of breast cancer cells (MDA-MB-231) from vitronectin in the presence of PAI-1, resulting in an increase in cellular adhesion. Therefore, the PAI-1 aptamer SM-20 demonstrates therapeutic potential as an antimetastatic agent and could possibly be used as an adjuvant to traditional chemotherapy for breast cancer.
Introduction
In the United States, the most prevalent cancer and second most common cause of death due to cancer in women is breast cancer (Cancer Statistics, 2007). Once detected, staging of the cancer ensues, which determines the therapeutic course of action. In addition to surgery, several classes of therapeutic agents can be utilized, depending on the nature of the cancer. However, many of these therapies, especially chemotherapeutic agents, cause side effects that greatly diminish the patient's quality of life. Moreover, if metastases are discovered, treatment goals are no longer curative but are largely palliative in nature, almost exclusively seeking to improve the patient's comfort as much as possible. Therefore, to prevent progression to this stage of disease, a nontoxic anticancer agent that represses metastasis could serve as an adjuvant to traditional therapeutic agents, possibly reducing the dose necessary for cytotoxicity while lessening the adverse effects associated with such therapies.
The serpin plasminogen activator inhibitor-1 (PAI-1; SERPINE1) is the main physiological inhibitor of the fibrinolysis system, as PAI-1 binds to and inhibits tissue-type plasminogen activator (uPA) and urokinase-type plasminogen activator (uPA). This results in inhibition of the conversion of plasminogen to plasmin. Plasmin cleaves pro-matrix metalloproteases (proMMPs), thereby permitting the invasion and migration of tumor cells through the MMP-mediated proteolytic breakdown of extracellular matrix proteins (for review see Dano et al., 2005; Beaulieu et al., 2007). Plasmin also directly cleaves components of the extracellular matrix. Plasminogen activation is greatly enhanced when uPA undergoes high affinity binding to the urokinase plasminogen activator receptor (uPAR). Since PAI-1 inhibits uPA activity, one would logically assume that an increase in PAI-1 would decrease tumor invasiveness. Surprisingly, this is not the case, as increases in expression levels of PAI-1, uPA, and uPAR are associated with a poor prognosis for several cancers including breast, brain, lung, bladder, liver, and pancreatic cancers (Dano et al., 2005; Lah et al., 2006). Many investigators have focused on blocking the interaction of selected components of the plasminogen activator (PA) system given its role in promoting tumor cell progression (Min et al., 1996; Ploug et al., 2001; Gondi et al., 2003; Pulukuri et al., 2005; Subramanian et al., 2006). For example, direct inhibition of uPA with a low-molecular-weight inhibitor retards tumor cell growth and metastasis (Zhu et al., 2007). Likewise, low-molecular-weight inhibitors of PAI-1 have successfully suppressed cancer cell invasion and angiogenesis (Leik et al., 2006). PAI-1 has also been proven responsible for the regulation of cellular adhesion and migration, and has been shown to interact with several extracellular matrix components (Chorostowska-Wynimko et al., 2004; Dellas and Loskutoff, 2005). Based on these qualities, many consider PAI-1 to be both a key regulator of tumor progression and an ideal target for cancer therapy.
In plasma, PAI-1 exists in two major forms, active and latent. While latent PAI-1 is functionally inactive, active PAI-1 effectively inhibits target proteases but is labile as it spontaneously converts to the latent form (Hekman and Loskutoff, 1985; Levin and Santell, 1987; Lindahl et al., 1989). To prevent this conversion, active PAI-1 binds to the extracellular matrix protein, vitronectin, which is unable to bind inactive PAI-1 or PAI-1 in complex with its target proteases (Declerck et al., 1988; Seiffert et al., 1994; Deng et al., 1996). Vitronectin also facilitates cell adhesion by binding to integrins and to surface-bound uPA (Wei et al., 1996; Chapman and WEI, 2001; Wei et al., 2001). PAI-1 competes with integrins and uPAR for vitronectin binding, resulting in the detachment of cells from the extracellular matrix (Deng et al., 1996; Stefansson and Lawrence, 1996; Kjoller et al., 1997). Therefore, the binding of PAI-1 to vitronectin prevents integrins from binding to vitronectin and inhibits cell adhesion in some cells, particularly breast cancer cells (Deng et al., 1996; Stefansson and Lawrence, 1996; Kjoller et al., 1997; Redmond et al., 2001; Palmieri et al., 2002). Therefore, the concentration of PAI-1, as well as the presence or absence of vitronectin, plasminogen activators, thrombin, and low-density lipoprotein receptor-related protein (LRP), all appear to play a role in the PAI-1-mediated effect on cell migration.
To shift the balance of the PA system toward cell adhesion, we exploited the properties of nucleic acid ligands termed aptamers. Aptamers are relatively small (~8–15 kDa) single-stranded nucleic acid ligands that bind to their targets with high affinity and specificity. In addition, they are nontoxic, have low to no immunogenicity, can be synthetically manufactured, and have adjustable bioavailability (Eyetech Study Group, 2002; Nimjee et al., 2005). Therefore, aptamers possess the affinity properties of monoclonal antibodies, while retaining the ease of synthesis of small molecules. For these reasons, aptamers are ideal agents for disrupting protein–protein interactions. Following the 2004 FDA approval of the first aptamer-based therapeutic agent, pegaptanib, a number of aptamers have entered clinical trials; for example, the aptamer targeting the interaction between Factor IXa and its substrate Factor X is currently in Phase 2 clinical trials as an antithrombotic agent with applications for percutaneous coronary intervention and generalized surgical anticoagulation (Eyetech Study Group, 2003; Dyke et al., 2006; Chan et al., 2008a, 2008b). In this study, we hypothesized that RNA aptamers that target the PAI-1/vitronectin interaction would increase cancer cell adhesion, which may translate to a clinical application in reducing breast cancer metastasis. To test this hypothesis, we generated aptamers that specifically bind to PAI-1 with high affinity (Kd < 4 nM) using SELEX (systematic evolution of ligands by exponential enrichment). Through functional assays including mutagenesis studies, competition binding, chromogenic assays, and cell adhesion experiments, we determined that aptamers WT-15 and SM-20 bind to PAI-1 in a region that spans the heparin- and vitronectin-binding domains, inhibiting the PAI-1/vitronectin interaction. Moreover, addition of SM-20 resulted in an increase in breast cancer cell adhesion. This aptamer molecule has therefore demonstrated potential as a novel antimetastatic therapeutic agent, which upon further development could potentially serve as adjuvant therapy for breast cancer.
Materials and Methods
Generation of aptamers using systematic evolution of ligands by exponential enrichment (SELEX)
The sequence of the starting RNA library (denoted Sel2 Library) was 5′-GGGAGGACGATGCGG-N40-CAGACG ACTCGCTGAGGATCC-3′, where “N40” denotes a random region 40-nucleotides in length. The RNA consisted of 2′-fluropyrimidines (2′-flurocytidine triphosphate and 2′-flurouridine triphosphate; Trilink Biotechnologies, San Diego, CA) in order to render the RNA nuclease resistant. Two separate selections against PAI-1 were performed: one selection used human wild-type PAI-1 (hWT PAI-1) protein, while the other used human stable mutant PAI-1 (hSM PAI-1), which contains four amino acid modifications (K154T, Q319L, M354I, and N150H) that hold PAI-1 in an active conformation (both kindly supplied by Dr. David Ginsburg, University of Michigan Medical Center) (Berkenpas et al., 1995). For both selections, the RNA library was incubated with its respective protein for 15 minutes at 37°C; unbound RNA was separated from RNA/PAI-1 complexes by passing the mixture over a nitrocellulose membrane. The selection against WT PAI-1 began in BSA binding buffer E (20 mM Hepes, pH 7.4, 50 mM NaCl, 2 mM CaCl2, and 0.1% bovine serum albumin [BSA]) and continued in this buffer for five rounds. Rounds 6c and 7c were performed in CHAPS binding buffer E (20 mM Hepes, pH 7.4, 50 mM NaCl, 2 mM CaCl2, and 0.05% CHAPS), and rounds 8c and 9c were performed in CHAPS binding buffer F (20 mM Hepes, pH 7.4, 150 mM NaCl, 2 mM CaCl2, and 0.05% CHAPS). The selection against hSM PAI-1 began in BSA binding buffer E and continued in this buffer for five rounds. Rounds 6c and 7c were performed in CHAPS binding buffer E/F (20 mM Hepes, pH 7.4, 100 mM NaCl, 2 mM CaCl2, and 0.05% CHAPS), and rounds 8c and 9c were performed in CHAPS binding buffer F.
Binding assays
Round and clone binding
Affinity constants (Kd values) were determined using nitrocellulose filter binding assays as previously described (Rusconi et al., 2000). Briefly, RNA was first dephosphorylated using bacterial alkaline phosphatase (Gibco BRL, Gatihersburg, MD) and 5′ end labeled with [γ-32P] ATP (PerkinElmer Life And Analytical Sciences, Inc., Waltham, MA) using T4 polynucleotide kinase (New England Biolabs, Beverly, MA). Round pools of RNA were incubated with increasing amounts of their respective PAI-1 protein in BSA binding buffer E, CHAPS binding buffer E, CHAPS binding buffer E/F, or CHAPS binding buffer F, depending on the initial round conditions. Following cloning to yield individual aptamer sequences, all binding was performed in BSA binding buffer F (20 mM Hepes, pH 7.4, 150 mM NaCl, 2 mM CaCl2, and 0.1% BSA). Radiolabeled RNA was incubated with PAI-1 protein for 5 minutes at 37°C; the mixture was then passed over a nitrocellulose membrane. The fraction of RNA that bound to the protein was quantified using a phosphoimager (Molecular Dynamics, Sunnyvale, CA). Nonspecific binding of the RNA was subtracted out such that only specific binding remained (denoted “corrected fraction bound”).
Competition binding
To determine if heparin and vitronectin were able to compete with aptamers WT-1, WT-15, and SM-20 for binding to PAI-1, the aptamers were first dephosphorylated and radiolabeled as described earlier. Briefly, a constant amount of PAI-1 (378 pM for WT-1, 888 pM for WT-15, and 3.4 nM for SM-20 when competing with heparin; 888 pM for WT-15 and 2.2 nM for SM-20 when competing with vitronectin) was incubated with increasing amounts of heparin or vitronectin at 37°C for 5 minutes. The PAI-1/competitor solution was then combined with a trace amount of radiolabeled aptamer and incubated at 37°C for 5 minutes before passing the mixture over a nitrocellulose membrane. The fraction of RNA bound to the protein was then quantified using a phosphoimager (Molecular Dynamics, Sunnyvale, CA).
PAI-1 complex formation
PAI-1 forms a high-molecular-weight, SDS-stable complex with tPA, uPA, and thrombin/heparin (Ehrlich et al., 1991; Perron et al., 2003) that can be visualized by SDS-PAGE. For that reason, we used SDS-PAGE analysis to evaluate the influence of the aptamers on the reaction products generated during the interaction of PAI-1 with its target proteases. The aptamers (500–1000 nM) were incubated with PAI-1 (500 nM; Molecular Innovations, Novi, MI) at 37°C for 10 minutes in HNPN buffer (20 mM Hepes, 150 mM NaCl, 0.01% PEG, 0.005% sodium azide) containing CaCl2 (2 nM), prior to the addition of proteases (tPA [Molecular Innovations, Novi, MI] or thrombin [Haematologic Technologies, Inc., Essex Junction, VT]). The aptamers were first heated to 65°C for 5 minutes before use. PAI-1/aptamer was then incubated with tPA (400 nM) for 15 minutes or with thrombin/heparin (200 nM and 10 μg/mL heparin) for 60 minutes at 37°C. The reactions were stopped through the addition of Laemmli buffer containing β-mercaptoethanol and were boiled for 5 minutes. The reactions were finally analyzed by SDS-PAGE followed by Coomassie staining to visualize PAI-1, thrombin, and tPA bands, as well as PAI-1/tPA and PAI-1/thrombin complexes.
Solid-phase vitronectin studies
Solid-phase PAI-1 binding studies were performed according to Lawrence et al., with slight modification (Lawrence et al., 1990). Briefly, 96-well plates used for immunochemistry (Nunc Immuno-module, Nalge Nunc International, Thermo Fisher Scientific, Rochester, NY) were coated with 150 μL of vitronectin (Molecular Innovations, Inc., Novi, MI) at 250 ng/mL and allowed to incubate overnight at 4°C. The wells were then washed three times with 300 μL PBST (phosphate buffered saline with 0.01% Tween 20). Following drying at room temperature (RT) for 15 minutes, the plate was blocked with 3% BSA for 1 hour at RT, followed by three washes with PBST. After heating an increasing amount of WT-15 and SM-20 (as well as the Sel2 library as a negative control) to 65°C for 5 minutes and allowing for cooling, the aptamers were added to 200 ng/mL PAI-1-A (active fraction of wild-type PAI-1, Molecular Innovations, Inc., Novi, MI) for a final volume of 200 μL. The RNA/PAI-1 mixture was incubated at RT for 10 minutes prior to its addition to the blocked plate, which then incubated at RT for 30 minutes. The plate was then washed three times with PBST and blotted on a paper towel, and 150 μL uPA (Molecular Innovations, Inc., Novi, MI) was added to the plate for a final concentration of 1 nM; this was incubated at RT for 30 minutes. Next, 50 μL S-2444 (DiaPharma, West Chester, OH), the chromogenic substrate for uPA activity, was added to the plate, which was then immediately placed into a microplate reader (BioTek EL311; BioTek Instruments, Inc., Winooski, VT) and set for kinetic readings every 35 seconds for 15 minutes at an absorbance of 405 nm. All RNA and protein were diluted in PBS with calcium and magnesium.
Thrombin activity assays
Heparin-mediated PAI-1 inhibition of thrombin
Increasing amounts of aptamer clones SM-20 and WT-15 (as well as the Sel2 library as a negative control) were heated to 65°C for 5 minutes and incubated with 50 nM WT PAI-1-A (active fraction of wild-type PAI-1; Molecular Innovations, Inc., Novi, MI) for 10 minutes at room temperature. Heparin (Diosynth Oss, the Netherlands; 10 μg/mL) and 1 nM α-thrombin (Haematologic Technologies, Inc., Essex Junction, VT) were then added to the RNA/PAI-1 mixture to bring the final volume to 100 μL and incubated at room temperature for 30 minutes. Residual thrombin activity was determined by cleavage of the chromogenic reagent specific for thrombin activity, S-2238 (DiaPharma, West Chester, OH) (150 μM). The plate was then immediately placed into a microplate reader (BioTek EL311; BioTek Instruments, Inc., Winooski, VT) and set for kinetic readings every 35 seconds for 15 minutes at an absorbance of 405 nm. All RNAs and proteins were diluted in HNPN buffer (20 mM Hepes, 150 mM NaCl, 0.01% PEG, 0.005% sodium azide).
Vitronectin-mediated PAI-1 inhibition of thrombin
When vitronectin was used to mediate PAI-1 inhibition of thrombin, the protocol remained unchanged with the exception that 10 μg/mL vitronectin (Molecular Innovations, Inc., Novi, MI) was used as opposed to heparin.
Cell adhesion assay
All cell adhesion assays were performed in 96-well microtiter plates that were coated with 150 μL vitronectin (Molecular Innovations, Inc., Novi, MI) at 7.5 μg/mL for 1 hour at 37°C. The plates were washed three times with warm PBS, and blocked for 1 hour at 37°C with 1% heat-inactivated BSA. The plates were then washed three times with PBS, air-dried, and either used immediately or wrapped in parafilm, and stored at 4°C. Aptamers WT-15 and SM-20 (as well as the Sel2 library as a negative control) (50–1000 nM) were heated to 65°C for 5 minutes prior to adding 50 nM PAI-1-A (Molecular Innovations, Inc., Novi, MI) for a final volume of 50 μL. The RNA/PAI-1 mixture was then incubated at 37°C for 15 minutes. MDA-MB-231 breast cancer cells were seeded at 5 × 104 cells/well in 200 μL serum-free medium onto vitronectin-coated plates and were allowed to attach for 1 hour at 37°C in the presence of PAI-1 or the RNA/PAI-1 complex. Following incubation, the nonadherent cells were removed with a multichannel pipette, and adhered cells were washed twice with warm PBS. To determine the quantity of adherent cells, an MTT assay was performed. Briefly, 20 μL of MTT (5 mg/mL) in 200 μL of serum-free media was added to each well and incubated for 3 hours at 37°C. Following incubation, the media was removed and 100 μL dimethyl sulfoxide (DMSO; Sigma, St. Louis, MO) was added to solubilize the formazan crystals for 15–30 minutes at room temperature. The absorbance was then measured at 570 nm in a VersaMax microtiter plate reader (Molecular Devices). The background absorbance was measured at 690 nm, which was subtracted from the absorbance at 570 nm.
Results
Nine rounds of SELEX yielded aptamers that bind to PAI-1 with high affinity
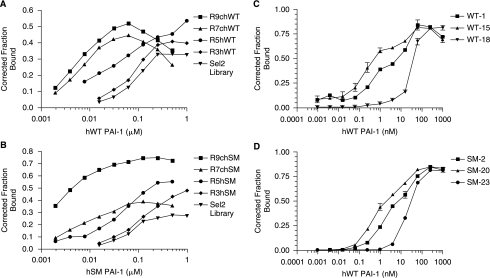
In order to generate aptamers that bound to the active form of PAI-1 with high affinity, SELEX was performed against two versions of human PAI-1. The native wild-type form of PAI-1 was used (hWT PAI-1), as well as a stable mutant version (hSM PAI-1) that contains the amino acid changes K154T, Q319L, M354I, and N150H that stabilize the otherwise labile protein in an active conformation (Berkenpas et al., 1995). First, a starting RNA library modified with 2′-fluoropyrimidines was bound with each protein; the RNA that bound to PAI-1 was separated from unbound RNA using a nitrocellulose membrane as described in Materials and Methods. By increasing the stringency in subsequent rounds, the binding affinity of each round increased (Fig. 1A and 1B). After nine rounds of SELEX, the affinity of the rounds reached low nanomolar levels. The round nine library (R9c) was then cloned and sequenced to generate individual aptamer sequences, as shown in Table 1. The WT selection produced only three aptamers, one of which, WT-1, dominated the pool of aptamers (Table 1). The SM selection, however, yielded a family of aptamers with similar sequences and several additional aptamers. Each WT aptamer (WT-1, WT-15, and WT-18) and selected aptamers from the SM selection were then chosen to undergo further characterization. As seen in Figure 1C and 1D, aptamers generated from both the WT and SM selections bound to WT PAI-1 with high affinity, as evidenced by their low nanomolar/high picomolar affinity constant (Kd) values (Table 1).
FIG. 1.
Systematic evolution of ligands by exponential enrichment (SELEX) yielded aptamers that bind to plasminogen activator inhibitor-1 (PAI-1) with high affinity. (A) The progress of the selection against the wild-type (WT) version of PAI-1 was followed using a nitrocellulose filter binding assay. Inverted triangles (▾) represent the starting RNA library (Sel2). Diamonds (♦) represent round 3, circles (•) represent round 5, triangles (▴) represent round 7, and squares (▪) represent round 9. The x-axis shows the human wild-type PAI-1 concentration and the y-axis shows the fraction of RNA bound to the protein. (B) The progress of the selection against the stable mutant (SM) version of PAI-1 was followed using a nitrocellulose filter binding assay. Inverted triangles (▾) represent the starting RNA library (Sel2). Diamonds (♦) represent round 3, circles (•) represent round 5, triangles (▴) represent round 7, and squares (▪) represent round 9. The x-axis shows the human stable mutant PAI-1 concentration and the y-axis shows the fraction of RNA bound to the protein. (C) Binding affinities of aptamers WT-1, WT-15, and WT-18 generated against WT PAI-1 were determined using a nitrocellulose filter binding assay. Squares (▪) represent WT-1, triangles (▴) represent WT-15, and inverted triangles (▾) represent WT-18. Each data point was performed in triplicate; error bars represent the standard error of the mean (SEM) of the data. (D) Binding affinities of aptamers SM-2, SM-20, and SM-23 generated against SM PAI-1 were determined using a nitrocellulose filter binding assay. Squares (▪) represent SM-2, triangles (▴) represent SM-20, and circles (•) represent SM-23. Each data point was performed in triplicate; error bars represent the SEM of the data.
Table 1.
PAI-1 Aptamer Sequences
| Clone ID | Variable region sequence | Frequency (%) | Kd |
|---|---|---|---|
| Selected against WT | |||
| R9chWT-1 | 5′-GTCCTAGCAGACACTCGGCCATCACGCCCATTGGTTTGCA-3′ | 89 | 1.2 nM |
| R9chWT-15 | 5′-ATCAACTCACCGTAGGTCTAGTGAGAACTTCAAGTCTACT-3′ | 5 | 177 pM |
| R9chWT-18 | 5′-ACTCCAACGATCCCACCGCGACAAGGGTCATCGGCACCGT-3′ | 5 | 35 nM |
| Selected against WT | |||
| R9chSM-20 | 5′-AGCGACTGACGATCTTGAGTAAACCGCTCATCCACGTAGT-3′ | 15 | 920 pM |
| R9chSM-14 | 5′-GTCCAGCTAAATCTCTACGAACCCCGCATTCCCCGTAACT-3′ | 10 | 37 nM |
| R9chSM-3 | 5′-GTCTCAATACGCACCCTGCATTCCCTTACTCGAGCCTAGC-3′ | 10 | 106 nM |
| R9chSM-23 | 5′-ATCTTTGGCCCTCCACAATATCCCTCCATGGGGTACCAG-3′ | 5 | 24 nM |
| R9chSM-2 | 5′-ATCCACTACGAACTCCGCATTCCCAGACACACTGGACCT-3′ | 5 | 3.3 nM |
| R9chSM-7 | 5′-GTCAAGNACGCGAACCCCGCATTCCCAAGAACGGCAACCCT-3′ | 5 | 120 nM |
| R9chSM-24 | 5′-TATCAACTAACGTACTGCCGCATTCCCACAATCCACAGGCTCC-3′ | 5 | 60 nM |
| R9chSM-6 | 5′-NTCAGTAAACGAACTCCGTATTCCCGATCATATGAGCACG-3′ | 5 | ND |
| R9chSM-16 | 5′-NTCACTCCAACGAACTCCGCATTCCCACACGCGACAATA-3′ | 5 | ND |
| R9chSM-8 | 5′-ATCAGGTACGAACTCTGCATTCCCAAAGCTCTCGGGGTGT-3′ | 5 | ND |
| R9chSM-5 | 5′-GTCAGGACTCAACTACGAACCCCGCATTCCCAACAGCACT-3′ | 5 | ND |
| R9chSM-9 | 5′-GTCAACCTCACGAACCCCGCATTCCCAGAGCCACTGCTCT-3′ | 5 | ND |
| R9chSM-17 | 5′-GTCAATTCAGCCACGTACCCCGCATTCCCAATGCCAGCAT-3′ | 5 | ND |
| R9chSM-12 | 5′-GTCAGCCTACGAACCCCGCATTCCCGATCGGGCCCGGCA-3′ | 5 | ND |
| R9chSM-18 | 5′-GTCATATCAACGAACCCCGCATTCCCGAGTAACTCCACCT-3′ | 5 | ND |
Abbreviation: ND, not determined.
Heparin and vitronectin compete with clones WT-15 and SM-20 for binding to PAI-1
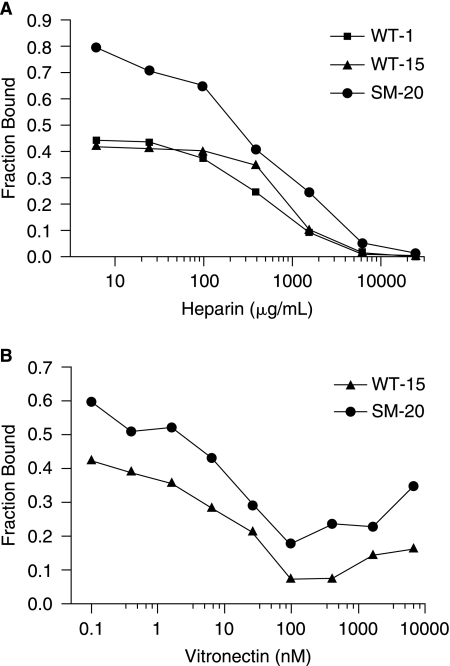
To determine the specific binding region of the lead aptamers, competition binding with heparin, given its similar charge distribution to RNA, and vitronectin, due to its proximity to the heparin-binding site, was performed. In this assay, a fixed amount of PAI-1 was incubated with increasing amounts of heparin or vitronectin, which was then combined with a trace amount of RNA. Protein competition with RNA for binding to PAI-1 is seen as a decrease in the fraction of RNA bound to PAI-1. As shown in Figure 2A, increasing amounts of heparin abolished binding of aptamer clones WT-1, WT-15, and SM-20 to PAI-1. In a like manner, competition binding was tested with vitronectin and similar results were attained with WT-15 and SM-20 (Fig. 2B). Therefore, heparin and vitronectin compete with WT-15 and SM-20 for binding to PAI-1 in solution, implying that these aptamers bind to PAI-1's heparin-/vitronectin-binding domains.
FIG. 2.
Heparin and vitronectin compete with aptamers for binding to PAI-1 in solution. (A) Increasing amounts of heparin compete with aptamers for binding to PAI-1; squares (▪) represent competition with clone WT-1 (PAI-1 concentration, 378 pM), triangles (▴) represent competition with WT-15 (PAI-1 concentration, 888 pM), and circles (•) represent competition with SM-20 (PAI-1 concentration, 3.4 nM). The x-axis shows the concentration of heparin, while the y-axis shows the fraction of RNA bound to PAI-1. (B) Increasing amounts of vitronectin compete with clones for binding to PAI-1; triangles (▴) represent competition with WT-15 (PAI-1 concentration, 888 pM), while circles (•) represent competition with SM-20 (PAI-1 concentration, 2.2 nM). The x-axis shows the concentration of vitronectin, while the y-axis shows the fraction of RNA bound to PAI-1.
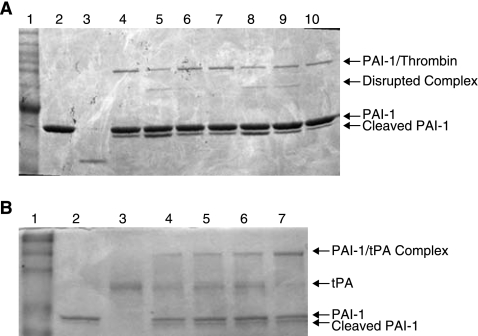
PAI-1 forms a SDS-stable complex with thrombin in the presence of heparin (Ehrlich et al., 1991). Therefore, if the aptamers prevent heparin binding to PAI-1, one would suspect that they will also disrupt the stable complex between PAI-1 and thrombin. Figure 3A shows that the aptamers (WT-15 and SM-20) are able to disrupt the complex in a concentration-dependent manner, as evidenced by the cleaved complex. The control aptamer library (Sel2) is unable to disrupt the complex (Fig. 3A). These data further suggest that the aptamers compete with heparin for binding to PAI-1.
FIG. 3.
Aptamers WT-15 and SM-20 disrupt the PAI-1/thrombin/heparin complex without affecting the PAI-1/tPA complex. (A) SM-20 and WT-15 disrupt the thrombin/PAI-1 complex in a concentration-dependent manner. PAI-1 (500 nM) was incubated with WT-15 (lanes 5 and 8), SM-20 (lanes 6 and 9), or Sel2 (lanes 7 and 10) (500–1000 nM) for 10 minutes before addition of thrombin (200 nM) and heparin (10 μg/mL). Lane 1: MW marker; lane 2: PAI-1 alone; lane 3: thrombin alone; lane 4: PAI-1/thrombin/heparin; lanes 5, 6, 7: aptamers at 500 nM; lanes 8, 9, 10: aptamers at 1000 nM. (B) The aptamers are unable to disrupt the PAI-1/tPA complex. PAI-1 (500 nM) was incubated with WT-15, SM-20, or Sel 2 (500 nM) prior to adding tPA (400 nM). Lane 1: MW marker; lane 2: PAI-1 alone; lane 3: tPA alone; lane 4: PAI-1/tPA; lane 5: PAI-1/tPA/WT-15; lane 6: PAI-1/tPA/SM-20; lane 7: PAI-1/tPA/Sel2.
The antiproteolytic activity of PAI-1 is dependent on its ability to form a stable complex with the plasminogen activators tPA and uPA. To determine if the aptamer clones were able to disrupt the PAI-1/tPA interaction, aptamers WT-15 and SM-20 were incubated with PAI-1, followed by addition of tPA. Figure 3B does not demonstrate any decrease in the level of PAI-1/tPA complex, suggesting that these aptamers do not interfere with the interaction of PAI-1 with tPA. These data suggest that the aptamers do not disrupt the antiproteolytic function of PAI-1.
Aptamers WT-15 and SM-20 prevent PAI-1 from binding to solid-phase vitronectin
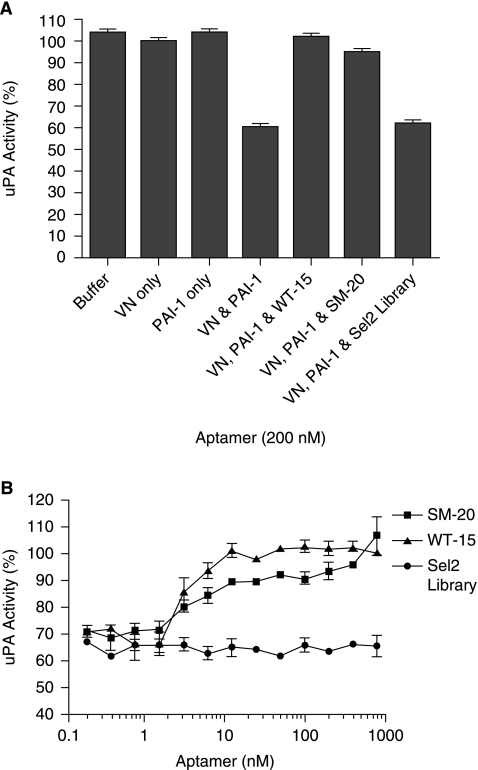
In vivo, vitronectin, an adhesive glycoprotein, binds to the extracellular matrix (PREISSNER, 1991). Therefore, although heparin and vitronectin competed with the clones for binding to PAI-1 in solution, we sought to determine if the clones could prevent PAI-1 from binding to vitronectin in a solid phase. In this assay, PAI-1 or PAI-1/RNA complex was added to vitronectin-coated plates and incubated as described in Materials and Methods. Following incubation, the plate was washed and incubated with uPA. If PAI-1 retains the ability to bind to vitronectin, it will inhibit uPA activity. As shown in Figure 4A, when only vitronectin is present, without PAI-1 or aptamers, uPA has maximal activity; the same is true when PAI-1, but no vitronectin, is present. As anticipated, the combination of vitronectin and PAI-1 decreases uPA activity ~60%. Aptamer WT-15 fully restores uPA activity and SM-20 restores uPA activity to greater than 90%, indicating that their binding to PAI-1 precludes binding to vitronectin. Figure 4B shows a dose–response curve of aptamer restoration of uPA activity; we see that near maximal levels of uPA activity, and thus inhibition of PAI-1 binding to vitronectin, occurs at concentrations as low as 12.5 nM.
FIG. 4.
Aptamers SM-20 and WT-15 prevent PAI-1 from binding to vitronectin that is bound in a solid phase. (A) Both 200 nM SM-20 and 200 nM WT-15 restore uPA activity, which indicates that they were able to free uPA from PAI-1 inhibition mediated by binding to vitronectin coated on a plate. Each data point was performed in duplicate in three separate experiments; error bars represent the standard error of the mean (SEM). (B) Clones SM-20 (represented by squares [▪]) and WT-15 (represented by triangles [▴]) restore uPA activity in a dose-dependent manner by preventing PAI-1 from binding to vitronectin and hence inhibiting uPA. Each data point was performed in duplicate in three separate experiments; error bars represent the SEM.
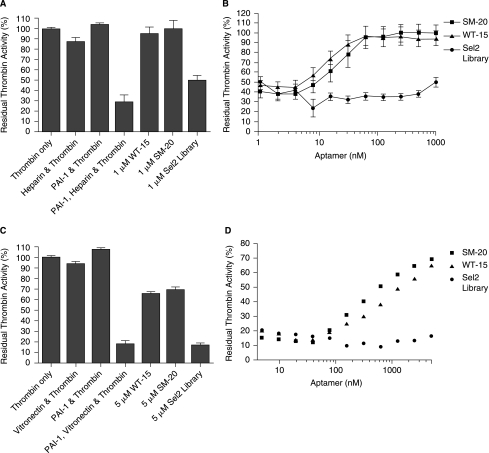
Aptamers WT-15 and SM-20 prevent heparin- and vitronectin-mediated PAI-1 inhibition of thrombin
In addition to preventing the binding of heparin and vitronectin to PAI-1, we sought to determine if this translated to an inhibition of PAI-1 function. To this end, we tested the ability of the aptamer clones to impede thrombin inhibition by PAI-1 that is mediated by the cofactors heparin or vitronectin. In this assay, the RNA/PAI-1 complex was incubated with either heparin or vitronectin prior to addition of thrombin. Residual thrombin activity was determined as described in Materials and Methods. In Figure 5A, we show that thrombin is inhibited by PAI-1/heparin, as evidenced by the decrease in thrombin activity. On the other hand, when PAI-1/heparin is incubated with 1 μM WT-15 or 1 μM SM-20, thrombin activity is completely restored. This rescue was also found to be dose-dependent, as seen in Figure 5B. Moreover, a decline in maximal restoration was not observed until the concentration of the clones reached below 62.5 nM. Similar results were found when in the presence of vitronectin (Fig. 5C and 5D). Here, the PAI-1/vitronectin had a more profound effect, decreasing thrombin activity to ~15% of its original value. As seen in Figure 5C, 5 μM WT-15, as well as 5 μM SM-20, restored ∼;65% of thrombin activity. Again, this occurred in a dose-dependent manner (Fig. 5D). Consequently, these data show that the aptamers WT-15 and SM-20 were able to prevent a functional reaction that stemmed from PAI-1 binding to heparin or vitronectin.
FIG. 5.
Aptamers SM-20 and WT-15 prevent heparin- and vitronectin-mediated PAI-1 inhibition of thrombin. (A) 1 μM SM-20, as well as 1 μM WT-15, restored thrombin activity despite the presence of PAI-1 and heparin, which in combination inhibited thrombin activity. Each data point was performed in duplicate in three separate experiments; error bars represent the standard error of the mean (SEM). (B) SM-20 (represented by squares [▪]) and WT-15 (represented by triangles [▴]) restore thrombin activity following PAI-1 and heparin inhibition in a dose-dependent manner. Each data point was performed in duplicate in three separate experiments; error bars represent the standard error of the mean (SEM). (C) 5 μM SM-20, as well as 5 μM WT-15, restored thrombin activity despite the presence of PAI-1 and vitronectin, which in combination inhibited thrombin activity. Each data point was performed in duplicate in three separate experiments; error bars represent the SEM. (D) Aptamers SM-20 (represented by squares [▪]) and WT-15 (represented by triangles [▴]) restore thrombin activity following PAI-1 and vitronectin inhibition in a dose-dependent manner. Each data point was performed in duplicate in three separate experiments; error bars represent the SEM.
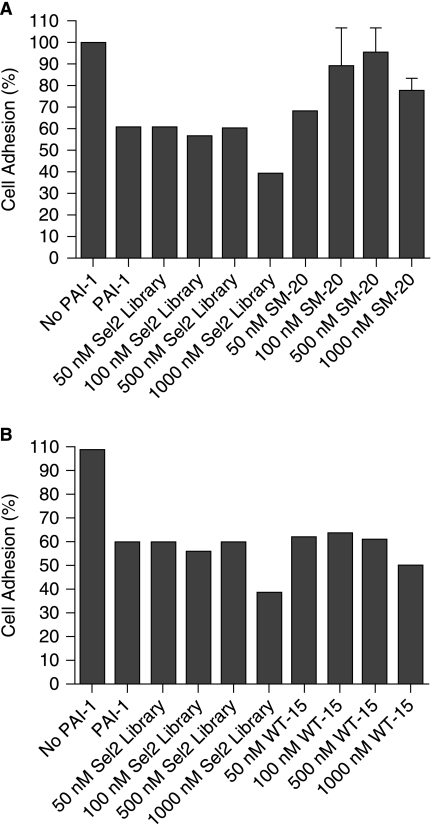
Aptamer SM-20, but not WT-15, restores breast cancer cell adhesion
Because both SM-20 and WT-15 prevented PAI-1 from binding to vitronectin bound to a solid surface and also disrupted a functional reaction involving PAI-1 and vitronectin, we sought to translate our results from in vitro to cell culture and test the PAI-1-specific aptamers' ability to disrupt a clinically relevant reaction, restoration of breast cancer cell adhesion. In this assay, breast cancer cells (MDA-MB-231) are incubated with PAI-1 or PAI-1/aptamer on vitronectin-coated plates. As seen in Figure 6, the addition of 50 nM PAI-1 decreases cell adhesion ~60%. However, the addition of increasing amounts of clone SM-20, with an optimal concentration at 500 nM, increases the percentage of cells that remain adherent to vitronectin (Fig. 6A). Although WT-15 demonstrated function in all previous assays, it was unable to increase breast cancer cell adhesion in the presence of PAI-1 at concentrations up to 1000 nM (Fig. 6B).
FIG. 6.
Aptamer SM-20, but not WT-15, restores breast cancer cell adhesion. (A) Breast cancer cells (MDA-MB-231) were incubated in the presence of SM-20 and PAI-1 in a vitronectin-coated plate. The SM-20 were performed in duplicate; error bars represent the standard error of the mean (SEM). (B) Breast cancer cells (MDA-MB-231) in the presence of WT-15 and PAI-1 in a vitronectin-coated plate did not show evidence of increased adhesion.
Discussion
PAI-1 is a multifunctional protein that interacts with several macromolecular substrates including plasminogen activators, vitronectin and thrombin. It contains three major domains, the active site region, the vitronectin-binding domain, and the lipoprotein-related protein-binding domain—all of which have been shown to contribute to the PAI-1-mediated pathogenesis of various diseases, including cancer (Wu and Zhao, 2002). In this study, we demonstrated that a PAI-1-specific RNA aptamer that binds to the vitronectin/heparin-binding sites of PAI-1 is able to restore breast cancer cell adhesion.
Considering the role of PAI-1 in cancer cell progression and metastasis, these data support the development of aptamers as a therapeutic option for cancer management. Aptamers have been shown to be nontoxic, well-tolerated molecules that cause low to no immunogenicity, can be synthetically manufactured, and have an adjustable bioavailability (Eyetech Study Group, 2002; Nimjee et al., 2005; Dyke et al., 2006; Chan et al., 2008a, 2008b). In addition to targeting PAI-1 for reductions in breast cancer metastasis, RNA aptamers have been selected that inhibit osteopontin, a secreted phosphoprotein that mediates tumorigenesis, local growth, and metastasis in several cancers (Mi et al., 2008). It has been shown that addition of OPN-R3 inhibits MDA-MB-231 in vitro adhesion, migration, and invasion, and decreases local progression and distant metastases in an in vivo xenograft model of breast cancer (Mi et al., 2008).
We describe the first RNA aptamer that binds PAI-1 while inhibiting the binding of PAI-1 to vitronectin. However, complete inhibition of all of PAI-1's activities might hinder its ability to regulate fibrinolysis, which could in principle engender bleeding. Therefore, elimination of the pathological functions of PAI-1 without hindering its physiological functions in hemostasis would be beneficial in a variety of disease settings. Here we demonstrated that aptamers WT-15 and SM-20 are able to disrupt vitronectin binding without compromising PAI-1's other functions, such as the inhibition of plasminogen activators.
The interaction between PAI-1 and vitronectin is critical to cancer progression. Given the importance of the PAI-1/vitronectin interaction in disrupting cellular adhesion, we hypothesized that an aptamer that inhibits this interaction would have a therapeutic application for breast cancer metastasis. To investigate this hypothesis, we first used SELEX technology targeting the human wild type (WT) and constitutively active stable mutant (SM) versions of PAI-1 to better ensure selection of an aptamer that bound the active version of PAI-1 (as opposed to the latent conformation). After nine rounds of SELEX, the enriched RNA pools were cloned and sequenced to generate individual aptamer sequences. The three aptamers that bound to WT or SM PAI-1 with the highest affinities were chosen for further characterization (WT-1, WT-15, and WT-18 from the WT selection, as well as SM-2, SM-20, and SM-23 from the SM selection). Following these assays, the two aptamers possessing binding affinities for WT PAI-1 in the picomolar range, WT-15 and SM-20, were found to have the greatest potential for disruption of the PAI-1/vitronectin interaction; hence, those clones were carried forward into all additional assays.
Aptamers tend to bind to highly positive regions on proteins such as heparin-binding sites. For example, the thrombin aptamer (TOG25) binds thrombin's heparin-binding site and has been shown to eliminate the heparin-accelerated inhibition of thrombin by various serpins (Jeter et al., 2004), and the VEGF aptamer that inhibits angiogenesis binds to the heparin-binding domain of VEGF 165 (Lee et al., 2005). Considering this, and the fact that PAI-1 is a heparin-binding serpin, we suspected that our aptamers would have a high probability of binding in the vicinity of this site. Also, PAI-1's vitronectin-binding site is located near the heparin-binding site. Consequently, we hypothesized that if our aptamer binds to the heparin site, it may also have an effect on vitronectin binding.
The first step in determining if the RNAs were able to disrupt the PAI-1/vitronectin interaction was to elucidate their binding location. When the aptamers were added to PAI-1 and uPA in a chromogenic assay used to detect uPA activity, no restoration of uPA function was found (data not shown). In addition, these aptamers were unable to disrupt the PAI-1/tPA high-molecular-weight complex (Fig. 3B). These observations led us to conclude that the aptamers were not binding to PAI-1 in a region (most likely the reactive center loop of PAI-1) that is important for its direct inhibition of uPA. Subsequently, we investigated the heparin-binding site of PAI-1. Binding studies with the PAI-1 R76E mutant (Stefansson et al., 1998), which does not bind heparin, suggested that the aptamers may bind in the heparin-binding region, as aptamer binding to this mutant protein was greatly reduced (data not shown). These results led us to hypothesize that the aptamers bind in the vicinity of PAI-1's heparin-binding site. To test this hypothesis, we investigated the ability of heparin to compete with aptamer binding to PAI-1. As seen in Figure 2A, increasing amounts of heparin prevented aptamer binding to PAI-1, suggesting that heparin binds in an overlapping site with the aptamers. When this assay was repeated with vitronectin as a competitor, similar results were achieved (Fig. 2B); however, at higher concentrations of vitronectin, binding somewhat increased. This was most likely due to nonspecific RNA interactions with high concentrations of vitronectin (data not shown). Recent studies have provided evidence that there are two vitronectin-binding sites on PAI-1, the somatomedin B (SMB) domain and another site outside of the SMB domain (Schar et al., 2008). Consequently, another plausible explanation is that the aptamers may also bind to the second vitronectin-binding site on PAI-1, although with reduced affinity. Nevertheless, collectively these data suggest that the clones bind to PAI-1 in a region spanning both the heparin- and vitronectin-binding domains (α-helices D and E, respectively).
Vitronectin is a main component of the extracellular matrix and is involved in several physiological processes including cell adhesion, migration, and proliferation, as well as fibrinolysis (PREISSNER, 1991). Whereas we were able to show that the aptamers bound to PAI-1 in a region overlapping the heparin- and vitronectin-binding sites, we did not know if this translated to the in vivo environment, that is, when vitronectin is bound to the extracellular matrix. Therefore, to further characterize the binding location and function of the clones, we performed an assay to determine if the clones were able to prevent PAI-1 from binding to vitronectin bound in a solid phase. We showed that both aptamers decreased PAI-1 inhibition of uPA (indicating that PAI-1 was not bound to vitronectin) in the presence of bound vitronectin (Fig. 4). Aptamer WT-15 fully attenuated PAI-1 inhibition of uPA, and such inhibition decreased only when its concentration became less than the concentration of PAI-1 in the assay (4.54 nM). This observation suggests that aptamer WT-15 binds to PAI-1 in a 1:1 stiochiometric ratio.
In addition to preventing PAI-1 from binding to vitronectin, aptamers WT-15 and SM-20 were found to disrupt the functional interaction between vitronectin and PAI-1 as it relates to thrombin inhibition (Fig. 5). Similar results were found when heparin was used as the cofactor, as both heparin and vitronectin accelerate PAI-1's inactivation of thrombin more than 200-fold, primarily by facilitating the formation of noncovalent complexes (Naski et al., 1993; REZAIE, 1999). Similar to the results noted for solid-phase vitronectin binding, aptamers WT-15 and SM-20 attenuated PAI-1/heparin and PAI-1/vitronectin inhibition of thrombin activity in a dose-dependent manner. Inhibition was particularly robust for the PAI-1/heparin interaction (Fig. 5B), as a decrease in maximal thrombin inhibition was again not apparent until the concentration of the aptamers was less than the concentration of PAI-1, further suggesting that the aptamers bind to PAI-1 in a 1:1 manner. On the other hand, the attenuation of the inhibition of PAI-1/vitronectin by the aptamers was not as robust as seen with heparin. One potential explanation is that at high concentrations of RNA, vitronectin non-specifically binds RNA, reducing the amount available to bind to PAI-1. These results corroborate the increase in binding noted during vitronectin's competition with the clones for binding to PAI-1. As vitronectin has heparin-binding domains itself that are positively charged, it is not unlikely that RNA would bind to those regions, whereas the negatively charged heparin would repel RNA due to its anionic properties. In spite of nonspecific binding, clones WT-15 and SM-20 are both able to attenuate PAI-1/cofactor inhibition of thrombin activity, further suggesting that they bind to a region on PAI-1 that overlaps the heparin- and vitronectin-binding sites.
We confirmed that the aptamers were specific for PAI-1 by assessing the ability of the structurally similar serpins, antithrombin III, heparin-cofactor II, and protein C inhibitor, to inhibit thrombin (in the presence of heparin and the aptamers) (REZAIE, 1999). The binding of the aptamers (WT-1, WT-15, SM-2, and SM-20) to these serpins was largely nonspecific; furthermore, they were unable to attenuate the inhibition of thrombin by these serpins (data not shown).
As mentioned earlier, the interaction between PAI-1 and vitronectin is important in the regulation of cell adhesion, cell migration, and fibrinolysis. In addition, vitronectin's interaction with PAI-1 has been implicated in several disease states (Stefansson and Lawrence, 1996; Dano et al., 2005), not only due to its function in keeping PAI-1 in an active conformation, but also due to PAI-1's competitive inhibition with the uPAR receptor for integrin binding, thus disrupting cellular adhesion (Stefansson and Lawrence, 1996; Kjoller et al., 1997). This interaction is particularly important for breast cancer cells, as increases in PAI-1 have been associated with a worsened prognosis for breast cancer patients (Beaulieu et al., 2007). In this case, inhibition of PAI-1's interaction with vitronectin would be beneficial and could lead to an increase in cellular adhesion, and therefore a decrease in cellular detachment and metastasis. For these reasons, we explored the potential of aptamers WT-15 and SM-20 in preventing breast cancer metastasis; this was achieved by determining if the aptamers could increase breast cancer cell adhesion in the setting of PAI-1-induced detachment. As seen in Figure 6, SM-20, but not WT-15, is able to restore cellular adhesion to ~90% its maximal value, calculated as the amount of adhesion noted in the absence of PAI-1. It is interesting that both aptamers SM-20 and WT-15 appear to bind to the overlapping heparin-/vitronectin-binding site on PAI-1, but only clone SM-20 is able to increase cellular adhesion in the presence of PAI-1. Differences between the sequences of the two aptamers likely impart distinct secondary and tertiary structures upon these RNAs; these structural variances, and how they alter PAI-1 binding, may be the basis behind the selectivity of one aptamer over the other in this instance.
Here, we have demonstrated the ability to isolate aptamers that bind to PAI-1 with high affinity. Furthermore, we have extensively functionally characterized two of these aptamers, mapping their binding location to an area on PAI-1 that spans α-helices D and E. In addition, we have shown that this binding is specific to PAI-1 and confers the ability to disrupt the PAI-1/vitronectin interaction, which has functional implications in preventing breast cancer metastasis. Plans for future work include further investigation of clone SM-20 in additional breast cancer cell lines in not only the cellular adhesion assay, but also assays of cellular migration. Also, further optimization of clone SM-20 through truncation and modification to increase its bioavailability would prepare the molecule for translation into an in vivo model of breast cancer metastasis. These developmental steps could lead to a novel antimetastatic agent that has low to no toxicity and is nonimmunogenic, making it an attractive agent to serve as an adjunct to traditional breast cancer therapy.
Acknowledgments
The authors would like to thank Dr. David Ginsburg for his generous donation of both hWT PAI-1 and hSM PAI-1, as well as Dr. Sabah Oney and Juliana Layzer for their technical advice. This work was supported by National Institutes of Health (NIH) grant #1F31NS058273-01 and the UNCF-Merck Graduate Science Research Dissertation Fellowship awarded to C.M.B., NIH grant #HL65222 awarded to B.A.S., and AHA grant #0765319U awarded to Y.M.F.
References
- BEAULIEU L.M. WHITLEY B.R. WIESNER T.F. REHAULT S.M. PALMIERI D. ELKAHLOUN A.G. CHURCH F.C. Breast cancer and metabolic syndrome linked through the plasminogen activator inhibitor-1 cycle. Bioessays. 2007;29:1029–1038. doi: 10.1002/bies.20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERKENPAS M.B. LAWRENCE D.A. GINSBURG D. Molecular evolution of plasminogen activator inhibitor-1 functional stability. EMBO J. 1995;14:2969–2977. doi: 10.1002/j.1460-2075.1995.tb07299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAN M.Y. COHEN M.G. DYKE C.K. MYLES S.K. ABERLE L.G. LIN M. WALDER J. STEINHUBL S.R. GILCHRIST I.C. KLEIMAN N.S. VORCHHEIMER D.A. CHRONOS N. MELLONI C. ALEXANDER J.H. HARRINGTON R.A. TONKENS R.M. BECKER R.C. RUSCONI C.P. Phase 1b randomized study of antidote-controlled modulation of factor IXa activity in patients with stable coronary artery disease. Circulation. 2008a;117:2865–2874. doi: 10.1161/CIRCULATIONAHA.107.745687. [DOI] [PubMed] [Google Scholar]
- CHAN M.Y. RUSCONI C.P. ALEXANDER J.H. TONKENS R.M. HARRINGTON R.A. BECKER R.C. A randomized, repeat-dose, pharmacodynamic and safety study of an antidote-controlled factor IXa inhibitor. J. Thromb. Haemost. 2008b;6:789–796. doi: 10.1111/j.1538-7836.2008.02932.x. [DOI] [PubMed] [Google Scholar]
- CHAPMAN H.A. WEI Y. Protease crosstalk with integrins: the urokinase receptor paradigm. Thromb. Haemost. 2001;86:124–129. [PubMed] [Google Scholar]
- CHOROSTOWSKA-WYNIMKO J. SKRZYPCZAK-JANKUN E. JANKUN J. Plasminogen activator inhibitor type-1: its structure, biological activity and role in tumorigenesis (Review) Int. J. Mol. Med. 2004;13:759–766. [PubMed] [Google Scholar]
- DANO K. BEHRENDT N. HOYER-HANSEN G. JOHNSEN M. LUND L.R. PLOUG M. ROMER J. Plasminogen activation and cancer. Thromb. Haemost. 2005;93:676–681. doi: 10.1160/TH05-01-0054. [DOI] [PubMed] [Google Scholar]
- DECLERCK P.J. DE MOL M. ALESSI M.C. BAUDNER S. PAQUES E.P. PREISSNER K.T. MULLER-BERGHAUS G. COLLEN D. Purification and characterization of a plasminogen activator inhibitor 1 binding protein from human plasma. Identification as a multimeric form of S protein (vitronectin) J. Biol. Chem. 1988;263:15454–15461. [PubMed] [Google Scholar]
- DELLAS C. LOSKUTOFF D.J. Historical analysis of PAI-1 from its discovery to its potential role in cell motility and disease. Thromb. Haemost. 2005;93:631–640. doi: 10.1160/TH05-01-0033. [DOI] [PubMed] [Google Scholar]
- DENG G. ROYLE G. WANG S. CRAIN K. LOSKUTOFF D.J. Structural and functional analysis of the plasminogen activator inhibitor-1 binding motif in the somatomedin B domain of vitronectin. J. Biol. Chem. 1996;271:12716–12723. doi: 10.1074/jbc.271.22.12716. [DOI] [PubMed] [Google Scholar]
- DYKE C.K. STEINHUBL S.R. KLEIMAN N.S. CANNON R.O. ABERLE L.G. LIN M. MYLES S.K. MELLONI C. HARRINGTON R.A. ALEXANDER J.H. BECKER R.C. RUSCONI C.P. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation. 2006;114:2490–2497. doi: 10.1161/CIRCULATIONAHA.106.668434. [DOI] [PubMed] [Google Scholar]
- EHRLICH H.J. GEBBINK R.K. PREISSNER K.T. KEIJER J. ESMON N.L. MERTENS K. PANNEKOEK H. Thrombin neutralizes plasminogen activator inhibitor 1 (PAI-1) that is complexed with vitronectin in the endothelial cell matrix. J. Cell. Biol. 1991;115:1773–1781. doi: 10.1083/jcb.115.6.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyetech Study Group. Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina. 2002;22:143–152. doi: 10.1097/00006982-200204000-00002. [DOI] [PubMed] [Google Scholar]
- Eyetech Study Group. Anti-vascular endothelial growth factor therapy for subfoveal choroidal neovascularization secondary to age-related macular degeneration: phase II study results. Ophthalmology. 2003;110:979–986. doi: 10.1016/S0161-6420(03)00085-X. [DOI] [PubMed] [Google Scholar]
- GONDI C.S. LAKKA S.S. YANAMANDRA N. SIDDIQUE K. DINH D.H. OLIVERO W.C. GUJRATI M. RAO J.S. Expression of antisense uPAR and antisense uPA from a bicistronic adenoviral construct inhibits glioma cell invasion, tumor growth, and angiogenesis. Oncogene. 2003;22:5967–5975. doi: 10.1038/sj.onc.1206535. [DOI] [PubMed] [Google Scholar]
- HEKMAN C.M. LOSKUTOFF D.J. Endothelial cells produce a latent inhibitor of plasminogen activators that can be activated by denaturants. J. Biol. Chem. 1985;260:11581–11587. [PubMed] [Google Scholar]
- JETER M.L. LY L.V. FORTENBERRY Y.M. WHINNA H.C. WHITE R.R. RUSCONI C.P. SULLENGER B.A. CHURCH F.C. RNA aptamer to thrombin binds anion-binding exosite-2 and alters protease inhibition by heparin-binding serpins. FEBS Lett. 2004;568:10–14. doi: 10.1016/j.febslet.2004.04.087. [DOI] [PubMed] [Google Scholar]
- KJOLLER L. KANSE S.M. KIRKEGAARD T. RODENBURG K.W. RONNE E. GOODMAN S.L. PREISSNER K.T. OSSOWSKI L. ANDREASEN P.A. Plasminogen activator inhibitor-1 represses integrin- and vitronectin-mediated cell migration independently of its function as an inhibitor of plasminogen activation. Exp. Cell Res. 1997;232:420–429. doi: 10.1006/excr.1997.3540. [DOI] [PubMed] [Google Scholar]
- LAH T.T. DURAN ALONSO M.B. VAN NOORDEN C.J. Antiprotease therapy in cancer: hot or not? Expert. Opin. Biol. Ther. 2006;6:257–279. doi: 10.1517/14712598.6.3.257. [DOI] [PubMed] [Google Scholar]
- LAWRENCE D.A. STRANDBERG L. ERICSON J. NY T. Structure-function studies of the SERPIN plasminogen activator inhibitor type 1. Analysis of chimeric strained loop mutants. J. Biol. Chem. 1990;265:20293–20301. [PubMed] [Google Scholar]
- LEE J.H. CANNY M.D. DE ERKENEZ A. KRILLEKE D. NG Y.S. SHIMA D.T. PARDI A. JUCKER F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc. Natl Acad. Sci. USA. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEIK C.E. SU E.J. NAMBI P. CRANDALL D.L. LAWRENCE D.A. Effect of pharmacologic plasminogen activator inhibitor-1 inhibition on cell motility and tumor angiogenesis. J. Thromb. Haemost. 2006;4:2710–2715. doi: 10.1111/j.1538-7836.2006.02244.x. [DOI] [PubMed] [Google Scholar]
- LEVIN E.G. SANTELL L. Conversion of the active to latent plasminogen activator inhibitor from human endothelial cells. Blood. 1987;70:1090–1098. [PubMed] [Google Scholar]
- LINDAHL T.L. SIGURDARDOTTIR O. WIMAN B. Stability of plasminogen activator inhibitor 1 (PAI-1) Thromb. Haemost. 1989;62:748–751. [PubMed] [Google Scholar]
- MI Z. GUO H. RUSSELL M.B. LIU Y. SULLENGER B.A. KUO P.C. RNA Aptamer Blockade of Osteopontin Inhibits Growth and Metastasis of MDA-MB231 Breast Cancer Cells. Mol. Ther. 2008 doi: 10.1038/mt.2008.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIN H.Y. DOYLE L.V. VITT C.R. ZANDONELLA C.L. STRATTON-THOMAS J.R. SHUMAN M.A. ROSENBERG S. Urokinase receptor antagonists inhibit angiogenesis and primary tumor growth in syngeneic mice. Cancer Res. 1996;56:2428–2433. [PubMed] [Google Scholar]
- NASKI M.C. LAWRENCE D.A. MOSHER D.F. PODOR T.J. GINSBURG D. Kinetics of inactivation of alpha-thrombin by plasminogen activator inhibitor-1. Comparison of the effects of native and urea-treated forms of vitronectin. J. Biol. Chem. 1993;268:12367–12372. [PubMed] [Google Scholar]
- NIMJEE S.M. RUSCONI C.P. SULLENGER B.A. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- PALMIERI D. LEE J.-W. JULIANO R.L. CHURCH F.C. Plasminogen Activator Inhibitor-1 and −3 Increase Cell Adhesion and Motility of MDA-MB-435 Breast Cancer Cells. J. Biol. Chem. 2002;277:40950–40957. doi: 10.1074/jbc.M202333200. [DOI] [PubMed] [Google Scholar]
- PERRON M.J. BLOUSE G.E. SHORE J.D. Distortion of the catalytic domain of tissue-type plasminogen activator by plasminogen activator inhibitor-1 coincides with the formation of stable serpin-proteinase complexes. J. Biol. Chem. 2003;278:48197–48203. doi: 10.1074/jbc.M306184200. [DOI] [PubMed] [Google Scholar]
- PLOUG M. OSTERGAARD S. GARDSVOLL H. KOVALSKI K. HOLST-HANSEN C. HOLM A. OSSOWSKI L. DANO K. Peptide-derived antagonists of the urokinase receptor. affinity maturation by combinatorial chemistry, identification of functional epitopes, and inhibitory effect on cancer cell intravasation. Biochemistry. 2001;40:12157–12168. doi: 10.1021/bi010662g. [DOI] [PubMed] [Google Scholar]
- PREISSNER K.T. Structure and biological role of vitronectin. Annu. Rev. Cell. Biol. 1991;7:275–310. doi: 10.1146/annurev.cb.07.110191.001423. [DOI] [PubMed] [Google Scholar]
- PULUKURI S.M. GONDI C.S. LAKKA S.S. JUTLA A. ESTES N. GUJRATI M. RAO J.S. RNA interference-directed knockdown of urokinase plasminogen activator and urokinase plasminogen activator receptor inhibits prostate cancer cell invasion, survival, and tumorigenicity in vivo. J. Biol. Chem. 2005;280:36529–36540. doi: 10.1074/jbc.M503111200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- REDMOND E.M. CULLEN J.P. CAHILL P.A. SITZMANN J.V. STEFANSSON S. LAWRENCE D.A. OKADA S.S. Endothelial cells inhibit flow-induced smooth muscle cell migration: role of plasminogen activator inhibitor-1. Circulation. 2001;103:597–603. doi: 10.1161/01.cir.103.4.597. [DOI] [PubMed] [Google Scholar]
- REZAIE A.R. Role of exosites 1 and 2 in thrombin reaction with plasminogen activator inhibitor-1 in the absence and presence of cofactors. Biochemistry. 1999;38:14592–14599. doi: 10.1021/bi9913303. [DOI] [PubMed] [Google Scholar]
- RUSCONI C.P. YEH A. LYERLY H.K. LAWSON J.H. SULLENGER B.A. Blocking the initiation of coagulation by RNA aptamers to factor VIIa. Thromb. Haemost. 2000;84:841–848. [PubMed] [Google Scholar]
- SCHAR C.R. JENSEN J.K. CHRISTENSEN A. BLOUSE G.E. ANDREASEN P.A. PETERSON C.B. Characterization of a site on PAI-1 that binds to vitronectin outside of the somatomedin B domain. J. Biol. Chem. 2008;283:28487–28496. doi: 10.1074/jbc.M804257200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEIFFERT D. CIAMBRONE G. WAGNER N.V. BINDER B.R. LOSKUTOFF D.J. The somatomedin B domain of vitronectin. Structural requirements for the binding and stabilization of active type 1 plasminogen activator inhibitor. J. Biol. Chem. 1994;269:2659–2666. [PubMed] [Google Scholar]
- STEFANSSON S. LAWRENCE D.A. The serpin PAI-1 inhibits cell migration by blocking integrin alpha V beta 3 binding to vitronectin. Nature. 1996;383:441–443. doi: 10.1038/383441a0. [DOI] [PubMed] [Google Scholar]
- STEFANSSON S. MUHAMMAD S. CHENG X.F. BATTEY F.D. STRICKLAND D.K. LAWRENCE D.A. Plasminogen activator inhibitor-1 contains a cryptic high affinity binding site for the low density lipoprotein receptor-related protein. J. Biol. Chem. 1998;273:6358–6366. doi: 10.1074/jbc.273.11.6358. [DOI] [PubMed] [Google Scholar]
- SUBRAMANIAN R. GONDI C.S. LAKKA S.S. JUTLA A. RAO J.S. siRNA-mediated simultaneous downregulation of uPA and its receptor inhibits angiogenesis and invasiveness triggering apoptosis in breast cancer cells. Int. J. Oncol. 2006;28:831–839. [PMC free article] [PubMed] [Google Scholar]
- U.S. Cancer Statistics Working Group. United States Cancer Statistics: 2004 Incidence and Mortality. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; 2007. [Google Scholar]
- WEI Y. EBLE J.A. WANG Z. KREIDBERG J.A. CHAPMAN H.A. Urokinase receptors promote beta1 integrin function through interactions with integrin alpha3beta1. Mol. Biol. Cell. 2001;12:2975–2986. doi: 10.1091/mbc.12.10.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEI Y. LUKASHEV M. SIMON D.I. BODARY S.C. ROSENBERG S. DOYLE M.V. CHAPMAN H.A. Regulation of integrin function by the urokinase receptor. Science. 1996;273:1551–1555. doi: 10.1126/science.273.5281.1551. [DOI] [PubMed] [Google Scholar]
- WU Q. ZHAO Z. Inhibition of PAI-1: a new antithrombotic approach. Curr. Drug Targets Cardiovasc. Haematol. Disord. 2002;2:27–42. doi: 10.2174/1568006023337727. [DOI] [PubMed] [Google Scholar]
- ZHU M. GOKHALE V.M. SZABO L. MUNOZ R.M. BAEK H. BASHYAM S. HURLEY L.H. VON HOFF D.D. HAN H. Identification of a novel inhibitor of urokinase-type plasminogen activator. Mol. Cancer Ther. 2007;6:1348–1356. doi: 10.1158/1535-7163.MCT-06-0520. [DOI] [PubMed] [Google Scholar]