Plants live in complex environments in which they intimately interact with a broad range of other organisms. Besides the plethora of deleterious interactions with pathogens and insect herbivores, relationships with beneficial microorganisms are frequent in nature as well, improving plant growth or helping the plant to overcome stress. The evolutionary arms race between plants and their enemies provided plants with a highly sophisticated defense system that, like the animal innate immune system, recognizes nonself molecules or signals from injured cells, and responds by activating an effective immune response against the invader encountered. Recent advances in plant immunity research underpin the pivotal role of cross-communicating hormones in the regulation of the plant’s defense signaling network (Spoel and Dong, 2008; Pieterse et al., 2009). Their powerful regulatory potential allows the plant to quickly adapt to its hostile environment and to utilize its resources in a cost-efficient manner. Plant enemies on the other hand, can hijack the plant’s defense signaling network for their own benefit by affecting hormone homeostasis to antagonize the host immune response (Grant and Jones, 2009). Similarly, beneficial microbes actively interfere with hormone-regulated immune responses to avoid being recognized as an alien organism (Van Wees et al., 2008). In nature, plants simultaneously or sequentially interact with multiple beneficial and antagonistic organisms with very different lifestyles. However, knowledge on how the hormone-regulated plant immune signaling network functions during multispecies interactions is still in its infancy. Bioinformatic and systems biology approaches will prove essential to crack this tough nut.
HORMONE CROSS TALK IN PLANT DEFENSE
Plant hormones are essential for the regulation of plant growth, development, reproduction, and survival. The importance of salicylic acid (SA), jasmonates (JA), and ethylene (ET) as primary signals in the regulation of plant defense is well established (Bari and Jones, 2009; Pieterse et al., 2009). Pathogens that require a living host (biotrophs) are commonly more sensitive to SA-mediated defense responses, whereas pathogens that kill the host and feed on the contents (necrotrophs) and herbivorous insects are generally affected by JA/ET-mediated defenses (Glazebrook, 2005; Howe and Jander, 2008). Changes in hormone concentration or sensitivity, such as triggered upon interactions with biotic agents, activates a bouquet of hormone signaling events that steers adaptive plant responses. The final outcome of the activated defense response is greatly influenced by the composition and kinetics of the hormonal blend produced (De Vos et al., 2005; Mur et al., 2006; Koornneef et al., 2008; Leon-Reyes et al., 2010).
Hormone cross talk is a process in which different hormone signaling pathways act antagonistically or synergistically, thereby providing a powerful regulatory potential to flexibly tailor the plant’s adaptive response to a variety of environmental cues. Cross talk between SA, JA, and ET signaling pathways emerged as an important regulatory mechanism of plant immunity (Spoel and Dong, 2008; Grant and Jones, 2009; Pieterse et al., 2009). Many studies have demonstrated that endogenously accumulating SA antagonizes JA-dependent defenses, thereby prioritizing SA-dependent resistance over JA-dependent defense (Koornneef and Pieterse, 2008). ET often plays a modulating role in this respect (Leon-Reyes et al., 2009, 2010; Zander et al., 2009). However, depending on the plant species and strategy of the attacker, JA can also antagonize the SA pathway. For instance, in wild tobacco (Nicotiana attenuata) it was demonstrated that the JA and ET bursts triggered by herbivore-derived elicitors, impede the SA burst, resulting in tuning of the SA-JA signal interaction and hence the defense responses of the plant to herbivory (Diezel et al., 2009). Recently, abscisic acid (ABA), auxins, gibberellins, and brassinosteroids emerged as forces on the battle field as well. In many cases, these hormones interact antagonistically or synergistically with the SA-JA-ET backbone of the plant immune signaling network, thereby redirecting its defense output (Wang et al., 2007; Navarro et al., 2008; Yasuda et al., 2008; Campos et al., 2009; de Torres-Zabala et al., 2009; Ton et al., 2009; De Vleesschauwer et al., 2010; Jiang et al., 2010).
HORMONE CROSS TALK IN A CHANGING ENVIRONMENT
While the importance of hormones in the hard wiring of the plant immune signaling circuitry is evident, little is known about how environmental factors feed into the regulatory network. Light signaling has been shown to strongly influence hormone-regulated defenses. For instance, Arabidopsis (Arabidopsis thaliana) plants display an altered sensitivity to JA-dependent defenses under conditions of crowding and shade, which is regulated via the photoreceptor phytochrome (Moreno et al., 2009; Robson et al., 2010). This results in a trade-off between resource investment in JA-dependent defense and increased growth to outcompete neighbors (Moreno et al., 2009). Cross talk between ABA-mediated drought stress and JA-dependent defense against necrotrophic pathogens in Arabidopsis was recently shown to be regulated by the transcription factor OVEREXPRESSOR OF CATIONIC PEROXIDASE3 (Ramirez et al., 2009). Interestingly, SA was shown to antagonize both adaptive responses (Spoel et al., 2007; Yasuda et al., 2008). However, in the adaptive response of Arabidopsis to heat stress, SA and JA seem to act together (Clarke et al., 2009), highlighting the complexity of the regulatory processes involved in the adaptive response of plants to their continuously changing environment.
DECOY TACTICS
While hormone cross talk may provide the plant with a powerful regulatory potential to finely tune its defense, cross talk is also a possible target for plant attackers to manipulate the immune signaling network for their own benefit. In recent years, several nice examples of pathogens and insect herbivores that hijack specific hormone-regulated signaling pathways to redirect the immune response have been described (for review, see Robert-Seilaniantz et al., 2007; López et al., 2008; Grant and Jones, 2009). For instance, the wilting pathogen Fusarium oxysporum has been shown to take control of the JA pathway in Arabidopsis, thereby enhancing Fusarium wilt-disease symptoms that lead to plant death (Thatcher et al., 2009). The bacterial pathogen Pseudomonas syringae was shown to promote disease by altering the host’s auxin and ABA physiology through the injection of specific virulence effectors into the host cell (Chen et al., 2007; de Torres-Zabala et al., 2007, 2009). P. syringae-induced ABA levels appeared to suppress SA biosynthesis and action, resulting in enhanced susceptibility to this pathogen (de Torres-Zabala et al., 2007, 2009). In addition, P. syringae produces the virulence factor coronatine, which acts as a molecular mimic of biologically the most active form of JA, jasmonoyl-Ile. Coronatine suppresses SA-dependent defenses, thereby also contributing to the suppression of the host’s immune response to this pathogen (Uppalapati et al., 2007; Katsir et al., 2008). Interestingly, coronatine directly binds to the JA receptor CORONATINE INSENSITIVE1 and is much more active than jasmonoyl-Ile (Katsir et al., 2008; Yan et al., 2009), exemplifying the ingenious ways by which pathogens can take control over the plant’s hormone signaling network to suppress host immunity.
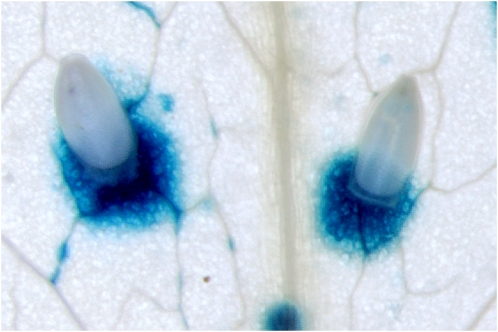
Insects have been demonstrated to exert similar decoy tactics. For instance, nymphs of the phloem-feeding silverleaf whitefly (Bremia tabaci) were shown to induce the SA pathway and suppress the JA pathway, resulting in a faster development of the nymphs (Zarate et al., 2007). A similar phenomenon was observed with caterpillars from the beet armyworm (Spodoptera exigua). Elicitors from salivary excretions of this herbivore suppressed effectual JA-dependent defenses through the activation of the SA pathway (Weech et al., 2008; Diezel et al., 2009). Recently, elicitors from insect eggs were found to activate the SA pathway in Arabidopsis at the site of oviposition. As a result, JA-dependent defenses were suppressed, resulting in an advantage for the newly hatched offspring that fed from the undefended tissue (Bruessow et al., 2010; Fig. 1).
Figure 1.
Eggs of the cabbage white butterfly P. rapae activate the SA-responsive PR1::GUS reporter gene at the site of oviposition. Activation of the SA pathway suppresses JA-dependent herbivore resistance, providing an advantage for newly hatched larvae of generalist herbivores that feed from this undefended tissue (Bruessow et al., 2010). Photo: Hans van Pelt.
MULTISPECIES INTERACTIONS: JUGGLING THE GOOD, THE BAD, AND THE UGLY
In nature, plants often deal with simultaneous or subsequent invasion by multiple pathogens and insects. In a field study with Brassica oleracea plants, it was demonstrated that early season herbivory significantly affected plant defense responses to secondary herbivores and the development of their populations (Poelman et al., 2008). Gene expression analyses implicated a dominant role for plant hormones in the regulation of this process. Also in Arabidopsis, pathogen and insect attack have been shown to affect secondary interactions with antagonists. For example, induction of the SA pathway by P. syringae suppressed JA signaling and rendered infected leaves more susceptible to the necrotrophic fungus Alternaria brassicicola (Spoel et al., 2007). Similarly, prior inoculation with the biotrophic downy mildew pathogen Hyaloperonospora arabidopsidis suppressed JA-mediated defenses that were activated upon feeding by caterpillars of the small cabbage white butterfly (Pieris rapae; Koornneef et al., 2008). In lima bean (Phaseolus lunatus) it was demonstrated that infestation with phloem-feeding whiteflies (Bemisia tabaci) negatively affected indirect plant defenses that are triggered by spider mites (Tetranychus urticae), resulting in a reduced attraction of predatory mites (Zhang et al., 2009). This effect could be mimicked by SA and involved suppression of JA biosynthesis, indicating that the effect of SA-JA cross talk extends to plant immune responses against organisms from different trophic levels.
In the soil, plant roots interact with an enormously diverse community of microbes, insects, and worms, which can influence the defense status of the aerial plant parts. For instance, belowground infestation by the Western corn rootworm (Diabrotica virgifera virgifera) was shown to affect the aboveground hormone signature of maize (Zea mays) and the level of aboveground resistance against herbivores and necrotrophs (Erb et al., 2009). Belowground interactions are frequently beneficial in nature. Because beneficial microbes are also recognized as alien organisms, active interference with the plant immune signaling network is fundamental for the establishment of intimate mutualistic relationships. In many cases, SA, JA, and ET emerged as the dominant regulators in this process (Van Wees et al., 2008; Gutjahr and Paszkowski, 2009; Van der Ent et al., 2009b; López-Ráez et al., 2010; Pineda et al., 2010). During the beneficial interaction of Arabidopsis roots with the endophytic fungus Piriformospora indica, components of the ET pathway were implicated in the balance between beneficial and nonbeneficial traits of this symbiosis (Camehl et al., 2010). Suppression of ET signaling was also observed in mycorrhizal tomato (Solanum lycopersicum) plants (López-Ráez et al., 2010). In addition, plant growth-promoting Pseudomonas fluorescens bacteria were shown to actively suppress ET-dependent microbe-associated molecular pattern responses in the roots of Arabidopsis (Millet et al., 2010), presumably to avoid being recognized as an uninvited guest. Colonization of roots by these beneficial rhizobacteria induces a systemic resistance in aerial plant parts that is effective against a broad spectrum of attackers. This induced systemic resistance is often based on priming for enhanced expression of JA-dependent defenses and boosts plant defenses that are activated upon subsequent attack by JA-inducing pathogens and insects (Pozo et al., 2008; Van Oosten et al., 2008; Van der Ent et al., 2009a). The list of examples by which beneficial microbes recruit hormone signaling pathways to establish a mutualistic interaction is growing rapidly. However, our understanding of how plants regulate their immune signaling network to maximize both profitable and protective functions during simultaneous interactions with the good, the bad, and the ugly is still limited.
UNRAVELING THE HOWS AND WHYS OF HORMONE CROSS TALK IN PLANT DEFENSE
In recent years, several players in the interactions between hormone-regulated defense signaling pathways have been described (for review, see Pieterse et al., 2009). These include transcriptional (co)regulators such as NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1, DELLAs, WRKYs, TGAs, MYC2, and APETALA2/ETHYLENE RESPONSE FACTOR. Unexpected connections between the gene regulatory networks of these factors start to emerge. For example, TGA transcription factors that were previously implicated in the regulation of SA-dependent defenses were also shown to be essential activators of induced defenses that are triggered by the combined action of JA and ET (Zander et al., 2009). However, upon simultaneous activation of the SA, JA, and ET pathways, TGAs act together with MYC2 to suppress JA/ET-dependent defenses (Zander et al., 2009), again demonstrating the delicate interrelationship between these hormones. While the players in hormone cross talk become more and more identified, it is clear that various posttranslational protein modifications add to the complexity of the hormone-regulated immune signaling network (for review, see Spoel et al., 2010). Exciting developments in hormone signaling research underpin the central role of the ubiquitin proteasome system in this respect, as this regulatory protein machinery is involved in hormone perception, derepression of hormone signaling pathways, degradation of hormone-specific transcription factors, and regulation of hormone biosynthesis (Santner and Estelle, 2010).
Recent genomics research has revealed that the signaling networks that are activated in response to mutualists, pathogens, and insects overlap, indicating that the regulation of the adaptive response to these organisms is finely balanced between protection against aggressors and acquisition of benefits. Major challenges for future research lie in understanding how complex multidimensional signal interactions are regulated during the interaction of plants with multiple organisms, and why they steer the immune response toward a particular defense output. Because of the high level of complexity, the bioinformatics and systems biology tools that become increasingly available (Volodarsky et al., 2009; Pitzschke and Hirt, 2010) will be extremely valuable in this respect. Recently, Wang et al. (2008) and Tsuda et al. (2009) provided nice examples of the power of a systems approach. Through gene expression profiling of a dedicated set of defense-related genes in wild-type and hormone signaling mutant backgrounds, they were able to pinpoint important regulators and interrelationships between SA, JA, and ET signaling sectors when plants express well-characterized immune responses, such as effector-triggered immunity and microbe-associated molecular pattern-triggered immunity. During interaction with multiple (a)biotic stresses, however, temporal changes in hormone signaling will add yet another layer of complexity to the gene regulatory network and the outcome of the defense response. Hence, tools to probe the dynamics of gene expression in microarray time series and to uncover underlying regulatory modules, such as described by Kiddle et al. (2010), will be highly valuable in future efforts to make sense out of hormone cross talk in the plant defense signaling network. Ultimately this will lead to an integrated view of how plants are able to simultaneously deal with mutualists, parasites, and environmental stresses, and maximize both beneficial and defensive functions.
References
- Bari R, Jones JDG. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Bruessow F, Gouhier-Darimont C, Buchala A, Métraux JP, Reymond P. (2010) Insect eggs suppress plant defence against chewing herbivores. Plant J 62: 876–885 [DOI] [PubMed] [Google Scholar]
- Camehl I, Sherameti I, Venus Y, Bethke G, Varma A, Lee J, Oelmüller R. (2010) Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytol 185: 1062–1073 [DOI] [PubMed] [Google Scholar]
- Campos ML, de Almeida M, Rossi ML, Martinelli AP, Litholdo CG, Figueira A, Rampelotti-Ferreira FT, Vendramim JD, Benedito VA, Peres LEP. (2009) Brassinosteroids interact negatively with jasmonates in the formation of anti-herbivory traits in tomato. J Exp Bot 60: 4346–4360 [DOI] [PubMed] [Google Scholar]
- Chen ZY, Agnew JL, Cohen JD, He P, Shan LB, Sheen J, Kunkel BN. (2007) Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA 104: 20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SM, Cristescu SM, Miersch O, Harren FJM, Wasternack C, Mur LAJ. (2009) Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol 182: 175–187 [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Bennett MH, Truman WH, Grant MR. (2009) Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J 59: 375–386 [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Egea PR, Bogre L, Grant M. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26: 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Yang Y, Cruz CV, Höfte M. (2010) Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol 152: 2036–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC, Dicke M, et al. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Diezel C, Von Dahl CC, Gaquerel E, Baldwin IT. (2009) Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol 150: 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M, Flors V, Karlen D, De Lange E, Planchamp C, D’Alessandro M, Turlings TCJ, Ton J. (2009) Signal signature of aboveground-induced resistance upon belowground herbivory in maize. Plant J 59: 292–302 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Grant MR, Jones JDG. (2009) Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Paszkowski U. (2009) Weights in the balance: jasmonic acid and salicylic acid signaling in root-biotroph interactions. Mol Plant Microbe Interact 22: 763–772 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Jiang CJ, Shimono M, Sugano S, Kojima M, Yazawa K, Yoshida R, Inoue H, Hayashi N, Sakakibara H, Takatsuji H. (2010) Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Mol Plant Microbe Interact 23: 791–798 [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiddle SJ, Windram OPF, McHattie S, Mead A, Beynon J, Buchanan-Wollaston V, Denby KJ, Mukherjee S. (2010) Temporal clustering by affinity propagation reveals transcriptional modules in Arabidopsis thaliana. Bioinformatics 26: 355–362 [DOI] [PubMed] [Google Scholar]
- Koornneef A, Leon-Reyes A, Ritsema T, Verhage A, Den Otter FC, Van Loon LC, Pieterse CMJ. (2008) Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol 147: 1358–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CMJ. (2008) Cross-talk in defense signaling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A, Du Y, Koornneef A, Proietti S, Körbes AP, Memelink J, Pieterse CMJ, Ritsema T. (2010) Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol Plant Microbe Interact 23: 187–197 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A, Spoel SH, De Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RAM, Ritsema T, Pieterse CMJ. (2009) Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol 149: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López MA, Bannenberg G, Castresana C. (2008) Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Curr Opin Plant Biol 11: 420–427 [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Verhage A, Fernández I, García JM, Azcón-Aguilar C, Flors V, Pozo MJ. (2010) Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway. J Exp Bot 61: 2589–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM. (2010) Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JE, Tao Y, Chory J, Ballare CL. (2009) Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc Natl Acad Sci USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C. (2006) The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol 140: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lison P, Nemri A, Harberd NP, Jones JDG. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. (2009) Networking by small-molecules hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Pineda A, Zheng SJ, Van Loon JJA, Pieterse CMJ, Dicke M. (2010) Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci 15: 507–514 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Hirt H. (2010) Bioinformatic and systems biology tools to generate testable models of signaling pathways and their targets. Plant Physiol 152: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelman EH, Broekgaarden C, Van Loon JJA, Dicke M. (2008) Early season herbivore differentially affects plant defence responses to subsequently colonizing herbivores and their abundance in the field. Mol Ecol 17: 3352–3365 [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Van der Ent S, Van Loon LC, Pieterse CMJ. (2008) Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol 180: 511–523 [DOI] [PubMed] [Google Scholar]
- Ramirez V, Coego A, Lopez A, Agorio A, Flors V, Vera P. (2009) Drought tolerance in Arabidopsis is controlled by the OCP3 disease resistance regulator. Plant J 58: 578–591 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JDG. (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10: 372–379 [DOI] [PubMed] [Google Scholar]
- Robson F, Okamoto H, Patrick E, Harris SR, Wasternack C, Brearley C, Turner JG. (2010) Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. (2010) The ubiquitin-proteasome system regulates plant hormone signaling. Plant J 61: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Tada Y, Loake GJ. (2010) Post-translational protein modification as a tool for transcription reprogramming. New Phytol 186: 333–339 [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Manners JM, Kazan K. (2009) Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J 58: 927–939 [DOI] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci 14: 310–317 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Wangdi T, Kunkel BN, Anand A, Mysore KS, Bender CL. (2007) The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 20: 955–965 [DOI] [PubMed] [Google Scholar]
- Van der Ent S, Van Hulten MHA, Pozo MJ, Czechowski T, Udvardi MK, Pieterse CMJ, Ton J. (2009a) Priming of plant innate immunity by rhizobacteria and β-aminobutyric acid: differences and similarities in regulation. New Phytol 183: 419–431 [DOI] [PubMed] [Google Scholar]
- Van der Ent S, Van Wees SCM, Pieterse CMJ. (2009b) Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Van Oosten VR, Bodenhausen N, Reymond P, Van Pelt JA, Van Loon LC, Dicke M, Pieterse CMJ. (2008) Differential effectiveness of microbially induced resistance against herbivorous insects in Arabidopsis. Mol Plant Microbe Interact 21: 919–930 [DOI] [PubMed] [Google Scholar]
- Van Wees SCM, Van der Ent S, Pieterse CMJ. (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11: 443–448 [DOI] [PubMed] [Google Scholar]
- Volodarsky D, Leviatan N, Otcheretianski A, Fluhr R. (2009) HORMONOMETER: a tool for discerning transcript signatures of hormone action in the Arabidopsis transcriptome. Plant Physiol 150: 1796–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Hendrickson Culler A, Dong X. (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17: 1784–1790 [DOI] [PubMed] [Google Scholar]
- Wang L, Mitra RM, Hasselmann KD, Sato M, Lenarz-Wyatt L, Cohen JD, Katagiri F, Glazebrook J. (2008) The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: roles of major regulators and the phytotoxin coronatine. Mol Plant Microbe Interact 21: 1408–1420 [DOI] [PubMed] [Google Scholar]
- Weech MH, Chapleau M, Pan L, Ide C, Bede JC. (2008) Caterpillar saliva interferes with induced Arabidopsis thaliana defence responses via the systemic acquired resistance pathway. J Exp Bot 59: 2437–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Maruyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S, et al. (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20: 1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander M, La Camera S, Lamotte O, Métraux JP, Gatz C. (2009) Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J 61: 200–210 [DOI] [PubMed] [Google Scholar]
- Zarate SI, Kempema LA, Walling LL. (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol 143: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PJ, Zheng SJ, Van Loon JJA, Boland W, David A, Mumm R, Dicke M. (2009) Whiteflies interfere with indirect plant defense against spider mites in Lima bean. Proc Natl Acad Sci USA 106: 21202–21207 [DOI] [PMC free article] [PubMed] [Google Scholar]