Abstract
Infectious diseases have been a prime testing ground for ecological theory. At the same time, the ecological perspective is increasingly recognized as essential in epidemiology. Long-term, spatially-resolved, reliable disease incidence data and the ability to confront them with mechanistic models have been critical in this cross-fertilization. Here, we review some of the key intellectual developments in epidemiology facilitated by long-term data. We proceed to identify research frontiers at the interface of ecology and epidemiology and their associated data needs.
Historical Background
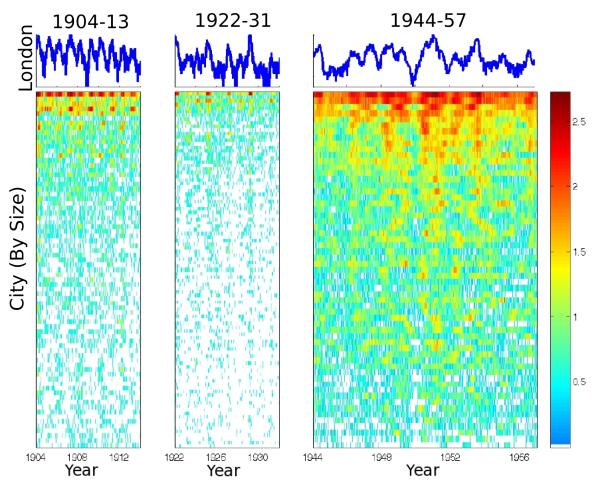
John Graunt, who in the 17th Century pioneered the collection of vital statistics, was a seminal figure in epidemiology [1]. Because he was interested in an early warning system for the spread of bubonic plague, his attention focused on disease mortality, but his systematic analyses of vital statistics were highly influential and led to programs for the regular documentation of demographic fluxes and causes of death [2]. One of Graunt’s most substantial legacies is a wealth of data on infectious disease morbidity and mortality systematically collected from the 16th Century. In England & Wales, for example, notifications of deaths attributable to several high-profile diseases (e.g., measles, whooping cough, diptheria, scarlet fever, plague) have been recorded since 1836 [3]. Fig. 1 illustrates spatially replicated data spanning different eras: we plot weekly notifications of whooping cough deaths (first two columns) and incidence (third column) for the largest population centres in England & Wales from the first years of the 20th century. Similarly, the United States Public Health Service has published the Weekly Abstract of Sanitary Reports since 1878 [4] and comparable collections are available in many other countries.
Figure 1.
Long-term data on whooping cough (pertussis) in the largest cities of England & Wales [12]. The first two panels depict weekly notifications of pertussis deaths from the largest 95 towns and cities in England & Wales for (a) 1904–1913 and (b) 1922–1931. Panel (c) depicts weekly case data for the 60 largest cities from 1944–1957. In the line graph at the top of each panel we present data from London, while the coloured panels present the spatial log-transformed data. The figures demonstrate changing epidemiological patterns of pertussis through the decades; in London, for example, pertussis outbreaks from 1904–1913 were annual, gave way to biennial cycles in 1922–1931 and were characterised by a mixture of annual and multiennial oscillations after World War II. The lower panels show that in between large outbreaks, pertussis becomes locally extinct (fades out) in small populations, as illustrated by regions of white.
Although long-term epidemiological data sets vary in reporting fidelity, frequency, and duration, relative to most other ecological time series they tend to be long and highly resolved. Examples include excellent data sets on cholera and malaria mortality in the former British India [5], recent dengue hemorrhagic fever incidence data from Thailand [6], raccoon rabies data from the eastern U.S. [7], and bubonic plague in gerbils in Kazakhstan [8]. This wealth of ecological data is perhaps rivalled only by fisheries data [9], small mammal trapping data[10], and forest insect outbreak data [11]. Another distinguishing feature of infectious disease data is the availability of parallel information, often including details of host demography, immunization practices, and societal and behavioral changes. These kinds of information have proved invaluable in placing observed epidemiological patterns within their ecological context (more on this below).
Long-term data were instrumental in the development of epidemiological ideas in the late 19th and early 20th Centuries, when a number of researchers explored the roles of seasonality, immunity, and competition in infectious disease dynamics [13, 14]. At the same time, fundamental theoretical insights led to the formulation of the classic mathematical models that underpin modern epidemiological research, including Hamer’s presentation of the so-called SIR (Susceptible-Infectious-Recovered) model [15], Ross’s development of the first malaria transmission model [16] and the influential work of Kermack & McKendrick [17] on the threshold properties of the SIR system. In the mid-20th Century, Bartlett’s ground-breaking analyses of measles epidemics and their extinction frequency led to the important concept of the Critical Community Size (the smallest host population size above which the pathogen persists; [18]) and the dynamical impact of demographic noise in amplifying fluctations and sustaining oscillations in SIR models [19]. Epidemiological theory was further boosted by the seminal contributions of Dietz [20] and Bailey [21]. In many ways, however, the true marriage of epidemiological theory and long-term datasets had to wait for the consummate work of Anderson & May.
Starting with their compelling 1979 treatise [22, 23], Anderson & May drew attention to the important parallels between ecological theory (especially predator-prey systems) and that of infectious disease. They subsequently published a series of elegant studies in which meaningful, policy-relevant conclusions were extracted from epidemiological data (summarized in [24]). Subsequently, infectious-disease ecology has burgeoned as a field, becoming a prime testing-ground for ecological concepts and theory [25, 26]. Currently, the cross-talk between ecology and epidemiology is exciting and productive: the examination of epidemiological data from an ecological perspective informs public health issues [27-30] and methodology developed for dealing with long-term epidemiological datasets are usefully applied in ecological contexts [eg, 31, 32]. The abundance of long-term data unquestionably continues to play a critical role in this blossoming.
In this opinion piece, we review the major epidemiological lessons learned from long-term data, outline some of the outstanding challenges to epidemiological theory, and identify an urgent need for new long-term datasets different in kind and scale.
Lessons learnt (so far)
To address issues of causality in natural systems, dynamical models are indispensable [33]. The most natural and rigorous means of evaluating such models is to confront them with long-term time-series data. In the ecology of many infectious diseases, two circumstances make such models relatively easy to formulate. The first is the pronounced separation between the generation times of micro-parasites (viruses, bacteria, protozoa) and those of their hosts. The second is the close ecological connection between many obligate specialist parasites and their hosts. Even given the relative simplicity of the ecology in such cases, it is remarkable that the very simplest models have proved surprisingly efficient at explanation of data. This is in stark contrast to the experience in ecology where, by and large, the simplest models are thought to be of limited use in explaining nature, with the notable exception of well studied laboratory systems (eg, [34-36]). This degree of success probably stems from a combination of factors. First, at the macroscopic scale (eg, long-term epidemic dynamics in a metropolitan centre), many infectious disease systems are characterised by well-understood biology and a reasonably simple natural history (host specificity, known durations of latency and infectiousness, and long-lasting immunity). Second, and intriguingly, many heterogeneities seem to average out in such infectious disease systems so that admittedly oversimplified descriptions often effectively capture prominent dynamical patterns [37].
Nonlinearity, Seasonality and Stochasticity
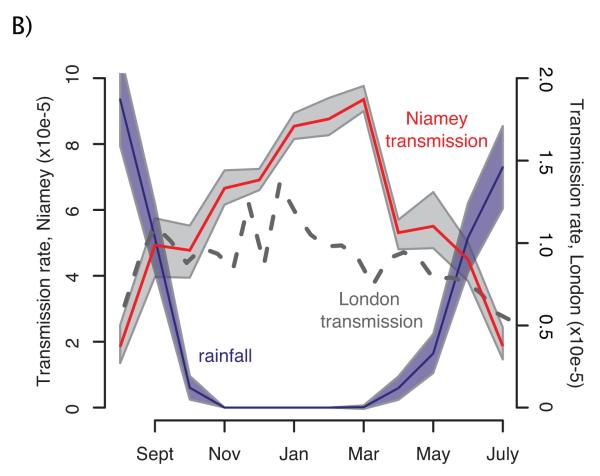
One of the earliest attempts to confront an epidemiological model with data was by Hamer [15], who noted the inconsisntecy between the constant prevalence predicted by the simplest transmission model and the violent, high amplitude oscillations observed in London measles case notifications. Hamer speculated that a missing component of the model was rhythmic variation in the number of susceptibles. This topic was re-examined by Soper [14], whose exploration of Glasgow measles data led him to suspect seasonal variation in transmission rates, attributable to the opening and closing of schools. This conclusion, that in large populations measles epidemics (and those of other childhood diseases) are driven by school-term driven seasonal changes in contact rates, has since been reaffirmed [38, 39]. Interestingly, a recent study of measles in sub-Saharan Africa has revisited the mechanism of transmission seasonality. Ferrari et al. [40] explored measles incidence in Niger and suggest that seasonal human migration associated with agricultural practices is a key driver there (see Fig. 2). In the data from Niger’s capital, the estimated amplitude of seasonality is much greater than it is in London (Fig. 2). This difference is thought to be largely responsible for the unpredictable and perhaps chaotic oscillations in measles in Niger.
Figure 2.
Dynamics of measles outbreaks in Niamey, Niger. Mean monthly rainfall from 1995 to 2004 (blue) are plotted together with 62 standard deviations (blue shading). In red, the estimated seasonal transmission rate for Niamey is depicted, with the shaded grey regions representing the 95% bayesian credible intervals; the dashed line depicts the seasonality (scaled for population size) for the pre-vaccine era in (1950-1968) London for comparison. Reproduced with permission from [40].
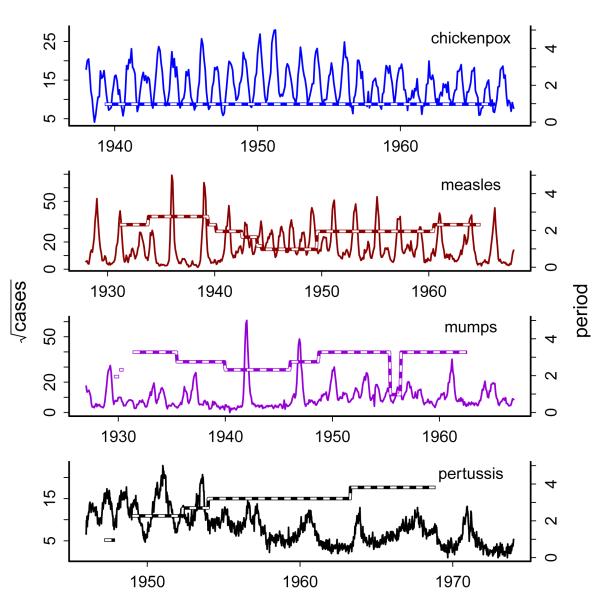
Compared with the pattern in Niger, measles data from England & Wales, Europe, and the US show a striking temporal regularity (eg, Fig. 3). In England & Wales, from 1950 until the introduction of national paediatric immunization in 1968, measles epidemics in larger towns and cities exhibited a predictable two-year cycle [24, 41]. The fact that the most basic SIR transmission model with school-term forcing reproduces this and other qualitative features of measles epidemics in large populations [42] has led some to comment on the essentially deterministic nature of these data [37]. As Fig. 3 shows, however, infectious diseases vary in their predictability and in the apparence of stochasticity and seasonality in their dynamics [43]. For example, epidemics of chickenpox (Figure 3a) are highly regular, with a constant inter-epidemic period. Similarly, while measles outbreaks (Figure 3b) exhibit distinct dynamical shifts, these are well explained by changes in birth rates (see below and [42]). In contrast, mumps (Figure 3c) and pertussis (Figures 3d and 1), display more unpredictable dynamics, an observation that presents both challenges and opportunities: challenges, inasmuch as increased noise levels obscure patterns that reveal the underlying ecology; opportunities, because increasing variability broadens the system’s dynamic range, thereby potentially revealing more about the mechanisms shaping the dynamics. From an ecological perspective, this observation is particularly interesting because of the historical, recurrent, and occasionally charged debate over the relative importance of exogenous (stochastic) and endogenous (density dependent) forces in shaping population dynamics [44, 45]. Analyses of childhood disease data have shed light on when stochasticity is dynamically important, identifying the epidemiological traits (eg, infectious pertiod and transmission rate) that determine the outcome of seasonality and demographic noise [46, 47]. Emerging theory on this front has very elegantly unpacked the necessary ingredients for noise amplification in such systems [48-50] and the accompanying response to seasonality [43].
Figure 3.
Depiction of long-term epidemiological data sets. We plot monthly case notifications from Copenhagen for (a) chickenpox, (b) measles, and (c) rubella. In (d), we plot weekly incidence of whooping cough in London (note that national immunization commenced in 1957 in England & Wales). For each panel, with the axis on the right hand side, we also plot the statistically significant dominant period through time as detected by wavelet spectral analysis.
Bifurcations, Chaos and Natural Experiments
One of the guises under which the noise vs. non-linearity controversy re-appeared was the 1980s and 1990s discussion surrounding the possibility of chaos in ecological systems [51]. The idea was that the nonlinearity inherent in pathogen transmission conjoined with seasonal forcing make childhood diseases prime candidates for chaotic dynamics. The high-profile work of Schaffer and colleagues [52, 53], Sugihara & May [54] and Ellner & Turchin [55] exploited long case notification time series for childhood diseases and novel theoretical approaches to identify the fingerprints of chaos. Ultimately, unequivocal evidence for chaos in these systems remains elusive, with perhaps the most likely example being that of measles in Niger discussed above [40]. In retrospect, the lasting impact of the hunt for chaos in ecology has been methodological. The question of whether any particular ecological system is chaotic has been eclipsed by a more basic question: What features must a mechanistic model have to explain ecological dynamics?
More recently, Earn et al. [42] argued that because measles is highly transmissibile and elicits long-lasting immunity, its epidemics are determined by the replenishment rate of the susceptible pool: ‘supply-side’ epidemiology. They pointed out that changes in the influx of susceptibles resulting from, for example, secular trends in birth rates or vaccination coverages might result in shifts in dynamical patterns and suggested that the observed recurrent annual epidemics of measles in developing nations [56] might be explained as a consequence of high fecundity, while the aperiodic dynamics observed in the vaccine era developed countries, previously considered to be an example of chaos, can be more parsimoniously attributed to the interaction between stochasticity and multiple attractors.
Direct experimental confirmation of the changes predicted by bifurcation analyses and stochastic simulations have here, as elsewhere in ecology, proven practically impossible. However, informative studies have exploited natural experiments of four types: (i) changes in host demography have afforded some of the most elegant and direct confirmations of predicted bifurcations [27, 42, 57], (ii) the commencement of mass vaccination campaigns [58], (iii) differential immunization strategies across countries, (iv) comparison of outbreak data among communities (towns, countries) of different sizes, which has allowed assessment of the relative importance of demographic stochasticity and extinction dynamics [37, 59-61].
Metapopulations, Spatial Synchrony, Travelling Waves & Transmission Networks
The systematically collected and spatially resolved UK measles and whooping cough incidence data represent a special, perhaps unique, resource. Recognising the significance of these data for long-standing questions in population ecology, Grenfell spearheaded a campaign to digitize such information [62]. Subsequent analysis revealed that the measles metapopulation in England & Wales in the pre-vaccine era was characterised by highly synchronous biennial outbreaks [37]. In the vaccination era, however, a significant reduction in spatial synchrony is observed [58, 62]. Phase differences among outbreaks in different populations have been mooted as a possible explanation for the paradoxical observation that the critical community size has not risen substantially as a result of vaccination. If correct, this is a prominent manifestation of the ecological concept of ‘rescue effect’ [63] and, importantly, suggests a strategy of spatially-targeted immunization programs [25].
Spatially-explicit epidemiological models make spatiotemporal predictions, and a good deal of attention has been focused on synchrony and traveling waves in disease systems [64]. Since Grenfell et al. [62] described such waves in measles incidence in England & Wales, they have been identified in a number of other systems, including spatially pulsed dengue outbreaks in Thailand, emanating from Bangkok [6]. A very active area of research has focused on the mechanisms of host and/or vector movement affecting such spatio-temporal patterns. It has been shown that “gravity” models generate patterns that are consistent with the waves of measles outbreaks in England & Wales [65]. Such models (borrowed from transportation theory) assume that the extent of epidemiological interaction (or ‘coupling’) between two centres is determined by the geographical distance between them and their respective population sizes. In contrast, the pronounced spatial waves of seasonal H3N2 influenza epidemics in the United States have been explained via coupling predicted by commuter movement between states [28].
Methodological development
Mechanistic models of epidemiological processes are nonlinear dynamical systems and as such are amenable to the tools of that field: most importantly numerical solution, stability and bifurcation analyses for deterministic models [24, 66] and simulation, computation of stationary distributions and stochastic resonance for probabilistic models [46, 48, 50]. To date, less attention has been focused on formal statistical inference in disease systems (estimating key parameters and evaluating competing hypotheses) than on analysis of models. The most widespread approach to formal statistical inference has used the “time-series SIR” (TSIR) approach [39], in which the dynamics of transmission are approximated by a simple discrete-time stochastic model that can be fit to time series data via nonlinear regression. Although this approach has been applied to a variety of diseases [43, 67] and has proven a rough-and-ready tool, the approximations it makes begin to break down the further one goes from the measles regime. Novel approaches based on the state-space framework have been applied to diseases such as influenza [68], cholera [29], and plant diseases [69] and show promise for dealing with strain dynamics, age structure, and environmental drivers. In a state-space framework, the underlying, not directly observable eco-epidemiological processes responsible for observable patterns are viewed as distinct from the observation process itself. Statistical inference on state-space models is computationally demanding but recent algorithmic breakthroughs have greatly improved the outlook for rigorous inference. Worthy of special note are tailored MCMC approaches [68, 69], indirect inference approaches based on nonlinear forecasting [70], and iterated filtering [29, 71, 72], the latter two of which enjoy the “plug-and-play” property: they require only model simulation, obviating the need for analytical tractability of model. These methods have enjoyed considerable success in infectious-disease settings and will likely lead to important insights in other ecological systems.
Theoretical Challenges & Data Needs
Here, we look to the future, outlining some of the research frontiers in disease ecology and advocating for new and different kinds of long-term data.
Strain evolution, phylodynamics, and the community perspective
The broader ecological stage on which infectious disease dynamics play out, their community context, is increasingly recognized as critical [12, 26, 73]. While the single host-single pathogen paradigm deepened our understanding of the epidemiology of measles and chickenpox, for example, there are many systems for which its explanatory power is limited. Obvious examples include strain-polymorphic pathogens, such as those responsible for malaria, influenza, dengue, and polio. There remain numerous open questions that can only be adequately answered by additional data. For instance, in disease systems with antigenic variability, much uncertainty surrounds the determinants of strain diversity [74], the limits to strain coexistence [75], the mechanisms responsible for the observed patterns of strain replacement and the strength, duration and impact of immunity [76, 77].
In the context of pathogens with limited diversity (eg, cholera and dengue), existing theory is complex but relatively straightforward [75, 78]. When genetic novelty continually arises, as in the case of influenza A, the theoretical challenges are greater. The so-called “phylodynamics” perspective attempts infer aspects of the ecology and evolution of hosts and pathogens from the shapes of pathogen phylogenies [74, 79, 80]. Finding ways of better integrating genetic and epidemiological data beyond visual or descriptive comparisons of phylogenies remains a challenge.
Beyond multi-strain systems, we now recognise “polymicrobial diseases”, in which transmission and pathogenicity involve interactions among distinct pathogens. Examples include opportunistic bacterial and viral infections (with numerous high-profile demonstrations in HIV/AIDS patients), periodontal diseases and some respiratory infections, including Haemophilus influenzae and Streptococcus pneumoniae [81]. Despite the recognised importance of multi-pathogen diseases in general, appropriate long-term data remain in very short supply. We believe breakthroughs in the understanding of these processes will require the collation of data of different kinds, especially serological cross-sectional information shedding light on the kinetics of population immunological profiles and interactions among infectious agents.
Within-host dynamics
Most epidemiological models that track the prevalence of an infectious disease within a population categorize individuals according to infection and immunity status, e.g., susceptible, infectious, or recovered and immune. Conceptually and mathematically, this resembles the Levins metapopulation model, in which habitat patches are either empty or fully colonized, irrespective of population densities [82]. This approach has limited value in a number of applications [83], including attempts to understand the epidemiological outcome of mixed infections [84], the evolutionary consequences of pathogen life-history traits [59, 85], and the evolution of drug-resistance [86]. In such cases, attempts to understand the underlying processes using mathematical models have been frustrated by the absence of long-term data at the scale of the individual infection [87]. Short-term or snapshot data for the initial stages of an infection are often available, but greater longitudinal information is likely to be the key to further progress for persistent infections such as HIV [88] and pathogens that can reinfect, such as influenza [89].
Another venue that increasingly calls for a finer-scale understanding is immunity dynamics. While infections by chickenpox, smallpox, and morbilliviruses (including measles, rinderpest, and canine and phocine distemper viruses) induce life-long immunity, this does not appear to be the norm. Population-level data have been used to infer the dynamics of immunity relating to cholera [29, 67], Haemophilus Influenzas Type B [90] and pertussis [30]. Ultimately, however, this question will need resolution via confrontation of better specific within-host models of infection with appropriate empirical information, to some extent obtainable from animal models.
Environmental drivers
From a public health and wildlife management perspective, research on the ecology and evolution of infectious diseases would ideally translate into the development of early-warning systems. This effort has for the most part focused on using climatological variables to inform epidemic predictions. This is largely because mechanisms linking environmental conditions, such as rainfall and temperature, with disease transmission are known [91]. For example, temperature determines the developmental rate of the malaria parasite Plasmodium falciparum [92], and the persistence of avian influenza viruses in aquatic environments [93], with qualitative impacts for transmission dynamics [94]. Perhaps the best-studied aspect has been the impact of climatic variables on disease vectors; ecological niche models have been used to predict the presence of vectors by reference to abiotic determinants of habitat suitability [95]. Though plausible, climatological determinants of epidemic risk and reliable early warning systems based on them require further empirical support. This has led a number of scientists to use long-term data to examine the statistical association between climate variation and the incidence and dynamics of infectious diseases [91], especially cholera [5] and lyme disease [96], with an eye to making predictions about the consequences of climate change [97]. These efforts are likely to be transformed by the increasingly abundant spatially highly-resolved satellite data on environmental drivers, while the acquisition of similarly resolved epidemiological data remains an active frontier.
Surveillance networks & policy
We have highlighted several epidemiological success stories made possible by long-term data sets accumulated through surveillance systems. Naturally, improved understanding of such systems leads to an expectation of reliable quantitative predictions. Indeed, epidemiological models are increasingly expected to quantify unobserved variables in an outbreak in progress (so-called ‘nowcasting’) and to make forecasts. For example, in the 2001 outbreak of foot-and-mouth disease in the UK, policy-makers and politicians relied heavily on mathematical modelling in their selection of epidemic control measures, with great success [98, 99]. The recent H1N1 pandemic, however, provides a sobering counter-example. Following the first wave of transmission in the northern hemisphere in the summer of 2009, epidemiological models were scrutinized for predictions about the severity and impact of the autumn influenza season. The models dramatically over-predicted the size of the winter ’flu outbreaks and numbers of likely fatalities. This prominent setback can be largely attributed to inadequate information, both as to the epidemiology of the virus (particularly the case fatality rate) and the true extent of the rst epidemic. The recent study by Miller et al. [100] goes a long way towards explaining why: they demonstrate that the first wave of the epidemic in the UK is likely to have ten times more children than was initially estimated. The resulting over-estimation of the number of susceptible individuals appears to have led directly to over-estimation of the second wave’s severity. This observation points to the need for systematic cross-sectional serological surveys as a prerequisite for better real-time modeling of the dynamics of emerging threats.
Increasing reliability of models for forecasting and nowcasting will depend on better data on the contact patterns and transmission networks within and between populations. Promising recent developments in this regard include detailed studies of contact networks in Portland, Oregon [101] and self-reported mixing-pattern data from European populations [102]. We are also better placed to understand the mechanistic basis of individual movements thanks to mobile phone geolocation data [103] and the geographical dynamics of monetary currency (the ‘where is George?’ project; [104]).
A universal challenge in the interpretation of incidence data is reporting bias that arises, for example, when subclinical infections play an important epidemiological role yet are less likely to be reported than severe disease [29]. More troubling are the potential dynamic interactions between reporting fidelity and epidemiological processes. For example, the 2009 H1N1 pandemic showed that sensationalism and fear can lead to increased clinic visits and thus higher reported incidence. Making the best use of long-term incidence data will require a better understanding of interactions between disease dynamics, transmission, behavioural changes, and the processes by which incidence data are recorded.
Data sharing policies
Policies and practices for systematic data sharing and access have yet to be formulated and adopted by the epidemiological research community. This leads to tension between those who have invested in the collection and digitization of data and those who have invested in the development of analytic tools. Whatever community-wide (or more likely funding agency-mandated) policies are eventually agreed upon must take care to adequately reward the initial investment in data mining and collection efforts.
Summary & Conclusions
Infectious disease ecology is a vibrant field of research. Long-term epidemiological data continue to feature prominently in the development and utility of the field. Epidemiology has furnished some of the most definitive tests of ecological principles and has proved an unrivalled testbed for ecological theory and method. In turn, epidemiology is beginning to benefit from an ecological perspective on complex multi-host and multi-pathogen systems. Continued progress will depend on our ability to gather new and different long-term data and effectively query them using more realistic models.
Acknowledgements
We thank Marc Choisy and two anynomous reviewers for comments on this manuscript. We are grateful to Matt Ferrari and Natalia Mantilla-Beniers for graciously sharing figures 2 and 3, respectively. PR & AAK are supported by the Research and Policy in Infectious Disease Dynamics program of the Science and Technology Directorate, Department of Homeland Security, and the Fogarty International Center, National Institutes of Health. PR was also supported by the Vaccine Modeling Initiative of the Bill & Melinda Gates Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Graunt J. Natural and Political Observations Made upon the Bills of Mortality. 1662.
- 2.Newsholme A. A national system of notification and registration of sickness. Journal of the Royal Statistical Society. 1896;59:1–37. [Google Scholar]
- 3.Creighton C. A History of Epidemics in Britain. Cambridge University Press; Cambridge: 1894. [Google Scholar]
- 4.Cliff A, Haggett P, Smallman-Raynor M. Cambridge Studies in Historical Geography. Cambridge University Press; Cambridge: 1998. Deciphering Global Epidemics: Analytical Approaches to the Disease Records of World Cities, 1888-1912. [Google Scholar]
- 5.Pascual M, Rodo X, Ellner SP, Colwell R, Bouma MJ. Cholera dynamics and El Niño-southern oscillation. Science. 2000;289:1766–1769. doi: 10.1126/science.289.5485.1766. [DOI] [PubMed] [Google Scholar]
- 6.Cummings DAT, Irizarry RA, Huang NE, Endy TP, Nisalak A, Ungchusak K, Burke DS. Travelling waves in the occurrence of dengue haemorrhagic fever in Thailand. Nature. 2004;427:344–7. doi: 10.1038/nature02225. [DOI] [PubMed] [Google Scholar]
- 7.Smith DL, Lucey B, Waller LA, Childs JE, Real LA. Predicting the spatial dynamics of rabies epidemics on heterogeneous landscapes. Proc Natl Acad Sci USA. 2002;99(6):3668–72. doi: 10.1073/pnas.042400799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis S, Begon M, De Bruyn L, Ageyev VS, Klassovskiy NL, Pole SB, Viljugrein H, Stenseth NC, Leirs H. Predictive thresholds for plague in Kazakhstan. Science. 2004;304:736–738. doi: 10.1126/science.1095854. [DOI] [PubMed] [Google Scholar]
- 9.Bjørnstad O, Fromentin J-M, Stenseth NC, Gjoaeter J. Cycles and trends in cod populations. Proceedings of the National Academy of Sciences USA. 1999;96:5066–5071. doi: 10.1073/pnas.96.9.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen T, Stenseth N, Henttonen H. Multiannual vole cycles and population regulation during long winters: An analysis of seasonal density dependence. American Naturalist. 1999;154:129–139. doi: 10.1086/303229. [DOI] [PubMed] [Google Scholar]
- 11.Esper J, Büntgen U, Frank DC, Nievergelt D, Liebhold A. 1200 years of regular outbreaks in Alpine insects. Proc. R. Soc. B. 2007;274:671–679. doi: 10.1098/rspb.2006.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohani P, Green CJ, Mantilla-Beniers NB, Grenfell BT. Ecological interference between fatal diseases. Nature. 2003;422:885–8. doi: 10.1038/nature01542. [DOI] [PubMed] [Google Scholar]
- 13.ransome A. On epidemics cycles. Proceedings of the Manchester Literary and Philosophical Society. 1880:75–96. [Google Scholar]
- 14.Soper H. The interpretation of periodicity in disease prevalence. Journal of the Royal Statistical Society. 1929;92(1):34–73. [Google Scholar]
- 15.Hamer W. Epidemic diseases in England – the evidence of variability and of persistency of type. Lancet. 1906;1:733–739. [Google Scholar]
- 16.Ross R. The prevention of malaria. John Murray; London: 1911. [Google Scholar]
- 17.Kermack W, McKendrick A. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. A. 1927;115:700–721. [Google Scholar]
- 18.Bartlett M. The critical community size for measles in the united states. (Series A (General)).Journal of the Royal Statistical Society. 1960;123(1):37–44. [Google Scholar]
- 19.Bartlett M. Stochastic processes or the statistics of change. (Series C (Applied Statistics)).Journal of the Royal Statistical Society. 1953;2(1):44–64. [Google Scholar]
- 20.Dietz K. The incidence of infectious diseases under the influence of seasonal uctuations. Lecture Notes in Biomathematics. 1976;11:1–15. [Google Scholar]
- 21.Bailey N. The Mathematical Theory of Infectious Diseases. Charles Griffin & Company Ltd; London: 1975. [Google Scholar]
- 22.Anderson R, May R. Population biology of infectious diseases: Part I. Nature. 1979;280:361–367. doi: 10.1038/280361a0. [DOI] [PubMed] [Google Scholar]
- 23.May R, Anderson R. Population biology of infectious diseases: Part II. Nature. 1979;280:455–161. doi: 10.1038/280455a0. [DOI] [PubMed] [Google Scholar]
- 24.Anderson R, May R. Infectious Diseases of Humans: Dynamics and Control. Oxford University Press; Oxford: 1991. [Google Scholar]
- 25.Earn D, Rohani P, Grenfell B. Persistence, chaos and synchrony in ecology and epidemiology. Proceedings of the Royal Society B: Biological Sciences. 1998;265:7. doi: 10.1098/rspb.1998.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostfeld R, Keesing F, Eviner V. Infectious Disease Ecology: Effects of Ecosystems on Disease and of Disease on Ecosystems. Princeton University Press; Princeton: 2008. [Google Scholar]
- 27.Cummings DAT, Iamsirithaworn S, Lessler JT, McDermott A, Prasanthong R, Nisalak A, Jarman RG, Burke DS, Gibbons RV. The impact of the demographic transition on dengue in Thailand: insights from a statistical analysis and mathematical modeling. PLoS Med. 2009;6:e1000139. doi: 10.1371/journal.pmed.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viboud C, Bjørnstad ON, Smith DL, Simonsen L, Miller MA, Grenfell BT. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science. 2006;312:447–51. doi: 10.1126/science.1125237. [DOI] [PubMed] [Google Scholar]
- 29.King A, Ionides E, Pascual M, Bouma M. Inapparent infections and cholera dynamics. Nature. 2008;454:877–880. doi: 10.1038/nature07084. [DOI] [PubMed] [Google Scholar]
- 30.Wearing H, Rohani P. Estimating the duration of pertussis immunity using epidemiological signatures. PLoS Pathogens. 2009;5:e1000647. doi: 10.1371/journal.ppat.1000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tidd C, Olsen L, Schaffer W. The case for chaos in childhood epidemics. i. predicting historical epidemics from mathematical models. Proceedings of the Royal Society of London B. 1993;254:257–273. doi: 10.1098/rspb.1993.0155. [DOI] [PubMed] [Google Scholar]
- 32.Cazelles B, Chavez M, Berteaux D, Ménard F, Vik O, Jenouvrier S, Stenseth NC. Wavelet analysis of ecological time series. Oecologia. 2008;156:287–304. doi: 10.1007/s00442-008-0993-2. [DOI] [PubMed] [Google Scholar]
- 33.Taper ML, Lele SR. The Nature of Scientific Evidence: Statistical, Philosophical, and Empirical Considerations. University of Chicago Press; Chicago: 2004. [Google Scholar]
- 34.Fussmann G, Ellner S, Sherzer K, Hairston N., Jr Crossing the hopf bifurcation in a live predator-prey system. Science. 2000;290:1358–1360. doi: 10.1126/science.290.5495.1358. [DOI] [PubMed] [Google Scholar]
- 35.McCauley E, Nelson W, Nisbet R. Small-amplitude cycles emerge from stage-structured interactions in Daphnia-algal systems. Nature. 2008;455:1240–1243. doi: 10.1038/nature07220. [DOI] [PubMed] [Google Scholar]
- 36.Costantino RF, Desharnais RA, Cushing JM, Dennis B, Henson SM, King AA. Nonlinear stochastic population dynamics: The flour beetle Tribolium as an effective tool of discovery. In: Desharnais RA, editor. Population Dynamics and Laboratory Ecology. Vol. 37 of Advances in Ecological Research. Academic Press; 2005. pp. 101–141. doi:10.1016/S0065-2504(04)37004-2. [Google Scholar]
- 37.Grenfell BT, Bjornstad O, Finkenstadt B. Dynamics of measles epidemics: Estimating scaling of transmission rates using a time series sir model. Ecological Monographs. 2002;72:169–184. [Google Scholar]
- 38.London W, Yorke JA. Recurrent outbreaks of measles, chickenpox and mumps. i. seasonal variations in contact rates. American Journal of Epidemiology. 1973;98:453–468. doi: 10.1093/oxfordjournals.aje.a121575. [DOI] [PubMed] [Google Scholar]
- 39.Finkenstädt B, Grenfell B. Time series modelling of childhood diseases: a dynamical systems approach. Applied Statistics. 2000;49:187–205. [Google Scholar]
- 40.Ferrari M, Grais R, Bharti N, Conlan A, Bjornstad O, Wolfson L, Guerin P, Djibo A, Grenfell B. The dynamics of measles in sub-saharan africa. Nature. 2008;451:679–684. doi: 10.1038/nature06509. [DOI] [PubMed] [Google Scholar]
- 41.Bolker B, Grenfell B. Chaos and biological complexity in measles dynamics. Proceedings: Biological Sciences. 1993:75–81. doi: 10.1098/rspb.1993.0011. [DOI] [PubMed] [Google Scholar]
- 42.Earn D, Rohani P, Bolker B, Grenfell B. A simple model for complex dynamical transitions in epidemics. Science. 2000;287:667–670. doi: 10.1126/science.287.5453.667. [DOI] [PubMed] [Google Scholar]
- 43.Metcalf C, Bjornstad O, Grenfell B, Andreasen V. Seasonality and comparative dynamics of six childhood infections in pre-vaccination copenhagen. Proceedings of the Royal Society of London B. 2009;276:4111–4118. doi: 10.1098/rspb.2009.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicholson A. The self-adjustment of populations to change. Cold Spring Harbour Symposium on Quantitative Biology. 1957;22:153–172. [Google Scholar]
- 45.Andrewartha H, Birch L. The Distribution and Abundance of Animals. Chicago University Press; Chicago: 1954. [Google Scholar]
- 46.Rohani P, Keeling M, Grenfell B. The interplay between determinism and stochasticity in childhood diseases. American Naturalist. 2002;159:469–481. 2128. doi: 10.1086/339467. [DOI] [PubMed] [Google Scholar]
- 47.Bauch CT, Earn DJD. Transients and attractors in epidemics. Proc Biol Sci. 2003;270:1573–8. doi: 10.1098/rspb.2003.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alonso D, McKane AJ, Pascual M. Stochastic amplification in epidemics. J R Soc Interface. 2007;4:575–82. doi: 10.1098/rsif.2006.0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuske R, Gordillo L, Greenwood P. Sustained oscillations via coherence resonance in SIR. Journal of theoretical biology. 2007;245:459–469. doi: 10.1016/j.jtbi.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 50.Black A, McKane A, Nunes A, Parisi A. Stochastic fluctuations in the susceptible-infective-recovered model with distributed infectious periods. Physical Review E. 2009;80:021922. doi: 10.1103/PhysRevE.80.021922. [DOI] [PubMed] [Google Scholar]
- 51.Hastings A, Hom C, Turchin P, Ellner S, Godfray H. Chaos in ecology: Is mother nature a strange attractor? Annual Review of Ecology and Systematics. 1993;24:1–33. [Google Scholar]
- 52.Schaffer W, Kot M. Chaos in ecological systems: the coals that newcastle forgot. Trends in Ecology & Evolution. 1986;1:58–63. doi: 10.1016/0169-5347(86)90018-2. [DOI] [PubMed] [Google Scholar]
- 53.Olsen L, Schaffer W. Chaos versus noisy periodicity: alternative hypotheses for childhood epidemics. Science. 1990;249:499. doi: 10.1126/science.2382131. [DOI] [PubMed] [Google Scholar]
- 54.Sugihara G, May R. Nonlinear forecasting as a way of distinguishing chaos from measurement error in time-series. Nature. 1990;344:734–741. doi: 10.1038/344734a0. [DOI] [PubMed] [Google Scholar]
- 55.Ellner S, Turchin P. Chaos in a noisy world: New methods and evidence from time-series analysis. American Naturalist. 1995;145:343–375. [Google Scholar]
- 56.McLean A, Anderson R. Measles in developing countries part I. Epidemiological parameters and patterns. Epidemiology and Infection. 1988;100:111–133. doi: 10.1017/s0950268800065614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pitzer VE, Viboud C, Simonsen L, Steiner C, Panozzo CA, Alonso WJ, Miller MA, Glass RI, Glasser JW, Parashar UD, Grenfell BT. Demographic variability, vaccination, and the spatiotemporal dynamics of rotavirus epidemics. Science. 2009;325:290–294. doi: 10.1126/science.1172330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rohani P, Earn DJ, Grenfell BT. Opposite patterns of synchrony in sympatric disease metapopulations. Science. 1999;286:968–71. doi: 10.1126/science.286.5441.968. [DOI] [PubMed] [Google Scholar]
- 59.King AA, Shrestha S, Harvill ET, Bjørnstad ON. Evolution of acute infections and the invasion-persistence trade-off. American Naturalist. 2009;173:446–455. doi: 10.1086/597217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Black F. Measles endemicity in insular populations: critical community size and its evolutionary implication. Journal of Theoretical Biology. 1966;11:207. doi: 10.1016/0022-5193(66)90161-5. [DOI] [PubMed] [Google Scholar]
- 61.Broutin H, Mantilla-Beniers N, Simondon F, Aaby P, Grenfell B, Guégan J-F, Rohani P. Epidemiological impact of vaccination on the dynamics of two childhood diseases in rural Senegal. Microbes & Infection. 2005;7:593–599. doi: 10.1016/j.micinf.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 62.Grenfell B, Bjornstad O, Kappey J. Travelling waves and spatial hierarchies in measles epidemics. Nature. 2001;414:716–723. doi: 10.1038/414716a. [DOI] [PubMed] [Google Scholar]
- 63.Brown J, Kodric-Brown A. Turnover rates in insular biogeography: effect of immigration on extinction. Ecology. 1977;58:445–449. [Google Scholar]
- 64.Zadokis J, Van den Bosch F. On the spread of plant disease - a theory on foci. Annual Review of Phytopathology. 1994;32:503–521. doi: 10.1146/annurev.py.32.090194.002443. [DOI] [PubMed] [Google Scholar]
- 65.Xia YC, Bjornstad ON, Grenfell BT. Measles metapopulation dynamics: A gravity model for epidemiological coupling and dynamics. American Naturalist. 2004;164:267–281. doi: 10.1086/422341. [DOI] [PubMed] [Google Scholar]
- 66.Diekmann O, Heesterbeek J. Mathenatical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation. Wiley; 2000. [Google Scholar]
- 67.Koelle K, Pascual M. Disentangling extrinsic from intrinsic factors in disease dynamics: A nonlinear time series approach with an application to cholera. American Naturalist. 2004;163:901–913. doi: 10.1086/420798. [DOI] [PubMed] [Google Scholar]
- 68.Cauchemez S, Carrat F, Viboud C, Valleron AJ, Boëlle PY. A Bayesian MCMC approach to study transmission of influenza: application to household longitudinal data. Statistics in Medicine. 2004;23:3469–3487. doi: 10.1002/sim.1912. [DOI] [PubMed] [Google Scholar]
- 69.Gibson GJ, Kleczkowski A, Gilligan CA. Bayesian analysis of botanical epidemics using stochastic compartmental models. Proceedings of the National Academy of Sciences of the U.S.A. 2004;101:12120–12124. doi: 10.1073/pnas.0400829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kendall B, Ellner S, McCauley E, Wood S, Briggs C, Murdoch W, Turchin P. Population cycles in the pine looper moth: Dynamical tests of mechanistic hypotheses. Ecological Monographs. 2005;75(2):259–276. [Google Scholar]
- 71.Ionides E, Breto C, King A. Inference for nonlinear dynamical systems. Proceedings of the National Academy of Sciences. 2006;103(49):18438. doi: 10.1073/pnas.0603181103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He D, Ionides EL, King AA. Plug-and-play inference for disease dynamics: measles in large and small populations as a case study. Journal of The Royal Society Interface. 2010;7:271–283. doi: 10.1098/rsif.2009.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fenton A, Pedersen A. Community epidemiology framework for classifying disease threats. Emerg Infect Dis. 2005;11:1815–1821. doi: 10.3201/eid1112.050306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferguson N, Galvani A, Bush R. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- 75.Gog J, Grenfell B. Dynamics and selection of many-strain pathogens. Proceedings of the National Academy of Sciences of the USA. 2002;99:17209–17214. doi: 10.1073/pnas.252512799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wearing H, Rohani P. Ecological and immunological determinants of dengue epidemics. Proceedings of the National Academy of Sciences of the USA. 2006;103:11802–11807. doi: 10.1073/pnas.0602960103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Adams B, Holmes EC, Zhang C, Mammen MP, Nimmannitya S, Kalayanarooj S, Boots M. Cross-protective immunity can account for the alternating epidemic pattern of dengue virus serotypes circulating in bangkok. Proc Natl Acad Sci USA. 2006;103(38):14234–9. doi: 10.1073/pnas.0602768103. doi:10.1073/pnas.0602768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta S, Ferguson N, Anderson R. Chaos, persistence and evolution of strain structure in antigenically diverse infectious agents. Science. 1998;280:912–915. doi: 10.1126/science.280.5365.912. [DOI] [PubMed] [Google Scholar]
- 79.Biek R, Drummond AJ, Poss M. A virus reveals population structure and recent demographic history of its carnivore host. Science. 2006;311:538–541. doi: 10.1126/science.1121360. [DOI] [PubMed] [Google Scholar]
- 80.Grenfell B, Pybus O, Gog J, Wood J, Daly J, Mumford J, Holmes EC. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 81.Brogden K, Guthmiller J. Polymicrobial Diseases. ASM Press; Washington DC: 2002. [PubMed] [Google Scholar]
- 82.Levins R. Some demographic and genetic consequences of environmental heterogeneity for biological control. Bulletin of the Entomological Society of America. 1969;15:237–240. [Google Scholar]
- 83.Mideo N, Alizon S, Day T. Linking within- and between-host dynamics in the evolutionary epidemiology of infectious diseases. Trends in Ecology & Evolution. 2008;23:511–517. doi: 10.1016/j.tree.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 84.Antia R, Yates A, Roode JD. The dynamics of acute malaria infections. I. Effect of the parasite’s red blood cell preference. Proceedings of the Royal Society B. 2008;275(1641):1449. doi: 10.1098/rspb.2008.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Read J, Keeling M. Disease evolution across a range of spatio-temporal scales. Theoretical Population Biology. 2006;70(2):201–213. doi: 10.1016/j.tpb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 86.Breban R, Napravnik S, Kahn J, Blower S. Quantifying the treatment efficacy of reverse transcriptase inhibitors: new analyses of clinical data based on within-host modeling. BMC Public Health. 2009;9:S11. doi: 10.1186/1471-2458-9-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Handel A, Longini I, Jr., Antia R. Neuraminidase inhibitor resistance in Influenza: Assessing the danger of its generation and spread. PLoS Computational Biology. 2007;3:e240. doi: 10.1371/journal.pcbi.0030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dixit N, Perelson A. Hiv dynamics with multiple infections of target cells. Proc. Natl. Acad. Sci. USA. 2005;102:8198–8203. doi: 10.1073/pnas.0407498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Handel A, Longini I, Jr., Antia R. Towards a quantitative understanding of the within-host dynamics of Influenza a infections. J. Roy. Soc. Interface. 2010;7:35–47. doi: 10.1098/rsif.2009.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Auranen K, Eichner M, Käyhty H, Takala AK, Arjas E. A hierarchical Bayesian model to predict the duration of immunity to haemophilus Influenzas type B. Biometrics. 2004:1306–1313. doi: 10.1111/j.0006-341x.1999.01306.x. [DOI] [PubMed] [Google Scholar]
- 91.Rogers L. The world incidence of leprosy in relation to meteorological conditions and its bearings on the probable mode of transmission. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1923;16:440–464. [Google Scholar]
- 92.MacDonald G. The epidemiology and control of malaria. Oxford University Press; London: 1957. [Google Scholar]
- 93.Brown J, Goekjian G, Poulson R, Valeika S, Stallknecht D. Avian influenza virus infectivity in water: Viral responses to varying pH, salinity, and temperature conditions. Veterinary Microbiology. 2009;136:20–26. doi: 10.1016/j.vetmic.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 94.Rohani P, Breban R, Stallknecht D, Drake J. Environmental transmission of low pathogenicity avian influenza viruses and its implications for pathogen invasion. Proceedings of the National Academy of Sciences. 2009;106:10365–10369. doi: 10.1073/pnas.0809026106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Craig M, Snow R, Le Sueur D. A climate-based distribution model of malaria transmission in sub-saharan africa. Parasitology Today. 1999;15:105–110. doi: 10.1016/s0169-4758(99)01396-4. [DOI] [PubMed] [Google Scholar]
- 96.Brownstein JS, Holford TR, Fish D. Effect of climate change on lyme disease risk in north america. EcoHealth. 2005;2:38–46. doi: 10.1007/s10393-004-0139-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rogers DJ, Randolph SE. The global spread of malaria in a future, warmer world. Science. 2000;289:1763–1766. doi: 10.1126/science.289.5485.1763. [DOI] [PubMed] [Google Scholar]
- 98.Keeling M, Woolhouse M, Shaw D, Matthews L, Chase-Topping M, Haydon D, Cornell S, Kappey J, Wilesmith J, Grenfell B. Dynamics of the 2001 uk foot and mouth epidemic: stochastic dispersal in a heterogeneous landscape. Science. 2001;294:813. doi: 10.1126/science.1065973. [DOI] [PubMed] [Google Scholar]
- 99.Ferguson N, Donnelly C, Anderson R. Transmission intensity and impact of control policies on the foot and mouth epidemic in great britain. Nature. 2001;413(6855):542–547. doi: 10.1038/35097116. [DOI] [PubMed] [Google Scholar]
- 100.Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. Incidence of 2009 pandemic Influenza A H1N1 infection in England: a cross-sectional serological study. The Lancet. doi: 10.1016/S0140-6736(09)62126-7. [DOI] [PubMed] [Google Scholar]
- 101.Eubank S, Guclu H, Kumar VA, Marathe M, Srinivasan A, Toroczkai Z, Wang N. Modelling disease outbreaks in realistic urban social networks. Nature. 2004;429(6988):180–184. doi: 10.1038/nature02541. [DOI] [PubMed] [Google Scholar]
- 102.Mossong J, Hens N, Jit M, Beutels P, Auranen K, Mikolajczyk R, Massari M, Salmaso S, Tomba GS, Wallinga J, Heijne J, Sadkowska-Todys M, Rosinska M, Edmunds WJ. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 2008;5:e74. doi: 10.1371/journal.pmed.0050074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.González M, Hidalgo C, Barabási A. Understanding individual human mobility patterns. Nature. 2008;453:779–82. doi: 10.1038/nature06958. [DOI] [PubMed] [Google Scholar]
- 104.Brockmann D, Hufnagel L, Geisel T. The scaling laws of human travel. Nature. 2006;439:462–5. doi: 10.1038/nature04292. [DOI] [PubMed] [Google Scholar]