Abstract
Activation-Induced Cytidine Deaminase (AID) is required for somatic hypermutation and immunoglobulin (Ig) class switch recombination in germinal center B cells. Occasionally, AID can target non-Ig genes and thereby promote GC B cell lymphomagenesis. We recently demonstrated that the oncogenic BCR-ABL1 kinase induces aberrant expression of AID in pre-B acute lymphoblastic leukemia (ALL) and lymphoid CML blast crisis. To elucidate the biological significance of aberrant AID expression, we studied loss of AID function in a murine model of BCR-ABL1 ALL. Mice transplanted with BCR-ABL1-transduced AID-/- bone marrow had prolonged survival as compared to mice transplanted with leukemia cells generated from AID+/+ bone marrow. Consistent with a causative role of AID in genetic instability, AID-/- leukemia had a lower frequency of amplifications, deletions and a lower frequency of mutations in non-Ig genes including Pax5 and Rhoh as compared to AID+/+ leukemias. AID-/- and AID+/+ ALL cells showed a markedly distinct gene expression pattern and AID-/- ALL cells failed to downregulate a number of tumor suppressor genes including Rhoh, Cdkn1a (p21), and Blnk (SLP65). We conclude that AID accelerates clonal evolution in BCR-ABL1 ALL by enhancing genetic instability, aberrant somatic hypermutation, and by negative regulation of tumor suppressor genes.
Introduction
Activation-Induced Cytidine Deaminase protein (AID) is essential for SHM and class switch recombination (CSR) in germinal center (GC) B cells (1, 2). AID introduces point mutations by converting cytidine into uridine followed by UNG1-mediated base excision repair (3). While mutations are largely confined to Ig variable region genes, occasionally AID targets non-Ig genes including BCL6, PIM1, BTG1, RHOH, and PAX5 (4). Hypermutation of non-Ig gene targets has been reported to occur in more than 50% of GC-derived diffuse large-cell lymphomas (DLBCL; 5, 6). In agreement with a role for AID in lymphomagenesis at the GC B cell stage, mice deficient in AID fail to develop GC-derived BCL6-dependent lymphoma (7). Likewise, AID enzymatic activity is required for the acquisition of Myc-IgH translocations (8), which drive malignant transformation in Burkitt's and DLBCL. In addition, recent work analyzing a database of over 1,700 breakpoints suggest that AID can synergize with the RAG1 and RAG2 enzymes to initiate chromosomal translocations at the pro-B/pre-B cell stage that are found in a wide range of B cell malignancies (9).
The BCR-ABL1 oncogene is found in a subset of patients with ALL carrying the so called Philadelphia chromosome. This translocation is the most common cytogenetic abnormality in adults with ALL occurring in approximately 25% of adult patients and approximately 3% of children with ALL (10). The BCR-ABL1 defines a high risk group and as such, patients receive intensive chemotherapy in combination with a tyrosine kinase inhibitor and are considered for bone marrow transplant. A recent study demonstrated dramatically improved outcome for children with BCR-ABL1 ALL, when they were treated with a combination of high dose tyrosine kinase inhibitors and intensive chemotherapy (11). BCR-ABL1 ALL arises from pre-B cells, a compartment that normally does not express AID. We and others recently reported aberrant expression of AID in BCR-ABL1 positive ALL (12, 13). In addition, our group recently showed that over-expression of AID promotes lymphoid blast crisis transformation in BCR-ABL1 chronic myeloid leukemia, an entity that appears clinically similar to de novo BCR-ABL1 ALL but carries multiple distinct molecular characteristics (14, 21). These studies collectively suggest a major role of AID in the clonal evolution of BCR-ABL1-driven leukemias. To formally test this hypothesis in a genetic experiment, we investigated the functional significance of aberrant AID expression in BCR-ABL1 ALL in a loss-of function study.
Materials and Methods
Mice, retroviral transductions and bone marrow transplants
6-8 week old Balb/cwere purchased from Jackson Laboratories (Bar Harbor, MN) and maintained in our animal care facility under standard conditions. A Balb/c AID-/- breeding pair was generously provided by Dr. Michel C. Nussenzweig (Rockefeller University). AID-/- mice were bred and maintained under standard conditions. AID-/- status was confirmed for all transplant donor mice by RT-PCR for Aid (Table S3). All experiments involving mice were reviewed and approved by the Institutional Animal Care and Use Committee. Retroviral transductions and bone marrow transplants were performed as described in Table S3.
Tyrosine kinase inhibitor treatment
Imatinib and Nilotinib were provided by Novartis (Basel, Switzerland). Treatments with Imatinib began 4 days post transplant by oral gavage with a dose of 200 mg/kg/day divided BID for a total of 30 days. In experiments using Nilotinib, 75 mg/kg/dose was administered orally every three days beginning 4 days post transplant and continuing until 60 days post transplant.
Agilent Comparative Genomic Hybridization Analysis
DNA was extracted from CD19 purified leukemia test samples using PureLink Genomic DNA Purification (Invitrogen). CD19 positive lymphocyte DNA obtained from 3 male Balb/c mice was pooled and used as a reference. CD19 purification was done using MACS microbead technology (Miltenyi Biotec). DNA was labeled and hybridized to the Agilent Mouse Genome CGH 244A chip according to manufacturer's protocol by MOgene LC (Saint Louis, MO). Data was analyzed with the use of DNA Analytics 4.0 software (Agilent Technologies, Santa Clara, California). Aberration detection method 2 (ADM-2) with centralization and fuzzy zero correction was used to define aberrant intervals. Default filter settings were applied.
Gene expression analysis
Total RNA was extracted from leukemia cells using Rneasy columns (Qiagen, Valencia, CA). Microarray was performed on mouse genome 430 2.0 arrays according to manufacturer instructions (Affymetrix, Santa Clara, CA). Analysis was done with Partek Genomic Suite software using 1 way ANOVA statistical analysis with a false discovery rate of less than 0.05 to detect differentially expressed genes (p<0.0026). RT-PCR verification was performed using primers and conditions as described in Table S3.
Western blot
Monoclonal antibody against p53 (IC12) was purchased from Cell Signaling Technologies (Danvers, MA). Polyclonal antibody against beta actin was purchased from Abcam (Cambridge, MA). Cells were lysed in the presence of protease inhibitors and the supernatant was run on NuPage 4-12% gradient Bis-Tris gels (Invitrogen). Protein was then transferred to a PVDF membrane and blocked with 5% nonfat milk prior to overnight incubation with primary antibodies at 4°C. Horseradish peroxidase conjugated secondary antibodies followed by incubation with substrate was used to detect protein bands.
Flow cytometry
Leukemia cells were resuspended in PBS and preincubated with anti-CD16/CD32 Fc-block (BD Pharmingen). Aliquots were stained for 15 minutes at 4°C with phycoerythrin-Cy5-conjugated monoclonal antibodies specific for murine Gr-1 and CD19, fluorescein isothiocyanate conjugated monoclonal antibody specific for murine CD19 or c-Kit, allophycocyanin conjugated monoclonal antibody specific for murine CD44 or the appropriate isotype control antibodies. All antibodies were purchased from BD Pharmingen. Cells were washed and resuspended in propidium iodide solution (50 μg/ml propidium iodide in PBS) for subsequent analysis using FACS Scan (BD Biosciences).
MTT assay
Cells were plated at a concentration of 2×105 cells per well in 96 well plates with a volume of 100ul media and incubated overnight at 37°C. The following day Imatinib or SU6656 was added at the indicated concentrations in a total volume of 50 μl and plates were kept at 37°C for 72 hours. Cell proliferation and viability were then determined using the TACS MTT Assay system according to manufacturer's instructions (R&D Systems, Minneapolis, MN).
Results
AID expression shortens leukemia latency in a mouse model of BCR-ABL1 leukemia
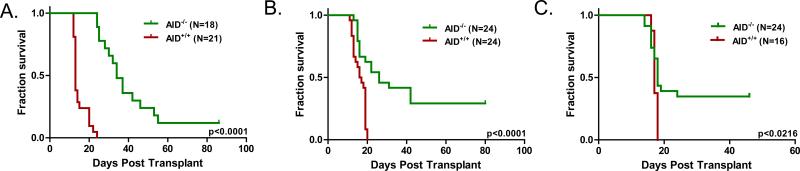
Our previous studies evaluated the role of AID in CML blast crisis progression in AID overexpression experiments (14). To measure the contribution of aberrant AID expression to the clonal evolution of p190 BCR-ABL1 ALL in a genetic experiment, we compared the course of disease of AID+/+ and AID-/- BCR-ABL1 ALL using a classical murine bone marrow transplant model (19; Figure S1). In this model, syngeneic lethally irradiated mice receiving AID-/- or AID+/+ bone marrow cells transduced with a retrovirus carrying the BCR-ABL1p190 oncogene, develop CD19+ pre-B cell leukemia within three weeks (Figure S2, Figure 1A, Table 1). AID mRNA expression was present by RT-PCR in BCR-ABL1 ALL cells and IL4/LPS-stimulated splenocytes from AID+/+ mice but not from AID-/- mice (Figure S2B). To study potential differences in biologic outcome of AID- deficient leukemia, we then evaluated survival in mice transplanted with AID-/- or AID+/+ BCR-ABL1-transformed bone marrow cells by Kaplan-Meier estimate (Figure 1, Table 1). Compared to AID+/+ leukemia, mice transplanted with AID-/- BCR-ABL1 transduced bone marrow had prolonged survival in the primary transplant setting (median 34 days vs. 13 days, p<0.0001 by logrank test). Transduction efficiencies of the retroviral BC-RABL1 oncogene were equivalent in both cohorts as demonstrated by quantitative PCR (Figure S2A). A difference in survival was also observed in a secondary transplant setting, in which leukemia cells from primary recipient mice were recovered from the spleen and purified by CD19+ MACS. From leukemic mice in primary transplant experiments, 104 leukemia cells were transplanted together with 106 non-transduced bone marrow cells into syngeneic lethally irradiated recipients (median survival 26 days vs. 16 days, p<0.0001 by logrank test; Figure 1B, Table 1).
Figure 1. AID-deficiency in acute lymphoblastic leukemia confers prolonged survival in primary and secondary transplants.
A. AID+/+ or AID-/- bone marrow was transduced with the BCR-ABL1 oncogene in vitro. Following transduction, 106 cells were injected into lethally irradiated Balb/c wild type recipients (curve compiled from 2 experiments). B. Kaplan-Meier overall survival of secondary transplantation. Leukemia cells from the spleen and peripheral blood of diseased mice sacrificed in primary transplant experiments were harvested, CD19 purified, and grown in vitro for one week. Following expansion, 104 cells from 6 different AID+/+ mice and 10 different AID-/- mice (~two recipient mice per clone) were injected into lethally irradiated Balb/c wild type recipients (curve compiled from 4 experiments). C. Kaplan-Meier overall survival of tertiary transplantation. 104 cells leukemia cells from secondary transplants were were injected into lethally irradiated Balb/c wild type recipients (curve compiled from 2 experiments). Statistical analysis of the serial transplantation experiments is presented in Table 1.
Table 1.
Statistical analysis of serial transplantation of AID+/+ and AID–/– leukemia cells
| Parameter | Primary AID+/+ | AID–/– | Secondary AID+/+ | AID–/– | Tertiary AID+/+ | AID–/– |
|---|---|---|---|---|---|---|
| Median survival | 13 days | 34 days | 16 days | 26 days | 17 days | 18 days |
| Number of mice | 21 | 18 | 24 | 24 | 16 | 24 |
| Hazard ratio | 25.56 | 5.08 | 2.92 | |||
| p-value | p<0.0001 | p<0.0001 | p=0.0216 |
In the secondary transplant experiments, we noted a narrowing in the median survival time between cohorts with a corresponding decrease in the hazard ratio (Figure 1A-B, Table 1). To determine if this trend continued with increasing passage of leukemia cells, we isolated AID-/- leukemia from secondary transplant recipients that died between 25 and 40 days. These were compared to AID+/+ leukemia isolated from mice in the primary transplant experiments (Figure 1C, Table 1). In this tertiary transplant setting, median survival became equivalent in the two cohorts and the hazard ratio decreased further. A one-sample t-test of the difference between survival times in secondary and tertiary transplants found an average difference of –5.9 days in mice that developed leukemia (standard error 4.05 days, z value –1.47, p=0.15; mice failing to develop leukemia were censored in this analysis). While median survival times narrowed over successive transplants, some clones failed to induce leukemia in both the secondary and tertiary setting. The acceleration of disease in sequential transplantations of AID-/- leukemia cells suggests that in that absence of AID, other factors are able to compensate in the development and clonal evolution of disease. A failure of this process to occur, however, leads to extinction of the clone as exemplified by decreased penetrance in secondary and tertiary recipients. We conclude that AID expression accelerates transformation and leukemic outgrowth in this model, leading to a more aggressive disease phenotype.
AID increases the frequency of amplifications and deletions in BCR-ABL1 leukemia
During somatic hypermutation and class switch recombination in germinal center B cells, deamination of cytosine residues leads to the generation of uracil residues which is subject to error-prone base excision repair and mismatch repair, leading to either mutations or double-strand DNA breaks (20). Deletion and amplification events require DNA double-strand breaks that can result from ubiquitous environmental (e.g. reactive oxygen intermediates) or endogenous factors (e.g. DNA replication errors as a consequence of a high proliferation rate). In the case of BCR-ABL1-driven ALL, two additional mechanisms of clonal evolution can be considered, namely (1) aberrant V(D)J recombination owing to deregulated Rag1 and Rag2 activity and (2) aberrant hypermutation and class-switching, which both depend on enzymatic activity of AID. Indeed, the majority of deletions at the IKZF1 (Ikaros) and CDKN2A (Arf, Ink4a) loci in BCR-ABL1 ALL are introduced exactly at recombination signal sequences that are recognized by the Rag1 and Rag2 enzymes during the process of V(D)J recombination (21, 22). On the other hand, in most cases of BCR-ABL1 ALL, the leukemia cells aberrantly express AID (12, 13), which leads to aberrant somatic hypermutation (Figure 2) and even class-switching in some cases (12). Based on these findings, we hypothesize that AID-dependent DNA breaks can result in amplification and deletion events in BCR-ABL1 ALL cells that aberrantly express AID (12, 13). Therefore we evaluated three AID-/- and three AID+/+ leukemia samples from diseased mice for genomic alterations by comparative genomic hybridization (CGH; Table 2, Table S1). AID-/- leukemia had a lower frequency of both amplifications and deletions in comparison to AID+/+ leukemia (17±2 vs. 45±7 amplifications p=0.0021 by student t-test; 11±2 vs. 40±7 deletions p=0.0025 by student t-test). In BCR-ABL1p190-transgenic mice, progression of leukemia is accompanied by increasing karyotypic abnormalities (23). The decreased frequency of amplification and deletions in our AID-/- leukemia demonstrates that AID indeed contributes to this genomic instability seen in BCR-ABL1-driven ALL. This is consistent with recent data in CML, where copy number alterations that are acquired during lymphoid blast crisis progression are often AID-dependent (14).
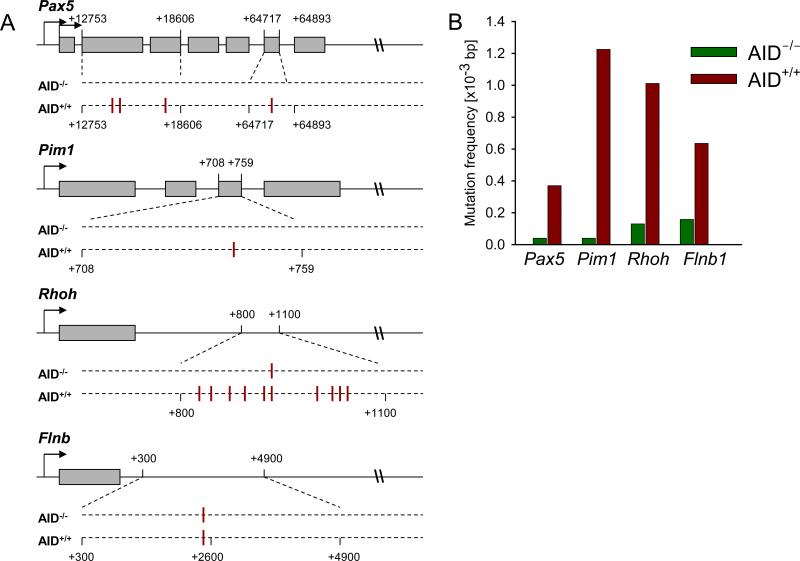
Figure 2. Mutation analysis of Rhoh, Pax5, Pim1, and Flnb.
In (A), genomic loci are shown with exon (boxes) and introns (lines), and are not drawn to scale. Location of mutations in both cohorts of mice are shown below loci with their positions. In AID+/+ leukemia, 4 clones had 4 separate mutations in the Pax5 gene, 2 clones had the same mutation in the Pim1 gene, and 8 clones had 10 different mutations in the Rhoh gene (2 clones had 2 mutations each, all of which were unique. In (B), The overall mutation frequencies [mutations/1,000 sequenced basepairs] are indicated for AID+/+ (red bars) and AID-/- (green bars) leukemia cells for Pax5, Pim1, Rhoh and Flnb genes.
Table 2.
Frequency of amplifications and deletions in murine BCR-ABL1 leukemias
| Cohort | N | Amplifications* (mean ± s.d.)** | Deletions* (mean ± s.d.)** | All Lesions* (mean ± s.d.)** |
|---|---|---|---|---|
| AID-/- | 3 | 17.0 ± 2.0 (15-19) | 10.7 ± 2.1 (9-13) | 27.7 ± 0.6 (27-28) |
| AID+/+ | 3 | 45.3 ± 6.7 (38-51) | 40.0 ± 7.2 (34-48) | 85.3 ± 11.9 (72-95) |
There are significant differences of amplifications (p=0.0021), deletions (p=0.0025) and all lesions (p=0.0011) between AID-/- leukemia and AID+/+ leukemia as determined by Student's t test.
Range shown in parentheses. Mean values were calculated based on the analysis of three tumor samples from three different mice per cohort.
AID induces aberrant somatic hypermutation in BCR-ABL1-driven ALL
Patients with diffuse large B cell lymphoma (DLCL), a germinal center-derived malignancy that expresses AID, are commonly found to have mutations in non-Ig targets of somatic hypermutation (5). In addition, CML cells often acquire somatic mutations in non-Ig loci (e.g. BCL6 and MYC) during progression into lymphoid blast crisis (14). Introduction of these mutations would be consistent with aberrant expression of AID in lymphoid blast crisis CML. For this reason, we studied whether somatic mutations of non-Ig genes are also acquired in BCR-ABL1 ALL and whether acquisition of these mutations is AID-dependent. To this end, we performed a comparative mutation analysis of three known AID target genes in AID+/+ and AID-/- BCR-ABL1 ALL cells (Pax5, Rhoh and Pim1; Figure 2). In AID+/+ but not AID-/- leukemia cells, we found a significant number of mutations within the first intron of Rhoh, similar to patients with DLBCL (5). Also the frequency of mutations in the Pax5 and Pim1 genes was higher in AID+/+ leukemia as compared to AID-/- leukemia, indicating that AID is active in BCR-ABL1 ALL cells. In summary with our CGH data, we conclude that AID contributes to genetic lesions in BCR-ABL1 ALL including amplifications, deletions, and point mutations.
AID-deficient leukemia has a distinct gene expression profile
Studies in primary human breast tumors have found that genetic lesions often result in far-reaching changes at the gene expression level (24). Additionally, lymphomas driven by deregulated expression of Myc and Bcl6 have distinct gene expression profiles in the presence and absence of AID (7). Therefore we investigated if the genomic alterations found in AID+/+ leukemia result in divergent gene expression patterns in BCR-ABL1 ALL as observed in Mycand Bcl6-driven B cell lymphomas.
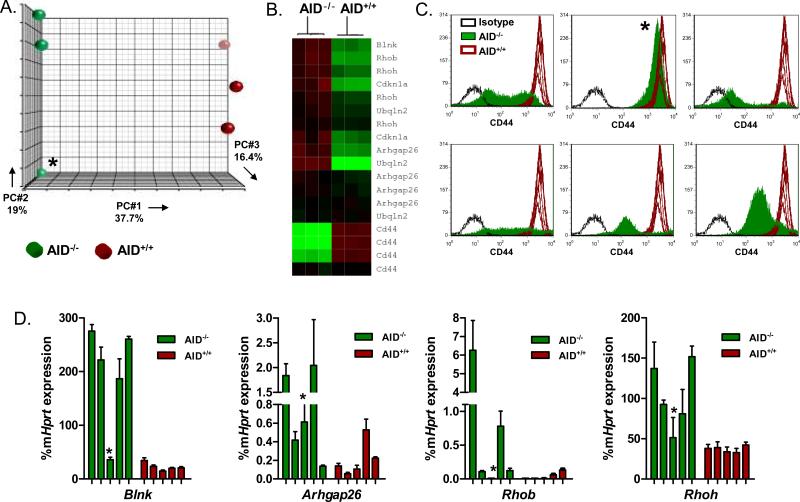
Three AID-/- leukemia samples were compared to three AID+/+ samples by Affymetrix gene expression arrays (Figure 3A, Table S2). Consistent with our hypothesis, we found that AID-/- leukemia was largely separate from AID+/+ leukemia by principle component analysis, supporting the concept that genomic alterations led to divergent gene expression patterns. One AID-/- leukemia sample was an outlier in relation to the other two, although it remains more closely related to AID-/- samples than AID+/+ leukemia (Figure 3A). This leukemia persisted to be unique in many of our analyses and is denoted by an asterisk in all of the figures. Using a one-way ANOVA for statistical analysis, we found 2,365 genes differentially expressed (p<0.0026 FDR 0.05).
Figure 3. Differential gene expression patterns in AID-/- and AID+/+ leukemia.
A. Principle component analysis. RNA was extracted from leukemia cells harvested from 6 mice; 3 mice with AID-/- leukemia and 3 mice with AID+/+ leukemia. The AID-/- outlier is designated by an asterisk in subsequent figures. Samples were hybridized to Affymetrix mouse genome 430A 2.0 arrays. 2,365 genes were found to be significantly differentially expressed (FDR 0.05, p<0.0026). B. Heatmap of genes chosen for validation by quantitative RT-PCR. C. CD44 expression in AID+/+ and AID-/- leukemia. 7 different AID+/+ samples comprise the wild type leukemia histogram (red lines). Shown are 6 different AID-/- leukemia samples in comparison (solid green overlay) and their isotype controls (dashed black line). D. Quantitative real-time RT-PCR of select differentially expressed genes. Data is shown as a percentage of murine Hprt gene expression. For each gene, 5 AID-/- samples and 5 AID+/+ samples from different mice were analyzed.
Inactive X specific transcripts (Xist) and Cd44 were among the genes with the highest gene expression ratio comparing AID+/+ and AID-/- leukemias (Table S2). These genes were also identified in a recent gene expression comparison between AID+/+ and AID-/- Myc- and Myc/Bcl6-driven B cell lymphomas (7). When evaluating the top 227 differentially expressed genes, we noted that several of the genes with higher expression levels in AID-/- leukemia have known tumor suppressor functions (Figure 3D, Figure 4C) including Blnk (SLP65; 28), Arhgap26 (GRAF; 25), Rhob (29), Cdkn1a (p21; 30), and Rhoh (31). Mutations in the first intron of Rhoh as observed in DLBCL and here in BCR-ABL1 ALL (Figure 2) have been implicated in downregulation of gene expression (32). Higher expression levels of Rhoh in AID-/- leukemia is therefore consistent with our mutation analysis of the Rhoh gene (Figure 2). Rhoh is a hematopoietic specific GTPase that is known to negatively regulate Rac-mediated signaling (31). Mice deficient in Rac GTPases have been demonstrated to have prolonged survival in a murine model of BCR-ABL1 disease (33). Mimicking Rhoh function by small molecule inhibition of Rac signaling attenuated leukemic growth in this model, underscoring the importance of Rac/Rhoh-interactions in BCR-ABL1 leukemia (33).
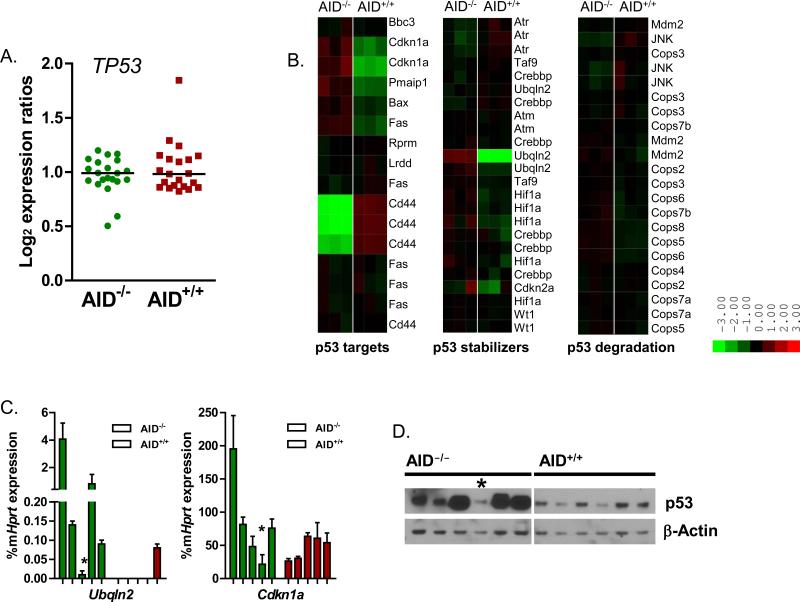
Figure 4. AID-/- leukemia cells fail to downregulate p53.
A. Shown is the log2 expression ratio for the seven different p53 probe sets in the mouse genome 430 2.0 array for each of the AID+/+ samples (N=3) and AID-/- samples (N=3). B. Heat map of downstream p53 targets, molecules involved in the p53 stabilization and p53 degradation. C. Confirmatory quantitative RT-PCR of Ubqln2 and Cdkn1a mRNA levels in AID-/- mice (percentage of Hprt mRNA levels). For each gene, 5 AID-/- samples and 5 AID+/+ samples from different mice were analyzed. D. Western blot analysis of p53 protein levels. Protein from 6 AID-/- leukemia samples and 6 AID+/+ leukemia samples was extracted and evaluated for p53 expression. The AID-/- outlier by PCA component analysis is designated by an asterisk.
In addition to multiple tumor suppressors, we found lower expression levels of Cd44 in AID-/- leukemia (Figure 3). The Cd44 gene encodes an adhesion molecule that is involved in migration and has been shown to be critical in the homing and engraftment of BCR-ABL1 leukemia cells (34). In this study, mice receiving Cd44-/- BCR-ABL1 transduced bone marrow had prolonged survival compared to Cd44+/+ bone marrow, an effect that was overcome by directly injecting cells into the femoral bone marrow of mice. Thus, AID-dependent upregulation of Cd44 may contribute to the shortened latency of AID+/+ compared to AID-/- leukemias and accelerate engraftment. Taken together, we conclude that differential expression patterns of multiple genes including Blnk (SLP65), Cdkn1a (p21), Rhoh, and Cd44 lead to more aggressive disease in AID+/+ leukemia.
AID deficient leukemia fails to downregulate p53
Cdkn1a and Cd44 are known downstream targets of the p53 tumor suppressor; with CD44 expression being suppressed by p53 in contrast to Cdkn1a expression which is enhanced by p53 (35, 36). Due to their differential expression in AID-/- leukemia, we evaluated other downstream targets of p53 and found higher expression levels of Pmaip1 (NOXA) and Bax in AID-/- leukemia as well, consistent with increased p53 levels in AID-/- leukemia (Figure 4). As p53 mRNA levels were similar in AID-/- and AID+/+ leukemia by Affymetrix gene array (Figure 4), we looked at transcript levels of multiple p53 protein stabilizers as well as proteins involved in p53 degradation. The majority of p53-related genes did not have a significant differential expression pattern with the exception of Ubqln2, a gene encoding an ubiquitin-like protein that affects in vivo protein degradation (Figure 4). Ubqln2 has been specifically shown to stabilize the p53 protein (37). In addition to the uniform downregulation of Ubqln2 in AID+/+ leukemia, two of the three AID+/+ samples demonstrated downregulation of Cdkn2a, a gene that encodes the Arf and Ink4a tumor suppressors. Arf inhibits the Mdm2 protein, a key regulator of p53 degradation, and has been reported to be deleted in ~50% of BCR-ABL1 ALL cases (35, 38). Consistent with the differential expression of downstream p53 targets, Cdkn2a and Ubqln2, p53 protein levels were lower in AID+/+ leukemia as determined by Western blot analysis (Figure 4D).
The importance of p53 in progression of multiple tumor types, including BCR-ABL1 leukemia, is well known (35, 39). Loss of p53 is thought to play an important role in disease progression from chronic phase CML to blast crisis (39). Recent studies have shown a loss of the CDKN2A gene in BCR-ABL1 ALL which encodes the Mdm2-inhibitor Arf (38). Therefore functional p53 inactivation by increased protein degradation is an important mechanism in this type of malignancy. p53 represents a DNA damage-response gene, expression of which is induced by DNA breaks that are acquired in GC B cells undergoing somatic hypermutation and class-switch recombination (40). Excessive upregulation of p53 in germinal center B cells however, is prevented by the BCL6 proto-oncogene (41). Outside of germinal centers, B cells lack BCL6 expression and hence do not tolerate AID activity, which potentially affords them protection from the deleterious accumulation of genetic lesions (42). AID-mediated activation of the DNA damage response in the absence of sufficient BCL6 expression would therefore constitute a setting, in which loss of p53 (and other DNA damage-response genes) would confer a critical selective advantage. In agreement with this hypothesis, p53 has been shown to be involved in the censoring of AID-dependent Myc-Igh translocations (43). We propose that specific downregulation of p53 in AID+/+ but not AID-/- leukemia clones is required to allow AID-dependent genetic lesions to occur and to prevent p53-induced apoptosis in response to AID-dependent DNA damage. Conversely, AID-/- leukemia clones only acquire a relatively small number of genetic lesions (Table 2) and therefore do not require downregulation of p53 to the extent that AID-expressing ALL cells do.
SRC kinase-mediated drug-resistance in murine BCR-ABL1 ALL is not AID-dependent
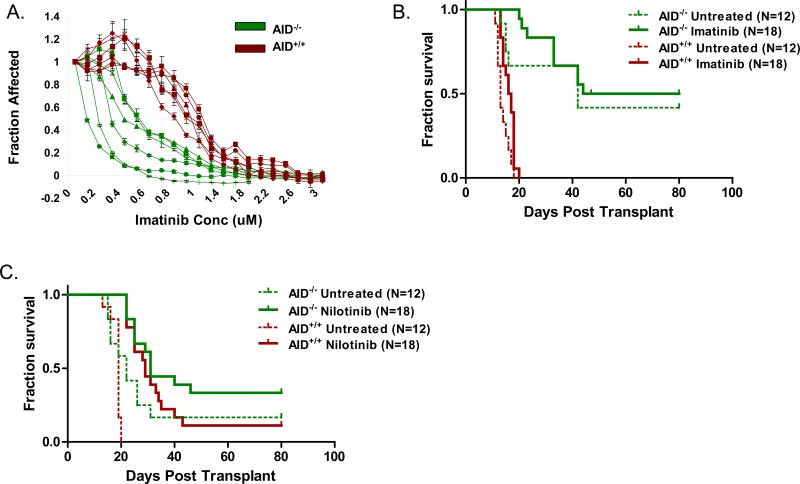
To determine if the differences in the clonal evolution of AID-/- and AID+/+ leukemia also involve the development of drug-resistance, we evaluated their sensitivity to tyrosine kinase inhibition. Fully transformed ALL cells were harvested from mice and subsequently cultured in vitro with Imatinib, a first generation BCR-ABL1 tyrosine kinase inhibitor. We found that AID-/- leukemia cells were more sensitive to treatment than AID+/+ leukemia in a standard cell proliferation assay (Figure 5A). To see if this finding extended to an in vivo setting, we treated secondary transplant recipients with either Imatinib or the second generation tyrosine kinase inhibitor Nilotinib beginning 4 days post transplant (Figure 5B, C). Imatinib failed to provide any benefit in either cohort (Figure 5B), however Nilotinib treatment led to prolonged survival in both AID+/+ and AID-/- leukemia (p=0.003 by Cox regression analysis; Figure 5C). Both AID-/-and AID+/+ cohorts had disease progression in a significant proportion of mice despite treatment with Nilotinib (Figure 5C). We therefore sought to determine if mechanisms of resistance were different.
Figure 5. Sensitivity of AID-/- leukemia to tyrosine kinase inhibition.
A. Leukemia cells were harvested from diseased mice and expanded in vitro. Subsequently cells were plated and treated with increasing concentrations with Imatinib as indicated. After 72 hours of incubation, proliferation was measured using a standard MTT assay. Data is compiled from two separate experiments, each experiment with three different AID-/- (green) and AID+/+ (red) clones. Each line represents an individual clone. B. Secondary transplant recipients were treated with Imatinib beginning 4 days post transplant until 30 days post transplant (data compiled from 2 experiments). C. Secondary transplant recipients were treated with Nilotinib beginning 4 days post transplant until 60 days post transplant as described in Materials and Methods (data compiled from 2 experiments).
Previously, we demonstrated that acquired kinase domain mutations represent a major cause of drug resistance in an in vitro model of p210 BCR-ABL1 B lymphoid blast crisis CML. Of note, the vast majority of these mutations are incorporated in an AID-dependent manner (14). Here we performed a similar experiment in our p190 BCR-ABL1 model for BCR-ABL1 ALL. Three AID-/- and three AID+/+ leukemia samples from tyrosine kinase inhibitor naive animals were grown in vitro with increasing concentrations of Imatinib to select for drug resistant cells. The kinase domain of multiple clones was sequenced from each sample to search for mutations that would confer drug-resistance (Figure S3). One AID+/+ sample was found to have a clinically relevant mutation in the kinase domain known to lead to drug resistance (L387F; 44). The other samples were found to have a wild type sequence, suggesting an alternative form of resistance in this experimental setting. Src kinases have been previously shown to play a significant role in this murine model of BCR-ABL1 driven leukemia, with an absence of disease when multiple Src kinases are deficient (45). Src-kinase-mediated drug-resistance, in which leukemia cells lack kinase domain mutations but have an increased dependence on the Src kinase LYN has also been described in patients (46). We reasoned that there may be differences between AID-/- and AID+/+ leukemia cells in their dependence on Src kinases, and thus a difference in the contribution of Src kinases to Imatinib resistance. To determine if our Imatinib resistant cells had a greater dependence on Src kinases due to BCR-ABL1 inhibition, we studied sensitivity to the Src kinase inhibitor SU6656 (47) and compared the response to Imatinib naïve cells (Figure S3). With the exception of one sample, both AID-/- and AID+/+ cells resistant to Imatinib had an increased sensitivity to Src kinase inhibition when compared to Imatinib naïve cells. We conclude that tyrosine kinase inhibition in both AID-/- and AID+/+ leukemia leads to an increased dependency on Src kinases in vitro. These data collectively indicate that AID expression in BCR-ABL1 ALL cells contributes to the acquisition of drug-resistance in vitro (Figure 5A). The mechanisms of drug-resistance in our experimental system, however, seem to differ from the pathways of drug-resistance most commonly encountered in patients, namely BCR-ABL1 kinase mutations.
Discussion
In a model of BCR-ABL1 driven ALL, we have shown that AID expression leads to a more aggressive phenotype, with a shorter median survival time as compared to AID deficient leukemia. In this model, AID+/+ leukemia is characterized by an increase in genetic lesions including amplifications, deletions, and evidence of aberrant somatic hypermutation. We propose that these genomic differences contribute to the divergent gene expression profiles found by gene expression profiling. As proof of principle, we demonstrated that mutations in the first intron of Rhoh occur at an increased frequency in AID+/+ leukemia consistent with decreased levels of transcript found in this cohort. Surprisingly, gene expression changes between AID+/+ and AID-/-leukemia were profoundly different. Given the function of AID as a cytosine deaminase, one would not expect gene expression changes of this magnitude. On the other hand, two recent reports (25, 26) show that AID also functions as a demethylase at CpG islands in a broad range of promoters throughout the genome. Based on these findings, the far-reaching differences in gene expression between AID-/- and AID+/+ leukemia cells that we observed here may also reflect changes of the methylation status of numerous promoter regions.
Many of the lesions found in our CGH and gene expression data were similar within AID-/- and AID+/+ cohorts, however there were also differences within the cohorts. In our murine model, introduction of the oncogene occurs in vitro followed by intravenous injection. The initial transformation thus happens outside of the mouse. Cell division of a few transformed clones may therefore lead to injection of these clones that carry many of the same lesions into several mice. Subsequent clonal evolution in vivo can then explain the differences found among the mice within a given cohort.
The significantly increased frequency of genetic lesions found in AID+/+ leukemia along with evidence of somatic hypermutation led us to question a possible contribution to tyrosine kinase inhibitor drug resistance. Sequencing of the BCR-ABL1 gene demonstrated a kinase domain mutation in only one of three resistant AID+/+ clones (Figure S3). In a previous study from our group (14), we had shown that AID contributes to the acquisition of BCR-ABL1 kinase domain mutations as a principal cause of drug-resistance. The fact that only one relevant BCRABL1 kinase domain mutation was found in our current experiment may be owing to the fact that leukemia cells acquired resistance to Imatinib by shifting their dependence from BCR-ABL1 to SRC kinases as demonstrated in Figure S3. Autonomous SRC kinase signaling was described previously as a major alternative mechanism of drug-resistance in BCR-ABL1 ALL (45, 46). Our experiments demonstrate that Imatinib resistant leukemia cells have shifted their dependence from BCR-ABL1 to SRC kinase signaling (Figure S3). These findings suggest that activation of SRC kinase signaling rather than acquisition of BCR-ABL1 tyrosine kinase mutations represents the predominant mechanism of drug-resistance in our mouse model of BCR-ABL1 ALL. In human BCR-ABL1 ALL, BCR-ABL1 kinase domain mutations are found in ~85% of Imatinib-resistant leukemia cases (14, 44). This difference may be owing to the fact that our mouse model of BCR-ABL1 ALL may not allow sufficient time for BCR-ABL1 kinase domain mutations to be acquired, or that it favors a resistance phenotype that is immediately available (i.e. SRC kinase signaling). Indeed, injection of BCR-ABL1-transformed B cell precursors into immunodeficient recipient mice leads to lethal disease with a median survival of 13 days (Figure 1, Table 1), whereas human BCR-ABL1 ALL is accociated with a median disease duration of 26 months (48) or longer (11).
In contrast to AID+/+ leukemia cells, their AID-/- counterparts failed to downregulate multiple tumor suppressors at the transcriptional level including Blnk (SLP65), Cdkn1a (p21), and Rhoh. Leukemia cells deficient in AID also failed to downregulate p53 protein levels as opposed to AID-expressing leukemia. Future studies will explore AID as a potential target for therapeutic approaches to prevent drug-resistance and relapse. For instance, AID activity can be suppressed by the cytidine deaminase inhibitor tetrahydrouridine (THU; 49), which is currently undergoing a Phase I trial in combination with 5-Fluoro-2’Deoxycytidine for adults with advanced solid tumors (50). Targeting of AID to hinder further clonal evolution may potentiate current treatment regimens and decrease the incidence of relapse in patients with ALL.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Paul Gaynon for critical discussions. Tanja A.Gruber is supported by NIH grant T32CA09659 and is a recipient of The Saban Research Career Development Fellowship. This work is supported by NIH (grants R01CA137060, R01CA139032, R21CA152497), the Leukemia and Lymphoma Society (Grants 6132-09 and 6097-10), the V Foundation for Cancer Research, the William Laurence and Blanche Hughes Foundation and a Stand Up To Cancer-American Association for Cancer Research (AACR) Innovative Research Grant (IRG00909). Markus Müschen is a Leukemia and Lymphoma Society Scholar.
References
- 1.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 2.Revy P, Muto T, Levy Y, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 3.Ramiro A, San-Martin BR, McBride K, et al. The role of activation-induced deaminase in antibody diversification and chromosome translocations. Adv. Immunol. 2007;94:75–107. doi: 10.1016/S0065-2776(06)94003-6. [DOI] [PubMed] [Google Scholar]
- 4.Liu M, Duke JL, Richter DJ, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 5.Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 6.Neuberger MS, Ehrenstein MR, Klix N, et al. Monitoring and interpreting the intrinsic features of somatic hypermutation. Immunol. Rev. 1998;162:107–116. doi: 10.1111/j.1600-065x.1998.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 7.Pasqualucci L, Bhagat G, Jankovic M, et al. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 8.Robbiani DF, Bothmer A, Callen E, et al. AID is required for chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh C-L, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pui C-H, Relling MV, Downing JR. Acute lymphoblastic leukemia. N. Engl. Med. 2004;350::1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 11.Schultz KR, Bowman WP, Aledo A, Slayton WB, Sather H, Devidas M, Wang C, Davies SM, Gaynon PS, Trigg M, Rutledge R, Burden L, Jorstad D, Carroll A, Heerema NA, Winick N, Borowitz MJ, Hunger SP, Carroll WL, Camitta B. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children's oncology group study. J Clin Oncol. 2009;27:5175–81. doi: 10.1200/JCO.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldhahn N, Henke N, Melchior K, et al. Activation-induced cytidine deaminase acts as a mutator in BCR-ABL1-transformed acute lymphoblastic leukemia cells. J. Exp. Med. 2007;204:1157–1166. doi: 10.1084/jem.20062662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iacobucci I, Lonetti A, Messa F, et al. Different isoforms of the B-cell mutator activation-induced cytidine deaminase are aberrantly expressed in BCR-ABL1-positive acute lymphoblastic leukemia patients. Leukemia. 2009 Sept 17; doi: 10.1038/leu.2009.197. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 14.Klemm L, Duy C, Iacobucci I, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16:232–45. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, Cohen CJ, Peng PD, et al. Development of optimal bicistronic lentiviral vectors facilitates high-level TCR gene expression and robust tumor cell recognition. Gene Ther. 2008;15:1411–1423. doi: 10.1038/gt.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–6. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jumaa H, Bossaller L, Portugal K, et al. Deficiency of the adaptor SLP-65 in pre-B-cell acute lymphoblastic leukaemia. Nature. 2003;423:452–6. doi: 10.1038/nature01608. [DOI] [PubMed] [Google Scholar]
- 18.Soneoka Y, Cannon PM, Ramsdale E, et al. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pear WS, Miller JP, Xu L, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving p210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 20.Chua KF, Alt FW, Manis JP. The function of AID in somatic mutation and class switch recombination: upstream or downstream of DNA breaks. J. Exp. Med. 2002;195:F37–F41. doi: 10.1084/jem.20020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, Relling MV, Shurtleff SA, Downing JR. BCR-ABL1 lymphoblastic leukemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 22.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5’-phosphorylated, RAG-dependent, and cell cycle regulated. Gene. Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 23.Voncken JW, Morris C, Pattengale P, et al. Clonal development and karyotype evolution during leukemogenesis of BCR-ABL transgenic mice. Blood. 1992;79:1029–1036. [PubMed] [Google Scholar]
- 24.Pollack JR, Sørlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. USA. 2002;2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–7. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–5. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jumaa H, Bossaller L, Portugal K, et al. Deficiency of the adapter SLP-65 in pre-B-cell acute lymphoblastic leukaemia. Nature. 2003;423:452–456. doi: 10.1038/nature01608. [DOI] [PubMed] [Google Scholar]
- 28.Borkhardt A, Bojesen S, Haas OA, et al. The human GRAF gene is fused to MLL in a unique t(5;11)(q31;q23) and both alleles are disrupted in three cases of myelodysplastic syndrome / acute myeloid leukemia with a deletion 5q. Proc. Nat. Acad. Sci. USA. 2000;97:9168–9173. doi: 10.1073/pnas.150079597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu A, Cerniglia GJ, Bernhard EJ, Prendergast GC. RhoB is required to mediate apoptosis in neoplastically transformed cells after DNA damage. Proc. Natl. Acad. Sci. USA. 2001;98:6192–6197. doi: 10.1073/pnas.111137198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z-Y, Perkins ND, Ohno T, Nabel EG, Nabel GJ. The p21 cyclin-dependent kinase inhibitor suppresses tumorigenicity in vivo. Nat. Med. 1995;1:1052–1056. doi: 10.1038/nm1095-1052. [DOI] [PubMed] [Google Scholar]
- 31.Gu Y, Jasti AC, Jansen M, Siefring JE. RhoH, a hematopoietic-specific Rho GTPase, regulates proliferation, survival, migration, and engraftment of hematopoietic progenitor cells. Blood. 2005;105:1467–1475. doi: 10.1182/blood-2004-04-1604. [DOI] [PubMed] [Google Scholar]
- 32.Williams DA, Zheng Y, Cancelas JA. Rho GTPases and regulation of hematopoietic stem cell localization. Method. Enzymol. 2008;439:365–393. doi: 10.1016/S0076-6879(07)00427-2. [DOI] [PubMed] [Google Scholar]
- 33.Thomas EK, Cancelas JA, Chae H-D, et al. Rac Guanosine Triphosphatases represent integrating molecular therapeutic targets for BCR-ABL-induced myeloproliferative disease. Cancer Cell. 2007;12:467–478. doi: 10.1016/j.ccr.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemia stem cells. Nat. Med. 2006;12:1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- 35.Whibley C, Pharoah PDP, Hollstein M. p53 polymorphisms: cancer and implications. Nat. Rev. Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 36.Godar S, Ince TA, Bell GW, et al. Growth-inhibitory and tumor suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleijnen MF, Shih AH, Zhou P, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 38.Trotta R, Vignudelli T, Candini O, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3:145–160. doi: 10.1016/s1535-6108(03)00020-5. [DOI] [PubMed] [Google Scholar]
- 39.Honda H, Ushijima T, Wakazono K, et al. Acquired loss of p53 induces blastic transformation in p210bcr/abl-expressing hematopoietic cells: a transgenic study for blast crisis of human CML. Blood. 2000;95:1144–1150. [PubMed] [Google Scholar]
- 40.Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-strand breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- 41.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 42.Muto T, Okazaki I, Yamada S, et al. Negative regulation of activation-induced cytidine deaminase in B cells. Proc. Natl. Acad. Sci. USA. 2006;103:2752–2757. doi: 10.1073/pnas.0510970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramiro AR, Jankovic M, Callen E, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soverini S, Colarossi S, Gnani A, et al. Contribution of ABL kinase domain mutations to imatinib resistance in different subsets of Philadelphia-positive patients: by the GIMEMA working party on chronic myeloid leukemia. Clin. Cancer Res. 2006;12:7374. doi: 10.1158/1078-0432.CCR-06-1516. [DOI] [PubMed] [Google Scholar]
- 45.Hu Y, Liu Y, Pelletier S, et al. Requirement of Src kinases Lyn, Hck, and Fgr for BCRABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat. Genet. 2004;36:453–461. doi: 10.1038/ng1343. [DOI] [PubMed] [Google Scholar]
- 46.Wu J, Meng F, Kong L-Y, et al. Association between Imatinib-resistant BCR-ABL mutation-negative leukemia and persistent activation of LYN kinase. J. Natl. Cancer I. 2008;100:926–939. doi: 10.1093/jnci/djn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blake RA, Broome MA, Liu X, et al. SU6656, a selective Src family kinase inhibitor, used to probe growth factor signaling. Mol. Cell. Biol. 2000;20:9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O'Brien S, Nicaise C, Bleickardt E, Blackwood-Chirchir MA, Iyer V, Chen TT, Huang F, Decillis AP, Sawyers CL. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–41. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 49.Muramatsu M, Sankaranand VS, Anant S, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18471–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 50.Beumer JH, Parise RA, Newman EM, et al. Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2’-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine (THU). Cancer Chemoth. Pharm. 2008;62:363–368. doi: 10.1007/s00280-007-0603-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.