The nature of time has long been an obsession for philosophers, scientists, and laymen. This nearly universal interest in time is encapsulated by the fact that the quotation “Time is nature’s way of keeping everything from happening at once” has been attributed to both Woody Allen and Albert Einstein! Although some argue that time is an illusion, ongoing work has revealed that most organisms possess an internal oscillator, the circadian clock, which has a pervasive influence on growth, development, and responses to the environment. Studies of circadian rhythms in plants reveal that because everything doesn’t happen at once, plants have evolved sophisticated mechanisms to partition physiological processes so that they occur at the most advantageous times, both on seasonal and daily scales.
A BRIEF HISTORY OF TIMING
It is apparent to even the most casual observer that plant physiology is strongly influenced by time: for example, plants generally fix carbon during the day but are carbon consumers at night, and plant reproduction is strongly tied to the seasons. Although these changes in physiology are clearly linked to changes in the environment, they are also strongly influenced by the plant circadian clock. Studies spanning the past 300 years have revealed that many features of plant physiology are affected by the circadian clock (McClung, 2006). Clock-influenced processes range from once-in-a-lifetime events such as germination and the transition from vegetative to reproductive growth, to annual events such as the onset of flowering or winter dormancy, to daily processes such as rhythmic movements of petals and leaves and emission of floral fragrance. Ongoing, intensive investigation into clock-regulated processes are revealing that plants are even more sophisticated time keepers than we previously thought, with most or all aspects of plant physiology influenced by the circadian system. The molecular nature of the plant oscillator is now becoming clear, and the development of mathematical models describing this clock are raising hopes that we will someday be able to predict how particular environmental conditions will interact with a given genotype to shape plant growth and development in the real world.
HOW DO PLANTS TELL TIME?
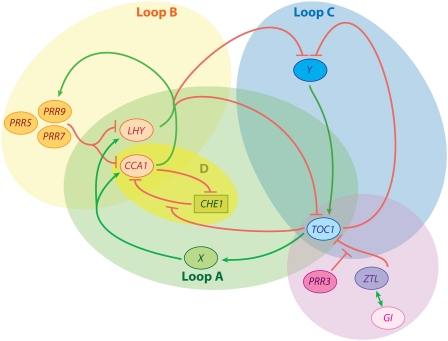
One hallmark of circadian rhythms is that these roughly 24-h cycles persist in constant environmental conditions; that is, although they can be reset by external cues such as changes in light or temperature, they do not depend upon these outside influences for their rhythmicity. In all organisms examined thus far, circadian rhythms are generated in a cell autonomous fashion. In eukaryotes as disparate as fungi, animals, and plants, the clock mechanism involves interlocked transcriptional feedback loops. However, despite these similarities, the clock genes involved in these transcriptional feedback loops are not conserved across higher taxa (Harmer, 2009). Extensive functional studies, carried out primarily in Arabidopsis (Arabidopsis thaliana), have begun to reveal the architecture of the circadian system in higher plants. The central transcriptional feedback loop consists of the Myb-like transcription factors CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) and the pseudo-response regulator protein TIMING OF CAB EXPRESSION1 (TOC1; Fig. 1, loop A). CCA1 and LHY repress TOC1 expression by binding to its promoter. TOC1 in turn indirectly promotes expression of CCA1 and LHY; in the case of CCA1, this is accomplished in part by inhibition of the transcription factor CHE (Fig. 1, loop D). Additional regulatory loops are also vital for clock function in Arabidopsis. The TOC1 homologs PSEUDO-RESPONSE REGULATOR5 (PRR5), PRR7, and PRR9, inhibit expression of LHY and CCA1. CCA1 and LHY in turn promote expression of PRR7 and PRR9, forming yet another negative feedback loop (Fig. 1, loop B). An additional transcriptional feedback loop has been proposed based on mathematical modeling, although the molecular identities of its components are currently unknown (Fig. 1, loop C). Additional genes such as EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRYTHMO/PHYTOCLOCK1 (LUX/PCL1) are known to function in the circadian network but have not yet been assigned specific functions.
Figure 1.
The plant clock is composed of multiple interacting transcriptional feedback loops (A–D). Posttranslational regulation is also key to clock function (pink circle). Figure adapted from Harmer (2009).
Posttranscriptional regulation is also essential for proper clock function. The F-box protein ZEITLUPE (ZTL) targets TOC1 for degradation in a manner modulated by PRR3. ZTL protein stability is in turn regulated by its interactions with GIGANTEA (GI; Fig. 1). Additional posttranscriptional mechanisms are also key (Somers et al., 2007).
How will unknown components of the plant clock (e.g. X and Y in Fig. 1) be identified? Recent genetic screens have identified multiple alleles of known genes, suggesting they are approaching saturation. However, high-throughput genomic approaches may reveal members of gene families with partially redundant functions in the circadian system (Pruneda-Paz et al., 2009). Careful genetic studies on higher-order mutants will then be required to determine their respective contributions to clock function.
Recent studies of the plant circadian clock indicate that its molecular composition can vary between different cell and organ types. For example, PRR3 appears to modulate TOC1 stability in plant vasculature but not in other cell types (Para et al., 2007). CCA1 and LHY are expressed but not able to inhibit TOC1 expression in dark-grown roots (James et al., 2008). It seems likely that as we continue to study cell- and organ-specific clock function we will find many additional examples of specializations in clock architecture in distinct tissues. The underlying reasons for these specializations may become apparent as we delve more deeply into the mechanisms connecting the circadian clock and plant physiology.
The Circadian Clock and Physiology—Complex Interacting Networks
Early representations of the circadian system consisted of three tidy categories: inputs, the core oscillator, and output pathways. Work in many model organisms has led to the disintegration of these boundaries, since it is now abundantly clear that key clock genes can play roles in input to the clock and that outputs can feed back to regulate clock function (Harmer, 2009).
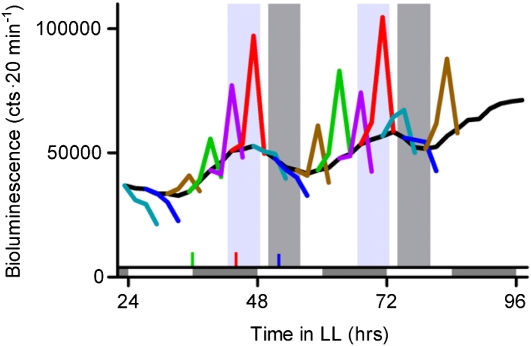
Recent studies have also shown that the clock is more thoroughly integrated with other signaling pathways than previously suspected. At least one-third of the plant genome is under circadian regulation. In addition, genes regulated by any one of the major plant hormones are more likely to be clock regulated than expected by chance, suggesting specific clock involvement in these pathways (Covington et al., 2008). Indeed, functional studies have shown that the circadian clock modulates plant sensitivity to exogenous auxin: even when plants are maintained in constant environmental conditions, responsiveness to auxin varies at different times of day (Fig. 2; Covington and Harmer, 2007). The clock also regulates responsiveness to abscisic acid (Legnaioli et al., 2009), and it seems likely that the clock will modulate other hormone pathways as well. Clock control of multiple signaling pathways may help to coordinate plant growth (Nozue et al., 2007; Michael et al., 2008). Intriguingly, exogenous hormone treatment can also influence circadian clock function (Robertson et al., 2009), suggesting that not only can the clock influence hormone signaling, but that hormone signaling may also influence the clock.
Figure 2.
Clock gating of auxin response. Plants expressing the auxin-sensitive reporter eDR5::LUC were treated with auxin at 4-h intervals. The black line indicates untreated control plants while the colored lines indicate plants treated at different circadian times. Data are from Covington and Harmer (2007). cts, counts; LL, constant light.
The circadian clock can also affect plant sensitivity to a variety of environmental stimuli. For example, chilling-sensitive plants show differential damage when the same cold treatment is applied at different circadian times (Rikin et al., 1993). Similarly, plant sensitivity to light or to drought stress varies in a circadian manner (Millar and Kay, 1996; Legnaioli et al., 2009). Responsiveness to biotic stimuli may also be influenced by the clock. For example, many genes regulated by mechanical wounding are also clock regulated, suggesting that plant defense responses may be modulated by the circadian system (Walley et al., 2007).
We are just beginning to appreciate the extensive connections between the circadian clock and plant metabolism. Clock genes influence nitrogen assimilation, tricarboxylic acid cycle intermediates, and general signaling molecules (Dodd et al., 2007; Gutierrez et al., 2008; Fukushima et al., 2009). The clock has long been known to influence photosynthesis, and recent work has demonstrated that the clock also controls the rate at which starch is utilized during the night (Graf et al., 2010). It now seems most appropriate to think of the circadian system as a network that is functionally interconnected with many (or perhaps all) signaling and metabolic networks within plant cells.
Why has evolution led to these pervasive interconnections? Recent studies suggest that consonance between the period of the circadian clock and environmental cycles enhances plant productivity and may provide an adaptive advantage (Dodd et al., 2005; Graf et al., 2010). That is, fitness is enhanced when the circadian system is able to appropriately time physiological processes so that they occur at the appropriate time in the day/night cycle. Modulation of clock function may also play a role in hybrid vigor (Ni et al., 2009).
The Beginning of Timing
An important outstanding question is how the plant clock evolved. Comparative genomic and functional studies suggest that the molecular nature of the plant clock is similar across the angiosperms and in the moss Physcomitrella patens (Okada et al., 2009; McClung, 2010). Studies in two green algae, Ostreococcus tauri and Chlamydomonas reinhardtii, paint a more complex picture, however. The O. tauri and C. reinhardtii genomes each contain single homologs of CCA1/LHY, TOC1, and LUX. Functional studies demonstrated a role for the CCA1/LHY-like and TOC1-like genes in the O. tauri circadian clock (Corellou et al., 2009) and for the CCA1/LHY-like and LUX-like genes in C. reinhardtii (Matsuo et al., 2008). These functional similarities suggest that the central transcriptional feedback loop between CCA1/LHY and TOC1 (Fig. 1, loop A) is conserved between green algae and land plants. But despite these similarities with angiosperm clocks, there must also be fundamental differences. The O. tauri and C. reinhardtii genomes lack additional PRR-like genes and do not have recognizable ELF3, ELF4, GI, or ZTL homologs (Matsuo et al., 2008; Corellou et al., 2009). In addition, forward genetic studies in C. reinhardtii have demonstrated that genes without obvious homologs in higher plants are essential for normal clock function in this species (Iliev et al., 2006; Matsuo et al., 2008). It will be fascinating to learn the mechanistic similarities and differences between the clocks of the green algae and those of the land plants.
The similarity between the overall architecture of circadian networks in higher eukaryotes coupled with the lack of conservation of clock genes make for a fascinating evolutionary puzzle. Are the similarities in circadian organization in animals, plants, and fungi due to convergent evolution, or evolution by descent? One possibility is that the last common ancestor to these eukaryotes possessed a circadian clock that relied upon metabolic oscillations, and that the clock genes that we now study evolved relatively recently to make this primordial biochemical clock more robust and more thoroughly integrated with environmental input and physiological output pathways.
Clocks in the Real World
As sessile organisms, plants are exquisitely well adapted to their local environments. However, as our global climate undergoes rapid warming, plants are changing their latitudinal ranges in an attempt to offset changes in the environment (Parmesan and Yohe, 2003). The resulting alterations in seasonal day lengths will likely necessitate evolutionary changes in circadian clock function for plants to maintain appropriate regulation of daily and seasonal events. Indeed, a recent study revealed that Arabidopsis plants grown in the field are acutely sensitive to seasonal influences, with small differences in time of sowing resulting in plants that either flower rapidly or overwinter before undergoing the transition from vegetative to reproductive growth (Wilczek et al., 2009). A relatively simple photothermal model of Arabidopsis development accurately predicted flowering time of multiple genotypes in this study. Mathematical models describing the plant circadian clock are currently being developed (Locke et al., 2006; Zeilinger et al., 2006). Future models may someday be used to inform breeding to optimize plant growth in diverse environments.
The above studies, and much research not discussed due to space constraints, give us a better appreciation of how plant physiology varies at different times of day. Further investigation of the mechanisms underlying the plant clockwork and its integration with pathways essential for growth and development will help us understand how plants are able to thrive in the ever-changing conditions of the real world.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CCA1, At2g46830; ELF3, At2g25930; ELF4, At2g40080; GI, At1g22770; LHY, At1g01060; LUX/PCL1, At3g46640; PRR3, At5g60100; PRR5, At5g24470; PRR7, At5g02810; PRR9, At2g46790; TOC1, At5g61380; and ZTL, At5g57360.
Acknowledgments
I would like to thank J.N. Maloof for helpful comments on the manuscript.
References
- Corellou F, Schwartz C, Motta JP, Djouani-Tahri el B, Sanchez F, Bouget FY. (2009) Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell 21: 3436–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Harmer SL. (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, Love J, Assie JM, Robertson FC, Jakobsen MK, Goncalves J, et al. (2007) The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science 318: 1789–1792 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K. (2009) Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci USA 106: 7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM. (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Iliev D, Voytsekh O, Schmidt EM, Fiedler M, Nykytenko A, Mittag M. (2006) A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol 142: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, Nimmo HG. (2008) The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322: 1832–1835 [DOI] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, Mas P. (2009) TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J 28: 3745–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M. (2008) A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev 22: 918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. (2006) Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. (2010) A modern circadian clock in the common angiosperm ancestor of monocots and eudicots. BMC Biol 8: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J. (2008) A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Kay SA. (1996) Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA 93: 15491–15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Okada R, Kondo S, Satbhai SB, Yamaguchi N, Tsukuda M, Aoki S. (2009) Functional characterization of CCA1/LHY homolog genes, PpCCA1a and PpCCA1b, in the moss Physcomitrella patens. Plant J 60: 551–563 [DOI] [PubMed] [Google Scholar]
- Para A, Farre EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA. (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmesan C, Yohe G. (2003) A globally coherent fingerprint of climate change impacts across natural systems. Nature 421: 37–42 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikin A, Dillwith JW, Bergman DK. (1993) Correlation between the circadian rhythm of resistance to extreme temperatures and changes in fatty acid composition in cotton seedlings. Plant Physiol 101: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson FC, Skeffington AW, Gardner MJ, Webb AA. (2009) Interactions between circadian and hormonal signalling in plants. Plant Mol Biol 69: 419–427 [DOI] [PubMed] [Google Scholar]
- Somers DE, Fujiwara S, Kim WY, Suh SS. (2007) Posttranslational photomodulation of circadian amplitude. Cold Spring Harb Symp Quant Biol 72: 193–200 [DOI] [PubMed] [Google Scholar]
- Walley J, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K. (2007) Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genetics 3: 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, Martin LJ, Muir CD, Sim S, Walker A, Anderson J, et al. (2009) Effects of genetic perturbation on seasonal life history plasticity. Science 323: 930–934 [DOI] [PubMed] [Google Scholar]
- Zeilinger MN, Farre EM, Taylor SR, Kay SA, Doyle FJ., III (2006) A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol 2: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]