Abstract
Noroviruses (NoV) are major agents of acute gastroenteritis in humans and the primary pathogens of shellfish-related outbreaks. Previous studies showed that some human strains bind to oyster tissues through carbohydrate ligands that are similar to their human receptors. Thus, based on presentation of shared norovirus carbohydrate ligands, oysters could selectively concentrate animal strains with increased ability to overcome species barriers. In comparison with human GI and GII strains, bovine GIII NoV strains, although frequently detected in bovine feces and waters of two estuaries of Brittany, were seldom detected in oysters grown in these estuaries. Characterization of the carbohydrate ligand from a new GIII strain indicated recognition of the alpha-galactosidase (α-Gal) epitope not expressed by humans, similar to the GIII.2 Newbury2 strain. This ligand was not detectable on oyster tissues, suggesting that oysters may not be able to accumulate substantial amounts of GIII strains due to the lack of shared carbohydrate ligand and that they should be unable to contribute to select GIII strains with an increased ability to recognize humans.
Environmental sources of animal pathogens and, most specifically, of RNA viruses may constitute substantial risk factors for cross-species transmission to humans (14). In this context, noroviruses (NoVs) infecting cattle could be of importance owing to the high densities of cows bred in areas of human activities. The ability of shellfish to concentrate pathogens released in seawater raises questions about the transmission of animal NoVs to humans through oyster consumption, but so far very few studies have compared water and shellfish contamination. One of the first such studies, conducted more than 30 years ago, comparing the presence of enterovirus by cell culture in water and oysters yielded about the same frequency of positive water (59%) and shellfish samples (35%) (12). More recently, phages of Bacteroides fragilis and Salmonella detected in sewage effluents were also detected in receiving waters and oysters (6). Human NoVs were detected in 75% of river water samples and in 60% of oyster beds (38). Only one study reported the detection of porcine norovirus in 15% of shellfish collected from the U.S. market but no information from the surrounding water was available (8).
NoVs are small nonenveloped viruses approximately 30 nm in diameter with a positive-sense, single-stranded RNA genome. They belong to the Caliciviridae family, and in humans they are the most frequent cause of diarrhea outbreaks in all age groups (11, 28). They are classified in five genogroups, with human strains belonging to genogroups I, II, and IV, GIII strains infecting cattle, and murine strains classified in GV (45). Recently, two new genogroups (VI and VII) infecting animals have been proposed (29). Based on analysis of the open reading frame 2 (ORF2) sequence encoding the capsid protein, high diversity has been observed, with the result that genogroups have been subdivided into clusters, including up to 19 for GII strains. Porcine NoVs have been classified into three clusters of GII (GII.11, GII.18, and GII.19) while all bovine strains of NoV described so far belong to GIII (25, 29, 41, 45). The first bovine strain, Bo/Newbury2/1976/UK (NB2), was isolated in the United Kingdom from calves with diarrhea (43). Later, another distinct genotype of bovine NoV, Bo/Jena/1978/GER, was identified in Germany (21). These two strains represent the prototypes of the GIII.2 and GIII.1 genotypes, respectively.
Although many gaps persist in our understanding of human NoV infections and pathogenesis, recent advances demonstrated a genetically determined host susceptibility based on histo-blood group antigen diversity. Various human NoV strains attach to distinct carbohydrates of the ABH and Lewis histo-blood group family, and evidence accumulated from volunteer studies and outbreaks indicates that binding to these carbohydrates is required for infection (19, 35). In addition, it was recently shown that the prototype bovine GIII.2 strain binds to a related carbohydrate structure which is absent from human tissues (44). Similarly, it was also demonstrated that some strains of either GI or GII specifically attach to oysters tissues through recognition of histo-blood group antigens (HBGAs) (17, 22, 36). This finding could help explain other observations, such as the rapid contamination of oysters, long persistence of viral particles, and, consequently, shellfish-borne outbreaks (3, 16). It additionally suggests that oysters may not merely act as passive filters randomly accumulating virus particles but, instead, may also act as selective filters specifically concentrating strains by recognition of carbohydrate epitopes shared with humans. As shellfish are grown in coastal waters frequently exposed to contamination from bovine in neighboring fields, they may be contaminated by these animal strains. This raises the issue of the potential role of oysters in the emergence of bovine NoVs into the human population.
The aim of our study is to provide quantitative data on the presence of GIII NoV strains in comparison with GI and GII strains in bovine feces, rivers, or estuarine waters as well as shellfish from an area of both high cattle density and high-density oyster breeding. The possibility of GIII strain-specific binding to carbohydrate ligands of oyster tissues that may be shared with cows and humans is additionally examined. The results are discussed in the context of the environmental data in order to provide a first appreciation of the risk of GIII NoV transmission to humans through oyster consumption.
MATERIALS AND METHODS
Reagents.
The lectin from Griffonia simplicifolia B4 isolectin 1 (GS1-B4), either peroxidase or fluorescein isothiocyanate (FITC) conjugated, which recognizes α1,3-linked terminal galactosyl residues, was purchased from Vector Laboratories (Burlingame, CA) and from EY Laboratories (San Mateo, CA), respectively. Alpha-galactosidase (α-Gal) from green coffee beans was purchased from Sigma (St. Louis, MO). The anti-B monoclonal antibody (MAb) ED3 was a kind gift from A. Martin (CRTS, Rennes, France). The anti-GIII rabbit polyclonal antiserum was prepared at the Veterinary School of Nantes by immunizing a rabbit with virus-like particles (VLPs) from the GIII.2 Bo/Newbury2/76/UK strain (44). This antiserum reacts nearly equally well with BEC28 VLPs (see below) and the immunizing Newbury2 VLPs. The anti-GI rabbit antiserum was prepared similarly using VLPs from the Norwalk virus (NV) strain. Synthetic oligosaccharides as polyacrylamide conjugates were kindly provided by N. Bovin (Moscow, Russia). Oligosaccharides coupled to human serum albumin (HSA) were obtained from IsoSep AB (Tulligen, Sweden). The structure of all oligosaccharides used is given in Table SA1 in the supplemental material.
Stool samples.
Bovine stool samples were collected from cattle in the surrounding area of the environmental sampling site in Brittany (northwestern France). For analysis, a 10% suspension was prepared, and then nucleic acids (NA) were extracted and purified with a QIAmp Viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions (1).
Environmental samples.
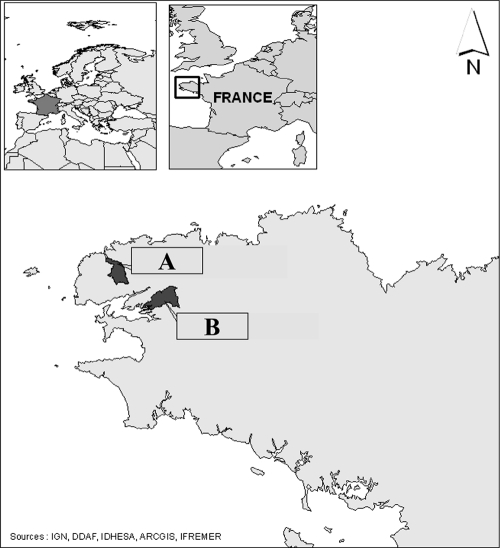
Two small estuaries, sites A and B, located in northwestern France (Brittany) were selected based on high-density bovine breeding since cows represent 8.86 × 106 and 1.84 × 106 inhabitant equivalents in terms of bacterial shedding in these two areas, respectively, and based on shellfish productions classified as B areas (European Regulation 54/2004/EC) (Fig. 1). Streams or small rivers impacted by farms and then flowing into the estuaries were identified and sampled during the study under a volume of 1 liter. Oyster samples, each one constituted by at least 12 individuals, located downstream of potentially contaminated water were collected.
FIG. 1.
Localization of the two sampling areas. The two sites (A and B) are located in Brittany, northwestern France. The dark gray areas represent the catchment basins impacting water quality at these sites.
Viral elution and concentration.
One-liter water samples without any pretreatment were concentrated to 40 ml by using an ultrafiltration membrane (Vivaflow 50; Sartorius) after addition of a known amount of mengovirus (see below) and then by polyethylene glycol 6000 (PEG 6000) precipitation (Sigma, St. Quentin, France) overnight at 4°C before centrifugation for 1.5 h at 1,500 × g (9). For oysters, all 12 individuals of a sample were dissected, and digestive tissues (DT) were pooled together to homogenize the sample and then frozen in 6-g portions. For analysis, one portion was thawed, and mengovirus was added as a viral extraction control. Then tissues were homogenized using beads (FastPrep), extracted by vortexing with an equal volume of chloroform-butanol for 30 s, and treated with Cat-Floc T (Calgon, Ellwood City, PA) for 5 min at room temperature before centrifugation for 15 min at 13,500 × g. The resulting suspension was precipitated with PEG 6000 for 1 h at 4°C and centrifuged for 20 min at 11,000 × g at 4°C (2).
Nucleic acid extraction.
For nucleic acid extraction and purification, a Nuclisens extraction kit (bioMérieux, Lyon France) was used according to the manufacturer's instructions, with minor modifications (18). PEG pellets from water or oyster samples were suspended in 1 ml of RNase-free water, mixed with the lysis buffer (2 ml), and incubated for 30 min at 56°C. After a brief centrifugation to eliminate particles (if needed), 50 μl of paramagnetic silica was added and incubated for 10 min at room temperature. All washes were performed using the magnetic ramp, and viral nucleic acids were recovered in 100 μl of elution buffer (bioMérieux, Lyon, France). Extracts were directly analyzed or frozen at −80°C until they were used.
Real-time RT-PCR.
All amplifications were conducted using a one-step real-time reverse transcription-PCR (rRT-PCR) kit (Ultrasens, Invitrogen) with previously described primers and probes for mengovirus (30) or NoV (18, 42). Briefly, they were as follows: for NoV GI, QNIF4 (5′-CGCTGGATGCGNTTCCAT-3′, where N is A, C, G or T), NV1LCR (5′-CCTTAGACGCCATCATCATTTAC-3′), and NV1LCpr (5′-TGGACAGGAGAYCGCRATCT-3′, where Y is C or T and R is A or G); for GII, QNIF2d (5′-ATGTTCAGRTGGATGAGRTTCTCWGA-3′, where R is A or G and W is A or T), COG2R (5′-TCGACGCCATCTTCATTCACA-3′), and QNIFs (5′-AGCACGTGGGAGGGCGATCG-3′); and for GIII, SW GIII forw (5′-CGCTCCATGTTYGCBTGG-3′, where Y is C or T), SW GIII rev (5′-TCAGTCATCTTCATTTACAAAATC-3′), and the probe SWGIII (5′-TGTGGGAAGGTAGTCGCGACRYC-3′, where R is A or G and Y is C or T).
All samples were analyzed in duplicate on 5 μl of undiluted or 10-fold diluted RNA extracts. Two negative amplification controls (water) were included in each amplification series, and no more than six samples were analyzed in an rRT-PCR assay. Precautions such as isolated rooms for various steps and the usage of filter tips were taken to prevent false-positive results.
The cycle threshold (CT) was defined as the cycle at which a significant increase in fluorescence occurred (i.e., when fluorescence became distinguishable from background). The number of NoV RNA copies present in positive samples was estimated using GI, GII, and GIII standard curves. To be included in the quantitative analysis, all wells had to yield a CT value of ≤41, which was considered the quantification threshold (QT). The final concentration was then determined based on the NA volume analyzed (5 μl of 100 μl of NA extract) and measured weight of DT (6 g analyzed) (18, 30).
The efficiency of virus extraction procedures was determined for each extraction by seeding 104 50% tissue culture infective doses of mengovirus prior to sample processing and determining mengovirus recovery by rRT-PCR, as previously described (7, 18). NoV concentrations were then corrected for virus loss during extraction by dividing the final norovirus concentration (uncorrected) by the mean mengovirus extraction efficiency.
Preparation of VLPs.
The entire capsid region was amplified from the intestinal content of a calf infected with strain BEC/28/03IT (accession number GQ397857) identified in a survey performed in Northern Italy, as described in the supplemental material. It was then cloned into the pFastBac vector, which was used to prepare the recombinant bacmid (BAC) BEC28, according to the manufacturer's instructions (Invitrogen). The sequence of the vector obtained confirmed the proper frame of the ORF2 and the close relationship (92% identity) of the Italian strain with a previously described Dutch BEC CH131-like strain (39). The bacmid was transfected into Sf9 insect cells to prepare the high-titer BAC BEC28 virus stock solution. VLPs were produced by infection of Sf9 cells with the stock baculovirus solution and purified by ultracentrifugation through a 30% (wt/vol) sucrose cushion, followed by a CsCl (1.362 g/cm3) density gradient (13). Quality of the purified BEC28 VLPs was analyzed by Western blotting and electron microscopy.
VLPs from the GIII.2 Bo/Newbury2/76/UK (NB2) strain prepared as previously described (44) were a kind gift from D. Poncet (CNRS, Gif/Yvette, France). VLPs from the GI.1 Norwalk virus strain were prepared as previously described (24).
Bioaccumulation of VLPs.
GI.1 (Norwalk) (0.8 μg) and GIII.2 (Newbury2) (1.28 μg) VLPs were added to 500 ml of clean seawater and homogenized for 5 min to obtain 3 × 107 and 7 × 107 VLPs/ml for GI and GIII, respectively. Two clean live oysters were added to the seawater and incubated for 24 h at 15°C (water temperature) under oxygenation. The bioaccumulation experiment was conducted twice.
Tissue samples and immunohistochemical analysis.
Bovine and porcine tissue samples from the gastroduodenal junction were obtained from healthy animals autopsied at the National Veterinary School of Nantes. Animals were handled in strict accordance with good animal practice as defined by the French national guidelines. Human gastroduodenal junction samples had been obtained from organ donors before the law 88-1138 of 20 December 1988 concerning resection of human tissues after death for scientific investigations. Oyster tissues were prepared from a batch of clean oysters (tested negative for NoV contamination) or from oysters harboring bioaccumulated VLPs so as to visualize all organs on a single section. Animal tissues were fixed in formalin, and human tissues were fixed in 95% ethanol for 48 h and paraffin embedded. Sections (5 μm) were rehydrated in graded ethanol and washed in phosphate-buffered saline (PBS). Endogenous peroxidase was inhibited by a 20-min incubation with methanol-0.3% H2O2. Sections were then washed in PBS-1% bovine serum albumin (BSA) for 30 min at room temperature. After sections were washed in PBS, they were incubated with the peroxidase-conjugated GS1-B4 lectin at 10 μg/ml at 4°C overnight. The attachment of VLPs to tissue sections was tested with BEC28 VLPs at 10 μg/ml or with NB2 VLPs at 1 μg/ml, followed by sequential incubations with a rabbit anti-GIII.2 antiserum, peroxidase-conjugated anti-rabbit IgG, and the 3-amino-9-ethylcarbazol peroxidase substrate, as previously described (45). Detection of Norwalk or Newbury2 bioaccumulated VLPs from oysters was performed directly by the anti-GI or anti-GIII rabbit antiserum. Periodate treatment at 1 and 10 mmol/liter for 30 min and alpha-galactosidase treatment were performed immediately after the endogenous peroxidase quenching, as previously described (17).
Enzyme-linked immunosorbent assay (ELISA)-based carbohydrate microtiter plate assays.
Oysters located in the same geographic area and under the same climatic conditions yet in a clean area were collected during the same period of time as those from the study field. Tissue extracts were prepared according to Maalouf et al. (22). For each tissue (digestive tissue, gills, and mantle), extracts were prepared from seven samples, each constituted by at least eight oysters collected throughout the year. Tissue extracts at 40 μg/ml were coated onto NUNC Maxisorp Immunoplates (ThermoFischer Scientific) in 100 mmol/liter carbonate buffer, pH 9.6, by overnight incubation at 4°C. After a blocking step with 10% nonfat dried cow's milk in PBS for 1 h, NB2 VLPs (1 μg/ml) or NV VLPs (1 μg/ml) in PBS-5% milk were added and incubated for 1 h at 37°C. Plates were then incubated with rabbit anti-GIII.2 serum or anti-GI.1 serum at a 1/1,000 dilution in PBS-5% milk for 1 h at 37°C. Then, peroxidase-conjugated anti-rabbit IgG (Uptima, Montluçon, France) at a 1/2,000 dilution in PBS-5% milk was added and incubated for 1 h at 37°C. After each step, the plates were washed three times with PBS-5% Tween 20. The enzyme signals were detected with TMB (3,3′,5,5′-tetramethylbenzidine) as a substrate (BD Bioscience, San Jose, CA) and then read at 450 nm.
Oligosaccharides as phosphonoacetic acid (PAA) and HSA conjugates were coated at 10 μg/ml at 37°C in a wet atmosphere. After samples were blocked with 5% nonfat dried cow's milk in PBS for 1 h, BEC28 VLPs (9.2 μg/ml) in PBS-5% milk were added. After incubation for 2 h at 4°C, binding of VLPs was detected as above using rabbit anti-GIII.2 serum and then peroxidase-conjugated anti-rabbit IgG with 1-h incubations at 4°C.
Cell transfection and flow cytometry analysis.
The complete coding sequence of the Ggta1 gene encoding the rat α1,3-galactosyltransferase was cloned as previously described (37) and inserted into the pCR3.1 eukaryotic expression vector (Invitrogen, Paisley, United Kingdom). It was used to transfect human embryonic kidney (HEK293) cells with Lipofectamine (Invitrogen). Forty-eight hours later, cells were labeled with either FITC-labeled GS1-B4 lectin at 10 μg/ml or BEC28 VLPs at 9.2 μg/ml as previously described (44). In the former case, fluorescence analysis was performed immediately following washes. In the latter case, cells were sequentially incubated with the rabbit anti-GIII.2 serum at a 1/1,000 dilution and with FITC-labeled anti-rabbit IgG (Sigma) for 30 min. After a final washing, fluorescence analysis was performed on a FACSCalibur (Becton Dickinson, Heidelberg, Germany) with the CellQuest program.
RESULTS
Detection of NoVs in feces and environmental samples.
GIII NoV was detected in 25 samples out of 136 stool samples analyzed (18%). A total of 70 water samples were collected (29 from site A and 41 from site B) in June, September, and November of 2008 and in January, March, April, May, July, August, and September of 2009 and tested for the presence of GI, GII, and GIII NoVs. The three genogroups were detected in water samples from both sites. GI NoVs were detected in two samples from site A and in three samples from site B. GII NoVs were detected in 4 samples from site A and in 13 samples from site B, and GIII NoVs were detected in 2 samples from site A and 8 samples from site B. A total of 47 samples of oysters (comprising at least 564 oysters) were collected over the study. Thirty-eight samples were collected from site A, and among these, one and seven were positive for GI and GII NoVs, respectively. From site B, over the nine oyster samples collected, one was found positive for GI, three were positive for GII, and one was positive for GIII NoVs. Multiple contaminations by GI and GII NoVs were detected in three water samples. Contaminations with both GII and GIII were detected in three additional water samples. The only oyster sample positive for GIII NoV was also found contaminated with a GII NoV. The frequencies of either water or oyster NoV contamination did not differ significantly between the two sites for any of the genogroups (P < 0.2, Fisher's exact test). The results from both areas were thus pooled for further analysis as shown in Table 1.
TABLE 1.
Detection and quantification of NoVs in water and oysters samples
| Genogroup | NoVs in water (n = 70)a |
NoVs in oysters (n = 47)a |
NoV concn in water/NoV concn in oystersc | ||||
|---|---|---|---|---|---|---|---|
| No. (%) of positive samples | Mean concn (no. of RNA copies/liter [range])b | Sum (no. of RNA copies/liter) | No. (%) of positive samples | Mean concn (no. of RNA copies/g of tissue [range])b | Sum (no. of NoV RNA copies/g of tissue) | ||
| GI | 5 (7) | 2,302 (120-4,900) | 11,510 | 2 (4) | 190 (71-310) | 381 | 30 |
| GII | 17 (24) | 19,149 (410-110,000) | 325,530 | 10 (21) | 45 (5-115) | 278 | 1,171 |
| GIII | 10 (14) | 14,222 (130-120,000) | 142,220 | 1 (2) | 90 | 90 | 1,580 |
n, number of samples.
Geometric mean. The means between the three NoV genogroups, either in water or in oyster samples, are not significantly different (Mann-Whitney two-tailed test).
Ratio of the sum of RNA copies detected in water to the sum of RNA copies detected in oyster samples during the study.
Quantitative analysis of NoV contamination of water samples indicated that far fewer GI than either GII or GIII RNA copies were detected during the study period. Yet the mean values of contaminated water samples did not differ between the three genogroups owing to the high variation in the number of detected RNA copies and to the lower number of GI-contaminated samples. Likewise, the mean numbers of RNA copies detected in contaminated oyster samples were not significantly different among genogroups. For both GI and GII NoVs, the percentages of contaminated water and oyster samples were quite similar (7% versus 4% for GI and 24% versus 21% for GII). Yet in the case of GIII NoV, the percentage of positive water samples was higher than that of oyster samples (14% and 2%, respectively), suggesting that GIII NoVs may not accumulate as efficiently as either GI or GII into oysters. However, owing to the small number of contaminated oyster samples, this difference did not reach statistical significance. When the ratio of total RNA copies detected in waters to copy numbers detected in oysters was calculated, a striking difference appeared between GI and either GII or GIII NoVs. Indeed, this ratio was 39- and 53-fold lower for GI than for GII and GIII, respectively, suggesting a highly efficient GI accumulation compared to GII and GIII accumulations in oysters (Table 1).
Characterization of the BEC28 VLPs ligand.
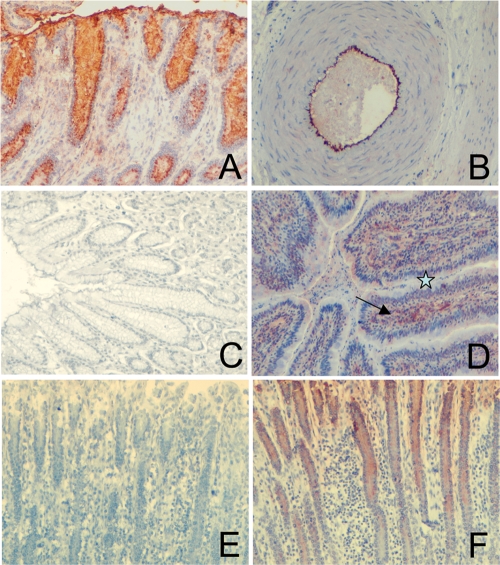
We previously used immunohistochemistry to show the binding of VLPs from several calicivirus strains, including the bovine GIII.2 Bo/Newbury2/76/UK (NB2) norovirus strain, to carbohydrates expressed on epithelial digestive cells (17, 24, 32, 44). Within genogroup, variations in carbohydrate specificity have been reported for human GII.4 strains (20). Thus, to determine if the ligand specificity of NB2 is also variable among GIII.2 strains, we sought to characterize a potential carbohydrate ligand of the newly described strain BEC28. This strain is clearly distinct from NB2, yet it can be classified as GIII.2 (see Fig. SA1 in the supplemental material). To this end, BEC28 VLPs were incubated on tissue sections from the gastroduodenal junction of bovine, porcine, and humans. Distinct results were obtained for each species. In bovine tissues, we observed staining on both endothelial and epithelial cells from the gut. In porcine tissues, the BEC28 capsids attached exclusively to nonepithelial cells of the digestive tract, mainly vascular endothelial cells, whereas staining was completely absent on human tissue sections tested in the same experiments (Fig. 2 A to D). Under similar experimental conditions, GI.1 and GII.4 VLPs readily stained human epithelial cells of serial tissue sections from the same individuals (data not shown). To determine if the binding involved carbohydrates, bovine tissue sections were pretreated with sodium periodate prior to incubation with the VLPs. At a 10 mmol/liter concentration, but not at 1 mmol/liter, the staining was completely lost, suggesting that BEC28 VLPs recognize a neutral glycan structure expressed both on bovine digestive surface epithelial cells and vascular endothelial cells. Since these results were reminiscent of what we previously observed with VLPs from the bovine NB2 strain which bind to the α-Gal epitope (see below), the effect of α-galactosidase treatment was tested. We observed that following treatment, the staining completely disappeared, indicating recognition of glycans with a terminal galactose residue in alpha linkage (Fig. 2E and F).
FIG. 2.
Analysis of BEC28 binding to tissues. VLPs were incubated on tissue sections and detected as described in the Materials and Methods section. Images show binding to bovine pyloric surface mucosa (A) and to the vascular endothelium of a bovine artery (B) and lack of binding to human pyloric surface mucosa (C). (D) Binding to porcine duodenal surface mucosa with lack of staining of epithelial cells is shown by a star, and staining of vascular endothelial cells of the lamina propria is shown by an arrow. (E) Effect of α-galactosidase treatment prior to VLP incubation on a bovine duodenum section. (F) Serial control section incubated under the same condition in the absence of enzyme.
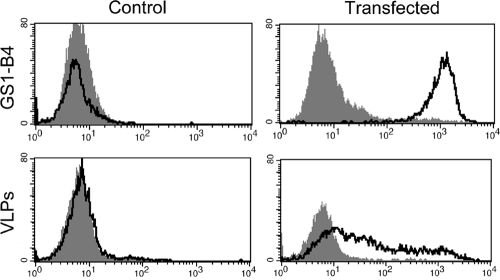
To define more precisely the carbohydrate specificity of BEC28 VLPS, binding was tested on a set of HBGA-related oligosaccharides by ELISA. A signal five times above background was observed on a single structure terminated by a galactose in α1,3 linkage, the Galα3Galβ4GlcNAc trisaccharide called the α-Gal or Galili epitope (Fig. 3). We next transfected human HEK293 cells with the rat Ggta1 cDNA to allow expression of the α-Gal antigen as detected by flow cytometry with the GS1-B4 lectin. Control HEK293 cells that lacked the α-Gal antigen were not recognized by BEC28 VLPs. In contrast, clear binding was observed on the Ggta1-transfected human cells (Fig. 4). Collectively, these results indicate that BEC28 VLPs have a carbohydrate specificity similar to that of NB2 VLPs (44), specifically recognizing the α-Gal antigen.
FIG. 3.
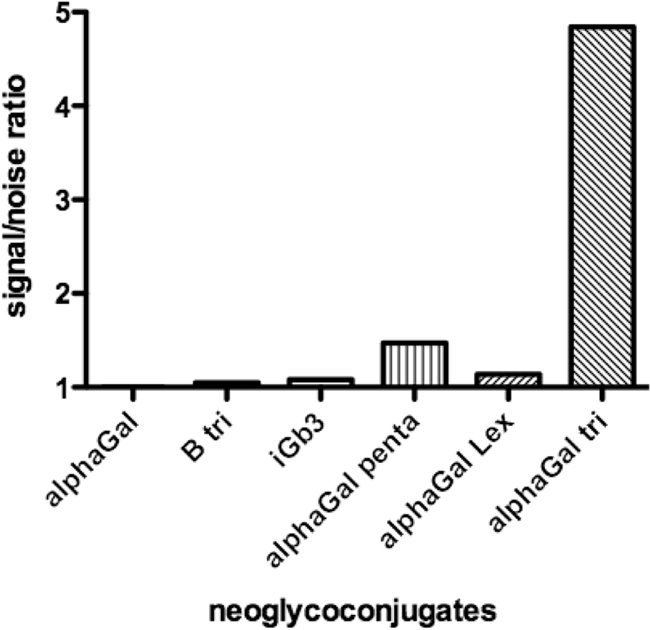
Binding of BEC28 VLPs to immobilized synthetic oligosaccharides. A panel of neoglycoconjugates was coated on ELISA plates, and the binding of BEC28 VLPs was detected as described in the Materials and Methods section. Results for a selected set of neoglycoconjugates with a terminal galactose in alpha linkage are shown as signal-to-noise ratios from one experiment representative of three independent ones. None of the other neoglycoconjugates tested showed a signal-to-noise ratio above 1 (see the list and structures in Table SA1 in the supplemental material). alphaGal, α-galactose monosaccharide; B tri, B trisaccharide; iGb3, isoglobotrihexosyl; alphaGal penta, α-Gal pentasaccharide; alphaGal Lex, α-Gal-Lewis x; alphaGal tri, α-Gal trisaccharide.
FIG. 4.
Flow cytometric analysis of the binding of BEC28 VLPs and the GS1-B4 isolectin to rat Ggta1-transfected or control mock-transfected human HEK293 cells. The respective negative controls correspond to either cells incubated in the absence of the lectin or to cells incubated in the presence of the VLPs followed by incubation with an irrelevant rabbit antiserum and FITC-labeled anti-rabbit IgG. The log of fluorescence intensities in arbitrary units is plotted against the cell number.
GIII.2 ligands from oyster tissues.
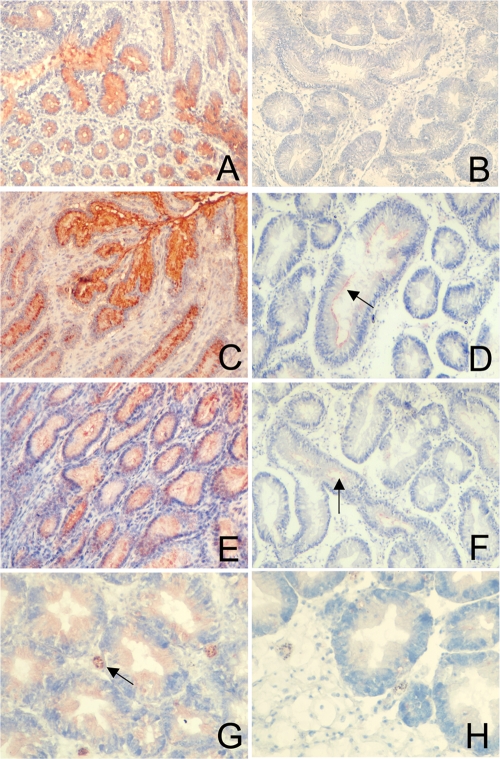
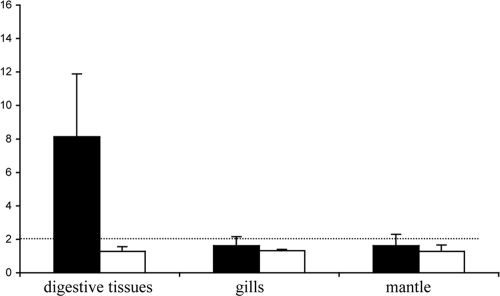
To search for the α-Gal epitope on oyster tissues, tissue sections allowing analysis of all organs were covered by the GS1-B4 lectin. No staining was observed, contrasting with the strong staining obtained on bovine gut tissue sections (Fig. 5 A and B). The α-Gal epitope is closely related to the B blood group antigen (compare α-Gal trisaccharide and B type 2 in Table SA1 in the supplemental material), we looked for the presence of the B antigen in oyster tissues. An anti-B MAb that strongly binds to human tissue sections from B blood group persons did not react with oyster tissues (data not shown). Since oysters may present a GIII ligand distinct from the α-Gal epitope that could be recognized by the VLPs, we tested the binding of the NB2 recombinant capsids to oyster soluble tissue extracts. No specific signal could be detected by ELISA on seven soluble extracts of three types of tissues (mantle, gills, and digestive tissues), each prepared from at least eight oysters, whereas in the same assays, these VLPs attached to the synthetic α-Gal epitope, and GI.1 VLPs used as positive controls readily attached to oyster digestive tissues soluble extracts (Fig. 6). Likewise, although clear binding of both NB2 and BEC28 VLPs was observed on bovine duodenum tissue sections used as positive controls in parallel assays, only slight, occasional labeling of the surface of some digestive tubules by either type of VLPs could be detected (Fig. 5C to F). In control experiments no staining could be observed in the absence of GIII VLPs, whereas strong unambiguous staining was obtained by using GI.1 VLPs detected with a specific antiserum, consistent with previous reports (data not shown). These results indicate that oyster tissues do not express the α-Gal epitope in detectable amounts but may nevertheless either express small amounts of another ligand yet to be characterized or bind small amounts of GIII VLPs in a nonspecific manner.
FIG. 5.
Expression of GIII ligands in oyster digestive tract and detection of bioaccumulated VLPs. Staining of bovine pyloric mucosa by the GS1-B4 isolectin (A), BEC28 VLPs (C), and NB2 VLPs (E) and comparison with oyster digestive tract are shown. Absence of staining of oyster tissues by GS1-B4 (B) and slight staining (arrows) of the lumen of oyster digestive tubules by BEC28 VLPs (D) and NB2 VLPs (F) are shown. (G) Presence of GI.1 (NV) VLPs in hemocytes and digestive epithelial cells following bioaccumulation. (H) Absence of GIII.2 (NB2) in oyster tissues following bioaccumulation.
FIG. 6.
Binding of recombinant GI.1 (NV) and GIII.2 (NB2) VLPs to oyster tissue extracts. Binding of GI.1 (black bars) and GIII.2 (white bars) VLPs to extracts of digestive tissues, gills, and mantle (x axis) is expressed as signal-to-noise ratio ± standard deviation (y axis). The positive threshold is indicated by a dashed line. Binding of NB2 VLPs to the α-Gal trisaccharide coupled to polyacrylamide and of NV VLPs to the H type 1 trisaccharide was used as a positive control on each plate (data not shown).
Next, the ability of GI.1 and GIII.2 VLPs to be bioaccumulated in oysters was compared. Both types of VLPs were seeded in water at similar concentrations (3 × 107 and 7 × 107 particles/ml, respectively). After 24 h oyster tissues were fixed, and the presence of VLPs was searched by immunohistology using anti-GI and anti-GIII antisera that have identical titers against their respective VLPs by ELISA. Under these conditions GI.1 VLPs, used as positive controls, were readily detected. In contrast, GIII VLPs could not be detected (Fig. 5G and H).
DISCUSSION
Tracing viral contamination in the open environment is quite difficult due to several factors such as source identification, dilution, currents, and, finally, sampling strategy (15, 31). Cows are bred at very high densities in some areas such as the west coast of France (Brittany), which is also a major area of shellfish production. The present study represents a first approach to evaluate the prevalence of bovine NoVs in environmental waters and in shellfish. Two sites from Brittany were selected based on their bovine population, which was at least a 100-fold greater than their human population in terms of sewage input (106 inhabitant equivalents for bovine compared with 104 and 103 inhabitant equivalents for humans at sites A and B, respectively). The shellfish-growing areas of these two sites (mainly oysters) are classified as B areas (European Community regulation 54/2004/EC). Eighteen percent of the bovine stool samples tested were positive for GIII NoV, a value comparable with results from other studies which range widely from 2% to 50% (25-27, 33, 39). Bovine NoVs were detected in a total of 14% of water samples. By comparison, GI and GII NoVs were detected in 7% and 24% of water samples, respectively. Moreover, the sum of GI RNA copies detected in water during the whole study period was 28 times lower than that of GII, in accordance with epidemiological data which show a large predominance of GII strains in the human population (34). The sum of GIII RNA copies detected was only two times lower than that of GII human NoVs, showing the high impact of bovine production on water contamination. In Brittany, bovines are bred in open fields most of the year, and thus feces may directly be dragged along toward small nearby rivers. Considering the substantial percentage of water-positive samples and the long persistence of human NoVs in contaminated oysters, we were surprised to find only one GIII shellfish-positive sample. Because extraction and inhibitor controls were used, we trust that negative samples were truly negative (18). The choice of primers and probe set used is another factor that could have influenced the results. However, the primers and probe used, which were selected from the literature, readily detected GIII viruses in bovine stools and water samples collected in the same area.
Specific binding of human NoV strains to oyster tissues was previously shown to occur through interaction with carbohydrate ligands shared with humans, suggesting that oysters may select NoV strains present in the environment, based on the presence of specific carbohydrate ligands (17, 36, 40). Contamination of seawater by animal NoVs could thus potentially lead to a selection of strains recognizing shared carbohydrate motifs, thereby facilitating cross-species transmission to humans. We show here that, similar to the GIII.2 prototype NB2 strain (44), BEC28 VLPs recognize the α-Gal epitope (Galα3Galβ4GlcNAc) expressed on surface epithelial cells of bovine stomach and duodenum. Specificity for this ligand therefore appears to be conserved among GIII.2 strains despite the genetic distance between the BEC28 and NB2 strains. Synthesis of the α-Gal epitope requires an α1,3-galactosyltransferase which is lacking in all human cells since the GGTA1 gene encoding this enzyme has been inactivated and is therefore completely absent from human tissues (23). In addition, the GIII ligand was not detectable on porcine digestive epithelial cells, making it unlikely that either humans or swine could be infected by these NoV strains since binding to HBGAs appears necessary for infection by various human strains (10, 19, 35). Moreover, we observed that the α-Gal epitope and the closely related B blood group antigen are not expressed on oyster tissues, suggesting that oysters should not be able to specifically concentrate bovine strains through binding to these carbohydrate structures, which share a terminal Galα3Gal-R motif. Nevertheless, weak binding of both NB2 and BEC28 VLPs to unidentified structures of oyster digestive epithelial cells was detectable, consistent with the rare and quantitatively weak detection of GIII sequences in oyster samples compared with the frequent water contamination. In bioaccumulation experiments no GIII VLPs were detected in oyster tissues, suggesting that the lack of specific ligand in these tissues may contribute to explain the rare occurrence of GIII oyster contamination despite frequent water contamination. In contrast, GI VLPs, which have a specific carbohydrate ligand in the oyster digestive tract, were efficiently bioaccumulated, as previously described (22). This is in accordance with the low ratio of GI RNA copies between water and oyster samples found in our environmental study. Alternatively, we cannot exclude the possibility that GIII strains, as well as GII strains, may not accumulate and persist in oysters as efficiently as GI strains because of a lower stability. Thus, GI NoVs could accumulate and persist in oysters because of high environmental stability and because of the presence of a specific ligand. In contrast, GII and GIII strains would accumulate and persist less well because of the absence of one of these two characteristics. Overall, our results indicate that GIII strains, while largely present in the coastal environment, are not so frequently found in oysters. Because of the absence of a specific carbohydrate ligand, selection of variants that would be able to cross-recognize human carbohydrate receptors would be unlikely to occur. Nevertheless, some contamination of oysters by GIII through nonspecific interactions may occur. Thus, it is unlikely that oyster consumption would facilitate GIII NoV infections in humans.
Human NoV strains of the GI and GII genogroups use distinct binding sites on the P2 domain of the capsid protein to attach to HBGAs (4, 5). The binding site of GIII strains and their mode of recognition of the α-Gal epitope are not known as yet, but to more precisely evaluate the risk of cross-species transmission, it will be important in future studies to determine the structural changes necessary for GIII strains to recognize HBGA motifs expressed on the human gut and potentially shared with cows. Likewise, bioaccumulation experiments with virions, rather than with VLPs, will be necessary to precisely determine the ability of oysters to accumulate and maintain GIII NoV strains. Stability in the environment of the different genogroups also needs to be compared. In conclusion, we demonstrated that a new approach combining in vitro studies and environmental sample analysis brings valuable information to investigate the possible transmission of animal viruses to humans through oyster consumption.
Supplementary Material
Acknowledgments
This work was supported in part by grant 2006 SEST 08 01 “Coquenpath” from the Agence Nationale pour la Recherche (ANR, 538), by a grant (CIMATH) from the Région des Pays de la Loire, and by the European Commission, DG Research Quality of 314 Life Program 6th Framework (EVENT; grants SP22-CT-2004-502571 and FP6-2002-SSP-315 1). H.M. was supported by a fellowship from Ifremer and the Conseil Régional des Pays de la Loire.
We are grateful to Didier Poncet and Nicolai Bovin for providing reagents and to the Alcyon laboratory (Morlaix), l'Ecole ONIRIS de Nantes, and the IDAC laboratory (Nantes) for providing bovine stools.
Footnotes
Published ahead of print on 13 August 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ambert-Balay, K., M. Lorrot, F. Bon, H. Giraudon, J. Kaplon, M. Wolfer, P. Lebon, D. Gendrel, and P. Pothier. 2008. Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients. J. Clin. Microbiol. 46:1252-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmar, R. L., F. H. Neill, J. L. Romalde, F. Le Guyader, C. M. Woodley, T. G. Metcalf, and M. K. Estes. 1995. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl. Environ. Microbiol. 61:3014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butt, A., K. E. Aldrige, and C. V. Sanders. 2004. Infections related to the ingestion of seafood. Part I: viral and bacterial infections. Lancet Infect. Dis. 4:201-212. [DOI] [PubMed] [Google Scholar]
- 4.Cao, S., Z. Lou, M. Tan, Y. Chen, Y. Liu, Z. Zhang, X. Zhang, X. Jiang, X. Li, and Z. Rao. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, J.-M., A. M. Hutson, M. K. Estes, and B. V. Prasad. 2008. Atomic resolution structural characterisation of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 105:9175-9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, H., L. A. Jaykus, G. Lovelace, and M. D. Sobsey. 1998. Bacteriophages and bacteria as indicators of enteric viruses in oysters and their harvest waters. Water Sci. Technol. 38:37-44. [Google Scholar]
- 7.Costafreda, M. I., A. Bosch, and R. M. Pinto. 2006. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 72:3846-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costantini, V., F. Loisy, L. Joens, F. S. Le Guyader, and L. J. Saif. 2006. Human and animal enteric caliciviruses in oysters from different coastal regions of the United States. Appl. Environ. Microbiol. 72:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Silva, A. K., F. S. Le Guyader, J. C. Le Saux, M. Pommepuy, M. A. Montgomery, and M. Elimelech. 2008. Norovirus removal and particle association in a waste stabilization pond. Environ. Sci. Technol. 42:9151-9157. [DOI] [PubMed] [Google Scholar]
- 10.Donaldson, E. F., L. Lindesmith, A. D. LoBue, and R. S. Baric. 2010. Viral shape-shifting: norovirus evasion of the human immune system. Nat. Rev. Microbiol. 8:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass, R. I., U. D. Parashar, and M. K. Estes. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyal, S. M., C. P. Gerba, and J. L. Melnick. 1979. Human enteroviruses in oysters and their overlaying waters. Appl. Environ. Microbiol. 37:572-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, M. G., Q. Wang, J. R. Smiley, K. O. Chang, and L. J. Saif. 2005. Self-assembly of the recombinant capsid protein of a bovine norovirus (BoNV) into virus-like particles and evaluation of cross-reactivity of BoNV with human noroviruses. J. Clin. Microbiol. 43:778-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heeney, J. L. 2006. Zoonotic viral diseases and the frontier of early diagnosis, control and prevention. J. Intern. Med. 260:399-408. [DOI] [PubMed] [Google Scholar]
- 15.Jimenez-Clavero, M. A., E. Escribano-Romero, C. Mansilla, N. Gomez, L. Cordoba, N. Roblas, F. Ponz, V. Ley, and J.-C. Saiz. 2005. Survey of bovine enterovirus in biological and environmental samples by a highly sensitive real-time reverse transcription-PCR. Appl. Environ. Microbiol. 71:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koopmans, M. 2008. Progress in understanding norovirus epidemiology. Curr. Opin. Infect. Dis. 21:544-552. [DOI] [PubMed] [Google Scholar]
- 17.Le Guyader, F. S., F. Loisy, R. L. Atmar, A. M. Hutson, M. K. Estes, N. Ruvöen-Clouet, M. Pommepuy, and J. Le Pendu. 2006. Norwalk-virus specific binding to oyster digestive tissues. Emerg. Infect. Dis. 12:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Guyader, F. S., S. Parnaudeau, J. Schaeffer, A. Bosch, F. Loisy, M. Pommepuy, and R. L. Atmar. 2009. Detection and quantification of noroviruses in shellfish. Appl. Environ. Microbiol. 75:618-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Pendu, J., N. Ruvöen-Clouet, E. Kindberg, and L. Svensson. 2006. Mendelian resistance to human norovirus infections. Semin. Immunol. 18:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindesmith, L., E. F. Donaldson, A. D. Lobue, J. L. Cannon, D. P. Zheng, J. Vinje, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maalouf, H., M. Zakhour, J. Le Pendu, J. C. Le Saux, R. L. Atmar, and F. S. Le Guyader. 2010. Distribution in tissue and seasonal variation of norovirus genogroup I and II ligands in oysters. Appl. Environ. Microbiol. 76:5621-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macher, B. A., and U. Galili. 2008. The Galα1,3Galβ1,4GlcNAc-R (α-Gal) epitope: a carbohydrate of unique evolution and clinical relevance. Biochim. Biophys. Acta 1780:75-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marionneau, S., N. Ruvöen-Clouet, B. Le Moullac-Vaidye, M. Clement, A. Cailleau-Thomas, G. Riuz-Palacios, P. W. Huang, X. Jiang, and J. Le Pendu. 2002. Norwalk virus binds to H types 1/3 histo-blood group antigens on gastro-duodenal epithelial cells of “secretor” individuals. Gastroenterology 122:1967-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattison, K., A. Shukla, A. Cook, F. Pollari, R. Friendship, D. Kelton, S. Bidawid, and J. M. Farber. 2007. Human noroviruses in swine and cattle. Emerg. Infect. Dis. 13:1184-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauroy, A., A. Scipioni, E. Mathijs, C. Saegerman, J. Mast, J. C. Briger, D. Ziant, C. Thys, and E. Thiry. 2009. Epidemiological study of bovine norovirus infection by RT-PCR and a VLP based antibody ELISA. Vet. Microbiol. 12:243-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, S. I., C. Jeong, H. H. Kim, S. H. Park, S. J. Park, B. H. Hyun, D. K. Yang, S. K. Kim, M. I. Kang, and K. O. Cho. 2007. Molecular epidemiology of bovine noroviruses in South Korea. Vet. Microbiol. 124:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel, M. N., M. A. Widdowson, R. I. Glass, K. Akazawa, J. Vinje, and U. D. Parashar. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phan, T. G., K. Kaneshi, Y. Ueda, S. Nakaya, S. Nishimura, A. Yamamoto, K. Sugita, S. Takanashi, S. Okitsu, and H. Ushijima. 2007. Genetic heterogeneity, evolution, and recombination in noroviruses. J. Med. Virol. 79:1388-1400. [DOI] [PubMed] [Google Scholar]
- 30.Pinto, R. M., M. I. Costafreda, and A. Bosch. 2009. Risk assessment in shellfish-borne outbreaks of hepatitis A. Appl. Environ. Microbiol. 75:7350-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riou, P., J. C. Le Saux, F. Dumas, M. P. Caprais, F. Le Guyader, and M. Pommepuy. 2007. Microbial impact of small tributaries on water and shellfish quality in shallow coastal areas. Water Res. 41:2274-2286. [DOI] [PubMed] [Google Scholar]
- 32.Ruvoën-Clouet, N., J. P. Ganière, G. André-Fontaine, D. Blanchard, and J. Le Pendu. 2000. Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J. Virol. 74:11950-11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scipioni, A., I. Bourgot, A. Mauroy, D. Ziant, C. Saegerman, G. Daube, and E. Thiry. 2008. Detection and quantification of human and bovine noroviruses by a TaqMan RT-PCR assay with a control for inhibition. Mol. Cell Probes 22:215-222. [DOI] [PubMed] [Google Scholar]
- 34.Siebenga, J. J., H. Vennema, D. P. Zheng, J. Vinje, B. E. Lee, X.-L. Pang, E. C. Ho, W. Lim, A. Choudekar, S. Broor, T. Halperin, N. B. Rasool, J. Hewitt, G. Greening, M. Jin, Z. J. Duan, Y. Lucero, M. O'Ryan, M. Hoehne, E. Schreier, R. M. Ratcliff, P. A. White, N. Iritani, G. Reuter, and M. Koopmans. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J. Infect. Dis. 200:802-812. [DOI] [PubMed] [Google Scholar]
- 35.Tan, M., and X. Jiang. 2007. Norovirus-host interaction: implications for disease control and prevention. Expert Rev. Mol. Med. 9:1-22. [DOI] [PubMed] [Google Scholar]
- 36.Tian, P., A. L. Engelbrektson, X. Jiang, W. Zhong, and R. E. Mandrell. 2007. Norovirus recognizes histo-blood group antigens on gastrointestinal cells of clams, mussels and oysters: a possible mechanism of bioaccumulation. J. Food Prot. 70:2140-2147. [DOI] [PubMed] [Google Scholar]
- 37.Turcot-Dubois, A. L., B. Le Moullac-Vaidye, S. Despiau, F. Roubinet, N. Bovin, J. Le Pendu, and A. Blancher. 2007. Long-term evolution of the CAZY glycosyltransferase 6 (ABO) gene family from fishes to mammals: a birth-and-death evolution model. Glycobiology 17:516-528. [DOI] [PubMed] [Google Scholar]
- 38.Ueki, Y., D. Sano, T. Watanabe, K. Akiyama, and T. Omura. 2005. Norovirus pathway in water environment estimated by genetic analysis of strains from patients of gastroenteritis, sewage, treated wastewater, river water and oysters. Water Res. 39:4271-4280. [DOI] [PubMed] [Google Scholar]
- 39.van der Poel, W. H. M., R. van der Heide, F. Verschoor, H. Gelderblom, J. Vinjé, and M. P. G. Koopmans. 2003. Epidemiology of Norwalk-like virus infections in cattle in The Netherlands. Vet. Microbiol. 92:297-309. [DOI] [PubMed] [Google Scholar]
- 40.Wang, D., Q. Wu, X. Kou, L. Yao, and J. Zhang. 2008. Distribution of norovirus in oyster tissues. J. Appl. Microbiol. 105:1966-1972. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Q. H., V. Costantini, and L. J. Saif. 2007. Porcine enteric caliciviruses: genetic and antigenic relatedness to human caliciviruses, diagnosis and epidemiology. Vaccine 25:5453-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf, S., W. M. Williamson, J. Hewitt, M. Rivera-Aban, S. Lin, A. Ball, P. Scholes, and G. E. Greening. 2007. Sensitive multiplex real-time reverse transcription-PCR assay for the detection of human and animal noroviruses in clinical and environmental samples. Appl. Environ. Microbiol. 73:5464-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woode, G. N., and J. C. Bridger. 1978. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J. Med. Microbiol. 11:441-452. [DOI] [PubMed] [Google Scholar]
- 44.Zakhour, M., N. Ruvöen-Clouet, A. Charpilienne, B. Langpap, D. Poncet, T. Peters, N. Bovin, and J. Le Pendu. 2009. The αGal epitope of the histo-blood group antigen family is a ligand for bovine norovirus Newbury2 expected to prevent cross-species prevention. PLoS Pathog. 5:e1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.