Abstract
The expression of virulence factors in Staphylococcus aureus is tightly coordinated by a vast network of regulatory molecules. In this report, we characterize a genetic locus unique to staphylococci called rsr that has a role in repressing two key virulence regulators, sarR and agr. Using strain SH1000, we showed that the transcription of virulence effectors, such as hla, sspA, and spa, is altered in an rsr mutant in a way consistent with agr upregulation. Analysis of RNAIII expression of the agr locus in rsr and rsr-sarR mutants indicated that rsr likely contributes to agr expression independently of SarR. We also provide evidence using a murine model of S. aureus skin infection that the effects mediated by rsr reduce disease progression.
Staphylococcus aureus is a leading cause of hospital- and community-acquired infections in the United States (8, 18). Its ability to colonize a variety of sites gives rise to a number of disease states ranging from superficial skin and soft tissue infections to pneumonia, endocarditis, osteomyelitis, and septic shock. Disease progression and severity are mediated by a diverse array of cell adhesion molecules, toxins, and immune system modulators which are tightly controlled by a large repertoire of regulatory molecules (5).
A basic model of S. aureus pathogenesis has virulence progressing in two discrete stages. The first stage is typified by the production of adhesins and surface proteins during the exponential phase of growth. The second stage of infection, analogous to the post-exponential and stationary phases of growth in vitro, is characterized by increased toxin production, which leads to increased tissue damage and bacterial spread. The regulatory events controlling this phenotypic switch center on the activities of the agr quorum-sensing system, which represses the expression of surface proteins, including protein A (spa), and upregulates the production of toxins, such as α-hemolysin (hla) and V8 protease (sspA), in transition from late exponential to post-exponential phases (22).
The agr system is comprised of two divergent transcripts, RNAII and RNAIII. RNAII carries four genes, agrDBCA, which are dedicated to the synthesis, sensing, and processing of the quorum-sensing autoinducing peptide (AIP) required for the activation of the RNAII promoter, primarily during the late exponential phase. The divergent transcript RNAIII, activated in response to RNAII via AgrA, is a regulatory RNA molecule expressed maximally during the post-exponential phase and controls virulence gene expression at the transcriptional and translational levels, the latter via an antisense mechanism (11). A number of regulators controlling agr expression (SarA, MgrA, ArlRS, SrrAB, SarX, and SarR) have been described, some with positive effects (e.g., SarA and MgrA) and others with negative effects (e.g., SrrAB and SarX) (7, 10, 15, 19, 20, 27).
Lying adjacent to sarR, which encodes a 13.6-kDa protein belonging to the SarA family of DNA binding global regulators in the S. aureus chromosome (6), is a gene we called rsr (for repressor of sarR) that is associated with sarR expression. We also confirmed that rsr is a repressor of the agr system. Additionally, an rsr mutant is hypervirulent in a murine model of skin infection. These data suggest that the rsr locus is a general inhibitor of virulence in S. aureus.
MATERIALS AND METHODS
Reagents, bacterial strains, and growth conditions.
Unless specified otherwise, all reagents were obtained from either Fisher (Pittsburgh, PA) or Sigma (St. Louis, MO). The bacterial strains used in this study are listed in Table 1. Escherichia coli strains were routinely grown on Luria-Bertani (LB) broth or agar containing 100 μg/ml of ampicillin where appropriate. S. aureus cultures were routinely cultured on either tryptic soy broth or agar. Strains carrying plasmids were selected and maintained on either 2.5 μg/ml erythromycin or 10 μg/ml chloramphenicol, as appropriate. Experiments investigating agr transcription were carried out in 03GL broth (23).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| S. aureus | ||
| RN4220 | Heavily mutagenized strain that accepts foreign DNA, agr mutant | 23 |
| SH1000 | 8325-4 with rsbU restored | 14 |
| COL | A methicillin-resistant strain, isolated from a human infection in the early 1960s | 13 |
| Newman | Methicillin-sensitive laboratory strain, isolated from a human infection in 1952 | 3 |
| RN6390 | Laboratory strain related to strain 8325-4 | 25 |
| ALC4657 | SH1000 Δrsr | This work |
| ALC4658 | COL Δrsr | This work |
| ALC4659 | Newman Δrsr | This work |
| ALC4660 | RN6390 Δrsr | This work |
| ALC6582 | SH1000 Δrsr/rsr complement | This work |
| ALC6583 | SH1000 ΔsarR | This work |
| ALC6584 | SH1000 ΔsarR Δrsr | This work |
| ALC2386 | SH1000 agr::tetM | 14 |
| ALC6585 | SH1000 agr::tetM Δrsr | This work |
| ALC6586 | SH1000 with pEPSA5::rsr | This work |
| E. coli | ||
| TOP10 | General cloning strain | Invitrogen |
| Plasmids | ||
| pMAD | E. coli-S. aureus shuttle vector containing a thermosensitive origin of replication bgaB, Emr, Apr | 2 |
| pEPSA5 | E. coli-S. aureus shuttle vector containing a xylose-inducible promoter, Apr, Cmr | 9 |
| pALC1484 | E. coli-S. aureus shuttle vector derived from pSK236 containing a multiple cloning site upstream of the gfpuvr gene (a gfp variant optimized for expression in S. aureus), Apr, Cmr | 16 |
| pALC1743 | agr P3 promoter driving the expression of gfpuvr in pALC1484 | 16 |
| pALC1741 | spa promoter driving the expression of gfpuvr in pALC1484 | 17 |
| pALC1740 | hla promoter driving the expression of gfpuvr in pALC1484 | 17 |
| pALC2831 | sspA promoter driving the expression of gfpuvr in pALC1484 | 26 |
Cultures for gene and protein expression analysis were grown in either glass sidearm flasks or 18-mm glass tubes in an Excella E24 shaking incubator set at 37°C and 250 rpm (New Brunswick Scientific, Edison, NJ). Growth was monitored by determining culture turbidity at 650 nm with an 18-mm borosilicate glass tube using a Spectronic 20D+ spectrophotometer (Spectronic Analytical Instruments, Garforth, England). Data collected at mid-, late, or post-exponential phases of growth corresponded to turbidity readings of 0.7, 1.1, and 1.7, respectively.
Genetic manipulations.
A list of oligonucleotides used in this study is available from the authors. Enzymes and reagents used for genetic manipulations were obtained from either Invitrogen (Irving, CA) or New England Biolabs (Ipswich, MA) and used according to the manufacturers' recommendations. Plasmids were originally propagated in E. coli and transformed by electroporation into RN4220, a heavily mutagenized, restriction-deficient strain of S. aureus capable of methylating foreign DNA. The methylated plasmid DNA was then electroporated into the appropriate S. aureus strains. All positive clones were initially identified by PCR and confirmed by DNA sequencing.
A markerless, in-frame deletion of rsr was constructed in four S. aureus strains using a PCR-based splicing technique followed by splicing overlap extension (SOEing), as described previously (2, 26). Briefly, the fragment was cloned into pMAD and introduced into S. aureus strains SH1000, Newman, RN6390, and COL (to obtain rsr) or ALC2386 (to obtain the rsr-agr mutant in SH1000) by electroporation, followed by the selection of blue colonies. Transformants were grown at 42°C, a nonpermissive temperature for pMAD replication, and then at 30°C to drive homologous recombination to yield white colonies (2, 26). A similar approach was used to construct the rsr-sarR double mutant in the SH1000 background, using primers flanking both sarR and rsr.
The mutation in the rsr mutant was restored by cloning the wild-type gene into pMAD and reintroducing it into the chromosome by homologous recombination, as described above. The rsr-overproducing strain was creating by cloning the rsr open reading frame behind the xylose-inducible promoter of the plasmid pEPSA5 (9).
Transcriptional analysis by Northern blotting.
Cells from either the mid-, late, or post-exponential phases of growth were harvested, resuspended in Trizol (Invitrogen), and lysed using 0.1-mm silica-zirconia beads in a reciprocating shaker. The RNA was extracted and resuspended in diethyl pyrocarbonate (DEPC)-treated water, according to the manufacturer's instructions. Ten micrograms of RNA was separated electrophoretically on 1.5% agarose-0.66 M formaldehyde gels in MOPS buffer (20 mM morpholinepropanesulfonic acid, 10 mM sodium acetate, 2 mM EDTA [pH 7]), transferred onto a Hybond XL nylon membrane (GE Healthcare) in 20× SSC (3 M NaCl, 0.3 M sodium citrate [pH 7]), and fixed to the membrane by cross-linking in a Stratalinker 1800 (Stratagene, La Jolla, CA).
The DNA probes, generated by either PCR or restriction digestion, were labeled with [α-32P]dCTP using the Roche random prime labeling kit (Hoffmann-La Roche Inc., Nutley, NJ) and added to the nylon membranes at 65°C. After hybridization overnight, the membranes were washed extensively and autoradiographed. Blots shown in Fig. 1, 2, 3, and 6 are representative of results from three separate experiments, and the intensities of the resulting bands were quantified using ImageJ (1).
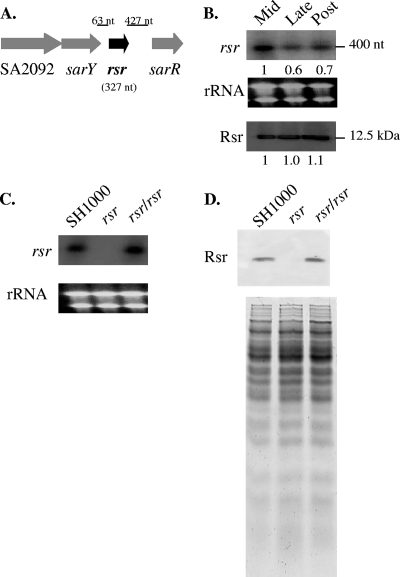
FIG. 1.
(A) Genomic context of the rsr gene. rsr lies in close proximity to three regulatory genes, sarR, sarY, and SA2092, which encodes a gene product with similarity to the AraC protein family. (B) Transcription (top) of rsr during the early, mid-, and late exponential phases of growth in strain SH1000. Numbers underneath the panels indicate relative intensities of the bands as determined by ImageJ. rRNA bands (middle) are shown as a loading control for the top panel. (C) Transcription (top) of rsr during the mid-exponential phase in the SH1000 parent, rsr mutant, and restored rsr mutant with a single copy of the rsr gene at its native chromosomal location. The intensities of the rRNA bands (bottom) serve as loading controls. (D) Translation of Rsr in the parent, mutant, and restored rsr mutant strains at late exponential phase as detected by a Western blot, with 20 μg of whole-cell lysate each and probed with a 1:1,000 dilution of murine anti-Rsr antibody (top). The bottom panel represents an equivalent SDS gel stained with Coomassie blue to demonstrate comparable loading among the lanes.
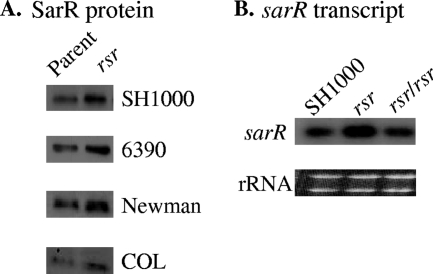
FIG. 2.
Effect of an rsr mutation on the translation (A) and transcription (B) of sarR. (A) Western blot analysis of SarR expression in rsr mutations in four S. aureus strains. Cell lysates were prepared from late-exponential-phase cells and probed with a murine anti-SarR monoclonal antibody. Expression of the SarR protein in the rsr mutants was 1.4, 1.6, 1.2, and 1.5 times that of the respective parents in SH1000, RN6390, Newman, and COL, as determined by ImageJ. (B) Northern blot analysis of sarR transcription during the late exponential growth phase in the SH1000 parent, the rsr mutant (expression at 1.7 times that of SH1000), and the restored rsr mutant. rRNA bands are shown as loading controls.
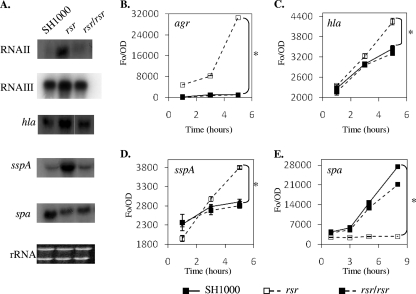
FIG. 3.
Effect of an rsr mutation on agr RNAII and RNAIII, hla, sspA, and spa transcript levels (A) and promoter activities (B to E). (A) Northern blot analysis of RNA isolated from postexponential-phase (RNAII, RNAIII, hla, and sspA) or late-exponential-phase (spa) cells. Late exponential and postexponential phases are defined as an OD650 of 1.1 and 1.7, respectively, using 18-mm borosilicate glass tubes in a Spectronic 20D+ spectrophotometer. (B to E) GFP-promoter fusion analysis of the agrP3, hla, sspA, and spa promoter activities, respectively, over the course of bacterial growth in the SH1000, rsr mutant, and restored rsr mutant strains. *, P < 0.01, as determined by the Student t test.
Real-time PCR analysis of gene transcription.
RNA, prepared as described above, was treated with RNase-free DNase to remove residual DNA, according to the manufacturer's instructions (Turbo-DNAfree; Applied Biosystems/Ambion, Austin, TX). Successful removal of the DNA was confirmed by PCR analysis. RNA (1 μg) was reverse transcribed using the Roche Transcriptor first-strand cDNA synthesis kit, and the resulting cDNA levels were quantified using the Maxima SYBR kit from Fermentas (Glen Burnie, MD) in a Roche LightCycler 1.5 according to the manufacturer's protocols. Reported levels of gene expression were normalized to those of gyrB.
Detection of SarR and Rsr by Western blotting.
Post-exponential-phase (optical density at 650 nm [OD650] of 1.7) or late-exponential-phase (OD650 of 1.1) cells were collected and lysed in 10 mM Tris, 1 mM EDTA (pH 8) with 0.1-mm silica-zirconia beads in a reciprocating shaker. Soluble cellular proteins were separated from debris by a high-speed spin at 12,000 × g for 5 min at 4°C and quantified using the BCA protein assay kit (Fisher). Equal amounts (20 or 75 μg) of protein were then separated on a 12.5% polyacrylamide-SDS gel and transferred onto a polyvinylidene difluoride membrane. The presence of SarR or Rsr was detected on the membranes using standard Western blotting procedures. The primary antibody was a murine-derived anti-SarR monoclonal antibody (clone 1D1) or murine anti-Rsr polyclonal antibody used at a concentration of 1:1,000, and the secondary antibody was donkey-derived anti-mouse immunoglobulin G conjugated with horseradish peroxidase (Jackson ImmunoResearch Laboratories, West Grove, PA) and used at a concentration of 1:10,000. After a series of extensive washes, an enhanced chemiluminescence kit (GE Healthcare) was used to detect binding of the antibodies to the membrane. Where applicable, ImageJ was used to quantify the relative intensities of the resulting bands. Results shown are representative of three separate experiments.
Transcriptional analysis using green fluorescent protein (GFP)-promoter fusions.
Shuttle plasmids containing the promoter regions of various genes (Table 1) driving the expression of GFPuvr (16) were transformed into SH1000, the rsr mutant, and the complemented mutant by electroporation. The resulting strains were grown overnight in tryptic soy broth (TSB), diluted 1:100 in fresh TSB the next morning, and incubated at 37°C with shaking at 250 rpm. Growth (OD600) and fluorescence (excitation/emission of 485/515 nm) of the cultures were monitored with an FL600 microplate fluorescence reader (BioTek Instruments, Winooski, VT). Experiments were performed three times using four independently isolated clones in triplicate. Error bars represent the standard errors of the means, and statistical significance was determined by the Student t test.
Subcutaneous murine model of skin infection.
SH1000 and its isogenic rsr mutant were grown in TSB to post-exponential phase (OD650 of 1.7), harvested, washed three times in sterile saline, and resuspended in sterile saline at 109 CFU/ml. CFU (108) in 100 μl of SH100 or the rsr mutant were injected subcutaneously into the shaved backs of six 12-week-old male C57BL/6 mice in the SH1000 group or in the rsr mutant group (Jackson Labs, Bar Harbor, ME). One mouse from the SH1000 group died upon handling. The mice were then monitored daily for subcutaneous abscess formation and other changes in general appearance (e.g., ruffled hair and activity). On the 8th day postinfection, the mice were sacrificed and 8-mm punch biopsy specimens of the skin were obtained from the infected sites. A portion of the skin biopsy specimen was fixed in 10% neutral buffered formalin for histological analysis; the remainder was processed in a glass tissue grinder and serially diluted to determine bacterial counts. Results reported are the means relative to the mass of the tissue section in grams. Statistical significance was determined using the Student t test.
Samples for histology were embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin according to standard histological procedures. Slides were viewed and images were captured using an Olympus digital light microscope (Olympus America Inc., Center Valley, PA).
RESULTS
Identification of rsr.
Recent complementation studies of the sarR mutant from our lab suggested that the genetic locus upstream of sarR may inhibit sarR expression. We have termed this locus rsr, for repressor of sarR. rsr is a unique gene bearing no homology to any gene outside the staphylococci, nor does it have any conserved motifs or domains. It is predicted to encode a 108-amino-acid protein with a molecular mass of ∼12.5 kDa and a theoretical pI of 4.54.
The rsr locus neighbors other regulatory genes (Fig. 1 A). Besides being upstream of sarR, rsr is also directly downstream of sarY, another member of the SarA protein family (5, 6). An uncharacterized gene, SA2092, lies upstream of sarY and is homologous to the AraC family of regulators. Despite its proximity to these regulatory genes, rsr appeared to be transcribed monocistronically with a transcript size of ∼400 nucleotides (nt), corresponding to the size of the rsr gene plus the intergenic region at the 5′ end (Fig. 1A and B). Analysis of rsr expression during different phases of S. aureus growth revealed that the gene was most highly transcribed during mid-exponential phase; however, the levels of the Rsr protein produced appeared to be stable throughout the growth period (Fig. 1B).
To investigate the phenotypic and genotypic effects of rsr, we first constructed an in-frame deletion of rsr in S. aureus strain SH1000, chosen for its similarity to the commonly published strains, including 8325-4 and RN6390, and its possession of an intact rsbU gene within the stress-induced sigB operon. As expected, the rsr mutant of SH1000 did not express the cognate transcript, while restoration of a single copy of rsr to the mutant chromosome reestablished the transcription of rsr (Fig. 1C). Immunoblot analysis demonstrated that the Rsr protein was absent in the rsr mutant but was expressed in the parent SH1000 strain and the restored rsr mutant strain (Fig. 1D).
Repression of SarR by rsr.
As described above, we initially suspected that rsr, a gene adjacent to sarR, may be a repressor of SarR expression in S. aureus. To verify the impact of rsr on sarR expression in a variety of prototypic S. aureus strains, including SH1000, we also deleted rsr in RN6390, Newman, and COL, thus encompassing both methicillin-susceptible (MSSA) and methicillin-resistant S. aureus (MRSA) strains. Lysates of rsr mutants from strains SH1000, RN6390, Newman, and COL prepared from post-exponential-phase cells (corresponding to the peak expression period for sarR) were immunoblotted and probed with an anti-SarR monoclonal antibody. In all four genetic backgrounds, expression of the SarR protein was increased in the mutant compared to that in the cognate parent, ranging from an ∼20% increase in the Newman mutant to ∼60% in the RN6390 mutant compared with the corresponding parent, thus demonstrating the repressive effect of rsr on SarR in a variety of strains (Fig. 2 A). Based on these trends, we decided to focus on SH1000, predicated upon the wealth of information we and others have generated on the regulatory pathway of agr and sarA in this strain (15, 20, 21, 29). Using the rsr mutant and the restored rsr mutant in which the mutated gene was replaced by the wild-type copy, we analyzed sarR transcription and showed that the sarR transcript level was increased in the rsr mutant compared with that in the cognate parent SH1000 strain but returned to the parental level in the restored rsr mutant, strongly suggesting that rsr was repressing sarR expression (Fig. 2B).
Role of rsr in virulence gene transcription.
Given the role of agr and sarA in modulating expression of many virulence genes (19, 21), we next examined the transcript levels of these two regulatory loci in the rsr mutant compared to those in the parent SH1000 strain and the restored rsr mutant strain. Curiously, the levels of sarA transcription were virtually identical among all 3 strains (data not shown), thus implying that rsr does not play a major role in controlling sarA expression. We then assessed agr expression by ascertaining the levels of RNAII (carrying the machinery of the agr system, agrDBCA), normally expressed maximally during the late exponential phase (OD650 of 1.1). As shown in Fig. 3 A, the transcription of RNAII was significantly increased in the rsr mutant compared to that in the parent and the restored rsr mutant. As RNAII augments RNAIII (the agr effector regulatory molecule), we also found that RNAIII expression was elevated in the rsr mutant compared to that in the cognate parent and restored mutant (Fig. 3A). GFP fusion analysis of the agr RNAIII promoter confirmed the Northern blot data (Fig. 3B). We also examined the expression of virulence genes controlled by agr. More specifically, the transcription of hla (encoding α-hemolysin), sspA (encoding V8 protease), and spa (encoding protein A) was also affected upon deletion of rsr, with both hla and sspA transcription upregulated by 2- to 3-fold in the rsr mutant compared to the that in the parent and the restored rsr mutant, as assessed by Northern blot and promoter fusion analyses (Fig. 3A, C, and D). In contrast, the transcription of spa was downregulated in the rsr mutant compared to that in the parent and the restored rsr mutant (Fig. 3A and E). The upregulation of hla and sspA and the downregulation of spa in the rsr mutant of SH1000 is in concordance with upregulation of RNAII and RNAIII in this strain.
Ascertaining the linkage between rsr and agr.
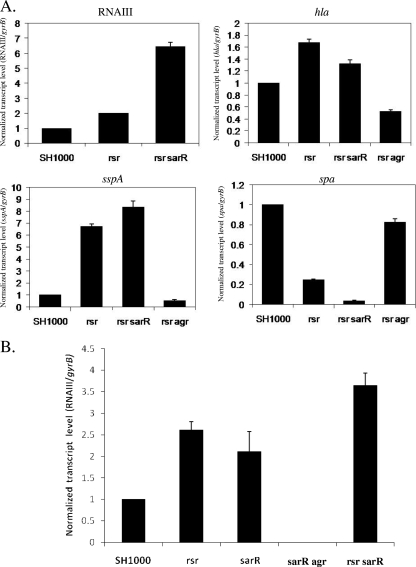
Based on the above transcriptional data, we wanted to determine if rsr requires SarR to impact agr and the ensuing downstream genes, such as hla, sspA, and spa. For this purpose, we first constructed an in-frame deletion mutant of both sarR and rsr to determine the role of sarR in mediating the effect of rsr on agr. Cognizant of the fact that the agr effector molecule RNAIII is maximally expressed during the post-exponential phase (OD650 of 1.7), we used real-time PCR transcription of RNAIII in the rsr-sarR double mutant during this growth phase. As shown in Fig. 4 A (top left), the transcription of RNAIII was more pronounced in the double rsr-sarR mutant than in the rsr single mutant by a factor of 3.5 and in the parent SH1000 by a factor of 6.7. As expected, the transcription of hla in the rsr and rsr-sarR mutants was increased compared with that in the parental SH1000 strain. However, hla expression in the rsr mutant was 1.3-fold higher than in the double rsr-sarR mutant (Fig. 4A, top right). With respect to sspA and spa transcription, the rsr-sarR double mutant demonstrated a phenotype consistent with a higher level of RNAIII than the single rsr mutant, with sspA transcript levels increased by 1.3-fold and spa transcription significantly decreased in the double mutant compared to that in the rsr single mutant (Fig. 4A, bottom). We also analyzed the effect of rsr on RNAIII during the late exponential phase (OD650 of 1.1). Similar to the findings during the post-exponential phase, the double rsr-sarR mutant exhibited a higher level of RNAIII expression than either the single rsr or the sarR mutant, as assessed by real-time PCR (Fig. 4B). More importantly, both rsr and sarR mutants demonstrated higher RNAIII expression than the parental SH1000 strain. Based on the above data with RNAIII, sspA, and spa, our results suggested that agr and its downstream effectors are repressed by rsr. This repressive effect of rsr is likely independent of sarR, because RNAIII expression is higher in the rsr-sarR double mutant than in the single rsr and sarR mutants.
FIG. 4.
(A) Transcriptional analysis of RNAIII, hla, sspA, and spa in an rsr-sarR and an rsr-agr double mutant from post-exponential-phase cells (OD650 of 1.7), treated with RNase-free DNase, reverse transcribed into cDNA, and analyzed by real-time PCR. (B) Real time-PCR of RNAIII of late-exponential-phase cells (OD650 of 1.1) from rsr, sarR, sarR-agr, and rsr-sarR mutants. Transcription levels in the mutants are normalized to the levels of gyrB and are reported relative to their transcription in the SH1000 parent (set to 1). The error bars represent individual standard deviations. The P values for SH1000 compared to the single rsr mutant, sarR mutants, and double mutants are <0.0008, <0.02, and <0.0006, respectively. The P values for the rsr-sarR double mutant compared to the rsr single mutant and the sarR mutant are <0.005 and <0.008, respectively.
To determine if the virulence genes controlled by rsr are totally dependent on agr repression, we constructed an rsr-agr double mutant, followed by transcriptional analysis of virulence genes in this strain by real-time PCR. In contrast to the rsr-sarR double mutant, the transcriptional patterns of the rsr-agr double mutant differed significantly from that of the single rsr mutant, with both hla and sspA transcription markedly reduced (over 50% reduction) compared to that in the parental strain, while spa transcription was almost fully restored to the parental level (Fig. 4A), again reaffirming the notion that the regulation of these effector genes by rsr is mediated via RNAIII of the agr locus (Fig. 4A).
Hypervirulence of an rsr mutant.
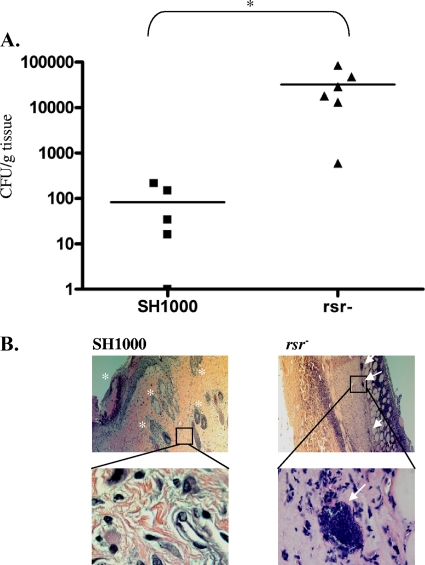
Based on the transcriptional profile of increased agr transcription in the rsr mutant, we reasoned that the mutant might be hypervirulent compared with the parent. To test this hypothesis, we employed a murine model of cutaneous infection. For this model, we injected 108 CFU/100 μl of SH1000 or the rsr mutant subcutaneously into the shaved backs of 6- to 12-week-old male C57BL/6 mice. On the first day postinfection, all of the mice in the mutant group had developed small areas of redness near the injection site, whereas none of the mice in the parental SH1000 group did. As the infection progressed, the mice infected with SH1000 did eventually develop small, red lesions on their backs that were indistinguishable from those of the mice in the rsr mutant group. The bacterial loads recovered from the tissue biopsy specimens were markedly different between the two groups of mice. On average, 83 and 32,500 CFU/gram of tissues were recovered from the mice in the SH1000 and rsr groups, respectively (Fig. 5 A). The histological profile of the tissue sections taken from the mice reflected their bacterial burden. Skin tissue samples taken from the mice infected with SH1000 had normal skin architecture with increased infiltration of phagocytic cells in the dermis (Fig. 5B, left) and a lack of tissue necrosis. Importantly, few, if any, bacteria were present in the infected tissue. Mice infected with the rsr mutant, in contrast, revealed abnormal skin histology; in particular, there was a massive infiltration of neutrophils and monocytes to the dermal and subdermal layers (Fig. 5B, right). The epidermal layer was absent, and several subcutaneous structures, such as hair shafts and sebaceous glands, were obliterated. Additionally, large clusters of bacteria were visible in the deep muscle tissue (Fig. 5B, bottom right).
FIG. 5.
Virulence of the rsr mutant. (A) A group of six C57BL/6 mice were each injected subcutaneously with 108 CFU of parent SH1000 or the rsr mutant and monitored daily. Eight days postinfection, the mice were sacrificed, and punch biopsies of the skin were done to determine bacterial load (*, P < 0.01, as determined by the Student t test) and to examine any histological differences between the mice from the parental SH1000 group (left) and the rsr mutant group (right). (B) Top, low magnification (100×) of the dermal and subdermal layers. Note the absence of the dermal layer and subcellular structures (*) in the biopsy specimen taken from a mouse infected with the rsr mutant and the presence of clusters of bacteria (arrows). Bottom, higher magnification (1,000×) of a representative bacterial cluster (right, arrow) present in the rsr mutant-infected tissue (right) and an equivalent region from the SH1000-infected tissue (left).
rsr as an inhibitor of agr.
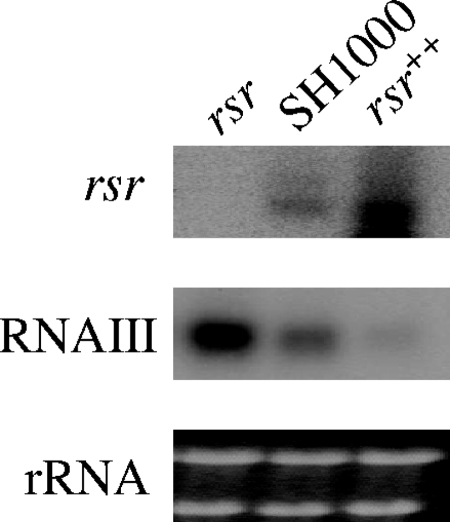
Both transcriptional analysis of virulence genes and the results obtained from the murine model of cutaneous infection suggested that the rsr mutant possessed an increased virulence phenotype. We thus considered the possibility that provision of rsr in multiple copies to S. aureus may decrease its virulence properties. To test this hypothesis, rsr was cloned downstream of the xylose-inducible promoter of the pEPSA5 plasmid and transformed into strain SH1000. The levels of rsr and RNAIII transcription were then assessed in the resultant strain, the rsr++ strain. As shown in Fig. 6, the level of rsr transcription was approximately 4-fold higher in rsr++ than in SH1000. The increased gene dosage of rsr in the rsr++ strain resulted in a dramatic reduction (∼80%) in agr RNAIII transcription compared with that in SH1000.
FIG. 6.
rsr as an inhibitor of virulence gene regulators. The rsr gene was overexpressed in SH1000 using a multicopy plasmid pEPSA5 containing rsr, and the transcript levels of rsr and agr RNAIII were assessed by Northern blotting using RNA extracted from post-exponential-phase cells (OD650 of 1.7). The expression of rsr and RNAIII in the rsr mutant expressing a high copy number of rsr was 4 times and 0.2 times that of the parental SH1000 strain, using ImageJ to determine the relative band intensities.
DISCUSSION
In this study, we report the identification of rsr, a gene unique to the staphylococci that bears no similarity to any known gene or conserved domain. Inactivation of rsr, residing in a chromosomal location directly adjacent to sarR, resulted in overproduction of the virulence gene regulator SarR in four S. aureus strains. Using SH1000 as the genetic backdrop, we found that the rsr mutant also exhibited augmented agr (RNAII and RNAIII), hla (α-hemolysin), and sspA (V8 protease) transcription as well as a decrease in the spa (protein A) transcript level, consistent with an increase in agr expression in the rsr mutant. As SarA is a positive regulator of agr, we also analyzed sarA transcription in the rsr mutant of S. aureus strain SH1000 and found comparable levels of sarA transcription among the mutant, parent, and rsr-restored mutant strains.
The rsr gene was transcribed most highly during the mid-exponential phase of growth; however, analysis of Rsr suggested that the protein level was relatively constant throughout the growth cycle. This observation suggested that the Rsr protein may not be involved in the temporal control of gene expression unlike several other S. aureus regulatory loci but may act throughout the growth cycle (4, 5). Indeed, changes in activity of both the agr and spa promoters, which exhibited the most striking changes in the rsr mutant, were sustained over the entire growth cycle (Fig. 3B and D). On the other hand, the promoter activities of hla and sspA, which are most active after the exponential phase, remained subdued in the mutant until the cells reached post-exponential phase (Fig. 3C and D), at which time these promoters became much more active than those in the parent, suggesting that regulatory factors other than rsr may have been at play in controlling these genes.
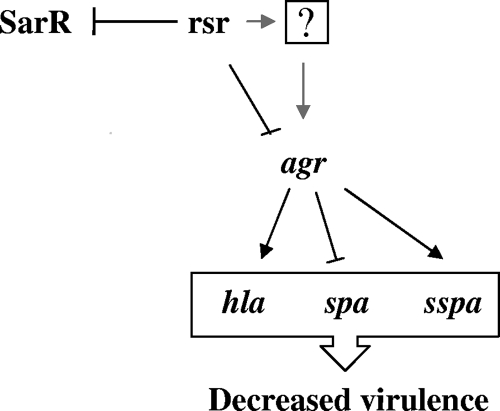
Although rsr is a repressor of sarR (Fig. 2) and agr (Fig. 3), we found that increased agr RNAIII expression in the rsr mutant is not attributable to enhanced sarR expression, because RNAIII expression in the double rsr-sarR mutant was higher than that in the single rsr and sarR mutants (Fig. 4B). More specifically, analysis of agr expression in the rsr-sarR double mutant indicated that the derepressive effects of rsr and sarR mutations on agr RNAII and subsequently on RNAIII appear to be additive (e.g., compare the RNAIII expression in rsr, sarR, and double rsr-sarR mutants in Fig. 4B). Consistent with this result was the observation that the effects of concomitant rsr and sarR mutations in SH1000 on sspA and spa expression also appeared to be culminative (Fig. 4A). These results indicated that either rsr directly represses agr transcription or does so via another regulatory molecule which acts as an intermediary between rsr and agr. In addition, the effect of rsr on agr expression is independent of SarR. The finding that the introduction of an agr mutation in an rsr mutant can downregulate hla and sspA as well as upregulate spa expression to near parental levels (Fig. 4) could lend support to the notion that rsr regulates virulence determinants primarily via an intact agr pathway (Fig. 7).
FIG. 7.
Proposed mechanism of rsr-mediated gene regulation. rsr represses both sarR and agr during the late exponential phase. The repressive effects on agr by rsr during the late exponential phase are shown with regard to hla, sspA, and spa transcription. It is unclear whether rsr represses agr directly or via an as-of-yet-unidentified regulator. The repression of agr contributes to the repression of both hla and sspA and to the activation of spa. Together, these regulatory changes are predicted to result in a net decrease in virulence. The genetic locus rsr thus acts independently of SarR to inhibit agr expression.
The transcriptional changes observed in the rsr mutant implied that its virulence phenotype may also have been enhanced. These phenotypic changes, however, were difficult to assess in SH1000 because this strain, by virtue of the restored rsbU gene, displays an enhanced sigB response and hence a more muted agr response (14), resulting in very low levels of hemolysis and proteolysis activities in vitro. We therefore decided to evaluate the clinical significance of rsr in vivo by comparing the virulence properties of the parent and mutant strains in a murine model of cutaneous infection. As expected, SH1000 was not proficient in causing subcutaneous abscesses in the mouse model, requiring up to 108 CFU to establish a mild abscess in the parent group. With the exception of a few erythematous lesions, most of the mice seemed unaffected initially by the subcutaneous injection of 108 CFU of strain SH1000. Histological studies also disclosed the integrity of the epidermis and dermis layers of the skin, thus corroborating with the clinical observation of these mice, which appeared relatively healthy. In contrast, subcutaneous injection of an equivalent dose of the rsr mutant offered dramatically different results, with on average almost 100-fold more bacteria recovered from the injection sites of the rsr group than from those of the parental group. Histological findings of mice infected with the rsr mutant also bore several classical hallmarks of infection/inflammation, with disruption of the epidermis accompanied by massive neutrophilic infiltration of the dermis layer.
Accordingly, this work extends our knowledge of the regulatory cascade controlling virulence in S. aureus. Our studies have identified rsr as the first known regulator of sarR. Besides repressing sarR expression, rsr also downregulates agr transcription. This reduction in agr expression was not due to changes in sarA expression. In addition, both rsr and SarR appear to repress agr expression in an additive manner during the late exponential phase, as exemplified by enhanced RNAII and RNAIII expression in the double rsr-sarR mutant over the single rsr and sarR mutants (Fig. 4). Whether the rsr-mediated effect on agr during the late exponential phase is direct or indirect via another regulator remains to be defined. However, we cannot directly infer from our studies here the role of rsr on stationary-phase expression of virulence determinants, because we have found that heightened expression of RNAIII during the stationary phase is less dependent on rsr (unpublished data). Nevertheless, it is clear that a mutation in rsr has led to an enhanced virulence response in our murine cutaneous model. Indeed, overexpression of rsr in the parental SH1000 strain, as expected, decreased the transcription of RNAIII of agr, and consequently, one would predict a corresponding decrease in virulence effectors and hence disease progression if rsr is upregulated in vivo.
One relevant question regarding our findings is the plausible mechanism by which a unique molecule like rsr, which is present only in staphylococcal species, could inhibit expression of agr-controlled virulence determinants. We have found that the Rsr protein does not bind to the agr promoter, thus ruling out direct protein-DNA binding as a mode of regulation. Additionally, pulldown studies showed that Rsr and SarR do not interact at the protein-protein level. Interestingly, model analysis of S. aureus mRNA structure revealed that the ∼400-nt rsr mRNA has features consistent with a small antisense RNA (12, 24). This mRNA, which is transcribed most highly during mid-exponential growth, may then regulate the target gene function early during the growth cycle. Work is ongoing to investigate this possibility.
Additional work in our lab is directed at determining whether these regulatory pathways are found in more clinically relevant strains, such as the community-acquired strains USA300 and USA400. If so, developing the means to either increase the levels of rsr or augment its activities may lead to the development of novel anti-infectives toward S. aureus.
Acknowledgments
This work was supported in part by NIH grant AI37142 to A.L.C.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 9 August 2010.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, T., T. Bae, O. Schneewind, F. Takeuchi, and K. Hiramatsu. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190:300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, A. L., K. A. Nishina, M. P. Trotonda, and S. Tamber. 2008. The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLeo, F. R., and H. F. Chambers. 2009. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J. Clin. Invest. 119:2464-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, G. C. Kedar, P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 10.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 11.Geisinger, E., R. P. Adhikari, R. Jin, H. F. Ross, and R. P. Novick. 2006. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 61:1038-1048. [DOI] [PubMed] [Google Scholar]
- 12.Geissmann, T., C. Chevalier, M. J. Cros, S. Boisset, P. Fechter, C. Noirot, J. Schrenzel, P. Francois, F. Vandenesch, C. Gaspin, and P. Romby. 2009. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 37:7239-7257. [DOI] [PMC free article] [PubMed]
- 13.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingavale, S., W. van Wamel, T. T. Luong, C. Y. Lee, and A. L. Cheung. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahl, B. C., M. Goulian, W. van Wamel, M. Herrmann, S. M. Simon, G. Kaplan, G. Peters, and A. L. Cheung. 2000. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect. Immun. 68:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kupferwasser, L. I., M. R. Yeaman, C. C. Nast, D. Kupferwasser, Y. Q. Xiong, M. Palma, A. L. Cheung, and A. S. Bayer. 2003. Salicylic acid attenuates virulence in endovascular infections by targeting global regulatory pathways in Staphylococcus aureus. J. Clin. Invest. 112:222-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liebowitz, L. D. 2009. MRSA burden and interventions. Int. J. Antimicrob. Agents 34(Suppl. 3):S11-S13. [DOI] [PubMed] [Google Scholar]
- 19.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manna, A. C., and A. L. Cheung. 2006. Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J. Bacteriol. 188:4288-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manna, A. C., and A. L. Cheung. 2006. Transcriptional regulation of the agr locus and the identification of DNA binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol. Microbiol. 60:1289-1301. [DOI] [PubMed] [Google Scholar]
- 22.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 23.Novick, R. P. 1990. The Staphylococcus as a molecular genetic system. VCH Publishers, New York, NY.
- 24.Pichon, C., and B. Felden. 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. U. S. A. 102:14249-14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity. W.B. Saunders Company, Philadelphia, PA.
- 26.Tamber, S., and A. L. Cheung. 2009. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect. Immun. 77:419-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]