Abstract
Immunoaffinity depletion with antibodies to the top 7 or top 14 high abundance plasma proteins is used to enhance detection of lower abundance proteins in both shotgun and targeted proteomic analyses. We evaluated the effects of top 7/top 14 immunodepletion on the shotgun proteomic analysis of human plasma. Our goal was to evaluate the impact of immunodepletion on detection of proteins across detectable ranges of abundance. The depletion columns afforded highly repeatable and efficient plasma protein fractionation. Relatively few nontargeted proteins were captured by the depletion columns. Analyses of unfractionated and immunodepleted plasma by peptide isoelectric focusing (IEF), followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) demonstrated enrichment of nontargeted plasma proteins by an average of 4-fold, as assessed by MS/MS spectral counting. Either top 7 or top 14 immunodepletion resulted in a 25% increase in identified proteins compared to unfractionated plasma. Although 23 low abundance (<10 ng mL−1) plasma proteins were detected, they accounted for only 5–6% of total protein identifications in immunodepleted plasma. In both unfractionated and immunodepleted plasma, the 50 most abundant plasma proteins accounted for 90% of cumulative spectral counts and precursor ion intensities, leaving little capacity to sample lower abundance proteins. Untargeted proteomic analyses using current LC-MS/MS platforms—even with immunodepletion—cannot be expected to efficiently discover low abundance, disease-specific biomarkers in plasma.
Keywords: plasma, high-abundance protein depletion, multiple affinity removal system, isoelectric focusing, shotgun proteomics
INTRODUCTION
The abundances of proteins in plasma proteomes exceeds 10 orders of magnitude and this wide dynamic range presents a barrier to detection of medium and low abundance proteins in proteomic analyses 1. The targeted depletion of abundant plasma proteins with antibody columns was first introduced in 2003 2 and has been employed both to increase the depth of proteome identifications in unbiased discovery and to increase sensitivity for targeted analyses of specific proteins. Columns containing immobilized antibodies to remove either 7 or 14 abundant plasma proteins (“top 7 or top 14 depletion”) are widely used 3, yet characterization of this approach has been limited. Studies based on two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis 2, 4–7 yielded small proteome inventories (<100 protein identifications) and provided at best a qualitative assessment of the impact of depletion on detection of plasma and serum proteins.
Two previous studies8, 9 employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) to generate larger proteome inventories (>200 protein identifications) and reported that immunodepletion increased the numbers of proteins detected. Gong et al.9 reported detection of 10 low-abundance proteins (concentrations <100 ng mL−1 based on literature annotations) only in depleted samples. The only quantitative assessment of enhanced detection of low abundance proteins as a result of immunodepletion was reported by Brand et al., who reported ELISA measurements on 11 proteins10. These results do not indicate whether immunodepletion broadly enhances detection of lower abundance plasma proteins. Moreover, previous reports did not assess the reproducibility of the depletion step or the degree to which nontargeted proteins were captured by these columns.
Studies of other immunodepletion media that target either 14 or more than 50 high abundance plasma proteins did address these questions 11–13 and the results suggested to us that a global LC-MS/MS inventory of the top7/top14-depleted plasma could provide much-improved characterization of this system.
Our interest in this problem is due in large part to the adoption of this approach for abundant plasma protein depletion in ongoing interlaboratory studies by the National Cancer Institute Clinical Proteomic Technologies Assessment for Cancer (CPTAC) program, in which we are participants. A major objective of the CPTAC program is to characterize the performance of proteome analysis platforms and sample preparation methods. In light of the widespread use of the top 7/top 14 system, the limited quantitative characterization of its performance in previous studies, and the planned use of these columns in CPTAC studies, we felt that a detailed characterization of this system was justified.
One striking characteristic of plasma proteome datasets is the relatively small number of proteins identified in comparison to analyses of cells or tissues. Application of one dimensional LC-MS/MS, together with commonly accepted criteria for protein identification (≥ 2 peptides at <5% false discovery rate (FDR)) yielded approximately 150–250 proteins 8, 13, whereas addition of multidimensional LC-MS/MS yielded 200–700 proteins 9, 13. Even the multilaboratory HUPO plasma proteome study yielded less than 900 high confidence protein identifications 14. These totals are approximately 2–5-fold less than have been reported for analyses of cell and tissue extracts using similar protein input amounts and similar LC-MS/MS platforms 15–19. Although this effect is commonly attributed to the overwhelming predominance of a small number of high abundance proteins, this relationship has not been quantitatively evaluated.
Here we present a detailed evaluation of top7/top14 immunodepletion in the analysis of human plasma by shotgun proteomics using LC-MS/MS. Our objective was to evaluate the impact of top7/top14 immunodepletion on the detection of plasma proteins across the broad range of detectable concentrations. The results document the high repeatability of target and nontargeted protein capture as well as an average 2 to 4-fold global enrichment of nontargeted proteins by immunodepletion. We also demonstrate a nearly 10-fold greater number of detected proteins in a human colon tumor cell line than in depleted plasma at the same level of protein input and show that the detection disparity is due to a steep protein abundance distribution in plasma versus cells. These findings demonstrate the consistent performance of abundant protein depletion with immunodepletion columns, but also illustrate the limitations of shotgun proteomics to detect disease biomarkers in plasma proteomes.
MATERIALS AND METHODS
Plasma and cell samples
Human blood plasma samples were obtained from three anonymous healthy volunteers through the Jim Ayers Institute for Precancer Detection and Diagnosis under an Institutional Review Board-approved protocol. Informed consent for research was obtained from all volunteers. Blood was drawn by venipuncture into 7 ml Vacutainer tubes using EDTA as anticoagulant (BD Worldwide, Franklin Lakes, NJ). Plasma was obtained by centrifugation of whole blood for 15 min at 380 × g. The initial plasma protein concentration was ~82 mg mL−1 as determined with the BCA protein assay (Pierce Chemicals, Rockford, IL). A human colon adenocarcinoma cell line (RKO) was obtained from ATCC (Manassas, VA) and cultured in 100 ml flasks in McCoy’s 5A media (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (Atlas Biologicals, Fort Collins, CO) at 37 °C in 5% CO2. RKO cells were grown to >90% confluence, then harvested in 5 ml of 0.25% trypsin-EDTA, washed with PBS and split into 1×107 cells per tube and frozen at −80 °C. Unless otherwise noted, all protein sample processing was performed at 4°C.
Immunoaffinity chromatography
Multiple Affinity Removal System™ (MARS‡) columns (4.6 × 100 mm) designed to deplete 7 abundant proteins (albumin, IgG, antitrypsin, IgA, transferrin, haptoglobin, fibrinogen) (MARS-7) or to deplete 14 abundant proteins (albumin, IgG, antitrypsin, IgA, transferrin, haptoglobin, fibrinogen, alpha2-macroglobulin, alpha1-acid glycoprotein, IgM, apolipoprotein AI, apolipoprotein AII, complement C3, and transthyretin) (MARS-14) were purchased from Agilent (Palo Alto, CA). Depletion was performed at room temperature with an Agilent 1100 series HPLC system. Plasma samples were diluted fourfold using the load/wash buffer supplied by the manufacture and remaining particulates in the diluted plasma were removed by centrifugation through a 0.22-μm spin filter 1 min at 16,000 × g.
After equilibration with the load/wash buffer, the MARS-7 column was loaded with 160 μL of the diluted plasma at a low flow rate (0.5 mL min−1) for 10 min. Flow-through fractions, representing depleted plasma, were collected and stored at −20 °C. The bound proteins were released with elution buffer at 1.0 mL min−1 for 7 min. The column was then washed with the load/wash buffer for 11 min at a flow rate of 1 mL min−1. Each depletion cycle took 28 min of total run time. The MARS-14 column was equilibrated with the load/wash buffer and 160 μL of the diluted plasma was loaded at a low flow rate (0.125 mL min−1) for 18 min and then for an additional 2 min at a flow rate of 1 mL min−1. The other binding and elution steps were identical to those used for the MARS-7 column. Thus, each depletion run cycle of the MARS-14 column took 38 min of total run time. Both depleted (flow-through fraction) and abundant plasma proteins (bound fraction) were collected and stored at −20 °C until further analysis.
Protein digestion
The depleted and bound protein fractions prepared from each sample were concentrated using Amicon 5 kDa molecular weight filters (Millipore, MA), followed by buffer exchange to 50 mM ammonium bicarbonate in the same unit. Protein concentration was then determined by the BCA protein assay (Pierce-Thermo Scientific, Rockford, IL). Proteins were denatured and reduced in 50 mM ammonium bicarbonate buffer, pH 8.0, 8 M urea, 10 mM dithiothreitol for 1 hour at 37 °C. Protein cysteines were alkylated with 50 mM iodoacetamide for 60 min at room temperature in the dark. The reduced and alkylated protein mixture was diluted 10-fold with 50 mM ammonium bicarbonate, pH 8.0, followed by the addition of trypsin (Promega, Cat#TB309, Madison, WI) at a trypsin/protein ratio of 1:50 (w/w). The digestion mixture was incubated overnight at 37 °C and then frozen at −80 °C and lyophilized. Samples were resuspended in 1 mL of deionized water and applied to SEP-Pak vac 1 cc (100 mg) C-18 cartridges (Waters Corp., Milford, MA), which were prewashed with 1 mL of acetonitrile and equilibrated with 2 mL of deionized water. The flow-through was discarded, the cartridges were washed with 1 mL of deionized water, the bound peptides were eluted with 80% acetonitrile in deionized water, and the eluate was evaporated in vacuo.
IEF of peptides
Tryptic peptides from 200 μg of protein were redissolved in 500 μL of 6 M urea and loaded in an IPGphor rehydration tray (GE Healthcare, Piscataway, NJ). Carrier ampholyte (2% (v/v) was included in the loading solution (IPG buffer pH 3–10 NL (GE catalog #17-6000-88)). IPG strips (24 cm, pH 3–10 NL) (GE Healthcare) were placed over the samples and allowed to rehydrate overnight at room temperature. The loaded strips were focused at 20 °C on an Ettan IPGPhor-III IEF system (GE Healthcare) using the following focusing program: 500 V for 1 hr; a gradient to 1000 V for 3 hr; a gradient to 10000 V for 3 hr; a step to 10000 V for 5 hr. The strips were then cut into 20 pieces and placed in separate wells of a 96-well Falcon flat bottom polystyrene ELISA plate (Fisher Scientific). Peptides were eluted from the strips with 200 μL of 0.1% formic acid for 15 min, followed by 200 μL of acetonitrile/0.1% formic acid (1:1, v/v) for 15 min, then with 200 μL of acetonitrile containing 0.1% formic acid for 15 min. The combined eluates for each IPG strip fraction were evaporated in vacuo and then redissolved in 0.1% trifluoroacetic acid (TFA) and applied to a 96 well C-18 Oasis HLB plate (30 μm particle size, 10 mg packing) (Waters Corp., Milford, MA) prewashed with 1 mL of acetonitrile and equilibrated with 2 mL of 0.1% TFA. The flow-through was discarded and the cartridges were washed with 1 mL of 0.1% TFA. The bound peptides were eluted with 0.3 mL each of 30% acetonitrile/0.1% TFA, 70% acetonitrile/0.1% TFA and 100% acetonitrile/0.1% TFA and the combined eluate was evaporated in vacuo and redissolved in 100 μL of 0.1% formic acid for LC-MS/MS analysis, as previously described 17.
Reverse-phase LC-MS/MS analysis
LC-MS/MS analyses were performed on a Thermo LTQ linear ion trap mass spectrometer equipped with a Thermo MicroAS autosampler and Thermo Surveyor HPLC pump, Nanospray source and Xcalibur 1.4 instrument control (Thermo Electron, San Jose, CA). Peptides were separated on a packed capillary tip (Polymicro Technologies, 100 μm × 11 cm) with Jupiter C18 resin (5 μm, 300 Å, Phenomenex) using an in-line solid-phase extraction column (100 μm × 6 cm) packed with the same C18 resin (using a frit generated with liquid silicate Kasil 120) similar to that previously described21. The flow from the HPLC pump was split prior to the injection valve to achieve flow-rates of 700 nL-1000 μL min-1 at the column tip. Mobile phase A consisted of 0.1 % formic acid and Mobile phase B consisted of 0.1% formic acid in acetonitrile. A 95 min gradient was performed with a 15 min washing period (100 % A for the first 10 min followed by a gradient to 98% A at 15 minutes) to allow for solid-phase extraction and removal of any residual salts. Following the washing period, the gradient was increased to 25% B by 50 min, followed by an increase to 90 % B by 65 min and held for 9 min before returning to the initial conditions. Liquid chromatography was carried out at ambient temperature at a flow rate of 0.6 μL min−1. Centroided MS/MS scans were acquired on the LTQ using an isolation width of 2 m/z, an activation time of 30 ms, an activation q of 0.250 and 30% normalized collision energy using 1 microscan with a max ion time of 100 ms for each MS/MS scan and 1 microscan with a max ion time of 500 ms for each full MS scan. The mass spectrometer was tuned prior to analysis using the synthetic peptide TpepK (AVAGKAGAR). Some parameters may have varied slightly from experiment to experiment, but typically the tune parameters were as follows: spray voltage of 2 kV, a capillary temperature of 150 °C, a capillary voltage of 50 V and tube lens of 120 V. The MS/MS spectra were collected using data-dependent scanning in which one full MS spectrum was followed by five MS-MS spectra. MS/MS spectra were recorded using dynamic exclusion of previously analyzed precursors for 60 s with a repeat count of 1 and a repeat duration of 1.
Data processing and analysis
The LC-MS/MS raw data were converted into mzData file format by ScanSifter v2.0, an in-house developed software, and the MyriMatch algorithm (version 2.1.11)22 was used to independently search all the MS/MS spectra against the human International Protein Index (IPI) database (version 3.37) with a total of 69,164 protein entries. Myrimatch employs a statistical model using the multivariate hypergeometric distribution to score peptide and places greater emphasis on matching intense peaks. The stratification of peak intensity in the scoring algorithm enables Myrimatch to outperform other scoring algorithms (Sequest, Mascot) that lack this feature. The search parameters used were as follows: 1.25 Da tolerance for precursor ion masses and 0.5 Da for fragment ion masses. Candidate peptides were permitted to feature semitryptic cleavages, which allow one non-tryptic end, and any number of missed cleavages was permitted. Carbamidomethylation of cysteines was specified as a fixed modification, variable modifications of methionine oxidation, N-terminal pyro-Glu from glutamine were allowed during the database search. The sequence database was doubled to contain each sequence in both forward and reversed orientations, enabling false discovery rate estimation.
The IDPicker algorithm23, 24 (version 2.1.5) filtered the identifications for each LC-MS/MS run to include the largest set for which a 5% peptide identification FDR could be maintained. IDPicker employs a bipartite graph analysis and efficient graph algorithms to identify protein clusters with shared peptides and to derive the minimal list of proteins. This bipartite parsimony technique simplifies protein lists by consolidating results that map to redundant database entries and also improves the accuracy of protein identification. This approach also groups functionally related proteins together and improves the comprehensibility of the results. These identifications from each LC-MS/MS run were pooled for each sample. IDPicker allows the user to specify a FDR threshold and then adjusts score threshold accordingly. For these studies a 5% peptide FDR was employed. Thus, peptide filtering employed reversed sequence database match information to determine Myrimatch score thresholds that yielded an estimated 5% peptide identification FDR for the identifications of each charge state, as calculated by the formula FDR = (2 × reverse)/(forward + reverse)25. Proteins were required to have at least two distinct peptide sequences observed in the analyses. Indistinguishable proteins were recognized and grouped. Parsimony rules were applied to generate a minimal list of proteins that explained all of the peptides that passed our entry criteria23. Further filtering of protein identification lists from IEF-LC-MS/MS analyses to achieve a protein FDR less than 5% is described under Results (see below).
Relative peptide/protein intensity analysis
Peptide and protein intensities for a subset of the identified plasma proteins were calculated from LC-MS/MS datafiles for analyses of plasma, MARS7/MARS-14 depleted plasma and cell lysates. MS/MS spectra were extracted to MGF files using ReAdWRaw4Mascot2.exe (version 20091016a) and identified by searching the NIST library of peptide fragmentation mass spectra (Human IT Rel. 3, 02/04/09) with MSPepSearch (Version 0.9, NIST), both were downloaded from http://peptide.nist.gov. Mass tolerances were set to 0.7 m/z and 0.8 m/z for precursor and fragment masses, respectively. Additionally, MSPepSearch was set to pre-search all spectra in fast, peptide mode (fiPv), and all top-ranked matches were filtered to a ‘Score’ threshold of >450 (<1% FDR according to previous decoy searches, data not shown). Then, for each identified peptide ion, the maximum abundance was estimated from extracted ion chromatograms (also with ReAdW4Mascot2.exe). Relative peptide intensity was then calculated as the fraction of all peptide intensity values for each peptide. To calculate relative protein intensity values, proteins were reassembled by first mapping the peptides to protein sequences. Only proteins with more than one distinct peptide sequence were counted and peptide intensities were assigned to only one protein. When several proteins contained a given sequence, assignment was made to the protein having the largest number of assigned peptides. Peptide ion intensities for each protein then were summed and the relative abundance for each protein was calculated as the fraction of all protein abundances.
RESULTS
Terminology
In this report, we refer to proteins against which antibodies on the MARS7 and MARS14 columns are directed as targeted proteins. Conversely, we refer to all other plasma proteins as nontargeted proteins, even though some of these may be retained by the columns (see below). Bound and flow-through fractions from MARS columns may contain both targeted and nontargeted proteins. We reserve the term low abundance proteins only for those proteins with reported concentrations of <10 ng mL−1 in human plasma (see Table S3 for references).
Sample recoveries and repeatability for immunodepletion column separations
Three separate samples of human plasma (3.3 mg plasma protein each) were fractionated with MARS-7 and MARS-14 columns and recoveries were determined for both the flow-through and bound fractions (Table 1). Ultraviolet detection (220 nm) traces for each run demonstrated consistent chromatography for both the MARS-7 column and MARS-14 column (Figure S1). An average of 0.40 mg (12% w/w) and 2.09 mg (63%) of applied protein was recovered from the MARS-7 flow-through and bound fractions, respectively. From the MARS-14, an average of 0.25 mg (8%) and 2.23 mg (68%) of the applied protein was recovered from the flow-through and bound fractions, respectively (Table 1). Recoveries of proteins from the flow-through and bound fractions across the three replicates were consistent, with standard deviations between 0 and 4%. Although the data indicate approximately a 25% sample loss of unknown nature, variability in sample recoveries is unlikely to be a major source of variation in analyses that incorporate MARS7 or MARS14 immunodepletion.
Table 1.
Sample yields and recoveries for MARS-7 and MARS-14 column separations.
| Method | Load volume (μL) | Load (mg) | Yield (μg) | % Recovery (±SD) |
|---|---|---|---|---|
| MARS-7 | 40 | 3.3 | 2487± 137 | 75 ± 4 |
| Flowthrough | 401 ± 6 | 12 ± 1 | ||
| Eluate | 2086 ± 132 | 63 ± 4 | ||
| MARS-14 | 40 | 3.3 | 2481± 83 | 75 ± 3 |
| Flowthrough | 253 ± 5 | 8 ± 1 | ||
| Eluate | 2228 ± 81 | 68 ± 3 |
Analysis of immunodepleted plasma protein fractions by LC-MS/MS
We evaluated the repeatability of immunodepletion using the same plasma sample and reverse phase LC-MS/MS. Separations with the MARS-7 and MARS-14 were done three times and the corresponding flow-through and bound fractions were individually analyzed by 1D LC-MS/MS, with database search by Myrimatch and peptide filtering and protein assembly by IDPicker (dataset A, Table S1). A total of 20 proteins were identified in all three replicates from the MARS-7 bound fractions and 107, 106 and 99 proteins were identified by a minimum of 2 distinct peptides from flow-through fractions, respectively. Not included in these totals are many spectral matches to a large number of different immunoglobulin sequences; the extensive sequence overlap between these database entries made it impossible to discern the total number of proteins present based on peptide identifications. A total of 19 proteins were identified in all three replicates from the MARS-14 bound fraction and 114, 118 and 113 proteins were identified from flow-through fraction, respectively. As for the MARS-7 analyses, spectral matches to immunoglobulins were excluded.
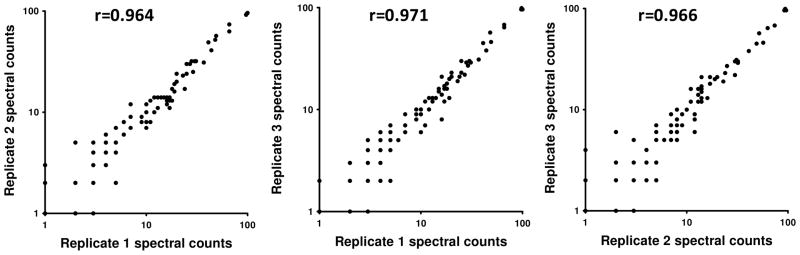
The numbers of proteins identified in the triplicate analyses were largely consistent in both fractions from each column. We used spectral count data for quantitative comparisons, as this method provides robust estimates of differential protein concentrations in shotgun proteomic analyses26, 27. An illustration of the consistency of proteome inventories based on spectral counting is shown in Figure 1. Nonzero spectral count measurements are plotted for all combinations of the 3 replicate analyses of MARS14 flow-through fractions. Spearman Rank correlation coefficients were 0.976 (1 versus 2), 0.971 (1 versus 3) and 0.977 (2 versus 3), respectively, indicating that spectral counts for the large majority of proteins proved highly reproducible across replicate depletion experiments using the MARS system.
Figure 1.
Linear correlations of the spectral counts between pairs LC-MS/MS analyses of three replicate MARS-14 FT fractions. Spearman correlation coefficient (r) is shown for each comparison. Similar results were obtained with MARS-7 depleted flow-through samples.
Efficiency of targeted protein depletion by MARS-7 and MARS-14 columns
The premise of immunodepletion is that enrichment of nontargeted proteins can be achieved through efficient extraction of targeted proteins by the column. We measured efficiency of targeted protein extraction on the basis of spectral counts from LC-MS/MS analyses of bound and flow-through fractions from three replicate depletions (dataset A). Although the bound and flow-through proteins are different in peptide composition, the spectral count data still allowed us to identify those proteins that are efficiently captured by the columns. The binding efficiency was calculated for each protein by dividing the spectral counts for each protein from the bound fraction (B) by the sum of spectral counts from the bound fraction and flow-through fraction (FT), i.e. B/(B+FT)12. In this study, albumin, IgG, antitrypsin, IgA, transferrin and haptoglobin were captured with >99% efficiency both by the MARS-7 and MARS-14 column, as shown in Table 2. Fibrinogen was captured with 71% and 76% efficiency by MARS-7 and MARS-14, respectively. Not all of the targeted proteins were captured efficiently by the MARS-14 column. For example, apolipoprotein AI and AII were captured only with 47% and 31% efficiency by the MARS-14 column.
Table 2.
Capture efficiency for MARS-7 and MARS-14 targeted proteins. Listed proteins 1–7 are targeted by the MARS-7 column; listed proteins 1–14 are targeted by the MARS-14 column.
| IPI no. | Protein name | MARS-7 | MARS 14 | Capture efficiency (%)c | ||||
|---|---|---|---|---|---|---|---|---|
| Ba | FTb | B | FT | MARS-7 | MARS-14 | |||
| #1 | IPI00553177 | Alpha-1-antitrypsin | 115 | 0 | 130 | 0 | >99 | >99 |
| #2 | IPI00641737 | Haptoglobin | 148 | 0 | 159 | 0 | >99 | >99 |
| #3 | IPI00744561 | IGHA1 protein | 69 | 0 | 56 | 0 | >99 | >99 |
| #4 | IPI00448925 | IGHG1 protein | 154 | 0 | 132 | 0 | >99 | >99 |
| #5 | IPI00021885 | Isoform 1 of fibrinogen alpha chain | 114 | 46 | 124 | 39 | 71 | 76 |
| #6 | IPI00745872 | Isoform 1 of serum albumin | 699 | 0 | 849 | 3 | >99 | >99 |
| #7 | IPI00022463 | Serotransferrin | 168 | 0 | 173 | 0 | >99 | >99 |
| #8 | IPI00022429 | Alpha-1-acid glycoprotein 1 | 21 | 48 | 48 | 8 | 86 | |
| #9 | IPI00478003 | Alpha-2-macroglobulin | 2 | 401 | 225 | 47 | 83 | |
| #10 | IPI00021841 | Apolipoprotein A-I | 25 | 154 | 83 | 92 | 47 | |
| #11 | IPI00021854 | Apolipoprotein A-II | 0 | 70 | 20 | 44 | 31 | |
| #12 | IPI00783987 | Complement C3 | 0 | 465 | 252 | 62 | 80 | |
| #13 | IPI00472610 | IGHM protein | 165 | 0 | 147 | 0 | >99 | |
| #14 | IPI00022432 | Transthyretin | 3 | 5 | 29 | 0 | >99 | |
B, bound fraction; listed values are MS/MS spectral counts for proteins detected
FT, flow-through fraction listed values are MS/MS spectral counts for proteins detected
Capture efficiency was calculated as B/B+FT; see text for discussion.
Binding of nontargeted plasma proteins to immunodepletion columns
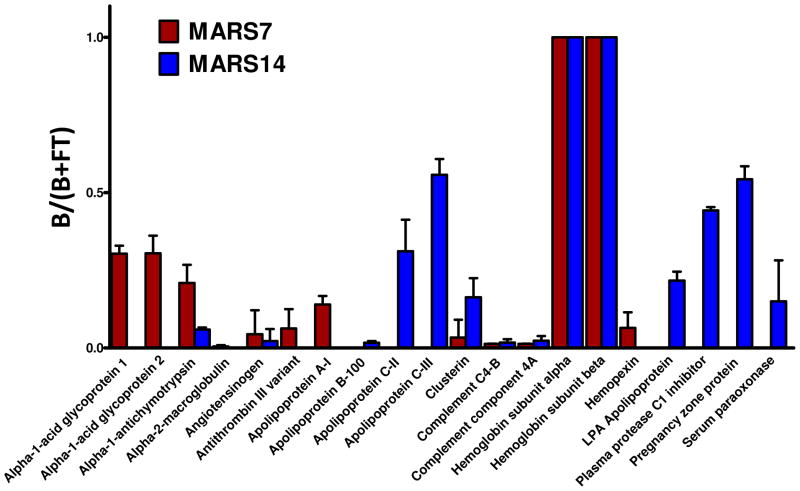
A potential problem with immunoaffinity protein depletion is that some nontargeted proteins might be removed along with the targeted proteins, either because they associate with targeted proteins or because they nonspecifically interact with the column. Capture of some nontargeted proteins by IgY14 and SuperMix was reported previously 13. To evaluate the potential binding of nontargeted proteins to the immunodepletion columns in more detail, we analyzed spectral count data from dataset A for evidence of nontargeted protein binding. Only proteins with two or more unique peptide identifications from all three experiments were considered and all immunoglobulin identifications were removed. Based on these criteria, a total of 20 nontargeted proteins were identified in LC-MS/MS analyses of the bound fractions from the MARS-7 and 19 proteins from the MARS-14 columns. (Spectral count data for all nontargeted proteins identified from the bound fraction are provided in Table S1.) To compare the extent of nonspecific binding in an individual high-abundance protein separation, a “nontargeted capture” value was calculated for each detected nontargeted protein as done for the binding efficiency of targeted proteins (B/(B + FT)). A larger value indicates that a high proportion of the protein is observed in the bound fraction. The capture values for each nontargeted protein with at least 5 spectral counts observed in the triplicate LC-MS/MS analyses of the bound fractions from MARS-7 and MARS-14 are listed in Figure 2. Several of the higher abundance nontargeted proteins bound to the MARS-7 column are target proteins for the MARS-14 column (α-2 macroglobulin, apolipoprotein AI, α1-acid glycoprotein 1, and α1-acid glycoprotein 2). Others were consistently detected only as nontargeted proteins on MARS-14 columns (pregnancy zone protein, apolipoprotein B-100, apoliproprotein CII, apolipoprotein CIII). Of the proteins we detected as having significant nontargeted binding to the MARS columns, CD5 antigen-like protein, zinc-α2–glycoprotein, hemoglobin, apolipoprotein C-III, apolipoprotein L1 and serum amyloid A-4 protein also were reported as nontargeted proteins for the IgY12 column12.
Figure 2.
Nontargeted human plasma proteins that bind to the MARS-7 and MARS-14 column in replicate LC-MS/MS analyses. The plotted values represent the fraction of the total counts for each protein that remain bound (i.e., B/(B + FT)).
Impact of targeted protein depletion on plasma proteome inventory
We used multidimensional LC-MS/MS to evaluate the extent to which lower abundance proteins could be detected in depleted versus undepleted plasma. The same quantities of peptides from unfractionated plasma, the MARS-7 FT fraction and the MARS-14 FT fraction were resolved by IEF into 20 fractions prior to LC-MS/MS (dataset B). Only those proteins identified with at least two unique peptides from the entire study set were considered confident identifications. The resulting IDPicker report summarizes single-analyses of crude plasma, MARS-7 and MARS-14 depleted plasma contained 651 IPI protein entries, 8 proteins included as contaminants and 93 proteins identified by reverse peptide matches for a total of 752 protein entries identified through 89,957 confident spectra-to-peptide matches (The IDPicker report, which contains an .html summary of all identified peptides and proteins is supplied as Supplementary Information. A spreadsheet summary of the identified proteins and associated spectral count data is provided in Table S2.) This original dataset had a unacceptably high protein FDR of 12.0% (2 × 45/752) and thus, we applied additional filtering steps to increase the quality of the protein identifications. We filtered out all proteins that had less than 10 spectra identified in the combined dataset (N=417) and collapsed an additional 19 proteins with more than one IPI identifier from protein groups that represented isoforms of the same protein. Our experience with large datasets is that removal of proteins with less than one spectral count per analytical run reduces false positive protein identifications to less than 5%. This filtering step has the additional benefit of removing proteins with insufficient spectral counts for statistically valid comparisons between datasets.
The final, filtered dataset contained 238 IPI entries with matching Human Genome Organization (HUGO) identifiers, 113 IPI entries without HUGO identifiers (mostly IgGs), 4 contaminant entries and 8 proteins identified by reverse peptide matches. The protein FDR for this filtered dataset was 4.6% (2 × 8/351). Removal of all 131 IPI entries clustering in IgG clusters (#2, 20, 155 and 242 in the IDPicker report) reduced the final dataset to 220 non-overlapping protein groups, of which 213 had matching HUGO identifiers. (The full dataset and the filtered versions are presented in Table S2.)
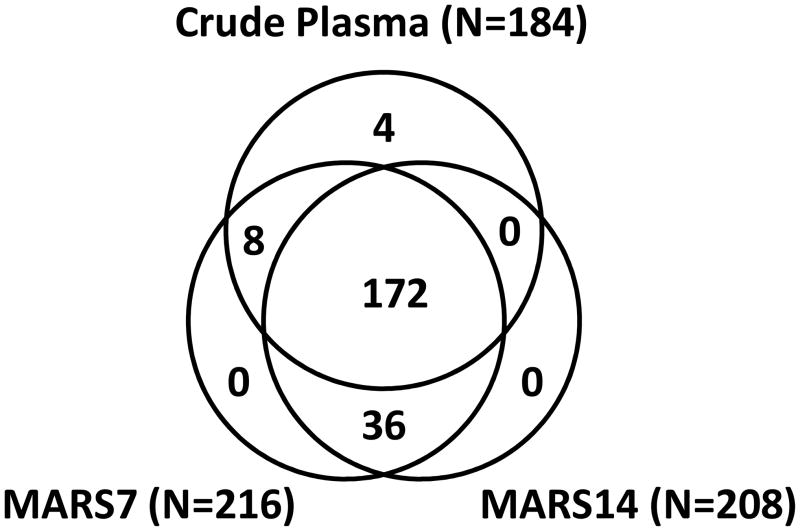
Figure 3 lists a VENN diagram of the identifications in crude plasma, MARS-7 FT and MARS-14 FT for this set of 220 proteins. The 4 proteins that were exclusively found in crude plasma had similarity to immunoglobulins but were sufficiently unique that they did not cluster based on shared sequence homology, while the 8 proteins that were observed in crude plasma and MARS7 included CETP, HP, HPR, ORM1, ORM2, SERPINA1, SLC9A11 and one unknown protein (IPI00877009.1). A total of 36 proteins listed in Figure 3 were found in the MARS-7FT and MARS-14FT fractions, but not in crude plasma. Of these, only four had reported plasma concentrations (isoform I of insulin-like growth factor II, IPI00001611, 380 ng mL−1; intercellular adhesion molecule 1, IPI00008494, 43 ng mL−1; macrophage colony stimulating factor receptor 1, IPI00011218, 26 ng mL−1; isoform D of proteoglycan 4, IPI00655676, 1 ng mL−1) (see Table S3).
Figure 3.
Comparison of proteome inventories from crude plasma, MARS-7 flow-through and MARS-14 flow-through analyzed by IEF-LC-MS/MS. A single IDpicker report was created from 3 separate single-dimensional MS/MS analyses and filtered for immunoglobulins, contaminant proteins and proteins identified by reverse sequences. A total of 220 proteins detectable by at least 10 spectral counts were included in this comparison.
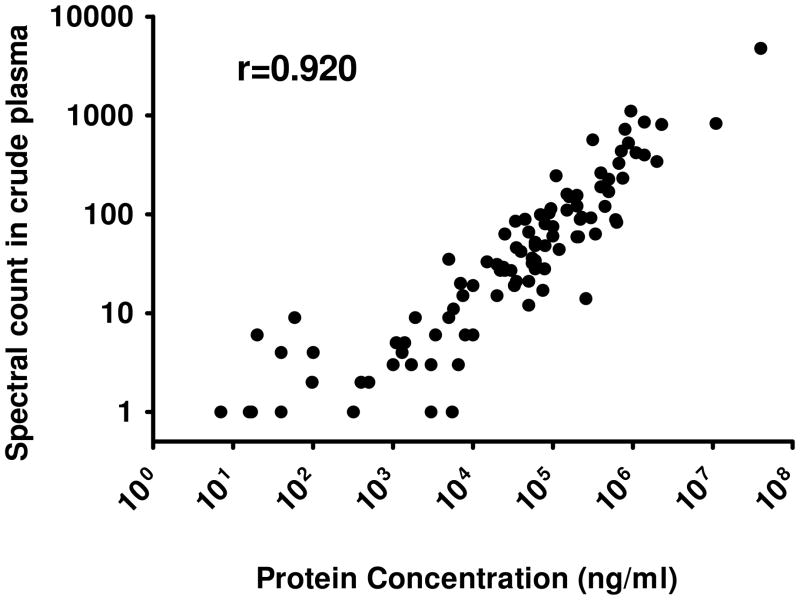
Spectral count data provide a robust means to estimate protein abundances from shotgun proteomic datasets26, 27. Figure 4 illustrates the rank correlation (Spearman r2 = 0.920) between the spectral counts for 100 proteins detected in crude plasma and with previously reported protein concentrations in plasma (a full list with references is provided as Table S3). We compared the spectral counts for proteins detected in the MARS FT fractions to the counts for the same proteins in crude plasma. Only the 100 proteins observed with at least 10 spectral counts in the entire study were used and these were ranked by spectral counts in the crude plasma. (The immunoglobulin identifications were excluded). To assess the level of enrichment for nontargeted proteins, the log2 of the ratio of counts in the MARS FT fraction to crude (unfractionated) plasma was used to estimate the increase (or decrease) in detection as a function of immunodepletion (Figure 5). For the 36 proteins detected only in MARS FT fractions, the denominator of the ratio is zero; thus, for these proteins a single count was used. A positive log2 ratio means that the spectral counts for the protein are higher in MARS flow-through than in crude plasma.
Figure 4.
Linear correlation between the spectral counts for each protein and previously reported concentrations of plasma proteins1 (Table S3.).
Figure 5.
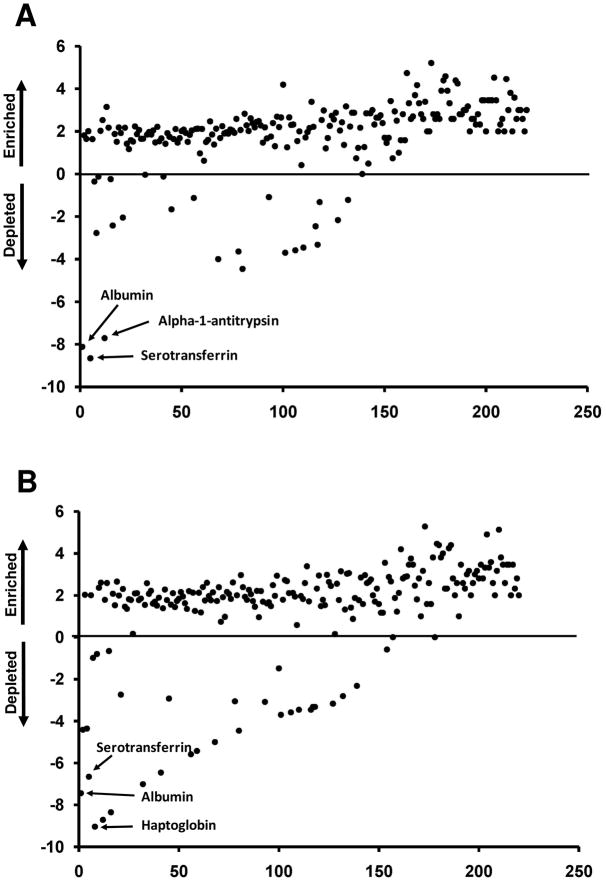
Global impact of MARS-7 (A) and MARS-14 (B) immunodepletion on detection of plasma proteins. Proteins observed with at least 10 spectral counts in the study are represented and are ranked by spectral counts in unfractionated (crude) plasma. Plotted values (y-axis) are the log2 ratios of detected spectral counts for each protein in depleted plasma relative to counts for the same protein in unfractionated plasma.
Analysis of the MARS-7 FT indicated that 188 proteins (85% of the total detected) increased in spectral counts by at least 2-fold compared to unfractionated plasma (Figure 5A). Similarly, in the MARS-14 FT, 180 proteins (82% of the total detected) increased the spectral counts by at least 2-fold (Figure 5B). Based on spectral counts, the median enrichment relative to undepleted plasma was 4.3-fold for the MARS-7 and 4.0-fold for the MARS-14 column. These data suggest that enrichment of nontargeted proteins by top7 and top14 immunodepletions is essentially equivalent. Figure 5 also illustrates the depletion of some proteins by the MARS-7 and MARS-14 columns; these include the targeted proteins and some nontargeted proteins described above and listed in Table 2 and Figure 2.
Among the proteins with enhanced detection in the MARS-7 and MARS-14 FT fractions were 23 proteins present at plasma concentrations of 10 ng mL−1 or lower (Table S3). These low abundance proteins accounted for only 5.6 and 6% of the proteins detected in both the MARS-7 and MARS-14 FT fractions, respectively. Concentrations for these proteins were obtained from previously reported data for plasma or serum levels in normal human subjects1. Because relatively few low abundance proteins (<10 ng mL−1) have been verified by clinical measurements, we are unable to estimate their enrichment.
Impact of protein abundance distribution on proteome analyses
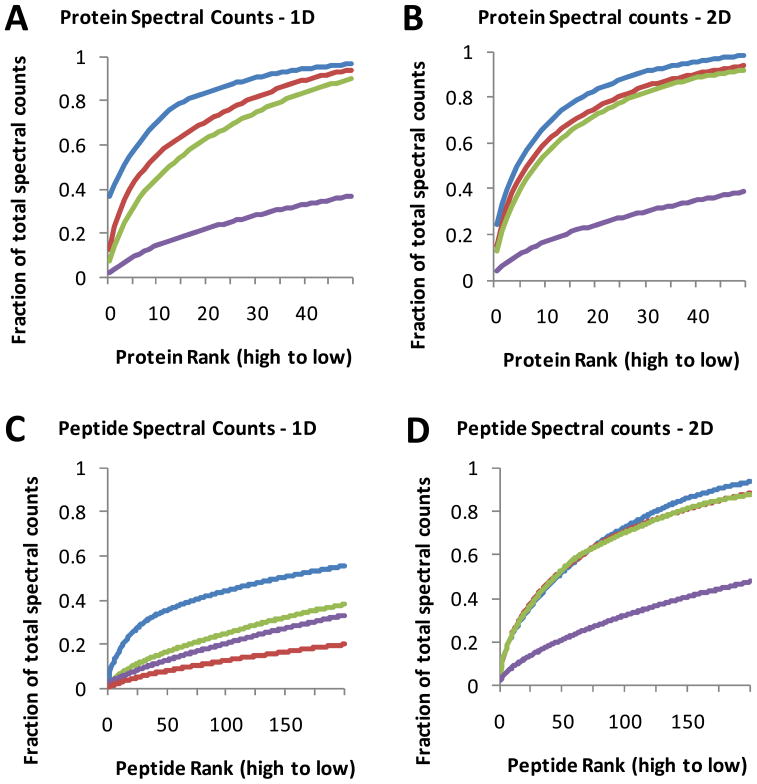
IEF-LC-MS/MS of crude and MARS-7 or MARS-14 immunodepleted plasma collectively yielded 220 high confidence protein identifications (see above). However, this total is only ~10% of the protein identifications generated from identical protein amounts of human colon adenomas and tumor cells with the same IEF-LC-MS/MS platform 17, 18. Although this disparity could be attributed to oversampling of peptide ion from abundant plasma proteins, we are unaware of published data that compare ion sampling in plasma and cell tryptic digests. Oversampling of highly abundant peptides should be evident in spectral count data for the detected peptides and proteins. Plots of protein rank as a function of spectral counts for human colon carcinoma RKO cells, undepleted plasma, and the MARS-7 FT and MARS-14 FT proteomes for a single LC-MS/MS run are shown in Figure 6 (Dataset C, supplemental data). In each plot, the spectral count curve for detected peptides and proteins is considerably steeper for plasma and the MARS FT fractions than for RKO cells. For example, the (blue) curve for plasma in Figure 6A shows that the top 10 proteins account for 70% of the spectral counts, whereas the curve for RKO cells shows that the top 10 proteins account for less than 205 of the total. These relationships are consistently observed in plots where spectral counts for multiple peptides are combined to generate protein spectral counts (Figures 6A and 6B) or when spectral counts for individual peptides are plotted (Figures 6C and 6D). These relationships held for both reverse phase LC-MS/MS analyses (Figures 6A and 6C) and for IEF-LC-MS/MS analyses (Figures 6B and 6D). In analyses of plasma, the MS instrument is presented with a selection of peptide ions that is most heavily biased in favor of the highest abundance proteins. This bias is lessened somewhat in the MARS FT fractions, but the curves for the MARS-7 and MARS-14 FT fractions are still considerably steeper than for RKO cells.
Figure 6.
The distribution of spectral counts for identified proteins (A and B) and peptides (C and D) in unfractionated plasma (blue line), MARS-7 FT (red line), MARS-14 FT (green line) and RKO cell lysate (purple line). Proteins and peptides are ranked on the x-axis in order of decreasing spectral counts in unfractionated plasma. The y-axis indicates cumulative spectral counts as a function of peptide or protein rank. Data are plotted for the top 50 proteins and the top 100 peptides. Results shown are for a single analysis by either reverse phase LC-MS/MS (A and C) or IEF-LC-MS/MS (B and D).
If bias in peptide ion sampling drives the plasma/cell disparity in protein identifications, then spectral counting provides an indirect measure of this effect. Measurement of peptide ion MS1 signal intensities would most directly represent the basis for dynamic sampling by the MS instrument. We therefore extracted MS1 signal intensities for peptides from the LC-MS/MS datasets for RKO cells, undepleted plasma, and the MARS-7 FT and MARS-14 FT fractions. We further calculated protein MS1 intensities from the combined peptide MS1 intensity data from reverse phase LC-MS/MS analyses (no IEF). This procedure utilizes a separate analysis pipeline, in which experimental MS/MS spectra are searched against spectral libraries to identify peptide sequences and their corresponding precursor MS1 signals. This approach thus represents a complementary analysis method to spectral counting based on database search results.
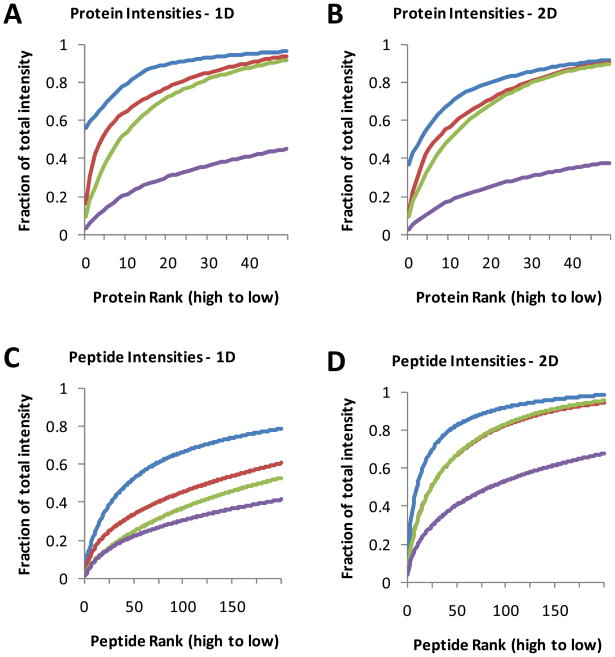
The distributions of the peptide and protein intensities for each sample type are shown in Figure 7. Relative abundance values were plotted according to intensity rank (from highest to lowest). Figure 7A and 7B show large differences in the fraction of total abundance accounted for by the same number of proteins and peptides. For example, in Figure 7A (reverse phase LC-MS/MS), the top protein and top 20 proteins account for 56% and 89% of the total MS1 signal intensity in crude plasma, whereas in RKO cells, the top protein and top 20 proteins account for only 3.7% and 30% of the total intensity. These plots also illustrate the relative effectiveness of the immunodepletion columns. Depletion of the top 14 proteins (with MARS-14) reduced the fractional intensity at protein rank 20 by about 19%, presumably allowing for the detection of additional lower abundance proteins. The protein and peptide signal distributions are similar for IEF-LC-MS/MS analyses (Figure 7C and 7D), with unfractionated plasma displaying the steepest signal intensity curve and RKO cells displaying the flattest curve. These data quantitatively support the hypothesis that the disparity in proteome inventories between plasma and cells is due to the steep protein abundance gradient for plasma.
Figure 7.
The distribution of MS1 signal intensities for identified proteins (A and B) and peptides (C and D) in unfractionated plasma (blue line), MARS-7 FT (red line), MARS-14 FT (green line) and RKO cell lysate (purple line). Proteins and peptides are ranked on the x-axis in order of decreasing signal intensity in unfractionated plasma. The y-axis indicates cumulative MS1 signal intensity as a function of peptide or protein rank. Data are plotted for the top 50 proteins and the top 100 peptides. Results shown are for a single analysis by either reverse phase LC-MS/MS (A and C) or IEF-LC-MS/MS (B and D).
DISCUSSION
We undertook our study to quantitatively assess the performance of top7/top14 plasma immunodepletion with a current generation LC-MS/MS analysis platform. Our results demonstrate that the immunodepletion columns provide specific and highly repeatable depletion of targeted proteins in human plasma and that they broadly enhance detection of nontargeted plasma proteins by a median of 4-fold. The immunodepletion columns displayed consistent performance in depleting targeted proteins, although not all targets were captured with equal efficiency. However, despite the enrichment for nontargeted proteins, analysis of depleted plasma with current multidimensional LC-MS/MS technology primarily detects medium to high abundance proteins. Low abundance proteins (<10 ng mL−1) comprised only 5–6% of the protein identifications in depleted plasma.
A potential concern regarding immunodepletion strategies is the removal of nontargeted proteins through non-specific association with the depletion columns or with captured proteins 28, 29. We observed that the bound fraction associated with immunodepletion columns contained 30 nontargeted proteins, most of which were captured at low levels based on spectral counts (Table S1). This finding is similar to the previous report of 38 nontargeted proteins captured by IgY-12 columns 12. For those nontargeted proteins captured at higher levels, binding appeared to be reproducible (Figure 2). We note that specific interaction with targeted proteins may explain at least a few of the nontargeted captures. For example, hemoglobin is reported to bind to haptoglobin and zinc-α2-glycoprotein may interact with the targeted apolipoproteins and interact directly with the Fc region of IgG 30, 31. Pregnancy zone protein, a relatively abundant pregnancy-associated plasma protein, has strong similarity to alpha-2-macroglobulin 32, which perhaps explained why pregnancy zone protein was only captured by the MARS-14 column (which captures alpha-2-macroglobulin), but not by the MARS-7 column.
Our observations regarding nontargeted protein capture are similar to those reported by Qian et al. 13, who performed similar evaluation of the IgY14 and SuperMix columns. We recognize that the single dimension LC-MS/MS analyses we used to characterize the bound fractions may not detect very low abundance proteins. Nevertheless, these analyses detected nontargeted capture of only a limited number of medium to high abundance proteins. We note that our shotgun proteomic analysis of the MARS-14 bound fraction detected two low abundance plasma proteins (paroxonase and serum amyloid), which suggests that even low abundance proteins would be detected in our bound fractions if they were present. It seems unlikely that the affinities of low abundance proteins for the capture media would be quite different from those of medium and high abundance proteins. Thus, inadvertent capture of nontargeted proteins is generally unlikely to interfere with the performance of analysis platforms that incorporate an abundant plasma protein depletion step.
In addition to consistently depleting targeted proteins and causing minimal depletion of nontargeted proteins, the immunodepletion columns broadly enhanced detection of nontargeted proteins, typically by about 4-fold, based on spectral counts. In our IEF-LC-MS/MS analyses, either MARS-7 or MARS-14 immunodepletion increased the number of detected plasma proteins by approximately 25% relative to non-depleted plasma. Although the MARS-14 column removed 7 additional proteins and a greater fraction of the plasma proteome mass, this conferred no real advantage over the MARS-7 column in enhancing global plasma protein identifications—indeed, the MARS-7 and MARS-14 FT fractions yielded equivalent numbers of identifications (208 vs. 216).
We used spectral counts and MS1 signal intensities to evaluate the sampling of peptide ion signals from plasma, depleted plasma and cell lysate digests. Both the spectral count plots (Figure 6) and the MS1 ion intensities plot (Figure 7) reveal the peptide mixture “seen” by the MS instrument and help to explain differences in identifications between sample types. For example, the profiles for the MARS-7 and MARS-14 fractions (compare the red and green curves) are nearly superimposable, which suggests that these two fractions are similarly complex. The 50 most abundant plasma proteins accounted for over 90% of the ions sampled, whereas in cell proteomes, only about 30% of the sampled ions map to the top 50 proteins. This leaves much greater bandwidth for detection of lower abundance proteins and yields a much larger protein inventory in cells than in plasma. Although these conclusions might have been considered predictable from a broad reading of the existing literature, our work provides the first quantitative documentation of this phenomenon. Moreover, the analytical approach demonstrated in Figures 6 and 7 could be used more generally to explain differences in the sizes of proteome inventories from different cell, tissue and biofluid types.
The principal rationale for using immunodepletion has been to enhance detection of low abundance plasma proteins. Immunodepletion by a top7/top14 approach certainly enhances detection of high and medium abundance nontargeted proteins, but does little to improve detection of low abundance proteins (<10 ng mL−1). Our analyses of the MARS-7 and MARS-14 FT fractions yielded only 23 proteins in this concentration range—approximately 5–6% of the total identifications. These findings confirm published results with IgY14 and SuperMix immunodepletion columns 13 and indicate that, even with immunodepletion, low abundance proteins are most likely to remain undetected by shotgun proteome analyses, even in immunodepleted plasma. Thus, untargeted proteomic analyses using current LC-MS/MS platforms—even with immunodepletion—cannot be expected to efficiently discover low abundance, disease-specific biomarkers in plasma.
Supplementary Material
Acknowledgments
This work was supported by a cooperative agreement award 5U24CA126479 from the National Cancer Institute through the Clinical Proteomic Technology Assessment for Cancer (CPTAC) program and by an interagency agreement between the National Cancer Institute and the National Institute of Standards and Technology.
Abbreviations
- CPTAC
Clinical Proteomic Technologies Assessment for Cancer
- CV
coefficient of variation
- FDR
false discovery rate
- IEF
isoelectric focusing
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MARS
multiple affinity removal system
- TFA
trifluoroacetic acid
Footnotes
MARS™ is a registered trademark of Agilent Technologies, Inc.
Disclaimer
Certain commercial equipment, instruments, or materials are identified in this document. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the products identified are necessarily the best available for the purpose.
References
- 1.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1 (11):845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Pieper R, Su Q, Gatlin CL, Huang ST, Anderson NL, Steiner S. Multi-component immunoaffinity subtraction chromatography: An innovative step towards a comprehensive survey of the human plasma proteome. Proteomics. 2003;3 (4):422–432. doi: 10.1002/pmic.200390057. [DOI] [PubMed] [Google Scholar]
- 3.Pernemalm M, Lewensohn R, Lehtio J. Affinity prefractionation for MS-based plasma proteomics. Proteomics. 2009;9 (6):1420–7. doi: 10.1002/pmic.200800377. [DOI] [PubMed] [Google Scholar]
- 4.Bjorhall K, Miliotis T, Davidsson P. Comparison of different depletion strategies for improved resolution in proteomic analysis of human serum samples. Proteomics. 2005;5 (1):307–17. doi: 10.1002/pmic.200400900. [DOI] [PubMed] [Google Scholar]
- 5.Desrosiers RR, Beaulieu E, Buchanan M, Beliveau R. Proteomic analysis of human plasma proteins by two-dimensional gel electrophoresis and by antibody arrays following depletion of high-abundance proteins. Cell Biochem Biophys. 2007;49 (3):182–95. doi: 10.1007/s12013-007-0048-z. [DOI] [PubMed] [Google Scholar]
- 6.Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5 (13):3292–303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 7.Roche S, Tiers L, Provansal M, Seveno M, Piva MT, Jouin P, Lehmann S. Depletion of one, six, twelve or twenty major blood proteins before proteomic analysis: the more the better? J Proteomics. 2009;72(6):945–51. doi: 10.1016/j.jprot.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Whiteaker JR, Zhang H, Eng JK, Fang R, Piening BD, Feng LC, Lorentzen TD, Schoenherr RM, Keane JF, Holzman T, Fitzgibbon M, Lin C, Zhang H, Cooke K, Liu T, Camp DG, 2nd, Anderson L, Watts J, Smith RD, McIntosh MW, Paulovich AG. Head-to-head comparison of serum fractionation techniques. J Proteome Res. 2007;6 (2):828–36. doi: 10.1021/pr0604920. [DOI] [PubMed] [Google Scholar]
- 9.Gong Y, Li X, Yang B, Ying W, Li D, Zhang Y, Dai S, Cai Y, Wang J, He F, Qian X. Different immunoaffinity fractionation strategies to characterize the human plasma proteome. J Proteome Res. 2006;5 (6):1379–87. doi: 10.1021/pr0600024. [DOI] [PubMed] [Google Scholar]
- 10.Brand J, Haslberger T, Zolg W, Pestlin G, Palme S. Depletion efficiency and recovery of trace markers from a multiparameter immunodepletion column. Proteomics. 2006;6 (11):3236–42. doi: 10.1002/pmic.200500864. [DOI] [PubMed] [Google Scholar]
- 11.Huang L, Harvie G, Feitelson JS, Gramatikoff K, Herold DA, Allen DL, Amunngama R, Hagler RA, Pisano MR, Zhang WW, Fang X. Immunoaffinity separation of plasma proteins by IgY microbeads: meeting the needs of proteomic sample preparation and analysis. Proteomics. 2005;5 (13):3314–28. doi: 10.1002/pmic.200401277. [DOI] [PubMed] [Google Scholar]
- 12.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, 2nd, Smith RD. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5 (11):2167–74. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian WJ, Kaleta DT, Petritis BO, Jiang H, Liu T, Zhang X, Mottaz HM, Varnum SM, Camp DG, 2nd, Huang L, Fang X, Zhang WW, Smith RD. Enhanced detection of low abundance human plasma proteins using a tandem IgY12-SuperMix immunoaffinity separation strategy. Mol Cell Proteomics. 2008;7 (10):1963–73. doi: 10.1074/mcp.M800008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.States DJ, Omenn GS, Blackwell TW, Fermin D, Eng J, Speicher DW, Hanash SM. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24 (3):333–8. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Qian WJ, Chin MH, Petyuk VA, Barry RC, Liu T, Gritsenko MA, Mottaz HM, Moore RJ, Camp DG, II, Khan AH, Smith DJ, Smith RD. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J Proteome Res. 2006;5 (2):361–9. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiteaker JR, Zhang H, Zhao L, Wang P, Kelly-Spratt KS, Ivey RG, Piening BD, Feng LC, Kasarda E, Gurley KE, Eng JK, Chodosh LA, Kemp CJ, McIntosh MW, Paulovich AG. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res. 2007;6 (10):3962–75. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 17.Slebos RJ, Brock JW, Winters NF, Stuart SR, Martinez MA, Li M, Chambers MC, Zimmerman LJ, Ham AJ, Tabb DL, Liebler DC. Evaluation of strong cation exchange versus isoelectric focusing of peptides for multidimensional liquid chromatography-tandem mass spectrometry. J Proteome Res. 2008;7 (12):5286–94. doi: 10.1021/pr8004666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprung RW, Jr, Brock JW, Tanksley JP, Li M, Washington MK, Slebos RJ, Liebler DC. Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis. Mol Cell Proteomics. 2009;8 (8):1988–98. doi: 10.1074/mcp.M800518-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009 doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 20.Cortes HJ, Pfeiffer CD, Richter BE, Stevens TS. Porous Ceramic Bed Supports for Fused-Silica Packed Capillary Columns Used in Liquid-Chromatography. Journal of High Resolution Chromatography & Chromatography Communications. 1987;10 (8):446–448. [Google Scholar]
- 21.Licklider LJ, Thoreen CC, Peng J, Gygi SP. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Analytical Chemistry. 2002;74 (13):3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- 22.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6 (2):654–61. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6 (9):3549–57. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma ZQ, Dasari S, Chambers MC, Litton MD, Sobecki SM, Zimmerman LJ, Halvey PJ, Schilling B, Drake PM, Gibson BW, Tabb DL. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 2009;8 (8):3872–81. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2 (9):667–75. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76 (14):4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 27.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77 (19):6218–24. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 28.Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin EF, 3rd, Liotta LA, Veenstra TD, Conrads TP. An investigation into the human serum “interactome”. Electrophoresis. 2004;25 (9):1289–98. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- 29.Zolotarjova N, Martosella J, Nicol G, Bailey J, Boyes BE, Barrett WC. Differences among techniques for high-abundant protein depletion. Proteomics. 2005;5 (13):3304–13. doi: 10.1002/pmic.200402021. [DOI] [PubMed] [Google Scholar]
- 30.Robert L, Bajic V, Jayle MF. The effect of certain glucides and polyelectrolytes on the combination of hemoglobin with serum haptoglobin and on its catalytic activity. C R Hebd Seances Acad Sci. 1956;242 (24):2868–70. [PubMed] [Google Scholar]
- 31.Kennedy MW, Heikema AP, Cooper A, Bjorkman PJ, Sanchez LM. Hydrophobic ligand binding by Zn-alpha 2-glycoprotein, a soluble fat-depleting factor related to major histocompatibility complex proteins. J Biol Chem. 2001;276 (37):35008–13. doi: 10.1074/jbc.C100301200. [DOI] [PubMed] [Google Scholar]
- 32.Philip A, Bostedt L, Stigbrand T, O’Connor-McCourt MD. Binding of transforming growth factor-beta (TGF-beta) to pregnancy zone protein (PZP). Comparison to the TGF-beta-alpha 2-macroglobulin interaction. Eur J Biochem. 1994;221 (2):687–93. doi: 10.1111/j.1432-1033.1994.tb18781.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.