Abstract
Naturally occurring poly(purine·pyrimidine) rich regions in the human genome are prone to adopt non-canonical DNA structures such as intramolecular triplexes (i.e. H-DNA). Such structure-forming sequences are abundant and can regulate the expression of several diseases-linked genes. In addition, the use of triplex-forming oligonucleotides (TFOs) to modulate gene structure and function has potential as an approach to targeted gene therapy. Previously, we found that endogenous H-DNA structures can induce DNA double-strand breaks and promote genomic rearrangements. Herein, we find that the DHX9 helicase co-immunoprecipitates with triplex DNA structures in mammalian cells, suggesting a role in the maintenance of genome stability. We tested this postulate by assessing the helicase activity of purified human DHX9 on various duplex and triplex DNA substrates in vitro. DHX9 displaced the third strand from a specific triplex DNA structure and catalyzed the unwinding with a 3′→5′ polarity with respect to the displaced third strand. Helicase activity required a 3′-single-stranded overhang on the third strand and was dependent on ATP hydrolysis. The reaction kinetics consisted of a pre-steady-state burst phase followed by a linear, steady-state pseudo-zero-order-reaction. In contrast, very little, if any helicase activity was detected on blunt triplexes, triplexes with 5′-overhangs, blunt duplexes, duplexes with overhangs, or forked duplex substrates. Thus, triplex structures containing a 3′-overhang represent preferred substrates for DHX9, where it removes the strand with Hoogsteen hydrogen-bonded bases. Our results suggest the involvement of DHX9 in maintaining genome integrity by unwinding mutagenic triplex DNA structures.
Alternative DNA conformations (i.e. non-B DNA), in addition to the canonical form of DNA consisting of a right-handed double helix (B-DNA), can form at repetitive DNA motifs, including left-handed Z-DNA adopted by alternating purine·pyrimidine sequences, cruciforms and hairpin/loops extruded from inverted and direct repeats, respectively, and multistranded triplex and quadruplex conformations assembled from poly(purine·pyrimidine) tracts with mirror repeat symmetry and G-rich sequences, respectively (1–7).
Bioinformatic analyses on the distribution of non-B DNA-forming sequences in the human genome revealed unexpected associations between the types of sequences, and hence their underlying non-B conformations, and specific classes of genes, gene locations and/or chromosomes (reviewed in (5)). Specifically, quadruplex-forming motifs were found to be enriched at telomeric ends and near transcription start sites, particularly in genes involved in growth and development (7–9). By contrast, long poly(purine·pyrimidine) tracts were clustered in the pseudoautosomal region (PAR1) of the sex chromosomes, essential for their meiotic segregation and recombination, and were enriched in genes involved in cell communication, particularly in the brain (10). Finally, large inverted repeats comprised most of the male-specific region of the Y-chromosome, where gene families essential to spermatogenesis are located (11).
Genome-wide analyses also revealed an unanticipated inter-individual variability in gene copy number variations (CNVs) (12) and the presence of highly homologous and modular low copy repeats [LCRs or segmental duplications (SDs)] (13). These analyses, together with studies of gene expression profiles (reviewed in (5)), the use of structure-specific binding probes (14, 15) and the sequencing of breakpoints underlying genomic rearrangements that occurred both during speciation (16, 17) and within the human population (18, 19), led to the view that non-B DNA structures are functional genomic elements that play pleiotropic roles in the cell. These include gene function and regulation, telomere and centromere function, and the generation of genomic diversity responsible for phenotypic variation.
At the same time, studies aimed at identifying the genomic elements underlying chromosomal rearrangements leading to human disease (reviewed in (6) and (20)), identified a common molecular mechanism of genetic instability based on the DNA structural features associated with repetitive DNA. The conclusion that such features coincide with non-B DNA conformations was supported by studies on model systems, ranging from bacteria (21, 22) to mammalian cell culture (23, 24) and the mouse (25), in which conditions that favor the structural transitions from B- to non-B DNA, including transcription and negative supercoiling, lead to genetic instability. Hence, non-B DNA conformations perform both a physiological role and potentiate genomic instability.
While the mechanisms that regulate the formation and resolution of non-B DNA structures remain to be elucidated, the use of exogenous small molecules that either form or stabilize non-B DNA represents an attractive means for therapeutic intervention aimed at compromising cancer cell function. For example, AS-1411, a 26 nucleotide aptamer, which folds into a quadruplex structure that inhibits nucleolin, is in phase II clinical trials for the treatment of acute myeloid leukemia and renal cancer (7). Similarly, the use of psoralen-conjugated triplex-forming oligonucleotides provides a means to deliver site-specific interstrand crosslinks (TFO-directed psoralen interstrand crosslinks or Tdp-ICLs) and thus inhibit gene expression and/or target damage to a specific site. For example, we reported a Tdp-ICL-based approach targeting the human c-MYC oncogene, a strategy that successfully inhibited c-MYC expression and that, in combination with gemcitabine, may provide an effective means to improve breast cancer therapy (26).
In this context, defining the cellular mechanisms that either promote or inhibit/resolve the formation of non-B DNA structures would greatly improve the design of effective drug molecules. The current data support a role for regulators of genomic stability, such as p53 (27) and the BRAFT super-complex (28), comprising the Fanconi’s anemia (FA) and Bloom (BLM) complexes, in the recognition and resolution of some non-B DNA structures, and point to a relevant role for helicases, such as BRIP1 and BLM (29, 30), in unwinding DNA secondary structures. In our search for components of the cellular machinery that are involved in the processing and resolution of triplex DNA structures, we used chromatin co-immunoprecipitation and MALDI-TOF analyses. We identified DHX9 as a component of the triplex DNA-protein complex in mammalian cells.
The DHX9 protein [unofficial HUGO nomenclature used; nuclear DNA helicase II (NDH II) or RNA helicase A (RHA)] belongs to the DEXH family of helicase superfamily 2, and was first isolated and characterized from calf thymus nuclei (31). The protein has a molecular weight of ~140 kDa and is characterized by 7 conserved core motifs, including two copies of a double stranded RNA-binding domain at the amino terminus, a helicase core domain in the central region, and an RGG-rich region at the carboxyl terminus (Figure 1) (32, 33), which confer both RNA and DNA helicase activities (34). The embryonic lethality of homozygous DHX9-knockout mice has been reported (35), which indicates an indispensable role for this helicase in mammalian cells (reviewed in (36)).
Figure 1. Schematic of the DHX9 helicase.
The schematic represents the N-terminal region of the protein (left) with the double-stranded RNA binding motifs I and II (dsRBMI and dsRBMII), followed by the helicase core domain (middle), comprising 7 evolutionarily conserved motifs, including the ATP binding and hydrolysis motif (boxes I, II and VI), the DNA binding motif (box Ia), the DEXH domain (box II), the nucleic acid unwinding domain (box III), and the RNA binding domain (box VI) and finally the C-terminal region (right), containing the conserved RGG box involved in single-stranded binding to nucleic acid substrates. Critical amino acids within the boxes are indicated. The relative positions of the motifs and spacing between motifs are not to scale. The numbers above the arrows are the typical range of amino acids spanning each region.
In this study, we tested the hypothesis that DHX9 unwinds triplex DNA structures as a part of its role in maintaining genomic integrity. The data demonstrate that the purified protein has the capacity to unwind intermolecular triplex DNA substrates in vitro with a specific 3′→5′ polarity with respect to the displaced third strand. This activity required a 3′-single-stranded overhang on the third strand and was dependent on ATP hydrolysis. In contrast, we detected comparatively low or no activity on a variety of duplex and forked duplex substrates, irrespective of the availability of 3′-single-stranded overhangs. The preference of DHX9 for triplex DNA structures, where it removes the strand with Hoogsteen hydrogen-bonded bases, suggests that it may be a part of the enzymatic repertoire of enzymes capable of resolving non-B DNA substrates. These findings support the notion that removal of such conformations may represent a major task, likely involving a larger number of enzymes than previously anticipated.
EXPERIMENTAL PROCEDURES
Expression and purification of recombinant DHX9 protein
Baculovirus stocks containing human DHX9 vectors were used to infect ~2 × 108 HiV cells at a multiplicity of infection of 1–5. Cells were harvested 40–48 h post-infection in TC100 medium. The following day fresh medium was added mixed with 1 ml of virus stock and incubated for 48 h at 27 °C. Cells were harvested by centrifugation at 1000 rpm for 5 min at 4 °C. Cells were washed with ice-cold phosphate-buffered saline (10 mM sodium phosphate, pH 7.4, 140 mM NaCl, 3 mM KCl, 1x PBS), further centrifuged at 1000 rpm for 5 min at 4 °C and stored at −70 °C. DHX9 was purified from whole cell extracts with Ni2+-NTA-Agarose (Qiagen) and with poly(rI·rC)-agarose, as previously described (32). DHX9 concentrations were determined by Bradford assays and polyacrylamide gel electrophoresis (PAGE).
Preparation of DNA substrates
PAGE-purified single-stranded oligo-deoxyribonucleotides used for the preparation of DNA substrates were obtained from Integrated DNA Technologies (Coralville, IA, USA) and are listed in Table 1A. The sequence of the triplex substrate was from the human rhodopsin gene (37). Prior to annealing the oligonucleotides to form the various DNA structures (Table 1B), 5′-ends were labeled with [γ-32P]ATP and T4 polynucleotide kinase for 1 h at 37 °C. The unincorporated nucleotides were removed by size-exclusion chromatography using MicroSpin G-25 columns (GE, Buckinghamshire, UK). To form the duplex and triplex substrates (Table 1B), the 5′-end-labeled oligonucleotides were mixed at molar ratios of 1:1 (duplex DNA) and 1:1:1 (triplex DNA), and annealed in triplex binding (TB) buffer [10 mM Tris·HCl (pH 7.6), 10 mM MgCl2, 10% glycerol (v/v)]. This high Mg2+ concentration was critical for triplex stability. Samples were heated to 95 °C for 10 min, and then cooled to 4 °C in a PCR apparatus (Bio-Rad) at the rate of 0.1 °C/min. The purity of the annealed substrates was assessed by 12% native PAGE (29:1, acrylamide/bis-acrylamide) in running (R) buffer (89 mM Tris-borate, pH 8.0, and 10 mM MgCl2). Electrophoretic separations were conducted for 4–5 h at 75 V (7.5 V/cm), and gels were exposed to a PhosphorScreen for visualization of the radiolabeled substrates by using a Storm PhosporImager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Table 1A.
Oligonucleotides used in this study.
| Oligo Name | Length | Sequence (5′→3′) |
|---|---|---|
| 21R | 21 | GGAAGC GGT AGG GGG AGG GGG |
| 21Y | 21 | GTA CAC GGT GAT CCT CTA GAG |
| 21R with 3′-overhang | 31 | CTC TAG AGG ATC ACC GTG TAC GTC ATA GTA T |
| 31R with 3′-overhang | 31 | GGA AGC GGT AGG GGG AGG GGG CAG TCG AGC G |
| 31R with 5′-overhang | 31 | ATA GCA GCT AGG AAG CGG TAG GGG GAG GGG G |
| 41R | 41 | AAA CAA CAC TGG GGG AGG GGG ACG GTG AAG GCC AAG TTC CC |
| 41Y | 41 | GGG AAC TTG GCC TTC ACC GTC CCC CTC CCC CAG TGT TGT TT |
| 45R | 45 | ACT CTA GAG GAT CCC CGG GTA CGT TAT TGC ATG AAA GCC CGG CTG |
| 45Y | 45 | ACT ATA ATA GCG ACG TAC CGC CAT TAG CCG GGG ATC CTC TAG AGT |
The triplex-forming oligonucleotide (TFO) binding site sequence is underlined
Table 1B.
Schematic structures of DNA substrates used in this study.
| Substrate name | Structure | Oligos used to make substrate |
|---|---|---|
| Blunt duplex DNA | 41R + 41Y | |
| Blunt triplex DNA |  |
41R + 41Y + 21R |
| 3′-overhang triplex DNA |  |
41R + 41Y + 31R with 3′-overhang |
| 5′-overhang triplex DNA |  |
41R + 41Y+ 31R with 5′-overhang |
| 3′-overhang duplex DNA |  |
21R with 3′-overhang + 21Y |
| Forked duplex DNA |  |
45R + 45Y |
Helicase/strand displacement assay
DHX9 activity was measured by the extent of strand displacement from the duplex or triplex DNA substrates (Table 1B) as follows; 10 μl reactions were carried out in helicase (H) buffer, consisting of TB buffer supplemented with 5 mM DTT, 5 mM ATP (unless stated otherwise) and 5 nM of the specific DNA substrate. The indicated amounts of DHX9 protein were added to the reaction mixtures and incubated in a temperature-controlled PCR apparatus at 32 °C for 20 min. This temperature was chosen based on previous experiments of helicase function on triplex structures (38, 39). The reactions were stopped by quick chilling on ice, followed by the addition of 2 μl loading buffer (40% sucrose, 0.1% bromophenol blue and 0.1% xylene cyanol). For the time-course kinetic experiments, 100 μl reactions were prepared as described above, from which 10 μl aliquots were withdrawn at the indicated time points, quick chilled on ice and mixed with 2 μl loading buffer. The reaction products were resolved by electrophoresis on 12% native polyacrylamide gels in ‘R’ buffer and visualized using a PhosphorImager. The images were quantified using the ImageQuant software and the percent of unwinding was determined from the total amount of the native DNA structures remaining at each time point.
RESULTS
DHX9 protein unwinds triplex DNA, but not blunt-end duplex DNA
In an effort to isolate the components of the enzymatic complex responsible for processing triplex DNA structures, we conducted co-immunoprecipitation experiments using anti-RPA antibodies in 293T cells to pull down protein complexes associated with plasmid substrates containing a triplex structure. MALDI-TOF analyses of the proteins resolved and eluted from denaturing polyacrylamide gels revealed the DHX9 helicase as a major component enriched with the triplex DNA structure (Suppl. Text, Suppl. Fig. 1 and Suppl. Table 1). This result prompted speculation that DHX9 may be involved in maintaining overall genome stability by resolving triplex DNA structures, a known source of DSBs leading to genomic rearrangements (21, 24, 25). Hence, to test the activity of DHX9 on triplex structures, substrates were prepared as described in “Experimental Procedures” (shown in Tables 1A and 1B). First, the annealing conditions for obtaining the relevant DNA substrates in high yield were optimized. These conditions included the use of gel-purified oligo-deoxyribonucleotides and a slow rate of cooling (0.1 °C/min) from 95 °C to 4 °C. Gel mobility-shift experiments indicated that the products (duplex and triplex) migrated at the expected positions according to their mass and structure on 12% native polyacrylamide gels. Specifically, the triplex DNA structures migrated slower than the duplex structures and single-stranded DNA. Also, the triplex structure containing a 5′-single-stranded overhang migrated slower than that containing a 3′-single-stranded overhang (Suppl. Text and Suppl. Fig. 2).
The helicase reaction assays were performed by incubating increasing amounts of purified human DHX9 protein (0, 50, 100, 200 nM) for 20 min at 32 °C with a constant amount of DNA substrate (5 nM). The enzyme displaced the third strand from a triplex substrate containing a 3′-single-stranded overhang in a concentration-dependent manner. Near complete displacement of the third strand was achieved at 200 nM, as evidenced from the products of the helicase reaction (Figure 2A), which migrated at the same position as the labeled single-strand and duplex DNA. In contrast, DHX9 (at 200 nM) failed to unwind a DNA duplex substrate of the same length and sequence composition as the duplex portion of the triplex substrate. Quantification of the activity of DHX9 helicase as a function of protein concentration on the triplex DNA substrate showed that at 50, 100, and 200 nM, 84, 57, and 8% triplex DNA remained on average after the reactions, respectively (Figure 2B) (r2 = 0.98). To determine whether this activity resulted from non-specific protein-DNA interactions, we tested the activity of bovine serum albumin (BSA) on the triplex DNA substrate. No detectable unwinding activity was observed (Figure 2A). Together, these results indicate that DHX9 possesses strand displacement activity on triplex DNA structures containing a 3′-single-stranded overhang, but undetectable activity on the blunt-end duplex DNA structures.
Figure 2. DHX9 unwinds triplex DNA.
Panel A. Five nM of triplex or double-stranded [γ32P]ATP-labeled DNA substrate (left) was incubated with various concentrations (0–200 nM) of DHX9 helicase or 150 nM BSA for 20 min at 32 °C (for details see “Experimental Procedures”). Helicase activity leading to strand-displacement was assessed by 12% native PAGE. Triangle; control lane in which triplex DNA was heat-denatured (5 min, 95 °C) to yield the duplex DNA plus the free third strand. Panel B. PhosphorImager quantification of DHX9 helicase activity on triplex DNA as a function of DHX9 concentration (mean ± SE from 3 independent experiments).
ATP is required for the strand displacement activity of DHX9 on triplex structures
Next, we explored whether hydrolysis of ATP was required for the unwinding activity of DHX9 on triplex DNA substrates. We incubated increasing amounts of protein (0, 50, 100, 200 nM) with triplex DNA substrates containing a 3′-single-stranded overhang either in the presence or absence of 5 mM ATP (Figure 3A) as well as in the presence of the non-hyrolyzable analogue of ATP, 5′-adenylyl-beta,gamma-imidodiphosphate (AMP-PNP) (Figure 3B). In the absence of ATP or in the presence of AMP-PNP, DHX9 failed to catalyze third strand displacement from the triplex substrate (Figure 3). In contrast, in the presence of hydrolyzable ATP (5 mM), the enzyme was able to displace the third strand from the triplex substrates. At 200 nM protein concentration, the third strand was entirely displaced from the triplex DNA substrate (Figure 3A), leaving duplex DNA intact. Hence, we conclude that ATP hydrolysis was indispensable for the helicase activity leading to strand displacement.
Figure 3. ATP is required for DHX9 helicase activity.
Panel A. Triplex DNA (see legend to Figure 1) was incubated with 0–200 nM DHX9 helicase in the presence (left) or in the absence (right) of 5 mM ATP. Panel B. Triplex DNA was incubated with 200 nM DHX9 in the presence of 5 mM ATP or in the presence 5 mM AMP-PNP, a nonhydrolyzable nucleotide analogue. Triangle; as specified in Figure 1.
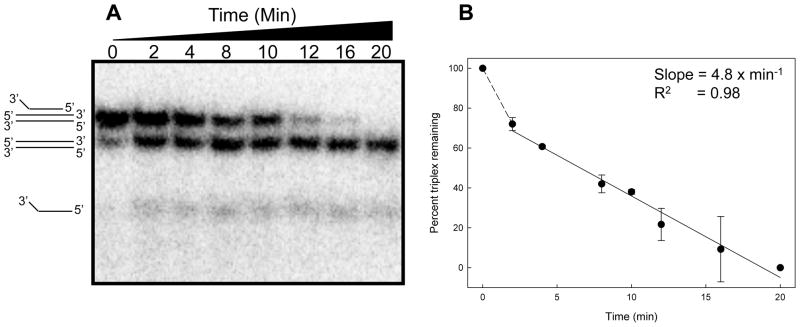
Kinetic analysis of DNA unwinding
Using the conditions described above, we then examined the rate of unwinding of DHX9 on the 3′-single-stranded overhang triplex substrate as a function of time. For this, 100 μl reaction mixtures were prepared in ‘H’ reaction buffer (“Material and Methods”), from which 10 μl aliquots were withdrawn at the indicated time points, quickly chilled on ice and mixed with 2 μl loading buffer to terminate the reaction. The reaction products were separated by 12% native polyacrylamide gel electrophoresis for analysis. DHX9 progressively unwound the third strand from the triplex DNA substrate (Figure 4A) with a biphasic kinetic behavior, which included a transient initial “burst” phase (~2 min) followed by a slower, linear phase (~18 min). This analysis confirmed that the DHX9 protein unwound the triplex substrate in a time-dependent manner (Figure 4B). Based on the slope of the reaction of ~4.8 min−1, we conclude that 4.8 Hoogsteen hydrogen-bonded bases were unwound per min. In summary, DHX9 displays a “bona fide” enzymatic behavior with respect to triplex DNA that follows pseudo-zero-order kinetics, in which the secondary DNA structure represents the substrate and the single-strand and duplex DNA species represent the reaction products.
Figure 4. Time-dependent DHX9 helicase activity.
Panel A. Triplex DNA was incubated with 200 nM DHX9 (Figure 1 and “Experimental Procedures”) and aliquots were withdrawn at the indicated time points and resolved by 12% native PAGE. Panel B. Graph of percent triplex remaining as a function of time (mean ± SE from 2 independent experiments). The rate of unwinding was calculated from the slope of the linear portion of the graph.
DHX9 polarity and substrate specificity
The strand to which helicases bind and migrate defines their polarity (40). To determine the polarity of DHX9 translocation, three different types of triplex DNA substrates were prepared; one blunt-end triplex, and two triplex DNA substrates containing a 10-nt ssDNA overhang at either the 3′- or 5′-end of the third strand (Table 1B). To avoid preferences in sequence composition, the core region of the triplex forming oligonucleotide (TFO) binding site (the sequence is underlined, Table 1A) contained the same nucleotide sequences. The activity of DHX9 on blunt-end triplexes and on the triplexes containing the 3′- or 5′-overhang is shown in Figure 5A and B. With varying concentrations of enzyme and under standard experimental conditions, the 3′-single-stranded overhang-containing triplex substrate was unwound efficiently, whereas little or no activity was detected on the 5′-single-stranded overhang and blunt substrates (Figure 5B). Indeed, no significant (>5%) unwinding activity on blunt-end triplexes was observed at any protein concentration tested (Figure 5A). These findings indicate that DHX9 has a strong preference for triplex DNA substrates with 3′-single-stranded overhang, where it translocates with a specific 3′→5′ polarity.
Figure 5. DHX9 activity requires a 3′-single-stranded overhang and displays 3′→5′ polarity.
Panel A. Lanes 1–2, mobility of the single-strands used to prepare the duplex and blunt-end triplex DNA structures. Helicase activity (see “Experimental Procedures”) in the presence of ATP was assessed on blunt-end duplex DNA (lanes 3–4) and blunt-end triplex DNA (lanes 5–8) by using 200 and 0–200 nM DHX9, respectively, whereas lane 9 shows the helicase activity of 200 nM DHX9 on blunt-end triplex in the absence of ATP. Triangle; as specified in Figure 1. Panel B. Helicase activity of DHX9 on various types of triplex structures. Lanes 1–3, mobility of the single-strands used to assemble triplex DNA structures with blunt-ends (lanes 5–6), 5′-single-stranded overhang (lanes 7–8) and 3′-single-stranded overhang (lanes 9–10) as depicted above the lanes, lane 4 shows the mobility of duplex DNA. A total of 200 nM DHX9 was used to determine the helicase activity on each type of triplex DNA structure.
Forked duplex DNA structures are weak substrates for DHX9 helicase activity
We first characterized DHX9 as a helicase active on duplex DNA substrates consisting of oligonucleotides hybridized to single-stranded (ss) M13mp18 (31, 34). This raised the question as to whether the enzyme would display stronger activity on triplex DNA, on 3′-single-stranded overhang on duplex substrates, or on other non-canonical DNA structures, such as forked duplexes. These structures are known to arise as intermediates during in vivo DNA metabolic events, such as replication. To address this question, we first repeated the studies conducted previously (34) on oligonucleotides bound to ssM13mp18 using the enzymatic preparations described herein (see Experimental Procedures). As expected, DHX9 displaced the oligonucleotides from ssM13mp18 (data not shown) (31, 34). Next, we prepared 10-nt 3′-single-stranded overhang duplexes and forked duplexes, using the sequences shown in Table 1A and described in “Experimental Procedures”. Five nM each of forked or 3′-single-stranded overhang duplex substrates were incubated with increasing amounts of DHX9 protein (50, 100, 200 nM) in ‘H’ reaction buffer (“Experimental Procedures”). The results showed little or no displacement of single strands from the duplex structures, even at the highest concentration (200 nM) of DHX9 used (Figure 6A and B). We also conducted similar experiments at 3.5 mM Mg2+ concentration, but still did not observe any detectable displacement activity on forked DNA substrates (data not shown). Thus, the ability of DHX9 to unwind duplex substrates was weaker than on triplex DNA, irrespective of the availability of a 3′-single-stranded overhang or a single strand-duplex junction.
Figure 6. Forked duplex DNA structures are weak substrates for DHX9 activity.
Panel A. Lanes 1–2, electrophoretic mobility of the participating single-strands (45R and 45Y) used to make the forked duplex structure. DNA helicase activity was determined (as described in “Experimental Procedures”) on a forked duplex structure in the presence (lanes 3–7) and in the absence (lanes 7–8) of ATP by using the indicated concentrations of DHX9 protein. Lane 9, the heat denatured (5 min, 95 °C) forked duplex structure without DHX9 to visualize the free participating single strands. Panel B. Helicase activity of DHX9 on various types of duplex and 3′-overhang triplex structures. Lanes 1–2, electrophoretic mobility of 21R with a 3′-overhang and 31R with a 3′-overhang single strand used to assemble the 3′-overhang duplex (lanes 5–6) and 3′-single-stranded overhang triplex (lanes 9–10) DNA structures. Lanes 3–4 and 5–6, represent blunt and forked duplex structures, respectively. A total of 200 nM DHX9 was used to determine the helicase activity on each type of DNA structure (5 nM). The structure of each substrate is shown on top of each lane.
DISCUSSION
The results presented in this study demonstrate that DHX9 has a marked preference for triplex DNA structures containing a 3′-single-stranded overhang over other triplex and duplex DNA substrates with or without 3′-tails, where it unwinds the Hoogsteen-bound bases by translocating with a 3′→5′ polarity. Unwinding required hydrolyzable ATP and the reaction was dependent on the amount of enzyme and incubation time. After 20 min incubation at 32 °C, 200 nM DHX9 completely unwound the triplex DNA substrate at a rate of ~24 bases min−1μM−1. Strand displacement was not observed when the non-hydrolysable ATP analog AMP-PNP was substituted for ATP. Previously, it was reported that ATP binding and/or ATP hydrolysis activities of DHX9 were required for cAMP-mediated transcriptional activation, since a point mutation (Lys to Asn) into the conserved ATP binding motif (Gly-Lys-Thr) of DHX9 led to the reduction in the level of transcription (41). DHX9 also required a 3′-single-stranded overhang on the third strand of the triplex substrate. Conversely, the enzyme failed to displace strands from a blunt-end DNA duplex, and displayed little or no activity on duplexes containing a 3′-single-stranded overhang, or forked duplex DNA molecules. These data support a model whereby DHX9 loads onto the 3′-ssDNA region of triplex substrates and subsequently unwinds the third strand with a 3′→5′ polarity. The requirement for a 3′-single-stranded overhang to load and translocate along triplex DNA is consistent with our earlier reports (31, 34) showing that DHX9 requires a pre-existing 3′-single-stranded overhang for unwinding. Herein, we show that this activity is ~17-fold higher on triplex than on duplex DNA substrates (42). Thus, DHX9 like other DNA helicases, may be activated by ssDNA regions where DNA unwinding is initiated and carried out by homo-oligomeric complexes (reviewed in (43)).
The unwinding activity of helicases on triplex DNA substrates is not without precedent. Mammalian WRN, BLM, (38) and BRIP1 (also known as BACH1 and FANCJ) (39), which belong to the helicase superfamily 2 and share sequence similarity with DHX9, can also unwind non-B DNA structures in vitro. These enzymes play important roles in maintaining chromosomal stability and as such, defects in their function can lead to genetic disorders, such as premature ageing and cancer (38, 44, 45). The WRN and BLM helicases unwind triplex substrates with the same polarity (i.e. from 3′→5′) as DHX9, whereas BRIP1 requires a 5′-single-stranded overhang to load onto triplex DNA structures (39). The WRN, BLM, and BRIP1 helicases can also unwind synthetic Holliday junctions, 12-nt bubble structures, G-quadruplex DNA, and forked structures (46–48). Moreover, WRN activity is potentiated by DHX9 on recombination-like DNA intermediates in vitro (42). In our assays, DHX9 was not able to unwind 3′-single-stranded overhang duplex DNA at 10 mM Mg2+ or forked DNA substrates at either 10 mM or 3.5 mM Mg2+ but displayed weak activity on oligodeoxynucleotides bound to ssM13mp18 when assayed at 3.5 mM Mg2+ (herein and (31, 34)). We showed previously that DHX9 activity may be suppressed by high (20 mM) Mg2+ concentrations (31). Since the stability of these DNA substrates increases with increasing Mg2+ concentration in the range of (0–10 mM) (49) and Hoogsteen-bound third strands in triplexes exhibit lower melting temperatures than their duplex counterparts (50, 51), it is possible that the DHX9 kinetic properties, such as Vmax, may depend on the thermal stability of the underlying DNA substrate. Previously, we have demonstrated that H-DNA-forming sequences induce mutations, largely by promoting the formation of DSBs (24). These sequences are abundant in the human genome (~1 in 50,000 bp in human) (52) and are enriched in introns and 5′- or 3′-untranslated regions of genes involved in cell communication and signaling (10). Analyses of chromosome translocation junctions support a model whereby non-B DNA sequences induce DSBs that are subsequently processed by non-homologous end-joining (NHEJ) (24, 53), which requires DNA-PKcs, Ku, and XRCC4-DNA ligase IV (54–57). The findings that DNA-PKcs can phosphorylate DHX9 and that DHX9 co-immuonoprecipitates with the Ku antigen (58), raises the possibility that the protein participates in NHEJ repair. This postulate is supported by the finding that, following induction of DSBs, the helicase co-localizes with gamma H2AX and Ku70 in the nucleus (59). Thus, DHX9 may be involved in resolving non-B DNA structures by interacting with Ku, thereby leading to an NHEJ-mediated processing of DNA secondary structures. DHX9 has also been implicated in assisting in the resolution of DNA metabolic intermediates, such as D-loops, that arise during recombination events (60), by facilitating degradation through the 3′→5′ exonuclease activity of WRN (60). Given its preference for RNA:DNA hybrids, DHX9 may also play a role in resolving R-loops during transcription and/or intermediates that might form at stalled replication forks. Finally, the enzyme has been found to interact with the C-terminal domain of BRCA1 in a complex with Ku80 at sites of DSBs (61–64), further supporting a role in genome stability.
In conclusion, our results demonstrate the ability of DHX9 to preferentially resolve mutagenic triplex DNA structures in an ATP-dependent fashion. The observation that DHX9 can unwind non-B DNA conformations suggests that the enzyme may belong to the class of DNA helicases that assist in maintaining genomic stability by functioning in DNA replication, recombination, and repair. The observations made in this study have in vivo implications since the triplex DNA substrates used herein are structurally very similar to the naturally occurring H-DNA conformations known to block replication fork progression and transcription (65–69), and to induce genetic instability (24, 25). Further investigations are warranted for a better understanding of the mechanism(s) involved in processing non-B DNA structures and/or the structural intermediates of DNA metabolism important for the maintenance of genomic stability.
Supplementary Material
Acknowledgments
We thank Ms. Sarah Henninger for manuscript preparation, and the members of the Vasquez laboratory and members of Richard Wood’s laboratory for helpful discussions.
ABBREVIATIONS
The abbreviations used are:
- AMP-PNP
5′-adenylyl-beta,gamma-imidodiphosphate
- bp
base pairs
- BSA
bovine serum albumin
- DSB
double strand break
- DTT
dithiothreitol
- PAGE
polyacrylamide gel electrophoresis
- ss
single-stranded
- Tdp-ICL
TFO-directed psoralen interstrand crosslink
- TFO
triplex-forming oligonucleotide
Footnotes
Funding was provided by NIH/NCI grants to K.M.V. (CA097175 and CA093729). Facility Core services were supported in part by the NIH/NIEHS Center Grant (ES007784) and M. D. Anderson’s Cancer Center Support Grant (CA016672).
This section describes the identification of DHX9 bound to triplex DNA by MALDI-TOF/TOF and the gel electrophoretic migration of duplex and triplex DNA species. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Mirkin SM. Discovery of alternative DNA structures: a heroic decade (1979–1989) Front Biosci. 2008;13:1064–1071. doi: 10.2741/2744. [DOI] [PubMed] [Google Scholar]
- 2.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 4.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr Opin Struct Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell Mol Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J Biol Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 7.Wong HM, Payet L, Huppert JL. Function and targeting of G-quadruplexes. Curr Opin Mol Ther. 2009;11:146–155. [PubMed] [Google Scholar]
- 8.Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008;18:233–241. doi: 10.1101/gr.6905408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacolla A, Collins JR, Gold B, Chuzhanova N, Yi M, Stephens RM, Stefanov S, Olsh A, Jakupciak JP, Dean M, Lempicki RA, Cooper DN, Wells RD. Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region. Nucleic Acids Res. 2006;34:2663–2675. doi: 10.1093/nar/gkl354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes JF, Skaletsky H, Pyntikova T, Minx PJ, Graves T, Rozen S, Wilson RK, Page DC. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature. 2005;437:100–103. doi: 10.1038/nature04101. [DOI] [PubMed] [Google Scholar]
- 12.Locke DP, Segraves R, Carbone L, Archidiacono N, Albertson DG, Pinkel D, Eichler EE. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. 2003;13:347–357. doi: 10.1101/gr.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marques-Bonet T, Girirajan S, Eichler EE. The origins and impact of primate segmental duplications. Trends Genet. 2009;25:443–454. doi: 10.1016/j.tig.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernando H, Sewitz S, Darot J, Tavare S, Huppert JL, Balasubramanian S. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009;37:6716–6722. doi: 10.1093/nar/gkp740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Xiao J, Li J, Lu L, Feng S, Droge P. Human genomic Z-DNA segments probed by the Z alpha domain of ADAR1. Nucleic Acids Res. 2009;37:2737–2746. doi: 10.1093/nar/gkp124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goidts V, Szamalek JM, de Jong PJ, Cooper DN, Chuzhanova N, Hameister H, Kehrer-Sawatzki H. Independent intrachromosomal recombination events underlie the pericentric inversions of chimpanzee and gorilla chromosomes homologous to human chromosome 16. Genome Res. 2005;15:1232–1242. doi: 10.1101/gr.3732505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolb J, Chuzhanova NA, Hogel J, Vasquez KM, Cooper DN, Bacolla A, Kehrer-Sawatzki H. Cruciform-forming inverted repeats appear to have mediated many of the microinversions that distinguish the human and chimpanzee genomes. Chromosome Res. 2009;17:469–483. doi: 10.1007/s10577-009-9039-9. [DOI] [PubMed] [Google Scholar]
- 18.Perry GH, Ben-Dor A, Tsalenko A, Sampas N, Rodriguez-Revenga L, Tran CW, Scheffer A, Steinfeld I, Tsang P, Yamada NA, Park HS, Kim JI, Seo JS, Yakhini Z, Laderman S, Bruhn L, Lee C. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupski JR. Genomic disorders ten years on. Genome Med. 2009;1:42. doi: 10.1186/gm42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bacolla A, Jaworski A, Larson JE, Jakupciak JP, Chuzhanova N, Abeysinghe SS, O’Connell CD, Cooper DN, Wells RD. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc Natl Acad Sci U S A. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bacolla A, Jaworski A, Connors TD, Wells RD. Pkd1 unusual DNA conformations are recognized by nucleotide excision repair. J Biol Chem. 2001;276:18597–18604. doi: 10.1074/jbc.M100845200. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc Natl Acad Sci U S A. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, Vasquez KM. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:13448–13453. doi: 10.1073/pnas.0405116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Carbajal S, Vijg J, DiGiovanni J, Vasquez KM. DNA structure-induced genomic instability in vivo. J Natl Cancer Inst. 2008;100:1815–1817. doi: 10.1093/jnci/djn385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen LA, Finch RA, Booker AJ, Vasquez KM. Targeting oncogenes to improve breast cancer chemotherapy. Cancer Res. 2006;66:4089–4094. doi: 10.1158/0008-5472.CAN-05-4288. [DOI] [PubMed] [Google Scholar]
- 27.Kim E, Deppert W. The versatile interactions of p53 with DNA: when flexibility serves specificity. Cell Death Differ. 2006;13:885–889. doi: 10.1038/sj.cdd.4401909. [DOI] [PubMed] [Google Scholar]
- 28.Brosh RM., Jr Molecular biology: The Bloom’s complex mousetrap. Nature. 2008;456:453–454. doi: 10.1038/456453a. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson LH, Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat Res. 2009;668:54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang SS, Grosse F. Purification and characterization of two DNA helicases from calf thymus nuclei. J Biol Chem. 1991;266:20483–20490. [PubMed] [Google Scholar]
- 32.Zhang S, Grosse F. Domain structure of human nuclear DNA helicase II (RNA helicase A) J Biol Chem. 1997;272:11487–11494. doi: 10.1074/jbc.272.17.11487. [DOI] [PubMed] [Google Scholar]
- 33.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S, Grosse F. Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry. 1994;33:3906–3912. doi: 10.1021/bi00179a016. [DOI] [PubMed] [Google Scholar]
- 35.Lee CG, da Costa Soares V, Newberger C, Manova K, Lacy E, Hurwitz J. RNA helicase A is essential for normal gastrulation. Proc Natl Acad Sci U S A. 1998;95:13709–13713. doi: 10.1073/pnas.95.23.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Grosse F. Multiple functions of nuclear DNA helicase II (RNA helicase A) in nucleic acid metabolism. Acta Biochim Biophys Sin. 2004;36:177–183. doi: 10.1093/abbs/36.3.177. [DOI] [PubMed] [Google Scholar]
- 37.Perkins BD, Wilson JH, Wensel TG, Vasquez KM. Triplex targets in the human rhodopsin gene. Biochemistry. 1998;37:11315–11322. doi: 10.1021/bi980525s. [DOI] [PubMed] [Google Scholar]
- 38.Brosh RM, Jr, Majumdar A, Desai S, Hickson ID, Bohr VA, Seidman MM. Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J Biol Chem. 2001;276:3024–3030. doi: 10.1074/jbc.M006784200. [DOI] [PubMed] [Google Scholar]
- 39.Sommers JA, Rawtani N, Gupta R, Bugreev DV, Mazin AV, Cantor SB, Brosh RM., Jr FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J Biol Chem. 2009;284:7505–7517. doi: 10.1074/jbc.M809019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 41.Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 42.Chakraborty P, Grosse F. WRN helicase unwinds Okazaki fragment-like hybrids in a reaction stimulated by the human DHX9 helicase. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq240. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 44.Martin GM. The Werner mutation: does it lead to a “public” or “private” mechanism of aging? Mol Med. 1997;3:356–358. [PMC free article] [PubMed] [Google Scholar]
- 45.Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 46.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 47.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 48.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Owczarzy R, Moreira BG, You Y, Behlke MA, Walder JA. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry. 2008;47:5336–5353. doi: 10.1021/bi702363u. [DOI] [PubMed] [Google Scholar]
- 50.Wu P, Kawamoto Y, Hara H, Sugimoto N. Effect of divalent cations and cytosine protonation on thermodynamic properties of intermolecular DNA double and triple helices. J Inorg Biochem. 2002;91:277–285. doi: 10.1016/s0162-0134(02)00444-0. [DOI] [PubMed] [Google Scholar]
- 51.Plum GE, Breslauer KJ. Thermodynamics of an intramolecular DNA triple helix: a calorimetric and spectroscopic study of the pH and salt dependence of thermally induced structural transitions. J Mol Biol. 1995;248:679–695. doi: 10.1006/jmbi.1995.0251. [DOI] [PubMed] [Google Scholar]
- 52.Schroth GP, Ho PS. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 1995;23:1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain A, Wang G, Vasquez KM. DNA triple helices: biological consequences and therapeutic potential. Biochimie. 2008;90:1117–1130. doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raghavan SC, Lieber MR. Chromosomal translocations and non–B DNA structures in the human genome. Cell Cycle. 2004;3:762–768. [PubMed] [Google Scholar]
- 55.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair. 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 56.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 57.Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 58.Zhang S, Schlott B, Gorlach M, Grosse F. DNA-dependent protein kinase (DNA-PK) phosphorylates nuclear DNA helicase II/RNA helicase A and hnRNP proteins in an RNA-dependent manner. Nucleic Acids Res. 2004;32:1–10. doi: 10.1093/nar/gkg933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mischo HE, Hemmerich P, Grosse F, Zhang S. Actinomycin D induces histone gamma-H2AX foci and complex formation of gamma-H2AX with Ku70 and nuclear DNA helicase II. J Biol Chem. 2005;280:9586–9594. doi: 10.1074/jbc.M411444200. [DOI] [PubMed] [Google Scholar]
- 60.Friedemann J, Grosse F, Zhang S. Nuclear DNA helicase II (RNA helicase A) interacts with Werner syndrome helicase and stimulates its exonuclease activity. J Biol Chem. 2005;280:31303–31313. doi: 10.1074/jbc.M503882200. [DOI] [PubMed] [Google Scholar]
- 61.Schlegel BP, Starita LM, Parvin JD. Overexpression of a protein fragment of RNA helicase A causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells. Oncogene. 2003;22:983–991. doi: 10.1038/sj.onc.1206195. [DOI] [PubMed] [Google Scholar]
- 62.Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 63.Bau DT, Mau YC, Shen CY. The role of BRCA1 in non-homologous end-joining. Cancer Lett. 2006;240:1–8. doi: 10.1016/j.canlet.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 64.Wei L, Lan L, Hong Z, Yasui A, Ishioka C, Chiba N. Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Mol Cell Biol. 2008;28:7380–7393. doi: 10.1128/MCB.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel HP, Lu L, Blaszak RT, Bissler JJ. PKD1 intron 21: triplex DNA formation and effect on replication. Nucleic Acids Res. 2004;32:1460–1468. doi: 10.1093/nar/gkh312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baran N, Lapidot A, Manor H. Formation of DNA triplexes accounts for arrests of DNA synthesis at d(TC)n and d(GA)n tracts. Proc Natl Acad Sci U S A. 1991;88:507–511. doi: 10.1073/pnas.88.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J Biol Chem. 1998;273:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 68.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol Cell Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J Biol Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.