Abstract
When oxygen is abundant, quiescent cells efficiently extract energy from glucose primarily by oxidative phosphorylation, whereas under the same conditions tumour cells consume glucose more avidly, converting it to lactate. This long-observed phenomenon is known as aerobic glycolysis1, and is important for cell growth2,3. Because aerobic glycolysis is only useful to growing cells, it is tightly regulated in a proliferation-linked manner4. Inmammals, this is partly achieved through control of pyruvate kinase isoform expression. The embryonic pyruvate kinase isoform, PKM2, is almost universally re-expressed in cancer2, and promotes aerobic glycolysis, whereas the adult isoform, PKM1, promotes oxidative phosphorylation2. These two isoforms result from mutually exclusive alternative splicing of the PKM pre-mRNA, reflecting inclusion of either exon 9 (PKM1) or exon 10 (PKM2). Here we show that three heterogeneous nuclear ribonucleoprotein (hnRNP) proteins, polypyrimidine tract binding protein (PTB, also known as hnRNPI), hnRNPA1 and hnRNPA2, bind repressively to sequences flanking exon 9, resulting in exon 10 inclusion. We also demonstrate that the oncogenic transcription factor c-Myc upregulates transcription of PTB, hnRNPA1 and hnRNPA2, ensuring a high PKM2/PKM1 ratio. Establishing a relevance to cancer, we show that human gliomas overexpress c-Myc, PTB, hnRNPA1 and hnRNPA2 in a manner that correlates with PKM2 expression. Our results thus define a pathway that regulates an alternative splicing event required for tumour cell proliferation.
Alternative splicing of PKM has an important role in determining the metabolic phenotype of mammalian cells. The single exon difference imparts the enzymes produced with important functional distinctions. For example, PKM2, but not PKM1, is regulated by the binding of tyrosine phosphorylated peptides, which results in release of the allosteric activator fructose-1-6-bisphosphate and inhibition of pyruvate kinase activity5, a property that might allow growth-factor-initiated signalling cascades to channel glycolytic intermediates into biosynthetic processes. The importance of tumour reversion to PKM2 was underscored by experiments in which replacement of PKM2 with PKM1 in tumour cells resulted in markedly reduced growth2. Consistent with a critical role in proliferation, re-expression of PKM2 in tumours is robust2, although little is known about the regulation of this process.
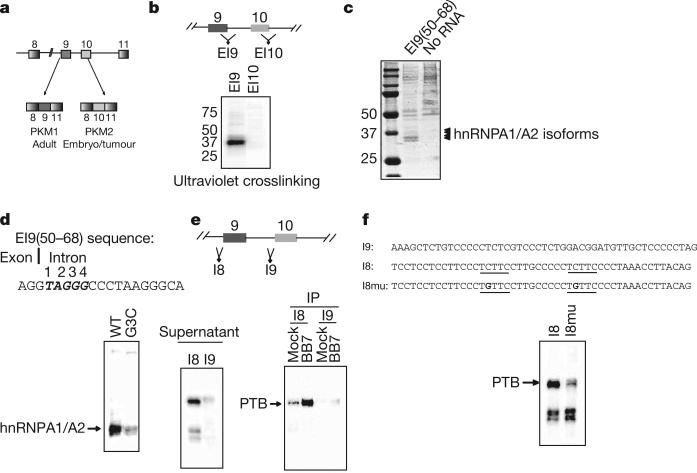
We set out to identify RNA binding proteins that might regulate PKM alternative splicing. To this end, we prepared an [α-32P]UTP-labelled 250-nucleotide RNA spanning the exon 9 (E9) 5′ splice site (EI9), previously identified as inhibitory to E9 inclusion6, as well as a labelled RNA from a corresponding region of E10 (EI10) (Fig. 1b), and performed ultraviolet crosslinking assays with HeLa nuclear extracts7. After separation by SDS–polyacrylamide gel electrophoresis (PAGE), multiple proteins from 35–40 kDa appeared using the EI9 substrate, whereas little binding was observed using the EI10 substrate (Fig. 1b). Strong binding was mapped to a 19-nucleotide region we named EI9(50–68) that spans the E9 5′ splice site (Supplementary Fig. 1). To identify the bound proteins, we performed RNA affinity chromatography using a 5′ biotin-labelled RNA corresponding to EI9(50–68). After SDS–PAGE and Coomassie staining, the pattern of specifically bound proteins closely matched that observed after ultraviolet crosslinking (Fig. 1c). The four indicated proteins between 35–40 kDa were excised and identified by mass spectrometry as isoforms of hnRNPA1 and hnRNPA2, RNA binding proteins with well established roles as sequence-specific repressors of splicing (for example, see refs 7, 8). This result was confirmed by immunoblotting with antibodies against hnRNPA1 (Supplementary Fig. 2).
Figure 1. hnRNP proteins bind specifically to sequences flanking E9.
a, Schematic diagram of PKM splicing. b, Position of probes spanning the E9 or E10 5′ splice sites (top). After ultraviolet crosslinking, proteins were detected by autoradiography (bottom). Position of molecular mass standards in kDa is indicated at left. c, Affinity chromatography using EI9(50–68). Bound proteins were separated by SDS–PAGE and Coomassie stained. Bands excised for mass spectrometry are indicated. d, Sequence of EI9(50–68); the putative hnRNPA1/A2 binding site is indicated in bold italics (top). Ultraviolet crosslinking with wild-type RNA, or RNA with a mutation in the putative hnRNPA1/A2 binding site, is shown in the bottom panel. e, Position of I8 and I9 (top). Ultraviolet crosslinking using I8 or I9 substrates is shown in the bottom left panel. Ultraviolet crosslinking reactions were immunoprecipitated with either anti-PTB (BB7) or anti-HA antibodies (bottom right panel). f, Ultraviolet crosslinking with I8 and the mutant derivative I8mu, sequences indicated above. Putative PTB binding sites in I8 are underlined.
The sequence immediately downstream of the E9 5′ splice site contains a UAGGGC sequence that is highly related to the consensus hnRNPA1 high affinity binding site identified by SELEX, UAGGG(A/U)9 (Fig. 1d). Consistent with previous mutational studies of an identical A1 binding site8, mutation of the G3 nucleotide of this motif to C led to a large decrease in hnRNPA1 and hnRNPA2 binding (Fig. 1d and Supplementary Fig. 3). The G3C mutation resulted in increased splicing in vitro when introduced into a splicing substrate containing E9 (Supplementary Fig. 4), and led to increased E9 inclusion in a minigene construct in vivo (Supplementary Fig. 5). These data confirm the presence of an inhibitory hnRNPA1/hnRNPA2 binding site immediately downstream of the E9 5′ splice site.
To explore the possibility that other splicing regulators bind upstream of E9 or E10, we constructed crosslinking substrates (48 nucleotides) that span the region upstream of each exon. Using these RNAs for ultraviolet crosslinking showed strong binding of a 55-kDa protein to the I8 RNA probe, but not to the I9 probe (Fig. 1e). Inspection of the polypyrimidine tract upstream of E9 revealed two potential PTB (polypyrimidine tract binding protein, or hnRNPI) binding sequences (UCUUC)10 within 35 nucleotides of the intron/exon boundary, whereas no such sequence exists in the E10 polypyrimidine tract. PTB frequently functions as a splicing repressor10, often by binding repressively to the polypyrimidine tract11. Immunoprecipitation confirmed that the 55-kDa crosslink observed using I8 RNA is PTB (Fig. 1e), and we observed strong binding of PTB to a biotinylated version of I8 (Supplementary Fig. 6). In addition, mutation of the two putative PTB binding sites from UCUUC to UGUUC significantly diminished binding (Fig. 1f and Supplementary Fig. 7). Our data indicate that the splicing repressor PTB binds specifically to the polypyrimidine tract of E9.
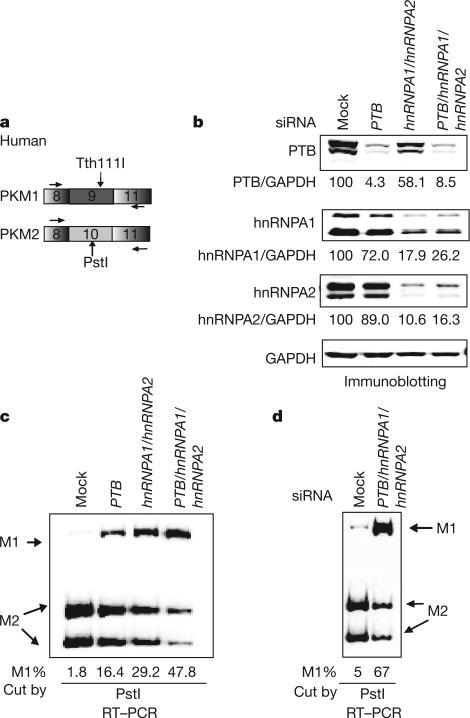
Because the locations of hnRNPA1/hnRNPA2 and PTB binding sites flanking E9 overlap elements critical to exon inclusion (the polypyrimidine tract for PTB11, the site of U1 snRNA–pre-mRNA base-pairing for hnRNPA1/hnRNPA212), we speculated that these proteins are inhibitors of E9 inclusion. To examine this possibility, we used short interfering RNA (siRNA) to deplete hnRNPA1, hnRNPA2 and/or PTB from HeLa cells. We assayed the PKM messenger RNA isoform ratio using RT–PCR followed by exon-specific restriction digestion (Fig. 2a). Knockdown of hnRNPA1 or hnRNPA2 in HeLa cells resulted in little change in splicing pattern (Supplementary Fig. 8). Because we have previously observed functional redundancy of hnRNPA1 and hnRNPA27, we next simultaneously depleted both proteins (Fig. 2b). This resulted in an increase in PKM1 mRNA, from 2% to 29%, and a concomitant decrease in PKM2 mRNA (Fig. 2c and Supplementary Fig. 8). PTB knockdown also increased the PKM1 isoform, to 16% (Fig. 2c; Supplementary Fig. 8), consistent with earlier observations13. Next, we simultaneously depleted all three factors, which further increased PKM1 levels to about 48% (Fig. 2c). Similar results were obtained using 293 cells, with the triple knockdown resulting in an increase from 5% to 67% PKM1 (Fig. 2d). Increases in PKM1 mRNA upon knockdown of hnRNPA1, hnRNPA2 and PTB were observed in all cell lines tested, including the breast cancer cell line MCF-7 and the glioblastoma cell line U87 (Supplementary Fig. 9). Knockdown of two other cancer-associated splicing factors in HeLa cells, the SR proteins ASF/SF2 and SRp20, while also resulting in slowed growth, failed to affect PKM1/PKM2 ratios significantly (Supplementary Fig. 8 and 10), indicating that the effects seen in PTB/hnRNPA1/hnRNPA2-depleted cells on PKM splicing are specific and not the result of pleiotropic effects due to changes in cell growth. Together, our results indicate that expression of PTB, hnRNPA1 and hnRNPA2 is the critical determinant of PKM isoform in transformed cells.
Figure 2. PTB, hnRNPA1 and hnRNPA2 are required for high PKM2/PKM1 mRNA ratios.
a, Scheme for assaying PKM1/PKM2 ratios in human cells. b, Immunoblots showing protein levels after the indicated siRNA treatment. Protein bands were quantified after LI-COR Odyssey scanning and normalized to GAPDH. c, The indicated splicing factors were depleted by siRNA, followed by PKM splicing assay outlined in a. Products corresponding to M1 and M2 isoforms are indicated with arrows. The PKM1 percentage is indicated below. d, PKM1 and PKM2 levels assayed after the indicated siRNA treatment in 293 cells.
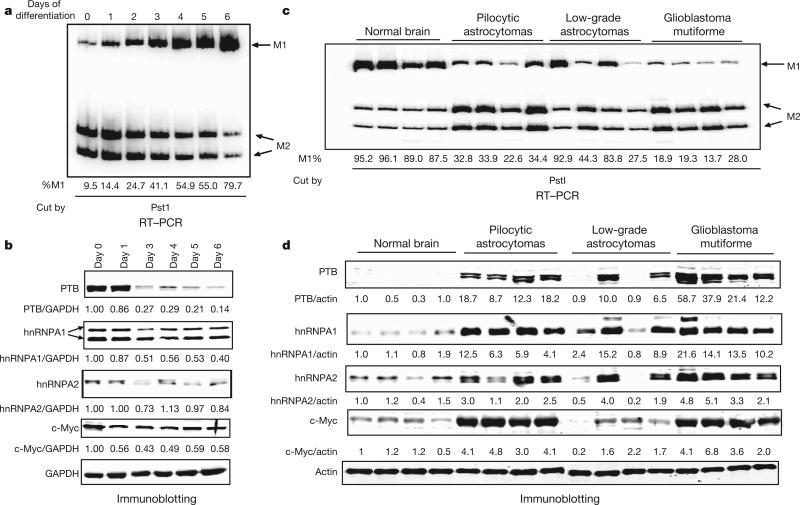
We next wished to determine whether expression levels of PTB, hnRNPA1 and hnRNPA2 and alternative splicing of PKM are correlated. We first examined whether changes in PTB, hnRNPA1 and hnRNPA2 levels correlate with changes in PKM splicing during switching from growth to quiescence. To this end, we used the mouse myoblast cell line C2C12, which, when grown to confluence and then switched to low-serum medium, undergoes myogenic differentiation, a process that includes PKM2 to PKM1 switching14. We differentiated C2C12 cells for 6 days, and used RT–PCR followed by restriction digestion to assess the PKM1/PKM2 ratio each day. We observed a large increase in Pkm1 and a corresponding decrease in Pkm2 mRNA during differentiation (Fig. 3a). We then prepared lysates of C2C12 cells at time points throughout differentiation and examined protein levels by immunoblotting (Fig. 3b). PTB expression dropped over 70% by day 3 of differentiation, after which it remained stable, consistent with previous studies15. We also observed an approximately 50% decrease in hnRNPA1 levels by day 3 of differentiation, although no significant changes were observed in the level of hnRNPA2. This result is consistent with a role for PTB and hnRNPA1 in maintaining high PKM2 levels in proliferating C2C12 cells.
Figure 3. Expression of PTB, hnRNPA1, hnRNPA2 and c-Myc correlates with PKM2 expression in C2C12 cells and tumours.
a, PKM splicing assay after the indicated number of days of C2C12 differentiation. b, Immunoblots for the indicated proteins were performed throughout differentiation, and normalized to GAPDH (day 0 5 1). c, RNA was extracted from brain tissue or tumour samples and assayed for PKM mRNA isoforms. d, Lysates were immunoblotted for PTB, hnRNPA1, hnRNPA2 or c-Myc and normalized to actin. Sample order is the same for RT–PCR and immunoblotting.
Because of the importance of the PKM2 isoform to the growth of cancer cells, we next examined human glioma tumour samples for a correlation between PTB, hnRNPA1 and hnRNPA2 expression and PKM splicing. We first assayed PKM1 and PKM2 mRNA levels as described earlier. Normal brain tissue ranged from 4% to 13% PKM2, pilocytic astrocytoma samples expressed approximately 66–77% PKM2, low-grade astrocytomas ranged from 7% to 73%, and glioblastoma multiforme samples expressed 72–86% PKM2 (Fig. 3c). To explore a potential correlation between elevated PKM2 mRNA levels and expression of the regulatory proteins we identified, we performed immunoblots for PTB, hnRNPA1 and hnRNPA2. Notably, all high-PKM2 tumours expressed elevated levels of PTB, hnRNPA1 and hnRNPA2, with the most striking overexpression in glioblastoma multiforme samples (Fig. 3d). Consistent with their uniformly high PKM2 expression, all four pilocytic astrocytoma samples also showed overexpression of PTB, hnRNPA1 and hnRNPA2. In low-grade astrocytomas the two high-PKM2 tumours showed elevated expression of the three proteins, whereas the two low-PKM2 tumours showed expression levels similar to normal brain. Immunoblotting for four other splicing factors (ASF/SF2, Tra2β, TLS (also called FUS) and hnRNPK) revealed no correlation with PKM2 expression (Supplementary Fig. 11), indicating that the correlation between an elevated PKM2/PKM1 mRNA ratio and overexpression of PTB, hnRNPA1 and hnRNPA2 is specific and not reflective of a general property of splicing factors.
The tight coupling of PKM2 expression to proliferation suggests that expression of the PKM splicing regulatory proteins we identified might be under the control of a proliferation-associated regulatory mechanism. A strong candidate to control this is the oncogenic transcription factor c-Myc, which, like PTB, hnRNPA1 and hnRNPA2, is upregulated in glioblastoma multiforme16, and has been shown to bind the PTB, hnRNPA1 and hnRNPA2 promoters17,18 and upregulate the expression of all three19,20. Consistent with a role for c-Myc in PTB, hnRNPA1 and hnRNPA2 regulation, we observed a near perfect correlation between the levels of c-Myc and those of PTB, hnRNPA1 and hnRNPA2 in gliomas and differentiating C2C12 cells (Fig. 3b, d). Furthermore, the transcription factor N-Myc, which is closely related to c-Myc21, was upregulated in pilocytic astrocytomas and to a lesser extent in glioblastoma multiforme samples (Supplementary Fig. 11), indicating that this protein may in some cases contribute to PTB, hnRNPA1 and hnRNPA2 upregulation.
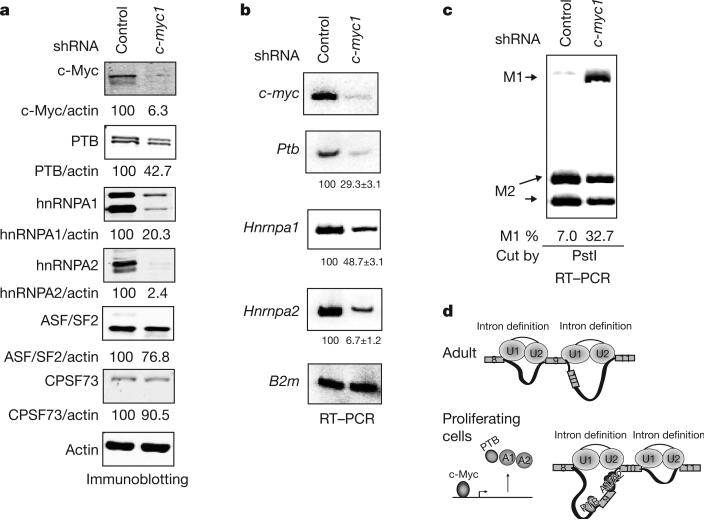
We next examined directly the involvement of c-Myc in expression of PTB, hnRNPA1 and hnRNPA2 and splicing regulation of PKM. We first asked whether decreasing c-Myc levels can affect PTB, hnRNPA1 and hnRNPA2 levels and the Pkm1/Pkm2 mRNA ratio. To this end, we stably transfected NIH-3T3 cells with vectors bearing a puromycin-resistance marker that express either a c-Myc-targeting short hairpin RNA (shRNA) or a control shRNA. Immunoblotting showed a reduction in c-Myc levels in cells stably transfected with c-myc shRNA compared to control cells (Fig. 4a). Protein levels of PTB, hnRNPA1 and hnRNPA2 were also significantly reduced after depletion of c-Myc, in contrast with two other RNA processing factors not implicated in PKM splicing regulation; that is, ASF/SF2 and CPSF73 (Fig. 4a). Ptb, Hnrnpa1 and Hnrnpa2 mRNA levels were also significantly reduced in the knockdown cells (Fig. 4b), supporting the idea that c-Myc regulates transcription of these genes. Importantly, the cells expressing the c-myc shRNA showed a pronounced increase in the Pkm1/Pkm2 ratio, expressing 33% Pkm1 mRNA compared to 7% in the control (Fig. 4c). A separate line stably expressing a second c-myc shRNA revealed a similarly elevated Pkm1/Pkm2 ratio, as well as reduced levels of PTB, hnRNPA1 and hnRNPA2, showing that the observed effects were not due to off-target effects of the c-myc shRNA (Supplementary Fig. 12). Additionally, we transiently co-transfected HeLa cells with an hnRNPA1 promoter-luciferase construct with a c-Myc expression vector22, which resulted in a dose- and c-Myc-binding-site-dependent increase in promoter activity (Supplementary Fig. 13).
Figure 4. c-Myc upregulates PTB, hnRNPA1 and hnRNPA2 and alters PKM splicing.
a, Immunoblotting using NIH-3T3 cells stably expressing control or c-Myc-targeting shRNAs. Signals were quantified and normalized to actin. b, RT–PCR using the same cell lines as in a, using B2m (β2-μglobulin) as a loading control. Real-time RT–PCR was performed separately to quantify the relative levels of Ptb, Hnrnpa1 and Hnrnpa2 mRNAs in control and c-Myc knockdown cells, using Rpl13a as a reference gene. Relative levels of each are shown below each panel, with s.d. indicated (n = 3). c, Pkm1/Pkm2 ratios in control and c-Myc knockdown cells determined as in Fig. 2a. d, A model for PKM splicing regulation. Top: in adult tissues, low expression of PTB, hnRNPA1 and hnRNPA2 allows for recognition of E9 by the splicing machinery and disrupts intronic structures favourable for E10 inclusion. Bottom: in embryonic and cancer cells, PTB, hnRNPA1 and hnRNPA2 are upregulated, bind to splicing signals flanking E9 and repress its inclusion. Binding of these proteins around E9 and possibly to other sites creates an intronic structure favourable to E10 inclusion.
The above results demonstrate a direct role for c-Myc in maintaining high PTB, hnRNPA1 and hnRNPA2 levels in NIH-3T3 cells. In contrast, c-Myc knockdown in HeLa cells revealed only a small decrease in PTB, hnRNPA1 and hnRNPA2 levels, and no change in the PKM1/PKM2 ratio (Supplementary Fig. 14), indicating that factors other than c-Myc might promote PTB, hnRNPA1 and hnRNPA2 expression in these cells. One possibility is the transcription factor E2F1, which, like c-Myc, binds upstream of all three genes18. However, knockdown of E2F1, or of Rb, a negative regulator of E2F family transcription factors23, resulted in little change in PTB, hnRNPA1 and hnRNPA2 levels (C.J.D., M.C. and J.L.M., unpublished data). However, because the E2F and Rb families exhibit redundancy, this result does not rule out involvement of the E2F/Rb pathway in regulation of PTB, hnRNPA1 and hnRNPA2. Indeed, because of their importance to proliferating cells, it is likely that PTB, hnRNPA1 and hnRNPA2 can be upregulated by proliferation-associated factors in addition to c-Myc.
The fact that PTB, hnRNPA1 and hnRNPA2 depletion results in switching to the PKM1 isoform suggests that RNA binding proteins can control the outcome of a mutually exclusive splicing event by simultaneously acting as repressors of one exon (E9) and activators of the other (E10) (Fig. 4d). Although it is easy to envision how these proteins exclude E9, how might PTB, hnRNPA1 and hnRNPA2 promote E10 inclusion? A variety of RNA binding proteins, including hnRNPA1 and hnRNPA2, have been shown to stimulate splicing of an adjacent exon through intronic binding sites24. One proposed mechanism for this is intron definition, in which intron-binding proteins induce intronic structures conducive to inclusion of the neighbouring exon24. We propose that, like many alternatively spliced exons, PKM E10 is poorly recognized by the splicing machinery in the absence of adjacent intron definition, and such a structure is promoted by PTB, hnRNPA1 and hnRNPA2 binding (Fig. 4d).
We have demonstrated a critical functional consequence for observations connecting PTB, hnRNPA1 and hnRNPA2 upregulation with cell proliferation25,26, transformation27,28 and a wide variety of cancers (for example, see refs 26, 27, 29, 30). Given the critical role of these proteins in promoting PKM2 production in tumours, overexpression of some combination of them is, like PKM2 expression, likely to be a general phenomenon in cancer. The fact that the proteins show some redundancy in promoting PKM2 splicing may ensure robust re-expression of PKM2 in tumours.
METHODS SUMMARY
Ultraviolet crosslinking substrates were cloned into the pcDNA3 vector (Invitrogen) and ultraviolet crosslinking was performed as previously described7. Mutations were introduced in EI9 by PCR-based site-directed muta-genesis. Biotinylated RNAs for affinity purification were purchased from Dharmacon, and RNA affinity chromatography was carried out as described7. Immunoprecipitations were carried out using protein A-agarose beads (Roche). RNA interference was performed as described7. We transfected 50 pmol of hnRNPA1 siRNA and 25 pmol of other siRNA duplex in a 24-well plate. After 72 h, we collected cells for RNA isolation and immunoblotting. C2C12 cells were grown in DMEM (Invitrogen) supplemented with 20% fetal bovine serum (FBS) (Hyclone) at 37°C in 5% CO2. For differentiation treatment, C2C12 cells were plated on gelatine-coated plates, allowed to reach confluence, and then switched to DMEM 2% donor equine serum (Hyclone). Human brain and glioma samples were obtained from the Bartoli Brain Tumour Bank at the Columbia University Medical Center. Samples were homogenized and used for Trizol RNA extraction and western blotting as described30. In all cases, immunoblots were scanned and quantified using the LI-COR Odyssey system. c-myc shRNA DNA sequences were purchased from Invitrogen and cloned into the pRS vector (Origene). shRNA constructs were transfected into NIH-3T3 cells and stable cell lines were selected with puromycin for RNA isolation and immunoblotting. PKM1/PKM2 ratio was analysed by extracting total RNA from cells and tissue samples and performing RT–PCR followed by PstI, Tth111I, or EcoNI digestion. qPCR for PTB, hnRNPA1 and hnRNPA2 in control and c-Myc knockdown cells was performed with SYBR green from Fermentas using the Applied Biosystems 7300 real-time PCR system. hnRNPA1 promoter sequence for dual luciferease reporter (DLR) assay was cloned into the PGL3-enhancer vector (Promega) and DLR assays were performed using the Dual Luciferase Reporter Assay System (Promega).
Supplementary Material
Supplementary Figure 1: Mapping of strong ~35-40 kDa crosslinking in the EI9 substrate. Truncations in EI9 were introduced with the indicated restriction enzymes (left). Strong crosslinking mapped to EI(1a) (not shown). Sections of EI9(1a) were cloned and used for UV crosslinking in HeLa NE.
Supplementary Figure 2: hnRNP A1 binds to EI9(50-68). After RNA affinity chromatography using biotinylated EI9(50-68), protein samples from EI9(50-68) (lane 1) and no RNA control (lane 2) were separated on 10% SDS-PAGE and analyzed by immunoblotting using an anti-hnRNP A1 antibody.
Supplementary Figure 3: hnRNP A1 binding to EI9(1a) is specifically decreased by G3C mutation. 32P-UTP labeled EI9(1a) RNA (see diagram in Supplementary Figure 1) was incubated with HeLa NE, in the presence of increasing amount (0, 1.5, 3, 7.5 μM) of cold WT EI9(1a) RNA (lanes 1-4) or cold EI9(1a) G3C mutant RNA (lanes 5-8), followed by UV crosslinking..
Supplementary Figure 4: Mutation of hnRNPA1/A2 binding site increases splicing of E9 containing in vitro splicing substrate. Schematic diagram of in vitro splicing constructs derived from AdML pre-mRNA substrate (top). In vitro splicing in HeLa nuclear extract using WT or G3C substrates.
Supplementary Figure 5: Minigene with mutated hnRNPA1/A2 binding site increases inclusion of E9 in HeLa cells. The mutation results in an increase in E9 and E10 double inclusion (left panel). Single inclusion band at left G3C lane contains a higher proportion of E9 containing transcripts as determined by Pst I digestion (right panel).
Supplementary Figure 6: a, PTB binds to biotinylated RNA oligo I8. After affinity chromatography using biotinylated I8 RNA supernatants (lanes 1 and 2) and protein samples bound to the beads from I8 (lane 4) and no RNA control (lane 3) were separated on 10% SDS-PAGE and analyzed by immunoblotting using an anti-PTB antibody (BB7). b, After RNA affinity chromatography using biotinylated I8, protein samples bound to the beads from I8 (lane 2) and no RNA control (lane 1) were separated on 10% SDS-PAGE and Coomassie stained. Positions of bands excised for mass spectrometry are indicated.
Supplementary Figure 7: PTB binding to I8 RNA is specifically abolished by a C to G mutation. 32P -UTP labeled I8 RNA (see sequence in Fig. 1) was incubated with HeLa NE in the presence of increasing amounts (0, 2, 3,3 6 μM) of cold I8 WT RNA (lanes 1–4) or cold I8mu RNA (lanes 5–8), followed by UV crosslinking.
Supplementary Figure 8: Knockdowns of splicing factors have differing effects on PKM splicing. The indicated splicing factors were depleted by siRNA, followed by PKM splicing assay outlined in Fig. 1a. Products corresponding to M1 and M2 are indicated with arrows. PCR products for the PKM amplicon and a load control, μglobulin, are displayed at left. The PKM amplicons after cutting with the indicated enzyme are shown in the middle and right panels.
Supplementary Figure 9: Assay of PKM splicing after siRNA depletion of ASF/SF2, or simultaneous knockdown of PTB/A1/A2. siRNAs were transfected into human breast cancer cell line MCF-7 and glioblastoma cell line U87. Cells were collected after three days. Total RNA was extracted and PKM isoform ratio was estimated as described in Fig. 2a.
Supplementary Figure 10: SRp20 knockdown does not affect the PKM1/2 mRNA ratio. a, Immunoblotting to detect SRp20 protein levels in HeLa cells treated with either control shRNA (lane 1) or SRp20-targeting shRNA (lane 2). b, PKM1/2 ratios in control and SRp20 knockdown cells determined as in Fig. 2a.
Supplementary Figure 11: Lysates of normal brain and glioma samples were immunoblotted for ASF/SF2, Tra2β, TLS, hnRNP K and n-Myc and normalized to actin. Quantitation of Tra2β, TLS and hnRNP K is indicated below each immunoblot (normal brain sample one=1). Sample order is the same as for RT-PCR and immunoblotting in Fig. 3c and 3d.
Supplementary Figure 12: A stable cell line expressing a second c-Myc-targeting shRNA also reduced PTB/A1/A2 and PKM2 mRNA levels. a, Immunoblotting using NIH-3T3 cells stably expressing control shRNA or c-Myc-targeting shRNA2. Signals were quantitated and normalized to actin. b, RT-PCR was used to determine relative mRNA levels in cells stably expressing control shRNA or c-Myc-targeting shRNA2. c, PKM1/2 ratios in control and c-Myc knockdown cells determined as in Fig. 2a.
Supplementary Figure 13: c-Myc upregulates transcription from the hnRNP A1 promoter via E boxes. a, Diagrams showing hnRNP A1 promoter-Luciferase reporter constructs. E boxes (CACGTG) are putative c-Myc binding sites, which are located within a ~700 nt hnRNP A1 promoter region cloned upstream of the luciferase gene. pGL3-A1p contains the wild-type promoter region. pGL3-A1pMu contains mutated E boxes (indicated in the diagram). b, Results of dual luciferase reporter assays showing relative luciferase activity (top) activated by overexpression of c-Myc in HeLa cells by co-transfection of c-Myc expression vector and pGL3-A1p (lanes 1-3) or pGL3-A1pMu (lanes 4-6). Luciferase activity was normalized to Renilla luciferase activity, and pGL3-A1p activity was set as 1. Immunoblotting using c-Myc antibody to show c-Myc overexpression in transfected cells (bottom).
Supplementary Figure 14: c-Myc knockdown in HeLa cells does not change PKM splicing. a, Immunoblotting using HeLa cells transiently expressing control or human c-Myc-targeting shRNAs. b, PKM1/2 ratios in control and c-Myc knockdown cells determined as in Fig. 2a.
Acknowledgements
We thank D. Black for BB7 antibody; T. Kashima for hnRNP A1/A2 siRNA; R. Prywes and E. Henckles for C2C12 cells; M. Sheetz and X. Zhang for NIH-3T3 cells; C. Prives, T. Barsotti and L. Biderman for MCF-7 cells, Rb and E2F1 siRNA; R. Dalla-Favera and Q. Shen for anti-c-Myc antibodies and c-Myc expression vector; R. Eisenmann for N-Myc antibody; and members of the Manley laboratory for discussions. This work was supported by grants from the Avon Foundation and the NIH.
METHODS
Plasmid constructs
Long ultraviolet crosslinking substrates (EI9, EI10) were prepared by amplifying fragments from HeLa genomic DNA using Pfu turbo (Stratagene), and cloning the products into pcDNA3 (Invitrogen). EI9(1–20), EI9(21–49) and EI9(50–68), and I8, I8mu and I9 DNA sequences were ordered from Invitrogen and cloned into pcDNA3. Primers used to amplify genomic DNA fragments were: EI9 forward, 5’-CGCGGATCCTTCTTATAAGTGTTTA GCAGCAGCT-3′, reverse, 5′-CGGAATTCACTGAGCCACAGGACCCTTTG-3′; EI10 forward, 5′-CGCGGATCCCTCCTTCAAGTGCTGCAGTG-3′, reverse, 5′-CGGAATCCTGGGCCCAGGGAAGGGG-3′; I8E9 forward, 5′-CCCAAGC TTAAATTCCCCATTCTGTCTTCCCATG-3′, reverse, 5′-CGGGATCCCTGC CAGACTCCGTCAGAACT-3′; I9E10 forward, 5′-CCCAAGCTTCTGTCCGG TGACTCTTCCCC-3′, reverse, 5′-CGGGATCCCTGCCAGACTTGGTGAGG ACG-3′. Mutations were introduced in EI9 by PCR-based site-directed mutagenesis. Mouse c-myc and control shRNA DNA sequences were ordered from Invitrogen and cloned into pRS vector (Origene) with BamHI and HindIII. The hnRNPA1 promoter region, either wild-type or the E box mutant, was cloned into PGL3-enhancer vector (Promega).
Antibodies
The following antibodies were used in this study: BB7 for human PTB immunoprecipitation (gift from D. Black), 3H8 for mouse/human PTB immunoblots (Sigma), MC3 for U2AF65 (Sigma), anti-HA (Covance) DP3D3 for hnRNPA2 (Abcam), N-262 for c-Myc (Santa Cruz), anti-actin (Sigma), anti-GAPDH (Sigma), 9H10 for hnRNPA1 (Sigma), monoclonal antibody 104 for SRp20.
Ultraviolet crosslinking, RNA affinity purification and immunoprecipitation assays
We carried out ultraviolet crosslinking as previously described7. Briefly, we linearized the ultraviolet crosslinking plasmids with an appropriate restriction enzyme and synthesized the RNAs with [32P]UTP or [32P]CTP. We incubated 1 × 105 c.p.m. RNAs with 10 μg HeLa or C2C12 NE in buffer D in a 20 μl reaction at 30°C for 15 min, then irradiated the samples with ultraviolet light in a Stratalinker 1800 (Stratagene), digested them with RNase A (10 μg ml−1) and resolved them by SDS–PAGE. The RNA affinity pull-down experiment and immunoprecipitation was preformed as described7. The 5′ biotinylated EI9(50–68) and I8 RNA oligonucleotides were purchased from Dharmacon. Antibodies were bound to protein A-agarose beads before immunoprecipitation. We used the following antibodies for immunoprecipitation: BB7 for PTB, and MC3 for U2AF65.
In vitro and in vivo splicing assays
A minigene containing PKM gene exon 8, exon 9, exon 10, exon 11 and flanking regions was cloned into pcDNA3 vector (Invitrogen). G to C mutation was introduced in the minigene by PCR-based site-directed mutagenesis11. Wild-type and mutated minigene vectors were transfected into HeLa cells. Twenty-four hours after transfection, cells were collected and PKM1/PKM2 ratio was analysed using RT–PCR followed by PstI digestion. In vitro splicing substrates were constructed by replacing the first exon and downstream intronic sequence of AdML pre-mRNA with PKM exon 9 and downstream intron 9 sequences or sequence with mutated hnRNPA1 binding site. Pre-mRNA substrates were synthesized by in vitro transcription using T7 RNA polymerase (Promega) following product protocol. In vitro splicing of the wild-type and mutated pre-mRNA was carried out using HeLa nuclear extract as described31.
RNA interference
We carried out RNA interference of PTB and hnRNPA1 and hnRNPA2 as described7. Briefly, we plated HeLa, 293, MCF-7, or U87 cells at 2.5–3 × 104 cells per well in 24-well plates. The next day, we mixed 50 pmol of hnRNPA1 duplex RNA and 25 pmol of the other duplex RNAs with 1.5 μl lipofectamine 2000 transfection reagent (Invitrogen) plus 100 μl of Opt-MEM medium and added this to cells after RNA duplex–lipid complex formation. For double and triple knockdowns in HeLa and 293 cells, RNA duplexes were transfected simultaneously. The control RNA duplex was used to ensure that parallel experiments had equal amounts of RNA. In MCF-7 and U87 cells, the second and third RNA duplexes were transfected 6 h after the previous transfection. Seventy-two hours after transfection, we collected cells for RNA isolation and immunoblotting. We used the following siRNAs (Dharmacon; the sense strand sequences are given): human hnRNPA1, 5′-CAGCUGAGGAAGCUCUUCA-3′; human hnRNPA2, 5′-GGAACAGUUCCGUAAGCUC-3′; human PTB, 5′-GCCUCAACGUCAAGUACAA-3′. ASF/SF2 depletion was performed as previously described7.
c-myc shRNA stable cell lines
Stable cell lines expressing c-myc shRNAs or control shRNA were obtained by transfecting pRS-shRNA vectors into NIH-3T3 cells followed by drug selection. Cells were plated in 10 cm plates. The next day, transfected cells were diluted and medium was replaced with medium containing a final concentration of 3 μg ml−1 puromycin. After 7–10 days, a mixture of fast- and slow-growing colonies appeared in cells transfected with c-myc shRNA, whereas only fast-growing colonies appeared in cells transfected with control shRNA. Single slow-growing colonies were isolated and cultured for c-Myc expressing cells. c-Myc expression was examined by immunoblotting. Positive colonies were collected for RT–PCR and western blotting. The following sense shRNA sequences were used: control, 5′-GAGGCTTCTTATAAGTGTTTACTCGAGTAAACACTTATAAGAAGCCTCTTTTT-3′; mouse c-myc shRNA1, 5′-CATCCTATGTTGCGGTCGCTACTCGAGTAGCGACCGCAACATAGGATGTTTTT-3′; mouse c-myc shRNA2, 5′-CGGACACACAACGTCTTGGAAC TCGAGTTCCAAGACGTTGTGTGTCCGTTTTT-3′; human c-Myc shRNA, 5′-CCATAATGTAAACTGCCTCAACTCGAGTTGAGGCAGTTTACATTATGGTTTTT-3′.
RT–PCR
Total RNA was extracted from tissue culture and human brain tumour samples using Trizol (Invitrogen) according to the manufacturer's instructions. Total RNA (2.5–5 μg) was used for each sample in a 20 μl reaction with 0.5 μl of SuperScript III RT (Invitrogen). One microlitre of the cDNA library was used in a 50 μl PCR reaction containing 3 μCi [32P]dCTP. Ten microlitres of the PCR products was digested by PstI and Tth111 I (human PKM) or EcoNI (mouse Pkm) and the products were resolved by 6% non-denaturing PAGE. Primers used in the PCR reactions were: human PKM exon 8 forward, 5′-CTGAAGGCAGTGATGTGGCC-3′; human PKM exon 11 reverse, 5′-ACCCGGAGGTCCACGTCCTC-3′; mouse Pkm exon 8 forward, 5′-CAAGGGGACTACCCTCTGG-3′; mouse Pkm exon 11 reverse, 5′-ACACGAAGGTCGACATCCTC-3′; B2M forward, 5′-GGCTATCCAGCGTACTCCAAA-3′, reverse, 5′-CGGCAGGCATACTCATCTTTTT-3′; B2m forward, 5′-TTCTGGTGCTTGTCTCACTGA-3′, reverse, 5′-CAGTATGTTCGGCTTCCCATTC-3′. qRT–PCR was performed using the following primers: mouse Hnrnpa1 forward, 5′-TGGAAGCAATTTTGGAGGTGG-3′, reverse, 5′-GGTTCCGTGGTTTAGCAAAGT-3′; mouse Hnrnpa2 forward, 5′-AAGAAATGCAGGAAGTCCAAAGT-3′, reverse, 5′-CTCCTCCATAACCAGGGCTAC-3′; mouse Ptb forward, 5′-AGCAGAGACTACACTCGACCT-3′, reverse, 5′-GCTCCTGCATACGGAGAGG-3′; mouse Rpl13a forward, 5′-GGGCAGGTTCTGGTATTGGAT-3′, reverse, 5′-GGCTCGGAAATGGTAGGGG-3′. Relative amounts of mRNA were calculated using the comparative Ct method.
Cell culture and differentiation
C2C12 cells were grown in DMEM (Invitrogen) supplemented with 20% fetal bovine serum (Hyclone) at 37°C in 5% CO2. For differentiation treatment, C2C12 were plated on gelatin-coated plates, allowed to reach confluence, and then switched to DMEM 2% donor equine serum (Hyclone). HeLa and 293 cells were grown in DMEM, 10% FBS. NIH-3T3 cells were grown in DMEM, 10% bovine calf serum (Hyclone).
Human brain tumour samples
De-identified brain and glioma samples were obtained from the Bartoli Brain Tumour Bank at the Columbia University Medical Center. Non-cancerous samples removed from epileptic patients were used for normal brain. Approximately 25–200 mg of each sample was obtained. Half of the homogenate was used for Trizol RNA extraction, the other half of each sample was processed for immunoblotting as described30.
Dual luciferase reporter (DLR) assay
c-Myc expression vector and hnRNPA1 promoter vector were co-transfected into HeLa cells. Twenty-four hours after transfection, cells were collected and DLR assays were preformed using the Dual Luciferase Reporter Assay System (Promega) following product protocol.
Footnotes
The authors declare no competing financial interests.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T, Marquardt C, Foker J. Aerobic glycolysis during lymphocyte proliferation. Nature. 1976;261:702–705. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- 5.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 6.Takenaka M, et al. Alternative splicing of the pyruvate kinase M gene in a minigene system. Eur. J. Biochem. 1996;235:366–371. doi: 10.1111/j.1432-1033.1996.00366.x. [DOI] [PubMed] [Google Scholar]
- 7.Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum. Mol. Genet. 2007;16:3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- 8.Del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spellman R, Smith CW. Novel modes of splicing repression by PTB. Trends Biochem. Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Sauliere J, Sureau A, Expert-Bezancon A, Marie J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the β-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol. Cell. Biol. 2006;26:8755–8769. doi: 10.1128/MCB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harada Y, Nakamura M, Asano A. Temporally distinctive changes of alternative splicing patterns during myogenic differentiation of C2C12 cells. J. Biochem. 1995;118:780–790. doi: 10.1093/oxfordjournals.jbchem.a124980. [DOI] [PubMed] [Google Scholar]
- 15.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 19.Shiio Y, et al. Quantitative proteomic analysis of Myc oncoprotein function. EMBO J. 2002;21:5088–5096. doi: 10.1093/emboj/cdf525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlosser I, et al. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene. 2005;24:520–524. doi: 10.1038/sj.onc.1208198. [DOI] [PubMed] [Google Scholar]
- 21.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu KJ, Mattioli M, Morse HC, III, Dalla-Favera R. c-MYC activates protein kinase A (PKA) by direct transcriptional activation of the PKA catalytic subunit beta (PKA-Cβ) gene. Oncogene. 2002;21:7872–7882. doi: 10.1038/sj.onc.1205986. [DOI] [PubMed] [Google Scholar]
- 23.Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Contreras R, et al. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biamonti G, et al. Human hnRNP protein A1 gene expression. Structural and functional characterization of the promoter. J. Mol. Biol. 1993;230:77–89. doi: 10.1006/jmbi.1993.1127. [DOI] [PubMed] [Google Scholar]
- 26.Zerbe LK, et al. Relative amounts of antagonistic splicing factors, hnRNP A1 and ASF/SF2, change during neoplastic lung growth: implications for pre-mRNA processing. Mol. Carcinog. 2004;41:187–196. doi: 10.1002/mc.20053. [DOI] [PubMed] [Google Scholar]
- 27.He X, et al. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26:4961–4968. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanamura A, Caceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J, et al. Differential expression of the early lung cancer detection marker, heterogeneous nuclear ribonucleoprotein-A2/B1 (hnRNP-A2/B1) in normal breast and neoplastic breast cancer. Breast Cancer Res. Treat. 2001;66:217–224. doi: 10.1023/a:1010631915831. [DOI] [PubMed] [Google Scholar]
- 30.Jin W, McCutcheon IE, Fuller GN, Huang ES, Cote GJ. Fibroblast growth factor receptor-1 α-exon exclusion and polypyrimidine tract-binding protein in glioblastoma multiforme tumors. Cancer Res. 2000;60:1221–1224. [PubMed] [Google Scholar]
- 31.Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Mapping of strong ~35-40 kDa crosslinking in the EI9 substrate. Truncations in EI9 were introduced with the indicated restriction enzymes (left). Strong crosslinking mapped to EI(1a) (not shown). Sections of EI9(1a) were cloned and used for UV crosslinking in HeLa NE.
Supplementary Figure 2: hnRNP A1 binds to EI9(50-68). After RNA affinity chromatography using biotinylated EI9(50-68), protein samples from EI9(50-68) (lane 1) and no RNA control (lane 2) were separated on 10% SDS-PAGE and analyzed by immunoblotting using an anti-hnRNP A1 antibody.
Supplementary Figure 3: hnRNP A1 binding to EI9(1a) is specifically decreased by G3C mutation. 32P-UTP labeled EI9(1a) RNA (see diagram in Supplementary Figure 1) was incubated with HeLa NE, in the presence of increasing amount (0, 1.5, 3, 7.5 μM) of cold WT EI9(1a) RNA (lanes 1-4) or cold EI9(1a) G3C mutant RNA (lanes 5-8), followed by UV crosslinking..
Supplementary Figure 4: Mutation of hnRNPA1/A2 binding site increases splicing of E9 containing in vitro splicing substrate. Schematic diagram of in vitro splicing constructs derived from AdML pre-mRNA substrate (top). In vitro splicing in HeLa nuclear extract using WT or G3C substrates.
Supplementary Figure 5: Minigene with mutated hnRNPA1/A2 binding site increases inclusion of E9 in HeLa cells. The mutation results in an increase in E9 and E10 double inclusion (left panel). Single inclusion band at left G3C lane contains a higher proportion of E9 containing transcripts as determined by Pst I digestion (right panel).
Supplementary Figure 6: a, PTB binds to biotinylated RNA oligo I8. After affinity chromatography using biotinylated I8 RNA supernatants (lanes 1 and 2) and protein samples bound to the beads from I8 (lane 4) and no RNA control (lane 3) were separated on 10% SDS-PAGE and analyzed by immunoblotting using an anti-PTB antibody (BB7). b, After RNA affinity chromatography using biotinylated I8, protein samples bound to the beads from I8 (lane 2) and no RNA control (lane 1) were separated on 10% SDS-PAGE and Coomassie stained. Positions of bands excised for mass spectrometry are indicated.
Supplementary Figure 7: PTB binding to I8 RNA is specifically abolished by a C to G mutation. 32P -UTP labeled I8 RNA (see sequence in Fig. 1) was incubated with HeLa NE in the presence of increasing amounts (0, 2, 3,3 6 μM) of cold I8 WT RNA (lanes 1–4) or cold I8mu RNA (lanes 5–8), followed by UV crosslinking.
Supplementary Figure 8: Knockdowns of splicing factors have differing effects on PKM splicing. The indicated splicing factors were depleted by siRNA, followed by PKM splicing assay outlined in Fig. 1a. Products corresponding to M1 and M2 are indicated with arrows. PCR products for the PKM amplicon and a load control, μglobulin, are displayed at left. The PKM amplicons after cutting with the indicated enzyme are shown in the middle and right panels.
Supplementary Figure 9: Assay of PKM splicing after siRNA depletion of ASF/SF2, or simultaneous knockdown of PTB/A1/A2. siRNAs were transfected into human breast cancer cell line MCF-7 and glioblastoma cell line U87. Cells were collected after three days. Total RNA was extracted and PKM isoform ratio was estimated as described in Fig. 2a.
Supplementary Figure 10: SRp20 knockdown does not affect the PKM1/2 mRNA ratio. a, Immunoblotting to detect SRp20 protein levels in HeLa cells treated with either control shRNA (lane 1) or SRp20-targeting shRNA (lane 2). b, PKM1/2 ratios in control and SRp20 knockdown cells determined as in Fig. 2a.
Supplementary Figure 11: Lysates of normal brain and glioma samples were immunoblotted for ASF/SF2, Tra2β, TLS, hnRNP K and n-Myc and normalized to actin. Quantitation of Tra2β, TLS and hnRNP K is indicated below each immunoblot (normal brain sample one=1). Sample order is the same as for RT-PCR and immunoblotting in Fig. 3c and 3d.
Supplementary Figure 12: A stable cell line expressing a second c-Myc-targeting shRNA also reduced PTB/A1/A2 and PKM2 mRNA levels. a, Immunoblotting using NIH-3T3 cells stably expressing control shRNA or c-Myc-targeting shRNA2. Signals were quantitated and normalized to actin. b, RT-PCR was used to determine relative mRNA levels in cells stably expressing control shRNA or c-Myc-targeting shRNA2. c, PKM1/2 ratios in control and c-Myc knockdown cells determined as in Fig. 2a.
Supplementary Figure 13: c-Myc upregulates transcription from the hnRNP A1 promoter via E boxes. a, Diagrams showing hnRNP A1 promoter-Luciferase reporter constructs. E boxes (CACGTG) are putative c-Myc binding sites, which are located within a ~700 nt hnRNP A1 promoter region cloned upstream of the luciferase gene. pGL3-A1p contains the wild-type promoter region. pGL3-A1pMu contains mutated E boxes (indicated in the diagram). b, Results of dual luciferase reporter assays showing relative luciferase activity (top) activated by overexpression of c-Myc in HeLa cells by co-transfection of c-Myc expression vector and pGL3-A1p (lanes 1-3) or pGL3-A1pMu (lanes 4-6). Luciferase activity was normalized to Renilla luciferase activity, and pGL3-A1p activity was set as 1. Immunoblotting using c-Myc antibody to show c-Myc overexpression in transfected cells (bottom).
Supplementary Figure 14: c-Myc knockdown in HeLa cells does not change PKM splicing. a, Immunoblotting using HeLa cells transiently expressing control or human c-Myc-targeting shRNAs. b, PKM1/2 ratios in control and c-Myc knockdown cells determined as in Fig. 2a.