Abstract
Histone chaperones regulate the density of incorporated histone proteins around DNA transcription sites and therefore constitute an important site-specific regulatory mechanism for the control of gene expression. At present, the targeting mechanism conferring this site specificity is unknown. We previously reported that the histone chaperone B23/nucleophosmin associates with rRNA chromatin (r-chromatin) to stimulate rRNA transcription. Here, we report on the mechanism for site-specific targeting of B23 to the r-chromatin. We observed that, during mitosis, B23 was released from chromatin upon inactivation of its RNA binding activity by cdc2 kinase-mediated phosphorylation. The phosphorylation status of B23 was also shown to strongly affect its chromatin binding activity. We further found that r-chromatin binding of B23 was a necessary condition for B23 histone chaperone activity in vivo. In addition, we found that depletion of upstream binding factor (UBF; an rRNA transcription factor) decreased the chromatin binding affinity of B23, which in turn led to an increase in histone density at the r-chromatin. These two major strands of evidence suggest a novel cell cycle-dependent mechanism for the site-specific regulation of histone density via joint RNA- and transcription factor-mediated recruitment of histone chaperones to specific chromosome loci.
Chromatin, the constituent substance of all eukaryotic chromosomes, is a highly compacted structure consisting mainly of genomic DNA in association with the four histone proteins, H2A, H2B, H3, and H4. Due to chromatin's occluded nature, significant chromatin remodeling is required to allow transcription factors to gain access to their DNA cognate sites. Thus, chromatin remodeling is capable of acting as a specific controlling mechanism, playing a role in a number of important biological events, including cell development, stress response, and cell cycle progression. Several studies have established that transcription factors bound to specific DNA sequences are capable of attracting histone-modifying and ATP-dependent chromatin-remodeling enzymes, which in turn act to promote chromatin's adoption of an open conformation (reviewed in reference 13). In addition to the action of these recruited enzymes, histone chaperones have been suggested to play a role in forming open chromatin structures via their direct association with histones. During transcription elongation phases, histones in nucleosome are dynamically evicted from, and then deposited back onto, DNA in concert with the progression of RNA polymerases. Several lines of evidence suggest that histone chaperones are involved in the regulation of histone density and thereby gene expression (29, 38). We also demonstrated that depletion of the histone chaperone template activating factor I (TAF-I) (16) changed the genetic expression profile of HeLa cells (10). Recent studies have suggested that a part of histone chaperones are recruited to the specific genes by interacting with certain DNA binding proteins (5, 7, 39). However, the molecular mechanism behind how histone chaperones achieve specific binding to a particular genomic region is not well understood.
rRNA synthesis is closely tied to cell growth and hence should, in theory, be tightly regulated in response to metabolic and environmental changes. The loading rate of RNA polymerase I (Pol I) onto the rRNA gene is a key regulatory step in controlling rRNA transcription levels (6). This step has been suggested to be regulated by the transcription factor upstream binding factor (UBF) that acts to recruit the Pol I complex (12). More recently, it was demonstrated that UBF plays roles in promoter escape (23) and transcription elongation (31) rather than preinitiation complex assembly. Recent research has also demonstrated that UBF associates with the entirety of the rRNA genes, including the intergenic region between rRNA coding regions (22). Therefore, it is likely that UBF plays a crucial role in defining rRNA gene loci. Another major factor determining rRNA expression levels is suggested to be the balance between active and inactive rRNA gene numbers. It was shown that only half of the rRNA genes are actively transcribed in exponentially growing cells (4). Epigenetic mechanisms are suggested to play a key role in regulating this active/inactive rRNA gene balance. The NoRC complex (33) has a reported involvement with rRNA transcription regulation. NoRC binds to the promoter region of rRNA genes by interacting with transcription termination factor I (TTF-I) and recruits the Sin3 corepressor complex (27, 42). It has also been reported that the SIRT1-Suv39h1-nucleomethylin complex mediates heterochromatin formation around rRNA genes in a manner sensitive to changing NAD+/NADH levels (15). These chromatin modification enzymes create and maintain an inactive chromatin structure around rRNA genes.
Histone chaperones, nucleolin and the FACT complex, also have the important role in rRNA chromatin (r-chromatin) regulation. Nucleolin was reported to play a role in enhancing chromatin remodeling by SWI/SNF and ATP-dependent chromatin assembly and remodeling factor (ACF) (1) and facilitate transcription by Pol I (25) in vitro. Nucleolin and the FACT complex have been shown to bind to r-chromatin, and their knockdown reduces the pre-rRNA transcription level (3, 25). We previously identified a nucleolar protein, B23, as a component of template activating factor III, the factor responsible for mediating the structural changes of adenovirus chromatin to stimulate DNA replication in vitro (18). B23 exists as two isoforms, B23.1 and B23.2. The C-terminal RNA binding domain of B23.1 is lacking in B23.2. Following our initial discovery, B23 was subsequently shown to function as a histone chaperone in vitro (20). Recently, we have reported that B23 binds to r-chromatin and regulates the histone density around rRNA genes (14). However, the mechanism by which B23 is recruited to and subsequently associated with r-chromatin remains unclear. Here, we clarify the molecular mechanism of how histone chaperone B23 is targeted to r-chromatin. Our findings suggest a novel mechanism for histone chaperone targeting to the specific chromosome loci.
MATERIALS AND METHODS
Cell culture, transfection, and cell cycle synchronization.
HeLa cells were maintained in modified Eagle medium (MEM) (Nissui) supplemented with 10% fetal bovine serum (FBS). 293T cells were maintained in Dulbecco's modified Eagle medium (DMEM) (Nissui) supplemented with 10% FBS.
Transient transfection of plasmid DNA was performed using GeneJuice (Novagen) according to the manufacturer's instructions. To establish stable cell lines, HeLa cells were transfected with pEGFP-Flag-B23.1, pEGFP-Flag-T4sA, and pEGFP-Flag-T4sD (see below). Neomycin-resistant cells were selected by G418.
Mitotic cells were collected following synchronization by two cycles of thymidine blockage following nocodazole arrest. Briefly, cells were treated with 3 mM thymidine (Sigma) in the culture medium for 16 h, released into a fresh culture medium without thymidine for 10 h, and then finally subjected to a second block with 3 mM thymidine for 14 h. After the double block, cells were released into a fresh culture medium for 8 h, after which 50 ng/ml nocodazole (Sigma) was added and cells were incubated for 4 to 10 h. Mitotic cells were collected by gentle shaking of the incubation dishes. For the experiment shown in Fig. 6, mitotic cells were released into culture medium in the absence or presence of 50 ng/ml of actinomycin D (Act D) (Sigma). To inhibit the RNA polymerase II activity, cells were incubated in the culture medium containing 5 μg/ml of α-amanitin (Sigma) for 24 h. To examine the effect of α-amanitin, U1 snRNA was amplified by reverse transcription-PCR (RT-PCR) using primer sets 5′- CGGGGTACCATACTTACCTGGCAGGGGAGA-3′ and 5′-CGGGGTACCGAATTCAGGGGAAAGCGCGAACG-3′.
siRNA transfection.
Small interfering RNA (siRNA) was transfected using Lipofectamine RNAi MAX (Invitrogen). Stealth RNAs for negative controls (Stealth RNAi negative-control duplex, catalog no. 12935-300, Invitrogen), for B23.1 (NPM1-HSS143154), and for UBF (UBTF-HSS111143, Invitrogen) were utilized.
Antibodies.
The following antibodies were used in this study: mouse monoclonal antibodies for B23.1 (Invitrogen), Flag tag (M2, Sigma), UBF (F9, Santa Cruz Biotechnology), nucleolin (MS-2, Santa Cruz Biotechnology), and cyclin B (GNS1, Santa Cruz Biotechnology) and rabbit polyclonal antibodies for histone H3 (Abcam) and phosphorylated histone H3 (S10, Millipore). Anti-H2A-H2B antibody was generated in rabbits by injecting recombinant His-tagged H2A-H2B (19). Specific antibodies against histones H2A and H2B were purified from rabbit serum by an H2A-H2B immobilized HiTrap NHS-activated HP column (GE Healthcare).
Immunoprecipitation.
Cells expressing Flag-tagged B23.1 or its mutants were lysed and sonicated in buffer A (50 mM Tris-HCl [pH 7.9], 0.1% Triton X-100) containing 100 mM NaCl. Anti-Flag-tag M2 affinity gel (Sigma) was added to the lysate, and the mixture was incubated at 4°C for 1 h. The gels were washed extensively with buffer A containing 200 mM NaCl, and the proteins bound to the gels were eluted with the buffer containing 0.1 mg/ml of FLAG peptide (Sigma). Eluted proteins were separate on a 13% SDS-polyacrylamide gel electrophoresis (PAGE) gel and analyzed by silver staining and Western blotting. For experiments involving immunoprecipitation for the subsequent detection of RNA, cells were incubated in a buffer (10 mM HEPES-NaOH [pH 7.9], 1.5 mM MgCl2, and 10 mM KCl) containing 0.1% Triton X-100, and the NaCl concentration was adjusted to 0.42 M. The extracts were recovered after extensive centrifugation (15 krpm for 20 min), and the NaCl concentration was diluted to 0.2 M. Immunoprecipitation experiments were carried out as described above. The RNAs bound to precipitated proteins were purified with phenol-chloroform extraction and ethanol precipitation. RNAs were separated on a 6% denaturing PAGE gel in 1× Tris-borate-EDTA (TBE) and visualized with GelRed (Biotium) staining.
Northern blotting.
RNAs were prepared by immunoprecipitation using anti-Flag antibody from nuclear extract, separated on a 6% denaturing PAGE gel, and transferred to nitrocellulose membranes. Northern blotting assays were carried out using Alkphos direct labeling reagents (GE Healthcare). The templates of probes were amplified by PCR using gene-specific primer sets.
ChIP assays.
Chromatin extracts were prepared from cells fixed with formaldehyde by extensive sonication. In all chromatin immunoprecipitation (ChIP) experiments, the lengths of DNA extracted from cell lysate were between 200 and 500 bp. ChIP assays were carried out according to the manual for the ChIP assay kit (Millipore). Cell lysates prepared from 2 × 106 cells were used for an immunoprecipitation assay, with 1% of the input cell lysate utilized as the input DNA sample. Precipitated DNAs were suspended in 50 μl of water and used as templates for PCR. The primer sets used to amplify A and B regions on the human rRNA gene were as follows: for region A, 5′-TGTGAATTGGATGGTGGCGTTTTTGGGGA-3′ and 5′-CAGGCGGCTCGAGCAGGAGC-3′; for region B, 5′-CGACTCTTAGCGGTGGATCACTC-3′ and 5′-AAGCGACGCTCAGACAGGCGT-3′. PCR products were separated using a 6% PAGE gel and were visualized by GelRed staining. Quantitative PCR (Q-PCR) was carried out using FastStart Universal Master (ROX) (Roche) and primer sets corresponding to the rRNA gene. For Fig. 7, HeLa cells were fixed and subjected to ChIP assays at 72 h after siRNA transfection. For Q-PCRs, the previously described primer sets (14) were used. For re-ChIP assays, precipitated protein-DNA complexes in the 1st ChIP assay were eluted in 20 μl of ChIP elution buffer (1% SDS, 10 mM Tris-HCl [pH 7.9], and 1 mM EDTA) containing 10 mM dithiothreitol (DTT), diluted with dilution buffer (0.1% SDS, 1.1% Triton X-100, 16.7 mM Tris-HCl [pH 7.9], 1.2 mM EDTA, and 167 mM NaCl), and used for 2nd ChIP assays.
Indirect immunofluorescence.
For immunofluorescence analyses, all procedures were carried out at room temperature. HeLa cells grown on coverslips were fixed with 1% or 3% paraformaldehyde for 10 min. Cells were then permeabilized for 5 to 10 min in a buffer (300 mM sucrose, 3 mM MgCl2 in phosphate-buffered saline [PBS]) containing 0.5% Triton X-100 and incubated in PBS containing 0.5% milk and 0.1% Triton X-100 for 30 min. The fixed and permeabilized cells were incubated with primary antibody for 1 h. The cells on coverslips were washed with PBS containing 0.1% Triton X-100 (PBST) and incubated with secondary antibodies (anti-mouse IgG-Alexa555; Invitrogen) for 30 min. Cells were washed extensively with PBST, and the DNA was stained with TO-PRO-3 (Invitrogen) for 5 min. All fluorescence images were captured by a confocal microscope (LSM 5 Exciter, Carl Zeiss) with a Plan-Apochromat 63× 1.4-numerical-aperture (NA) oil immersion objective lens. Images were cropped, sized, and arranged into panels using Adobe Photoshop CS3 (Adobe Systems).
Cell fractionation.
Cells washed with PBS were incubated in a hypotonic buffer (10 mM HEPES-NaOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) on ice for 10 min and centrifuged at 2,000 rpm for 3 min, and the supernatant was removed. Cell pellets were suspended in hypotonic buffer containing 0.1% (vol/vol) Triton X-100 and incubated on ice for 2 min, following which the soluble proteins were recovered. The cell pellets were suspended in an SDS sample buffer as insoluble fractions. Equal amounts of total, soluble, and insoluble fractions were separated by SDS-PAGE and analyzed by Western blotting.
Plasmid construction.
Plasmids pET-14b-B23.1, -B23.2, and -T4sA have been described previously (18, 21). To construct pET-14b-T199D, the cDNA fragments containing a mutation at T199 were generated using a two-step PCR. In the first PCR, two DNA fragments containing the mutation were amplified using T7 promoter primer and 5′-ATCTATACGAGATGATCCAGCCAAAAATG-3′ and the T3 promoter primer and 5′-CATTTTTGGCTGGATCATCTCGTATAGAT-3′, with pBS-Flag-B23.1 as a template. These DNA fragments were then separated and purified from an agarose gel and used in the second PCR as templates. The second PCR was performed with T7 and T3 promoter primers. Full-length mutated cDNA was digested with BamHI and cloned into a BamHI-digested pBS-Flag vector. pBS-Flag-T199D was digested with NdeI and BamHI and then subcloned into NdeI- and BamHI-digested pET-14b. T219D and T234/237D expression vectors were constructed with the same procedure. Primer sets for T219D and T234/237D were 5′-AAAACCATCATCAGATCCAAGATCAAAAG-3′ and 5′-CTTTTGATCTTGGATCTGATGATGGTTTT-3′ and 5′-CAGGAAAAAGATCCTAAAGATCCAAAAGGA-3′ and 5′-TCCTTTTGGATCTTTAGGATCTTTTTCCTG-3′, respectively. T219/234/237D and T4sD expression vectors were prepared by the same method using the appropriate primers (described above) and plasmid vectors as templates. To construct pET-14b-T95D, the primer set 5′-AAATAGATCCACCAGTGGTCTTAAGG-3′ and 5′-GGTGGATCTATTTCAAAGCCCCCAAG-3′ was used along with pET-14b-B23.1 as a template. To construct S125D, S125A, S70D, and S254D expression vectors, primer sets 5′-CAGAGGATGAAGATGAAGAGGAGGAG-3′ and 5′-TCTTCATCCTCTGCATCTTCCTCCAC-3′, 5′-CAGAGGCAGAAGATGAAGAGGAGGAG-3′ and 5′-TCTTCTGCCTCTGCATCTTCCTCCAC-3′, 5′-AAGGCGATCCAATTAAAGTAACA-3′ and 5′-ATTGGATCGCCTTCGTAATTCAT-3′, and 5′-AAGCAGATATAGAAAAAGGTGGT-3′ and 5′-TCTATATCTGCTTGCATTTTTGC-3′ were used, respectively. The plasmids for expressing S70/125D and S70/125/254D (S3sD) were constructed using the identical protocol with the appropriate primer sets (described above) and plasmid DNA templates. For expression of EGFP-Flag and Flag-tagged proteins in HeLa cells, each cDNA attached by Flag-tag at its 5′ terminus was cut out from a pBS-Flag vector by digestion with BamHI and then subcloned into a BamHI-digested pEGFP-C1 vector (CLONTECH) or a BglII-digested pCAGGS vector. The sequences of all plasmids were confirmed using an ABI Prism BigDye Terminator version 3.1 cycle sequencing kit (PE Applied Biosystems) with the appropriate primers.
Purification of recombinant proteins and biochemical analyses.
For expression and purification of recombinant proteins, BL21(DE3) was transformed with pET-14b plasmids containing B23.1 mutant cDNAs. B23 proteins were expressed and purified as previously described (18). For histone transfer assays, core histones were preincubated with His-tagged proteins in a buffer (20 mM HEPES-NaOH [pH 7.9], 10% glycerol, 0.4 mg/ml bovine serum albumin [BSA], and 50 mM NaCl) at room temperature for 10 min, and then 20 ng of 147-bp-long DNA fragment containing the 5S rRNA gene was added. The mixture was incubated at 37°C for 1 h and analyzed by 6% PAGE in 0.5× TBE buffer. The gel was run at 14.3 V/cm for 90 min, and DNA was visualized with GelRed staining. Filter binding assays and supercoiling assays were carried out as described previously (19, 20).
Examination of rRNA transcription levels in siRNA-treated cells.
rRNA transcription levels were examined as described previously (14). At 24 h after siRNA transfection, plasmid DNA for expression of Flag-tagged B23.1 proteins was transfected using GeneJuice (Novagen). Cells were collected 72 h after siRNA transfection, and RNA was purified from transfected cells with an RNeasy kit (Qiagen). Reverse transcription was carried out with ReverTraAce (Toyobo). The synthesized cDNA was used as a template for quantitative PCR using FastStart Universal Master (ROX) (Roche). Q-PCRs were carried out using the Applied Biosystems 7500 Fast Real-Time PCR system. Primer sequences used for RT-PCR were described previously (14).
RESULTS
B23.1 is released from chromatin during mitosis.
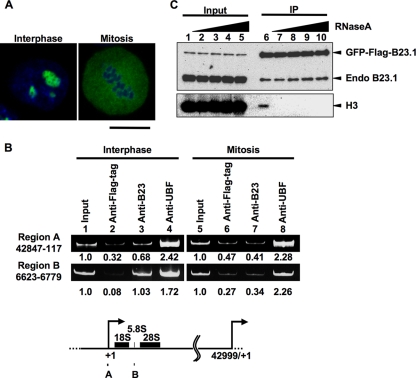
We previously demonstrated that B23 is involved in the regulation of rRNA transcription through its histone chaperone activity (14); however, how B23 is targeted to r-chromatin was unknown. As shown in Fig. 1 A, B23.1 is localized at the nucleoli in interphase cells, whereas it is randomly located during mitosis. We first examined whether B23.1 is associated with r-chromatin during mitosis. Asynchronous and mitotic HeLa cells were fixed, and ChIP assays were performed (Fig. 1B). In agreement with previous findings (14), we found that in asynchronous cells, B23.1 and UBF were associated with both the promoter (region A) and coding (region B) regions of the rRNA gene. In mitotic cell extracts, rRNA gene DNA fragments precipitated with anti-UBF were clearly observable (lane 8), whereas those precipitated with anti-B23 were not (lane 7), indicating that B23 was released from r-chromatin during mitosis. Considering that B23 is phosphorylated during mitosis by the cdc2-cyclin B complex and that this phosphorylation inactivates the RNA binding activity (21), we tested whether the RNA binding activity of B23.1 was related to its chromatin binding activity. Extracts treated without or with increasing concentrations of RNase A were prepared from HeLa cells stably expressing green fluorescent protein (GFP)-Flag-B23.1 and subjected to immunoprecipitation with anti-Flag-tag antibody (Fig. 1C). GFP-Flag-B23.1 coimmunoprecipitated with chromosomal histone H3 (Fig. 1C, lane 6); however, the level of coprecipitated histone H3 was significantly diminished when the extracts were treated with low concentrations of RNase A (lanes 7 to 10). These results suggest that the RNA binding activity of B23.1 is important for its association with chromatin.
FIG. 1.
B23.1 is released from chromatin during mitosis. (A) Localization of GFP-Flag-B23.1. HeLa cells stably expressing GFP-Flag-B23.1 grown on coverslips were fixed with 3% paraformaldehyde, and the localization of the protein was observed under a confocal microscope. DNA was counterstained with TO-PRO-3. Bar, 10 μm. (B) ChIP assays of endogenous B23. Asynchronous and mitotic HeLa cells were subjected to ChIP assays using anti-Flag-tag as a control, anti-B23, and anti-UBF antibodies. The input (lanes 1 and 5) and coimmunoprecipitated (lanes 2 to 4 and 6 to 8) DNA were purified, and the DNA fragment harboring the rRNA gene promoter (region A; nucleotide positions between 42847 (−153) and 117 relative to the transcription start site [+1]) and coding (region B; nucleotide positions 6623 to 6779) regions were amplified by PCR. PCR products were analyzed by 6% PAGE and visualized with GelRed staining. The amounts of PCR products amplified from input DNA were set as 1.0, and the enrichment is shown at the bottom of the panel. The positions of primers on the rRNA gene are schematically represented at the bottom. (C) Immunoprecipitation of GFP-Flag-B23.1 with RNase-treated cell extracts. HeLa cells expressing GFP-Flag-B23.1 were treated without (lanes 1 and 6) or with 1, 10, 100, or 1,000 μg/ml of RNase A (lanes 2 to 5 and 7 to 10). GFP-Flag-B23.1 was immunoprecipitated with anti-Flag-tag antibody and separated by SDS-PAGE followed by Western blotting with anti-B23.1 and anti-histone H3 antibodies.
The RNA binding activity of B23.1 is required for its association with r-chromatin.
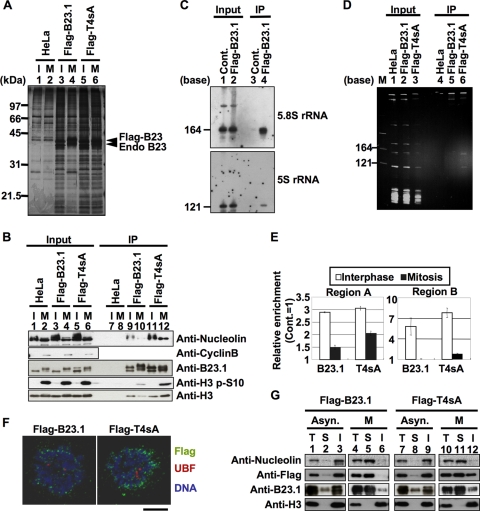
To test the above notion, we next examined whether the B23.1 mutant T4sA associated with RNA and chromatin in mitotic cells. T4sA is incapable of being phosphorylated due to the fact that its four cdc2 target sites, T199, T219, T234, and T237 (21), are replaced with alanine. We previously demonstrated that T4sA was associated with RNA regardless of whether it was pretreated with cdc2-cyclin B kinase in vitro (21). If the RNA binding activity of B23.1 is required for its association with chromatin, we supposed that T4sA would remain associated with chromatin during mitosis. Cell extracts derived from asynchronous or mitotic cells stably expressing either Flag-B23.1 or -T4sA were subjected to immunoprecipitation (Fig. 2 A and B). From asynchronous cell extracts, it can be seen that B23.1 and T4sA coprecipitated with a variety of proteins, including nucleolin and histone H3 (Fig. 2A, lanes 3 and 5, and B, lanes 9 and 11). Due to the fact that the mobility of endogenous and Flag-tagged B23.1 in mitotic extracts was significantly retarded due to hyperphosphorylation and the amount of histone H3 phosphorylated at serine 10 was increased (Fig. 2B, lanes 2, 4, and 6), we took care to make sure that extracts were taken from cells properly synchronized at prometaphase. The mobility shift of mitotic Flag-T4sA was minimal (Fig. 2B, compare lanes 5 and 6 and 11 and 12), indicating that this mutant was not efficiently phosphorylated during mitosis. Interestingly, we found that nucleolin and histone H3 proteins efficiently coprecipitated with Flag-T4sA from mitotic extracts (lane 12). In parallel, we analyzed RNAs coprecipitated with Flag-tagged proteins from mitotic extracts (Fig. 2C and D). Flag-B23.1 coprecipitated with 5.8S and 5S rRNAs from asynchronous cell extracts (Fig. 2C) as previously reported (2, 41). However, Flag-B23.1 did not efficiently precipitate the RNAs from mitotic extracts (Fig. 2D, lane 5). In contrast, two small rRNAs were coprecipitated with Flag-T4sA even from mitotic extracts (lane 6). These results strongly support the idea that the RNA binding activity of B23.1 is regulated by cdc2-mediated phosphorylation in vivo.
FIG. 2.
Mitotic phosphorylation of B23.1 is required for the inactivation of its chromatin and RNA binding activities. (A and B) Immunoprecipitation of Flag-tagged proteins. Cell extracts were prepared from asynchronous (I) or mitotic (M) HeLa cells and HeLa cells expressing either Flag-B23.1 or -T4sA, and immunoprecipitation using anti-Flag-tag antibody was carried out. Immunoprecipitated proteins were separated by SDS-PAGE and detected by silver staining (A) and Western blotting (B). Input and immunoprecipitated proteins (A, lanes 1 to 6 and 7 to 12, respectively) were analyzed by Western blotting using anti-B23, antinucleolin, anti-histone H3 S10P (histone H3 phosphorylated at serine 10), and anti-histone H3 antibodies. Western blotting with anti-cyclin B antibody was used as a mitotic marker (B, second panel). (C) RNAs coprecipitated with Flag-B23.1. Nuclear extracts were prepared from control and Flag-B23.1-expressing HeLa cells, and immunoprecipitation with anti-Flag-tag antibody was carried out. The input and coimmunoprecipitated RNA was purified, separated on a 6% denaturing PAGE gel, and analyzed by Northern blotting with 5S and 5.8S rRNA gene probes. (D) RNA binding activity of B23.1 and T4sA during mitosis. Cell extracts were prepared from mitotic HeLa cells expressing either Flag-B23.1 or -T4sA, as described in Materials and Methods, and immunoprecipitation with anti-Flag-tag antibody was carried out. Coimmunoprecipitated RNAs were purified, separated on a 6% denaturing PAGE gel, and detected with GelRed staining. Lane M indicates RNA molecular markers. (E) Cell cycle-dependent association of B23.1 and T4sA with the rRNA gene. Asynchronous and mitotic HeLa cells expressing either Flag-B23.1 or -T4sA were subjected to ChIP assays using control IgG and anti-Flag-tag antibody. Coimmunoprecipitated and input DNA were used as templates for Q-PCR using the primer sets for both regions A and B. The amount of amplified DNA precipitated with control IgG was set as 1.0, and the relative enrichment level of the DNA fragments by anti-Flag-tag immunoprecipitation are shown. White and black bars in the graph show the results of ChIP assays with asynchronous and mitotic cells, respectively. Q-PCRs were carried out in triplicate. (F) Localization of B23.1 and T4sA in mitotic cells. HeLa cells expressing either Flag-B23.1 or -T4sA grown on coverslips (left and right panels, respectively) were fixed in 1% paraformaldehyde, and Flag-tagged proteins (green) and UBF (red) were detected by specific antibodies. DNA was counterstained with TO-PRO-3 (blue). Localization of proteins was observed under a confocal microscope. Bar, 10 μm. (G) Extractability of B23.1 and T4sA in asynchronous and mitotic cells. Asynchronous or mitotic (lanes 1 to 3 and 7 to 9 or 4 to 6 and 10 to 12, respectively) HeLa cells expressing either Flag-B23.1 or -T4sA were fractionated to soluble and insoluble fractions. Total cell extracts and soluble and insoluble fractions (indicated by T, S, and I, respectively) were separated by SDS-PAGE and analyzed by Western blotting using antibodies against nucleolin, Flag tag, B23.1, and histone H3.
We next examined whether Flag-T4sA remains associated with r-chromatin during mitosis by using ChIP assays (Fig. 2E). In asynchronous cells, Flag-B23.1 and Flag-T4sA bound to r-chromatin (white bars). During mitosis, the affinity of both Flag-B23.1 and Flag-T4sA with r-chromatin was markedly decreased as endogenous B23 did so (Fig. 1B), although the association level of T4sA was slightly higher than that of the wild-type B23.1.
The level of association between T4sA and r-chromatin detected in mitotic cells was decreased in comparison to that measured in asynchronous cells (Fig. 2E), whereas the degree of association between T4sA and chromosomal histone H3 in asynchronous and mitotic cells was not significantly altered (Fig. 2B, lanes 11 and 12). To explore this apparent inconsistency, we examined the localization of Flag-T4sA in mitotic cells (Fig. 2F). Under the assay conditions employed, soluble proteins were extracted, and proteins associating with the nuclear structure were retained. As previously reported (28) in prometaphase cells, Flag-B23.1 was detected around the chromosome periphery (Fig. 2F). In comparison with wild-type B23.1, T4sA was found to be more concentrated around the chromosome periphery. To address this point more quantitatively, cell fractionation experiments were carried out (Fig. 2G). Cells expressing either Flag-B23.1 or -T4sA were fractionated into soluble and insoluble fractions, and the proteins existing in each fraction were analyzed by Western blotting. Endogenous B23 and nucleolin appeared mainly in the insoluble fraction in asynchronous cells (Fig. 2G, lanes 3 and 9). In mitotic cells, however, these proteins were mainly in the soluble fraction (lanes 5 and 11). Throughout the cell cycle, histone H3 was observed only in the insoluble fraction. The fractionation pattern of Flag-B23.1 was similar to that of endogenous B23 (lanes 1 to 6), whereas Flag-T4sA was present in both the soluble and insoluble fractions in mitotic cells (lanes 11 and 12). The r-chromatin termed NOR in mitotic cells was visualized by UBF staining; however, the colocalization of NOR with Flag-B23.1 or -T4sA was not clear (Fig. 2F). Thus, it is quite likely that the chromosomal histone H3, which coprecipitates with Flag-T4sA from mitotic extracts, is itself derived from the entire chromosome. These results suggest that the RNA binding activity of B23.1 is a necessary condition for its association with chromatin but is, by itself, not sufficient for maintaining an association with r-chromatin.
A B23.1 mutant mimicking mitotic phosphorylation does not efficiently associate with RNA and chromatin in asynchronous cells.
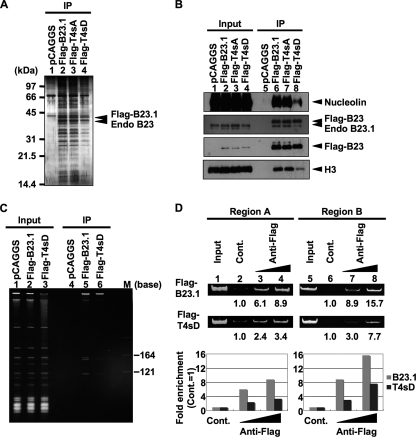
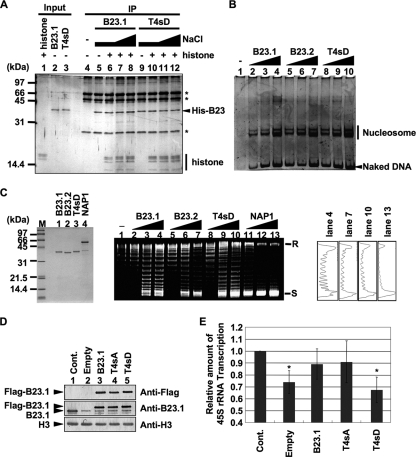
We next examined the chromatin binding activity of a phosphomimetic mutant, termed T4sD, in which four cdc2 target sites were replaced with aspartic acids in asynchronous cells (Fig. 3). Flag-tagged B23.1, T4sA, and T4sD were transiently expressed in 293T cells and immunoprecipitation was carried out (Fig. 3A to C). We found that Flag-B23.1 and Flag-T4sA bound to a variety of proteins, including nucleolin and histone H3 (Fig. 3A and B) and Flag-B23.1 also bound to 5.8S and 5S rRNAs (Fig. 3C). Conversely, chromosomal histone H3 and nucleolin did not efficiently coprecipitate with T4sD (Fig. 3B, lane 8). Additionally, the amount of 5S and 5.8S rRNAs coprecipitated with Flag-T4sD was less than half of that coprecipitated with wild-type B23.1 (Fig. 3C, lane 6). We noted that a low but distinct level of RNA was coprecipitated with T4sD from cell extracts, although T4sD does not associate with RNAs in vitro (Fig. 4). This may be due to the fact that T4sD forms an oligomer with endogenous B23 (Fig. 3B). Indeed, B23.1 and B23.2 were equally precipitated with Flag-tagged B23.1, T4sA, and T4sD (data not shown). To investigate the association of T4sD with r-chromatin, ChIP assays were performed (Fig. 3D). The results clearly demonstrated that the association level of T4sD with r-chromatin was lower than that with B23.1. Taken in total, we concluded from these data that the RNA binding activity of B23.1 is crucial for its chromatin binding activity.
FIG. 3.
The phosphomimetic B23.1 mutant does not efficiently associate with chromatin. (A and B) Immunoprecipitation of Flag-tagged B23.1 proteins. 293T cells were transfected with empty vector or vectors for expression of Flag-B23.1, -T4sA, and -T4sD (lanes 1 to 4, respectively), and immunoprecipitation was carried out with anti-Flag-tag antibody. Precipitated proteins were separated by SDS-PAGE and detected with silver staining (A) or Western blotting (B). Western blotting was carried out with anti-Flag-tag, -nucleolin, -B23, and -histone H3 antibodies. (C) RNA binding activity of T4sD in 293T cells. Cell extracts were prepared as described in Materials and Methods. RNAs coimmunoprecipitated with Flag-tagged proteins were separated on a 6% denaturing PAGE gel and visualized with staining by GelRed. Lane M indicates RNA molecular markers. (D) r-chromatin association of B23.1 and T4sD. Flag-B23.1 or -T4sD was transiently expressed in 293T cells, and ChIP assays using anti-Flag-tag antibody (0.5 and 1.0 μg) were carried out. Anti-Myc-tag antibody (1 μg) was used as a control. Precipitated and input DNA were used as templates for PCR with specific primer sets (regions A and B). PCR product from control immunoprecipitation was set as 1.0, and the relative enrichment levels of DNA fragments by anti-Flag-tag immunoprecipitation are indicated at the bottom and graphically represented. Gray and black bars in the graphs show the results of ChIP assays with Flag-B23.1 and -T4sD, respectively.
FIG. 4.
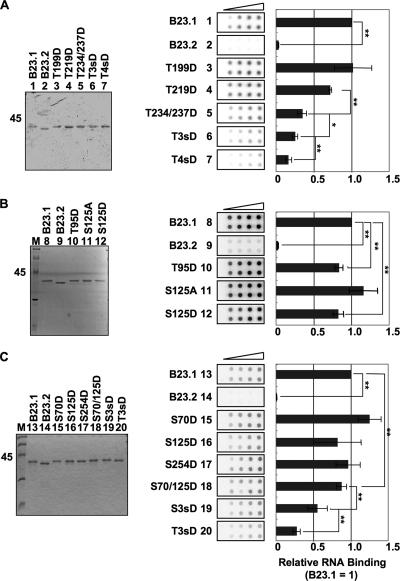
Phosphorylation of four cdc2 consensus sites of B23 is crucial for efficient suppression of its RNA binding activity. (A) RNA binding activity of B23 proteins containing mutations at cdc2 consensus sites. B23.1, B23.2, T199D, T219D, T234/237D, T3sD, and T4sD proteins (lanes 1 to 7, respectively) (200 ng) were separated by SDS-PAGE and visualized by Coomassie brilliant blue (CBB) staining (left). T3sD and T4sD are B23.1 mutants in which T219, T234, and T237 and all four threonines (T), respectively, are replaced with aspartic acids (D). Purified proteins (50, 100, 200, and 400 ng) were mixed with 32P-labeled HeLa cell total RNA (10 ng). The mixture was incubated and then filtered though nitrocellulose membranes. The membranes were extensively washed, and RNA retained on the membrane was detected with an image analyzer (middle). The intensity of each spot was analyzed, and the RNA binding activity obtained with the same amount of B23 proteins was first calculated relative to that of B23.1 (1.0). Then the relative RNA binding activity at each protein amount was averaged. Means ± standard deviations (SD) obtained from two duplicate independent experiments are shown (center and right). (B) The effects of T95 and S125 phosphorylation on the RNA binding activity of B23.1. Purified proteins B23.1, B23.2, T95D, S125A, and S125D (200 ng) were separated by SDS-PAGE (lanes 8 to 12, respectively) and visualized with CBB staining. The RNA binding activity of B23 mutant proteins was examined by filter binding assays as described for panel A. (C) The effects of the position and number of phosphorylation sites on the RNA binding activity of B23.1. Purified proteins B23.1, B23.2, S70D, S125D, S254D, S70/125D, S3sD (S70/125/254D), and T3sD (T219/234/237D) (200 ng) were separated by SDS-PAGE (lanes 13 to 20, respectively) and visualized with CBB staining. The RNA binding activity of each protein was examined as described for panel A. For all experiments, statistical P values were calculated by t tests and indicated with * for P < 0.05 and ** for P < 0.01.
Cdc2-mediated phosphorylation at four consensus sites of B23 inactivates its RNA binding activity.
We next tried to identify critical phosphorylation site(s) affecting the RNA binding activity of B23.1. To this end, we constructed a series of phosphomimetic mutants of B23.1. Since four threonine residues were known to be phosphorylated by cdc2 kinase in mitosis, one to four threonine residues were substituted by aspartic acid (designated T199D, T219D, T234/237D, T3sD [T219/234/237D], and T4sD). The RNA binding activities of these mutant proteins were examined by filter binding assay (Fig. 4A). RNAs alone or RNAs mixed with B23.2, lacking the RNA binding domain of B23.1, were not retained on the membrane, whereas RNAs mixed with B23.1 were retained on the membrane in a B23.1 dose-dependent manner. The RNA binding activity of T199D was similar to that of wild-type B23.1, whereas that of T219D was slightly lower. The RNA binding activity of B23.1 was gradually reduced in response to an increasing number of phosphomimetic mutations. When all four threonine residues were replaced with aspartic acids, the RNA binding activity was dramatically reduced. Since B23 is known to be phosphorylated at several different sites, we also examined the effect of other phosphorylation sites on the RNA binding activity of B23.1. One putative site, threonine 95 (T95), within the nuclear export signal sequence has been suggested as a likely phosphorylation site (40). Another potential phosphorylation regulatory site, serine 125 (S125), has been previously shown to be phosphorylated by CK2 kinase (37). Filter binding assays demonstrated that the RNA binding activities of T95D and S125D were slightly decreased (Fig. 4B). Since the number of phosphorylation sites was likely to be important for affecting the RNA binding activity of B23.1, we constructed mutant proteins having two or three phosphomimetic mutations. Because B23 at the mitotic spindle pole was found to be phosphorylated at three serine residues (S70, S125, S254) (17), we constructed phosphomimetic mutants S70D, S254D, S70/125D, and S3sD (S70/125/254D) and compared their RNA binding activities with that of T3sD containing three mutations at the cdc2 consensus sites (Fig. 4C). The RNA binding activity of the serine mutants was gradually decreased upon an increasing number of mutations, as was seen in the case of cdc2 consensus sites (Fig. 4A and C). Importantly, S3sD showed about a 2-fold higher activity than T3sD (P = 0.0097, t test). From these data, we concluded that the inactivation of the RNA binding activity depends on the number and position of phosphorylation sites.
Chromatin and RNA binding activities of B23.1 are required for its maximal stimulatory activity of rRNA transcription.
So far, we have demonstrated that the RNA binding activity of B23.1 was required for its association with chromatin. To examine the biological significance of this finding, we next examined whether the chromatin binding activity of B23.1 was necessary for its stimulatory effect upon rRNA transcription. To address this point, we first examined the histone binding activity of wild-type and T4sD mutant B23.1 by immunoprecipitation (Fig. 5 A). Both wild-type and T4sD B23.1 proteins were bound to histones similarly even in the increasing concentrations of NaCl (Fig. 5A, lanes 6 to 8 and 10 to 12, respectively). We next examined the histone chaperone function of the T4sD mutant via histone transfer assay and supercoiling assay (Fig. 5B and C). His-tagged B23.1, B23.2, and T4sD similarly transfer histones to a 147-bp-long DNA fragment, and the assembled nucleosome core particles were observed (Fig. 5B). In supercoiling assays, when nucleosome is assembled on relaxed circular DNA, supercoil is introduced in the plasmid DNA. NAP1, a well-characterized histone chaperone, efficiently assembled nucleosome, and supercoil was introduced into the plasmid DNA (Fig. 5C, lanes 11 to 13). Nucleosome was assembled on the plasmid DNA with increasing amounts of His-B23.1 and His-B23.2 (lanes 1 to 7) in agreement with previous studies (20), although the activity of B23 proteins was lower than that of NAP1. T4sD also showed a similar level of nucleosome assembly activity (lanes 8 to 10; see also the densitometric analysis). These results indicated that phosphorylation at the four cdc2 consensus sites did not affect the histone chaperone activity of B23.1. We next examined the histone chaperone activity of T4sD in vivo. HeLa cells were treated with control or B23.1 siRNA, and then Flag-tagged B23.1 and its mutants were produced by transient transfection. Under these conditions, the expression level of B23.1 in cells treated with B23.1 siRNA was decreased by approximately one-third of that in control cells (compare lanes 1 and 2 in Fig. 5D). Total RNA was extracted, and the amount of 45S pre-rRNA was quantified by Q-PCR using the primer set specific for the 5′ external transcribed sequence (5′-ETS) (Fig. 5E). Since the 5′-ETS region of pre-rRNA is quickly processed in vivo, the amount of 45S pre-rRNA should be taken as reflecting the ongoing transcription activity of rRNA. When endogenous B23.1 was depleted, the amount of 45S pre-rRNA was decreased to 70% of that in the control cells. We also found that the amounts of UBF and the p194 subunit of RNA polymerase I were not significantly decreased by B23 depletion (data not shown). Therefore, it is likely that transcription inhibition by B23 depletion is due to reduced RNA polymerase processivity, because we have demonstrated that B23 depletion increases histone density along the r-chromatin (Fig. 7) (14). However, the pre-rRNA transcription level was recovered by the expression of Flag-B23.1 and -T4sA (Fig. 5E). In contrast, the expression of T4sD did not rescue the rRNA transcription, although T4sD showed the potential histone chaperone activity in vitro (Fig. 5A to C). Therefore, we concluded that the RNA binding activity of B23.1 is required for its correct targeting to r-chromatin and that association of B23.1 with r-chromatin plays a crucial role in achieving maximal stimulatory activity of rRNA transcription in vivo.
FIG. 5.
RNA binding activity of B23.1 is required to facilitate rRNA transcription. (A) Histone binding activity of recombinant proteins. Recombinant B23.1 or T4sD (1 μg) was preincubated without (lanes 5 and 9) or with core histones (300 ng, lanes 6 to 8 and 10 to 12), and immunoprecipitation was carried out with anti-B23.1 antibody. Bound proteins were washed in a buffer containing 150 mM (lanes 4 to 6 and 9 to 10), 250 mM (lanes 7 and 11), or 400 mM (lanes 8 and 12) NaCl and were separated by SDS-PAGE followed by silver staining. The asterisks indicate bands derived from the antibody. (B) The histone transfer activity of B23 proteins. Core histones (72 ng) were preincubated without (lane 1) or with 50 (lanes 2, 5, and 8), 150 (lanes 3, 6, and 9), or 450 (lanes 4, 7, and 10) ng of recombinant B23.1, B23.2, or T4sD (lanes 2 to 4, 5 to 7, and 8 to 10, respectively) and then mixed with the 147-bp-long DNA fragment and further incubated. The mixtures were separated on a 6% PAGE gel, and DNA was visualized with GelRed staining. (C) Supercoiling assays. B23.1, B23.2, T4sD, and NAP1 proteins were separated by SDS-PAGE and visualized by CBB staining (left). Core histones (1.8 pmol) were preincubated without (lane 1) or with 1.8 (lanes 2, 5, 8, and 11), 5.4 (lanes 3, 6, 9, and 12), or 16.2 (lanes 4, 7, 10, and 13) pmol of recombinant B23.1, B23.2, T4sD, or NAP1 (lanes 2 to 4, 5 to 7, 8 to 10, or 11 to 13, respectively) and then mixed with plasmid DNA preincubated with topoisomerase I and further incubated. DNA was purified and separated on a 1% agarose gel and visualized with GelRed staining. Positions of relaxed (R) or supercoiled (S) circular plasmid DNA are indicated. The band intensities of lanes 4, 7, 10, and 13 were quantified and plotted (right). (D) Expression level of endogenous and exogenous B23 proteins. HeLa cells were transfected with control or B23.1 siRNAs. The cells were supertransfected 24 h after siRNA transfection with empty vector or vectors expressing either Flag-B23.1, -T4sA, or -T4sD. An equal number of cells was collected 72 h after siRNA transfection and subjected to Western blotting with anti-Flag-tag, -B23.1, and -histone H3 antibodies. (E) rRNA transcription level of siRNA-treated cells. Total RNA was isolated from 7 × 105 cells prepared as described for panel D, and the rRNA transcription level was examined by quantitative RT-PCR using a 5′-ETS-specific primer set. The amount of pre-rRNA was normalized by that of β-actin mRNA. Results are means ± SD obtained from three independent experiments. Statistical P values were calculated by t tests and are indicated with * for P < 0.05.
Nascent RNAs are dispensable for the recruitment of B23 on r-chromatin.
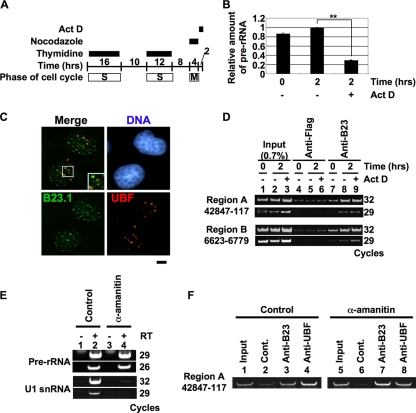
We next explored which molecule(s) was involved in the recruitment of B23.1 to r-chromatin. Because B23 associates with mature rRNAs (Fig. 2C), we hypothesized that B23 is recruited to r-chromatin via its association with nascent pre-rRNAs. In fact, we found that B23 also bound to pre-rRNA in cells (data not shown). Therefore, we examined whether the association of B23.1 with nascent pre-rRNA was required for its recruitment to r-chromatin. B23.1 is released from r-chromatin during mitosis, with the association being restored 2 h after exit from mitosis (Fig. 6 D, lanes 5 and 6). We examined the effect of Pol I transcription initiation following mitosis on the recruitment of B23.1 to r-chromatin. Mitotic HeLa cells were released for 2 h with or without low concentrations of Act D, and the extent of B23 association with r-chromatin was examined using a ChIP assay. Low concentrations of Act D specifically inhibit the Pol I transcription (Fig. 6B). In addition, the nucleolar structure was not properly assembled, and B23 was distributed throughout the nucleus with forming small foci (Fig. 6C). It should also be noted that distinct amounts of pre-rRNA remained during mitosis and the pre-rRNA level was decreased when cells underwent entry into G1 phase, suggesting that pre-rRNA processing was not inhibited by Act D (Fig. 6B). ChIP assays demonstrated that the association of B23.1 with r-chromatin was recovered even in the presence of Act D (Fig. 6D, lane 9). Thus, we concluded that the newly synthesized pre-rRNA is not required for the recruitment of B23.1 to r-chromatin, although we cannot exclude the possibility that pre-rRNAs synthesized during the previous cell cycle and located at r-chromatin may play a role in recruitment of B23.1 to r-chromatin after reentry into the new G1 phase. Next, we examined whether B23 is recruited to r-chromatin with RNAs transcribed by RNA polymerase II. HeLa cells were cultured for 24 h in the absence or presence of α-amanitin, and ChIP assays were carried out. RT-PCR demonstrated that the amount of U1 snRNA transcribed by Pol II was specifically reduced (Fig. 6E). Under this condition, ChIP assays demonstrated that both UBF and B23 remained associated with r-chromatin (Fig. 6F, lanes 3, 4, 7, and 8). These results suggest that the nascent RNAs are not required for the recruitment of B23 to r-chromatin.
FIG. 6.
Nascent rRNA is not crucial for the recruitment of the B23-RNA complex to r-chromatin. (A to C) Effect of Act D on rRNA transcription. HeLa cells were synchronized at mitosis and released for 2 h in growth medium in the absence or presence of 50 ng/ml Act D (A). Total RNAs were purified, and the rRNA transcription level was examined by quantitative RT-PCR using a 5′-ETS-specific primer set (B). The amount of pre-rRNA was normalized by that of β-actin mRNA. Results are means ± SD obtained from three independent experiments. Statistical P values are indicated with ** for P < 0.01. HeLa cells prepared as described for panel A were fixed and subjected to immunofluorescence analysis with anti-B23.1 and anti-UBF antibodies (C). DNA was counterstained with TO-PRO-3. Bar, 5 μm. (D) Effect of rRNA transcription initiation on the recruitment of B23 to r-chromatin. HeLa cells prepared as described for panel A were subjected to ChIP assays using anti-Flag-tag and -B23 (lanes 4 to 6 and 7 to 9, respectively) antibodies. Precipitated (lanes 4 to 9) and input (lanes 1 to 3) DNA were used for PCR with region A and B primer sets. PCR products were analyzed on a 6% PAGE gel and visualized with GelRed staining. (E) Effect of α-amanitin on transcription. HeLa cells were cultured in the absence or presence of 5 μg/ml of α-amanitin for 24 h, and total RNA was isolated from 4.5 × 105 cells. The amounts of pre-rRNA and U1 snRNA were examined by RT-PCR. PCR products were analyzed on a 6% PAGE gel and visualized with GelRed staining. (F) Effect of Pol II inhibition on the association of B23 with r-chromatin. HeLa cells prepared as described for panel E were subjected to ChIP assays using control IgG, anti-B23, and anti-UBF (lanes 2 and 6, 3 and 7, and 4 and 8, respectively) antibodies. Precipitated and input DNA was amplified by PCR with the region A primer set.
UBF is involved in the recruitment of B23 on r-chromatin.
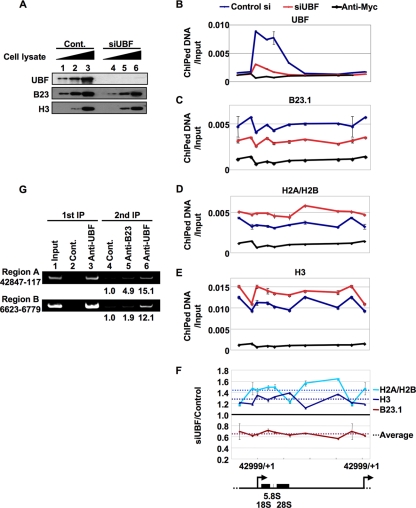
Since UBF was found to be associated with the entire rRNA gene (22) and has previously been reported to be necessary for recruiting factors involved in linking rRNA transcription and processing (24), we next focused on the function of UBF in the recruitment of the B23-RNA complex to r-chromatin. To examine this point, ChIP assays were performed with control or UBF siRNA-treated HeLa cells (Fig. 7). The expression level of UBF protein in cells treated with UBF siRNA was reduced to less than 25% of the control siRNA-treated cells, while those of B23 and histone H3 were not changed (Fig. 7A). The enrichment of DNA fragments containing the rRNA genes was quantitatively analyzed by Q-PCR, with several primer sets extending across the entire rRNA gene being employed (Fig. 7B to F). UBF was apparently enriched around the promoter and coding regions of the rRNA gene in control siRNA-treated cells. UBF siRNA treatment significantly decreased the association level of UBF to r-chromatin (Fig. 7B). B23 bound to the entire rRNA genes in control siRNA-treated cells, whereas the B23 association was decreased by UBF siRNA treatment (Fig. 7C). This result indicated that UBF is involved in the recruitment of B23 to r-chromatin. Additionally, we examined the effect of UBF siRNA treatment on the histone density around r-chromatin that was shown to be regulated by B23 (14). Histone H2A-H2B and H3 were distributed evenly across the r-chromatin in control siRNA-treated cells. In contrast to the B23 association level, histone density on r-chromatin was evenly increased by UBF siRNA treatment (Fig. 7D and E). The effect of UBF knockdown on the distribution pattern of B23 and histones around r-chromatin is shown in Fig. 7F. These results suggest that UBF plays an important role in the recruitment of B23 to r-chromatin. In order to confirm this notion, we performed sequential ChIP assays (Fig. 7G). ChIP assays were first carried out with anti-UBF antibody, and the UBF-containing complex was subsequently subjected to the second ChIP assays using anti-B23 and anti-UBF antibodies. Both on the regions A and B, we detected the colocalization of B23 and UBF (Fig. 7G, lane 5). The colocalization level of B23 and UBF on region A was higher than that on region B (lanes 5 and 6). These results strongly suggest that B23 is recruited to r-chromatin through the joint actions of its own RNA binding activity and UBF's ability to regulate the histone density and thereby rRNA transcription level.
FIG. 7.
UBF plays a crucial role in the recruitment of the B23-RNA complex to r-chromatin. (A) Expression level of UBF, B23.1, and histone H3 proteins in siRNA-treated HeLa cells. HeLa cells were transfected with control and UBF siRNAs (lanes 1 to 3 and 4 to 6, respectively). Cells were fixed 72 h after siRNA transfection and cell extracts were prepared. Increasing amounts of cell extracts were subjected to Western blotting with anti-UBF, -B23, and -histone H3 antibodies. (B to F) Effect of UBF knockdown on the distributions of B23.1 and histones along the r-chromatin. Cell lysates prepared as described for panel A were subjected to ChIP assays using anti-UBF (B), -B23 (C), -histone H2A/H2B (D), and -histone H3 (E) antibodies. Anti-Myc-tag antibody was used as a control. Precipitated DNA and input DNA were used as templates for Q-PCR using primer sets for the rRNA gene. The enrichment level of amplified DNA is shown as the relative amount to input DNA (B to E). The x axes of the graphs correspond to the position along the 43-kbp-long rRNA genes shown schematically at the bottom. Blue and red lines in the graphs (B to E) show the results of ChIP assays with control and UBF siRNA-treated cells, respectively. Q-PCRs were carried out in triplicate. The amounts of B23.1, histone H2A-H2B, and histone H3 along the r-chromatin in UBF siRNA-treated cells were estimated relative to those in control siRNA-treated cells (F). Dotted lines show the average of change. (G) Colocalization of UBF and B23 on r-chromatin. Fixed HeLa cells were subjected to ChIP assays using control IgG and anti-UBF antibody (primary ChIP, lanes 2 and 3). The UBF-containing complex was subjected to a second immunoprecipitation with control IgG, anti-B23, and anti-UBF (lanes 4 to 6) antibodies. Precipitated (lanes 2 to 6) and input (lane 1) DNA was amplified by PCR. PCR products were separated by 6% PAGE and visualized with GelRed staining.
DISCUSSION
Here, we have shown that the r-chromatin binding of histone chaperone B23 is dependent on B23's RNA binding activity and is required for its stimulatory function of rRNA transcription. This conclusion was drawn from the following results: (i) the chromatin binding activity of B23.1 in cell extracts is sensitive to RNase treatment (Fig. 1C), (ii) T4sA, a mutant of B23.1 in which the RNA binding activity is not influenced by cdc2-mediated phosphorylation, remains bound to chromatin during mitosis (Fig. 2), and (iii) T4sD, a mutant mimicking the mitotic phosphorylated state of B23.1, does not efficiently associate with RNA and r-chromatin (Fig. 3). The chromatin association of B23 was shown to be necessary for it to exert its histone chaperone activity in cells (Fig. 5). In addition, we demonstrated that recruitment of B23.1 to r-chromatin depends not only upon its RNA binding activity but also on the presence of UBF. Our results suggest a novel regulatory mechanism for conferring target gene specificity to histone chaperones. The other important conclusion in this study is that the histone chaperone activity of B23.1 is indirectly regulated by cdc2-mediated phosphorylation during the cell cycle.
RNA molecules required for B23 recruitment to r-chromatin.
We demonstrated that B23.1 is recruited to r-chromatin in an RNA binding activity-dependent manner. We found that nascent pre-rRNA was dispensable for the recruitment of B23 to r-chromatin (Fig. 6). In mitotic cells, since not only Pol I transcription but also pre-rRNA processing is suggested to be inactivated, we cannot exclude the possibility that pre-rRNA synthesized during the previous cell cycle helps to recruit B23.1 to r-chromatin. Another possibility is that mature rRNAs function as B23.1-recruiting RNAs. Several lines of evidence support this idea: (i) B23.1 associates with 5.8S and 28S rRNAs in vitro (21), (ii) Flag-tagged B23.1 was coimmunoprecipitated with mature rRNAs from cell extracts (Fig. 2C and 3C), and (iii) endogenous B23 was coprecipitated with rRNAs from cell extracts (2, 35, 41). It is also possible that a minor population of non-rRNA is involved in the recruitment of B23.1. In this context, we examined the effect of Pol II transcription inhibition on the association of B23 with r-chromatin (Fig. 6). However, the association level of B23 with r-chromatin was not significantly reduced after at least 24 h of Pol II transcription inhibition. Clarification of the RNA binding specificity of B23.1 will provide a clue to determine which RNA molecule is required to recruit the B23 histone chaperone to r-chromatin.
We have previously demonstrated that purified recombinant B23.1 did not show efficient nucleosome disassembly activity in vitro (19). However, the depletion of B23.1 increased the histone density around the rRNA gene in a histone chaperone activity-dependent manner (14). Thus, complex formation with RNA molecules may be required not only for r-chromatin binding but also for stimulating the B23 histone chaperone function. In addition, given that acetylation of B23 enhances its histone chaperone activity (36), it would be interesting to examine the chromatin remodeling function of acetylated B23 in combination with its RNA binding activity.
Cell cycle-dependent regulation of histone chaperones.
In addition to the inactivation of the pre-rRNA processing pathway, an important consequence of mitotic phosphorylation of B23 may be the inactivation of rRNA transcription through r-chromatin regulation. During mitosis, most of genes are silenced and chromatin structure is highly condensed to ensure proper chromosome segregation. Silencing of rRNA transcription during mitosis by the phosphorylation-mediated inactivation of Pol I machinery could be crucial for repetitive rRNA gene segregation (8). The inactivation of the histone chaperone function of B23 may be in part a mechanism for inactivation of rRNA transcription during mitosis. It was recently reported that rRNA gene clusters distributed throughout 10 chromosomes in diploid human cells are recombination hot spots for a variety of solid tumors (34). Thus, the rigorous regulation of B23-RNA complex formation in mitosis may be very important for maintaining the genome stability.
UBF function in recruiting the B23-RNA complex to r-chromatin.
We demonstrated that the RNA binding activity of B23.1 is essential but not sufficient for its r-chromatin binding, as shown in Fig. 2. UBF was found to be required for the recruitment of B23 to r-chromatin to colocalize preferentially at the promoter region of rRNA genes (Fig. 7). However, the distribution patterns of UBF and B23 along r-chromatin were different (Fig. 7B and C), and the direct interaction between UBF and B23 was not detected in vitro (data not shown). Thus, it is suggested that UBF recruits the B23 complex indirectly. Three possibilities exist for UBF's role in the recruitment of B23.1 to r-chromatin. The first possibility is that the r-chromatin structure attracts the B23-RNA complex. It has been established that UBF binds to and then subsequently bends r-chromatin via its HMG boxes to create a nucleosome-like structure (32). The r-chromatin structure associated with UBF may recruit the B23-RNA complex. The second possibility is that UBF removes the inhibitory factor for the B23-RNA complex assembly from r-chromatin. It was recently reported that UBF depletion increased the level of linker histone H1 on r-chromatin (11, 26). Since HMG box proteins and histone H1 are suggested to associate with chromatin in a mutually exclusive manner (9), histone H1-bound chromatin may restrict the access of the B23-RNA complex. The third possibility is that UBF is needed for tethering the factor(s) that recruits B23 to r-chromatin. Further analysis is required to distinguish these different possibilities.
In summary, here we have demonstrated that the histone chaperone B23.1 is recruited to r-chromatin through its RNA binding activity and B23 recruitment is regulated by its cell cycle-dependent phosphorylation. This discovery represents a novel regulatory mechanism for targeting histone chaperones to specific genes. It was recently reported that B23.1 is recruited to the specific gene transcribed by RNA polymerase II outside the nucleolus (30). These observations prompted us to speculate that B23.1 is differentially recruited to the different chromosome loci by changing the partner RNA molecules, implying that RNA molecules may specify the chromosome loci to be remodeled by B23.1. We feel that this hypothesis is noteworthy, as it represents a new role for RNA participation in chromatin regulation.
Acknowledgments
We thank D. Hall (University of Tsukuba) for a critical reading of the manuscript.
This work was supported by PRESTO from Japan Science and Technology Agency (to M.O.), Special Coordination Funds for Promoting Science and Technology (to M.O.), Grants-in-Aid for Scientific Research (grant numbers 19038003, 20052005, 21113005, and 17013018) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M.O. and K.N.), the Bioarchitect research Program from RIKEN (to K.N.), and a grant from the Ichiro Kanehara Foundation (to M.O.).
Footnotes
Published ahead of print on 16 August 2010.
REFERENCES
- 1.Angelov, D., V. A. Bondarenko, S. Almagro, H. Menoni, F. Mongelard, F. Hans, F. Mietton, V. M. Studitsky, A. Hamiche, S. Dimitrov, and P. Bouvet. 2006. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 25:1669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertwistle, D., M. Sugimoto, and C. J. Sherr. 2004. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birch, J. L., B. C. Tan, K. I. Panov, T. B. Panova, J. S. Andersen, T. A. Owen-Hughes, J. Russell, S. C. Lee, and J. C. Zomerdijk. 2009. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 28:854-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conconi, A., R. M. Widmer, T. Koller, and J. M. Sogo. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57:753-761. [DOI] [PubMed] [Google Scholar]
- 5.Eckey, M., W. Hong, M. Papaioannou, and A. Baniahmad. 2007. The nucleosome assembly activity of NAP1 is enhanced by Alien. Mol. Cell. Biol. 27:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.French, S. L., Y. N. Osheim, F. Cioci, M. Nomura, and A. L. Beyer. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodfellow, H., A. Krejci, Y. Moshkin, C. P. Verrijzer, F. Karch, and S. J. Bray. 2007. Gene-specific targeting of the histone chaperone asf1 to mediate silencing. Dev. Cell 13:593-600. [DOI] [PubMed] [Google Scholar]
- 8.Heix, J., A. Vente, R. Voit, A. Budde, T. M. Michaelidis, and I. Grummt. 1998. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 17:7373-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson, J. B., J. M. Pollock, Jr., and R. L. Rill. 1979. Chromatin fractionation procedure that yields nucleosomes containing near-stoichiometric amounts of high mobility group nonhistone chromosomal proteins. Biochemistry 18:3739-3748. [DOI] [PubMed] [Google Scholar]
- 10.Kato, K., M. Miyaji-Yamaguchi, M. Okuwaki, and K. Nagata. 2007. Histone acetylation-independent transcription stimulation by a histone chaperone. Nucleic Acids Res. 35:705-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kermekchiev, M., J. L. Workman, and C. S. Pikaard. 1997. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol. Cell. Biol. 17:5833-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McStay, B., M. W. Frazier, and R. H. Reeder. 1991. xUBF contains a novel dimerization domain essential for RNA polymerase I transcription. Genes Dev. 5:1957-1968. [DOI] [PubMed] [Google Scholar]
- 13.Montecino, M., J. L. Stein, G. S. Stein, J. B. Lian, A. J. van Wijnen, F. Cruzat, S. Gutierrez, J. Olate, S. Marcellini, and J. L. Gutierrez. 2007. Nucleosome organization and targeting of SWI/SNF chromatin-remodeling complexes: contributions of the DNA sequence. Biochem. Cell Biol. 85:419-425. [DOI] [PubMed] [Google Scholar]
- 14.Murano, K., M. Okuwaki, M. Hisaoka, and K. Nagata. 2008. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol. Cell. Biol. 28:3114-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murayama, A., K. Ohmori, A. Fujimura, H. Minami, K. Yasuzawa-Tanaka, T. Kuroda, S. Oie, H. Daitoku, M. Okuwaki, K. Nagata, A. Fukamizu, K. Kimura, T. Shimizu, and J. Yanagisawa. 2008. Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133:627-639. [DOI] [PubMed] [Google Scholar]
- 16.Nagata, K., H. Kawase, H. Handa, K. Yano, M. Yamasaki, Y. Ishimi, A. Okuda, A. Kikuchi, and K. Matsumoto. 1995. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl. Acad. Sci. U. S. A. 92:4279-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nousiainen, M., H. H. Sillje, G. Sauer, E. A. Nigg, and R. Korner. 2006. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. U. S. A. 103:5391-5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okuwaki, M., A. Iwamatsu, M. Tsujimoto, and K. Nagata. 2001. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 311:41-55. [DOI] [PubMed] [Google Scholar]
- 19.Okuwaki, M., K. Kato, H. Shimahara, S. Tate, and K. Nagata. 2005. Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I. Mol. Cell. Biol. 25:10639-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuwaki, M., K. Matsumoto, M. Tsujimoto, and K. Nagata. 2001. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 506:272-276. [DOI] [PubMed] [Google Scholar]
- 21.Okuwaki, M., M. Tsujimoto, and K. Nagata. 2002. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell 13:2016-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Sullivan, A. C., G. J. Sullivan, and B. McStay. 2002. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panov, K. I., J. K. Friedrich, J. Russell, and J. C. Zomerdijk. 2006. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 25:3310-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prieto, J. L., and B. McStay. 2007. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 21:2041-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rickards, B., S. J. Flint, M. D. Cole, and G. LeRoy. 2007. Nucleolin is required for RNA polymerase I transcription in vivo. Mol. Cell. Biol. 27:937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanij, E., G. Poortinga, K. Sharkey, S. Hung, T. P. Holloway, J. Quin, E. Robb, L. H. Wong, W. G. Thomas, V. Stefanovsky, T. Moss, L. Rothblum, K. M. Hannan, G. A. McArthur, R. B. Pearson, and R. D. Hannan. 2008. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 183:1259-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santoro, R., J. Li, and I. Grummt. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32:393-396. [DOI] [PubMed] [Google Scholar]
- 28.Savino, T. M., J. Gebrane-Younes, J. De Mey, J. B. Sibarita, and D. Hernandez-Verdun. 2001. Nucleolar assembly of the rRNA processing machinery in living cells. J. Cell Biol. 153:1097-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwabish, M. A., and K. Struhl. 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22:415-422. [DOI] [PubMed] [Google Scholar]
- 30.Shandilya, J., V. Swaminathan, S. S. Gadad, R. Choudhari, G. S. Kodaganur, and T. K. Kundu. 2009. Acetylated NPM1 localizes in the nucleoplasm and regulates transcriptional activation of genes implicated in oral cancer manifestation. Mol. Cell. Biol. 29:5115-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stefanovsky, V., F. Langlois, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2006. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21:629-639. [DOI] [PubMed] [Google Scholar]
- 32.Stefanovsky, V. Y., G. Pelletier, D. P. Bazett-Jones, C. Crane-Robinson, and T. Moss. 2001. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 29:3241-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strohner, R., A. Nemeth, P. Jansa, U. Hofmann-Rohrer, R. Santoro, G. Langst, and I. Grummt. 2001. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20:4892-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stults, D. M., M. W. Killen, E. P. Williamson, J. S. Hourigan, H. D. Vargas, S. M. Arnold, J. A. Moscow, and A. J. Pierce. 2009. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 69:9096-9104. [DOI] [PubMed] [Google Scholar]
- 35.Sugimoto, M., M. L. Kuo, M. F. Roussel, and C. J. Sherr. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11:415-424. [DOI] [PubMed] [Google Scholar]
- 36.Swaminathan, V., A. H. Kishore, K. K. Febitha, and T. K. Kundu. 2005. Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol. Cell. Biol. 25:7534-7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szebeni, A., K. Hingorani, S. Negi, and M. O. Olson. 2003. Role of protein kinase CK2 phosphorylation in the molecular chaperone activity of nucleolar protein b23. J. Biol. Chem. 278:9107-9115. [DOI] [PubMed] [Google Scholar]
- 38.Takahata, S., Y. Yu, and D. J. Stillman. 2009. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol. Cell 34:405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Telese, F., P. Bruni, A. Donizetti, D. Gianni, C. D'Ambrosio, A. Scaloni, N. Zambrano, M. G. Rosenfeld, and T. Russo. 2005. Transcription regulation by the adaptor protein Fe65 and the nucleosome assembly factor SET. EMBO Rep. 6:77-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, W., A. Budhu, M. Forgues, and X. W. Wang. 2005. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat. Cell Biol. 7:823-830. [DOI] [PubMed] [Google Scholar]
- 41.Yu, Y., L. B. Maggi, Jr., S. N. Brady, A. J. Apicelli, M. S. Dai, H. Lu, and J. D. Weber. 2006. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 26:3798-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, Y., R. Santoro, and I. Grummt. 2002. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 21:4632-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]