Abstract
Within the Diptera and outside the suborder Brachycera, the blood feeding habit occurred at least twice, producing the present day sand flies, and the Culicomorpha, including the mosquitoes (Culicidae), black flies (Simulidae), biting midges (Ceratopogonidae) and frog feeding flies (Corethrellidae). Alternatives to this scenario are also discussed. Successful blood feeding requires adaptations to antagonize the vertebrate's mechanisms of blood clotting, platelet aggregation, vasoconstriction, pain and itching, which are triggered by tissue destruction and immune reactions to insect products. Saliva of these insects provides a complex pharmacological armamentarium to block these vertebrate reactions. With the advent of transcriptomics, the sialomes (from the Greek word sialo=saliva) of at least two species of each of these families have been studied (except for the frog feeders), allowing an insight into the diverse pathways leading to today's salivary composition within the Culicomorpha, having the sand flies as an outgroup. This review catalogs 1,288 salivary proteins in 10 generic classes comprising over 150 different protein families, most of which we have no functional knowledge. These proteins and many sequence comparisons are displayed in a hyperlinked spreadsheet that hopefully will stimulate and facilitate the task of functional characterization of these proteins, and their possible use as novel pharmacological agents and epidemiological markers of insect vector exposure.
Introduction
The blood feeding habit evolved independently several times among different insect orders, and even within insect orders, such as the Diptera (Ribeiro, 1995). Among the several challenges associated with this peculiar diet, the vertebrate response against blood loss, the hemostasis process, represents a formidable barrier to efficient blood feeding. Within seconds of vessel laceration, mammalian platelets adhere to each other, forming a plug, and produce or expose pro-clotting and vasoconstrictory substances. Immune reactions can lead to mast cell degranulation and the release of biogenic amines (mainly histamine and serotonin) and eicosanoids (mainly leukotriene C4 and prostaglandin D2) that induce host itching reactions and edema that can prevent feeding or even kill the micropredator. Perhaps for these reasons, insects evolved a salivary concoction that disarms their host's hemostasis and inflammation. Because vertebrate hemostasis and inflammation is complex and redundant, hematophagous insect saliva is also complex and redundant, containing dozens of active compounds (Ribeiro and Arca, 2009). Because this feeding mode evolved independently in several insect orders and families, the salivary composition among insects is typical of a convergent evolutionary scenario (Mans and Francischetti, 2010). On the other hand, the diversity among different genera within a same family has been also found to be large possibly due to the vertebrate host immune pressure over the salivary products, or due to the appearance of new hemostatic challenges that are posed by the evolving host genomes, such as the appearance of platelets in mammals, much more efficient than, for example, bird thrombocytes (Didisheim et al., 1959).
The Diptera were classically divided into two suborders, Nematocera, and Brachycera both of which have hematophagous flies, such as the mosquitoes, sand flies and black flies in the Nematocera, and horse flies, tsetse and stable flies, for example, in the Brachycera. This traditional division in two suborders is challenged because the Nematocera is considered paraphyletic regarding Brachycera (Yeates and Wiegmann, 1999), being replaced by several infra-orders, including the the Psycodomorpha that includes the family Psychodidae (sand flies), and the Culicomorpha on the other, thus postulating two independent events of blood feeding in the non-Brachycera flies (Null hypothesis). Notice that the common usage of Nematocera in this paper should be understood as “non-Brachycera” according to the current phylogenetic view. The Culicomorpha clade may have originated from a single blood feeding ancestor during the Triassic, over 200 million years ago (MYA). This ancestor gave rise to the 11 extant families, 4 of which retain blood feeding, namely the Culicidae (mosquitoes), Simuliidae (black flies), Ceratopogonidae (biting midges) and Corethrellidae (frog biting midges) (Grimaldi and Engel, 2005). One family, the Blephariceridae, (the net-winged midges) actually feeds on insect hemolymph, not vertebrate blood, and the remaining families (Chaoboridae, Dixidae, Niphomyiidae, Chironomidae, Thaumaleidae and Deuterophlebiidae) lost the blood feeding habit (Grimaldi and Engel, 2005). Within the Nematocera hematophagy is restricted solely to the adult stage, and in most cases solely to the adult female stage, such as in mosquitoes and sand flies, where it is essential for egg development. These insects, in addition to blood, will also take sugar meals, which energize flight and basal metabolic needs; accordingly, their saliva will reflect adaptations to sugar feeding as well, including the presence of antimicrobial compounds.
On the other hand, given the extensive losses proposed in blood-feeding lifestyles of the Culicomorpha an even more parsimonious scenario would be that hematophagy evolved in the last common ancestor of the Psychodomorpha and the Culicomorpha, with extensive losses in hematophagous behavior across a number of lineages. Alternatively, blood-feeding behavior could have evolved independently in each major family of the Nematocera (Mans and Francischetti, 2010; Pawlowski et al., 1996; Ribeiro, 1995).
Methodologies developed in the past 10 years created an unprecedented window from which the transcription repertoire of particular organs can be brought to light. In the case of blood feeding Nematocera, representative sialotranscriptomes (from the Greek sialo=saliva) exist from 4 of the 5 blood feeding families, excluding the Corethrellidae (Table I). With the exception of the frog biting flies, there are two or more sialotranscriptomes available for analysis for all other families of blood sucking Nematocera (Psychodidae, Culicidae, Ceratopogonidae and Simuliidae) allowing for an insight into the evolution of blood feeding within the Nematocera.
Table I.
Salivary transcriptomes and transcripts from Nematocera compiled in this review
The available data
Using the information from the sialotranscriptomes indicated in Table I, which includes the non-blood sucking mosquito Toxorhynchites amboinensis, a total of 1,280 proteins were retrieved from GenBank and from our previous annotation. These proteins were grouped either (i) by comparing their primary sequence against each other by the tool Blast and producing groups of sequences with a particular threshold of identity over 40% of their length (note that this relatively small value was chosen because many sequences are fragments or truncated) (columns CL to DH of table S1); or (ii) grouped following Psiblast at different levels of the switches −h and −j determining the e value threshold for inclusion of matches and number of iterations, respectively (columns DI to DN of table S1) (Altschul et al., 1997). The sequences obtained were automatically aligned with the program ClustalX (Thompson et al., 1997) if less than 100 sequences were found in a particular group. Sequences were also submitted to several servers of the Danish Center for Biological Sequence Analysis to obtain indications of secretory signals (signalP, secretomeP and targetP servers) (Bendtsen et al., 2004; Emanuelsson et al., 2000; Nielsen et al., 1997; Sonnhammer et al., 1998), transmembrane domains (tmhmm server) (Sonnhammer et al., 1998) and mucin-type galactosylation (NetOglyc server) (Julenius et al., 2005). The data was transferred to an Excel spreadsheet and manually annotated to help classification of the various protein families (Supplemental table S1). Notice that table S1 may be somewhat redundant by including proteins that are >95% similar among themselves, but on the other hand may provide indication of alleles. These can be easily picked up on columns DE or DG of table S1.
The salivary composition of blood sucking Nematocera
As mentioned above, the salivary composition of blood feeding insects is quite complex and varied, containing various enzymes, proteins with various protease inhibitor domains, mucins, kratagonists (chelators of agonists) (Ribeiro and Arca, 2009) and a large number of proteins that are only found in blood sucking insects, as indicated by comparisons of their primary structure to the non-redundant protein database (NR) of the National Center for Biotechnology Information (NCBI). Most proteins of this last group have unknown function, as will be indicated below.
Annotation of table S1 grouped the 1,280 proteins in 10 main classes representing 155 different protein families (excluding the 106 orphan proteins), summarized in Table II. These classes are organized in a gradient regarding a possible functional perspective, starting with enzymes (170 proteins in 23 families) and proteins with typical protease inhibitor domains, such as serpins or Kunitz domain containing polypeptides to proteins with unknown function, many of which are uniquely found within this 1,280 set.
Table II.
Summary classification of the protein families possibly associated with a salivary function in blood sucking Nematocera.
| Class | Total proteins | Total families |
|---|---|---|
| I – Ubiquitous protein families existing outside Diptera, function known or presumed | ||
| Enzymes | 170 | 23 |
| Ubiquitous protease inhibitor domains | 52 | 6 |
| Immunity-related proteins | 75 | 12 |
| Mucins | 106 | 12 |
| OBP superfamily | 186 | 14 |
| Other ubiquitous families | 22 | 5 |
| II – Ubiquitous protein families existing outside Insects, function unknown | 40 | 1 |
| III– Ubiquitous insect protein families existing outside Nematocera, function unknown | 31 | 6 |
| IV - Protein families exclusive of blood sucking Nematocera | 56 | 2 |
| V - Protein families specific of mosquitoes | ||
| Found in both culicines and anophelines | 83 | 12 |
| Uniquely found in culicines | 40 | 7 |
| Uniquely found in anophelines | 63 | 10 |
| Uniquely found in Aedes | 21 | 3 |
| Uniquely found in Culex | 33 | 1 |
| VI - Protein families specific of black flies | 123 | 24 |
| VII - Protein families specific of sand flies | 32 | 9 |
| VIII - Protein families specific of Culicoides | 41 | 8 |
| IX - Salivary-orphan proteins of conserved secreted families | 43 | 43 |
| X - Orphan proteins of unique standing | 63 | 63 |
| Total | 1280 | 261 |
A breakdown of the classes shown in Table II indicates the inner complexity of each class, and how much is known about the function of each subclass (Table III). Most of the protein families have no known function; only 19 of the 155 families have been studied so far with at least one published work available. Below follows a description of the different families, which should serve as a guide for browsing supplemental file S1.
Table III.
Classification and functional status of protein families putatively assigned a salivary function in blood feeding Nematocera.
| Class |
Number | Function known? | Reference |
|---|---|---|---|
| Subclass | |||
| Name | |||
| I – Ubiquitous protein families existing outside Diptera, function known or presumed | |||
|
| |||
| Enzymes | |||
|
| |||
| Phlebotomine Cimex-type apyrase | 11 | Y | (Valenzuela et al., 2001b) |
| 5'nucleotidas/Apyrase family | 17 | Y | (Champagne et al., 1995; Sun et al., 2006) |
| Adenosine deaminase family | 7 | Y | (Ribeiro et al., 2001) |
| Purine hydrolase | 3 | Y | (Ribeiro and Valenzuela, 2003) |
| Pyrophosphatase/Phosphodiesterase - Phlebotomine specific | 3 | ||
| Endonuclease | 11 | Y | (Calvo and Ribeiro, 2006) |
| Hyaluronidase | 5 | Y | (Cerna et al., 2002; Ribeiro et al., 2000) |
| Ribonucleases | 3 | ||
| Alkaline phosphatase | 2 | ||
| Serine proteases | 48 | ||
| Metalloprotease - may be housekeeping | 1 | ||
| Dipeptidyl peptidase | 1 | ||
| Cathepsins - maybe housekeeping | 2 | ||
| Glutamate carboxypeptidase - may be housekeeping | 1 | ||
| Phlebotomus phospholipase A2 | 3 | ||
| Mosquito lipase | 2 | ||
| Triacylglycerol lipase | 1 | ||
| Carboxylesterase | 1 | ||
| Destabilase family | 3 | ||
| Anopheline peroxidases | 4 | Y | (Ribeiro and Valenzuela, 1999) |
| Peroxiredoxin | 3 | ||
| Glycosidases | 34 | Y | (James et al., 1989; Marinotti et al., 1996; Marinotti et al., 1990; Marinotti and James, 1990) |
| Chitinase | 4 | ||
|
| |||
| Ubiquitous protease inhibitor domains | |||
|
| |||
| Serpin family | 10 | Y | (Stark and James, 1998) |
| Simulium Kunitz proteins | 6 | ||
| Culicoides Kunitz proteins | 15 | ||
| Cystatins - may be housekeeping | 2 | ||
| TIL domain family found in mosquitoes | 8 | ||
| Kazal domain containing peptides | 11 | ||
|
| |||
| Immunity-related proteins | |||
|
| |||
| Lysozyme | 13 | Y | (Moreira-Ferro et al., 1998; Rossignol and Lueders, 1986) |
| Defensin | 9 | ||
| Diptericin | 1 | ||
| Gambicin | 4 | Y | (Vizioli et al., 2001) |
| Cecropins | 7 | ||
| GGY peptide | 3 | ||
| Fred/Ficolin domain containing proteins | 7 | ||
| Gram negative binding protein | 7 | ||
| Peptidoglycan recognition protein | 2 | ||
| Leucine rich protein | 2 | ||
| Galectin - maybe housekeeping | 2 | ||
| C type lectins | 18 | ||
|
| |||
| Mucins | |||
|
| |||
| Widespread mucin family | 3 | ||
| Virus induced mucin | 6 | ||
| Mucin I mosquito family | 29 | ||
| gSG5 mucin protein family | 5 | ||
| SG3 mucin family | 11 | ||
| Aedes-specific mucin | 4 | ||
| Simulium mucin | 5 | ||
| 30 kDa Phlebotomine mucin family | 13 | ||
| Culicoides 10 kDa mucin | 3 | ||
| Other mucins | 12 | ||
| Peritrophin /chitin binding | 6 | ||
|
| |||
| OBP superfamily | |||
|
| |||
| Long D7 mosquito family | 23 | Y | (Calvo et al., 2006a; Calvo et al., 2009a; Mans et al., 2007) |
| Anopheline short D7 family | 27 | Y | (Calvo et al., 2006a; Isawa et al., 2002) |
| Culicine short D7 proteins | 15 | ||
| Salivary mosquito OBP | 6 | ||
| Phlebotomine long D7 family | 22 | ||
| Phlebotomine SP-15 family | 33 | ||
| Lutzomyia short D7 | 3 | ||
| Simulium specific long D7 family with 2 OBP domains | 3 | ||
| Simulium nigrimanum long D7 with only 1 OBP domain | 1 | ||
| Simulium OBP | 3 | ||
| Simulium nigrimanum OBP | 3 | ||
| Simulium short D7 family | 28 | Y | (Cupp and Cupp, 2000) |
| Culicoides OBP family | 12 | ||
| Culicoides OBP family II | 7 | ||
|
| |||
| Other ubiquitous families | |||
|
| |||
| Aedes sialokinin - neurokinin family | 2 | Y | (Beerntsen et al., 1999; Champagne and Ribeiro, 1994; Zeidner et al., 1999) |
| Selenoprotein | 1 | ||
| Phosphatidylethanolamine binding protein | 3 | ||
| Aedes Phosphatidylethanolamine-binding protein | 2 | ||
| Yellow phlebotomine family | 14 | ||
|
| |||
| II – Ubiquitous protein families existing outside Nematocera, function unknown | |||
|
| |||
| Antigen-5 family | 40 | ||
|
| |||
| III– Ubiquitous insect protein families existing outside Nematocera, function unknown | |||
|
| |||
| 15–17 kDa Insect family | 7 | ||
| Simulium/Culicoides insect conserved secreted protein family | 9 | ||
| 12–14 kDa mosquito family | 8 | ||
| Hypothetical conserved secreted protein | 3 | ||
| Culex/Drosophila WAP subfamily | 3 | ||
| Aedes hypothetical secreted conserved protein | 1 | ||
|
| |||
| IV - Protein families exclusive of blood sucking Nematocera | |||
|
| |||
| 30 kDa/Aegyptin family | 55 | Y | (Boppana et al., 2009; Calvo et al., 2007b; Calvo al.; Yoshida et al., 2008) |
| 41 kDa family | 18 | ||
|
| |||
| V - Protein families specific of mosquitoes | |||
|
| |||
| Found in both culicines and anophelines | |||
|
| |||
| 56 kDa mosquito family | 7 | ||
| 37.7 kDa mosquito family | 3 | ||
| Basic tail mosquito family | 13 | ||
| 4.3 kDa mosquito peptide | 3 | ||
| HHH peptide family | 10 | ||
| Glycine-rich mosquito family | 3 | ||
| Salivary protein 16 family | 2 | ||
| Aedes/darlingi 14–15family | 2 | ||
| gSG8 family | 3 | ||
| Hyp6.2 family | 5 | ||
|
| |||
| SG1/62kDa superfamily | |||
|
| |||
| Anopheline SG1 family | 27 | ||
| Aedes 62 kDa kDa family | 5 | ||
|
| |||
| Uniquely found in Culicines | |||
|
| |||
| 9.7 kDa | 8 | ||
| hyp8.2 culicine family | 5 | ||
| 30.5 kDa protein | 12 | ||
| 23.5 kDa culicine family | 4 | ||
| KKK circle family | 2 | ||
| GQ rich culicine family | 7 | ||
| Culex/Toxorhynchites family | 2 | ||
|
| |||
| Uniquely found in anophelines | |||
|
| |||
| cE5/Anophelin family | 7 | Y | (Francischetti et al., 1999; Valenzuela et al., 1999) |
| gSG7 family/Anophensins | 9 | Y | (Isawa et al., 2007) |
| gSG6 family | 3 | ||
| Hyp8.2 family | 6 | ||
| hypl5–17 family | 7 | ||
| SG2 family | 8 | ||
| hyp10/hyp12 family | 7 | ||
| 4.2 kDa family | 4 | ||
| Anopheles acidic protein | 7 | ||
| An. darlingi GGGG peptide family | 5 | ||
|
| |||
| Uniquely found in Aedes | |||
|
| |||
| Aedes 6.5–8.5 protein family | 4 | ||
| Aedes W rich peptides | 8 | ||
| 34 kDa Aedes family | 9 | ||
|
| |||
| Uniquely found in Culex | |||
|
| |||
| Culex WRP/16 kDa family | 33 | ||
|
| |||
| VI - Protein families specific of black flies | |||
|
| |||
| Simulium SVEP family | 16 | Y | (Cupp et al., 1998) |
| Simulium collagen-like family | 20 | ||
| Sv7.0 kDa family | 7 | ||
| Simulium Q-rich proteins | 8 | ||
| Basic 7–13 Simulium family | 4 | ||
| Sv7.8 kDa family | 4 | ||
| Basic 7–13 Simulium family | 2 | ||
| Simulium Basic 13 kDa family | 4 | ||
| Simulium Basic 28 kDa family | 4 | ||
| Simulium Acid 28kDa family | 4 | ||
| Simulium Sv7.8 kDa family | 4 | ||
| Simulium nigrimanum 8–10CysW family | 5 | ||
| Simulium nigrimanum conserved hypothetical family | 3 | ||
| Simulium 5-Cys secreted protein | 3 | ||
| Simulium nigrimanum 8–10Cys family | 3 | ||
| Simulium family of conserved secreted proteins | 3 | ||
| Simulium disintegrin similar to Phenoloxidase inhibitor peptide | 2 | ||
| Simulium basic 7.4 kDa family | 2 | ||
| Simulium basic 13 kDa family | 2 | ||
| Sv7.8 kDa family | 2 | ||
| Simulium 4.8 kDa family | 5 | ||
| Simulium Basic 7–13 Simulium family | 2 | ||
| H-rich, acidic proteins of Simulium | 13 | ||
| Simulium nigrimanum Sn Actinohivin | 1 | ||
|
| |||
| VII - Protein families specific of sand flies | |||
|
| |||
| Phlebotomine hyp13 family | 5 | ||
| SP13 Phlebotomine family | 3 | ||
| 37–46 kDa phlebotomine family | 5 | ||
| 33 kDa Phlebotomine family of Xa inhibitors | 7 | ||
| 27–30 kDa phlebotomine family | 4 | ||
| Lutzomyia 9–10kDa family | 2 | ||
| 16 kDa Phlebotomus family | 3 | ||
| 10 kDa Phlebotomine peptide - also found in midgut | 2 | ||
| Maxadilan | 1 | Y | (Lerner et al., 1991; Lerner and Shoemaker, 1992) |
|
| |||
| VIII - Protein families specific of Culicoides | |||
|
| |||
| H-rich, acidic proteins of Culicoides | 3 | ||
| 15 kDa Culicoides protein family | 2 | ||
| 19–21 kDa Culicoides protein family | 2 | ||
| Culicoides 15 kDa family | 20 | ||
| Culicoides proteins distantly similar to Culex 16.7 kDa protein | 6 | ||
| Culicoides 18.5 kDa family | 3 | ||
| Culicoides 20 kDa family | 2 | ||
| Culicoides nebuculosus His rich peptides | 3 | ||
|
| |||
| IX - Salivary-orphan proteins of conserved secreted families | |||
|
| |||
| Various | 43 | ||
|
| |||
| X - Orphan proteins of unique standing | |||
|
| |||
| Various | 63 | ||
I – Ubiquitous protein families existing outside Diptera, function known or presumed
Enzymes
Apyrase
Blood sucking Nematocera were known to hydrolyze ATP and ADP (Cupp et al., 1994; Cupp et al., 1995; Marinotti et al., 1996; Perez de Leon and Tabachnick, 1996; Reno and Novak, 2005; Ribeiro et al., 1989; Ribeiro et al., 1985; Ribeiro et al., 1986; Ribeiro et al., 1984), which are important hemostasis and inflammatory agonists released by platelets and broken cells following tissue injury. ADP is a potent inducer of platelet aggregation, and ATP induces neutrophil aggregation and granule release (Francischetti et al., 2009; Ribeiro and Arca, 2009). In Aedes aegypti a protein coding for the salivary apyrase was identified as a member of the 5'nucleotidase family (Champagne et al., 1995), and the recombinant protein shown to indeed hydrolyze ATP and ADP (Sun et al., 2006). 5' nucleotidases are ubiquitous enzymes normally associated with the extracellular side of cell membranes, through a phosphatidylinositol lipid anchor, attached to their carboxyterminus. Aedes apyrase was first shown to lack the typical carboxyterminus where the inositol phosphate lipid is anchored, making this enzyme a soluble secreted protein (Champagne et al., 1995). Transcripts for all 5' nucleotidases indicated in Table I also lack the carboxyterminal region typical of phospholipid anchor binding. As far as mosquitoes are concerned the presence of a salivary apyrase/5'nucleotidase activity seems to have a different importance in Anopheles and Aedes as compared to Culex. Transcripts encoding 5'nucleotidase family members were not found in C. tarsalis whereas in Culex quinquefasciatus the expression levels were quite low compared to Aedes and Anopheles mosquitoes. In contrast, as first shown for Anopheles gambiae (Arcà et al, 1999; Lombardo et al, 2000), Aedes and Anopheles mosquitoes carry in their saliva two different members of the 5'nucleotidase family, although still remain to be determined if they both act as apyrases or if one is an apyrase and the other a true 5'nucleotidase. This difference may be due to the bird feeding preferences of Culex mosquitoes, which do not face the platelet barrier of mammal feeding mosquitoes (Ribeiro, 2000). In sand flies the apyrase activity results from a completely different protein family (Charlab et al., 1999; Valenzuela et al., 2001b), which was first discovered in bed bug salivary glands, constituting a new family of nucleotide binding enzymes (Valenzuela et al., 1998). In the sand fly Lutzomyia longipalpis there also exists a 5'-nucleotidase enzyme that was shown to hydrolyze not only AMP, but also UDP-Glucose, thus acting as a phosphodiesterase (Ribeiro et al., 2000b).
5'-nucleotidase/apyrase-coding transcripts are found in mosquitoes and black flies, but are not shown for Culicoides in our supplemental table. However, search of the NCBI database allows for identification of a 88 aa truncated protein from C. sonorensis (gi|51557826), derived from a salivary transcriptome, that has similarity to several 5'-nucleotidase/apyrases previously described in sialotranscriptomes, the matches of which can be verified at http://www.ncbi.nlm.nih.gov/sutils/blink.cgi?pid=51557826.
Adenosine deaminase (ADA) and purine hydrolase (PH)
Following the first sialotranscriptome analysis of Ae.aegypti (Valenzuela et al., 2002b), transcripts were found coding for ADA and PH, indicating that adenosine could be hydrolyzed to inosine, and then to hypoxantine plus ribose. Following up on these transcriptomic leads, the enzymatic activities were found in the insect saliva and salivary gland homogenates (Ribeiro et al., 2001; Ribeiro and Valenzuela, 2003). These activities may decrease the concentration of adenosine and inosine at the site of the bite, thus decreasing these agonists that induce mast cell degranulation (Tilley et al., 2000) and trigger the itching reaction. Transcripts coding for adenosine deaminase were found in culicines and anophelines, as well as in L. longipalpis (Charlab et al., 2000; Charlab et al., 2001; Ribeiro et al., 2001) and Phlebotomus duboscqi, but not on black flies or Culicoides. The recombinant enzyme from P. duboscqi was expressed and its activity confirmed (Kato et al., 2007). Interestingly, P. papatasi lacks transcripts coding for ADA, and has large amounts of adenosine and AMP in its saliva, which are vasodilators and platelet aggregation antagonists (Katz et al., 2000; Ribeiro et al., 1999; Ribeiro and Modi, 2001), underlining the large differences in salivary strategy within a single genus of fly, as is the case of P. duboscqi (which does not have salivary adenosine and AMP – unpublished) and P. papatasi. PH transcripts have been found so far only in culicine mosquitoes.
Pyrophosphatase/Phosphodiesterase
Transcripts coding for these enzymes have been found only on the genus Phlebotomus. They may be important to hydrolyze dinucleotides such as the diadenosine polyphosphates Ap4A and Ap5A, important inflammatory mediators released by platelets (Gasmi et al., 1996; Schluter et al., 1996), a function that might have been taking over in Lutzomyia by the 5'-nucleotidase/phosphodiesterase mentioned above (Ribeiro et al., 2000b), emphasizing convergent evolution scenarios within the sand flies.
Endonucleases and Hyaluronidases
These two activities may be related to a common function of decreasing the skin matrix viscosity around the feeding site. Endonuclease transcripts were found in C. quinquefasciatus, and the activity confirmed by probing mosquitoes and by expression of a functional recombinant enzyme (Calvo and Ribeiro, 2006). Transcripts coding for endonucleases were also found in Ochlerotatus triseriatus, but not in Aedes or anopheline mosquitoes. They were found in sand flies and in Simulium. Hyaluronidase transcripts were first found in L. longipalpis (Charlab et al., 1999), and the salivary activity detected in various sand flies and black flies (Cerna et al., 2002; Ribeiro et al., 2000a; Volfova et al., 2008).
Ribonucleases
Transcripts coding for secreted ribonucleases were found solely in mosquitoes. Their function is unknown, and they could be playing a housekeeping function.
Alkaline phosphatase
This activity has been determined in salivary glands and in probes of Ae. aegypti and appears to be found in both males and female glands (Argentine and James, 1995). Their function is unknown. Transcripts coding for alkaline phosphatase have been found in Ae. aegypti and C. tarsalis.
Serine proteases, metalloproteases, and other peptidases
Transcripts coding for trypsin-like enzymes abound in all Culicomorpha. Several gene products are found expressed in the salivary glands of individual mosquito species, as well as in black flies and biting midges. In sand fly sialomes only one example of this family is found, in P. ariasi indicating that it may be underrepresented in Psychodiidae. The function of this family, none of which has been biochemically characterized so far, is unknown, but presumably it may function in immunity, serving as prophenoloxidase activators, or in blood feeding, by digesting the host matrix or affecting hemostasis, as in a fibrinogenolytic function known to exist in ticks and tabanid flies (Francischetti et al., 2003; Xu et al., 2008) or activating host plasminogen. Not as widespread as serine proteases coding transcripts are those coding for metalloproteases (singly found in Simulium. nigrimanum), dipeptidyl peptidase (singly found in L. longipalpis), cathepsins (S. nigrimanum and C. tarsalis) and glutamate carboxypeptidase (C. tarsalis). None of these have been functionally characterized and they may be associated to housekeeping functions.
Lipases/esterases
This group of enzymes are scattered throughout the Nematocera sialomes. A typical secretory phospholipase A2 is found in 3 species of the genus Phlebotomus. Additionally, triacylglycerol lipases are also found in mosquitoes (C. quinquefasciatus and Anopheles stephensi) and in the sand fly P. argentipes. Transcripts coding for a carboxylesterase were found in Ae. aegypti. None have been biochemically characterized. The mosquito C. quinquefasciatus has a potent phospholipase C activity that hydrolyzes Platelet Aggregation Factor (PAF) but its molecular nature remains unknown (Ribeiro and Francischetti, 2001).
Destabilase
This enzyme is related to lysozyme (Zavalova et al., 2000) and was first discovered in the salivary glands of leeches (Baskova and Nikonov, 1991; Fradkov et al., 1996), where it works as epsilon-(gamma-Glu)-Lys isopeptidase. Transcripts coding for this type of enzyme were found in Simulium transcriptomes, but the biochemical activity was not characterized.
Anopheline peroxidases
The salivary vasodilator of Anopheles albimanus was shown to be a peroxidase with catecol oxidase activity which destroyed vasoconstrictory amines such as norepinephrine (Ribeiro and Valenzuela, 1999; Ribeiro and Nussenzveig, 1993). Transcripts coding for these enzymes were found in An. gambiae and An. darlingi.
Peroxiredoxin-coding transcripts were found in S. vittatum, C. tarsalis and A. aegypti. They may play a housekeeping function, or be related to detoxifying host peroxinitrites (Ferrer-Sueta and Radi, 2009; Trujillo et al., 2007). None have been biochemically characterized.
Glycosidases
Maltase and amylase transcripts are represented in all Nematocera sialomes done so far, and are most probably associated with a sugar feeding function, as indicated by these activities to exist in both male and female mosquito salivary glands (James et al., 1989a). These enzymes represent one of the few families common to all blood feeding Nematocera, possibly because sugar feeding is an earlier trait that was present in the common ancestor of both Culicomorpha and Psychodidae.
Chitinase-coding transcripts were identified in P. arabicus and A. aegypti. They may play a role in immunity, as similar genes in An. gambiae were found to be immunoresponsive (Shi and Paskewitz, 2004).
Ubiquitous protease inhibitor domains
Several protein domains associated with an inhibitory function of proteases are known, and possibly have a primeval function of regulating housekeeping (for example, lysosomal enzymes), immune (regulating clotting or phenololoxidase activating cascades) or digestive proteases. Since many host's hemostatic and inflammatory processes involve proteolytic cascades or processing, it is not surprising that many such domains were recruited to function in the saliva of blood feeding insects.
Serpin stands for Serine Protease Inhibitor, a ubiquitous family regulating serine protease cascades in mammals and invertebrates. In Nematocera blood feeders, the family is found in the sialome of Culicine, but not Anopheline mosquitoes, and there is one known serpin described in the sand fly L. longipalpis. The salivary serpin of Ae. aegypti was identified as a factor Xa inhibitor and is the main salivary anti clotting in this mosquito (Stark and James, 1995; Stark and James, 1998). However, Aedes and Culex have two different salivary serpins; the specificity of this second serpin is still unknown, as is the serpin from the sand fly.
The Kunitz domain is ubiquitously found and associated with protease inhibitors (Ascenzi et al., 2003), but also with ion channel blockers (Castaneda and Harvey, 2009; Harvey, 1997; Paesen et al., 2009), which could impose a vasodilatory function. Only black flies and biting midges have this protein family represented in their transcriptomes, several different proteins being present in a single organism, indicating gene duplication events.
Cystatins are inhibitors of cysteine proteinases (Abrahamson et al., 2003). Transcripts coding for these proteins were found only in Ae. aegypti and Ae. albopictus, but their function in blood feeding, is unknown. Ticks secrete multiple cystatins that have an anti inflammatory and immunosuppressive function (Kotsyfakis et al., 2008; Kotsyfakis et al., 2007; Kotsyfakis et al., 2006).
The TIL domain refers to Trypsin Inhibitor Like domain containing 5 disulphide bonds and are ubiquitously found in animals (Rawlings et al., 2004). In sialotranscriptomes it is found restricted to mosquitoes, including those of male An. gambiae, suggesting an antimicrobial role (Kanost, 1999). None of these mosquito peptides have been biochemically or microbiologically characterized.
The Kazal domain is also associated with serine protease inhibitors and antimicrobial activity (Kanost, 1999; Rawlings et al., 2004). Members of this family have been found exclusively in anopheline and culicine mosquitoes, except for one additional example in Culicoides sonorensis.
Immunity related proteins
Antimicrobial peptides (AMP) and microbial pattern recognition molecules (MPRM) are commonly found in sialotranscriptomes of blood feeding insects and ticks. Some AMP families are ubiquitous, such as the lysozymes and defensins, found in vertebrates and invertebrates (Torres and Kuchel, 2004), others are quite restricted, such as the mosquito specific gambicin family. The GGY peptide listed in supplemental file S1 has similarities to a nematode peptide characterized to have an AMP function (Couillault et al., 2004). The pattern recognition molecules include families associated with activation of the complement system, such as the ficolins and C-type lectins (Fujita et al., 2004; Thiel, 2007), as well as the peptidoglycan recognition proteins (PRPs) and Gram-negative binding proteins (GNBPs) (Ferrandon et al., 2004), also associated with the initiation and regulation of immune reactions. It is possible that some of these proteins play a role in blood-feeding, instead of antimicrobial. For example, many snake venom C-type lectins are known to have anti-clotting and anti-platelet functions (Jennings et al., 2005; Morita, 2005; Polgar et al., 1997). Supplemental file S1 lists 12 families of proteins in this category, only two of which had at least a member characterized to some extent: Lysozyme activity was detected in salivary extracts of male and female mosquitoes (Moreira-Ferro et al., 1998; Rossignol and Lueders, 1986), and the gambicin peptide (a family exclusive to mosquitoes) was microbiologically characterized (Vizioli et al., 2001).
Mucins and peritrophins
Mucins are highly glycosylated proteins containing serine or threonine residues that are O-linked to N-acetyl-galactosamine containing oligosaccharides, and are ubiquitously found in salivary and gut epithelia of vertebrates and invertebrates (Korayem et al., 2004; Rayms-Keller et al., 2000; Sarauer et al., 2003). Many of these proteins are immune regulated and function in the entrapment of virus and bacteria, as we are reminded during an upper respiratory tract viral infection. Mucins have an unusual large amount of Ser and Thr and their glycosylation potential can be recognized by the NetOGlyc server (Julenius et al., 2005). This class of proteins is commonly found in sialotranscriptomes, and may be associated with the coating of the chitinous lining of the salivary channels, as indicated by many members having a chitin binding domain. Supplemental file S1 indicates 10 different families recognized by their primary amino acid sequence similarity, plus 12 proteins with indication of heavy glycosylation from different insects that have no similarities to other sialotranscriptome proteins. Six additional proteins are added to this group that have chitin binding domains and are annotated as peritrophins; these do not have indication of abundant glycosylation, but are grouped with mucins due to possible similarity of function. No protein listed in this section has been characterized. It is to be noted that recombinant expression and purification of mucins is difficult due to their glycosylation requirement and stickiness. However, RNAi experiments may produce interesting results.
Odorant binding protein/D7 superfamily
The protein named D7 was one of the first deducted from a salivary cDNA cloned from a mosquito salivary gland (James et al., 1991). Later this protein was found to be a member of the Odorant Binding Protein (OBP) superfamily (Hekmat-Scafe et al., 2000), to be organized as a multi gene family in the mosquito An. gambiae (Arca et al., 1999; Arca et al., 2002) and to be widespread within blood sucking Nematocera (Valenzuela et al., 2002a). More recently, mosquito members of this family were shown to be strong binders, or kratagonists, of biogenic amines and leukotrienes exerting their effects by antagonizing inflammation and hemostasis agonists (Calvo et al., 2006a; Calvo et al., 2009a). Proteins with one or two OBP domains characterize the short and long D7 protein subfamilies, which were found so far in all sialotranscriptomes of mosquitoes. Short and long mosquito proteins were crystallized, showing 2 additional helices per OBP domain when compared to the canonical OBP domain. In An. gambiae there are 3 genes coding for the long proteins and 5 genes coding for the short D7 proteins, organized as tandem repeats in chromosome 3R (Arca et al., 2005). The last gene of each long or short series is poorly transcribed and it was proposed they may be turning into pseudogenes (Calvo et al., 2006a; Calvo et al., 2009a; Mans et al., 2007). Transcripts coding for canonical OBP proteins are also found in mosquito sialotranscriptomes, albeit at a much lower frequency than those coding for classical D7 proteins. Abundant transcripts of mRNA coding for sequences with the OBP domain are also found in black flies, biting midges, and sand flies, however, none have their crystal structure resolved.
Alignment of 147 selected members of the OBP/D7 family shown in supplemental file S1 (excluding the few true OBPa from mosquitoes, excessively truncated proteins, and proteins of the same species having more than 95% identity) allows the creation of the condensed bootstrapped phylogram shown in supplemental file S2. Although these sequences have indications of OBP domains by RPSblast to the CDD or PFAM databases (Marchler-Bauer et al., 2002), no strong bootstrap support exists indicating a common ancestor for all proteins. However, most of the proteins group into strong clades and subclades that might help to categorize functions of these proteins as they become known.
The mosquito D7 proteins organize themselves into several clades with strong bootstrap support, including a clade (81 % bootstrap support) with all Long D7 proteins, except one resulting from the poorly transcribed third gene coding for a D7 protein from An. gambiae, and a long D7 sequence of C. tarsalis. The Culicidae Long D7 clade (supplemental file S2) contains subclades that organize themselves neatly into genera, including two clades for the genus Culex. The strong bootstrap support for this clade points to the existence of this family in the common ancestor of anophelines and culicines, over 150 million years ago (MYA) (Krzywinski et al., 2006). The mosquito short D7 proteins organize into several clades, containing those for Aedes and Ochlerotatus (3 clades and one singleton), one for Culex and another for the Anopheles genera. The Anopheline subclade contains the 5 known gene products of the An. gambiae short D7 proteins, which are arranged in tandem directly opposite and in the reverse direction of the long D7 gene cassette in chromosome arm 3R (Arca et al., 2005). The subclades containing these proteins are numbered I–V in supplemental file S2. Subclade I contains the An. stephensi protein named Hamadarin (gi|21314941) which is an inhibitor of bradykinin formation, a peptide inducing pain and produced during blood clotting (Isawa et al., 2002); it contains also the An. gambiae protein gi|4538887 which was shown, together with those marked with a circle in clades II, III and IV, to strongly bind biogenic amines (BABP, Biogenic Amines Binding Proteins), thus having anti-hemostatic and anti-inflammatory properties (Calvo et al., 2006a; Mans et al., 2007). The An. gambiae protein of subclade V is poorly transcribed and does not bind biogenic amines, and is possibly in the process of becoming a pseudogene (Calvo et al., 2006a). Interestingly, an An. darlingi homolog exists, indicating this gene is active in this South American mosquito. These subclades should help assigning functions to the orthologous members in different genera and subgenera when the function of one member is known.
The phlebotomine OBP's organize themselves in 3 groups, containing (1) the classical phlebotomine D7 protein clade (100 % bootstrap support), which have ~27 kDa mature molecular mass; (2) the shorter (12–16 kDa) proteins similar to the SP-15 antigen of P. papatasi that confers protection to Leishmaniasis (Valenzuela et al., 2001a), also organizes under strong (99 %) bootstrap support, plus (3) two orphan short Lutzomyia sequences. Notice that each of the two large clades is composed of strong subclades indicative of gene duplications. None of these proteins have been characterized functionally, but most of the long D7 proteins of this group have the motif [ED]-[EQ]-x(7)-C-x(12,17)-W-x(2)-W-x(7,9)-[TS]-x-C-[YF]-x-[KR]-C-x(8,22)-Q-x(22,32)-C-x(2)-[VLI] which is found in the cysteinyl leukotriene binding D7 proteins of mosquitoes, suggesting a potential role of this proteins in phlebotomines and indicated in the column labeled “Salivary gland family motifs” of supplemental spreadsheet S1, suggesting a potential role of this proteins in phlebotomines (see column Z labeled “Salivary motifs” of supplemental spreadsheet S1). Since it is not believed that sand flies and mosquitoes share a common blood feeding ancestor, the evolution of a multigene family expressing salivary gland OBP's in both families is a remarkable product of convergent evolution. Alternatively, these findings support a common hematophagous ancestor between sand flies and Culicomorpha.
The black fly proteins organize themselves into 4 independent clades each with strong bootstrap support, plus two singleton proteins. The protein gi|197260866 has been described by the Cupp's as an anti-thrombin in a patent (Cupp and Cupp, 2000), and may be responsible for the anti-thrombin activity of S. vittatum salivary gland homogenates (Abebe et al., 1995). Clade IV (supplemental file S2) contains one protein sequence from S. vittatum and one from S. nigrimanum, which are unique among non-mosquito proteins for having, like the long D7 proteins of mosquitoes, two OBP domains. Except for gi|197260866, no other protein in this group has been characterized functionally.
The biting midge salivary OBP's fall into two well supported super-clades that are more closely related to each other than any other D7-clades. Each clade contains members of C. sonorensis and C. nubeculosus and suggests a gene duplication event occurred in the last common ancestor to these species giving rise to two ancestral OBP-like molecules found in biting midge salivary glands. Within these clades at least five orthologous clades exist that suggest that extensive gene duplication events occurred in the last ancestor. Of interest is the fact that although the Culicoides D7 proteins seems to be only distantly related to the biogenic amine binding proteins of mosquitoes, they also exist as highly abundant and antigenic proteins in biting midge saliva (Wilson et al., 2008). This could as such, be another example of convergent evolution to target biogenic amines.
Aedes sialokinin
This peptide was identified as the vasodilator found in Ae. aegypti salivary homogenates; it was found expressed in the medial lobe of adult female salivary glands and the gene was later cloned, and found expressed in the same tissue location (Beerntsen et al., 1999; Champagne and Ribeiro, 1994). The peptide is a typical member of the vertebrate neurokinin family, by its common end of Phe-X-Gly-Leu-Met where X is any hydrophobic peptide. The Gly following the Met produces a final peptide that is amidated. Sialotranscriptomes of Ae. albopictus and O. triseriatus failed to retrieve homologues, possibly due to the relatively small size of the transcript, which tends to be lost in the cDNA library construction. Sialokinins were shown to affect virus transmission and immunomodulate their vertebrate hosts (Zeidner et al., 1999).
Selenoprotein
A typical selenoprotein with signal peptide indicative of secretion was found in the sialotranscriptome of An. gambiae and putative homologs are found in C. quinquefasciatus and Ae. aegypti. This protein may be of housekeeping role in the ER or Golgi, or participate in the detoxification of oxygen radicals in the bite wound during feeding.
Phosphatidylethanolamine-binding protein
This is a ubiquitous protein family, members of which have been found expressed in sialotranscriptomes of black flies and mosquitoes. These proteins have a signal peptide indicative of secretion, but there is no evidence that they are found in saliva. Their function is unknown, but could be playing a role as kratagonists of lipid mediators, if any.
Yellow phlebotomine family
This is a protein family found exclusively in insects. The family is so named because mutation of a gene in Drosophila produced a yellow phenotype indicative of disruption of the melanization process (Albert et al., 1999; Geyer et al., 1986), leading to characterization of its product as a novel protein. The major royal jelly protein of bees was found also to be part of this family (Albert et al., 1999). Later, mosquitoes and Drosophila family members were shown to have dopachrome convertase activity (Han et al., 2002; Johnson et al., 2001). Yellow proteins are abundant in phlebotomine sialomes, producing strong bands in salivary gland homogenate protein chromatograms, where more than one gene for the family exists (Anderson et al., 2006; Hostomska et al., 2009; Valenzuela et al., 2004). Members of the Yellow family of proteins appear to be good markers of vector exposure (Bahia et al., 2007). No dopachrome convertase activity could be detected in P. papatasi or L. longipalpis salivary homogenates (Ribeiro, unpublished). It is possible that these proteins in phlebotomines lost the catalytic function and work as binders of biogenic amines (Ribeiro and Arca, 2009).
II – Ubiquitous protein families existing outside Nematocera, function unknown
Antigen 5 family
This is a ubiquitous protein family belonging to the wider CAP superfamily (Gibbs et al., 2008). Its members are associated with host defenses in plants, and various functions in animals, such as toxins in snake and lizard venoms (Nobile et al., 1996; Yamazaki et al., 2002), proteolytic activity in Conus snails (Milne et al., 2003), and platelet aggregation inhibition in a tabanid salivary protein (Xu et al., 2008), although this latter function results from the novel incorporation of an RGD domain that functions as a disintegrin. A protein member of this family, found expressed in the salivary glands of the stable fly Stomoxys calcitrans binds immunoglobulin (Ameri et al., 2008) and may function as an inhibitor of the classic pathway of complement (Wang et al., 2009). The variety of functions that this family is endowed with precludes assigning a function to any of the proteins listed in supplemental file S1, none of which has been characterized functionally.
III– Ubiquitous insect protein families existing outside Nematocera, function unknown
This group of proteins possibly contains housekeeping proteins that could be performing a function in the ER or Golgi, or may have been recruited to perform a salivary function.
15–17 kDa family
Several mosquito species and also Phlebotomus and Culicoides presented ESTs in their sialotranscriptomes coding for secreted proteins of MW ranging from 15–17 kDa also found in Drosophila, and in the midgut of phlebotomines. Psiblast (results on supplemental file S3) of a representative protein from P. duboscqi indicates that the family is multigenic in Drosophilids, and retrieves salivary expressed proteins from triatomines and the stable fly Stomoxys calcitrans.
12–14 kDa family similar to Drosophila proteins
Psiblast for this mosquito family represented by 8 proteins of mature mass ranging from 11 to 16 kDa retrieves Drosophila proteins of the same size. Expression analysis of representative members of this family from Aedes and Anopheles indicate expression in male mosquitoes in addition to salivary gland of female mosquitoes, suggesting a function unrelated to blood feeding.
Simulium/Culicoides insect conserved secreted protein family
The sialotranscriptomes of Simulium and Culicoides revealed transcripts coding for putative secreted proteins that are exclusive to insects, as indicated by 4 iterations of Psiblast (Supplemental file S4). The function of these proteins is unknown.
Hypothetical conserved secreted protein
The sialotranscriptome of An. darlingi and O. triseriatus revealed transcripts coding for secreted proteins matching solely insect proteins by Psiblast (Supplemental file S5).
Culex/Drosophila WAP family
Culex sialotranscriptomes indicate the expression of proteins with mature MW ~ 10 kDa containing the non-specific hit Pfam WAP domain which includes a four disulphide core motif. These proteins are similar to other insect proteins of the same size found in the NR database. The high sequence identity (93 %) between the C. tarsalis and C. pipiens sequences are indicative of a housekeeping role, because salivary proteins known to be secreted in saliva have lesser identity between these two species than housekeeping proteins (Calvo et al., 2010a).
IV– Protein families specific to blood sucking Nematocera
Found in more than one insect family
30 kDa antigen/Aegyptin
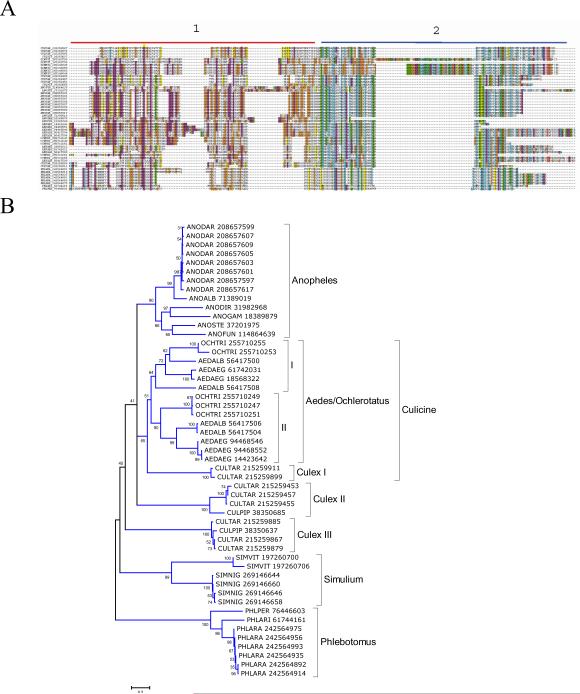
Members of this protein family are found in the sialotranscriptomes of mosquitoes and black flies where they are abundantly expressed, and also in sand flies albeit at lower expression levels. They were first identified as a 30 kDa antigen in Aedes mosquitoes (Docena et al., 1999; Simons and Peng, 2001). Proteins of this family from An. stephensi and Ae. aegypti were shown more recently to inhibit platelet aggregation by interfering with collagen recognition (Calvo et al., 2007b; Yoshida et al., 2008). Aegyptin was the name given to the Aedes inhibitor. The promoter region of the anopheline gene was identified and used for robust salivary gland specific expression of foreign genes in adult female mosquitoes (Yoshida and Watanabe, 2006). The family has 2 distinct domains, in addition to the secretory signal peptide, characterized by an acidic, low complexity domain rich in Pro, Gly, Glu and Asp residues (for this reason some proteins were named as GE rich proteins in anophelines), marked 1 in figure 1A, and a more complex carboxyterminal domain, marked with 2 in figure 1A. The richness of acidic residues in domain 1 confers a pI for this protein family in the range of 3.9–4.6 The bootstrapped phylogram (Fig 1B) indicates robust clades, as expected, for the Anopheline, Culicine, Simulium and Phlebotomus sequences. Notice also that anophelines and Simulium appear to have a single gene coding for this family (those from An. darlingi appearing to be alleles from a single gene), while Culicines have multiple genes. The Aedes/Ochlerotatus genes fall with strong bootstrap support into two subclades, marked I and II in Fig 1B. Aedes have shorter versions of Aegyptin, and recently a member of this short Aegyptin (gi|18568322) was shown to program host CD4 T cells to express IL-4 in mice (Boppana et al., 2009). Whether this effect is due do interaction of the protein with host collagen-like proteins is unknown.
Figure 1.
The 30 kDa antigen/Aegyptin family of blood sucking Nematocera. A) Clustal alignment showing the 2 domain regions (first 20 amino acids excluded). B) Bootstrapped phylogram of the alignment in A. The number near the branches indicates percent bootstrap support after 10,000 trials. The bar at the bottom of the figure indicates 20% amino acid sequence divergence. The sequence names are represented by the first 3 letters of the genus name, followed by two letters of the species name, followed by the GenBank accession number.
41.9 kDa superfamily
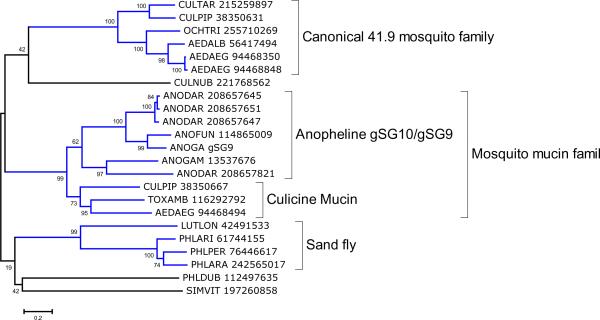
The first member of this protein family was identified in the sialotranscriptome of C. quinquefasciatus (Ribeiro et al., 2004) when it did not provide significant matches to any known protein. Additional sialotranscriptomes identified related proteins in the mosquitoes Ae. aegypti, Ae. albopictus, C. tarsalis, Oc. triseriatus and An. darlingi and in the black fly S. vittatum and biting midge C. nubeculosus, as well as the 39kDa family of the phlebotomines L. longipalpis, P. arabicus, P. ariasi, P. dubosci and P. perniciosus, indicating this family to be an ancient. Interestingly, this family was not found in members of the Cellia anopheline subgenera so far investigated (An. gambiae, An. funestus and An. stephensi), but was found in the New World anopheline An. darlingi. Psiblast of the An. darlingi sequence against the NR protein database after 6 iterations retrieves additional mosquito proteins, including the shorter gSG10 mucin family of Cellia anophelines (~ 16 kDa proteins), and Culex and Toxorhynchites proteins annotated as mucins (Fig 2 and supplemental file S6). The gSG9 protein of An. gambiae is clearly derived from a further truncation of the gSG10 gene, producing a 6 kDa peptide. These results indicate that the gSG10/gSG9 family in the Cellia subgenus may have originated from the canonical 41.9 kDa ancestors. This mucin subfamily has 10–20 putative N-acetyl-galactosamine glycosylation sites (Julenius et al., 2005), while the larger members of the superfamily have less than 8 sites. Expression analysis in Ae. aegypti and Ae. albopictus showed that the transcripts coding for members of this family, gi|94468350 and gi|56417494 respectively, are found in adult female salivary glands and in male mosquitoes (Ribeiro et al., 2007; Arcà et al., 2007), the same pattern found in An. gambiae for the gSG10 and gSG9 proteins (Arca et al., 2005), suggesting a role in sugar feeding either as a glycolytic enzyme or as an antimicrobial, or in salivary duct maintenance as a mucin.
Figure 2.
Phylogram of the 41 kDa family of mosquitoes, biting midges, black flies and biting midges. The number near the branches indicates percent bootstrap support after 10,000 trials. The bar at the bottom of the figure indicates 20% amino acid sequence divergence. The sequence names are represented by the first 3 letters of the genus name, followed by two letters of the species name, followed by the GenBank accession number, except for some An. gambiae sequences.
V - Mosquito specific salivary proteins
Found in both culicines and anophelines
The 56 kDa protein family is expressed in adult mosquito salivary glands, including male An. gambiae mosquitoes (Calvo et al., 2006b). RT-PCR experiments in Ae. aegypti and Ae. albopictus also revealed this transcript in female salivary glands and in males, indicating a function associated with sugar feeding or as an antimicrobial. Four iterations of psi-blast (Altschul et al., 1997) retrieves bacterial proteins in addition of mosquito proteins (Arca et al., 2007; Arca et al., 2005; Ribeiro et al., 2007). In Ae. aegypti and An. gambiae, the genes coding for these proteins are uniexonic. These observations lead to the suggestion that this protein family was acquired by mosquitoes via horizontal transfer from a bacterial genome.
Supplemental file S1 shows 11 additional protein families that are mosquito specific and represented in both anophelines and culicines. The protein families named 37.7 kDa, basic tail, hyp6.2 and Aedes/darlingi 14–15 family and SG1 were found expressed exclusively, or enriched, on adult female salivary glands, while the families HHH peptide family, Glycine rich mosquito family and salivary protein 16 family were found to be expressed also in male mosquitoes and other tissues, suggesting a possible antimicrobial or housekeeping function. The Ae. aegypti member of the 4.3 kDa family was found preferentially expressed in adult female salivary glands, while the Ae. albopictus homolog was ubiquitously expressed. Within the gSG8 family, the An. gambiae member was found expressed exclusively in adult female salivary glands, while the Ae. aegypti member was found expressed also in male mosquitoes (Arca et al., 2007; Arca et al., 2005; Ribeiro et al., 2007). No members of this group have their function characterized.
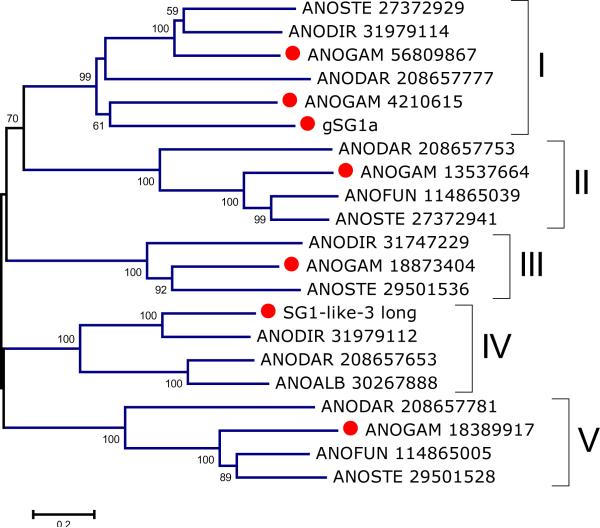
The SG1 family has been previously indicated as unique to anophelines (Arca et al., 2005). Its remarkable gene expansion in An. gambiae was indicated by both transcriptome and genome analysis, with 6 genes previously described, five of which occur in tandem in chromosome X (Arca et al., 2005). Further analysis of supplemental table S1 indicates one additional SG1 gene in chromosome X (worksheet named SG1 on supplemental file S1), located more than 1 Mb away from the previously identified cluster. All these genes are uniexonic, despite the protein products being relatively large, on the order of 45 kDa. Phylogenetic analysis following alignment of a selected non-redundant group of the available proteins shows the presence of 5 robust clades (Fig 3). Clades I and II merge with relatively good bootstrap support (70 %), indicating they are most related. Clade I contains 3 An. gambiae members, the remaining 4 clades one member each. Except for clade III, all others contain one member from the New World An. darlingi mosquito, indicating these clades were formed prior to the breakup of Gondwanaland (Krzywinski and Besansky, 2003), and that the SG1 gene expansion occurred early in Anopheles evolution.
Figure 3.
Bootstrapped phylogram of the SG1 family of Anophelines. The number near the branches indicates percent bootstrap support after 10,000 trials. The bar at the bottom of the figure indicates 20% amino acid sequence divergence. The sequence names are represented by the first 3 letters of the genus name, followed by two letters of the species name, followed by the GenBank accession number. The An. gambiae sequences are marked with a filled circle.
Psiblast of the An. gambiae SG1 protein against the NR database retrieves all proteins listed in supplemental file S1 under the SG1 group, but additionally retrieves after 6 iterations the salivary proteins of Aedes named 62 kDa proteins which are roughly of the same size. Alignment and phylogenetic analysis of the SG1 and 62 kDa family does not support a common ancestry between these two families, for lack of bootstrap support (results not shown); indeed only a common tyrosine is found in the alignment of the SG1 and 62 kDa set.
Proteins so far found only in culicine mosquitoes
Seven protein families are found exclusive to culicines. Some members have been studied with regards to their tissue specific expression (Arca et al., 2007; Ribeiro et al., 2007), indicating that the families named 9.7 kDa and hyp8.2 culicine family are enriched or selectively expressed in adult female glands, suggesting some role connected to blood feeding. Members of the families named 30.5 kDa and 23.5 kDa were found mainly expressed in female glands and in adult males, a pattern that appears compatible with possible functions in sugar feeding or as antimicrobial. The Ae. albopictus member of the KKK circle family has ubiquitous expression whereas the families named GQ rich and Culex/Toxorhynchites family do not have any members with known tissue expression. No members of this group have their function characterized.
Proteins so far found only in anopheline mosquitoes
Ten protein families are found exclusive to anophelines, only two of which have members that were functionally characterized, the anophelins and the anophensins. Anophelin is a thrombin inhibitor found in salivary gland homogenates of the New World mosquito An. albimanus. The An. gambiae anophelin primary amino acid sequence is only 60 % and 49 % identical to An. funestus and An. stephensi which are, as An. gambiae is, members of the Cellia subgenus, and only 42 and 43 % identical to An. albimanus and An. darlingi, which are both from the Nyssorhynchus subgenus only found in the New World. The New World anophelins are also shorter than their Old World homologues. The An. gambiae homolog of anophelin, was found ubiquitously expressed (Arca et al., 2005), suggesting it may have additional functions beyond salivary anticlotting activity.
Anophensin was the name given to the An. stephensi homolog of the An. gambiae protein previously named gSG7 (Arca et al., 1999). Anophensin prevents kallikrein activation thus preventing formation of the pain-inducing peptide bradykinin (Isawa et al., 2007; Poole et al., 1999). Two gSG7 genes, arranged in tandem and named gSG7 and gSG7-2 respectively, are present in An. gambiae and homologues are found in An. funestus, An. darlingi and An. stephensi (Calvo et al., 2007a). Only one member of the family, the An. stephensi anophensin, which is an orthologue of the An. gambiae gSG7, has been functionally characterized. Given the divergence between the duplicated genes in the different species (~40% identity and ~55% similarity) it would not be surprising if the gSG7 protein will be found to have acquired some different specificity.
The families named gSG6, hyp8.2 and hyp15–17 were found expressed exclusively, or enriched, in adult female salivary glands, suggesting a role in blood feeding. Actually, reduction of gSG6 protein levels by RNAi affected blood feeding in An. gambiae (Lombardo et al., 2009), although its specific function is still unknown. The families named SG2 and hyp10/hyp12 were found in adult female salivary glands and in males, but no in other female tissues, suggesting a possible role in sugar feeding or as an antimicrobial. The family named 4.2 kDa was ubiquitously expressed, and the families named Anopheles acidic protein and An. darlingi GGGG peptide family have not been studied regarding tissue expression.
Peptides designed on the gSG6 protein from An. gambiae has been recently used as a marker of vector exposure in an epidemiological context (Poinsignon et al., 2009; Poinsignon et al., 2008), a trend initiated with whole salivary homogenates from vector arthropods (Barral et al., 2000; Cornelie et al., 2007; Remoue et al., 2006; Schwartz et al., 1991; Schwartz et al., 1990). The use of recombinant antigens increases the amount of available protein for large epidemiological studies, improves reproducibility and decreases the amount of possible cross reactivity by careful selection of the candidate antigens. In this context recombinant An. gambiae gSG6 has also been tested as marker of exposure to malaria vector bites in a large epidemiological survey in an area with high level of transmission/exposure of Burkina Faso giving very encouraging results (Arcà, in preparation).
Proteins so far found only in the Aedes/Ochlerotatus mosquito genera
Three protein families were found exclusively in Aedes and the closely related Ochlerotatus genus, consisting of the families named 6.5–8.5, W-rich and 34 kDa. Expression analysis of these transcripts indicate the two first families to be expressed in female salivary glands and also in males, but not female carcasses, indicating a salivary function in both males and females, while the 34 kDa family is uniquely expressed, or enriched, in adult female salivary glands in both Ae. aegypti and Ae. albopictus. No functions have been attributed to any of these proteins.
Proteins so far found only in the Culex genus
A single expanded family is unique to the Culex genus, namely the 16.7 kDa family, found in both C. tarsalis and C. quinquefasciatus (Calvo et al., 2010a; Ribeiro et al., 2004). As recently indicated (Calvo et al., 2010a), this family consists of 30 genes in C. quinquefasciatus, 28 of which are uniexonic. Psiblast analysis retrieves bacterial proteins with sugar binding domains, lectins and hemolysins, the crystal structure of which have a trefoil structure (Calvo et al., 2010a). This family is by far the most transcribed in Culex sialotranscriptomes, dwarfing the expression of the D7 family, and for this reason it was suggested that this family may have acquired the serotonin-binding function found in the D7 of other mosquito genera so far studied.
VI - Proteins so far found only in black flies
Two sialotranscriptomes were done so far for black flies, one with the North American autogenous species S. vittatum (Andersen et al., 2009) and the other with the South American S. nigrimanum (In press). From these 2 transcriptomes, 24 protein families were identified as exclusively found in black flies. Only one of these proteins has a known characterized function, the salivary vasodilator named SVEP for S. vittatum erythema protein (Cupp et al., 1998). These proteins families were described in detail in a recent publication (submitted) and will not be further described.
VII - Proteins so far found only in sand flies
Eight protein families were found unique to sand flies. Two of the families have been functionally characterized, including the vasodilatory peptide Maxadilan, specific of the genus Lutzomyia and the 33 kDa family, found in both New World and Old World genera and identified as a FXa clotting inhibitor (J. Valenzuela, unpublished). The remaining families have no known function. Interestingly, the 27–30 kDa family of P. argentipes, consisting of 4 related proteins (Anderson et al., 2006), produces no significant matches to any protein in the NR database, but provides a Rpsblast match to the CDD ETX_MTX2 motif spanning the whole 226 amino acids of the motif with a low e value (1e-17). This motif is named Clostridium epsilon toxin ETX/Bacillus mosquitocidal toxin MTX2, suggesting an acquisition by horizontal transfer.
VIII - Proteins so far found only in biting midges
Eight protein families are exclusive to biting midges, including a quite expanded family of proteins in the range of 14–15 kDa, found in both C. sonorensis and C. nubeculosus. No member of any of the families has been characterized functionally.
IX - Orphan proteins of conserved families of unknown function
Spreadsheet S1 provides links to 43 protein sequences found in sialotranscriptomes that have no match to other proteins described in sialotranscriptomes, but match proteins deposited in the NR database. These proteins may have been uniquely co-opted for a salivary function in the named insects, or may be poorly expressed housekeeping transcripts that were revealed by chance in the transcriptomes.
X - Orphan proteins of unique standing
An additional 63 proteins from various insects and containing a signal peptide indicative of secretion are described in spreadsheet S1 which have neither matches to any sialotranscriptome, nor matches to known proteins in the NR database.
Evolutionary considerations
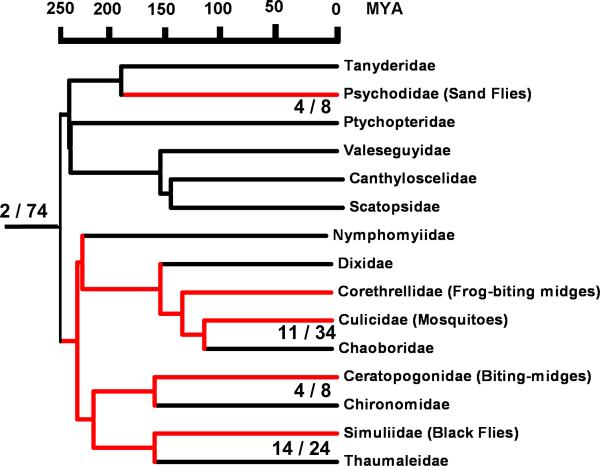
The evolution of the blood sucking habit in the Nematocera may have occurred at least twice, over two hundred million years ago, producing today's sand flies and all the families within the Culicomorpha (the null evolutionary hypothesis) (Fig. 4). The salivary glands of these insects incorporate the evolutionary process toward this adaptation, a trajectory that included a menu initially composed mainly by dinosaurs and reptiles, changing to mostly birds and mammals after the extinction of the dinosaurs and irradiation of mammal 65 MYA. Mammals “invented” the blood platelet, creating a major barrier to blood feeding, or at least creating the opportunity for insects to develop new pharmacological agents to disarm platelet function that increase their fitness by increasing blood uptake success. Comparative sialotranscriptome analysis allows a snapshot of this evolutionary process over a long evolutionary time, and allows some insight into the Culicomorpha “Eve”, with sand flies as an outgroup.
Fig. 4.
Blood-feeding evolution in the Nematocera. Indicated in red are the Null hypothesis (independent evolution of blood-feeding behavior in the Psychomorpha and Culicomorpha) presented in this study, according to Grimaldi and Engel (2005). Numbers at the basal branch indicate the total number of unique sialome protein families not recognizable outside Nematocera that are common to all Nematocera lineages (30 kDa antigen and 41 kDa protein family), followed by the number of all protein families (excluding the orphan proteins) unique to any Nematocera. Below the family branches with available sialome data, the numbers represents the number of protein families unique to the lineage followed by the unique families found in all members (excluding orphan proteins). Notice that the denominators shown at the end of the branches (8, 34, 8 and 24) add to the 74 unique salivary protein families. 4/8 in the sand fly branch indicate that of the 8 unique sand fly protein families, only 4 are shared between Lutzomyia and Phlebotomus. Phylogenetic and divergence date data were obtained from Grimaldi and Engel (2005).
But before we launch into this exercise, here are some difficulties: First, when attempting to visualize the evolutionary scenario to hematophagy starting with the Culicomorpha or Psycomorpha “Eve”, we contemplate that the majority of the proteins discovered so far have no known function, and many have no similarities to any other known proteins. It is fair to say at this point in time that the overwhelming majority of salivary proteins in the Nematocera have no known function, as indicated in Table III. Even if the protein family is well known, we may not “a priori” determine its function; for example, Kazal-domain containing proteins may function as a serine protease inhibitor, and thus could have anti-clotting function, but can also be antimicrobial. For this reason, in species where only females blood-feed, for example in mosquitoes, comparative transcriptomes analysis between male and females can indicate those that are adaptive to sugar feeding, if they are found in male salivary glands. It is thus important to determine whether a protein functions within a blood feeding or a sugar feeding context to ascribe evolutionary pathways associated with hematophagy. Secondly, large expanded families may have multiple functions. For example, a few years ago we were completely ignorant on the function of the D7 family in mosquitoes. We learned that they mainly function as kratagonists, in some cases antagonizing serotonin, in other cases histamine and in other cases leukotrienes or thromboxane A2 (Calvo et al., 2009a; Mans et al., 2007); but it can also function as an inhibitor of bradykinin formation (Isawa et al., 2002). Identification of orthologs helps in assigning functions, and from their alignment, models can be built for functional identification in new organisms, but we cannot a priori determine the specificity of the more distant members, where novel functions may be acquired. Thirdly, it appears that the salivary gland genes are at a relatively rapid pace of evolution, leading to the appearance of protein families that are unique even at the genus level of some families, or even at the subgenus level, as is the gSG7 protein of anophelines. This fast evolution is probably a response to the immune pressure posed by their hosts. Perhaps for this last consideration, very few salivary protein families are conserved in all Culicomorpha, allowing for only a few unique genes common to all members that resisted the passage of time. And these few genes could have been the product of convergent evolution and not common ancestry.
A most probable common mode of feeding of the adult Nematocera “Eve,” from which both sand flies and Culicomorpha were derived, must have been a plant feeding behavior, surely not on flower nectar, because these did not exist at 200 MYA. This may explain the ubiquitous presence of multiple sugar hydrolyzing enzymes such as amylase and maltase, common to sand flies and Culicomorpha. The same can be stated for antimicrobial peptides such as the ubiquitous lysozyme and other common antimicrobial peptide domains, as they might have controlled microbial growth in the insect's crop. This set of salivary proteins should be common to all Nematocera, blood feeding or not.
Genes coding for ubiquitous proteins, such as antigen 5 or trypsins, are difficult to assign an evolutionary scenario without a clear characterization of their orthologous function in different families. For example, if salivary antigen 5 proteins across the different families are shown to retain a immunoglobulin binding function, or if trypsins are found to have a conserved fibrinolytic function, these orthologous functions could make a strong point on their association with the evolution of blood feeding. But these genes might otherwise be associated with innate immunity and not related to hematophagy. Their functional characterization is important in this evolutionary context, in addition to their possible function in immunity and blood feeding.
From an evolutionary (not biochemical) viewpoint, the assignment of proteins to a specific protein family obviously holds the implication that all members of such a family are homologous. That is, that all members derived from a common ancestral fold that originated once in the distant past. In the case where such families occur universally throughout the animal kingdom, their presence in sialomes per se cannot be taken as evidence of common blood-feeding origins. These protein families reside in most genomes and could have been recruited to the sialome at any given time in the evolution of blood-feeding behavior. This would certainly be the case for the majority of Class I families such as the enzymes, protease inhibitor domains and anti-microbials. Results do indicate, however, that different arthropod lineages and families possess unique sialome compositions (Mans and Francischetti, 2010). This corresponds to lineage specific innovations directly related to adaptation to a blood-feeding lifestyle and do indicate that specific families responded in unique ways to the challenge of the blood-feeding interface. What is of interest though, is that the various lineages share very few of the protein families common outside the Nematocera and no indication can be found that there is a lineage specific association of protein families as might have been expected if the various lineages share common blood-feeding histories (Fig. 4). The fact that common protein families are found in closely related lineages is certainly indicative of a shared history, but the ways these protein folds have been exploited to evolve novel functions are what is remarkable and significant (Mans and Francischetti, 2010).
In the elucidation of molecular origins, the accurate identification of orthologous proteins, rather than protein folds is crucial. Orthologous proteins are related by vertical descent and generally possess similar functions and mechanisms that are conserved over time due to negative selection. While immune pressures and gene losses can certainly account for some of the variations observed in the association of protein folds and function, an analysis of the data clearly show that orthologous proteins (with conserved function) are found within the various nematocerous families and that within these lineages immune pressure may play a relatively small role in determining functional switches (but could lead to gene function loss and gene elimination). In many cases, these functions have been conserved for more than 100–200 million years. An example is the biogenic amine binding D7-fold found in mosquitoes with conserved binding mechanisms, but known to be highly immunogenic (Calvo et al., 2009a; Mans et al., 2007).
The review thus far considered the sialomes from the perspective of the distribution of protein families in and outside the Nematocera and their possible functions. As such, there are a number of protein families shared between all Nematocera that is also found in other arthropods and a number of 74 families that are specifically nematocerous (Fig. 4, Table 3). Only two of these 74 unique families are shared between the Culicomorpha and sand flies, the 41 kDa superfamily, possibly associated with sugar feeding, and the 30 kDa/Aegyptin family, associated with blood feeding (Fig 4). As we move into the Nematocera lineages, the number of shared families increase, but even so it is only near 50% of the totality of unique families (Fig 4) indicating many recent acquisitions of novel families, or evolution of old families to such a degree that they are not recognizable by sequence similarity analysis. As such, the argument could have been made that blood-feeding evolved in the last common ancestor of the Culicomorpha and Psycomorpha, with subsequent lineage specific innovations after speciation of the main families. However, most of the protein families that are shared between the nematocerous families are ubiquitous and found widely in other blood-feeding and toxic arthropods as well (Fry et al., 2009; Mans et al., 2008; Mans and Francischetti, 2010). Those that have been exapted from house-keeping functions to function at the host-vector feeding interface could have been recruited at any stage and their use as indicators of common blood-feeding origins seems tenuous. A nice example of such proteins are the 5'-nucleotidases, which have been recruited several times in different unrelated blood-feeding arthropods that include kissing bugs, mosquitoes and ticks (Champagne et al., 1995; Faudry et al., 2004; Stutzer et al., 2009); accordingly, the common finding of 5'-nucleotidases serving an apyrase function in Culicomorpha could have been acquired by independent recruitment of this gene family by the different lineages. In the same line, the recruitment of the OBP superfamily by both Culicomorpha and sand flies could have been the result of convergent evolution. Elucidation of the function and crystal structure of these sand fly proteins may throw more light into this issue.
The data obtained, even if extensive gene losses and continuous recruitment of new folds are considered, does not give overwhelming support for the Null hypothesis. In the light of the above exposition, the possibility that the various blood-feeding Nematoceran families evolved blood-feeding behavior independently should be considered seriously, even if this seems to be the least likely parsimonious scenario (Mans and Francischetti, 2010). Of interest, would be the fact that this is the third group of closely related organisms (the other two being ticks and triatomine bugs) that posses similar folds, but on closer analysis would seem to indicate independent evolution of functions involved at the blood-feeding interface. Previous observations with triatomine bugs from the genera Rhodnius and Triatoma, as well as soft and hard tick families, suggest that these organisms evolved hematophagous behavior independently (Mans et al. 2008; Mans and Francischetti, 2010). The question is raised why closely related organisms are prone to evolve blood-feeding behavior independently. In this regard, previous similar lifestyles that would predispose towards blood-feeding (plant feeding, predation of insects, scavenging of arthropod hemolymph), in which birds and mammals (and possibly dinosaurs) suddenly occupies the same environmental niche in which non-hematophagous arthropods were living, could have precipitated an association of vertebrates and the previous food niche, thus causing an initial exploration of vertebrates as a potential food source.
Within this discussion, it is puzzling that there is no common salivary anticlotting protein in the Culicomorpha. Blood clotting is relatively conserved in vertebrates, all having thrombin as a final enzyme that processes fibrinogen to fibrin. This clotting system is quite ancient and must have posed a barrier to blood feeding by the ancestral hematophagous Culicomorpha. Nonetheless, Culicoides and black flies abound with Kunitz-domain proteins (yet to be demonstrated to be anti-clotting), a black fly anti-thrombin is member of the OBP family, Culicine mosquitoes have a serpin that inhibits factor Xa, and anopheline mosquitoes have a unique anti thrombin named anophelin (see references in Table III). No conservation of an anti-clotting is verified even at the Culicidae level. It has been observed (Ribeiro, unpublished) that salivary gland homogenates (SGH) of Ae. aegypti failed to increase “in vitro” recalcification clotting time of plasma derived from mosquito exposed Guinea pigs, and also the plasma of one of the authors (JMCR), who used to feed mosquitoes on himself at the time; however, elevated recalcification time was observed with SGH-treated plasma derived from unexposed mammals. It appears that the value of the Aedes serpin is clearly under pressure to either be less immunogenic or for Aedes to “find out” something else better suited to the job. Better documented is the case of the salivary vasodilatory peptide from L. longipalpis named maxadilan, shown to be immunogenic; host immunity was associated with less fly blood ingestion and egg output (Milleron et al., 2004a; Milleron et al., 2004b). Perhaps for this reason, maxadilan genes from wild populations are very variable at the amino acid level, with up to 23% amino acid divergence between alleles (Lanzaro et al., 1999). Interestingly these alleles have different cross reactivity to antibodies, but equal vasodilatory activity (Milleron et al., 2004b) indicating the adaptive value of this polymorphism which reflects antigenic variation, not functional improvement. A detailed population genetics work on salivary genes involved in blood feeding will certainly enhance our vision on the evolutionary paths leading toward this peculiar diet. The lack of a common salivary anti-clotting agent among the Culicomorpha could thus support either the renunciation of the null hypothesis, or represent fast gene turnover within a commonly derived hematophagous lineage.
Concluding remarks
Due possibly to host immune pressure, the evolutionary speed of salivary gland genes is at a fast pace. As such, it is remarkable that some common protein families involved in blood feeding and secreted in large amounts still exist in organisms that diverged over 150 MYA, as represented by the D7 protein family of mosquitoes, or the 5'-nucleotidase family of apyrases in the Culicomorpha. To the extent that these genes cannot mutate fast enough or keep a state of balanced polymorphism, they should move into extinction thus creating a new opportunity for recruitment of a new gene family. Perhaps for these reasons it is difficult to use salivary gland-specific genes to trace the evolutionary history of hematophagous Nematocera, but this at the same time stresses the evolutionary creativity of novel pharmacological compounds needed for successful blood feeding found in the saliva of such animals.
The organization of the available data hopefully will provide a stimulus for the tremendous task ahead for functional characterization of the proteins here listed, and may provide a platform for helping in the definition of candidate proteins to be used as genus- or species-specific antigens to be used as markers of insect exposure.
Supplementary Material
Acknowledgements
This work was supported in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA. Because J.M.C.R. is a government employee and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abebe M, Cupp MS, Champagne D, Cupp EW. Simulidin: a black fly (Simulium vittatum) salivary gland protein with anti-thrombin activity. J. Insect Physiol. 1995;41:1001–1006. [Google Scholar]
- Abrahamson M, Alvarez-Fernandez M, Nathanson CM. Cystatins. Biochem Soc Symp. 2003:179–99. doi: 10.1042/bss0700179. [DOI] [PubMed] [Google Scholar]
- Albert S, Bhattacharya D, Klaudiny J, Schmitzova J, Simuth J. The family of major royal jelly proteins and its evolution. J Mol Evol. 1999;49:290–7. doi: 10.1007/pl00006551. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameri M, Wang X, Wilkerson MJ, Kanost MR, Broce AB. An immunoglobulin binding protein (antigen 5) of the stable fly (Diptera: Muscidae) salivary gland stimulates bovine immune responses. J Med Entomol. 2008;45:94–101. doi: 10.1603/0022-2585(2008)45[94:aibpao]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JF, Pham VM, Meng Z, Champagne DE, Ribeiro JM. Insight into the Sialome of the Black Fly, Simulium vittatum. J Proteome Res. 2009 doi: 10.1021/pr8008429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Oliveira F, Kamhawi S, Mans BJ, Reynoso D, Seitz AE, Lawyer P, Garfield M, Pham M, Valenzuela JG. Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC Genomics. 2006;7:52. doi: 10.1186/1471-2164-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca B, Lombardo F, de Lara Capurro M, della Torre A, Dimopoulos G, James AA, Coluzzi M. Trapping cDNAs encoding secreted proteins from the salivary glands of the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. U S A. 1999;96:1516–21. doi: 10.1073/pnas.96.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Francischetti IM, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JM. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem Mol Biol. 2007;37:107–27. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Lanfrancotti A, Spanos L, Veneri M, Louis C, Coluzzi M. A cluster of four D7-related genes is expressed in the salivary glands of the African malaria vector Anopheles gambiae. Insect Mol. Biol. 2002;11:47–55. doi: 10.1046/j.0962-1075.2001.00309.x. [DOI] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JM. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J. Exp. Biol. 2005;208:3971–86. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Argentine JA, James AA. Characterization of a salivary gland-specific esterase in the vector mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 1995;25:621–30. doi: 10.1016/0965-1748(94)00103-o. [DOI] [PubMed] [Google Scholar]
- Ascenzi P, Bocedi A, Bolognesi M, Spallarossa A, Coletta M, De Cristofaro R, Menegatti E. The bovine basic pancreatic trypsin inhibitor (Kunitz inhibitor): a milestone protein. Curr Protein Pept Sci. 2003;4:231–51. doi: 10.2174/1389203033487180. [DOI] [PubMed] [Google Scholar]
- Bahia D, Gontijo NF, Leon IR, Perales J, Pereira MH, Oliveira G, Correa-Oliveira R, Reis AB. Antibodies from dogs with canine visceral leishmaniasis recognise two proteins from the saliva of Lutzomyia longipalpis. Parasitol Res. 2007;100:449–54. doi: 10.1007/s00436-006-0307-8. [DOI] [PubMed] [Google Scholar]
- Barral A, Honda E, Caldas A, Costa J, Vinhas V, Rowton ED, Valenzuela JG, Charlab R, Barral-Netto M, Ribeiro JM. Human immune response to sand fly salivary gland antigens: a useful epidemiological marker? Am. J. Trop. Med. Hyg. 2000;62:740–5. doi: 10.4269/ajtmh.2000.62.740. [DOI] [PubMed] [Google Scholar]
- Baskova IP, Nikonov GI. Destabilase, the novel epsilon-(gamma-Glu)-Lys isopeptidase with thrombolytic activity. Blood Coagul Fibrinolysis. 1991;2:167–72. doi: 10.1097/00001721-199102000-00025. [DOI] [PubMed] [Google Scholar]
- Beerntsen BT, Champagne DE, Coleman JL, Campos YA, James AA. Characterization of the Sialokinin I gene encoding the salivary vasodilator of the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 1999;8:459–67. doi: 10.1046/j.1365-2583.1999.00141.x. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17:349–56. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- Boppana VD, Thangamani S, Adler AJ, Wikel SK. SAAG-4 is a novel mosquito salivary protein that programmes host CD4 T cells to express IL-4. Parasite Immunol. 2009;31:287–95. doi: 10.1111/j.1365-3024.2009.01096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Andersen J, Francischetti IM, de LCM, deBianchi AG, James AA, Ribeiro JM, Marinotti O. The transcriptome of adult female Anopheles darlingi salivary glands. Insect Mol. Biol. 2004;13:73–88. doi: 10.1111/j.1365-2583.2004.00463.x. [DOI] [PubMed] [Google Scholar]
- Calvo E, Dao A, Pham VM, Ribeiro JM. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect Biochem Mol Biol. 2007a;37:164–75. doi: 10.1016/j.ibmb.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J. Biol. Chem. 2006a;281:1935–42. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Ribeiro JM, Andersen JF. Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc Natl Acad Sci U S A. 2009a;106:3728–33. doi: 10.1073/pnas.0813190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Pham VM, Lombardo F, Arca B, Ribeiro JM. The sialotranscriptome of adult male Anopheles gambiae mosquitoes. Insect Biochem. Mol. Biol. 2006b;36:570–5. doi: 10.1016/j.ibmb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Calvo E, Pham VM, Marinotti O, Andersen JF, Ribeiro JM. The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC Genomics. 2009b;10:57. doi: 10.1186/1471-2164-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Pham VM, Ribeiro JM. An insight into the sialotranscriptome of the non-blood feeding Toxorhynchites amboinensis mosquito. Insect Biochem Mol Biol. 2008;38:499–507. doi: 10.1016/j.ibmb.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Ribeiro JM. A novel secreted endonuclease from Culex quinquefasciatus salivary glands. J. Exp. Biol. 2006;209:2651–9. doi: 10.1242/jeb.02267. [DOI] [PubMed] [Google Scholar]
- Calvo E, Sanchez-Vargas I, Favreau AJ, Barbian KD, Pham VM, Olson KE, Ribeiro JM. An insight into the sialotranscriptome of the West Nile mosquito vector, Culex tarsalis. BMC Genomics. 2010a;11:51. doi: 10.1186/1471-2164-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Sanchez-Vargas I, Kotsyfakis M, Favreau AJ, Barbian KD, Pham VM, Olson KE, Ribeiro JM. The salivary gland transcriptome of the eastern tree hole mosquito, Ochlerotatus triseriatus. J Med Entomol. 2010b;47:376–86. doi: 10.1603/me09226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Tokumasu F, Marinotti O, Villeval JL, Ribeiro JM, Francischetti IM. Aegyptin, a novel mosquito salivary gland protein, specifically binds to collagen and prevents its interaction with platelet glycoprotein VI, integrin alpha2beta1, and von Willebrand factor. J Biol Chem. 2007b;282:26928–38. doi: 10.1074/jbc.M705669200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Tokumasu F, Mizurini DM, McPhie P, Narum DL, Ribeiro JM, Monteiro RQ, Francischetti IM. Aegyptin displays high-affinity for the von Willebrand factor binding site (RGQOGVMGF) in collagen and inhibits carotid thrombus formation in vivo. Febs J. 277:413–27. doi: 10.1111/j.1742-4658.2009.07494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Vandyke KA, Letchworth GJ, Drolet BS, Hanekamp T, Wilson WC. Midgut and salivary gland transcriptomes of the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae) Insect Mol. Biol. 2005;14:121–36. doi: 10.1111/j.1365-2583.2004.00537.x. [DOI] [PubMed] [Google Scholar]
- Castaneda O, Harvey AL. Discovery and characterization of cnidarian peptide toxins that affect neuronal potassium ion channels. Toxicon. 2009 doi: 10.1016/j.toxicon.2009.02.032. [DOI] [PubMed] [Google Scholar]
- Cazares-Raga FE, Gonzalez-Lazaro M, Montero-Solis C, Gonzalez-Ceron L, Zamudio F, Martinez-Barnetche J, Torres-Monzon JA, Ovilla-Munoz M, Aguilar-Fuentes J, Rodriguez MH, et al. GP35 ANOAL, an abundant acidic glycoprotein of female Anopheles albimanus saliva. Insect Mol Biol. 2007;16:187–98. doi: 10.1111/j.1365-2583.2006.00712.x. [DOI] [PubMed] [Google Scholar]
- Cerna P, Mikes L, Volf P. Salivary gland hyaluronidase in various species of phlebotomine sand flies (Diptera: psychodidae) Insect Biochem. Mol. Biol. 2002;32:1691–7. doi: 10.1016/s0965-1748(02)00109-1. [DOI] [PubMed] [Google Scholar]
- Champagne D, Ribeiro JMC. Sialokinins I and II: Two salivary tachykinins from the Yellow Fever mosquito, Aedes aegypti. Proc. Natl. Acad. Sci. (U.S.A.) 1994;91:138–142. doi: 10.1073/pnas.91.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne DE, Smartt CT, Ribeiro JM, James AA. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5'-nucleotidase family. Proc. Natl. Acad. Sci. U S A. 1995;92:694–8. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlab R, Rowton ED, Ribeiro JM. The salivary adenosine deaminase from the sand fly Lutzomyia longipalpis. Exp. Parasitol. 2000;95:45–53. doi: 10.1006/expr.2000.4503. [DOI] [PubMed] [Google Scholar]
- Charlab R, Valenzuela JG, Andersen J, Ribeiro JM. The invertebrate growth factor/CECR1 subfamily of adenosine deaminase proteins. Gene. 2001;267:13–22. doi: 10.1016/s0378-1119(01)00393-6. [DOI] [PubMed] [Google Scholar]
- Charlab R, Valenzuela JG, Rowton ED, Ribeiro JM. Toward an understanding of the biochemical and pharmacological complexity of the saliva of a hematophagous sand fly Lutzomyia longipalpis. Proc. Natl. Acad. Sci. U S A. 1999;96:15155–60. doi: 10.1073/pnas.96.26.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelie S, Remoue F, Doucoure S, Ndiaye T, Sauvage FX, Boulanger D, Simondon F. An insight into immunogenic salivary proteins of Anopheles gambiae in African children. Malar J. 2007;6:75. doi: 10.1186/1475-2875-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 2004;5:488–94. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Cupp EW, Cupp MS, Ramberg FB. Salivary apyrase in african and New-World vectors of Plasmodium species and its relationship to malaria transmission. Am. J. Trop. Med. Hyg. 1994;50:235–240. doi: 10.4269/ajtmh.1994.50.235. [DOI] [PubMed] [Google Scholar]
- Cupp MS, Cupp EW. Antithrombin protein and DNA sequences from black fly. USA. 2000 [Google Scholar]
- Cupp MS, Cupp EW, Ochoa AJ, Moulton JK. Salivary apyrase in New World blackflies (Diptera: Simuliidae) and its relationship to onchocerciasis vector status. Med Vet Entomol. 1995;9:325–30. doi: 10.1111/j.1365-2915.1995.tb00141.x. [DOI] [PubMed] [Google Scholar]
- Cupp MS, Ribeiro JM, Champagne DE, Cupp EW. Analyses of cDNA and recombinant protein for a potent vasoactive protein in saliva of a blood-feeding black fly, Simulium vittatum. J Exp Biol. 1998;201:1553–61. doi: 10.1242/jeb.201.10.1553. [DOI] [PubMed] [Google Scholar]
- Didisheim P, Hattori K, Lewis JH. Hematologic and coagulation studies in various animal species. J Lab Clin Med. 1959;53:866–75. [PubMed] [Google Scholar]
- Docena GH, Benitez P, Campos RE, Macia A, Fernandez R, Fossati CA. Detection of allergens in Aedes albifasciatus mosquito (Diptera: Culicidae) extracts by immunological methods. J Investig Allergol Clin Immunol. 1999;9:165–71. [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Faudry E, Lozzi SP, Santana JM, D'Souza-Ault M, Kieffer S, Felix CR, Ricart CA, Sousa MV, Vernet T, Teixeira AR. Triatoma infestans apyrases belong to the 5'-nucleotidase family. J. Biol. Chem. 2004;279:19607–13. doi: 10.1074/jbc.M401681200. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hoffmann JA. Sensing infection in Drosophila: Toll and beyond. Semin Immunol. 2004;16:43–53. doi: 10.1016/j.smim.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Ferrer-Sueta G, Radi R. Chemical biology of peroxynitrite: kinetics, diffusion, and radicals. ACS Chem Biol. 2009;4:161–77. doi: 10.1021/cb800279q. [DOI] [PubMed] [Google Scholar]
- Fradkov A, Berezhnoy S, Barsova E, Zavalova L, Lukyanov S, Baskova I, Sverdlov ED. Enzyme from the medicinal leech (Hirudo medicinalis) that specifically splits endo-epsilon(-gamma-Glu)-Lys isopeptide bonds: cDNA cloning and protein primary structure. FEBS Lett. 1996;390:145–8. doi: 10.1016/0014-5793(96)00644-8. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Mather TN, Ribeiro JM. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochem Biophys Res Commun. 2003;305:869–75. doi: 10.1016/s0006-291x(03)00857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J. Exp. Biol. 2002;205:2429–51. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Ribeiro JM. Anophelin: kinetics and mechanism of thrombin inhibition. Biochemistry. 1999;38:16678–85. doi: 10.1021/bi991231p. [DOI] [PubMed] [Google Scholar]
- Francischetti IMB, Sá-Nunes A, Mans BJ, Santos IM, Ribeiro JMC. The role of saliva in tick feeding. Frontiers in Biosciences. 2009;14:2051–2088. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JD, King GF, Nevalainen TJ, Norman JA, Lewis RJ, Norton RS, et al. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- Fujita T, Matsushita M, Endo Y. The lectin-complement pathway--its role in innate immunity and evolution. Immunol Rev. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- Gasmi L, McLennan AG, Edwards SW. Neutrophil apoptosis is delayed by the diadenosine polyphosphates, Ap5A and Ap6A: synergism with granulocyte-macrophage colony-stimulating factor. Br J Haematol. 1996;95:637–9. doi: 10.1046/j.1365-2141.1996.d01-1960.x. [DOI] [PubMed] [Google Scholar]
- Geyer PK, Spana C, Corces VG. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. Embo J. 1986;5:2657–62. doi: 10.1002/j.1460-2075.1986.tb04548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs GM, Roelants K, O'Bryan MK. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins--roles in reproduction, cancer, and immune defense. Endocr Rev. 2008;29:865–97. doi: 10.1210/er.2008-0032. [DOI] [PubMed] [Google Scholar]
- Grimaldi D, Engel M. Evolution of the insects. Cambridge University Press; New York: 2005. [Google Scholar]
- Han Q, Fang J, Ding H, Johnson JK, Christensen BM, Li J. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem. J. 2002;368:333–40. doi: 10.1042/BJ20020272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey AL. Recent studies on dendrotoxins and potassium ion channels. Gen Pharmacol. 1997;28:7–12. doi: 10.1016/s0306-3623(96)00173-5. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Dorit RL, Carlson JR. Molecular evolution of odorant-binding protein genes OS-E and OS-F in Drosophila. Genetics. 2000;155:117–27. doi: 10.1093/genetics/155.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostomska J, Volfova V, Mu J, Garfield M, Rohousova I, Volf P, Valenzuela JG, Jochim RC. Analysis of salivary transcripts and antigens of the sand fly Phlebotomus arabicus. BMC Genomics. 2009;10:282. doi: 10.1186/1471-2164-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isawa H, Orito Y, Iwanaga S, Jingushi N, Morita A, Chinzei Y, Yuda M. Identification and characterization of a new kallikrein-kinin system inhibitor from the salivary glands of the malaria vector mosquito Anopheles stephensi. Insect Biochem Mol Biol. 2007;37:466–77. doi: 10.1016/j.ibmb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Isawa H, Yuda M, Orito Y, Chinzei Y. A mosquito salivary protein inhibits activation of the plasma contact system by binding to factor XII and high molecular weight kininogen. J. Biol. Chem. 2002;13:13. doi: 10.1074/jbc.M203505200. [DOI] [PubMed] [Google Scholar]
- James AA, Blackmer K, Marinotti O, Ghosn CR, Racioppi JV. Isolation and characterization of the gene expressing the major salivary gland protein of the female mosquito, Aedes aegypti. Mol. Biochem. Parasitol. 1991;44:245–254. doi: 10.1016/0166-6851(91)90010-4. [DOI] [PubMed] [Google Scholar]
- James AA, Blackmer K, Racioppi JV. A salivary gland-specific, maltase-like gene of the vector mosquito, Aedes aegypti. Gene. 1989a;75:73–83. doi: 10.1016/0378-1119(89)90384-3. [DOI] [PubMed] [Google Scholar]
- James AA, Blackmer K, Racioppi JV. A salivary gland-specific, maltase-like gene of the vector mosquito, Aedes aegypti. Gene. 1989b;75:73–83. doi: 10.1016/0378-1119(89)90384-3. [DOI] [PubMed] [Google Scholar]
- Jariyapan N, Choochote W, Jitpakdi A, Harnnoi T, Siriyasatein P, Wilkinson MC, Bates PA. A glycine- and glutamate-rich protein is female salivary gland-specific and abundant in the malaria vector Anopheles dirus B (Diptera: Culicidae) J Med Entomol. 2006;43:867–74. doi: 10.1603/0022-2585(2006)43[867:agagpi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Jennings B, Spearman W, Shephard E. A novel 25 kDa protein from the venom of Bitis arietans with similarity to C-type lectins causes fibrinogen-dependent platelet agglutination. Toxicon. 2005;46:687–98. doi: 10.1016/j.toxicon.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Johnson JK, Li J, Christensen BM. Cloning and characterization of a dopachrome conversion enzyme from the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2001;31:1125–35. doi: 10.1016/s0965-1748(01)00072-8. [DOI] [PubMed] [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–64. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Kanost MR. Serine proteinase inhibitors in arthropod immunity. Dev Comp Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- Kato H, Anderson JM, Kamhawi S, Oliveira F, Lawyer PG, Pham VM, Sangare CS, Samake S, Sissoko I, Garfield M, et al. High degree of conservancy among secreted salivary gland proteins from two geographically distant Phlebotomus duboscqi sandflies populations (Mali and Kenya) BMC Genomics. 2006;7:226. doi: 10.1186/1471-2164-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jochim RC, Lawyer PG, Valenzuela JG. Identification and characterization of a salivary adenosine deaminase from the sand fly Phlebotomus duboscqi, the vector of Leishmania major in sub-Saharan Africa. J Exp Biol. 2007;210:733–40. doi: 10.1242/jeb.001289. [DOI] [PubMed] [Google Scholar]
- Katz O, Waitumbi JN, Zer R, Warburg A. Adenosine, AMP, and protein phosphatase activity in sandfly saliva. Am J.Trop Med Hyg. 2000;62:145–50. doi: 10.4269/ajtmh.2000.62.145. [DOI] [PubMed] [Google Scholar]
- Korayem AM, Fabbri M, Takahashi K, Scherfer C, Lindgren M, Schmidt O, Ueda R, Dushay MS, Theopold U. A Drosophila salivary gland mucin is also expressed in immune tissues: evidence for a function in coagulation and the entrapment of bacteria. Insect Biochem Mol Biol. 2004;34:1297–304. doi: 10.1016/j.ibmb.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Kotsyfakis M, Anderson JM, Andersen JF, Calvo E, Francischetti IM, Mather TN, Valenzuela JG, Ribeiro JM. Cutting edge: Immunity against a “silent” salivary antigen of the Lyme vector Ixodes scapularis impairs its ability to feed. J Immunol. 2008;181:5209–12. doi: 10.4049/jimmunol.181.8.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsyfakis M, Karim S, Andersen JF, Mather TN, Ribeiro JM. Selective cysteine protease inhibition contributes to blood-feeding success of the tick Ixodes scapularis. J Biol Chem. 2007;282:29256–63. doi: 10.1074/jbc.M703143200. [DOI] [PubMed] [Google Scholar]
- Kotsyfakis M, Sa-Nunes A, Francischetti IM, Mather TN, Andersen JF, Ribeiro JM. Antiinflammatory and immunosuppressive activity of sialostatin L, a salivary cystatin from the tick Ixodes scapularis. J. Biol. Chem. 2006;281:26298–307. doi: 10.1074/jbc.M513010200. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Besansky NJ. Molecular systematics of Anopheles: from subgenera to subpopulations. Annu Rev Entomol. 2003;48:111–39. doi: 10.1146/annurev.ento.48.091801.112647. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phylogenet Evol. 2006;39:417–23. doi: 10.1016/j.ympev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Lanzaro GC, Lopes AH, Ribeiro JM, Shoemaker CB, Warburg A, Soares M, Titus RG. Variation in the salivary peptide, maxadilan, from species in the Lutzomyia longipalpis complex. Insect Mol Biol. 1999;8:267–75. doi: 10.1046/j.1365-2583.1999.820267.x. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Ribeiro JMC, Nelson RJ, Lerner MR. Isolation of maxadilan, a potent vasodilatory peptide from the salivary glands of the sand fly Lutzomyia longipalpis. J. Biol. Chem. 1991;266:11234–11236. [PubMed] [Google Scholar]
- Lerner EA, Shoemaker CB. Maxadilan: Cloning and functional expression of the gene encoding this potent vasodilator peptide. J. Biol. Chem. 1992;267:1062–1066. [PubMed] [Google Scholar]
- Lombardo F, Ronca R, Rizzo C, Mestres-Simon M, Lanfrancotti A, Curra C, Fiorentino G, Bourgouin C, Ribeiro JM, Petrarca V, et al. The Anopheles gambiae salivary protein gSG6: an anopheline-specific protein with a blood-feeding role. Insect Biochem Mol Biol. 2009;39:457–66. doi: 10.1016/j.ibmb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, Andersen JF, Francischetti IM, Valenzuela JG, Schwan TG, Pham VM, Garfield MK, Hammer CH, Ribeiro JM. Comparative sialomics between hard and soft ticks: Implications for the evolution of blood-feeding behavior. Insect Biochem Mol Biol. 2008;38:42–58. doi: 10.1016/j.ibmb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, Calvo E, Ribeiro JM, Andersen JF. The crystal structure of D7r4, a salivary biogenic amine-binding protein from the malaria mosquito Anopheles gambiae. J Biol Chem. 2007;282:36626–33. doi: 10.1074/jbc.M706410200. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Francischetti IMB. Sialomic perspectives on the evolution of blood-feeding behavior in Arthropods: Future therapeutics by natural design. In: Kini RM, Clemetson KJ, Markland FS, McLane MA, Morita T, editors. Toxins and Hemostasis: From Bench to Bed-side. Springer; 2010. [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–3. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinotti O, de Brito M, Moreira CK. Apyrase and alpha-glucosidase in the salivary glands of Aedes albopictus. Comp. Biochem. Physiol. 1996;113B:675–679. doi: 10.1016/0305-0491(95)02035-7. [DOI] [PubMed] [Google Scholar]
- Marinotti O, James A, Ribeiro JMC. Diet and salivation in female Aedes aegypti mosquitoes. J. Insect Physiol. 1990;36:545–548. [Google Scholar]
- Marinotti O, James AA. An alpha-glucosidase in the salivary glands of the vector mosquito, Aedes aegypti. Insect Biochem. 1990;20:619–623. [Google Scholar]
- Milleron RS, Mutebi JP, Valle S, Montoya A, Yin H, Soong L, Lanzaro GC. Antigenic diversity in maxadilan, a salivary protein from the sand fly vector of American visceral leishmaniasis. Am J Trop Med Hyg. 2004a;70:286–93. [PubMed] [Google Scholar]
- Milleron RS, Ribeiro JM, Elnaime D, Soong L, Lanzaro GC. Negative effect of antibodies against maxadilan on the fitness of the sand fly vector of American visceral leishmaniasis. Am J Trop Med Hyg. 2004b;70:278–85. [PubMed] [Google Scholar]
- Milne TJ, Abbenante G, Tyndall JD, Halliday J, Lewis RJ. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J Biol. Chem. 2003;278:31105–10. doi: 10.1074/jbc.M304843200. [DOI] [PubMed] [Google Scholar]
- Montero-Solis C, Gonzalez-Ceron L, Rodriguez MH, Cirerol BE, Zamudio F, Possanni LD, James AA, de la Cruz Hernandez-Hernandez F. Identification and characterization of gp65, a salivary-gland-specific molecule expressed in the malaria vector Anopheles albimanus. Insect Mol Biol. 2004;13:155–64. doi: 10.1111/j.0962-1075.2004.00473.x. [DOI] [PubMed] [Google Scholar]
- Moreira-Ferro CK, Daffre S, James AA, Marinotti O. A lysozyme in the salivary glands of the malaria vector Anopheles darlingi. Insect Mol. Biol. 1998;7:257–64. doi: 10.1111/j.1365-2583.1998.00067.x. [DOI] [PubMed] [Google Scholar]
- Morita T. Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon. 2005;45:1099–114. doi: 10.1016/j.toxicon.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nobile M, Noceti F, Prestipino G, Possani LD. Helothermine, a lizard venom toxin, inhibits calcium current in cerebellar granules. Exp Brain Res. 1996;110:15–20. doi: 10.1007/BF00241369. [DOI] [PubMed] [Google Scholar]
- Oliveira F, Kamhawi S, Seitz AE, Pham VM, Guigal PM, Fischer L, Ward J, Valenzuela JG. From transcriptome to immunome: identification of DTH inducing proteins from a Phlebotomus ariasi salivary gland cDNA library. Vaccine. 2006;24:374–90. doi: 10.1016/j.vaccine.2005.07.085. [DOI] [PubMed] [Google Scholar]
- Paesen GC, Siebold C, Dallas ML, Peers C, Harlos K, Nuttall PA, Nunn MA, Stuart DI, Esnouf RM. An ion-channel modulator from the saliva of the brown ear tick has a highly modified Kunitz/BPTI structure. J Mol Biol. 2009;389:734–47. doi: 10.1016/j.jmb.2009.04.045. [DOI] [PubMed] [Google Scholar]
- Pawlowski J, Szadziewski R, Kmieciak D, Fahrni J, Bittar G. Phylogeny of the infraorder Culicomorpha (Diptera: Nematocera) based on the 28S RNA gene sequences. Sys. Entomol. 1996;21 [Google Scholar]
- Perez de Leon A, Tabachnick WJ. Apyrase activity and adenosine diphosphate induced platelet aggregation inhibition by the salivary gland proteins of Culicoides variipennis, the North American vector of bluetongue viruses. Vet. Parasitol. 1996;61:327–338. doi: 10.1016/0304-4017(95)00828-4. [DOI] [PubMed] [Google Scholar]
- Poinsignon A, Cornelie S, Ba F, Boulanger D, Sow C, Rossignol M, Sokhna C, Cisse B, Simondon F, Remoue F. Human IgG response to a salivary peptide, gSG6-P1, as a new immuno-epidemiological tool for evaluating low-level exposure to Anopheles bites. Malar J. 2009;8:198. doi: 10.1186/1475-2875-8-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinsignon A, Cornelie S, Mestres-Simon M, Lanfrancotti A, Rossignol M, Boulanger D, Cisse B, Sokhna C, Arca B, Simondon F, et al. Novel peptide marker corresponding to salivary protein gSG6 potentially identifies exposure to Anopheles bites. PLoS ONE. 2008;3:e2472. doi: 10.1371/journal.pone.0002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgar J, Clemetson JM, Kehrel BE, Wiedemann M, Magnenat EM, Wells TNC, Clemetson KJ. Platelet activation and signal transduction by convulxin, a C-type lectin from Crotalus durissus terrificus (tropical rattlesnake) venom via the p62/GPVI collagen receptor. J. Biol. Chem. 1997;272:13576–83. doi: 10.1074/jbc.272.21.13576. [DOI] [PubMed] [Google Scholar]
- Poole S, Lorenzetti BB, Cunha JM, Cunha FQ, Ferreira SH. Bradykinin B1 and B2 receptors, tumour necrosis factor alpha and inflammatory hyperalgesia. Br J.Pharmacol. 1999;126:649–56. doi: 10.1038/sj.bjp.0702347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Tolle DP, Barrett AJ. Evolutionary families of peptidase inhibitors. Biochem J. 2004;378:705–16. doi: 10.1042/BJ20031825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayms-Keller A, McGaw M, Oray C, Carlson JO, Beaty BJ. Molecular cloning and characterization of a metal responsive Aedes aegypti intestinal mucin cDNA. Insect Mol Biol. 2000;9:419–26. doi: 10.1046/j.1365-2583.2000.00202.x. [DOI] [PubMed] [Google Scholar]
- Remoue F, Cisse B, Ba F, Sokhna C, Herve JP, Boulanger D, Simondon F. Evaluation of the antibody response to Anopheles salivary antigens as a potential marker of risk of malaria. Trans R Soc Trop Med Hyg. 2006;100:363–70. doi: 10.1016/j.trstmh.2005.06.032. [DOI] [PubMed] [Google Scholar]
- Reno HE, Novak RJ. Characterization of apyrase-like activity in Ochlerotatus triseriatus, Ochlerotatus hendersoni, and Aedes aegypti. Am J Trop Med Hyg. 2005;73:541–5. [PubMed] [Google Scholar]
- Ribeiro JM. Blood-feeding in mosquitoes: probing time and salivary gland anti-haemostatic activities in representatives of three genera (Aedes, Anopheles, Culex) Med. Vet. Entomol. 2000;14:142–8. doi: 10.1046/j.1365-2915.2000.00227.x. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel SK. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem. Mol. Biol. 2004;34:543–63. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Charlab R, Rowton ED, Cupp EW. Simulium vittatum (Diptera: Simuliidae) and Lutzomyia longipalpis (Diptera: Psychodidae) salivary gland hyaluronidase activity. J. Med. Entomol. 2000a;37:743–7. doi: 10.1603/0022-2585-37.5.743. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Charlab R, Valenzuela JG. The salivary adenosine deaminase activity of the mosquitoes Culex quinquefasciatus and Aedes aegypti. J. Exp. Biol. 2001;204:2001–10. doi: 10.1242/jeb.204.11.2001. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Francischetti IM. Platelet-activating-factor-hydrolyzing phospholipase C in the salivary glands and saliva of the mosquito Culex quinquefasciatus. J. Exp. Biol. 2001;204:3887–94. doi: 10.1242/jeb.204.22.3887. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Katz O, Pannell LK, Waitumbi J, Warburg A. Salivary glands of the sand fly Phlebotomus papatasi contain pharmacologically active amounts of adenosine and 5'-AMP. J. Exp. Biol. 1999;202:1551–9. doi: 10.1242/jeb.202.11.1551. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Modi G. The salivary adenosine/AMP content of Phlebotomus argentipes Annandale and Brunetti, the main vector of human kala-azar. J. Parasitol. 2001;87:915–7. doi: 10.1645/0022-3395(2001)087[0915:TSAACO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Rowton ED, Charlab R. The salivary 5'-nucleotidase/phosphodiesterase of the hematophagous sand fly, Lutzomyia longipalpis. Insect Biochem. Mol. Biol. 2000b;30:279–85. doi: 10.1016/s0965-1748(99)00123-x. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Valenzuela JG. Purification and cloning of the salivary peroxidase/catechol oxidase of the mosquito Anopheles albimanus. J. Exp. Biol. 1999;202:809–16. doi: 10.1242/jeb.202.7.809. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Valenzuela JG. The salivary purine nucleosidase of the mosquito, Aedes aegypti. Insect Biochem Mol. Biol. 2003;33:13–22. doi: 10.1016/s0965-1748(02)00078-4. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Valenzuela JG, Pham VM, Kleeman L, Barbian KD, Favreau AJ, Eaton DP, Aoki V, Hans-Filho G, Rivitti EA, et al. An insight into the sialotranscriptome of Simulium nigrimanum, a black fly associated with fogo selvagem in South America. Am J Trop Med Hyg. 82:1060–75. doi: 10.4269/ajtmh.2010.09-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC. Blood-feeding arthropods: Live syringes or invertebrate pharmacologists? Infect. Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- Ribeiro JMC, Arca B. From sialomes to the sialoverse: An insight into the salivary potion of blood feeding insects. Adv Insect Physiol. 2009;37:59–118. [Google Scholar]
- Ribeiro JMC, Modi GB, Resh RB. Salivary apyrase activity of some old world phlebotomine sand flies. Insect Biochem. 1989;19:409–412. [Google Scholar]
- Ribeiro JMC, Nussenzveig RH. The salivary catechol oxidase/peroxidase activities of the mosquito, Anopheles albimanus. J. Exp. Biol. 1993;179:273–287. doi: 10.1242/jeb.179.1.273. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Rossignol PA, Spielman A. Salivary gland apyrase determines probing time in anopheline mosquitoes. J. Insect Physiol. 1985;9:551–560. [Google Scholar]
- Ribeiro JMC, Rossignol PA, Spielman A. Blood finding strategy of a capillary feeding sandfly, Lutzomyia longipalpis. Comp. Biochem. Physiol. 1986;83A:683–686. doi: 10.1016/0300-9629(86)90709-7. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Sarkis JJF, Rossignol PA, Spielman A. Salivary apyrase of Aedes aegypti: Characterization and secretory fate. Comp. Biochem. Physiol. 1984;79B:81–86. doi: 10.1016/0305-0491(84)90081-6. [DOI] [PubMed] [Google Scholar]
- Rossignol PA, Lueders AM. Bacteriolytic factor in the salivary glands of Aedes aegypti. Comp. Biochem. Physiol. 1986;83B:819–822. doi: 10.1016/0305-0491(86)90153-7. [DOI] [PubMed] [Google Scholar]
- Russell CL, Heesom KJ, Arthur CJ, Helps CR, Mellor PS, Day MJ, Torsteinsdottir S, Bjornsdottir TS, Wilson AD. Identification and isolation of cDNA clones encoding the abundant secreted proteins in the saliva proteome of Culicoides nubeculosus. Insect Mol Biol. 2009;18:383–93. doi: 10.1111/j.1365-2583.2009.00882.x. [DOI] [PubMed] [Google Scholar]
- Sarauer BL, Gillott C, Hegedus D. Characterization of an intestinal mucin from the peritrophic matrix of the diamondback moth, Plutella xylostella. Insect Mol Biol. 2003;12:333–43. doi: 10.1046/j.1365-2583.2003.00420.x. [DOI] [PubMed] [Google Scholar]
- Schluter H, Tepel M, Zidek W. Vascular actions of diadenosine phosphates. J Auton Pharmacol. 1996;16:357–62. doi: 10.1111/j.1474-8673.1996.tb00053.x. [DOI] [PubMed] [Google Scholar]
- Schwartz BS, Ford DP, Childs JE, Thomas RJ. Anti-tick saliva antibody: A biologic marker of tick exposure that is a risk factor for Lyme disease seropositivity. Am. J. Epidemiol. 1991;134:86–95. doi: 10.1093/oxfordjournals.aje.a115996. [DOI] [PubMed] [Google Scholar]
- Schwartz BS, Ribeiro JM, Goldstein MD. Anti-tick antibodies: an epidemiologic tool in Lyme disease research. Am J.Epidemiol. 1990;132:58–66. doi: 10.1093/oxfordjournals.aje.a115643. [DOI] [PubMed] [Google Scholar]
- Shi L, Paskewitz SM. Identification and molecular characterization of two immune-responsive chitinase-like proteins from Anopheles gambiae. Insect Mol. Biol. 2004;13:387–98. doi: 10.1111/j.0962-1075.2004.00496.x. [DOI] [PubMed] [Google Scholar]
- Simons FE, Peng Z. Mosquito allergy: recombinant mosquito salivary antigens for new diagnostic tests. Int. Arch. Allergy Immunol. 2001;124:403–5. doi: 10.1159/000053771. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- Stark KR, James AA. A factor Xa-directed anticoagulant from the salivary glands of the yellow fever mosquito Aedes aegypti. Exp. Parasitol. 1995;81:321–331. doi: 10.1006/expr.1995.1123. [DOI] [PubMed] [Google Scholar]
- Stark KR, James AA. Isolation and characterization of the gene encoding a novel factor Xa-directed anticoagulant from the yellow fever mosquito, Aedes aegypti. J. Biol. Chem. 1998;273:20802–9. doi: 10.1074/jbc.273.33.20802. [DOI] [PubMed] [Google Scholar]
- Stutzer C, Mans BJ, Gaspar AR, Neitz AW, Maritz-Olivier C. Ornithodoros savignyi: soft tick apyrase belongs to the 5'-nucleotidase family. Exp Parasitol. 2009;122:318–27. doi: 10.1016/j.exppara.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Sun D, McNicol A, James AA, Peng Z. Expression of functional recombinant mosquito salivary apyrase: A potential therapeutic platelet aggregation inhibitor. Platelets. 2006;17:178–84. doi: 10.1080/09537100500460234. [DOI] [PubMed] [Google Scholar]
- Thiel S. Complement activating soluble pattern recognition molecules with collagen-like regions, mannan-binding lectin, ficolins and associated proteins. Mol Immunol. 2007;44:3875–88. doi: 10.1016/j.molimm.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley SL, Wagoner VA, Salvatore CA, Jacobson MA, Koller BH. Adenosine and inosine increase cutaneous vasopermeability by activating A(3) receptors on mast cells. J. Clin. Invest. 2000;105:361–7. doi: 10.1172/JCI8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AM, Kuchel PW. The beta-defensin-fold family of polypeptides. Toxicon. 2004;44:581–8. doi: 10.1016/j.toxicon.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ferrer-Sueta G, Thomson L, Flohe L, Radi R. Kinetics of peroxiredoxins and their role in the decomposition of peroxynitrite. Subcell Biochem. 2007;44:83–113. doi: 10.1007/978-1-4020-6051-9_5. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Belkaid Y, Garfield MK, Mendez S, Kamhawi S, Rowton ED, Sacks DL, Ribeiro JM. Toward a defined anti-Leishmania vaccine targeting vector antigens: characterization of a protective salivary protein. J. Exp. Med. 2001a;194:331–42. doi: 10.1084/jem.194.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JG, Belkaid Y, Rowton E, Ribeiro JM. The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J. Exp. Biol. 2001b;204:229–37. doi: 10.1242/jeb.204.2.229. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Charlab R, Galperin MY, Ribeiro JM. Purification, cloning, and expression of an apyrase from the bed bug Cimex lectularius. A new type of nucleotide-binding enzyme. J. Biol. Chem. 1998;273:30583–90. doi: 10.1074/jbc.273.46.30583. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Charlab R, Gonzalez EC, Miranda-Santos IKF, Marinotti O, Francischetti IM, Ribeiro JMC. The D7 family of salivary proteins in blood sucking Diptera. Insect Mol. Biol. 2002a;11:149–55. doi: 10.1046/j.1365-2583.2002.00319.x. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem. Mol. Biol. 2003;33:717–32. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Ribeiro JM. Purification, cloning, and synthesis of a novel salivary anti-thrombin from the mosquito Anopheles albimanus. Biochemistry. 1999;38:11209–11215. doi: 10.1021/bi990761i. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Garfield M, Rowton ED, Pham VM. Identification of the most abundant secreted proteins from the salivary glands of the sand fly Lutzomyia longipalpis, vector of Leishmania chagasi. J Exp Biol. 2004;207:3717–29. doi: 10.1242/jeb.01185. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Pham VM, Garfield MK, Francischetti IM, Ribeiro JMC. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2002b;32:1101–1122. doi: 10.1016/s0965-1748(02)00047-4. [DOI] [PubMed] [Google Scholar]
- Vizioli J, Bulet P, Hoffmann JA, Kafatos FC, Muller HM, Dimopoulos G. Gambicin: a novel immune responsive antimicrobial peptide from the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. U S A. 2001;98:12630–5. doi: 10.1073/pnas.221466798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volfova V, Hostomska J, Cerny M, Votypka J, Volf P. Hyaluronidase of bloodsucking insects and its enhancing effect on leishmania infection in mice. PLoS Negl Trop Dis. 2008;2:e294. doi: 10.1371/journal.pntd.0000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ribeiro JM, Broce AB, Wilkerson MJ, Kanost MR. An insight into the transcriptome and proteome of the salivary gland of the stable fly, Stomoxys calcitrans. Insect Biochem Mol Biol. 2009 doi: 10.1016/j.ibmb.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AD, Heesom KJ, Mawby WJ, Mellor PS, Russell CL. Identification of abundant proteins and potential allergens in Culicoides nubeculosus salivary glands. Vet Immunol Immunopathol. 2008;122:94–103. doi: 10.1016/j.vetimm.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Xu X, Yang H, Ma D, Wu J, Wang Y, Song Y, Wang X, Lu Y, Yang J, Lai R. Toward an understanding of the molecular mechanism for successful blood feeding by coupling proteomics analysis with pharmacological testing of horsefly salivary glands. Mol Cell Proteomics. 2008;7:582–90. doi: 10.1074/mcp.M700497-MCP200. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Koike H, Sugiyama Y, Motoyoshi K, Wada T, Hishinuma S, Mita M, Morita T. Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur J.Biochem. 2002;269:2708–15. doi: 10.1046/j.1432-1033.2002.02940.x. [DOI] [PubMed] [Google Scholar]
- Yeates DK, Wiegmann BM. Congruence and controversy: toward a higher-level phylogeny of Diptera. Annu Rev Entomol. 1999;44:397–428. doi: 10.1146/annurev.ento.44.1.397. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sudo T, Niimi M, Tao L, Sun B, Kambayashi J, Watanabe H, Luo E, Matsuoka H. Inhibition of collagen-induced platelet aggregation by anopheline antiplatelet protein, a saliva protein from a malaria vector mosquito. Blood. 2008;111:2007–14. doi: 10.1182/blood-2007-06-097824. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Watanabe H. Robust salivary gland-specific transgene expression in Anopheles stephensi mosquito. Insect Mol Biol. 2006;15:403–10. doi: 10.1111/j.1365-2583.2006.00645.x. [DOI] [PubMed] [Google Scholar]
- Zavalova LL, Baskova IP, Lukyanov SA, Sass AV, Snezhkov EV, Akopov SB, Artamonova, Archipova VS, Nesmeyanov VA, Kozlov DG, et al. Destabilase from the medicinal leech is a representative of a novel family of lysozymes. Biochim Biophys Acta. 2000;1478:69–77. doi: 10.1016/s0167-4838(00)00006-6. [DOI] [PubMed] [Google Scholar]
- Zeidner NS, Higgs S, Happ CM, Beaty BJ, Miller BR. Mosquito feeding modulates Th1 and Th2 cytokines in flavivirus susceptible mice: an effect mimicked by injection of sialokinins, but not demonstrated in flavivirus resistant mice. Parasite Immunol. 1999;21:35–44. doi: 10.1046/j.1365-3024.1999.00199.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.