Abstract
Intrinsic and acquired cellular resistance factors limit the efficacy of most targeted cancer therapeutics. Synthetic lethal screens in lower eukaryotes suggest that networks of genes closely linked to therapeutic targets would be enriched for determinants of drug resistance. We developed a protein network centered on the epidermal growth factor receptor (EGFR), which is a validated cancer therapeutic target, and used siRNA screening to comparatively probe this network for proteins that regulate the effectiveness of both EGFR-targeted agents and nonspecific cytotoxic agents. We identified subnetworks of proteins influencing resistance, with putative resistance determinants enriched among proteins that interacted with proteins at the core of the network. We found that EGFR antagonists and clinically relevant drugs targeting proteins connected in the EGFR network, such as the kinases protein kinase C or Aurora kinase A, or the transcriptional regulator STAT3, synergized to reduce cell viability and tumor size, suggesting the potential for a direct path to clinical exploitation. Such a focused approach can potentially improve the coherent design of combination cancer therapies.
Introduction
A central premise driving the development of targeted cancer therapies has been that agents directed against specific proteins that promote tumorigenesis or maintain the malignant phenotype will have greater efficacy and less toxicity than untargeted cytotoxic agents. Although small molecule and antibody drugs directed against well-validated cancer targets, such as epidermal growth factor receptor (EGFR), the Philadelphia chromosome-associated chimeric oncoprotein BCR-ABL, vascular endothelial growth factor (VEGF), mammalian target of rapamycin (mTOR), and other proteins are clinically useful, many tumors fail to respond because of intrinsic or acquired resistance. In some cases, a clear and unique determinant of resistance can be identified, for example when mutational activation of the EGFR downstream effector K-RAS limits response to EGFR-targeting drugs (1, 2). However, for most tumors, heterogeneous resistance to oncogene-targeting therapies appears to arise from partial contributions by multiple proteins.
This result is compatible with the paradigm of a robust signaling network (3), which is gradually replacing the idea of minimally branching signaling pathways marked by hierarchical signaling relationships. Network models (4–6) emphasize dense connections among signaling proteins, lack of hierarchy, feedback signaling loops, and tendencies towards protective redundancy due to the existence of paralogous proteins with overlapping functionality (3). A robust network paradigm has critical implications for targeted cancer therapies, predicting that in cells treated with therapies inhibiting an oncogenic node, rescue signaling can be provided by modifying signaling output from any of a number of distinct proteins that are enriched among the components of the web of interactions centered on the target of inhibition. This concept is reinforced by studies in model organisms demonstrating that quantitatively significant signal-modulating relationships commonly involve proteins that have closely linked functions (7). The goal of this study was to use siRNA libraries targeting the EGFR signaling network to identify potential regulators of resistance to EGFR-targeted therapies, and to provide leads for overcoming therapeutic resistance.
Results
Integration of orthogonal data sets allows construction of an EGFR-centered signaling network for targeted RNAi screening
To construct a network-based library, genes encoding proteins with evidence of functional interactions with EGFR were collected from multiple databases (Fig. 1A, and Materials and Methods). We used two members of the EGFR family, EGFR (also known as ERBB1) and HER2 (also known as ERBB2), as seed nodes to select first- and second-order binary protein-protein interactions (PPIs). We mined non-PPI functional linkages relevant to the EGFR pathway from five pathway databases. From BOND (8) and EBI (9), we identified proteins that associated with the seed proteins in purified complexes. We included genes that were transcriptionally responsive to inhibition or stimulation of EGFR that we identified from the NIH GEO resource (10). We added human orthologs for genes identified in other species (predominantly Drosophila) that genetically interacted with evolutionarily conserved EGFR orthologs. Together, these data nominated 2689 genes encoding proteins linked by at least one criterion to the initial seed list. We chose 638 genes to target in the siRNA library (Table S1) predominantly on the basis of representation in at least two overlapping orthogonal sources. Also included in the 638 genes were those of the 2689 genes that exhibited a physical interaction with the EGFR adaptor protein SHC, or close signaling connections to the nonreceptor tyrosine kinase SRC and transforming growth factor β (TGF-β) pathways that interact with ERBB family proteins to promote tumor aggressiveness (11, 12).
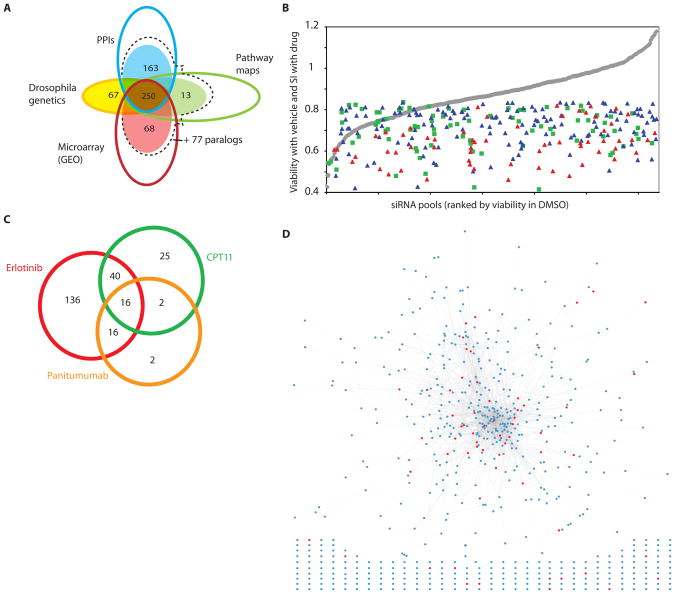
Fig. 1. Design and screening of a targeted library.
(A) Genes targeted in the library were identified from a preliminary large set (open circles) of genes engaged in protein-protein interactions (PPIs) with one of the seeds, or interacting with one of the direct interactors; collected from 5 curated pathway resources for the EGFR signaling network (Pathway maps); shown by microarray experiments to be rapidly transcriptionally responsive to stimulation or inhibition of EGFR; or human orthologs of genes known to genetically interact with EGFR in Drosophila (Drosophila genetics). Genes from this larger set were prioritized by a series of collection criteria aimed at finding genes connected to EGFR by more than one criterion (detailed in table S1); final numbers of genes added to the library from each group are shown in filled circles. At the intersection of the circles is the number of genes added to the library, because they belong to at least two sets. For the PPI, pathway map, and microarray sets, additional 77 genes were identified as closely related in sequence and potentially paralogous in function (dashed line). (B) Distribution of hits as a factor of overall viability reduction with the siRNA. siRNAs in library are listed in order of intrinsic impact on viability of A431 cells treated with DMSO (gray line). Blue triangles, sensitization index (SI) for primary hits with erlotinib; red triangles, SI for validated hits with erlotinib; green squares, SI for primary hits with CPT11. (C) Degree of overlap between primary hits obtained for erlotinib, panitumumab, and CPT11. (D) Network of validated (red circles) hits sensitizing to EGFR-targeting agents, in the context of the full library. Lines (edges) represent connections based on direct protein-protein interactions or genetic interactions in Drosophila. Hits and genes from the starting set that were not connected in the network are shown below the network.
siRNA screening defines subsets of genes that sensitize cells to EGFR inhibition
The A431 cervical adenocarcinoma cell line is dependent on EGFR signaling for proliferation and survival. We reiteratively screened this cell line with the targeted siRNA library in combination with DMSO (vehicle), or small molecule inhibitors of EGFR, or function-blocking EGFR antibodies, or with the non-EGFR-targeted cytotoxic and DNA-damaging agent camptothecin (CPT11) applied at IC25–IC35 concentrations (fig. S1A). Viability was measured with Alamar blue, a metabolic indicator of the number of viable cells. Primary hits were defined as genes that when targeted with siRNAs reduced negative control-normalized viability by at least 15% in the presence of a drug compared to the viability in the presence of DMSO [defined as the Sensitization Index (SI) <0.85], with a false discovery rate (FDR) < 20% (fig. S1B, S2). (In the absence of drug treatment, knockdown of (247/638) of genes in the library reduced the viability of DMSO-treated A431 cells by at least 15%, including 45 that reduced viability more than 30%. The distribution of primary hits was independent of the tendency of a siRNA to affect cell viability in the absence of drug treatment (Fig. 1B), indicating the action of hits was not merely a reflection of accumulated injury to hit-depleted cells. The majority of hits obtained by treating the cells with the EGFR-targeted antibody panitumumab were included within the larger set of genes identified as hits in the cells exposed to the EGFR-targeted small molecule inhibitor erlotinib (Fig. 1C). Knockdown of 212 primary hits, including 95 hits with an SI <0.7, sensitized to cells to one or both EGFR-targeting agents. In contrast, knockdown of only 83 primary hits, including 30 hits with an SI <0.7, sensitized cells to CPT11 (Fig. 1C).
Performance of additional validation testing (fig. S2, S3) identified a set of 61 genes (Table 1) for which 2 or more independent gene-targeted siRNAs both efficiently knocked down their target gene and sensitized cells to EGFR-targeting agents. The majority of the sensitizing genes (48/61) encoded proteins that were connected in a physically interacting network (Fig. 1D). The remaining 13 encoded proteins that are not known to interact physically with EGFR or its direct partners, but instead are linked to EGFR on the basis of rapid changes in the abundance of their mRNA transcripts in response to pathway activation, inhibition, or both.
Table 1. Validated EGFR-sensitizing genes.
Validated screen hits. Symbol is the official Entrez gene name, ID is Entrez gene ID.
| Symbol | ID | Gene Description | Origin | Clinical agent |
|---|---|---|---|---|
| ABL1 | 25 | v-abl Abelson murine leukemia viral oncogene homolog 1 | PPI1 | Imatinib, dasatinib |
| AKT2 | 208 | v-akt murine thymoma viral oncogene homolog 2 | PG | Perifosine, triciribine, GSK690693 |
| ANXA6 | 309 | annexin A6 | C, PPI2 | |
| ARF4 | 378 | ADP-ribosylation factor 4 | PM, PPI1 | |
| ARF5 | 381 | ADP-ribosylation factor 5 | PG | |
| ASCL2 | 430 | achaete-scute complex homolog 2 (Drosophila) | PPI2, DG | |
| BCAR1 | 9564 | breast cancer anti-estrogen resistance 1 | PM, C, PPI1 | |
| CALM1 | 801 | calmodulin 1 (phosphorylase kinase, delta) | C PPI1 | phenothiazines |
| CBLC | 23624 | Cas-Br-M (murine) ecotropic retroviral transforming sequence c | PM PPI1 | |
| CCND1 | 595 | cyclin D1 | PM, PPI2 | |
| CD59 | 966 | CD59 molecule, complement regulatory protein | C, PPI2 | Roche, Preclinical |
| CDH3 | 1001 | cadherin 3, type 1, P-cadherin (placental) | PG | |
| CXCL12 | 6387 | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | MA | |
| DCN | 1634 | decorin | PPI1 | |
| DDR2 | 4921 | discoidin domain receptor family, member 2 | PPI1 | |
| DIXDC1 | 85458 | DIX domain containing 1 | MA | |
| DLG4 | 1742 | discs, large homolog 4 (Drosophila) | PPI1 | |
| DUSP4 | 1846 | dual specificity phosphatase 4 | MA | |
| DUSP6 | 1848 | dual specificity phosphatase 6 | DG | |
| DUSP7 | 1849 | dual specificity phosphatase 7 | DG | |
| EPHA5 | 2044 | EPH receptor A5 | PG | |
| ERBB3 | 2065 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | PM, PPI1, DG | antibodies, for example MM-121 |
| FER | 2241 | fer (fps/fes related) tyrosine kinase (phosphoprotein NCP94) | PPI1 | |
| FGFR2 | 2263 | fibroblast growth factor receptor 2 (bacteria-expressed kinase, keratinocyte growth factor receptor, craniofacial dysostosis 1, Crouzon syndrome, Pfeiffer syndrome, Jackson-Weiss syndrome) | DG | For example, brivanib, masitinib, TKI258, PHA-739358 |
| FLNA | 2316 | filamin PG, alpha (actin binding protein 280) | C, PPI2 | |
| GRB7 | 2886 | growth factor receptor-bound protein 7 | PM, PPI1 | |
| HSPA9 | 3313 | heat shock 70kDa protein 9 (mortalin) | C, PPI2 | |
| INPPL1 | 3636 | inositol polyphosphate phosphatase-like 1 | PM, PPI1 | |
| KLF10 | 7071 | Kruppel-like factor 10 | MA | |
| LOC284393 | 284393 | similar to ribosomal protein L10 | PG | |
| LOC63920 | 63920 | transposon-derived Buster3 transposase-like | MA | |
| LTK | 4058 | leukocyte tyrosine kinase | PPI1 | |
| MAP3K1 | 4214 | mitogen-activated protein kinase kinase kinase 1 | PM, PPI2 | |
| MAPK1 | 5594 | mitogen-activated protein kinase 1 | PM, PPI1 | |
| MATK | 4145 | megakaryocyte-associated tyrosine kinase | PM, PPI1 | |
| NEDD9 | 4739 | neural precursor cell expressed, developmentally down-regulated 9 | PPI1 | |
| NOTCH2 | 4853 | notch 2 | DG | |
| PIK3R1 | 5295 | phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | PM, PPI1 | For example, PX-866, BGT226, GDC-0941, XL765 |
| PIK3R2 | 5296 | phosphoinositide-3-kinase, regulatory subunit 2 (p85 beta) | PM, PPI1 | |
| PIN1 | 5300 | protein (peptidylprolyl cis/trans isomerase) NIMA-interacting 1 | PM | |
| PKN2 | 5586 | protein kinase N2 | PM, PPI2 | |
| PLSCR1 | 5359 | phospholipid scramblase 1 | PM, PPI1, MA | |
| PPIA | 5478 | peptidylprolyl isomerase PG (cyclophilin PG) | C, PPI2 | |
| PRKACB | 5567 | protein kinase, cAMP-dependent, catalytic, beta | PG | |
| PRKCD | 5580 | protein kinase C, delta | PM, PPI1 | ruboxistaurin, enzastaurin, tamoxifen |
| PRKCE | 5581 | protein kinase C, epsilon | PM, PPI2 | |
| PRKCZ | 5590 | protein kinase C, zeta | PM, PPI2 | |
| PTPRF | 5792 | protein tyrosine phosphatase, receptor type, F nexin-II, Alzheimer disease) | PPI1 | |
| RAC1 | 5879 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | PM, PPI2 | |
| RAPGEF1 | 2889 | Rap guanine nucleotide exchange factor (GEF) 1 | PPI1 | |
| RASA3 | 22821 | RAS p21 protein activator 3 | DG | |
| RET | 5979 | ret proto-oncogene | PPI1, M2 | valdetanib |
| RPS6KA5 | 9252 | ribosomal protein S6 kinase, 90kDa, polypeptide 5 | PM | ruboxistaurin |
| SC4MOL | 6307 | sterol-C4-methyl oxidase-like | MA | |
| SH2D3C | 10044 | SH2 domain containing 3C | PM, PPI1 | |
| SHC1 | 6464 | SHC (Src homology 2 domain containing) transforming protein 1 | PM, C, PPI1, DG | |
| SMAD2 | 4087 | SMAD family member 2 | PM, PPI1 | peptide 144 targets TGFβ1RIII |
| SOS2 | 6655 | son of sevenless homolog 2 (Drosophila) | PM, PPI1, DG | |
| STAT3 | 6774 | signal transducer and activator of transcription | PM, C, PPI1 | STAT 3 decoy oligonucleotide |
| TBL1Y | 90665 | 3 (acute-phase response factor) transducin (beta)-like 1Y-linked | DG | |
| VAV3 | 10451 | vav 3 oncogene | PM, C, PPI1 |
Hits are listed alphabetically by Gene Symbol (Symbol). Entrez Gene ID (ID), description (including alternative common names), reason for inclusion in the library (Origin), and existence of any targeted small molecule agent (Clinical Agent) are also provided. C, found in a protein complex with seeds; DG, identified from Drosophila genetics; MA, microarray indicates transcriptionally responsive to EGFR; PG, paralog; PM, present in pathway maps; PPI1, direct protein interactions with seeds; PPI2, direct protein interactions with a protein within the PPI1 group.
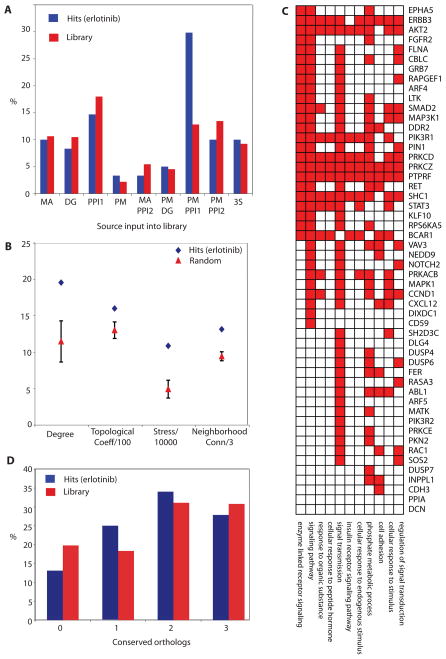
Relative to the overall properties of the 638-gene library, the erlotinib-sensitizing hits were significantly enriched for genes that were first-order PPIs of the seeds and were also present in the pathway maps (Fig. 2A). When examined within the context of the EGFR-centered network, the erlotinib-sensitizing hits encoded proteins that exhibited topology parameters distinct from those of the overall network, such as increased degree, which reflected the number of edges linked to it; topological coefficient, which provided an estimate for the trend of nodes in the network to have shared neighbors; stress, which reflected how frequently a node was in the shortest path connecting other nodes; and neighborhood connectivity, which represented the average number of neighbors for each direct interactor of the node. Together these properties suggest that these genes encode proteins that serve as network “hubs” and connect with many other proteins in the network (Fig. 2B). On the basis of their Gene Ontology (GO) function, erlotinib-sensitizing hits encoded proteins that were significantly enriched for involvement in phosphate metabolism (kinases or phosphatases) and signaling (represented by several GO categories) relative to the overall composition of the siRNA library (Fig. 2C). We observed a weak trend for hits to be evolutionarily conserved, as reflected by the increased number of orthologs in lower eukaryotes among hits relative to the overall library (Fig. 2D).
Fig. 2. Network properties of hits.
(A) Hits by source of input in library. MA, microarray indicates transcriptionally responsive to EGFR; DG, Drosophila genetics; PPI1, direct protein interactions with seeds; PM, pathway maps; PPI2, direct protein interactions with a protein within the PPI1 group, or found in a complex with seed proteins; 3S, any 3 sources combined. (B) Topological analysis of erlotinib hit network identified in A431 cells. Data shown represents difference between properties of the set of 61 validated hits and the average for 20 randomly generated sets of 61 genes from the library. Measures of degree, topological coefficient (Topological Coeff/100), stress (Stress/1000), and neighborhood connectivity (Neighborhood Conn/3) show significant enrichment for hits validated with erlotinib, with the error bars for the random set data reflecting a 99% confidence interval. (C) Enriched GO classifications (for each category shown, p<0.01). among hits. Enrichment is significant for proteins annotated as involved in phosphate metabolism (kinases and phosphatases) and in multiple signal transduction-related categories. (D) Percentage of hits versus library proteins having a recognized ortholog in S. cerevisiae, C. elegans or D. melanogaster. X axis, the number of species (among listed) having a recognized ortholog.
A subgroup of validated genes is active in multiple cell lines and promotes drug-induced apoptosis
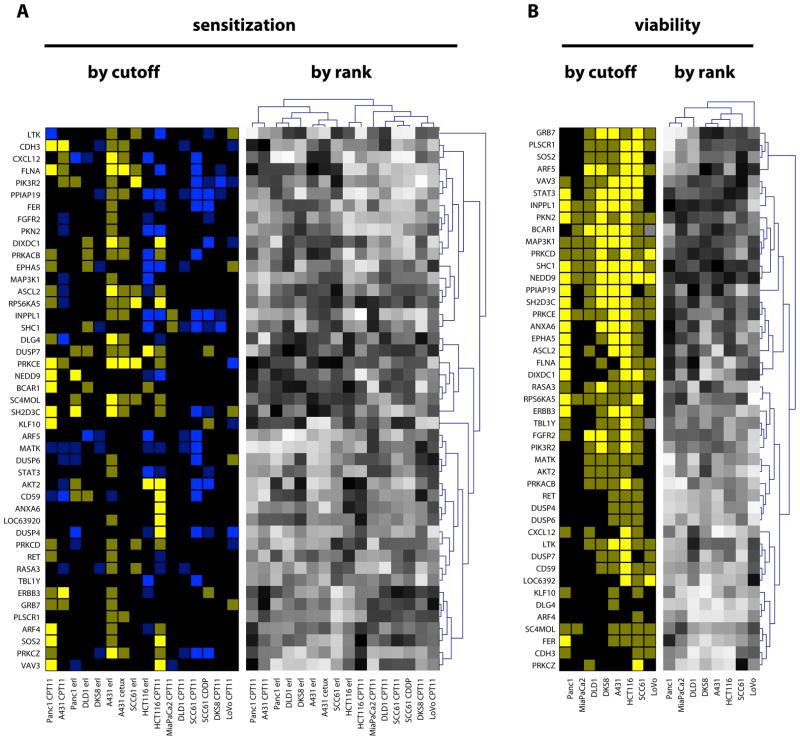
To assess if the genes that sensitized A431 cells to EGFR inhibitors or non-EGFR-targeted cytotoxic agents also influenced the sensitivity of other cancer cell lines to these drugs, we profiled the efficacy of siRNAs targeting 45 of these genes in sensitizing 7 other cell lines to erlotinib, cetuximab (an EGFR function-blocking antibody), or CPT11. These lines included A431, the colorectal adenocarcinoma cell lines HCT116, DLD-1, DKS-8, and LoVo, the head and neck squamous cell carcinoma cell line SCC61, and the pancreatic adenocarcinoma cell lines PANC-1 and MIA PaCa-2 (Fig. 3A). Cell lines with mutations in genes encoding proteins that are known to produce drug resistance (for example, in K-Ras or p53, or both) had more noise in their sensitization responses, with the result that lines containing such mutations (DLD-1, DKS-8, LoVo, MIA PaCa-2) yielded many fewer sensitizing hits than we found in the A431 cells, as judged by a strict FDR-based statistical criteria. One contributing factor to the reduced number of hits was an increase in the stochastic “noise”, which caused greater standard deviation in experimental repetitions. To compensate for this factor, we analyzed the data in two ways-- not only by statistically stringent conventional threshold analysis (Fig. 3A, left) but also by assessing the rank order of sensitization phenotype, using relaxed statistical criteria (compare to Fig. 3A, right; see Materials and Methods). This analysis indicated a subset of sensitizing genes were consistently most sensitizing among the group analyzed.
Fig. 3. Sensitization profile of hits.
(A) Left, SI values for erlotinib and CPT11, calculated as (test siRNA)/(GL2) for cells grown in drug, divided by (test siRNA/GL2) for cells grown in DMSO: dim yellow, SI≤0.85; bright yellow, SI≤0.7; dim blue, SI≥1.15; bright blue, SI≥1.3. Right, gradient of relative ranking of the efficacy of hits to sensitize cells to drugs indicated: black, most sensitizing; white, least sensitizing or in some cases antagonizing (compare to left panels). For rank order analysis, cluster analysis was performed to identify genes with similar profiles of sensitization (dendrogram, Y axis), and also to cluster cell lines by similarity of response (dendrogram, X axis): these clustered patterns are used to organize the display of genes selected by threshold. Two cell lines, MIA-Paca2 and LoVo, yielded no reproducible hits sensitizing to erlotinib; BCAR1 and TBL1Y were not tested in LoVo. Note, not all hits initially obtained and validated in A431 score as positive by rigorous statistical criteria with A431/erlotinib in this experiment because of the intrinsic false negative rate of the assay. (B) Left, siRNA-induced viability reduction below 0.85 (dim yellow), or 0.7 (bright yellow) that of control siRNA-treated cells, calculated based on the formula (test siRNA)/(GL2 control siRNA) for cells grown in DMSO. Right, relative ranking of hit efficacy in reducing cell line viability.
None of the 45 genes when knocked down sensitized all tested cell lines to erlotinib. On the basis of the threshold analysis (Fig. 3A, left), knockdown of the 45 genes originally identified in the A431 cells, most consistently sensitized this cell line to erlotinib, with many in this group also sensitizing A431 cells to cetuximab (Table 1). Knockdown of a subset of these genes (including those targeting RPS6KA5, FLNA, DUSP7, PRKCE, PRKACB, SC4MOL, and ASCL2) sensitized cells to erlotinib, CPT11, or both, in 3 to 5 cell lines, suggesting a broader action in resistance, but less specificity for EGFR-targeting agents. This overlap in CPT11 sensitizing genes with erlotinib sensitizing genes may indicate general roles for some of the genes in general cell survival pathways, or alternatively, reflect the important role of genes closely linked to EGFR in supporting general cell survival. Surprisingly, we also observed that a small number of genes originally identified as sensitizing in A431 cells treated with erlotinib actually antagonized the effects of this or other drugs in other cell lines (for example, PPIAP19 and INPPL1).
Reanalyzing the same set of 45 genes on the basis of sensitization ranking (Fig. 3A, right), all genes detected on the basis of strict thresholds were again identified, but additional genes of interest were now detected (Table 2). For example, in the ranking analysis, PRKCE was one of the most sensitizing genes in 11/16 conditions assessed, whereas in the threshold analysis it only scored as significantly sensitizing in 6/16 conditions.
Table 2.
Genes that when knocked down sensitized multiple cell lines to drugs. Threshold analysis provides a set of target genes based on strict statistical criteria. Rank order analysis provides a different view of the target gene set by compensating for stochastic noise in the responsiveness of cell lines (including DLD-1, DKS-8, LoVO, MIA PaCa-2, Panc1, and HCT116) with nonspecific drug-resistance mutations, such as in the genes encoding K-RAS or p53. For rank analysis, genes indicated represent those found in the highest and lowest quartiles based on absolute activity for each of the categories specified.
| Cutoff Analysis Only | Rank Analysis Only | |
|---|---|---|
| Genes that sensitize A431 cells to erlotinib and cetuximab | ASCL2 CDH3 CXCL12 DIXDC1 DLG4 DUSP7 FLNA INPPL1 PLSCR1 PPIAP19 PRKACB PRKCE PRKCZ RPS6KA5 SC4MOL SH2D3C |
CDH3 CXCL12 FLNA INPPL1 PRKACB PRKCE RPS6KA5 SC4MOL SH2D3C |
| Genes that sensitize two or more cell lines to erlotinib | AKT2 ASCL2 BCAR1 CD59 DIXDC1 DUSP7 FLNA LTK PIK3R2 PRKACB PRKCD PRKCE RPS6KA5 SC4MOL SH2D3C |
AKT2 ASCL2 BCAR1 CD59 DIXDC1 DLG4 DUSP7 FLNA KLF10 LTK NEDD9 PRKACB PRKCE SC4MOL SH2D3C SHC1 |
| Genes that sensitize one or more resistant cell lines to erlotinib | AKT2 BCAR1 CD59 DIXDC1 DUSP7 EPHA5 NEDD9 PIK3R2 PRKACB SC4MOL SH2D3C SHC1 |
AKT2 ANXA6 ASCL2 BCAR1 CD59 DIXDC1 DLG4 DUSP6 DUSP7 EPHA5 FGFR2 KLF10 LOC63920 LTK NEDD9 PKN2 PPIAP19 PRKACB RET SC4MOL SH2D3C SHC1 |
| Genes that sensitized cells to both erlotinib and CPT11 in >3 cell lines, | none | BCAR1 CD59 DLG4 |
| Genes that sensitized cells to erlotinib and CPT11, but did not reduce viability in the absence of drug | CXCL12 (only in A431) | DLG4 DUSP6 |
| Genes that affected both the viability and sensitization to erlotinib or CPT11 in ≥ 2 tumor cells lines | ASCL2 BCAR1 DUSP7 ERBB3 FLNA INPPL1 LTK PIK3R2 PRKACB PRKCD PRKCE RASA3 RPS6KA5 SC4MOL SH2D3C SHC1 SOS2 VAV3 |
DIXDC1 FLNA PRKCE SH2D3C SHC1 |
| Genes that reduced viability of at least 2 RAS mutant cell lines but not more than one wild-type cell line | AKT2 ARF4 CDH3 CXCL12 DUSP7 FLNA LOC63920 PPIAP19 |
ANXA6 FLNA PPIAP19 |
The effects of inhibiting a selected target gene reflect not only drug-related sensitizing activity, but also an intrinsic effect on cell growth due to loss of the gene product, which may cumulatively result in an altered rank order of target genes in influencing cell viability. We therefore also established the baseline intrinsic activity of the validated siRNAs in reducing cell viability in DMSO-treated cells (Fig. 3B). In multiple cell lines in the presence of vehicle alone, targeting of some genes, such as RPS6KA5 and SHC1, significantly reduced cell viability; whereas targeting of others, including DUSP7 and DLG4, had relatively little effect on cell viability in the absence of drug treatment (Table 2).
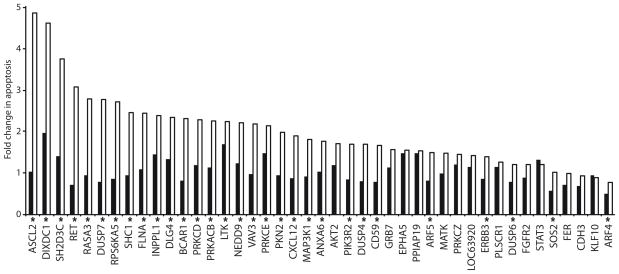
On the basis of the combination of intrinsic and sensitizing effects, knock down of many genes (including PRKCE, DUSP7, SH2D3C, SHC1, SC4MOL, FLNA, and NEDD9) strongly reduced the viability of multiple tumor cell lines treated with EGFR-targeting agents. Further, depletion of 30 of the hits showed statistically significant drug-gene interactions by selectively enhancing apoptosis in the presence of erlotinib versus GL2-targeted control siRNA A431 cells, including 9 of the hits that selectively enhanced apoptosis >2-fold in erlotinib- versus DMSO-treated cells (Fig. 4).) These genes may be particularly useful targets for cancer therapy, because of their ability to induce cell death rather than only cytostasis.
Figure 4. siRNA hits enhance apoptosis in the presence of EGFR inhibitor.
Apoptosis was determined by annexin-V labeling and normalized to negative control siRNA. Composite results from two independent experiments are shown as odds ratio columns; black, DMSO-treated; white, erlotinib-treated. Asterisks indicate statistically significant (FDR ≤ 0.05) erlotinib-gene interactions from two independent experiments.
Many strongly sensitizing hits populate a protein network connected to EGFR
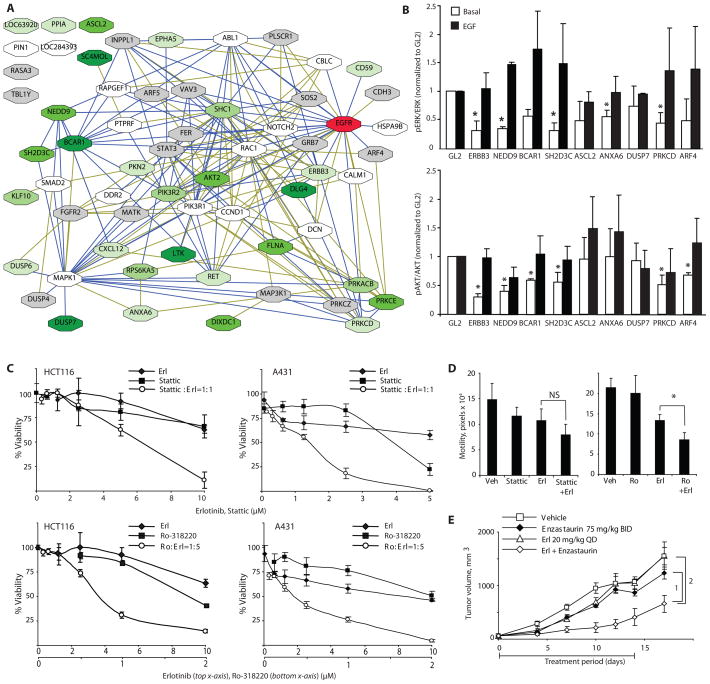
These findings support the idea that a cogently designed network focused around a core cancer target, such as EGFR, would provide a rich source of genes that modulate resistance to EGFR pathway-targeted agents. In general, we observed a greater effect on the core viability of cell lines containing wild-type versus mutant RAS (for example, SCC61 and DKS8 in A431 cells), although the stronger hits (SI <0.7) were typically active in both (Fig. 3, Table 2); in contrast, no meaningful correlation was detected between sensitization profile and RAS mutational status, suggesting that sensitizing activity occurred downstream or independently from core RAS signaling outputs. We investigated the relative interactions of the stronger hits within the overall topology of the EGFR signaling network (Fig. 5A). We could place the majority of hits in a connected subnetwork defined by direct physical interactions. We identified genes encoding 2 members of the protein kinase C (PRKC) family as sensitizing in multiple cell lines (PRKCD and PRKCE), with a third PRKC encoding gene PRKCE also directly connecting to another sensitizer, PRKACB (encoding the catalytic subunit of cAMP-dependent protein kinase). A second cluster included SH2D3C, BCAR1, and NEDD9 (in each case encoding a scaffolding protein involved in integrin-dependent signaling), which on the basis of rank-order analysis (Fig. 3A) sensitized cells preferentially to erlotinib and cetuximab relative to non-EGFR targeted agents, and were all connected by direct physical interactions. Many of these most sensitizing hits were directly connected to MAPK1 (encoding mitogen-activated protein kinase 1, also known as extracellular signal-regulated protein kinase ERK2), PIK3R (encoding the regulatory subunit of phosphoinositide 3-kinase), STAT3 (encoding signal transducer and activator of transcription 3), SHC1 (encoding a scaffolding protein intermediate between receptor tyrosine kinases and RAS), and EGFR itself, supporting the idea that these proteins modulated core outputs of the central EGFR signaling pathway.
Fig. 5. Functional classification of hits.
(A) Network of validated genes that sensitize cells to EGFR-targeting agents. Blue lines represent connections based on direct PPIs or genetic interactions in Drosophila; green lines represent connections generated by pathway maps and text mining. Green shadowing on boxes indicates genes that are in the top quartile by rank order of those strongly sensitizing at least 1 (lightest green) to at least 5 (darkest green) cancer cell lines to erlotinib. Gray boxes, genes that were never in the top quartile; white boxes, validated genes not tested for sensitization. (B) Analysis of ERK (top) and AKT (bottom) status in cell lysates from A431 cells following siRNA transfection, under basal medium conditions and following EGF stimulation. Average results of three independent Western blots are shown as ratios of amounts of phosphorylated and total proteins. Results were normalized to corresponding ratios in GL2 siRNA-transfected control cells; error bars are standard deviations. Asterisks indicate statistically significant difference with negative control; t-test p<0.05. (C) Viability curves for erlotinib and Stattic used as single agents, or combined at 1:1 molar ratio in A431 (left) and HCT116 (right) cells, or for erlotinib and Ro-318220 used as single agents, or combined at 1:5 molar ratio in A431 (left) and HCT116 (right) cells. (D) Motility was measured by wound healing assay in A431 cells cells treated with 0.5 μM erlotinib alone or in combination with 0.5 μM Stattic (left) or 0.25 μM Ro-318220 (right), and assessed over 18 hours. NS, not significant. *, FDR=3.57*10−5. (E) A431 xenografts implanted in SCID mice were treated with drugs as indicated when palpable tumors appeared (day zero) Drug treatment was administered from days 0–14, and tumor measurement continued until day 18. The generalized estimating equations approach (with an autoregressive correlation structure) was used to model tumor growth. A linear time-effect was included in the model for the logarithm of tumor volume and interacted with the treatments in each comparison. P-values are 1, 0.042 and 2, 0.032, n=5 mice per group.
We next tested the ability of a number of the hits in this network to directly modulate both basal and EGF-stimulated activation of the core pathway effectors MAPK1 and AKT, which is activated by PI3K (Fig. 5B and fig. S4). Knockdown of ERBB3, ANXA6 (encoding annexin VI), PRKCD, NEDD9, BCAR1, or SH2D3C reduced basal activation of MAPK1 or AKT, or both, implying the encoded proteins could influence activity of these canonical effectors of EGFR-RAS signaling. However, knockdown of none of these genes reduced EGF-stimulated activation of AKT or MAPK1, indicating that EGF signaling to MAPK1 and AKT does not require these components of the network.
By contrast, a small number of the hits, including TBL1Y [encoding transducin (beta)-like, an adaptor protein], PIN1 (encoding peptidyl-prolyl cis/trans isomerase), NIMA-interacting 1 protein), SC4MOL (encoding sterol-C4-methyl oxidase-like protein, involved in sterol biosynthesis), and ASCL2 (encoding achaete-scute complex homolog 2, a transcription factor), were not connected by direct protein-protein interactions to the core network (Fig. 5A), suggesting either a different mode of action or previously undetected connections.
Direct testing of knockdown of ASCL2 showed that a reduction of the encoded protein failed to statistically significantly affect MAPK1 or AKT activation under basal or EGF-stimulated conditions, although it potently sensitized erlotinib-treated cells to apoptosis (Fig. 4). ASCL2 is a target of Wnt signaling that is increased in abundance in a subset of colon carcinomas (13), and that also controls the expansion of epithelial stem cells (14). Together, these observations suggest that inhibition of ASCL2 may be promising as a direction for therapeutic development.
Chemical inhibition of proteins encoded by or associated with hit genes synergizes with erlotinib in reducing cell viability and tumor growth
We wanted to gain insights that could be rapidly translated into the clinic. Although the clinical use of RNAi is a topic of intense current research, small molecules and monoclonal antibodies remain the most broadly applicable therapy platforms. Further, given that siRNA rarely depletes targeted genes more than 90%, whereas small molecule inhibitors can completely block the functions of targeted gene products, they may produce more robust effects relative to RNAi. For some sensitizing hits, targeted small molecules exist, including Stattic [a small molecule inhibitor of STAT3 activation and dimerization (15)], enzastaurin and Ro-318220 [both targeting the PRKC family (16), including members represented among the hits].
Stattic synergized with erlotinib in inhibiting the viability of both A431 and HCT116 cells (Fig. 5C and fig. S5A) in keeping with the reported dependency of EGFR-driven autocrine growth on STAT3 activation in cancer (17, 18), but showed no statistically significant synergy in reducing cell motility (Fig. 5D, left panel). Both Ro-318220 and enzastaurin synergized with erlotinib in A431 and HCT116 cells (Fig. 5C and fig. S5B), at multiple ratios of drug combination. Combined application of erlotinib and Ro-318220 also significantly reduced tumor cell motility (Fig. 5D, right panel), and reduced tumor growth in a xenograft assay (Fig. 5E). We analyzed the effect of drug combinations on the activation state of a series of benchmark signaling proteins relevant to proliferation and apoptosis, including AKT, ERK, MDM2 (an E3 ubiquitin ligase), and p53 (Fig. S6). Erlotinib used as a single drug reduced basal ERK activation, and basal and EGF-stimulated AKT signaling, but did not affect MDM2 or p53. None of these proteins exhibited changes in amount of phosphorylated (active) species as a consequence of combined application of two drugs, with the exception of AKT, which consistently trended towards reduced phosphorylation on S473 in cells treated with erlotinib in combination with either stattic or enzastaurin. S473 phosphorylation of AKT has been described as dependent on integrated signaling by PRKC, EGFR, and mTOR (19), which may be a pathway by which the enzastaurin-erlotinib combination reduced cell viability.
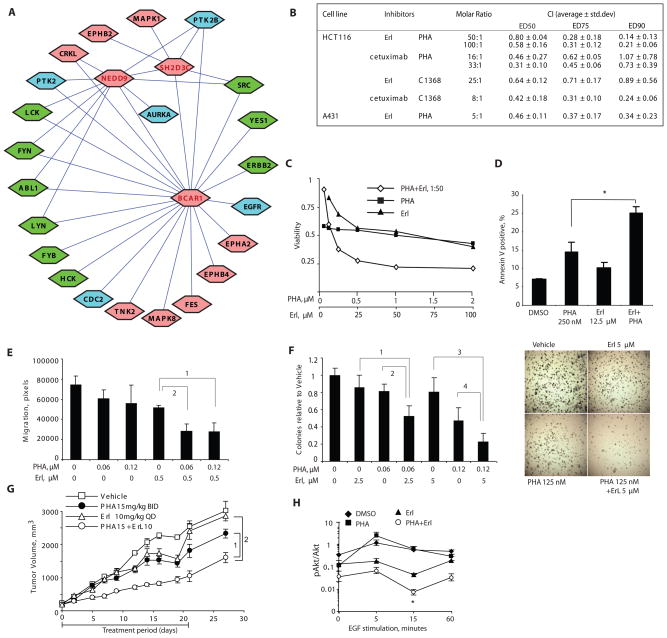
The proteins of the sensitizing BCAR1-SH3D2C-NEDD9 cluster have been linked to control of cell survival in the context of integrin-mediated signaling cascades that are frequently active in advanced and metastatic tumors (20–24), suggesting this cluster may be of particular interest for therapeutic exploitation. However, these proteins are scaffolding proteins and not catalytic, and in contrast to STAT3, have not been targeted by existing small molecule agents. Given the results suggesting the enrichment of sensitizing genes among gene encoding proteins closely linked to core hits, we hypothesized that small molecules targeting kinases closely linked to this cluster by physical interactions might similarly provide a source of synergizing agents for combination with erlotinib. We identified more than 20 kinases as direct interaction neighbors around BCAR1, SH3D3C, and NEDD9 (Fig. 6A). Ten of these kinases (either uniquely, or as one member of a protein family) are targeted by drugs that are in pre-clinical or clinical development, or approved agents, and some of these drugs have indeed been combined productively with EGFR-directed therapeutics, for example dasatinib, targeting Src family kinases (25). Among these, the NEDD9-interacting (26) kinase AURKA (known as Aurora A kinase or STK6) also stimulates the EGFR effector RALA (a guanosine trisphosphatase) (27), and when overexpressed in tumors is associated with increased amounts of phosphorylated AKT (28). Moreover, drugs targeting AURKA are currently undergoing clinical evaluation (29–31).
Fig. 6. Synergy between inhibitors of AURKA and inhibitors of EGFR.
(A) Kinases directly associated with the BCAR1-NEDD9-SH2D3C cluster. Kinases for which no clinical small molecule inhibitor is available are indicated in pink; kinases for which small molecule inhibitors are available are indicated in blue or in green, if inhibitor has previously been tested for synergy with EGFR-targeting agents. (B) Inhibitors of EGFR and inhibitors of AURKA synergize to reduce viability of cancer cells in vitro. Summary results of drug interactions calculated as Chou-Talalay combination index (CI) based on Cell Titer blue viability determinations. CI values <1 indicate synergy, and <0.5 strong synergy, between the two agents in producing cytotoxic effect.. (C) Dose-dependent inhibition of HCT116 cell viability by combination of PHA-680632 and erlotinib. (D) Combination of PHA-680632 and erlotinib treatment increases apoptosis in HCT116 cells at 72 hours; *, t-test p=0.001. (E) Cell motility was measured by wound healing assay in cells treated with drugs at concentrations indicated, and assessed over 18 hours. FDR is <10−5 for 1 and 2. (F) Left, relative soft agar colony formation of cells grown for 2 to 3 weeks in drugs at the concentrations indicated. FDR is equal to 1, 0.0003; 2, 0.0006; 3, 0.0003; and 4, 0.004. Right, results from typical experiment. (G) Tumor xenografts were implanted in SCID mice treated with drugs as indicated. The generalized estimating equations approach (with an autoregressive correlation structure) was used to model tumor growth. A linear time-effect was included in the model for the logarithm of tumor volume and interacted with the treatments in each comparison. P-values are 1: 0.005; 2: 0.008. (H) Quantitation of 3 independent Western analyses of protein lysates of cells treated with erlotinib and PHA-680632 for 3 hrs followed by EGF stimulation. Error bars represent standard error of the mean (SEM) of three independent experiments; *, t-test comparing erlotinib to erlotinib + PHA-680632 yielded P = 0.013.
Analysis on the basis of the Chou-Talalay coefficient of interaction showed that the small molecule AURKA inhibitor PHA-680632 (29) synergized with erlotinib in reducing cell viability of both A431 and HCT116 cells (Fig. 6B). In HCT116 cells, we found strong synergy (coefficient of interaction values <0.5) between cetuximab and either PHA-680632 or another AURKA inhibitor C1368 (32) (Fig. 6B). Erlotinib exhibited strong synergy with PHA-680832 (combination index <0.5) and a slightly less strong interaction with C1368. Combination of AURKA and EGFR-targeting agents did not merely produce cytostasis, but resulted in cell death, increasing the frequency of apoptosis nearly two-fold (Fig. 6C, 6D). In addition, combination of these drugs significantly reduced cell motility (Fig. 6E), colony growth in soft agar (Fig. 6F), and the growth of tumor xenografts implanted in SCID mice (Fig. 6G).
Co-inhibition of EGFR and AURKA reduces SRC family kinase activity
We explored the signaling changes underlying the synergy between EGFR inhibition with erlotinib and the AURKA inhibitor PHA-680632. Treatment of cells with PHA-680632 alone did not reduce the abundance of EGFR or alter EGFR autophosphorylation, and activation when compared to DMSO-treated cells (fig. S7). Furthermore, inhibition of AURKA alone with PHA-680632 had little effect on ERK1/2 or AKT phosphorylation in response to transient EGF stimulation (Fig. 6H and fig. S7). However, in combination with erlotinib treatment, PHA-680632 significantly reduced Ser473-AKT phosphorylation below the amounts seen in cells treated with either agent alone (Fig. 6H), which is consistent with the reduced survival of cells treated with the drug combination, despite not significantly influencing other EGFR-dependent signaling benchmarks (fig. S7).
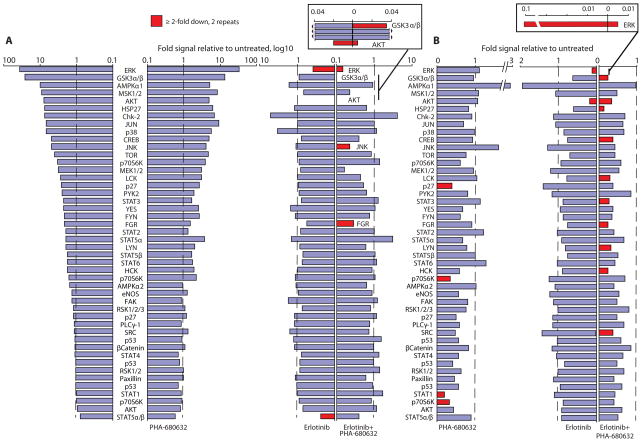
To explore signaling consequences of co-inhibition of AURKA and EGFR in greater depth, we performed a more comprehensive phosphoproteomic analysis of 46 signaling proteins linked to cell proliferation or survival responses, or both, following treatment of A431 cells with erlotinib, PHA-680632, or both. Analysis of two independently performed Western-based screens with phosphorylation-directed antibodies (Fig. 7A) established that erlotinib blocked EGF-induced activation of multiple signaling pathways (reducing AKT and ERK phosphorylation below the amount in unstimulated cells), and PHA-680632 had little effect on EGF-mediated phosphorylation events when used as single agent. In contrast, the combination of drugs led to specific inhibition of a subset of proteins, including greater inhibition of ERK and AKT, as well as inhibition of GSK3β (glycogen synthase kinase 3β, a known functional partner of AUKRA (33)), JNK (c-Jun-N-terminal kinase), and the SRC family kinase FGR.
Fig. 7. Dual inhibition of Aurora-A and EGFR suppresses activation of multiple signaling nodes.
Ranked fold increase in phosphorylation signal of 46 proteins in A431 cells stimulated with EGF (15 ng/mL, 15 min) (A) or grown in 1% serum media (B) and treated with indicated drugs. Stimulation in cells exposed to EGF in the absence of drugs is shown in the far left of panel A. In panel A, the proteins are listed in the same order in the right and left sides, with all proteins labeled on the left and those that are substantially reduced by drug treatment labeled on the right. In panel A, cells were exposed to 0.5 μM erlotinib or 1 μM PHA-680632, or both. In panel B, cells were exposed to 1 μM erlotinib or 0.5 μM PHA-680632, or 0.5 μM erlotinib plus 1 μM PHA-680632. Inset, graphs illustrate magnified scale of indicated phosphoproteins. Proteins showed in red have consistent decrease of >2-fold in signal intensity in independent biological replicates, as indicated.
We performed similar experiments to analyze signaling changes under the steady-state growth conditions in the presence of serum (when the activation state of pathways was not strictly dependent on EGF), which we used to assess synergistic killing of cells (Fig. 7B). Strikingly, this analysis re-identified the same targets for the drug combination as those seen with EGF-dependent signaling (Fig. 7A), but in addition showed significant reduction in the phosphorylation of STAT3 and a group of SRC kinases, including FGR, HCK, LYN, SRC, and LCK. These last hits in particular are intriguing, because the BCAR1-NEDD9-SH2D3C proteins that led us to consider AURKA are direct activators and substrates of these same kinases of SRC family (Fig. 6A) (34). AURKA inhibitors may weaken this resistance cluster in the network.
Discussion
Another potential use of this data set is for the nomination of new biomarkers for selecting patient responsiveness. However, extensive analysis of the expression of siRNA targets in cell lines used for functional analysis (Fig. S8) showed no statistically significant correlation between expression level and role in modulating resistance, whereas analysis of Oncomine profiles (Fig. S9) did not reveal specific trends of altered expression in tumors. Large sequencing projects, including among others the Cancer Gene Census, have noted mutations with some frequency for RET, FLNA, FGFR2, SMAD2, PIK3R1, ABL1, CCND1, and AKT2 [(35) and http://www.sanger.ac.uk/CGP/]; however, most of the genes we identified are not common targets for mutations. These observations have potentially important translational implications, because much effort has gone into analyzing gene expression or mutational status to predict drug resistance. This cumulative lack of a clear pattern of expression or mutation likely reflects the complexity of cancer-associated signaling networks. For many solid tumors, no unique oncogenic driver has been yet identified, but instead, tumor cells undergo multiple, sequential process-oriented oncogenic alterations that together reprogram multiple yet discrete aspects of tumor functionality. In such a scenario, fitness of a cancer cell is determined by the robustness of its signaling network as a whole. The resistance-mediating genes that we have identified should undergo scrutiny as alternative EGFR modulators, joining with proteins such as KRAS, BRAF (a MAPKKK), c-MET (a receptor tyrosine kinase), IGF1 (insulin growth factor 1), and others (36).
A major goal of systems-level bioinformatics analyses is to nominate critical nodes to target in combination to enhance therapy in the clinic and successes are beginning to emerge from this information-driven strategy (37). Separately, screening of siRNA libraries has emerged as an approach to identify genes that when knocked down can kill cancer cells or sensitize them to cytotoxic agents. To date, such screening has typically employed either full genome screens, or screens of small libraries targeting limited groups of proteins, such as the kinome or phosphatome (38, 39). A genome-wide screen to identify sensitizers to the microtubule-targeting agent paclitaxel identified a number of hits that clustered into coherent groups of genes associated with the proteasome or mitotic spindle (40), which had been linked to paclitaxel activity on the basis of existing pathway knowledge.
In the current study, we employed bioinformatics design and direct screening and found that many proteins influencing cellular resistance to EGFR-targeting agents clustered in connection-dense, highly interactive portions of the EGFR signaling network, thus supporting our core hypothesis that these characteristics would be enriched for synthetic lethal interactions. These sensitizing protein clusters were useful for predicting the efficacy of combining protein-targeted drugs with inhibitors of EGFR, suggesting the potential of this approach for speeding the translation of results to the clinic. We believe this targeted approach has several advantages in comparison to a full genome screen. Beyond the pragmatic factors of convenience, speed, and cost, all hits arising from a targeted screen already have at least some defined functional relationships to the signaling pathway under investigation, which should accelerate validation and mechanistic analysis. Further, the limited size of the library allowed the use of more relaxed statistical criteria in nominating hits for validation than would be necessary in a full genome screen, which allowed us to repeat the primary screen multiple times. Given the intrinsic noise in siRNA screening, these are important advantages. Although the use of focused screening approaches (for instance, a kinome library) overcomes a number of these problems, it is notable that only 25/61 of our hits were kinases, and some of the most potent, such as the BCAR1-SH2D3C-NEDD9 cluster, are entirely noncatalytic. Together with our observation that the single greatest source of enrichment for hits (Figure 2A) is among the proteins with both direct physical interactions and literature-based pathway connections to the library seeds, these observations provide guidance for future library optimization.
Materials and Methods
Cell lines, compounds, and antibodies
The A431 cervical adenocarcinoma, (wild type for the gene encoding KRAS and mutation in the gene encoding p53 (41), HCT116 and LoVo,(both of which have mutations in the gene encoding KRAS and are p53 wild type) colorectal carcinoma, and the PANC-1 (mutations in the genes encoding KRAS and p53) and MIA PaCa-2 (mutations in the genes encoding KRAS, p53, and the cell cycle inhibitor p16) pancreatic adenocarcinoma (42) cell lines were obtained from the ATCC (USA). The DLD-1 (mutations in the genes encoding KRAS and p53) and DKS-8 (a cell line derived from DLD-1 in which the activated KRAS allele is disrupted, mutation in the gene encoding p53) (43) were a gift of Robert J. Coffey (Vanderbilt University, TN). SCC61 cells (wild type for KRAS, mutation in the gene encoding p53), derived from squamous cell carcinomas of the head and neck, were provided by Dr. Tanguy Y. Seiwert (University of Chicago). All cell lines were maintained in DMEM supplemented with 10% v/v fetal bovine serum (FBS) and L-glutamine without antibiotics. Cetuximab, panitumumab, and erlotinib were purchased from the Fox Chase Cancer Center pharmacy; CPT11 and C1368 from Sigma-Aldrich (USA); Stattic and Ro-318220 from EMD Chemicals (Gibbstown, NJ, USA). PHA-680632 was obtained from Nerviano Medical Sciences (Nerviano, Italy), as a gift of Dr. Jurgen Moll. Enzastaurin was provided by the Elli Lilly Company (Indianapolis, IN). All antibodies used in Western blot experiments were purchased from Cell Signaling (Danvers, MA, USA), except the mouse monoclonal antibody against p53, which was from Calbiochem (EMD Biosciences, USA).
EGFR network construction
Four sources of information were used, including (i) published EGFR pathway maps, (ii) human PPI data from mulitple databases, (iii) human orthologs of PPIs and genetic interactions modeled from Drosophila, and (iv) microarray data obtained at brief intervals after treatment of cells with stimulators or inhibitors of EGFR or ERBB2. Following initial assembly of a larger gene list, genes were parsed into high confidence (“core”, denoted as “1” after the corresponding letter code) versus lower confidence sets (denoted as “2”), on the basis of the confidence criteria outlined for each section below. For each category of information, all “core” components were included in the final library, as were genes noted as lower confidence but that were included in at least two categories of search criteria (for example, second order protein-protein interaction and microarray linkage). Finally, for the assembled set of EGFR interactors, multiple paralogous genes were identified in humans with the KEGG Sequence Similarity DataBase (SSDB) resource (http://www.genome.jp/kegg/ssdb/) (44). 77 paralogs of the best-characterized and biologically significant genes were added to the set. All data storage, handling, and analysis were done primarily in Cytoscape (http://www.cytoscape.org/)(45).
For the data from pathway map sources, protein names for were extracted from the following pathway maps focused on EGFR: Science Signaling Database of Cell Signaling (http://stke.sciencemag.org/cgi/cm/stkecm;CMP_14987) (46); Biocarta (http://www.biocarta.com/pathfiles/PathwayProteinList.asp?showPFID=102); the Systems Biology model repository (http://www.systems-biology.org/001/005.html) (47); NetPath (http://www.netpath.org/pathways?path_id=NetPath_4) (48); and from Protein Lounge (http://www.proteinlounge.com/pop_pathways1.asp?id=EGF+Pathway). Protein names were individually inspected and, where necessary, converted to the corresponding official (NCBI or EMBL) symbols. Proteins mentioned on at least two EGFR-centered pathways were designated as “pathway core”; substantial divergence was seen among different interpretations of the “EGFR pathway” by the 5 sources.
EGFR (ERBB1) and ERBB2 were used as seeds for PPI searches. Curated information regarding human PPIs of these seeds was collected from the following databases: Biomolecular Object Network Databank (BOND) (http://bond.unleashedinformatics.com/), which derives from the Biomolecular Interaction Network Database (8); General Repository for Interaction Datasets (http://thebiogrid.org/) (49); EMBL_EBI IntAct (http://www.ebi.ac.uk/intact/site/index.jsf) (9); The Human Protein Reference Database (http://www.hprd.org/) (50); Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/about_genomenet/service.html) (44); and Prolinks Database 2.0 (http://mysql5.mbi.ucla.edu/cgi-bin/functionator/pronav) (51). Data for first rank (direct) interactors were collected both by export from the corresponding database and subsequent import into Cytoscape, and by directly querying those databases with the BioNetBuilder plugin (http://err.bio.nyu.edu/cytoscape/bionetbuilder/) (52)), and then cross-comparing retrieved results. Data for the second order interactions were obtained by using EGFR and ERBB2 first rank interactors as seeds for an additional round of query with the BioNetBuilder plugin. Finally, an orthogonal set of second rank interactors was obtained by analysis of protein complexes with more than 2 subunits, which included EGFR. Information for complexes was obtained from BOND and IntAct (XX), and manually compared to the lists in the corresponding publications. We also used the SHC1 and SHC3 adaptors, which bridge between EGFR and downstream signaling effectors, and the CAS (EFS, BCAR1, and NEDD9) scaffolding proteins, which connect EGFR to the SRC and TGF-β core signaling cascades (34, 53), as seeds for first order PPI searches. Second order PPIs with EGFR and ERBB2 were ranked higher (as a P1) if they were also first order interactors of SHC or CAS proteins
To extract a set of EGFR-centered interactions potentially conserved between humans and D. melanogaster, we used information assembled by the Michigan Proteomics Consortium in the Drosophila Interactions Database (DroID) (http://www.droidb.org) (54). Briefly, this database integrates genetic and or protein interaction data from various nonmammalian species (yeast, worms, and flies). Of the 105 EGFR interactors (almost exclusively from Drosophila genetic interactions), 65 had 1 or 2 conserved human orthologs (encoded by 117 genes).
Microarray data were obtained from The Gene Expression Omnibus (GEO, release date Dec 15, 2006) (10). In the selected dataset (GSE6521; raw data available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6521), MCF-7 human breast cancer cells were incubated with the growth hormone heregulin (HRG), or AG1478 (an EGFR kinase inhibitor), or both growth hormone and AG1478. Controls were set as cells that were not treated with growth hormone or inhibitor. A total of 348 genes with a >1.5 fold change (+ or −) upon AG1478 treatment was identified. In this group, the core set included 89 genes that showed a >2-fold change in expression upon AG1478 treatment, or which were inducible by HRG (>1.5 fold) and repressible by AG1478 (>1.5 fold).
High-throughput siRNA screening methods
IC values for erlotinib, panitumumab, and CPT11 were established (fig. S1A). The custom siRNA library targeting 638 human genes was designed and synthesized with two siRNA duplexes for each gene target (Qiagen, Valencia, CA). Transfection conditions were established for the A431 cervical adenocarcinoma ((41) cell line (Fig. S1B) using PLK1 & GL2 siRNA controls to achieve Z′ values (55) of 0.5 or greater. Details of establishment of Z′ factor for transfections (fig S1), and statistical consideration for selection of preliminary positive candidates graphically outlined in fig. S2 and based on standard approaches described in detail in (40). For each gene targeted, two independent siRNA duplexes were combined and arrayed in 96-well plates with a layout that systematically placed positive control siRNA (targeting PLK1) and negative control siRNA (targeting insect luciferase GL2, Thermo Fisher Scientific, USA) amongst the test siRNAs. We used a reverse transfection protocol in which siRNA at a final concentration of 50 nM was mixed with Dharmafect 1 transfection reagent according to the manufacturer’s instructions (Thermo Fisher, USA). Cells (3500 per well, resuspended in 1% FBS/DMEM) were added directly to wells with an automated liquid dispenser. At 24 hr following transfection, two replica plates were treated with drugs at previously established IC30 or 0.02% DMSO diluted in culture media. We assessed viability 96 hr post-transfection with Alamar blue (CellTiter Blue Viability Assay, Promega, USA). Dose-responses for each drug and cell line were retested in parallel with each screen.
For screening, A431 cells were transfected with siRNA followed by exposure to vehicle (0.02% DMSO), or drug used at inhibitory concentrations of 30% (IC30). Viability was determined for each target gene and normalized to the averaged GL2 viability on each plate. Sensitization index (SI) was calculated for each individual well on a 96-well plate as SI=(Vdrug/GL2drug)/(VDMSO/GL2DMSO), where V was viability in wells transfected with targeting duplexes and GL2 was the averaged viability of 4 wells with non-targeting negative control siRNA on the same plate. All calculations were automated using cellHTS package within open source Bioconductor Package (http://www.bioconductor.org)(56). The effect of drug treatment on viability was measured based on the normalized viabilities in the drug-treated and vehicle wells using Limma (57). Limma borrows strength across genes on the basis of an empirical Bayes approach and identifies statistically significant changes in viability by combining information from a set of gene-specific tests. Hits were identified based on statistical significance, as well as biological significance. Statistical significance was determined by p-value controlled for the false discovery rate (FDR) using the Benjamini-Hochberg step-up method (58) to account for multiple testing. Hits showing an FDR of less than 20% were considered statistically significant. Biological significance was arbitrarily defined as an increase or decrease in SI greater than 15%. Hits identified as statistically and biologically significant were further validated.
Primary sensitizing hits obtained with erlotinib, cetuximab, or both were further tested with erlotinib and DMSO in the A431 cell line with 4 siRNA individual duplexes (the two originally used in the screen, plus two new nonoverlapping RNA oligoribonucleotides), to confirm the sensitization phenotype at 10 nM and 50 nM concentrations. Hits were considered as validated by this method if at least 2 out of 4 siRNA reproduced the sensitization phenotype with SI≤0.85, FDR≤20% for each individual siRNA sequence in at least two independent experiments. For a number of hits, we additionally confirmed that sensitizing siRNAs reduced mRNA abundance for the targeted genes, using qRT-PCR; and confirmed reduction in protein abundance by Western analysis (fig. S3).
Cell line and drug specificity of candidate sensitizing genes
Of the confirmed set of 61 siRNA targets identified as causing erlotinib sensitivity in A431 cells, 45 were further tested for sensitization to erlotinib, cetuximab and CPT11 in A431 versus refractory adenocarcinoma cell lines for which optimal transfection conditions and drug sensitivity had been established. In this analysis, for each target, the two most active siRNA duplexes identified during the validation stage were pooled in a 96-well format, cells were transfected with these siRNA pools and drug-treated under conditions similar to those described above for the initial A431 screen. SI and statistical significance were calculated as in the validation experiments. All experiments were performed at least three times independently.
We used two approaches in subsequent data analysis. For the relative ranking approach, for each experiment, SI values (regardless of FDR) for each siRNA pool were ranked from the strongest (assigned a value of 0) to the weakest (assigned as 1). For all experiments performed with a given cell:drug combination (for example, A431:erlotinib, or HCT116:CPT11) averages were determined on the basis of at least three experimental runs. The averaged data were imported and clustered in MultiExperiment Viewer (MeV_4_3) software (59), and dendrograms were created using HCL Support Trees (using Euclidian Distance as a metric, and bootstrapping with 100 iterations). For the absolute threshold approach, specific SI thresholds were applied for each data point, considering only data with an FDR ≤20% in each independent experiment. Data were visualized in MultiExperiment Viewer using color assignments to indicate SI cutoffs obtained in at least two independent experiments, as described in figure legends. The resulting output of both analytic strategies was processed using the graphic software package Canvas (ACD Systems International, Canada) to improve visualization of data.
Quantitative RT-PCR
For evaluation of expression of validated target genes, each of the cell lines was grown to 70% confluency in DMEM media with 10% FBS, then total RNA was extracted with RNeasy Minikit (Qiagen, Valencia, CA). To confirm mRNA depletion by siRNA, 48 hrs after transfection of A431 cells grown in 96-well plates, total RNA was extracted with a Cell-to-Ct kit from Applied Biosystems, Foster City, CA. Quantitative RT-PCR reactions were performed with TaqMan probes and primers designed by the manufacturer of the Cell-to-Ct kit, using an ABI PRISM 7700 detection system (Applied Biosystems, Foster City, CA). The results were analyzed with the comparative Ct method to establish relative expression curves.
To assess whether gene expression correlated with the ability of gene-targeted siRNAs to inhibit intrinsic cell growth, we used a Pearson correlation of the mean values of gene expression relative to that obtained in A431 cells measured by RT-PCR, against the mean growth observed in DMSO-treated cells in all experiments. To test significance, we permuted the labels on the cell lines in the RT-PCR measurements, which created a series of 100 data sets that should show only chance correlation, and generated Pearson correlation values on this permuted set. Significance was defined as an FDR of 5%, setting Pearson correlation greater than 0.745 or less than −0.71 for positive correlated or negative-correlated, respectively. Positive correlation indicates that higher expression (lower number of RT-PCR cycles) correlated with greater growth inhibition, whereas negative correlation indicates higher expression is correlated with lower inhibition.
Network analysis with hits
For all genes in the library, the String search engine (60) was used in subsequent analysis to augment information on PPIs in human cells, PPIs between homologous genes in model organisms, database or pathway links, and text mining (coappearance of gene names in PubMed). Data regarding experimentally proven interactions in human and model organisms were merged. Topological properties of the library network were assessed with the NetworkAnalyzer plugin for Cytoscape (61), on the basis of STRING-expanded defined interactions among genes in the library (based only on experimental data). In this analysis, for each node, degree, stress, and neighborhood connectivity were separately assessed. The topological coefficient was calculated to provide an estimate for the trend of the nodes in the network to have shared neighbors. To provide additional context in some analyses (Fig. 5) STRING-extracted information from pathway databases and text-mining data were merged and displayed using Cytoscape as indicated in figure legends.
Apoptosis assays
Apoptosis was measured with the Annexin V assay (Guava Technologies, Hayward, CA). Annexin V-positive A431 cells were counted using Guava flow cytometry 72 hours post transfection, 48 hours after treatment. Statistical significance versus cells transfected with the control GL2 siRNA was determined by logistic regression models to identify genes that when knocked down increased apoptosis in the presence of erlotinib relative to vehicle.
Pathway analysis
To measure the effect of siRNAs on the activity of EGFR effectors, cells were transfected with siRNA and the culture media was replaced with glutamine-supplemented serum-free DMEM at 24 hrs post-transfection. After overnight incubation, cells were treated with DMSO, erlotinib, or PHA-680632 for 2 hrs, then either left untreated or stimulated with EGF at 15 ng/ml for 15 minutes. Cell extracts were prepared using M-PER™ mammalian protein extraction buffer (Thermo Scientific, Rockford, IL) supplemented with the Halt™ phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL) and the Complete Mini™ protease inhibitor cocktail (Roche Diagnostics Gmbh, Manheim, Germany). Extracts were centrifuged at 15,000g for 10 min at 4°C. Western signal detection was performed using antibodies to indicated proteins with LiCor technology (Lincoln, NE, USA) or standard X-ray film.
For phosphoproteomic analysis, we used the Proteome Profiler™ array (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s protocol. In brief, A431 cells were grown for 24 hours in DMEM supplemented with L-glutamine and 1% FBS to 70% confluency. Cells were either then serum-starved overnight or maintained in the same media. Serum-starved and cells incubated in 1% serum were either left untreated or incubated with IC30 concentrations of inhibitors for 3 hours. For a subset of phosphoproteins, phosphorylation status was confirmed by Western blot. Quantification was done with ImageJ software (62).
Drug synergy testing
The combination index (CI) between pharmacological inhibitors was established by the Chou-Talalay method (63). We used the software package CalcuSyn (BioSoft, UK) to automate calculations. Briefly, for each drug tested, an IC50 curve was established in each cell line, and used to select combination doses of drugs for subsequent synergy tests. 3500 cells were plated per well in 96-well plates. After 24 hours, cells were treated with serial dilutions of individual inhibitors or combinations of two inhibitors maintained at a constant molar ratio. After 72 hours incubation, cell viability was measured using either CellTiter Blue (Promega, USA) or a WST1 assay (Roche Applied Science, Indianapolis, IN). The CI values for each dose and corresponding cytotoxicity were expressed as the fraction affected (Fa) and were calculated using CalcuSyn computer software and presented as Fa-CI plots.
Anchorage-independent growth and cell motility
Soft agar assays were done essentially as described (64). Cells were seeded at 2000 cells per well and grown for 2 to 3 weeks. Colonies were stained with thiazolyl blue tetrazolium bromide, and scored with a Nikon SMZ1500 microscope coupled with Cool Snap charge-coupled device camera (Roper Scientific, Inc., Tucson, AZ) with Image Pro-Plus software (Media Cybernetics, Silver Spring, MD). Survival curves were based on at least two concentration points, with values determined in at least two separate experiments, with each assay done in duplicate. Drug interactions were calculated as above using CalcuSyn software. For motility assays, movement of A431 cells grown in 1% FCS into a scratched area of the monolayer was monitored with a phase contrast 10x objective using an inverted microscope (Nikon TE2000). Images were obtained every 20 min for 18 hours. Areas of migration were estimated using MetaMorph software. For both studies, analysis of variance was used to determine the treatment effect for each comparison. The logarithm of normalized ratios (to vehicle control) was used in the analysis. Multiple hypothesis testing performed with the FDR method of Benjamini & Hochberg (58).
Tumor formation in vivo
Male CB.17/scid mice aged 6 to 8 weeks were obtained from the Fox Chase Cancer Center breeding colony. All experiments were performed according to protocols approved by the institutional animal use committee. Mice were injected with 3 × 106 A431 cells subcutaneously into the flanks. Palpable tumors appeared in all animals in 10 to 14 days, and were measured 3 times a week in two dimension and volume calculated by modified ellipsoidal formula as Length × Width2 × 0.52. Mice were randomized and treatments commenced when tumor volume exceeded 65 mm3. Erlotinib at doses 10 to 20 mg/kg was given by oral gavage in 10% DMSO/saline. Enzastaurin was suspended in 5% dextrose in water and dosed at 75 mg/kg by gavage twice daily. PHA-680632 was freshly dissolved in acidified 5% dextrose in water and administered intraperitoneally twice daily at 15 mg/kg dose. The generalized estimating equations approach (with an autoregressive correlation structure) was used to model tumor growth. A linear time-effect was included in the model for the logarithm of tumor volume and interacted with the treatments in each comparison.
Supplementary Material
Fig. S1. Optimization assays to establish critical assay parameters for screening.
Fig. S2. Schematic of screen strategy and hit selection
Fig. S3. Efficiency of gene knockdown for a representative set of siRNAs used.
Fig. S4. Representative Western blots showing the overall abundance and phosphorylation status of EGFR pathway members in A431 cells transfected with siRNAs targeting the indicated genes.
Fig. S5. Synergistic in vitro activity of signaling inhibitors.
Fig. S6. Phosphoprotein changes with signaling inhibitors.
Fig. S7. PHA-680632 and erlotinib combine to reduce AKT phosphorylation.
Fig. S8. Expression profile of hits in cell lines.
Fig. S9. Expression profile of hits in Oncomine.
Table S1. List of genes included in the targeted library, including official gene name, gene symbol, and reason(s) for inclusion in the library.
Table S2. Sequences of the siRNAs for the 61 validated target genes.
Acknowledgments
We thank M. F. Ochs (Johns Hopkins School of Public Health) for consultation on statistics, and M. Brown (Lombardi Cancer Center), J. Moll (Nerviano Medical Sciences), R. Coffey (Vanderbilt University), T. Y. Seiwert (University of Chicago), and V. Khazak (NexusPharma) for reagents and help with design of xenograft studies. We particularly thank the Fox Chase Cancer Center Biostatistics Service, and High Throughput Screening Facilities.
Funding: This work was supported by NIH core grant CA06927, by the Pew Charitable Fund, and by a generous gift from Mrs. C Greenberg, to Fox Chase Cancer Center; by NIH R01 CA50633, by the Jeannik Littlefield Award from the AACR and by research funds from Amgen (to LW); by NIH R01 CA63366 and R01 CA113342, and by Pennsylvania Tobacco Settlement funding (to EAG); by the Fox Chase Cancer Center Head and Neck Keystone Program; and by a career development award from Genentech (to IA).
References
- 1.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS medicine. 2005 Jan;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006 Apr 15;66:3992. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 3.Friedman A, Perrimon N. Genetic screening for signal transduction in the era of network biology. Cell. 2007 Jan 26;128:225. doi: 10.1016/j.cell.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999 Oct 15;286:509. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 5.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999 Dec 2;402:C47. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 6.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003 Oct 14;100:12123. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy SR. Genetics. Total information awareness for worm genetics. Science. 2006 Mar 10;311:1381. doi: 10.1126/science.1125655. [DOI] [PubMed] [Google Scholar]
- 8.Bader GD, Donaldson I, Wolting C, Ouellette BF, Pawson T, Hogue CW. BIND--The Biomolecular Interaction Network Database. Nucleic Acids Res. 2001 Jan 1;29:242. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, Kerssemakers J, Leroy C, Menden M, Michaut M, Montecchi-Palazzi L, Neuhauser SN, Orchard S, Perreau V, Roechert B, van Eijk K, Hermjakob H. The IntAct molecular interaction database in 2010. Nucleic Acids Res. Jan;38:D525. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Edgar R. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009 Jan;37:D885. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimri M, Naramura M, Duan L, Chen J, Ortega-Cava C, Chen G, Goswami R, Fernandes N, Gao Q, Dimri GP, Band V, Band H. Modeling breast cancer-associated c-Src and EGFR overexpression in human MECs: c-Src and EGFR cooperatively promote aberrant three-dimensional acinar structure and invasive behavior. Cancer Res. 2007 May 1;67:4164. doi: 10.1158/0008-5472.CAN-06-2580. [DOI] [PubMed] [Google Scholar]
- 12.Seton-Rogers SE, Lu Y, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, Brugge JS. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc Natl Acad Sci U S A. 2004 Feb 3;101:1257. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jubb AM, Chalasani S, Frantz GD, Smits R, Grabsch HI, Kavi V, Maughan NJ, Hillan KJ, Quirke P, Koeppen H. Achaete-scute like 2 (ascl2) is a target of Wnt signalling and is upregulated in intestinal neoplasia. Oncogene. 2006 Jun 8;25:3445. doi: 10.1038/sj.onc.1209382. [DOI] [PubMed] [Google Scholar]
- 14.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, van Es JH, Barker N, Peters PJ, van de Wetering M, Clevers H. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009 Mar 6;136:903. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 15.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006 Nov;13:1235. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 16.Dieter P, Fitzke E. RO 31-8220 and RO 31-7549 show improved selectivity for protein kinase C over staurosporine in macrophages. Biochem Biophys Res Commun. 1991 Nov 27;181:396. doi: 10.1016/s0006-291x(05)81432-9. [DOI] [PubMed] [Google Scholar]
- 17.Grandis JR, Drenning SD, Chakraborty A, Zhou MY, Zeng Q, Pitt AS, Tweardy DJ. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor- mediated cell growth In vitro. J Clin Invest. 1998 Oct 1;102:1385. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehm AL, Sen M, Seethala R, Gooding WE, Freilino M, Wong SM, Wang S, Johnson DE, Grandis JR. Combined targeting of epidermal growth factor receptor, signal transducer and activator of transcription-3, and Bcl-X(L) enhances antitumor effects in squamous cell carcinoma of the head and neck. Mol Pharmacol. 2008 Jun;73:1632. doi: 10.1124/mol.107.044636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan QW, Cheng C, Knight ZA, Haas-Kogan D, Stokoe D, James CD, McCormick F, Shokat KM, Weiss WA. EGFR signals to mTOR through PKC and independently of Akt in glioma. Science signaling. 2009;2:ra4. doi: 10.1126/scisignal.2000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almeida EA, Ilic D, Han Q, Hauck CR, Jin F, Kawakatsu H, Schlaepfer DD, Damsky CH. Matrix survival signaling: from fibronectin via focal adhesion kinase to c-Jun NH(2)-terminal kinase. J Cell Biol. 2000;149:741. doi: 10.1083/jcb.149.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Law SF, O’Neill GM, Fashena SJ, Einarson MB, Golemis EA. The docking protein HEF1 is an apoptotic mediator at focal adhesion sites. Mol Cell Biol. 2000;20:5184. doi: 10.1128/mcb.20.14.5184-5195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Neill GM, Golemis EA. Proteolysis of the docking protein HEF1 and implications for focal adhesion dynamics. Mol Cell Biol. 2001;21:5094. doi: 10.1128/MCB.21.15.5094-5108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Y, Brush J, Stewart T. NSP1 Defines a Novel Family of Adaptor Proteins Linking Integrin and Tyrosine Kinase Receptors to the c-Jun N-terminal Kinase/Stress-activated Protein Kinase Signaling Pathway. J Biol Chem. 1999;274:10047. doi: 10.1074/jbc.274.15.10047. [DOI] [PubMed] [Google Scholar]
- 24.Hartmann TN, Burger JA, Glodek A, Fujii N, Burger M. CXCR4 chemokine receptor and integrin signaling co-operate in mediating adhesion and chemoresistance in small cell lung cancer (SCLC) cells. Oncogene. 2005 Jun 23;24:4462. doi: 10.1038/sj.onc.1208621. [DOI] [PubMed] [Google Scholar]
- 25.Song L, Morris M, Bagui T, Lee FY, Jove R, Haura EB. Dasatinib (BMS-354825) selectively induces apoptosis in lung cancer cells dependent on epidermal growth factor receptor signaling for survival. Cancer Res. 2006 Jun 1;66:5542. doi: 10.1158/0008-5472.CAN-05-4620. [DOI] [PubMed] [Google Scholar]
- 26.Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005 Oct;7:937. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu JC, Chen TY, Yu CT, Tsai SJ, Hsu JM, Tang MJ, Chou CK, Lin WJ, Yuan CJ, Huang CY. Identification of V23RalA-Ser194 as a critical mediator for Aurora-A-induced cellular motility and transformation by small pool expression screening. J Biol Chem. 2005 Mar 11;280:9013. doi: 10.1074/jbc.M411068200. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T, Deng CX. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006 Nov 16;25:7148. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- 29.Soncini C, Carpinelli P, Gianellini L, Fancelli D, Vianello P, Rusconi L, Storici P, Zugnoni P, Pesenti E, Croci V, Ceruti R, Giorgini ML, Cappella P, Ballinari D, Sola F, Varasi M, Bravo R, Moll J. PHA-680632, a novel Aurora kinase inhibitor with potent antitumoral activity. Clin Cancer Res. 2006 Jul 1;12:4080. doi: 10.1158/1078-0432.CCR-05-1964. [DOI] [PubMed] [Google Scholar]
- 30.Carpinelli P, Ceruti R, Giorgini ML, Cappella P, Gianellini L, Croci V, Degrassi A, Texido G, Rocchetti M, Vianello P, Rusconi L, Storici P, Zugnoni P, Arrigoni C, Soncini C, Alli C, Patton V, Marsiglio A, Ballinari D, Pesenti E, Fancelli D, Moll J. PHA-739358, a potent inhibitor of Aurora kinases with a selective target inhibition profile relevant to cancer. Mol Cancer Ther. 2007 Dec;6:3158. doi: 10.1158/1535-7163.MCT-07-0444. [DOI] [PubMed] [Google Scholar]
- 31.Manfredi MG, Ecsedy JA, Meetze KA, Balani SK, Burenkova O, Chen W, Galvin KM, Hoar KM, Huck JJ, LeRoy PJ, Ray ET, Sells TB, Stringer B, Stroud SG, Vos TJ, Weatherhead GS, Wysong DR, Zhang M, Bolen JB, Claiborne CF. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proc Natl Acad Sci U S A. 2007 Mar 6;104:4106. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tari LW, Hoffman ID, Bensen DC, Hunter MJ, Nix J, Nelson KJ, McRee DE, Swanson RV. Structural basis for the inhibition of Aurora A kinase by a novel class of high affinity disubstituted pyrimidine inhibitors. Bioorg Med Chem Lett. 2007 Feb 1;17:688. doi: 10.1016/j.bmcl.2006.10.086. [DOI] [PubMed] [Google Scholar]
- 33.Fumoto K, Lee PC, Saya H, Kikuchi A. AIP regulates stability of Aurora-A at early mitotic phase coordinately with GSK-3beta. Oncogene. 2008 Jul 24;27:4478. doi: 10.1038/onc.2008.92. [DOI] [PubMed] [Google Scholar]
- 34.Defilippi P, Di Stefano P, Cabodi S. p130Cas: a versatile scaffold in signaling networks. Trends Cell Biol. 2006 May;16:257. doi: 10.1016/j.tcb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004 Mar;4:177. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hopper-Borge EA, Nasto RE, Ratushny V, Weiner LM, Golemis EA, Astsaturov I. Mechanisms of tumor resistance to EGFR-targeted therapies. Expert opinion on therapeutic targets. 2009 Mar;13:339. doi: 10.1517/14712590902735795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pritchard JR, Cosgrove BD, Hemann MT, Griffith LG, Wands JR, Lauffenburger DA. Three-kinase inhibitor combination recreates multipathway effects of a geldanamycin analogue on hepatocellular carcinoma cell death. Mol Cancer Ther. 2009 Aug;8:2183. doi: 10.1158/1535-7163.MCT-08-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanton C, Marani M, Pardo O, Warne PH, Kelly G, Sahai E, Elustondo F, Chang J, Temple J, Ahmed AA, Brenton JD, Downward J, Nicke B. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007 Jun;11:498. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Iorns E, Turner NC, Elliott R, Syed N, Garrone O, Gasco M, Tutt AN, Crook T, Lord CJ, Ashworth A. Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell. 2008 Feb;13:91. doi: 10.1016/j.ccr.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG, Xie XJ, White MA. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007 Apr 12;446:815. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 41.Kwok TT, Mok CH, Menton-Brennan L. Up-regulation of a mutant form of p53 by doxorubicin in human squamous carcinoma cells. Cancer Res. 1994 Jun 1;54:2834. [PubMed] [Google Scholar]
- 42.Brunner TB, Cengel KA, Hahn SM, Wu J, Fraker DL, McKenna WG, Bernhard EJ. Pancreatic cancer cell radiation survival and prenyltransferase inhibition: the role of K-Ras. Cancer Res. 2005 Sep 15;65:8433. doi: 10.1158/0008-5472.CAN-05-0158. [DOI] [PubMed] [Google Scholar]
- 43.Sarthy AV, Morgan-Lappe SE, Zakula D, Vernetti L, Schurdak M, Packer JC, Anderson MG, Shirasawa S, Sasazuki T, Fesik SW. Survivin depletion preferentially reduces the survival of activated K-Ras-transformed cells. Mol Cancer Ther. 2007 Jan;6:269. doi: 10.1158/1535-7163.MCT-06-0560. [DOI] [PubMed] [Google Scholar]
- 44.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002 Jan 1;30:42. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003 Nov;13:2498. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlessinger J. Epidermal Growth Factor Receptor Pathway. Sci Signal. (Connections Map in the Database of Cell Signaling, as seen January 2008), http://stke.sciencemag.org/cgi/cm/stkecm;CMP_14987.
- 47.Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Molecular systems biology. 2005;1:2005 0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD, Mathivanan S, Pecquet C, Gollapudi SK, Tattikota SG, Mohan S, Padhukasahasram H, Subbannayya Y, Goel R, Jacob HK, Zhong J, Sekhar R, Nanjappa V, Balakrishnan L, Subbaiah R, Ramachandra YL, Rahiman BA, Prasad TS, Lin JX, Houtman JC, Desiderio S, Renauld JC, Constantinescu SN, Ohara O, Hirano T, Kubo M, Singh S, Khatri P, Draghici S, Bader GD, Sander C, Leonard WJ, Pandey A. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, Dolinski K, Tyers M. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008 Jan;36:D637. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, Menon S, Hanumanthu G, Gupta M, Upendran S, Gupta S, Mahesh M, Jacob B, Mathew P, Chatterjee P, Arun KS, Sharma S, Chandrika KN, Deshpande N, Palvankar K, Raghavnath R, Krishnakanth R, Karathia H, Rekha B, Nayak R, Vishnupriya G, Kumar HG, Nagini M, Kumar GS, Jose R, Deepthi P, Mohan SS, Gandhi TK, Harsha HC, Deshpande KS, Sarker M, Prasad TS, Pandey A. Human protein reference database--2006 update. Nucleic Acids Res. 2006 Jan 1;34:D411. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowers PM, Pellegrini M, Thompson MJ, Fierro J, Yeates TO, Eisenberg D. Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol. 2004;5:R35. doi: 10.1186/gb-2004-5-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avila-Campillo I, Drew K, Lin J, Reiss DJ, Bonneau R. BioNetBuilder: automatic integration of biological networks. Bioinformatics. 2007 Feb 1;23:392. doi: 10.1093/bioinformatics/btl604. [DOI] [PubMed] [Google Scholar]
- 53.O’Neill GM, Fashena SJ, Golemis EA. Integrin signaling: a new Cas(t) of characters enters the stage. Trends Cell Biol. 2000;10:111. doi: 10.1016/s0962-8924(99)01714-6. [DOI] [PubMed] [Google Scholar]
- 54.Yu J, Pacifico S, Liu G, Finley RL., Jr DroID: the Drosophila Interactions Database, a comprehensive resource for annotated gene and protein interactions. BMC Genomics. 2008;9:461. doi: 10.1186/1471-2164-9-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 56.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 58.Benjamini Y, Hochhberg Y. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J Royal Stat Soc B. 1995;57:289. [Google Scholar]
- 59.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003 Feb;34:374. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 60.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009 Jan;37:D412. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008 Jan 15;24:282. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 62.Rasband WS. ImageJ. National Institutes of Health; Bethesda, Maryland, USA: 1997–2009. http://rsb.info.nih.gov/ij/ [Google Scholar]
- 63.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 64.Skobeleva N, Menon S, Weber L, Golemis EA, Khazak V. In vitro and in vivo synergy of MCP compounds with mitogen-activated protein kinase pathway- and microtubule-targeting inhibitors. Mol Cancer Ther. 2007 Mar;6:898. doi: 10.1158/1535-7163.MCT-06-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Optimization assays to establish critical assay parameters for screening.
Fig. S2. Schematic of screen strategy and hit selection
Fig. S3. Efficiency of gene knockdown for a representative set of siRNAs used.
Fig. S4. Representative Western blots showing the overall abundance and phosphorylation status of EGFR pathway members in A431 cells transfected with siRNAs targeting the indicated genes.
Fig. S5. Synergistic in vitro activity of signaling inhibitors.
Fig. S6. Phosphoprotein changes with signaling inhibitors.
Fig. S7. PHA-680632 and erlotinib combine to reduce AKT phosphorylation.
Fig. S8. Expression profile of hits in cell lines.
Fig. S9. Expression profile of hits in Oncomine.
Table S1. List of genes included in the targeted library, including official gene name, gene symbol, and reason(s) for inclusion in the library.
Table S2. Sequences of the siRNAs for the 61 validated target genes.