Abstract
ToxR-dependent recruitment of TcpP to the toxT promoter facilitates toxT transcription in Vibrio cholerae, initiating a regulatory cascade that culminates in cholera toxin expression and secretion. Although TcpP usually requires ToxR to activate the toxT promoter, TcpP overexpression can circumvent the requirement for ToxR in this process. To define nucleotides critical for TcpP-dependent promoter recognition and activation, a series of toxT promoter derivatives with single-base-pair transversions spanning the TcpP-binding site were generated and used as plasmid-borne toxT-lacZ fusions, as DNA mobility shift targets, and as allelic replacements of the chromosomal toxT promoter. When present in ΔtoxR V. cholerae overexpressing TcpP, several transversions affecting nucleotides within two direct repeats present in the TcpP-binding region (TGTAA-N6-TGTAA) caused defects in TcpP-dependent toxT-lacZ fusion activation and toxin production. Electrophoretic mobility shift assays demonstrated that these same transversions reduced the affinity of the toxT promoter for TcpP. The presence of ToxR suppressed transcription activation defects associated with most, but not all, transversions. Particularly, the central thymine nucleotide of both pentameric repeats was essential for efficient toxT activation, even in the presence of ToxR. These results suggest that the toxT promoter recognition function provided by ToxR can facilitate the interaction of TcpP with the toxT promoter but is insufficient for promoter activation when the TcpP-binding site has been severely compromised by mutation. Thus, the interaction of TcpP with nucleotides of the direct repeat sequences appears to be a prerequisite for toxT promoter activation.
The gastrointestinal disease cholera is primarily due to secretion of cholera toxin (CT) by ingested Vibrio cholerae. Cholera toxin expression is regulated by a cascade that requires ToxR-dependent recruitment of TcpP to the promoter immediately upstream of the toxT gene (following tcpPH activation by AphA and AphB) (9, 23), where TcpP activates toxT transcription and initiates ToxT expression (4, 11, 20; reviewed in reference 17). Although ToxT directly activates the transcription of the ctxAB operon encoding cholera toxin, to fully activate ctxAB expression, ToxT must also activate the tcpA promoter. The tcpABQCRDSTEF operon has the same polarity as toxT but is positioned several kilobases upstream. Due to the presence of an inefficient transcriptional terminator between the tcpABQCRDSTEF operon and toxT, a portion of these transcripts extend into toxT (5, 6, 26), resulting in a positive feedback loop whereby ToxT further activates its own expression. Once primed by TcpP-dependent toxT expression, this positive feedback loop elevates ToxT expression to levels sufficient to fully activate ctxAB transcription, leading to maximal expression and secretion of cholera toxin (26).
The tcpP and toxT genes lay within the Vibrio pathogenicity island (VPI) on V. cholerae chromosome I (8). Transcriptional profiling using V. cholerae genomic microarrays has suggested that TcpP may be involved in the regulation of several open reading frames both within and outside the VPI (1). To date, the toxT promoter is the only promoter experimentally verified to be bound and directly regulated by TcpP (11). DNase I protection analysis indicates that the TcpP footprint covers the region from at least position −51 to −32 relative to the toxT transcription start site. However, ToxR can provide a TcpP derivative deficient in DNA binding (TcpP-H93L) with the promoter recognition function required for promoter activation (11). This observation led to the hypothesis that the ability of TcpP to bind the toxT promoter was unnecessary for promoter activation as long as ToxR provided promoter recognition function (10). This culminated in a model in which ToxR recruits TcpP to the toxT promoter region via a protein-protein interaction independent of TcpP-DNA interactions, appropriately positioning TcpP for productive interactions with the transcriptional apparatus (10).
TcpP and ToxR are integral inner membrane proteins with amino-terminal cytoplasmic domains that display significant homology to the PhoB/OmpR family of winged helix-turn-helix transcription activators (16). Modeling based on the PhoB-DNA cocrystal structure (2) or nuclear magnetic resonance (NMR) structural analysis (25) and the phenotypes of TcpP mutant derivatives has identified several amino acid residues that are important for the interaction of TcpP with its binding site at the toxT promoter, with ToxR, and possibly with RNA polymerase (10, 11).
Activation of the toxT promoter and cholera toxin production can be rendered ToxR independent if TcpP is artificially overexpressed (4, 11, 20). This observation suggests that the ability of TcpP to productively interact with the toxT promoter in the absence of ToxR is limited by the low affinity of TcpP for its binding site and/or the limited expression level of TcpP. This ability of TcpP to activate the toxT promoter in the absence of ToxR has been exploited to identify TcpP positive-control and DNA-binding mutants (10). Overexpression of one such DNA-binding mutant, TcpP-H93L, activated the toxT promoter only in the presence of ToxR (11). These results suggest that TcpP can be recruited to the toxT promoter by two different mechanisms, depending upon the availability of ToxR. We hypothesized that when ToxR is present, TcpP recruitment to the toxT promoter is less dependent on DNA binding by TcpP (promoter recognition being provided by ToxR). However, when ToxR is absent, overexpression of TcpP can compensate by raising the concentration of TcpP to a level whereby it can efficiently recognize the toxT promoter without the assistance of ToxR. In the latter case, TcpP promoter binding is predicted to be critical for toxT activation.
In this study, nucleotides within the TcpP-binding site at the toxT promoter constituting a direct pentameric repeat of TGTAA-N6-TGTAA that influence toxT promoter activation and cholera toxin production in both ToxR+ and ΔtoxR strains were identified. These same nucleotides are shown to influence the affinity of the toxT promoter for TcpP in vitro, thus defining a DNA sequence motif bound by TcpP.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All V. cholerae strains used in this study are derived from the O1 serotype classical biotype strain O395. V. cholerae and Escherichia coli strains and plasmids used in this study are listed in Table 1. Strains were routinely grown in LB medium containing 10 g/liter NaCl at 37°C. Unless otherwise indicated, antibiotics were used at the following concentrations: streptomycin, 100 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; and kanamycin, 40 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| V. cholerae strains | ||
| O395 | Wild type, Strr | Laboratory strain |
| RY1 | O395 ΔtcpP | 26 |
| RY6 | O395 toxTΔpro | 26 |
| EK307 | O395 ΔtoxR | 11 |
| EK459 | O395 ΔtoxR ΔtcpP | 11 |
| EK1647 | O395 ΔtcpP toxTΔpro | This study |
| EK1654 | O395 ΔtoxR ΔtcpP toxTΔpro | This study |
| TG128 | EK459/pBluescript SK(+)/pMMB207 | This study |
| TG129 | EK459/pSK-toxR-HA/pMMB207 | This study |
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (F80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| DH5α λpir | supE44 ΔlacU169 (F80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 (λpir) | Laboratory collection |
| SM10 λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr (λpir) | Laboratory collection |
| TG389 | DH5α/pREP4/pBluescript KS(−) | This study |
| TG387 | DH5α/pREP4/pTG64 | This study |
| Plasmids | ||
| pBluescript SK(+) | Apr; high-copy-no. cloning vector | Stratagene |
| pBluescript KS(−) | Apr; high-copy-no. cloning vector | Stratagene |
| pSK-toxR-HA | pBluescript SK(+)-toxR-HA | this study |
| pMMB207 | IPTG-inducible expression vector; CmrlacIqtacP | 19 |
| pTL61T | Apr; promoterless lacZ fusion vector | 13 |
| pKAS32 | Suicide plasmid; Apr Sms | 24 |
| pKAS32-tcpF-toxTΔpro-toxT | For assembly of the parental toxT promoter | 26 |
| pKAS32-tcpF-toxTpro-toxT | For introducing transversion mutations onto the chromosome | This study |
| pEK41 | pMMB207-tcpP-HSV | 11 |
| pREP4 | p15A, lacIq Kmr | Qiagen |
| pRK2013 | ColE1::RK2 tra+ Kmr | 3 |
| pTG24 | Modified form of pTL61T with NotI site inserted into the XhoI, XhoI site disrupted | This study |
| pTG49 | pBluescript SK(+) with tacPtcpP-HSV inserted as a PstI/EcoRV fragment | This study |
| pTG64 | pBluescript KS(−)-tacP-tcpP-HSV | This study |
| pBluescript SK(+)-toxTpro | Vector for generation of transversion mutations and cloning into pTL61T | This study |
| pBluescript SK(+)-tcpF-toxTpro-toxT | Vector for generation of transversion mutations and cloning into pKAS32 | This study |
DNA manipulations.
Cloning procedures and transformation of E. coli strains were carried out using standard protocols (22). PCR products were generated using the Expand high-fidelity PCR system (Roche) or Pfu Turbo (Stratagene). Linear PCR products and DNA fragments were purified by agarose gel electrophoresis followed by gel extraction using the QIAquick gel extraction kit (Qiagen). The DNA sequences of cloned PCR products were verified at the University of Michigan Sequencing Core. DNA was transferred into V. cholerae by either electroporation (2.2 kV, 200 W) using an E. coli Pulsor (Bio-Rad), filter conjugation using the mobilization functions of E. coli SM10 λpir, or triparental mating using mobilization plasmid pRK2013 (3).
Transversion mutagenesis.
Nucleotide substitutions within the TcpP-binding region of the toxT promoter were generated by mutagenic PCR followed by DpnI digestion (14), using a template containing the toxT promoter region that was constructed sequentially using a starting plasmid containing most of the toxT region of O395 but lacking nucleotides −112 to +1 relative to the start site of transcription (26) (Table 1). This plasmid generates an NruI site at the position of the toxT promoter deletion. An annealed double-stranded oligonucleotide corresponding to the basal promoter elements (−10 and −35 RNA polymerase recognition elements) and TcpP-binding site was then ligated into the vector following NruI digestion. The primers were designed to regenerate a new NruI site between the TcpP- and ToxR-binding sites (see Table S1 in the supplemental material). The ToxR-binding site was then introduced by ligating an annealed double-stranded oligonucleotide containing the toxT promoter sequence from position −112 to −60 into the NruI-digested intermediate plasmid containing the TcpP-binding site plus basal promoter elements. This reconstructed toxT promoter construct is referred to as the parental toxT promoter vector in these studies and contains a TTC-to-CGA substitution at nucleotides −59 to −57 due to a remaining NruI half-site. The parental toxT promoter or mutant derivatives were used to clone toxT promoter mutants into the β-galactosidase reporter vector, pTL61T (13), or the allelic replacement suicide plasmid, pKAS32 (24). All PCR-generated V. cholerae DNA fragments were sequenced to verify the absence of additional nucleotide changes.
Construction of promoter-lacZ fusion plasmids.
DNA fragments corresponding to the region from position −172 to +45 relative to the toxT transcription start site (5), carrying the parental and transversion-substituted toxT promoters, were PCR amplified from the pBluescript SK(+)-toxTpro parental construct and recloned into pBluescript SK(+) via BamHI and EcoRI restriction sites. Alternatively, the region from position −172 to +45 was amplified from the chromosome of a previously constructed recombinant strain harboring the mutation of interest and ligated into pBluescript SK(+) following BamHI and EcoRI digestion as described above. Parental or mutant toxT promoters were then excised from pBluescript-based constructs with SalI and NotI, gel purified, and religated into pTG24 (Table 1), a modified version of pTL61T (13), generating lacZ transcriptional fusions. The pTL61T derivatives were electroporated into derivatives of V. cholerae, and the resulting strains were further modified by triparental matings to introduce pMMB207 (19) or pEK41 (11).
Measurement of lacZ fusion activity.
Cultures of strains carrying both pTG24 and pMMB207 or their derivatives were grown overnight in Luria-Bertani broth containing 5 g/liter NaCl at 30°C and diluted 1:50 in Luria-Bertani broth which had been adjusted to an initial pH of 6.5 (classically referred to as ToxR-inducing conditions) supplemented with chloramphenicol, ampicillin, streptomycin, and the indicated concentrations of isopropyl-β-d-thiogalactopyranoside (IPTG). After incubation for 3.5 h at 30°C, the optical densities at 600 nm (OD600) of the cultures were determined, and 5- to 100-μl samples were used in a standard β-galactosidase assay (18).
Construction of modified chromosomal toxT promoters.
The construction of V. cholerae strains carrying modified toxT promoters at the toxT locus was carried out in strains carrying toxTΔpro, a chromosomal deletion of the region from position −112 to +1 (26) relative to the toxT transcription start site, using the published procedure for allelic replacement. Mutations were made as described above using PCR mutagenesis (14) in either the pBluescript SK(+)-tcpF-toxTpro-toxT vector or pKAS32-tcpF-toxTpro-toxT. The parental construct and its transversion derivatives were religated into pKAS32 (24) as an EcoRI fragment or a NotI/EcoRV fragment when necessary and transformed into E. coli strain SM10 λpir, following confirmation of insertions in E. coli DH5α λpir. SM10 λpir carrying the various toxT promoter mutants was conjugated into derivatives of V. cholerae O395 toxTΔpro, followed by verification of toxTΔpro allele replacement with the parental promoter or its transversion derivatives as previously described (26). Finally, pMMB207 (19) or pEK41 (11) was introduced into these strains by triparental mating.
Cholera toxin enzyme-linked immunosorbent assays (ELISAs).
Measurement of cholera toxin production was carried out essentially as described earlier (11), inducing virulence gene expression in V. cholerae strains grown for 20 h at 30°C in Luria-Bertani broth containing 5 g/liter NaCl that had been adjusted to an initial pH of 6.5, which was supplemented with 1 mM IPTG and appropriate antibiotics (ToxR-inducing conditions).
Construction of a pSK-toxR-HA epitope-tagged expression plasmid.
To construct an expression plasmid carrying toxR harboring a C-terminal influenza virus hemagglutinin (HA) epitope tag, toxR from strain O395 was amplified by PCR using a 5′ primer with a BamHI site and 3′ primer with an XhoI site (see Table S1 in the supplemental material), and BamHI/XhoI-digested PCR products were ligated into BamHI/XhoI-digested epitope-tagging vector pcDNA3-HA (7). Clones with inserts were then digested with BamHI and ApaI to liberate the toxR-HA gene fusion, and that fragment was ligated into the pBluescript SK(+) vector (Stratagene) digested with BamHI and ApaI. The pSK-toxR-HA construct was confirmed by sequencing.
Construction of an E. coli strain overexpressing TcpP-HSV.
To obtain high levels of TcpP-HSV expression for gel shift analysis, the 2-kb PstI/EcoRV fragment carrying tacP-tcpP-HSV, in which the tac promoter directs the transcription of tcpP-HSV, was excised from the intermediate-copy-number plasmid pEK41 with PstI/EcoRV and ligated into PstI/EcoRV-digested Bluescript pSK(+), yielding pTG49 (Table 1). After verifying the DNA sequence of the insert, the fragment was recloned in PstI/EcoRV-digested pBluescript KS(−) in DH5α/pREP4, resulting in pTG64.
Mobility shift assays.
DNA gel mobility shift assays were performed essentially as previously described (11) using membrane preparations obtained either from E. coli strains TG389 [DH5α + pREP4 + pBluescript KS(−)] and TG387 (DH5α + pREP4 + pTG64) grown in LB broth supplemented with 1 mM IPTG, kanamycin, and ampicillin or from TG128 [O395 ΔtcpP ΔtoxR + pBluescript SK(+) + pMMB207] and TG129 (O395 ΔtcpP ΔtoxR + pSK-toxR-HA + pMMB207) grown in LB broth supplemented with 1 mM IPTG, streptomycin, chloramphenicol, and ampicillin, essentially as previously described (11) (Table 1). Membrane protein concentration was determined using the quick-start Bradford dye reagent (Bio-Rad) with bovine serum albumin (BSA) as a standard. DNA fragments carrying the region from position −172 to +45 relative to the toxT transcription start site were excised from pBluescript clones carrying the parental promoter derivatives using EcoRI and BamHI, gel purified, and end-labeled with Klenow DNA polymerase (Invitrogen) in the presence of 2 mM dGTP, 2 mM dCTP, 2 mM dTTP, and 0.25 μl [α-32P]dATP (MP Biomedicals). After a 15-min incubation at 37°C, labeled promoter fragments were ethanol precipitated, resuspended in distilled water, and diluted to approximately 2,000 cpm/μl. Various amounts of membrane preparations were mixed with the end-labeled DNA targets in a solution containing 10 mM Tris (pH 7.4), 1 mM EDTA, 5 mM NaCl, 50 mM KCl, 50 μg/ml BSA, and 10 μg/ml sheared salmon sperm DNA. Binding reactions were performed at 30°C for 30 min, and the free and membrane-associated DNA targets samples were separated by electrophoresis on a 6% polyacrylamide-Tris-borate-EDTA (TBE) gel prerun with 5% thioglycolic acid as previously described (11). After electrophoresis, the gels were dried, the extent of DNA migration was recorded by autoradiography, and the relative intensities of the recorded signals were determined using a Biospectrum image analyzer (UVP, LLC). A composite image for Fig. 3 was generated from a single piece of autoradiography film displaying migration patterns from five different gel runs (Fig. 3A to E), with each run consisting of a set of four individual gels run simultaneously in a single gel box.
RESULTS
Nucleotide transversions within the TcpP-binding site affect promoter activation in ΔtoxR V. cholerae.
The TcpP-binding site within the toxT promoter has been defined previously by DNase I footprinting analysis (11); however, the nucleotides within the TcpP-protected region that are required for TcpP binding and TcpP-dependent activation have not yet been determined. To identify these nucleotides, a collection of toxT promoter derivatives were constructed that carry transversion substitutions at each base pair in the region from position −56 to −38, corresponding to the TcpP-binding site (Fig. 1) (11). DNA fragments corresponding to the region from position −172 to +45 relative to the toxT transcription start site (5), carrying the parental promoter or its transversion derivatives, were ligated upstream of the promoterless lacZ reporter gene in a derivative of plasmid pTL61T (13), pTG24. β-Galactosidase levels were assessed in a V. cholerae O395 ΔtoxR ΔtcpP background containing either the empty expression vector pMMB207 (19) or pEK41 (an IPTG-inducible pMMB207-tcpP-HSV-carrying plasmid) (11).
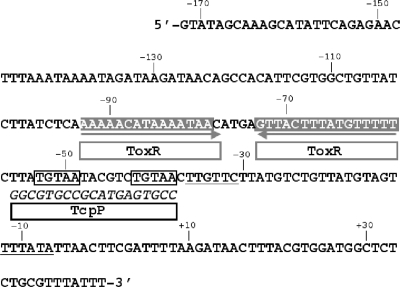
FIG. 1.
DNA sequence of the V. cholerae classical strain O395 toxT promoter region from position −172 to +45, numbered relative to the start of transcription (5). Nucleotides corresponding to the −35 and −10 hexamers are underlined, and gray horizontal arrows below the sequence and the gray boxed regions indicate the locations of inverted repeat sequences containing ToxR binding sites (5). The region of the promoter protected by TcpP from DNase I digestion (11) and subjected to transversion mutagenesis is indicated below the DNA sequence, with the substituted nucleotides in italic. The pentameric repeats identified as the refined TcpP-binding sites are boxed in black.
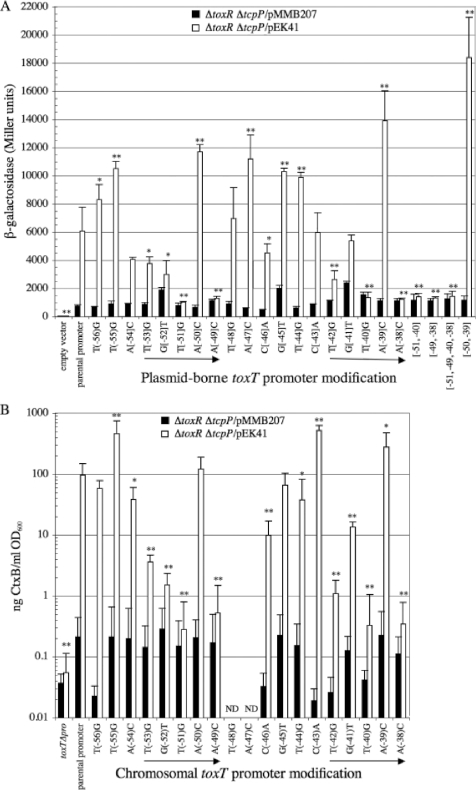
In the ΔtoxR ΔtcpP/pEK41 background, the activity of the parental toxT-lacZ fusion was about 6,000 Miller units, nearly 8-fold above its activity in the ΔtoxR ΔtcpP/pMMB207 background lacking TcpP. Thus, TcpP can give strong ToxR-independent activation of the toxT promoter when TcpP-HSV is expressed from pEK41. The −51, −49, −40, and −38 transversions reduced TcpP-dependent activity dramatically, to levels similar to that observed in the absence of TcpP (Fig. 2 A). In addition, the −52 and −42 transversions decreased parental fusion activity more than 50%, and the transversions at positions −53 and −46 caused a less severe defect of about a 30% decrease in β-galactosidase activity (Fig. 2A). The transversion at position −54 also caused about a 30% decrease in β-galactosidase activity, but that defect did not reach statistical significance (P = 0.056).
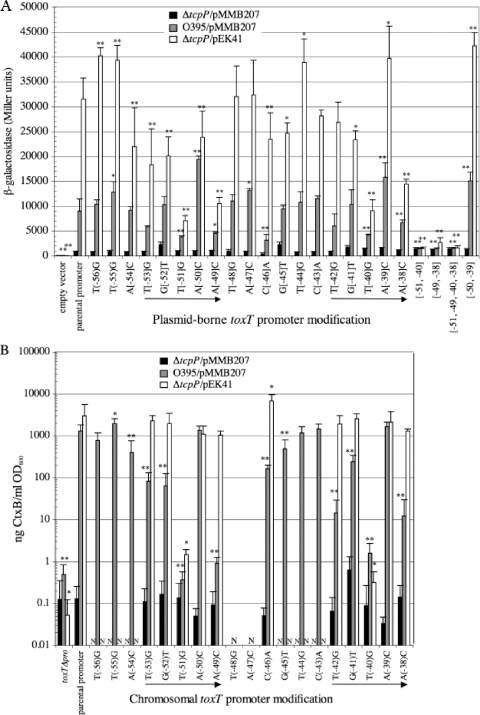
FIG. 2.
Effects of TcpP-binding site mutations on toxT-lacZ and CT reporter activities in a ΔtoxR ΔtcpP background with TcpP-HSV overexpression. The positions of single- and multiple-base-pair transversions in the parental promoter are indicated relative to the toxT transcription start site. Black bars, values for ΔtoxR ΔtcpP/pMMB207 strains; white bars, values for ΔtoxR ΔtcpP/pEK41 strains overexpressing TcpP-HSV. (A) β-Galactosidase activities for 3.5-h cultures of strains carrying either pTG24 alone (empty vector) or toxT-lacZ fusions in which the parental promoter or mutant derivatives drive the expression of the lacZ reporter. Values represent the averages from at least three independent cultures. (B) CtxB levels secreted by 20-h cultures from strains carrying various chromosomal toxT promoter derivatives. toxTΔpro is a chromosomal deletion of the toxT promoter region from position −112 to +1. Values represent the averages from at least six independent cultures. ND, not determined. Strains were grown under ToxR-inducing conditions in the presence of 1 mM IPTG. Error bars represent the standard deviation for each data set. *, P < 0.05; **, P < 0.005 (relative to the parental toxT promoter).
Analysis of the toxT promoter transversion mutations that most strongly altered parental fusion activity revealed a pentameric direct repeat element, TGTAA-N6-TGTAA, containing nucleotides critical for toxT transcription (Fig. 1). The −51, −49, −40, and −38 transversions had the strongest effects on TcpP-mediated activation of the toxT promoter. These transversions replaced the middle thymine base (−51 and −40) or the final adenine base (−49 and −38), suggesting that those nucleotides are the most critical for TcpP/toxT promoter interactions. Moreover, the −53, −52, and −42 transversions, which had less pronounced negative effects on toxT-lacZ fusion activity, also replaced bases found within the direct repeat motifs. As these two direct repeat sequences are centered 11 bp apart, one turn of the DNA helix (Fig. 1), these transversions alter nucleotides that may directly interact with TcpP, possibly bound in a head-to-tail dimer arrangement.
Some transversions resulted in notable increases in β-galactosidase activity (Fig. 2A). Those at positions −50 and −39 gave the highest increase in toxT-lacZ activation, and both alter the penultimate position of the TGTAA-N6-TGTAA pentameric repeat element within the toxT promoter. Combining both mutations in a single promoter gave an even higher level of toxT-lacZ activation, suggesting that the effects are cumulative (Fig. 2A). These results indicate that the existing TcpP-binding site within the toxT promoter is suboptimal.
Effects of chromosomal transversions on TcpP-dependent CT production in the absence of ToxR.
As the effects of the transversions on toxT-lacZ fusion activity were determined in a plasmid context, their influences on cholera toxin (CT) production, a downstream reporter of total toxT expression, were determined. Selected transversion derivatives were moved into the chromosomal toxT locus of strain EK1654 (O395 ΔtoxR ΔtcpP toxTΔpro [Table 1]). The resulting strains then received pMMB207 (empty vector) or pEK41 (pMMB207-tcpP-HSV) to allow TcpP-dependent CT production in the absence of ToxR.
In the ΔtoxR ΔtcpP/pMMB207 background, all strains secreted less than 1 ng CT/ml/OD600 unit, comparable to that of the ΔtoxR ΔtcpP toxTΔpro (EK1654 parent)/pMMB207 negative control lacking the toxT promoter region from position −112 to +1 (Fig. 2B). In the ΔtoxR ΔtcpP/pEK41 background, the chromosomal parental promoter directed toxin production to more than 100 ng CT/ml/OD600 unit, elevated more than 100-fold from that obtained in either the ΔtoxR ΔtcpP/pMMB207 background or the toxTΔpro ΔtoxR ΔtcpP/pEK41 strain (Fig. 2B). The −51, −49, −40, and −38 transversions reduced toxin production more than 100-fold, essentially rendering toxin production unresponsive to TcpP-HSV overexpression. The −53, −52, and −42 transversions decreased toxin production between 10-fold and 100-fold. All other transversions tested altered toxin production less than 10-fold (Fig. 2B). Thus, all transversions that altered toxin production 10-fold or greater were located within the TGTAA-N6-TGTAA pentameric repeat motif.
Effects of toxT promoter mutations on TcpP binding.
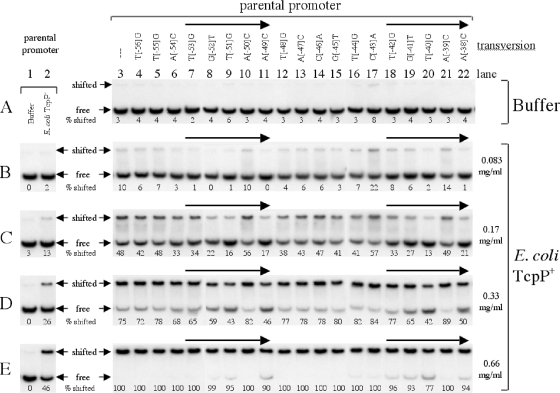
One likely explanation for why certain transversion mutations within the toxT promoter disrupt TcpP-mediated activation is that they disrupt promoter recognition by TcpP. To determine if changes in the affinity of the promoter-TcpP interaction underlie transversion-dependent changes in the ability of overexpressed TcpP-HSV to activate the toxT promoter, electrophoretic mobility shift assays were performed. These studies use membrane preparations containing overexpressed TcpP-HSV. DNA fragments carrying the parental toxT promoter and its derivatives were used as targets in mobility shift assays with either membrane buffer alone (Fig. 3 A, lanes 3 to 22, and B to E, lanes 1) or dilutions of membrane preparations obtained from an E. coli strain carrying either the empty vector pBluescript KS(−) (E. coli TcpP−) (Fig. 3B to E, lanes 2) or pTG64 expressing TcpP-HSV (Fig. 3B to E, lanes 3 to 22).
FIG. 3.
Electrophoretic mobility shift assay for the binding of toxT promoter derivatives to TcpP-HSV in purified membrane preparations. The 32P-end-labeled toxT promoter targets used in each lane in all panels carry the parental promoter (lanes 1, 2, and 3) or single-transversion derivatives with mutations at positions −56 to −38 (lanes 4 to 22, respectively), as indicated at the top of the figure. The horizontal arrows above each panel indicate the positions of the pentameric repeat motif (TGTAA-N6-TGTAA) within the TcpP-footprinted region of the toxT promoter. For lanes 3 to 22, the DNA-binding solutions containing end-labeled DNA targets were mixed with membrane buffer only (A) or TcpP-containing E. coli membrane preparation at either 0.083 mg/ml (B), 0.17 mg/ml (C), 0.33 mg/ml (D), or 0.66 mg/ml (E), as indicated on the right. The positions of free and shifted end-labeled DNA target migration through the gel are indicated in the column between lanes 2 and 3. The percent shifted values given below the unshifted target band for each sample indicate the percentage of shifted signal, relative to the sum of free and shifted signals, as quantified by densitometry. Control samples containing the parental target mixed with either membrane buffer only (lane 1) or negative-control E. coli TcpP− membrane preparation (lane 2), present at the same protein concentration as the experimental membrane preparation used for lanes 3 to 22 as indicated on the right side of the figure, were included in each gel run where appropriate.
In the presence of TcpP-HSV-containing membranes, the parental toxT promoter was shifted in a concentration-dependent fashion by increasing amounts of membrane (Fig. 3B to E, lanes 3). Binding curve analysis indicated that the parental toxT promoter fragment would be shifted 50% with 0.177 mg/ml TcpP-containing membrane protein (Table 2; see Fig. S1 in the supplemental material). This is approximately 3-fold less protein than was required for a 50% shift using membranes lacking TcpP (Table 2). In addition, many transversion-bearing targets have 50% shifting values of between 0.177 and 0.208 mg/ml TcpP-containing membrane protein (Table 2), altering the affinity of the toxT promoter for TcpP by less than 20%. In contrast, almost all transversion mutations in the pentameric repeat regions resulted in a 35 to 225% increase in the 50% shifting value (Table 2). The most dramatic defects were observed with the −52, −51, −49, −40, and −38 transversion mutations, which resulted in toxT promoters clearly defective for TcpP binding at various protein concentrations (Fig. 3). The 50% shifting values for these mutations were between 0.332 and 0.400 mg/ml TcpP-containing membrane protein (Table 2), reducing the affinity for the toxT promoter approximately 2-fold. Four of these transversion mutations affect the center positions (−51 and −40) and final positions (−49 and −38) of the pentameric repeats (TGTAA-N6-TGTAA) (Fig. 1) and were some of the least active promoters for toxT activation in the absence of ToxR (Fig. 2). toxT promoter fragments carrying combinations of either the −51 and −40 transversions (center positions of the pentameric repeats) or the −49 and −38 transversions (final position of the pentameric repeats) required 0.533 and 0.527 mg/ml TcpP-containing membrane protein, respectively, to be shifted 50% (Table 2). Thus, these pairs of transversions reduced the affinity of the toxT promoter for TcpP-HSV approximately 3-fold. Other mutations in the pentameric repeat region (−53, −42, and −41) had less dramatic effects on toxT promoter binding but still had 50% shifting values of 0.220 to 0.252 mg/ml TcpP-containing membranes (a 24 to 42% increase relative to the parental toxT promoter) (Table 2).
TABLE 2.
Relative binding of DNA targets for TcpP+ and TcpP− membrane preparations
| Targeta | mg protein/ml required for a 50% shift |
|
|---|---|---|
| TcpP− (TG389) | TcpP+ (TG387) | |
| Parental | 0.526 | 0.177 |
| −56 | 0.548 | 0.182 |
| −55 | 0.526 | 0.179 |
| −54 | 0.578 | 0.233 |
| −53 | 0.589 | 0.239 |
| −52 | 0.589 | 0.376 |
| −51 | 0.570 | 0.373 |
| −50 | 0.620 | 0.154 |
| −49 | 0.685 | 0.358 |
| −48 | 0.586 | 0.208 |
| −47 | 0.636 | 0.193 |
| −46 | 0.649 | 0.182 |
| −45 | 0.637 | 0.197 |
| −44 | 0.585 | 0.193 |
| −43 | 0.610 | 0.148 |
| −42 | 0.648 | 0.220 |
| −41 | 0.617 | 0.252 |
| −40 | 0.622 | 0.400 |
| −39 | 0.677 | 0.173 |
| −38 | 0.610 | 0.332 |
| −51, −40 | 0.692 | 0.533 |
| −49, −38 | 0.698 | 0.527 |
| −51, −49, −40, −38 | 0.676 | 0.515 |
| −50, −39 | 0.676 | 0.167 |
Numbers represent the positions of transversion substitutions, relative to the toxT transcription start site. Direct repeat elements (wild-type nucleotides TGTAA) are from positions −53 to −49 and −42 to −38.
The −50 and −39 transversions (located at the penultimate position of the pentameric repeats), individually or together on the same toxT promoter fragment, appeared to lead to slightly enhanced TcpP binding relative to the parental toxT promoter (Fig. 3). For the −50 transversion this was reflected in a 13% lower concentration of TcpP-containing membrane required for 50% shifting, while for the −39 transversion the 50% shifting concentration was nearly the same as for the parental toxT promoter (0.173 versus 0.177 mg/ml, respectively) (Table 2). This may explain why the −50 and −50/−39 double mutant promoters activate a toxT-lacZ fusion better than the parental promoter (Fig. 2A).
The −43 transversion also bound TcpP slightly better than the parental toxT promoter (Fig. 3; Table 2). While this mutant did not show increased toxT-lacZ activation relative to the parental toxT promoter (Fig. 2A), it did lead to enhanced toxin production in the absence of ToxR (Fig. 2B).
When considered along with the toxT-lacZ activation and toxin production data, these results suggest that in the absence of ToxR, the strong defects in TcpP-dependent promoter activation associated with the −52, −51, −49, −40, and −38 transversions can be largely attributed to reductions in affinity of the toxT promoter for TcpP. Mutations at several other positions (−53, −42, and −41) within the pentameric repeats which also affected toxT-lacZ activation and toxin production led to defects in TcpP binding as well, albeit a less dramatic defect than mutations at positions −52, −51, −49, −40, and −38. Conversely, the increases in TcpP-dependent promoter activation associated with the −50 and −39 transversions (also within the pentameric repeats) may be attributed to slight increases in the affinity of these mutant promoters for TcpP (alternatively, the increased activation associated with these transversions may be due to transversion-induced changes in the conformation of promoter-bound TcpP). These results define nucleotides within the TcpP-footprinted region of the toxT promoter that are important for TcpP-dependent promoter activation and TcpP binding in the absence of ToxR.
ToxR restores TcpP-dependent activation to several toxT promoter mutants.
Given that ToxR enhances the ability of TcpP to activate the toxT promoter (4, 11, 20), ToxR can restore toxT activation to a form of TcpP (TcpP-H93L) that has poor DNA binding activity (11), and ToxR can interact with TcpP via a protein-protein interaction (10), we hypothesized that the presence of ToxR may allow TcpP to activate toxT promoters harboring mutations that affect DNA binding by TcpP.
Using the plasmid-borne toxT-lacZ constructs, the presence of ToxR in the ΔtcpP/pEK41 background elevated TcpP-HSV-dependent activation of the toxT promoter approximately 5-fold relative to that in the ΔtoxR ΔtcpP background (∼30,000 Miller units [Fig. 4 A] compared to 6,000 Miller units [Fig. 2A]). Those mutations in the toxT promoter that rendered the promoter insensitive to TcpP activation in the absence of ToxR (−51, −49, −40, and −38) (Fig. 2A), showed intermediate but detectable activation in the presence of ToxR (Fig. 4A). However, upon combining the two most defective mutations (−51 and −40, the two center positions of the pentameric repeat) no TcpP-mediated toxT activation was detected, even in the presence of ToxR (Fig. 4A). Thus, while ToxR can facilitate partial activation of a toxT promoter mutated for either pentameric repeat of the TcpP-binding site, if both sites are mutated, ToxR is no longer able to facilitate activation. This suggests that although ToxR facilitates TcpP-mediated activation and can restore activation of TcpP mutants defective for DNA binding (TcpP-H93L) (11), DNA binding by TcpP to the toxT promoter is still required for toxT activation. Combining the −49 and −38 toxT promoter mutations (at the last position of each repeat) results in severe attenuation of toxT activation, but some activation was still detectable (Fig. 4A), indicating that the central position of the pentameric repeat is the most critical for TcpP promoter activation.
FIG. 4.
Effects of TcpP-binding site mutations on toxT-lacZ and CT reporter activities in a ΔtcpP background (toxR+) with TcpP-HSV overexpression. The positions of single- and multiple-base-pair transversions in the parental promoter are indicated relative to the toxT transcription start site. Black bars, values for ΔtcpP/pMMB207 strains; gray bars, values for O395/pMMB207 strains; white bars, values for ΔtcpP/pEK41 strains overexpressing TcpP-HSV. (A) β-Galactosidase activities for 3.5-h cultures of strains carrying either pTG24 alone (empty vector) or derivatives in which the parental promoter or its transversion derivatives drive the expression of the lacZ reporter. Values represent the averages from at least three independent cultures (n ≥ 3). (B) CtxB levels secreted by 20-h cultures of strains carrying modified chromosomal toxT promoters. toxTΔpro is a chromosomal deletion of the toxT promoter region from position −112 to +1. Values represent the averages from at least six independent cultures (n ≥ 6). N, not determined. Error bars represent the standard deviation for each data set. Statistical variance (Student's t test) from parental promoter values: *, P ≤ 0.05; **, P ≤ 0.005. Strains were grown under ToxR-inducing conditions in the presence of 1 mM IPTG.
To determine the impact of toxT promoter mutations in nucleotides most defective for toxT activation on CT production in the presence of ToxR, we tested a number of chromosomal toxT promoter mutants in a ΔtcpP background with TcpP-HSV provided from the plasmid pEK41. In this assay almost all toxT promoter mutants were completely rescued for toxin production by the presence of ToxR (compare Fig. 4B to Fig. 2B). Thus, for six out of eight toxT promoter mutations within the pentameric repeat that gave reduced CT production in the absence of ToxR, the presence of ToxR restored CT levels to that of the parental toxT promoter. Reflecting their poor levels of toxT-lacZ activation even in the presence of ToxR (Fig. 4A), transversion mutations at the center of the pentameric repeats (−51 and −40 positions) caused a strong defect in CT production, even in the presence of ToxR (>1,000-fold decreased) (Fig. 4B). This lends further support to the hypothesis that the center position of these two repeats is the most critical for DNA binding and TcpP-mediated activation.
Given that our studies so far had been performed using ΔtcpP mutants overexpressing a TcpP-HSV derivative from plasmid pEK41, we determined the effect of toxT promoter transversion mutations in the O395 background expressing wild-type alleles of tcpP and toxR. To make these studies directly comparable to our previous experiments, O395 derivatives were tested in the presence of the empty cloning vector pMMB207, thus removing any effects of growth in chloramphenicol on the assay.
In the O395 background, parental toxT-lacZ fusion activity was nearly 9,000 Miller units of β-galactosidase, more than 10-fold higher than in the ΔtcpP/pMMB207 background but about 3-fold less than in the ΔtcpP/pEK41 background. Thus, endogenous levels of TcpP can activate the toxT promoter in the presence of ToxR, but not as efficiently as TcpP-HSV overexpressed from a plasmid.
In the O395 background, four transversion mutations resulted in significant decreases in toxT activation of about 2-fold (−51, −49, −46, and −40) (Fig. 4A, gray bars), while the −38 mutation resulted in a slightly less dramatic but significant decrease in toxT activation (25% reduction). As was the case when TcpP was overexpressed, pairwise combinations of transversions comparably positioned within the pentameric repeats ([−51, −40] and [−49, −38]) reduced toxT-lacZ fusion activity further, now to undetectable levels when TcpP was expressed at endogenous levels.
Mutations with a tendency to increase toxT promoter activation by TcpP (−50 and −39) maintained this trend at endogenous levels of TcpP, with significant increases in toxT-lacZ activation when alone or in the −50, −39 double mutant (Fig. 4A).
The −46 transversion mutation was somewhat unexpected in its defects in toxT-lacZ activation. It should be noted that the −46 transversion did not alter binding affinity for the parental toxT promoter (Table 2) but did decrease background activation of the toxT-lacZ fusion in the absence of TcpP by more than 2-fold (Fig. 4A, black bars) (P < 0.005). Thus, although the −46 transversion reduced toxT-lacZ fusion activity about 3-fold in the O395 background, some of that defect may reflect a general deficiency in transcription potential.
When present on the chromosome in the O395 background, the parental toxT promoter facilitated toxin production to over 1,200 ng CT/ml/OD600 unit (Fig. 4B), comparable to that obtained in the ΔtcpP/pEK41 background. This suggests that although the activity of the toxT-lacZ fusion was not saturated by the endogenous levels of TcpP expression in the O395 background, endogenous TcpP expression fully saturated toxin expression and the ToxT-dependent positive feedback loop that primes additional toxT expression. Again, analysis of CT production focused on transversions in the pentameric repeats or those that had a major affect on toxT-lacZ expression in the O395 background. In the O395 background, the chromosomal −53, −52, −42, and −38 transversion mutations reduced toxin production 10 to 100-fold, while the −51, −49, and −40 transversions reduced production about 1,000-fold, to levels similar to a strain deleted for the toxT promoter (toxTΔpro) (Fig. 4B). Thus, in the presence of endogenous levels of TcpP, the center position of the pentameric repeats emerged again as among those most critical for toxT promoter activity. Positions −49 and −38 (at the final positions of the repeats) were also of particular importance. It appears that the promoter-distal direct repeat element is more essential than the promoter-proximal TGTAA repeat, as the −51 and −49 transversions are more defective that the −40 and −38 transversions for toxin production in the O395 background.
The fact that the −46 transversion mutant directed toxin production to within 10-fold of the parental toxT promoter but reduced toxT-lacZ fusion activity to the lowest level for any single-transversion mutant tested was surprising. This may reflect the fact that in the toxT-lacZ fusion experiments, the target promoter is carried on a multicopy plasmid, and in the CT assay, the toxT promoter is present on the chromosome. Upon reducing the number of promoter targets, the endogenous levels of TcpP may now be able to sufficiently bind and activate the −46 transversion mutant to initiate moderately robust toxT expression. However, additional factors must be at play, since other promoter mutants slightly less defective for toxT-lacZ activation (−51, −49, −40, and −38) were not as competent for toxin production in the O395 background.
toxT promoter mutations minimally affect ToxR binding.
While most toxT promoter mutations affecting TcpP-mediated activation were compensated for by ToxR coexpression, two notable mutations were not, i.e., the mutations at positions −51 and −40 at the centers of the pentameric repeats (Fig. 4A and B). This could indicate that even in the presence of ToxR, TcpP must bind the toxT promoter to activate transcription, or that certain transversion mutations affect ToxR binding to the promoter even though they lie adjacent to the ToxR-binding site. To determine whether toxT promoter transversion mutations affect ToxR binding to the promoter, electrophoretic mobility shift assays were carried out using V. cholerae membrane preparations (from a ΔtoxR ΔtcpP derivative) containing overexpressed ToxR-HA (encoding a carboxy-terminal, HA epitope-tagged derivative of the protein). Buffer alone or dilutions of membrane preparations containing or lacking ToxR-HA were used. Binding curve analysis following a series of ToxR gel shifts in the presence of various concentrations of membranes was used to calculate the concentration at which each promoter derivative would be 50% shifted (Table 3; see Fig. S2 and S3 in the supplemental material). In the absence of ToxR, all single-transversion mutants displayed comparable 50% binding curves with the parental toxT vector (Table 3). In the presence of the ToxR, the parental toxT promoter and single-transversion-bearing toxT promoter targets could be 50% shifted with between 0.532 and 0.659 mg/ml membrane protein (about 10-fold less membrane than is required to shift the toxT promoter in the absence of ToxR). Thus, none of the transversions altered the affinity of the toxT promoter for ToxR by greater than 20% (Table 3). Furthermore, there was no correlation between the level of increased ToxR-containing membrane concentration required for promoter binding and the effect of the transversion on the ability of ToxR to facilitate TcpP-mediated toxT promoter activation. Thus, the inability of ToxR to restore activation to the −51 and −40 toxT promoter mutants at the center positions of the pentameric repeats is not due to lack of ToxR binding to these mutant promoters.
TABLE 3.
Relative binding of DNA targets for ToxR+ and ToxR− membrane preparations
| Targeta | mg/ml protein required for a 50% shift |
|
|---|---|---|
| TG128 (ToxR−) | TG129 (ToxR+) | |
| Parental | 5.43 | 0.548 |
| −56 | 5.32 | 0.539 |
| −55 | 5.41 | 0.552 |
| −54 | 5.18 | 0.575 |
| −53 | 5.83 | 0.532 |
| −52 | 4.96 | 0.641 |
| −51 | 5.60 | 0.618 |
| −50 | 5.62 | 0.630 |
| −49 | 5.48 | 0.624 |
| −48 | 6.18 | 0.659 |
| −47 | 5.70 | 0.630 |
| −46 | 5.12 | 0.629 |
| −45 | 5.24 | 0.591 |
| −44 | 5.08 | 0.620 |
| −43 | 4.86 | 0.605 |
| −42 | 5.14 | 0.642 |
| −41 | 5.34 | 0.605 |
| −40 | 5.42 | 0.629 |
| −39 | 5.13 | 0.615 |
| −38 | 5.38 | 0.658 |
| −51, −40 | >7.0 | 0.718 |
| −49, −38 | 6.21 | 0.690 |
| −51, −49, −40, −38 | 5.90 | 0.714 |
| −50, −39 | 5.40 | 0.665 |
Numbers represent the positions of transversion substitutions, relative to the toxT transcription start site. Direct repeat elements (wild-type nucleotides TGTAA) are from positions −53 to −49 and −42 to −38.
DISCUSSION
In this study we defined nucleotides within the TcpP-binding site of the toxT promoter that are critical for TcpP-dependent transcription activation in the absence and presence of ToxR. Previous studies have shown that endogenous levels of TcpP require coexpression of ToxR to activate the toxT promoter (5, 11). However, TcpP can activate the toxT promoter when overexpressed in the absence of ToxR (4, 11, 20). TcpP overexpression in the absence of ToxR likely compensates for weak toxT promoter-binding activity of TcpP. In this study, toxT promoter mutagenesis defined nucleotides within the TcpP-binding region that are unconditionally required, conditionally required, or not required for toxT promoter activation. Our studies revealed that transversions with either unconditional or conditional effects alter nucleotides within the pentameric direct repeat element, TGTAA-N6-TGTAA (Fig. 1), required for TcpP-dependent activation. Several nucleotides in the repeat element were important for toxT promoter activation and CT production in the absence of ToxR, whereas only the center positions of the repeat element (−51 and −40) were critical in the presence of ToxR (Fig. 2 and 4). These data indicate that the center position is the most important for TcpP-dependent promoter activation. Consistent with that interpretation, mutagenesis of either the −51 or −40 position reduced the affinity of toxT promoter DNA for TcpP-HSV more than 2-fold (Fig. 3; Table 2). The final positions in the repeat element, −49 and −38, were also particularly important for plasmid-based toxT-lacZ activation in the absence or presence of ToxR (Fig. 2A and 4A). Mutations at these positions also affected CT production when TcpP was expressed at endogenous levels in the presence of ToxR (Fig. 4B). However, when TcpP was overexpressed from plasmid pEK41, the defects of the −49 and −38 transversions were overcome for CT production (Fig. 4B). toxT promoter mutations at positions −49 and −38 also resulted in a 2-fold or nearly 2-fold decrease in binding affinity for TcpP, respectively (Fig. 3; Table 2). Taking the results together, the difference in CT production by the center-position (−51 and −40) mutants and last-position (−49 and −38) mutants suggests that the −51 and −40 mutations affect something in addition to binding affinity for TcpP, since the −49 and −38 mutants have comparable DNA-binding defects. Perhaps the −51 and −40 mutations result in TcpP molecules oriented on the toxT promoter in a way that is especially poor for productive RNA polymerase interaction.
Activation of the toxT promoter by TcpP and ToxR leads to priming of an autoregulatory positive feedback loop where ToxT protein activates the tcpA promoter and an elongated tcp transcript extends through tcpF and into the toxT gene, leading to further enhancement of toxT expression (21, 26). In this study we assessed direct activation of the toxT promoter by TcpP in the presence or absence of ToxR using plasmid-based toxT-lacZ fusions. We also assessed production of a downstream product of toxT activation, CT. The ctxAB operon is directly activated by ToxT (27). Thus, expression from the toxT-lacZ fusion represents a simplified picture of toxT promoter activation, whereas measurement of CT reflects all transcription activity leading to ToxT production.
The results of this study are consistent with the binding of two TcpP monomers in a head-to-tail configuration to the two pentameric direct repeat motifs within the TcpP-footprinted region, analogous to the structure determined for the E. coli PhoB-DNA cocrystal (2). The fact that mutating a critical nucleotide in only one of the pentameric repeats, −51 (promoter distal) or −40 (promoter proximal), dramatically affects DNA-binding activity indicates that productive toxT promoter binding may require a dimer of TcpP (Fig. 3). Based on the fact that mutations in the promoter-distal TcpP-binding site (centered at position −51) have a more dramatic effect on toxT-lacZ activation and CT production in the presence of ToxR than mutations in the promoter-proximal TcpP-binding site (centered at −40) (Fig. 4), the promoter-distal TcpP-binding site may be the initiation site for TcpP binding, perhaps with the assistance of ToxR (Fig. 5) (10). It is interesting to note that TcpP is a member of the PhoB/OmpR family of winged-helix-turn-helix transcription activators and that the pentameric repeat identified for TcpP binding to the toxT promoter differs by only one nucleotide from the previously characterized pho box bound by PhoB in E. coli (TGTAA versus TGTCA) (2, 15). In the course of our toxT promoter analysis, we identified one pair of mutations that resulted in the ability of TcpP to activate transcription even better than the parental toxT promoter. Those mutations were at the penultimate position of the pentameric repeat and made the TcpP-binding site into a consensus pho box (TGTCA). This may indicate that the toxT promoter is not optimized for TcpP binding and that TcpP binds DNA much like PhoB. It is possible that when the Vibrio pathogenicity island containing tcpP and toxT was acquired by horizontal transfer, TcpP was capable of directly activating the toxT promoter in the absence of ToxR. Since that time, ToxR has become an essential component of toxT promoter activation, and alteration of the TcpP-binding site to a less well recognized sequence for TcpP may have facilitated this occurrence.
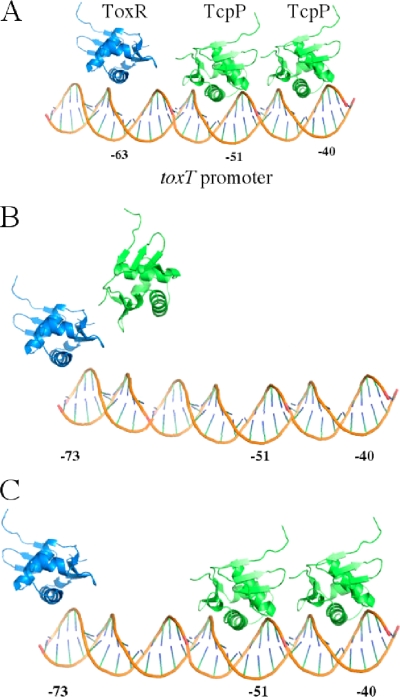
FIG. 5.
Models of toxT promoter activation by TcpP and ToxR. (A) Binding of ToxR to its upstream binding site (potentially centered at nucleotide −63 [12]) brings TcpP to the toxT promoter via a wing-wing interaction between ToxR and TcpP (10; E. S. Krukonis et al., submitted for publication). TcpP is then released to bind its promoter-binding site centered at nucleotides −51 and −40 (the critical nucleotides of the TGTAA-N6-TGTAA repeat). In this model, wing-wing interactions may be maintained even while ToxR and TcpP engage the DNA. (B) Based on putative assignments of the precise ToxR-binding site (12), ToxR may bind to a position further upstream, centered at nucleotide −73. In this case, ToxR could recruit TcpP to the toxT promoter via a protein-protein interaction(s), but it would then release TcpP so that TcpP could then bind its more promoter-proximal binding site (C). In both cases, TcpP binding to the center position of the TGTAA-N6-TGTAA direct repeat (nucleotides −51 and −40) is required for toxT activation.
In the solved structures of PhoB interacting with DNA, the wing domain of PhoB binds to the minor groove at a position neighboring the pho box (2, 25). Our studies also identified one nucleotide, −46, outside the pentameric repeat TcpP-binding site that affected toxT-lacZ activation and CT production in the absence of ToxR and toxT-lacZ expression in the presence of ToxR (Fig. 2 and 4A). We hypothesize that this position in the toxT promoter may be bound by the wing domain of TcpP, although this position is a CG base pair in the toxT promoter and an AT base pair in the pho box (2, 25). The analogous position in the promoter-distal pentameric repeat, −57, was originally considered to be outside the TcpP-binding site based on footprinting analysis (11) and contains part of a residual NruI site remaining from construction of the parental toxT promoter used for these studies (the region from −59 to −57 has a TTC-to-CGA change [see Materials and Methods]). Further analysis of the triplet substitution revealed that the TTC-to-CGA change results in a 3.5-fold decrease in TcpP-mediated activation of the toxT-lacZ fusion, and most of this defect can be attributed to the C-to-A substitution at position −57. The triplet substitution affects toxT-lacZ expression only in the absence of ToxR (data not shown). This suggests that the −57 and −46 positions may play analogous roles for the two bound TcpP-molecules at the toxT promoter. We hypothesize that the wing domains of TcpP, bound as a head-to-tail dimer, may interact in the minor groove with base pairs at positions −57 and −46 of the toxT promoter. Previous studies have suggested a role for the wing domain of TcpP in interaction with ToxR. Specifically, several mutations in the wing domain of TcpP disrupted the ability of ToxR and TcpP to interact in a cross-linking assay (10). Our proposal that the wing domain of TcpP may also interact with the minor groove of the toxT promoter does not exclude this model. In fact, ToxR may bind to the wing domain of TcpP and bring TcpP to the toxT promoter by the interaction of ToxR with its upstream ToxR-binding site, and then ToxR may release TcpP to engage the toxT promoter using the DNA recognition helix (helix 3) of TcpP to bind in the major groove and wing domain in the minor groove (Fig. 5). One might think of ToxR as an escort, delivering a TcpP molecule with poor DNA-binding affinity for the toxT promoter to its binding site with close enough proximity to facilitate TcpP-toxT promoter interactions. Consistent with this hypothesis is the fact that all TcpP mutations that affected ToxR binding were in the wing domain of TcpP, and all affected the DNA-binding activity of TcpP (10). Alternatively, the promoter-proximal TcpP molecule may interact with nucleotide −46 via its wing domain, but the promoter-distal TcpP molecule may maintain interaction with ToxR for the duration of the activation process. This “hand-holding” model for ToxR assistance of TcpP-mediated toxT promoter activation (Fig. 5) may explain why mutation of the promoter-distal potential wing-binding nucleotide, position −57, affects toxT promoter activation by TcpP in the absence of ToxR but not the in the presence of ToxR. According to the “hand-holding” model, the wing of the promoter-distal TcpP molecule would be engaged by interacting with ToxR and thus would not need to interact with the nucleotide at position −57. This model does not explain why mutations at the other putative wing interaction nucleotide, −46, are also more affected when TcpP is overexpressed in the absence of ToxR than in the presence of ToxR. One possibility is that by binding just upstream of the TcpP-binding site, ToxR induces DNA bending that alters the architecture of the toxT promoter and facilitates stronger TcpP interaction with the toxT promoter, thus overcoming binding defects caused by the position −46 transversion mutation.
As the current pandemic of V. cholerae is caused by the El Tor biotype and these studies were performed with the classical biotype strain O395, it is important to note that the toxT promoter region in these biotypes is identical for the ToxR-binding site (inverted repeat) and the TcpP-binding site (direct repeats) (Fig. 1). Furthermore, the DNA-binding helices and wing domains of TcpP from both biotypes are identical. Thus, we anticipate that the El Tor biotype would respond to toxT promoter mutations similarly to as we describe here for O395.
In searching the genome of V. cholerae strain N16961 for the TcpP-binding site TGTAA-N6-TGTAA, three sites in addition to the toxT promoter were identified. Two were within genes, VC1542 (DNA ligase) and VC1732 (3-phosphoshikimate 1-carboxyvinyltransferase), whereas one was between the hypothetical open reading frames (ORFs) VC2472 and VC2473. Microarray analysis of gene expression in V. cholerae N16961 grown under AKI (virulence-inducing) conditions indicated that the levels of several transcripts may be altered greater than 2-fold in a TcpP-dependent, ToxT-independent manner (1). Of those, three were regulated independently of ToxR: VC1865 (encoding a hypothetical protein), VC2033 (encoding alcohol dehydrogenase), and VC2368 (fexA/arcA). If we change the consensus binding site to the preferred sequence TGTCA-N6-TGTCA identified in our toxT promoter mutant analysis (Fig. 2 and 3), we are able to identify such a site upstream of the 5′ end of the VC2033 coding strand, suggesting that it may in fact be directly regulated by TcpP. This preferred TcpP-binding site is actually downstream of the neighboring hypothetical ORF VC2034, but that ORF is only 35 amino acids long and has just three homologs (all in V. cholerae), and thus it may not represent a true ORF.
Supplementary Material
Acknowledgments
We thank Victor DiRita for many helpful discussions throughout this work and for providing the pKAS32-toxTΔpro plasmid (26). We thank Nicole Koropatkin for help in assembling molecular models of ToxR and TcpP based on the PhoB crystal structure (2).
This work was supported by grant R01-AI075087 to E.S.K. from the NIH NIAID.
Editor: A. Camilli
Footnotes
Published ahead of print on 2 August 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Bina, J., J. Zhu, M. Dziejman, S. Faruque, S. Calderwood, and J. Mekalanos. 2003. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc. Natl. Acad. Sci. U. S. A. 100:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco, A. G., M. Sola, F. X. Gomis-Ruth, and M. Coll. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10:701-713. [DOI] [PubMed] [Google Scholar]
- 3.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Häse, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgins, D. E., and V. J. DiRita. 1994. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol. Microbiol. 14:17-29. [DOI] [PubMed] [Google Scholar]
- 6.Higgins, D. E., E. Nazareno, and V. J. DiRita. 1992. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J. Bacteriol. 174:6974-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inohara, N., L. del Peso, T. Koseki, S. Chen, and G. Nunez. 1998. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J. Biol. Chem. 273:12296-12300. [DOI] [PubMed] [Google Scholar]
- 8.Karaolis, D. K., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. U. S. A. 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovacikova, G., and K. Skorupski. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 181:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krukonis, E. S., and V. J. DiRita. 2003. DNA binding and ToxR responsiveness by the wing domain of TcpP, an activator of virulence gene expression in Vibrio cholerae. Mol. Cell 12:157-165. [DOI] [PubMed] [Google Scholar]
- 11.Krukonis, E. S., R. R. Yu, and V. J. Dirita. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38:67-84. [DOI] [PubMed] [Google Scholar]
- 12.Li, C. C., J. A. Crawford, V. J. DiRita, and J. B. Kaper. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 35:189-203. [DOI] [PubMed] [Google Scholar]
- 13.Linn, T., and R. S. Pierre. 1990. Improved vector system for constructing transcriptional fusions that ensure independent translation of lacZ. J. Bacteriol. 172:1077-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makarova, O., E. Kamberov, and B. Margolis. 2000. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29:970-972. [DOI] [PubMed] [Google Scholar]
- 15.Makino, K., M. Amemura, S. Kim, A. Nakata, and H. Shinagawa. 1993. Role of the sigma 70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev. 7:149-160. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301-312. [DOI] [PubMed] [Google Scholar]
- 17.Matson, J. S., J. H. Withey, and V. J. DiRita. 2007. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 75:5542-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 20.Murley, Y. M., P. A. Carroll, K. Skorupski, R. K. Taylor, and S. B. Calderwood. 1999. Differential transcription of the tcpPH operon confers biotype-specific control of the Vibrio cholerae ToxR virulence regulon. Infect. Immun. 67:5117-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nye, M. B., J. D. Pfau, K. Skorupski, and R. K. Taylor. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 182:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Skorupski, K., and R. K. Taylor. 1999. A new level in the Vibrio cholerae virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31:763-771. [DOI] [PubMed] [Google Scholar]
- 24.Skorupski, K., and R. K. Taylor. 1996. Broad-host-range positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 25.Yamane, T., H. Okamura, M. Ikeguchi, Y. Nishimura, and A. Kidera. 2008. Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment. Proteins 71:1970-1983. [DOI] [PubMed] [Google Scholar]
- 26.Yu, R. R., and V. J. DiRita. 1999. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J. Bacteriol. 181:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43:119-134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.