Abstract
The Polarity protein complex Par6/aPKC/Cdc42 regulates polarization processes during epithelial morphogenesis, astrocyte migration and axon specification. Others and we have demonstrated that this complex is also required for disruption of apical-basal polarity during the oncogene ErbB2-induced transformation and TGFβ-induced epithelial mesenchymal transition of mammary epithelial cells. Here, we report that expression of Par6 by itself in mammary epithelial cells induces epidermal growth factor independent cell proliferation and development of hyperplastic three-dimensional acini without affecting apical-basal polarity. This is dependent on the ability of Par6 to interact with aPKC and Cdc42 but not Lgl and Par3 and its ability to promote sustained activation of MEK/Erk signaling. Downregulation of Cdc42 or aPKC expression suppresses the ability of Par6 to induce proliferation, demonstrating that Par6 promotes cell proliferation by interacting with aPKC and Cdc42. We also show that Par6 is overexpressed in breast cancer derived cell lines and in both precancerous and advanced primary human breast cancers suggesting that Par6 overexpression regulates tumor initiation and progression. Thus, in addition to regulating cell polarization processes, Par6 is an inducer of cell proliferation in breast epithelial cells.
Introduction
Par6 is a scaffolding molecule identified in C. elegans as a regulator of asymmetric cell division during embryonic development (1). In mammals, Par6 localizes to tight junctions (2, 3), axon tips (4), and nuclear speckles (5) and regulates diverse cellular processes. Par6 regulates establishment of apical-basal polarity (2, 3), inhibition of cell death (6), directional migration of astrocytes and keratinocytes (7, 8), and axon specification in neurons (4, 9). Thus, Par6 regulates diverse biological processes likely in a cell type and context-specific manner.
Par6 is a scaffolding molecule that is involved in multiple protein-protein interactions (10, 11). The most prominent interactions are those made with the members of the Par polarity complex, which consists of Par3, an additional scaffolding molecule, atypical protein kinase (aPKC) and the GTP binding proteins Cdc42/Rac (12–15). These interactions are required for Par6 regulation of cell biological processes. For instance, Par6 regulates aPKC induced phosphorylation of Lgl (16), Par1 (17) and Numb proteins (18) during establishment of apical-basal polarity in epithelial cells. Likewise, Par6 associated aPKC regulates glycogen synthase kinase 3β (GSK3β) activity to induce polarized migration of astrocytes (19) and to promote cell death during 3D epithelial morphogenesis (6). Thus, the Par6 complex can serve as a signaling node that regulates diverse biological processes by controlling activation of downstream pathways.
In addition to its role during normal cell biology, Par6 also regulates initiation and progression of cell transformation. For example, Par6 cooperates with Rac1/Cdc42 to transform fibroblasts (12). We recently found that Par6 interacts with the oncogene receptor tyrosine kinase ErbB2 and this interaction is required for ErbB2-induced disruption of cell polarity and inhibition of cell death in three-dimensional mammary epithelial structures (20). Par6 also cooperates with transforming growth factor beta (TGFβ) receptor to promote TGFβ-induced epithelial to mesenchymal transition (21). Interestingly, among the three isoforms of Par6 (gene name Pard6), Pard6a, Pard6b, and Pard6c the Pard6b isoform is located in a region of the genome that is frequently amplified and overexpressed in breast cancer (22–25). It is likely that Par6 not only cooperates with regulators of cell transformation but may also directly regulate cell transformation processes.
Here we show that expression of Par6 induces epidermal growth factor independent proliferation of normal mammary epithelial cells by promoting activation of mitogen activated protein kinase (MAPK) signaling. This function of Par6 was dependent on its ability to interact with aPKC/Cdc42, demonstrating that Par6 regulates cell proliferation pathways, in addition to its previously demonstrated role as a regulator of cell polarity and cell migration. Furthermore, we show that Par6 is overexpressed both in human precancerous breast lesions, and in estrogen receptor positive breast cancers suggesting that Par6 pathways are likely to play critical roles during initiation and progression of breast cancer. Thus, the polarity protein Par6 promotes transformation of epithelial cells not only by regulating cell polarity pathways but also by inducing cell proliferation.
Materials and Methods
Cell culture and stable cell line generation
MCF-10A cells were maintained as previously described (44) Comma-1D β geo cells were kindly provided by Daniel Medina (Baylor College of Medicine) and were maintained in DMEM/F12 supplemented with 2% FBS, 10 µg/mL insulin and 5 ng/mL EGF (45). HC11 cells were obtained from the Rosen laboratory and cultured according to (46). Preparation of virus and infection were performed as previously described (47). All cells lines were selected with 1µg/ml of puromyocin.
Cell growth and S-phase assays
MCF-10A (Vector, Par6α,ΔK19A, Pro136, M235W and Par6β) cells were grown to confluency under normal growth conditions and placed in assay medium (DMEM/F12 supplemented with 2% horse serum, mg/ml Insulin, mg/ml hydrocortisone and mg/ml cholera toxin). Cells were then trypsinized and 5×104 (growth curve) or 2.5×105 (S-phase) cells were plated in assay medium. For growth curve assays cells were trypsinized and counted by hemacytometer on day 1,3,5,7,9 and 11. For S-phase assay 30% of the assay media was changed on day 1 and cells were imaged by phase micrscopy before harvesting on day 3. Cells were collected for flow cytometery by trypsinization and subsequent ethanol (70%) fixation. Cells were rehydrated in PBS with 1% Calf serum and stained with 20mg/ml Propidium Iodide (Sigma) containing 100ug/ml RNAseA. Samples were analyzed using an LSRII flow cytometer (Becton Dickinson, San Jose, CA) at least 10,000 cells per sample were collected. The Data from three independent experiments were analyzed using ModFit software (Verity, Topsham, ME). Comma1D cells (Vector, Par6α) were grown to confluency under normal growth conditions and then placed in assay medium (DMEM/F12 supplemented with 0.5% Fetal Bovine Serum, 5µg/ml Insulin). 1×105 cells were plated and cells numbers were counted on day 3 using a hemacytometer.
3D morphogeneis assay
MCF-10A stable cell lines (Vector, Par6α, K19A, and Par6β) were trypsinized and 4,000 single cells/well in an 8 well chamber slide (BD) coated with Matrigel. Cells were grown in assay medium with various amount of EGF (5, 0.5, 0.1 and 0ng/ml). Media was changed and acini were imaged every four days. Day 12 acini structures were analyzed using using AxioVison 4.5 (Zeiss, Thornwood, NY). The acini size distribution of ~600 acini from 3 independent experiments was represented in a box plot. Each box represents 50% of the data within the inter-quartile range. The blue line represents the median value and the spread represents 1.5 times the inter-quartile range and outliers are shown as circles. Statistical analyses were performed using GraphPad Prism software and Mann-Whitney test. HC11 stable cell lines (Vector, Par6α) were grown on Matrigel with 0.5ng/ml EGF and day 12 structures were analyzed as done for MCF-10A cells.
Immunofluorescence
All Immunofluorescence procedures were performed as previously described 44. Microscopy was performed on Zeiss Axiovert 200M using AxioVison 4.5 and ApoTome imaging system (Zeiss, Thornwood, NY).
Biochemistry and immunoprecipitation
MCF-10A cells (Vector, Par6α, K19A, ΔPro136, M235W and Par6β) were plated 2×106 in growth media and cells were stimulated on day 5 with 2ug/ml EGF. Cells were lysed in TNE buffer and immunoprecipitation of with Flag or Par6 antibodies as described previously (20).
DNA constructs
Carboxy or amino-terminal Flagepitope tag mouse par6α was generated by PCR amplification of mPar6α from pFlag-CMV-mpar6α (14) and cloned into MSCV-PURO-IRES-GFP (kindly provided by S. Lowe, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). Point mutations of mPar6α were generated using site directed mutagenesis. Amino-terminal Flag epitope-tag human Par6β was generated by PCR amplification from pBluescriptR-hPar6β (ATCC) and cloned in to MSCV-PURO-IRES-GFP. Two PCKι short-hairpin vectors were obtained from Open Biosystems, Huntsville, AL and subcloned into MSCV-LTR-PURO-IRES-GFP (48). Targeting sequence for PKCι 1: CACAGACAGTAA TTCCATATTAG and PKCι 2: GATTATCTCTTCCAAGTTATTAG. Cdc42 Short-hairpin vector number 3 was obtained from Open Biosystems, Huntsville, AL and subcloned into MSCV-LTR-HYGRO-IRES-GFP (48). Cdc42 Short-hairpin vector number 5 was generated by PCR and subcloned into MSCV-LTR-HYGRO-IRES-GFP. Targeting sequence for Cdc42-3 CGGCCTAAAGAATGTATTTGAT and Cdc42-5: CAAGAATGTATTTGACGAAGCA.
Quantitative PCR
25 breast tumor samples were obtained from the Wigler laboratory (CSHL). RNA was isolated using a Versagene RNA tissue kit (Gentra Systems). Normal breast tissue RNA was a gift from David Mu (CSHL). Breast cancer cell line RNA was a gift from Adrian Krainer (CSHL). MCF-10A control and Par6β overexpressing RNA were obtained from trizol lysis (Invitrogen, Carlsbad, CA) according to the manufactures protocol. cDNA was generated using a Taqman reverse transcriptase kit (Roche). Quantitative real-time PCR was performed using SYBR® green master mix (Applied Biosystems). The following primer sequences were used for Par6β, 5’- GTGAAGAGCAAGTTTGGAGC-3’, and 5’-GATGTCTGATAGCCTACCA-3’ and, for GAPHDH, 5’-CGACAGTCAGCCGCATCTT-3’ and 5’-CGTTGACTCCGA CCTTCA-3’. Samples were run on a Peltier thermalcycler (PTC-200) from MJ Research and data was collected using an Opticon monitor chromo4 continuous fluorescence detector. Cycles were normalized to GAPDH and compared to MCF-10A or normal breast tissue Par6β levels.
Antibodies
Antibodies used in this study were against: Ki67 (Zymed, San Francisco, CA); PKCι, GM130, Ccdc42, Erk2 (BD Biosciences, San Jose, CA); hPar6c antibody was previously described (20); mPar3 (Upstate Biotechnology, Lake Placid, NY); PKCζ (Santa Cruz Biotechnology, Santa Cruz, CA) cleaved caspase-3, p-ERM, p-AKT (Cell Signaling Technology, Danvers, MA); Flag M2, b-Actin (Sigma, St Louis, MO); Laminin (Chemicon, Temecula, CA); p-ERK 1 and 2, p-PKCi, Alexa-Fluor-conjugated secondary antibodies (Invitrogen, Carlsbad, CA) were used.
Analysis of human breast cancers
Gene expression was performed on 112 breast tumors samples and data was generously provided by Therese Sorlei and colleagues. The expression values are median-polished log ratios of expression in tumor cells to that in a reference cell mixture. A kolmogorov-Smirnov null hypothesis test was used to calculate the p-value.
Results
Overexpression of Par6 does not disrupt 3D acini morphogenesis
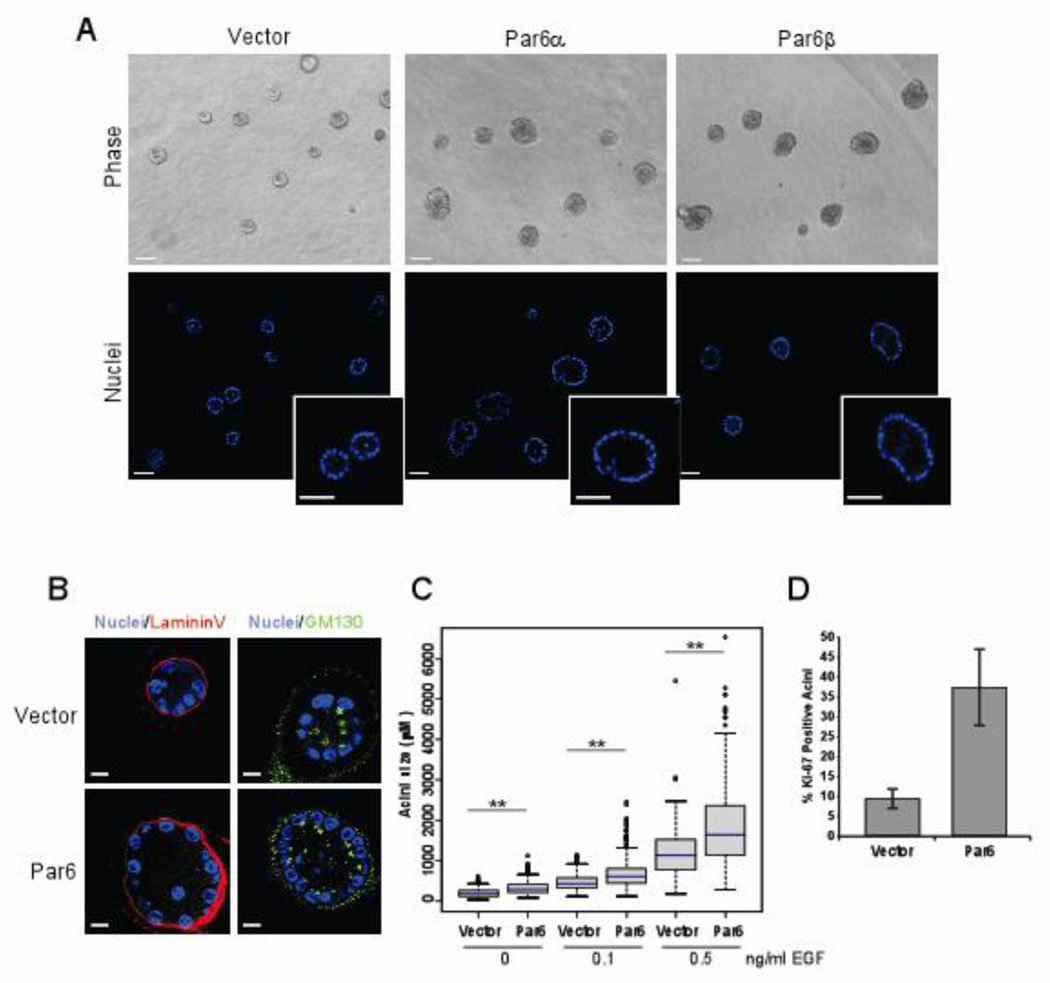
Although Pard6b is amplified in breast cancer, it is not known what role, if any, overexpression of Par6 plays during transformation on breast epithelial cells. To investigate if overexpression of Par6 affects polarization and morphogenesis of mammary epithelial cells in three-dimensional (3D) culture, we expressed Pard6a (protein referred to as Par6α) and Pard6b (protein referred to as Par6β) in an EGF-dependent, non-transformed human mammary epithelial cell line, MCF-10A. When plated on Matrigel™ in the presence of EGF, MCF-10A cells form 3D acini-like structures comprised of a single layer of polarized, proliferation-arrested epithelial cells surrounding a hollow central lumen (26). Overexpression of either Par6 isoform did not affect the ability of MCF-10A cells to form acini with hollow lumens (Figure 1A). In addition, these acini had no detectable loss of apical-basal polarity as determined by using the apical polarity marker GM130, a basal polarity marker Laminin V (Figure 1B and Supp. Figure 1) and a basolateral marker E-cadherin (data not shown). Thus, overexpression of Par6 did not affect polarization and morphogenesis of MCF-10A cells on Matrigel™.
Figure 1. Overexpression of Par6 promotes proliferation without disrupting 3D acini morphogenesis.
(A) Phase contrast (top) and optical sections of acini stained with DAPI (bottom) images of Day 20 acini grown in 0.5 ng/ml EGF, insets show detail of hollow lumen. Scale bar represents 50 µm. (B) Immunofluorescence staining of Day 12 acini structures grown in 0.5 ng/ml EGF with the apical marker GM130 (green and basal marker Laminin V (Red) and co-stained with DAPI to monitor nuclei. Scale bar represents 10 µm. (C) Distribution of acini size (circumferential area) of Day 12 acini structures grown in 0, 0.1 and 0.5 ng/ml EGF. Area of each acini was measured using Zeiss Axiovision 4.5 software and plotted as box plots. The blue line represents the median value and the spread represents 1.5 times the inter-quartile range and outliers are shown as circles. Each condition represents approximately 800 acini structures from three dependent experiments. The P-value between Vector and Par6, for each EGF concentration was less than 0.0001 calculated by Mann-Whitney test. (D) Quantitation of Day 12 acini with at least one positive Ki-67 positive nuclei on Day 12 acini structures grown in 0.5ng/ml EGF Data is represented as percent Ki-67 positive structures +/− s.d. Each bar represents approximately 700 acini from three independent experiments.
Overexpression of Par6 promotes cell proliferation
Although we did not detect any difference in the organization of the acinar structures, we did observe that Par6 expressing acini were larger than the control acini (Figure 1A). To test the hypothesis that Par6 overexpression promotes acinar growth, acini size was quantitated by measuring the area occupied by each acinus. Par6α and β overexpressing cells formed acini that were significantly larger than those formed by control cells, irrespective of the dose of EGF (Figure 1C and Supp. Figure 2B). To determine if the increase in size was due to an increase in cell number or cell size, we counted the number of cells per structure. Acini derived from Par6 overexpressing cells had significantly more cells than those derived from control cells (Supp. Figure 3).
To determine if the increase in cell number is due to changes in cell proliferation rates, we monitored the presence of the proliferation marker, Ki-67 during 3D morphogenesis. Consistent with our previous results (26), acini derived from control cells had low proliferation rates on day 12 (Figure 1D). However, acini derived from Par6 expressing cells had higher proliferation rates until day 20, when compared to acini derived from control cells (Supp. Figure 4A). Furthermore we did not observe any evidence for inhibition of cell death pathways in acini derived from Par6 overexpressing cells (Supp. Figure 4B and C). Thus, Par6 overexpression induces development of hyperplastic acini by inhibiting proliferation arrest during 3D morphogenesis without disrupting polarity.
The ability of Par6 to promote proliferation was observed in multiple, independent populations of MCF-10A cell and in a mouse mammary epithelial cell line demonstrating that there was no clonal or cell line bias to the phenotype (Supp. Figure 5).
Par6 induced cell proliferation requires interaction with aPKC and Cdc42
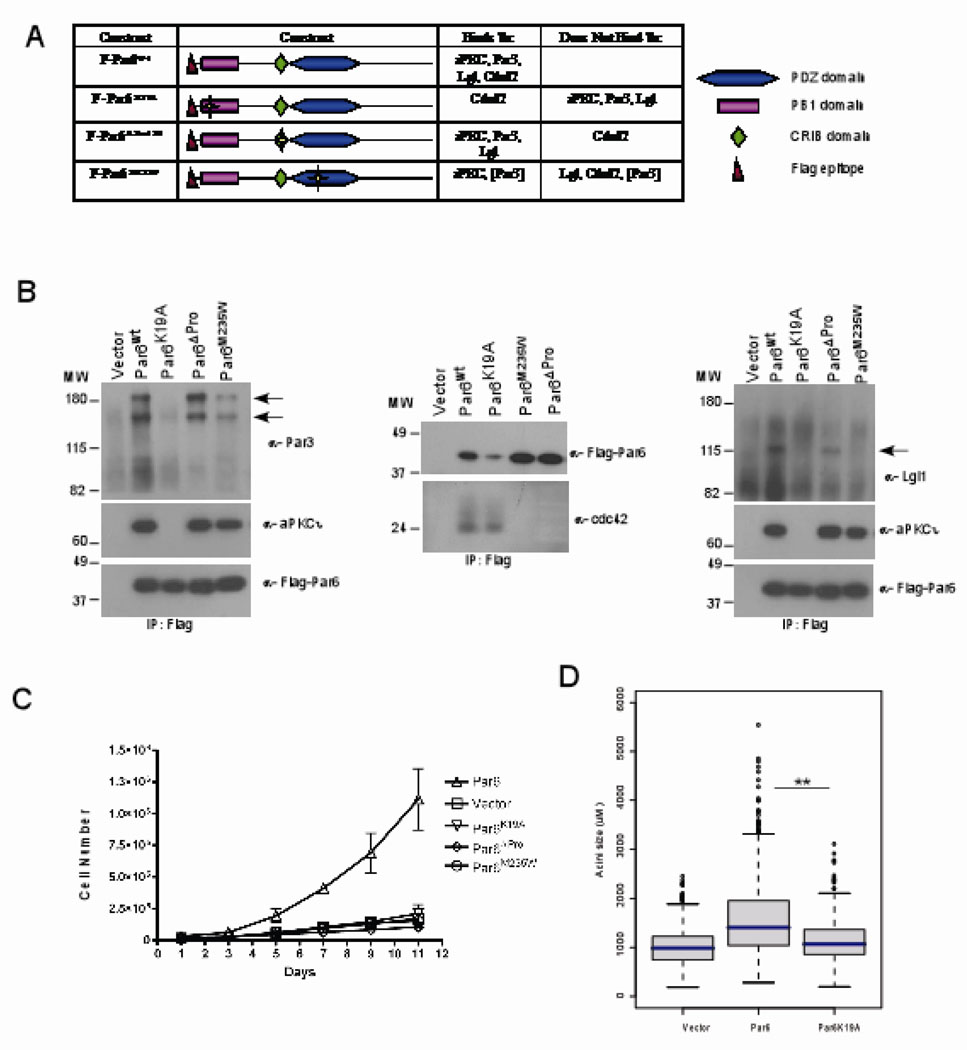
In order to gain insight into the mechanisms by which Par6 induces cell proliferation, we generated Par6 mutants that are defective in binding to members of the Par6 complex such as Par3, Cdc42, Lgl and aPKC. We generated a Lysine to Alanine mutation in the PB1 (Phox/Bem1p) domain (K19A) to abolish binding to aPKC, deleted Proline 136 in the semi-CRIB binding domain (ΔPro136) to disrupt binding to Cdc42, and substituted a Methione with a Tryptophan in the PDZ domain (M235W) to disrupt binding to Lgl (14, 27–30) (Figure 2A). Each mutant was expressed in MCF-10A cells and the ability of the mutants to interact with components of the Par complex was determined by co-immunoprecipitation analysis (Fig 2B, C, D). As expected, K19A failed to associate with aPKC and retained its ability to associate with Cdc42 (Figure 2B). In addition we found that this mutant also lost its ability to interact with Par3 and Lgl (Figure 2B). The ΔPro136 mutant was defective in its ability to bind Cdc42, but associated with Par3, aPKC and Lgl. The M235W mutant did not bind Cdc42, Lgl and was defective in binding Par3 but still associated with aPKC (Figure 2B).
Figure 2. Par6 induced cell proliferation requires interaction with aPKC and Cdc4.
(A) Table summary of the Par6 mutants and their different binding partners. (B) Immunoprecipitaion of cell extracts with Par6 (anti-flag antibodies) and immunoblotted with anti-aPKCi, anti-Flag, anti-Par3, anti-Cdc42 and anti-lgl antibodies. (C) Growth curve of vector control cells compared to Par6wt and mutant Par6 cells over a period of 11 days in EGF free media. Data are means +/− s.d. of estimated cell numbers from three independent experiments. (D)= Distribution of acini size (circumferential area) of Day 12 structures grown in 0.5ng/ml EGF. Each condition represents approximately 800 acini structures from three dependent experiments. The P-value between Vector and Par6wt or Par6wt and K19A is less than 0.0001 calculated by Mann-Whitney test.
Stable populations of MCF-10A cells expressing comparable levels of the Par6 mutants were generated and analyzed for EGF-independent cell proliferation. None of the mutants induced EGF-independent cell proliferation in monolayer cultures (Figure 2C) or enhance cell proliferation during acini formation (Figure 2D and data not shown). The inability of Par6K19A to promote proliferation demonstrated that aPKC binding was necessary and that Cdc42 binding is not sufficient. Conversely, the Par6ΔPro136 mutant demonstrated that interaction with Cdc42 was necessary while aPKC, Par3 and Lgl were not sufficient to promote proliferation. The inability of Par6M235W to promote proliferation confirmed our conclusion that interaction with Cdc42 was necessary and that an interaction with aPKC was not sufficient to induce EGF-independent proliferation. Because the binding of Cdc42 to Par6 induces aPKC kinase activity in the Par6 complex (3, 7) it is likely that Par6-aPKC-Cdc42 forms a core complex to promote EGF-independent cell proliferation of mammary epithelial cells.
Par6 induced cell proliferation requires both aPKC and Cdc42
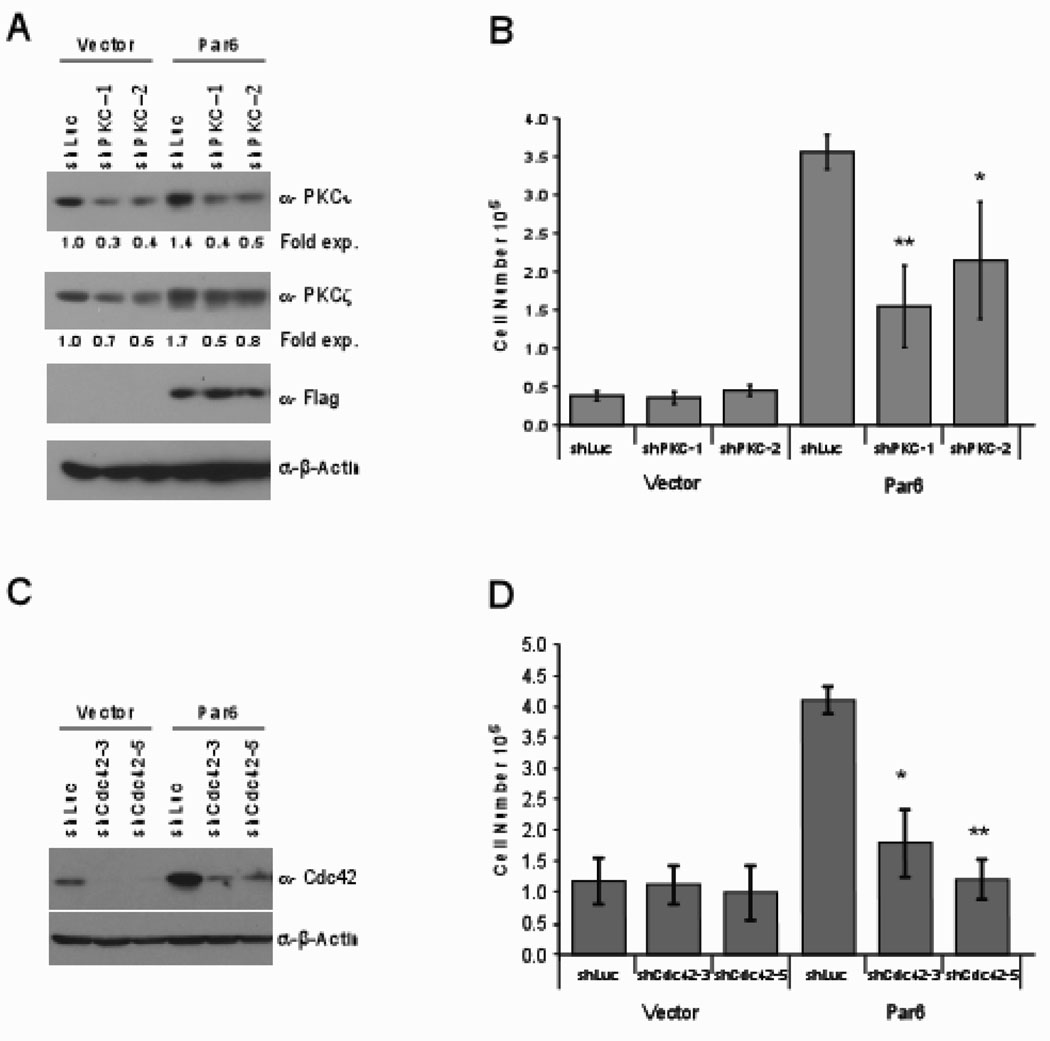
To determine if aPKC and Cdc42 play a role during Par6 induced cell proliferation, we knocked down expression of aPKC and Cdc42 in both control and Par6 expressing cell lines using two independent short-hairpin RNAs for each aPKC and Cdc42. (Figure 3A). The aPKC RNAi vectors significantly downregulated the expression level of PKCι and had a modest effect on the levels of PKCζ (Figure 3A). Decreased expression of PKCi significantly impaired the ability of Par6 overexpressing cells to proliferate in the absence of EGF, while having no significant effect on the basal proliferation observed in vector control cells (Figure 3B).
Figure 3. Par6 induced cell proliferation requires aPKC and Cdc4.
(A) Cell extracts from PKC short-hairpin infected MCF-10A cells immunobloted with indicated antibodies. The numbers below the bands refer to fold change in PKCι and PKCζ protein expression normalized to β-actin as determined by densometric analysis. (B) The indicated cells were grown in EGF free media for seven days and cell number was determined. Data are means +/− s.d. of quantitated cell numbers from three independent experiments. Student t-test were performed showing statistical significance between Par6wtshLuc and Par6wtshaPKC1 (P=0.004) or Par6wtshaPKC2 (P=0.04). (C) Cell extracts from Cdc42 short-hairpin infected MCF-10A cells immunobloted with indicated antibodies. (D) The indicated cells were grown in EGF free media for seven days and cell number was determined. Data are means +/− s.d. of quantitated cell numbers from three independent experiments. Student t-test were performed showing statistical significance between Par6wtshLuc and Par6wtshCdc42-3 (P=0.009) or Par6wtCdc42-5(P=0.0005).
We identified two independent short-hairpins that target Cdc42 (Figure 3C). Both RNAi vectors significantly downregulated Cdc42 in both control and Par6 overexpressing MCF-10A cells (Figure 3C). As observed for aPKC, downregulation of Cdc42 inhibited the ability of Par6 to induce proliferation in the absence of EGF, whereas loss of Cdc42 did not have a significant effect on the basal proliferation observed in vector control cells (Figure 3D). Together these data demonstrates that aPKC and Cdc42 play a critical role during Par6 induced proliferation in MCF-10A cells.
Par6 overexpression activates MAPK signaling
To understand how Par6 expression promotes cell proliferation, we tested if Par6 induced autocrine production of growth factors or juxtacrine activation of cell-cell signaling pathways. Conditioned media from Par6 expressing cells failed to promote proliferation of vector control MCF-10A cells (data not shown). In addition, co-culturing experiments demonstrated that Par6 expressing cells did not induce EGF-independent proliferation of control cells (data not shown). These observations suggest that Par6 promotes proliferation in a cell autonomous manner.
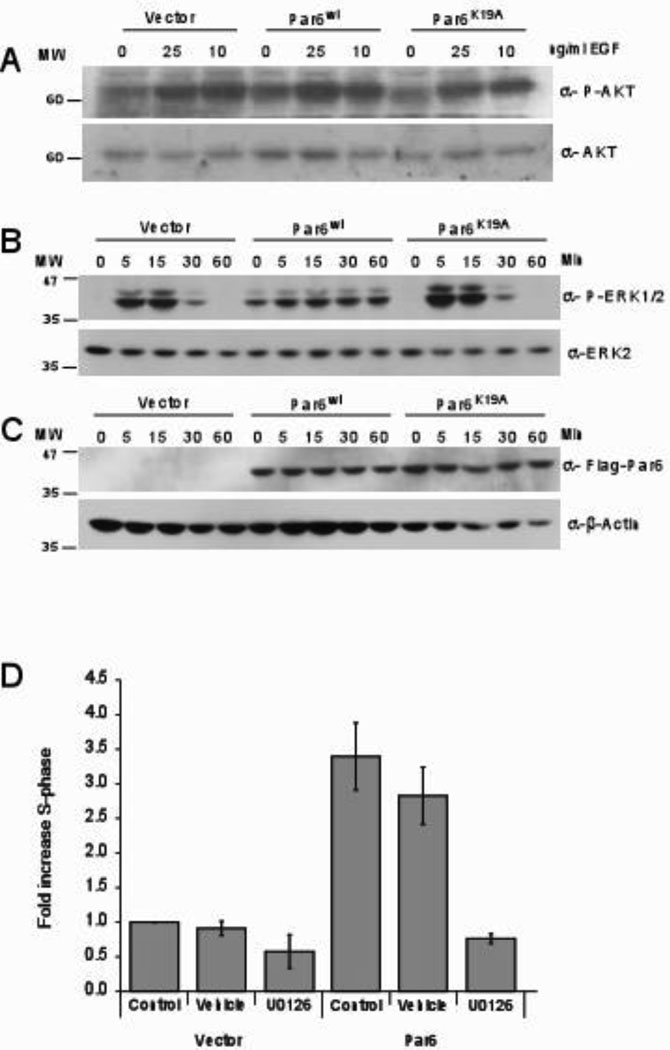
We therefore investigated cell autonomous mechanisms by which Par6 promotes cell proliferation. Activation of the EGFR pathway is an attractive possibility because our data showed that Par6 induces EGF independent proliferation of MCF-10A cells (Figure 2D and Supp. Figure 2C and 5A). However, we did not observe EGF stimulation independent phosphorylation of EGFR in Par6 overexpressing cells (data not shown), suggesting that Par6 overexpression does not directly induce EGFR phosphorylation. To determine if Par6 activates pathways downstream of EGFR, we investigated activation of PLCγ, PI3K, Ras/MAPK and JNK, both in the presence and in the absence of EGF stimulation. Par6 overexpression had no effect on activation of PLCγ, PI3K or JNK pathways either in the presence or in the absence of EGF. (Figure 4A and data not shown). However, Par6 overexpressing cells had a significant increase in ERK phosphorylation in the absence of EGF stimulation. In the presence of EGF stimulation, ERK phosphorylation returned to basal levels by 30 minutes in vector control cells, whereas in Par6 overexpressing cells it was prolonged for 60 (Figure 4B) and 120 minutes (Supp. Figure 2D). In contrast to Par6, the Par6 mutants neither induced phosphorylation of ERK in the absence of EGF nor promoted sustained ERK phosphorylation in the presence of EGF (Figure 4B and Supp. Figure 6).
Figure 4. Par6 overexpression activates MAPK signaling.
(A) Cell extracts from cells that were stimulated with 0, 10 and 25 ng/ml EGF for 15 minutes were immunoblotted with anti-phospho specific AKT and reprobed for total AKT. (B) Cell extracts from cells that were stimulated for 0,5,15, 30 and 60 minutes with 2 ng/ml EGF were immunoblotted with antibodies specific for phosphorylated ERK1/2 and reprobed with total ERK2. (C) Cell extracts were immunobloted with anti-flag antibodies and reprobed with anti-β-actin. (D) Cells were grown in EGF free media for three days with no inhibitors (Control), DMSO (vehicle) or Mek inhibitor (10 µM UO126) and analyzed by flow cytometry. Data are fold increase in S-phase of Par6 expressing cells compared to control cells and are means +/− s.d. of three independent experiments.
To confirm that the effect of Par6 on proliferation is directly due to its effect on ERK activation, we inhibited ERK activation using the MEK kinase inhibitor U0126. Treatment of Par6 overexpressing cells with U0126 blocked both basal and EGF-induced sustained ERK phosphorylation (data not shown). In addition, the inhibitor also blocked EGF-independent cell proliferation (Figure 4D) suggesting a role for MEK kinase in Par6 induced cell proliferation. Thus, we conclude that Par6 overexpression promotes EGF independent cell proliferation by activating the MEK/ERK signaling pathway.
Pard6b is overexpressed in estrogen receptor positive human breast tumors
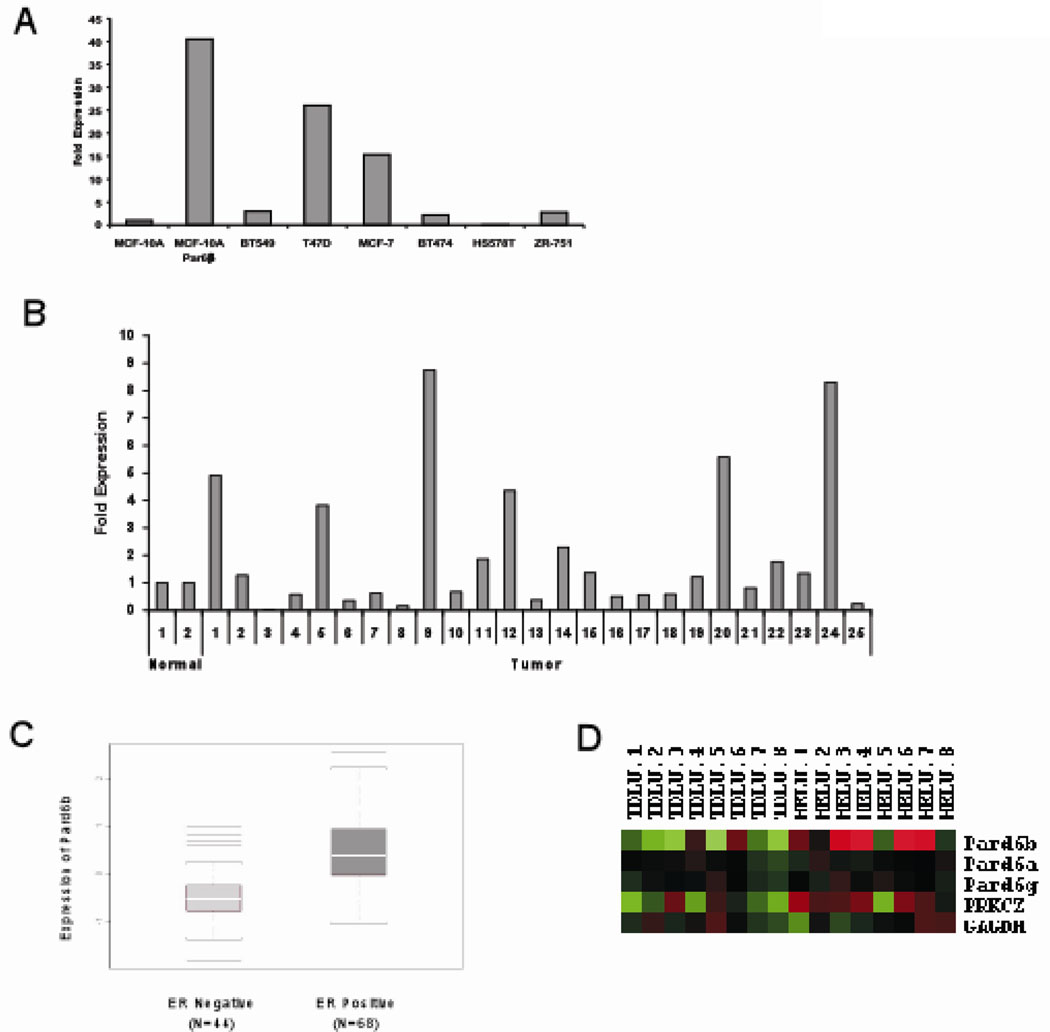
Next we investigated if the genes encoding for the Par6 family of proteins (Pard6) are targeted for overexpression in breast cancer. Interestingly, Pard6b (Chr. 20q13.13) but not Pard6a (Chr. 16q22.1) or Pard6g (Chr. 18q23) is located in a region of the genome that is frequently amplified in breast cancer (22–25) and this amplification correlated with increased Pard6b gene expression (23, 24). To directly test if Pard6b is overexpressed in primary human breast cancers, we performed quantitative PCR analysis from 25 primary breast tumors, two normal breast tissue samples, six breast cancer cell lines and MCF-10A cells. We found that 6/25 primary tumors and 2/6 breast cancer cells express mRNA at least two fold more than the levels observed in their respective controls (Figure 5A and B). Thus, among the Pard6 family members, Pard6b is amplified genomically and transcriptionally upregulated in breast cancer.
Figure 5. Pard6b is overexpressed in estrogen receptor positive human breast tumors.
(A) Quantitative PCR analysis of Par6β gene expression using cDNA from breast cancer cell lines and normalized to GAPDH gene expression. Data is represented as fold increase over MCF-10A control cells. (B) Quantitative PCR analysis of Par6 β gene expression using cDNA generated from primary breast tumors and normalized to GAPDH gene expression. Data is represented as fold increase over the average levels expressed in normal breast tissue. (C) A box plot of the microarray data comparing Pard6b gene expression in ER− verses ER+ tumor samples. A Kolmogorov-Smirnov null hypothesis test was used to calculate the p-value (P = 2.9 × 10−7) (D) Hierarchical clustering from a supervised comparison between paired samples of normal TDLUs and HELUs. Pard6b is expressed 2.5 fold (P = 0.02) and PKCZ 1.46 fold (P = 0.029) in HELU compared to TDLU (40).
Among the breast cancer cell lines tested, the two cell lines (MCF-7 and T47D) that overexpressed Par6b are also estrogen receptor (ER) positive, suggesting a relationship between Par6b overexpression and ER status. To determine if there is a relationship between ER status and Pard6b expression, we compared Pard6b mRNA levels in 68 of ER positive and 44 of ER negative breast tumors. Overexpression of Pard6b showed a significant positive correlation with ER+ status (Figure 5C). We did not observe any relationship between Par6 overexpression and ErbB2 amplification in human breast tumors (data not shown), demonstrating that Par6db overexpression is specific to the ER positive subtype of breast cancer. In addition, analysis of a public gene expression database Oncomine (31) showed that Pard6b expression positively correlated with ER in four independent studies (Supp. Figure 7) (23, 32–34). Inhibition of ER signaling in MCF-7 cells by treatment with Tamoxifen did not result in a significant decrease in the levels of Par6db mRNA (Supp. Figure 8), suggesting that Pard6b levels may not be directly regulated by estrogen. Thus, it is likely that overexpression of Par6 cooperates with EGF to provide a proliferative advantage to ER positive breast cancers.
Almost all precancerous breast lesions are ER positive (35). Furthermore, we recently showed that hyperlastic enlarged lobular units (HELU), the earliest histologically identifiable precursor of breast cancer, show high proliferation rates and an increased expression of members of both EGF and ER signaling modules (35, 36). Interestingly, like the 3D acini derived from Par6 overexpressing MCF-10A cells, HELUs are hyperplastic structures where the acini are enlarged in size due to increase in cell number with no apparent loss of acinar architecture. Given the architectural similarity, we tested if Par6 was overexpressed in HELUs. RNA isolated from microdissected HELU and adjacent TDLU were analyzed by microarray (36). Pard6b but not Pard6a or Pard6g, was overexpressed by 2.5 fold (p=0.002) in HELUs compared to adjacent TDLUs. The gene encoding for PKCζ (PRKCZ), a component of Par6 complex, was also upregulated by 1.46 fold (p=0.029) compared to TDLUs (36) (Figure 5D). Thus our results demonstrate that overexpression of Par6/aPKC is observed early in precancerous lesions and retained during development of ER positive cancers.
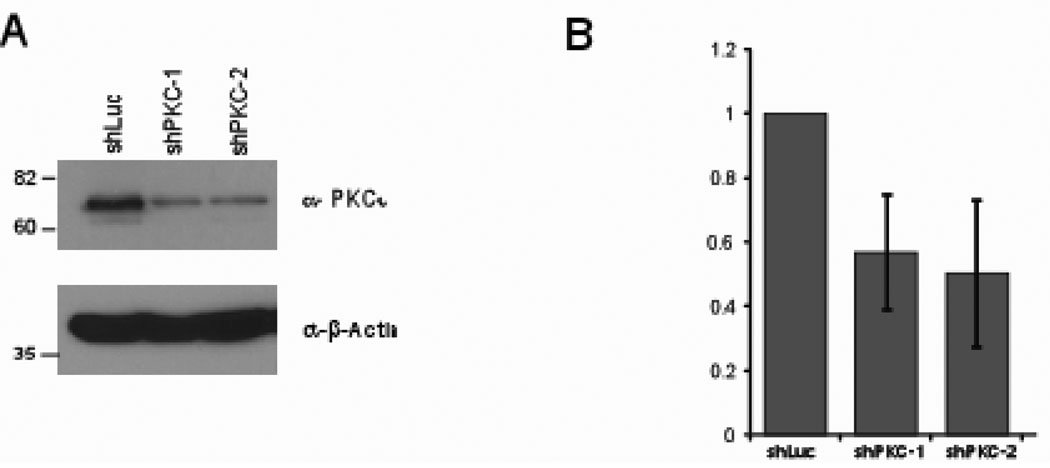
It is possible that the Par6/aPKC/Cdc42 complex is a novel target for therapeutic intervention for both early and late stage breast cancers. To test this possibility, we downregulated expression of aPKC in a breast cancer-derived cell line, MCF7. Downregulation of PKCi decreased cell proliferation (Figure 6A and B) suggesting that inhibiting aPKC is likely to have to important therapeutic relevance. Taken together our results demonstrate that Par6/Cdc42/aPKC is a regulator of cell proliferation and suggests that the Par6 complex as novel therapeutic target for breast cancer.
Figure 6. aPKC regulates MCF-7 cell proliferation.
(A) Cell extract from PKC short-hairpin infected MCF-10A cells immunobloted with indicated antibodies. (B) The indicated cells were grown growth media containing 10% serum for seven days and cell number was determined. Data are fold decrease in cell number of MCF-7shPKC expressing cells compared to vector control cells and are means +/− s.d. of three independent experiments.
Discussion
We demonstrate that overexpression of Par6 in mammary epithelial cells promotes activation of MAPK and induces cell proliferation. In addition, we show that Par6 is overexpressed in both precancerous human breast lesions and in advanced breast cancers. Thus our results identify a novel role for the polarity protein Par6 as an inducer of cell proliferation, which is likely to play an important role during initiation and progression of breast cancer.
Our results show that overexpression of Par6α and β in non-transformed mammary epithelial cells does not affect establishment of apical-basal polarity during acinar morphogenesis. While, several studies have reported that altered expression of both Par6α and β delays establishment of apical polarity (2,3), they also find that the cells recover from the effects of Par6 overexpression and eventually develop cell polarity. This suggests that compensatory mechanisms act redundantly to ensure apical polarity formation in the presence of altered Par6 expression. Since MCF-10A acinar morphogenesis takes several days, it is likely that Par6 overexpressing cells use these compensatory mechanisms to establish proper apical polarity. We found that ectopic expression of Par6 promotes proliferation of multiple mammary epithelial cell lines, demonstrating that in addition to its known role as a polarity regulator, Par6 can also induce cell proliferation.
Both aPKC and Cdc42 are required for Par6 to stimulate of cell proliferation. This is consistent with the established role of aPKC and Cdc42 in Par6 mediated regulation of tight junction biogenesis, polarized cell migration, and cell death (3,6,19,37). Although previous studies have shown that Par6 induced modulation of GSK3β activity is required for directed cell migration and apoptosis, GSK3β is unlikely to play a role in Par6 induced cell proliferation, because we did not see any differential phosphorylation of GSK3β in Par6 overexpressing cells (data not shown). Instead we found that Par6 overexpression induces ERK phosphorylation in the absence of EGF, identifying MAPK pathway as an effector of the Par6-aPKC-Cdc42 complex. MAPK signaling can be activated by ligand-independent phosphorylation of EGFR (38). However, we did not observe phosphorylation of EGFR in the absence of EGF ruling out a role for EGFR phosphorylation as a mechanism to activate MAPK pathway in Par6 overexpressing cells. Previous studies have shown that atypical PKC is not only sufficient but is also required for activation of ERK in a Mek-dependent manner in response to serum stimulation (39–42). Similarly, we find that Par6 uses aPKC to activate ERK in a MEK-dependent manner. The precise mechanism by which aPKC activates MEK remains to be understood. Thus, in addition to its known effectors that regulate cell polarity, the Par6-aPKC-Cdc42 complex also regulates molecules that induce cell proliferation.
Our results show that Par6β is overexpressed both in HELUs, and in ER positive advanced carcinomas. HELUs are characterized by high rates of cell proliferation, ER positivity and increased expression of EGF family of growth factors. It is possible that HELUs arise from cooperation between overexpressed Par6 and EGF ligands, similar to how Par6 overexpression induced hyperplastic acini in our MCF-10A 3D cultures. Considering that Par6 overexpression promoted hyperplastic acini in MCF-10A 3D culture in cooperation with EGF, it is possible that these two factors could cooperate during development of premalignant lesions in the breast. Our data combined with other studies that show that Par6 is amplified genomically and overexpressed in ER positive advanced carcinoma suggest that there is a genetic advantage associates with increased Par6 expression during breast cancer progression.
Par6 is also known to cooperate with oncogenes associated to breast cancer progression. For example Par6 plays a critical role during ErbB2 induced transformation of organized epithelia (20) and ErbB2 amplification is associated with the premalignant breast disease, DCIS (43). We did not observe either a positive or negative correlation between Par6 overexpression and ErbB2 status. Therefore, ErbB2 requires expression of Par6 but not overexpression. In addition, Par6 is required for TGFβ induced epithelial to mesenchymal transition, a cellular process that is associated with invasion. The ability of Par6 to interact with oncogenes in distinct cellular processes suggests that Par6 has multiple functions. The results presented in this manuscript, taken together with those above, suggest that the polarity signaling pathways regulated by Par6 plays a cooperative role during the initiation and progression to breast cancer. Thus, a further understanding of the alterations in the Par6 signaling pathways could identify both novel drug targets and predictive biomarkers for breast cancer progression.
Supplementary Material
Acknowledgements
The authors would like to thank Tony Pawson for Par6a cDNA, and Therese Sorlei and Anne Lisa Bornestein for sharing the microarray data. We would like to thank members of the Muthuswamy laboratory, Dan Nolan and Cathy Courmier for helpful suggestions. This work was supported by US Army predoctoral fellowship, DAMD 17-03-1-0193 (MEN), and funding support to SKM that include grants from NCI CA098830 and CA105388, and grants from Rita Allen Foundation, FACT foundation, Glen Cove Cares, and Long Islanders Against Breast Cancer.
References
- 1.Watts JL, Etemad-Moghadam B, Guo S, et al. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development. 1996;122:3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- 2.Gao L, Joberty G, Macara IG. Assembly of epithelial tight junctions is negatively regulated by Par6. Curr Biol. 2002;12:221–225. doi: 10.1016/s0960-9822(01)00663-7. [DOI] [PubMed] [Google Scholar]
- 3.Yamanaka T, Horikoshi Y, Suzuki A, et al. PAR-6 regulates aPKC activity in a novel way and mediates cell-cell contact-induced formation of the epithelial junctional complex. Genes Cells. 2001;6:721–731. doi: 10.1046/j.1365-2443.2001.00453.x. [DOI] [PubMed] [Google Scholar]
- 4.Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- 5.Cline EG, Nelson WJ. Characterization of mammalian Par 6 as a dual-location protein. Mol Cell Biol. 2007;27:4431–4443. doi: 10.1128/MCB.02235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim M, Datta A, Brakeman P, Yu W, Mostov KE. Polarity proteins PAR6 and aPKC regulate cell death through GSK-3beta in 3D epithelial morphogenesis. J Cell Sci. 2007;120:2309–2317. doi: 10.1242/jcs.007443. [DOI] [PubMed] [Google Scholar]
- 7.Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 8.Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. ACF7: an essential integrator of microtubule dynamics. Cell. 2003;115:343–354. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- 9.Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- 10.Macara IG. Parsing the polarity code. Nat Rev Mol Cell Biol. 2004;5:220–231. doi: 10.1038/nrm1332. [DOI] [PubMed] [Google Scholar]
- 11.Brajenovic M, Joberty G, Kuster B, Bouwmeester T, Drewes G. Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J Biol Chem. 2004;279:12804–12811. doi: 10.1074/jbc.M312171200. [DOI] [PubMed] [Google Scholar]
- 12.Qiu RG, Abo A, Steven Martin G. A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCzeta signaling and cell transformation. Curr Biol. 2000;10:697–707. doi: 10.1016/s0960-9822(00)00535-2. [DOI] [PubMed] [Google Scholar]
- 13.Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 14.Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol. 2000;2:540–547. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- 15.Johansson A, Driessens M, Aspenstrom P. The mammalian homologue of the Caenorhabditis elegans polarity protein PAR-6 is a binding partner for the Rho GTPases Cdc42 and Rac1. J Cell Sci. 2000;113:3267–3275. doi: 10.1242/jcs.113.18.3267. [DOI] [PubMed] [Google Scholar]
- 16.Plant PJ, Fawcett JP, Lin DC, et al. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat Cell Biol. 2003;5:301–308. doi: 10.1038/ncb948. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki A, Hirata M, Kamimura K, et al. aPKC Acts Upstream of PAR-1b in Both the Establishment and Maintenance of Mammalian Epithelial Polarity. Current Biology. 2004;14:1425–1435. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 18.Smith CA, Lau KM, Rahmani Z, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO Journal. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- 20.Aranda V, Haire T, Nolan ME, et al. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol. 2006;8:1235–1245. doi: 10.1038/ncb1485. [DOI] [PubMed] [Google Scholar]
- 21.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 22.Hicks J, Krasnitz A, Lakshmi B, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginestier C, Cervera N, Finetti P, et al. Prognosis and Gene Expression Profiling of 20q13-Amplified Breast Cancers. Clin Cancer Res. 2006;12:4533–4544. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- 24.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 26.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamanaka T, Horikoshi Y, Sugiyama Y, et al. Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr Biol. 2003;13:734–743. doi: 10.1016/s0960-9822(03)00244-6. [DOI] [PubMed] [Google Scholar]
- 28.Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12:39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Hurd TW, Margolis B. Tight junction protein Par6 interacts with an evolutionarily conserved region in the amino terminus of PALS1/stardust. J Biol Chem. 2004;279:30715–30721. doi: 10.1074/jbc.M401930200. [DOI] [PubMed] [Google Scholar]
- 30.Noda Y, Kohjima M, Izaki T, et al. Molecular recognition in dimerization between PB1 domains. J Biol Chem. 2003;278:43516–43524. doi: 10.1074/jbc.M306330200. [DOI] [PubMed] [Google Scholar]
- 31.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perou CM, Jeffrey SS, van de Rijn M, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proceedings of the National Academy of Sciences. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Vijver MJ, He YD, van 't Veer LJ, et al. A Gene-Expression Signature as a Predictor of Survival in Breast Cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 34.Zhao H, Langerod A, Ji Y, et al. Different Gene Expression Patterns in Invasive Lobular and Ductal Carcinomas of the Breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S, Mohsin SK, Mao S, Hilsenbeck SG, Medina D, Allred DC. Hormones, receptors, and growth in hyperplastic enlarged lobular units: early potential precursors of breast cancer. Breast Cancer Res. 2006;8:R6. doi: 10.1186/bcr1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, Medina D, Tsimelzon A, et al. Alterations of Gene Expression in the Development of Early Hyperplastic Precursors of Breast Cancer. Am J Pathol. 2007 doi: 10.2353/ajpath.2007.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao L, Macara IG, Joberty G. Multiple splice variants of Par3 and of a novel related gene, Par3L, produce proteins with different binding properties. Gene. 2002;294:99–107. doi: 10.1016/s0378-1119(02)00681-9. [DOI] [PubMed] [Google Scholar]
- 38.Miranti CK, Brugge JS. Sensing the environment: a historical perspective on integrin signal transduction. 2002;4:E83–E90. doi: 10.1038/ncb0402-e83. [DOI] [PubMed] [Google Scholar]
- 39.Berra E, Diaz-Meco MT, Dominguez I, et al. Protein kinase C ? isoform is critical for mitogenic signal transduction. Cell. 1993;74:555–563. doi: 10.1016/0092-8674(93)80056-k. [DOI] [PubMed] [Google Scholar]
- 40.Berra E, Diaz-Meco MT, Lozano J, et al. Evidence for a role of MEK and MAPK during signal transduction by protein kinase C zeta. Embo J. 1995;14:6157–6163. doi: 10.1002/j.1460-2075.1995.tb00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bjorkoy G, Perander M, Overvatn A, Johansen T. Reversion of Ras- and phosphatidylcholine-hydrolyzing phospholipase C-mediated transformation of NIH 3T3 cells by a dominant interfering mutant of protein kinase C lambda is accompanied by the loss of constitutive nuclear mitogen-activated protein kinase/extracellular signal-regulated kinase activity. J Biol Chem. 1997;272:11557–11565. doi: 10.1074/jbc.272.17.11557. [DOI] [PubMed] [Google Scholar]
- 42.Schonwasser DC, Marais RM, Marshall CJ, Parker PJ. Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol Cell Biol. 1998;18:790–798. doi: 10.1128/mcb.18.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allred DC, Clark GM, Molina R, et al. Overexpression of HER-2/neu and its relationship with other prognostic factors change during the progression of in situ to invasive breast cancer. Hum Pathol. 1992;23:974–979. doi: 10.1016/0046-8177(92)90257-4. [DOI] [PubMed] [Google Scholar]
- 44.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 45.Danielson KG, Oborn CJ, Durban EM, Butel JS, Medina D. Epithelial Mouse Mammary Cell Line Exhibiting Normal Morphogenesis in vivo and Functional Differentiation in vitro. PNAS. 1984;81:3756–3760. doi: 10.1073/pnas.81.12.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xian W, Schwertfeger KL, Vargo-Gogola T, Rosen JM. Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J Cell Biol. 2005;171:663–673. doi: 10.1083/jcb.200505098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci U S A. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dickins RA, Hemann MT, Zilfou JT, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.