Abstract
A novel assay was developed for Daudi cells in which the antiviral (AV) and antiproliferative (AP) activities of interferon (IFN) can be measured simultaneously. Using this novel assay, conditions allowing IFN AV protection but no growth inhibition were identified and selected. Daudi cells were treated under these conditions, and gene expression microarray analyses were performed. The results of the analysis identified 25 genes associated with IFN-α AV activity. Upregulation of 23 IFN-induced genes was confirmed by using reverse transcription-PCR. Of 25 gene products, 17 were detected by Western blotting at 24 h. Of the 25 genes, 10 have not been previously linked to AV activity of IFN-α. The most upregulated gene was IFIT3 (for IFN-induced protein with tetratricopeptide repeats 3). The results from antibody neutralizing experiments suggested an association of the identified genes with IFN-α AV activity. This association was strengthened by results from IFIT3-small interfering RNA transfection experiments showing decreased expression of IFIT3 and a reduction in the AV activity induced by IFN-α. Overexpression of IFIT3 resulted in a decrease of virus titer. Transcription of AV genes after the treatment of cells with higher concentrations of IFN having an AP effect on Daudi cells suggested pleiotropic functions of identified gene products.
Over the past 50 years, type I interferons (IFNs) have emerged as major components of the innate immune system and are recognized for their ability to combat viral infections. The major type I human IFNs include the IFN-α subtypes, IFN-β, and IFN-ω. In addition to their antiviral (AV) function, these proteins also have antiproliferative (AP) and immunomodulatory effects on cells (10, 41). Therapeutic approaches have exploited the multiple effects of IFN, with human IFN-α2 approved for the treatment of hepatitis B and C, as well as certain forms of cancer. Identification of genes important for AV function holds considerable potential for the development of novel prophylactics and therapeutics. However, delineating genes associated specifically with AV function from genes involved in other IFN functions has been challenging.
To activate IFN-stimulated gene transcription, the type I IFNs bind to their receptors and initiate intracellular signaling through the JAK/STAT pathway (15, 17). IFN gene expression profiles have been previously studied by using microarray analysis (5, 13, 16, 23, 28, 34, 40), but this work has been unable to identify IFN-induced genes under conditions that allow for AV protection without growth inhibition. Moreover, although it is possible to perform AP or AV assays individually (1, 3, 37, 30), no assay has been described in the literature that measures both activities simultaneously.
Effective delineation of genes and proteins associated with AP and AV responses is hindered by the lack of a single platform approach. Using human Burkitt's lymphoma (Daudi) cells as the cell model, we modified an MTT assay to identify conditions that allowed for AV protection but not AP activities. Treatments with IFN-α2c and IFN hybrid 2 [HY-2, IFN-α21b(1-95)/IFN-α2c(96-165)] were compared, and conditions for AV protection without AP activity were identified. Gene expression analysis under conditions of AV protection revealed a set of 25 genes that are important for AV activity, with the most upregulated gene identified as IFN-induced protein with tetratricopeptide repeats 3 (IFIT3).
IFIT3 was recently described as a key mediator of AP activity of IFN-α (36). The AP effect of IFIT3 correlated with increased p21 and p27, two factors that negatively regulate progression of the cell cycle from G1 phase into S phase (36). The transcription of IFIT3 is mediated by the formation of the ISGF3 complex containing pSTAT1, pSTAT2, and IRF9, but it also can be mediated by IRF1 through IRF9/STAT2-dependent or -independent mechanisms (18). Different mechanisms inducing IFIT3 highlight the importance of this protein in IFN-α functions. However, IFIT3 was not previously shown to be associated with IFN AV activity.
In summary, this research provides a novel assay that distinguishes between AV and AP activities of type I IFNs on a Daudi cell line. Using conditions from this assay, in conjunction with gene expression analysis, enabled us to identify a number of genes not previously associated with the AV effect of IFN. IFIT3 was the most highly upregulated gene at 6 and 24 h after IFN treatment. Reduction of IFIT3 expression by small interfering RNA (siRNA) in human A549 cells resulted in decreased AV activity, suggesting an AV function for IFIT3. Overexpression of IFIT3 led to a decrease in virus titer. We illustrate here the importance of the application of biologically significant concentrations of IFN-α for the identification of its AV genes. This is also the first study identifying AV-associated genes of the Daudi cell line treated with IFN-α. Demonstration of IFIT3 as an IFN-α-induced protein associated with AV activity in Daudi cells is particularly significant given the importance of this protein in IFN-α functions and the lack of previous evidence linking this gene or protein to IFN AV activity.
MATERIALS AND METHODS
Cell lines.
Suspension Daudi cells were selected as the human cell model based on their sensitivity to type I IFNs and their wide use in measuring IFN AP activity (10-12). The Daudi cells were obtained from P. Grimley (Department of Pathology, Uniformed Services University of the Health Sciences, Bethesda, MD). Adherent Madin-Darby bovine kidney (MDBK) cells, green monkey kidney epithelial cells (Vero), human alveolar basal epithelial cells (A549), human cervical carcinoma cells (HeLa), human monocytoid cells (U937), and human T lymphocytes (Jurkat) were obtained from the American Type Cell Culture Collection (ATCC; Manassas, VA). Human ovary carcinoma cells (OVCAR-3) were obtained from NCI-60 collection, human hepatoma cells (Huh-7) were a generous gift from B. Rehermann (NIDDK, NIH), human B-lymphoblastoid cells (B-JAB) were a generous gift from M. Lenardo (NIAID, NIH), and human fibrosarcoma cells (2fTGH) were provided by G. Stark (The Cleveland Clinic Foundation). Human monocytes from healthy volunteers were obtained from the NIH Blood Bank. Cell lines were cultured as previously described (31).
Viruses.
Vesicular stomatitis virus (VSV; single-strand, negative-sense RNA virus, Indiana strain) was a gift from R. Friedman (Department of Pathology, Uniformed Services University of the Health Sciences, Bethesda, MD). Murine encephalomyelitis virus (EMCV; single-strand, positive-sense RNA virus) was obtained from the ATCC.
To estimate the quantity of infectious virus, harvested supernatants were evaluated by using a plaque assay on MDBK cells. Cells were seeded at 2 × 105 per well in 24-well plates (Falcon; Becton Dickinson Labware, Franklin Lakes, NJ) and incubated for 24 h prior to virus infection. Subsequently, 5-fold dilutions of the supernatants (three pooled wells for each concentration of IFN) were transferred onto MDBK cell layers in 24-well plates and then serially diluted in 10-fold increments. After 1 h of incubation, the supernatants were removed, and MDBK cells were overlaid with fresh Dulbecco modified Eagle medium containing 2% fetal bovine serum (FBS) and methylcellulose. Cell sheets were fixed after 24 h and stained with crystal violet. Virus titers were calculated by first multiplying the plaque count by the dilution factor and then multiplying the result by the dilution factor of 5. The virus titers for VSV were 1.5 × 107 PFU/ml on MDBK cells, 2.5 × 105 PFU/ml on A549 cells, and 1.3 × 106 PFU/ml on Vero cells. The virus titers for EMCV were 3.0 × 106 PFU/ml on A549 cells and 2.3 × 108 PFU/ml on Vero cells.
To study the influence of VSV on IFN-α2-induced gene products, Daudi cells (3 × 106 cells in 10 ml of RPMI) were incubated with 2.5 IU of human IFN-α2a/ml and VSV (1.5 × 104 PFU/ml). To study the time course of gene products induced by endogenous IFN, Daudi cells (3 × 106 cells in 10 ml of RPMI) were incubated with EMCV (3.0 × 105 PFU/ml). All samples were harvested at the indicated time points. Cell lysates were prepared and analyzed as describe above.
IFNs.
IFN-α2c and HY-2 [HY-2, IFN-α21b (1-95)/IFN-α2c (96-165)] were prepared and analyzed as previously described (11, 37, 41). IFN hybrid HY-2 was selected based on our earlier observations of induced AP activity on Daudi cells, compared to IFN-α21b and IFN-α2c (11). The specific AV activities of IFNs on Daudi cells were 3.5 ×108 IU/mg for IFN-α2c and 3.5 ×107 IU/mg for HY-2. The antiproliferative activities (50% inhibition of proliferation in 48 h after IFN treatment) were 36 ng/ml for HY-2 and 0.072 ng/ml for IFN-α2c. All IFN AV units were expressed with reference to the NIH standard Gxa01-901-535 (human recombinant IFN-α2a).
Antiproliferative assays and neutralization of IFN antiproliferative activity.
For the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma-Aldrich, Inc., St. Louis, MO] AP assay, Daudi cells (3 × 104 cells/well) were treated with IFN-α2c or HY-2 at an initial concentration of 360 ng/ml and 10-fold serial dilutions in 10% FBS-RPMI as previously described (10).
For neutralization of AP activity, Daudi cells were mock treated or treated with 10-fold dilutions from an initial concentration of 360 ng of IFN-α2c or HY-2/ml with or without 10 μg of anti-IFN-α2c monoclonal antibody (MAb) N54 (a generous gift from P. Kontsek, Institute of Neuroimmunology, SAS, Bratislava, Slovakia)/ml. Concentrations of IFNs that inhibit cell growth by 50% (IC50) were calculated at 48 h after IFN treatment (32).
Antiviral assays and neutralization of antiviral activities of IFNs.
For the MTT antiviral assay, Daudi cells (3 × 104 cells/well) were treated with IFN-α2c or HY-2 for 24 h as described above, with the exception of using 2% FBS-RPMI. Subsequently, 50 μl (1.5 × 103 PFU/ml) of VSV was added per well, followed by incubation for 24 h. When a cytopathic effect (CPE) was observed on ca. 50% of cells, a standard MTT assay was performed. The percentage of cells protected by IFN from virus was calculated by using the following equation: {[(ODIFN sample treated well − ODvirus control)/ODcell control] − ODvirus control} × 100.
To select an optimal time point for gene expression analysis, neutralizing experiments were performed. To achieve neutralization of IFN-α AV activity and to study the kinetics of signal transduction for AV effect, 1 μg of anti-IFNAR1 MAb 64.10 (a generous gift from M. Tovey, GenOdyssee S.A., Courtabaeuf, France)/ml was added at 6, 15, and 24 h after the IFN treatments.
Each IFN AV assay was simultaneously performed on two 96-well plates, with one used for colorimetric MTT assay and the other used for cell and cell supernatant harvest to evaluate cell viability and VSV titer. Cell viability was measured microscopically after staining cells with trypan blue in a hemocytometer (Cellometer; Nexcelom Bioscience, LLC, Lawrence, MA) according to the manufacturer's instructions.
RNA isolation.
RNA was isolated by using an RNeasy minikit (Qiagen, Stanford, CA). Cells were homogenized by using QIA shredder columns, and samples were eluted in 50 μl of RNase-free water. To ensure that there was no residual DNA contamination, samples were treated with RNase-free DNase (Qiagen) as described in the RNeasy protocol.
Gene expression microarray.
Gene expression was initially measured in four Daudi cell groups (3 × 106 in 10 ml of RPMI) treated for 24 h at IFN concentrations selected to allow comparison of gene activity in AV activity environments with environments in which no AV activity was observed. These treatment groups were: (i) 0.0036 ng of IFN-α2c/ml (allowing AV activity only); (i) 0.00036 ng of IFN-α2c/ml (no AV observed); (iii) 0.036 ng of HY-2/ml (allowing AV activity); and (iv) 0.0036 ng of HY-2/ml (no AV observed) (see Tables 3 and 4).
To refine the list of genes associated with AV activity, samples showing AP phenotype (achieved by treatment with IFN-α2c [0.036 ng/ml] or HY-2 [36 ng/ml]) were compared to identically treated samples manipulated to display the AV phenotype through subsequent neutralization with anti-IFNAR1 MAb 64.10 (1 μg/ml). Gene expression was also measured in cells treated with 2.5 IU of IFN standard Gxa01-901-535/ml, a concentration allowing AV protection. AV protection of IFN standard (2.5 IU/ml) was completely neutralized using the same concentration of antibody. Untreated cells were included in each experiment.
Two-color spotted oligonucleotide arrays (Microarray Research Facility NIAID, NIH, Bethesda, MD) were utilized for transcriptional expression data acquisition. Total RNA was purified by using an RNeasy kit (Qiagen, Valencia, CA). A 5-μg portion of total RNA was labeled by using a modified aminoallyl-labeling method, followed by hybridization as previously published by Han et al. (8). After washing, arrays were dried using a Labnet Spectrafuge Mini (Labnet, Edison, NJ) and scanned using an Axon GenePix 4200A microarray scanner with GenePix Pro 5.1 software (Molecular Devices, Sunnyvale, CA). Raw microarray data was uploaded to the mAdb microarray database (http://nciarray.nci.nih.gov/). Array data were normalized by 50th percentile normalization, and the spot intensity was calculated by subtracting the local median background from the mean foreground intensity. Array replicates were averaged across treatments, and gene expression fold values were obtained from the normalized Cy5/Cy3 ratios.
Genes that were upregulated at 24 h after IFN treatment were selected by comparison of IFN-treated and untreated samples. Differences in gene expression were tested for statistical significance by using t tests, and all genes for which differential expression in the AV samples was found significant at the <0.05 P value were noted. The genes commonly induced by both IFN-α2c and HY-2 were reported. Further analysis of gene expression profiles after 6 h of incubation helped indicate genes associated with early roles in IFN-induced AV mechanisms.
Quantitative real-time RT-PCR.
To confirm gene expression data from microarray experiments, quantitative reverse transcription-PCR (qRT-PCR) was performed on samples identical to those used for microarray analysis.
SYBR green qRT-PCR was performed by using a Brilliant II SYBR green qRT-PCR master mix kit (Stratagene, La Jolla, CA). The final 25-μl reaction mix consisted of a SYBR green qRT-PCR master mix, reference dye (2 μM), RT/RNase block enzyme mix (0.0625 μl per 25-μl reaction), forward and reverse gene specific primers (400 nM each), and template RNA (20 ng per reaction). qRT-PCRs were set up in 96-well ABgene PCR plates (Thermo Scientific, Waltham, MA) and run on MX3000P or MX2005P real-time thermocyclers (Stratagene).
Primers of ∼20mer length (Table 1) were designed, and a three-step cycling protocol was performed as previously described (35). The results were analyzed by using MxPro software (version 4.01). The data were expressed as the mean fold increase relative to baseline levels. All qRT-PCR data were normalized to the level of housekeeping gene GAPDH.
TABLE 1.
Forward and reverse primers used in qRT-PCRa
| Gene |
GenBank accession no. | Primerb |
||
|---|---|---|---|---|
| Name | Description | Name | Primer sequence (5′-3′) or Qiagen catalog no. (QT) | |
| BST2 | Bone marrow stromal cell antigen 2 | NM_004335 | BST2#1 F | TGC TGG GGA TAG GAA TTC TG |
| BST2#1 R | TCA GCT CTT GTT GCA GGA GA | |||
| GOT1 | Glutamic-oxaloacetic transaminase 1 | NM_002079 | GOT1#1 F | GGC CAT TCG CTA TTT TGT GT |
| GOT1#1 R | GAC CAA GTA ATC CGC ACG AT | |||
| IFI27 | IFN, alpha-inducible protein 27 | NM_005532 | IFI27#1 F | TCT GGC TCT GCC GTA GTT TT |
| IFI27#1 R | GAA CTT GGT CAA TCC GGA GA | |||
| IFI35 | IFN-induced protein 35 | NM_005533 | IFI35#1 F | CAG GGC CTA GCA GTC TTC AC |
| IFI35#1 R | GGC ATG CAG GCT CTT TTT AC | |||
| IFI44 | IFN-induced protein 44 | NM_006417 | IFI44#1 F | TTC GAT GCG AAG ATT CAC TG |
| IFI44#1 R | CCC TTG GAA AAC AGA CCT CA | |||
| IFI44L | IFN-induced protein 44-like | NM_006820 | IFI44L#1 F | TAT GTG TGT TGG CTG GGA GA |
| IFI44L#1 R | GGG CCT GCA TAC CTC ATA GA | |||
| IFIT3 | IFN-induced protein with tetratricopeptide repeats 3 | NM_001549 | IFIT3#1 F | GAA CAT GCT GAC CAA GCA GA |
| IFIT3#1 R | CAG TTG TGT CCA CCC TTC CT | |||
| IFITM1 | IFN-induced transmembrane protein 1 | NM_003641 | IFITM1#1 F | CAA CAC TTC CTT CCC CAA AG |
| IFITM1#1 R | GAA CAG GGA CCA GAC GAC AT | |||
| IRF7 | IFN regulatory factor 7 | NM_001572 | IRF7#1 F | GAA CTG TGA CAC CCC CAT CT |
| IRF7#1 R | TGC TGC TAT CCA GGG AAG AC | |||
| IRF9 (ISGF3G) | IFN regulatory factor 9 | M87503 | IRF9#1 F | AGG TCCA GCT GTC TGG AAG A |
| IRF9#1 R | ATG GCA TCC TCT TCC TCC TT | |||
| KCNJ1 | Potassium inwardly rectifying channel, subfamily J, member 1 | NM_000220 | KCNJ1#2 F | GTG GCT TTT CAA CGG GAG TA |
| KCNJ1#2 R | ATG CAT GTC TTG TGG GAT CA | |||
| LY6E | Lymphocyte antigen 6 complex, locus E | NM_001127213 | LY6E#1 F | TGA TGT GCT TCT CCT GCT TG |
| LY6E#1 R | ACA GGT CTT GCT CAG GCT GT | |||
| MX1 | Myxovirus (influenza virus) resistance 1, IFN-inducible protein | NM_002462 | MX1#2 F | ACC ACA GAG GCT CTC AGC AT |
| MX1#2 R | CTC AGC TGG TCC TGG ATC TC | |||
| OAS1 | 2′-5′-Oligoadenylate synthetase 1, 40/46 kDa | X04371 | OAS1#1 F | ACA GGC AGA AGA GGA CTG GA |
| OAS1#1 R | TAG AAG GCC AGG AGT CAG GA | |||
| OAS2 | 2′-5′-Oligoadenylate synthetase 2, 69/71 kDa | M87284 | OAS2#1 F | ACA GCT GAA AGC CTT TTG GA |
| OAS2#1 R | GCA TTA AAG GCA GGA AGC AC | |||
| PARP12 | Poly(ADP-ribose) polymerase family, member 12 | NM_022750 | PARP12#1 F | GCA GTG CAT CAA GCT CCA TA |
| PARP12#1 R | CCT GTG GGA CAA AAA GAG GA | |||
| PKR (EIF2AK2) | Eukaryotic translation initiation factor 2-alpha kinase 2 | NM_002759 | PKR#1 F | ACG CTT TGG GGC TAA TTC TT |
| PKR#1 R | TTC TCT GGG CTT TTC TTC CA | |||
| RSAD2 | Radical S-adenosyl methionine domain containing 2 | NM_080657 | RSAD2#2 F | TTC TGA AGC GAG GAG GAA AA |
| RSAD2#2 R | TGG GAA ATA CCA ACG GGA TA | |||
| SP100 | SP100 nuclear antigen | NM_003113 | SP100#1 F | TCC CTC CTA AAG GGG AGA AA |
| SP100#1 R | CTT CAG CTT TGC AGC CTT CT | |||
| STAT1 | Signal transducer and activator of transcription 1, 91 kDa | NM_007315 | STAT1 F | CCG TTT TCA TGA CCT CCT GT |
| STAT1 R | TGA ATA TTC CCC GAC TGA GC | |||
| TREX1 | Three prime repair exonuclease 1 | NM_016381 | TREX1#1 F | GCT CAG CAT CTG TCA GTG GA |
| TREX1#1 R | ATC CTT GGT ACC CCT GCT CT | |||
| UBE2L6 | Ubiquitin-conjugating enzyme E2L 6 | NM_004223 | UBE2L6#1 F | CAA CCT CCC TAC CAC CTG AA |
| UBE2L6#1 R | GCA AGG CTT CCA GTT CTC AC | |||
| IFI6 | IFN, alpha-inducible protein 6 | NM_022873 | Hs_IFI6_1_SG | QT 00244503 |
| HSH2D | Hematopoietic SH2 domain containing | NM_032855 | Hs_HSH2D_1_SG | QT 00087185 |
| FLJ11286 | Chromosome 19 open reading frame 66 | NM_018381 | Hs_C19orf66_1_SG | QT 00031535 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | NM_002046 | Hs_GAPDH_2_SG | QT 01192646 |
qRT-PCR primers were designed using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) or obtained from Qiagen.
Primer sequences are given or the Qiagen QuantiTect primer assay catalog number is provided.
Western blot analysis.
Daudi cells (3 × 106 cells in 10 ml of RPMI) were incubated with various concentrations of IFNs and harvested at the indicated time points as described under Gene Expression Microarray. To compare the presence of IFIT3 and HSH2D in different cell lines, 3 × 106 A549, OVCAR-3, HeLa, HuH7, 2fTGH, B-JAB, Jurkat, and U937 cells and monocytes were treated with the lowest IFN-α2c concentrations (as indicated in the figure legends), allowing AV properties, and harvested 24 h after treatment. Cell lysates were prepared by using mammalian protein extraction reagent (M-PER) with protease and phosphatase inhibitor cocktails (Pierce, Rockford, IL). Protein concentrations were determined by measuring the absorbance at 280 nm, using a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE). Samples (50 μg) were analyzed by SDS-PAGE using 10 to 20% Tris-glycine gels, (Invitrogen Corp., Gaithersburg, MD) under reducing conditions, followed by Western blotting (WB) (32). Antibodies for the detection of STAT1 (immunogen 592-731), pSTAT1 (pY701), IFIT3 (RIG-G), and IRF-9 were obtained from BD Transduction Lab (San Jose, CA). Antibodies for detection of STAT2 (immunogen 671-806) and pSTAT2 (pY689) were obtained from Upstate (Charlottesville, VA), monoclonal antibodies for detection of actin, UBE2L6, IRF7 (C-term), and SP100 were obtained from Abcam (Cambridge, MA). The anti-MxA antibody was a generous gift from O. Haller, University of Freiburg, Freiburg, Germany; anti-OAS1 antibody was a gift from S. Sarkar, Lerner Research Institute, Cleveland, OH; anti-IFI44 antibody was a gift from U. Certa, Hoffmann-LaRoche, Basel, Switzerland; and anti-BST2 antibody was a gift from K. Strebel, NIAID, NIH, Bethesda, MD. The anti-IFI44L and anti-HSH2D antibodies were purchased from Novus Biological, Inc. (Littleton, CO). Antibodies to PKR, TREX-1, OAS2, and CD225-17 (IFITM1) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to RSAD2 were obtained from the Proteintech Group (Chicago, IL), and anti-IFI35 antibodies were obtained from Abnova (Walnut, CA). Biotinylated protein ladder (10 to 200 kDa; Cell Signaling Technology) was used as a molecular weight marker. The nitrocellulose membranes were developed by using a SuperSignal West Femto maximum sensitivity kit (Pierce, Rockford, IL) and visualized with an LAS-3000 charge-coupled device camera system (Fujifilm Medical System, Stamford, CT).
RNA interference antiviral assay.
A549 cells were seeded at 2.5 × 105 cells/well on 24-well plates and immediately transfected for 24 h with 20 nM 25-bp specific (IFIT3, sense, 5′-AUUCGAAUAGUCCAUAGCAUAUUGC-3′) or negative control (low GC) Stealth siRNA oligonucleotides (Invitrogen, Carlsbad, CA), using 5 μg of Lipofectamine 2000 (Invitrogen) diluted in Opti-MEM (Gibco/Invitrogen Corp., Grand Island, NY). Cells were then treated with IFN for 24 h as indicated. After IFN treatment, the culture medium was removed, and EMCV, at a multiplicity of infection (MOI) of 0.01, was added in medium containing 2% FBS. Each experiment was performed simultaneously on two 24-well plates. At 48 h postinfection, supernatants were harvested from one plate to evaluate virus titers, and corresponding cell layers were stained with crystal violet. The absorbance at 570 nm (A570) of each well was determined, and the percentage of cells protected by IFN from virus was calculated by using the above-mentioned equation. The effect of IFIT3-siRNA transfection on virus titer was examined by plaque assay on Vero cells as described above, using the harvested supernatants. Cells from the second plate were harvested after IFN treatment and subjected to WB analysis as previously described to analyze the efficiency of IFIT3-siRNA transfection. The results were confirmed by using 19-bp specific (IFIT3, sense, 5′-GCA AUA UGC UAU GGA CUA U-3′) or negative control siGenome nontargeting #1 siRNA oligonucleotides (Dharmacon, Inc., Lafayette, CO).
Overexpression of IFIT3.
To construct the IFIT3 expression vector, a 1,473-bp fragment of the IFIT3 open reading frame from genomic DNA of the A549 cell line (ATCC) was amplified using a forward primer containing an EcoRV site (underlined) with the sequence 5′-ATCGATATCATGAGTGAGGTCACCAAGAATTC-3′ (start codon in boldface) and a reverse primer containing a BamHI site (underlined) with the sequence 5′-GATGGATCCTCAGTTCAGTTGCTCTGAGTTAG-3′ (stop codon in boldface). PCR amplification was performed using a DNA thermal cycler. Amplified products were digested with EcoRV and BamHI, purified from agarose gel, and then cloned into vector pIRESpuro3 (Clontech Laboratories, Inc., Mountain View, CA). The generated plasmid pIRESpuro3-IFIT3 contains a puromycin resistance gene for the selection of stable clones in human cell lines. The Escherichia coli strain DH5α competent cell (Invitrogen) was used for the production of pIRESpuro3-IFIT3 plasmid. Individual ampicillin-resistant colonies were isolated and grown overnight at 37°C in LB medium supplemented with ampicillin. The plasmid DNA was purified by using Qiagen plasmid kits, and the quality of the preparation was verified by agarose gel analysis and restriction mapping (data not shown). The transient cell line containing the IFIT3 gene was created by transfecting Vero cells with the plasmid containing IFIT3 using FuGENE HD (Roche Diagnostic Corp., Indianapolis, IN), as recommended by the supplier. Expression of IFIT3 was confirmed by WB analysis. To estimate the effect of IFIT3 on the virus titer, Vero cells were infected with either VSV or EMCV at an MOI of 0.0001. The supernatant and cell layer were collected at 15 h postinfection. The virus titer (TCID50) was determined on Vero cells as described elsewhere (1).
RESULTS
Establishment of conditions for gene expression analysis.
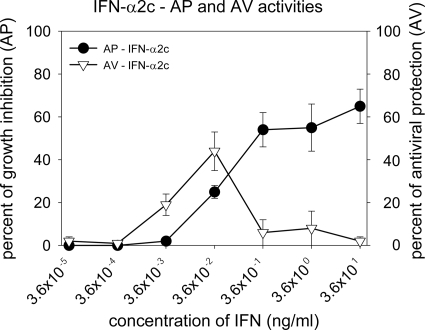
An MTT assay was developed that distinguishes between the AP and AV activities of IFN-α in a single assay. Testing of IFNs in this novel assay identified conditions that limited IFN-induced activity to AV protection. Treatment of Daudi cells with 0.0036 ng of IFN-α2c/ml resulted in 19% AV protection, and treatment of cells with 0.036 ng of HY-2/ml yielded 23% AV protection. Fifty-four percent AV protection was observed with international standard (Gxa1-901-535) at a concentration of 2.5 IU/ml (0.0125 ng/ml). Treatment of Daudi cells with higher concentrations of IFN-α2c resulted in overlapping AP/AV phenotype or AP phenotype only (Fig. 1).
FIG. 1.
AP and AV effects of IFN-α2c on Daudi cells. Daudi cells were treated with IFN-α2c in the presence or absence of VSV. Plates were developed 48 h after IFN treatment.
The results of cell viability and virus titer evaluation after treatment with the selected concentrations confirmed that the concentration required for AV protection had no effect on cell proliferation but resulted in increased cell viability in comparison to virus-infected cells only (data not shown). The same concentration of IFN-α2c was able to reduce the virus titer in comparison to an untreated control (Table 2). The concentrations associated with AV protection but not AP activity were utilized in the gene microarray analysis. All experiments were performed with HY-2 and IFN-α2a standard, and similar results were obtained (data not shown).
TABLE 2.
Effect of IFN treatment on virus titera
| IFN-α2c amt (ng/ml) | Virus titer (PFU/ml) |
|---|---|
| 0.036 | (1.2 ± 1.0) × 102 |
| 0.0036 | (4.3 ± 2.5) × 104 |
| 0.00036 | (1.3 ± 0.6) × 105 |
| None (control) | (1.3 ± 0.6) × 105 |
Daudi cells were incubated with IFN-α2c under conditions that resulted in AV protection but no growth inhibition at 24 h, followed by challenge with VSV. When a CPE was observed on ca. 50% of cells, supernatants from the duplicate plate were harvested and used for estimation of the VSV titer. Mean numbers are derived from three independent assays. Control, cells infected with virus and no IFN.
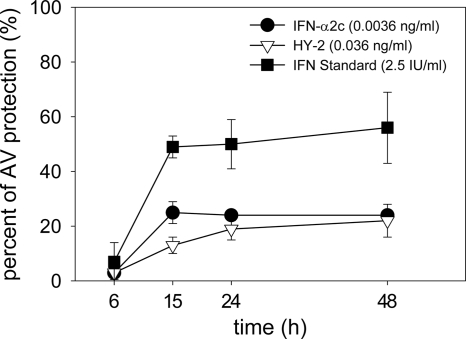
Results from neutralizing experiments showed achievement of maximal AV protection at 15 h of incubation with IFN (Fig. 2). No significant difference in AV protection was observed between 15 and 24 h. The 24-h time point was selected to monitor the effect of IFN on gene expression by microarray, qRT-PCR, and WB experiments. The results of neutralizing AP experiments showed that treatment of Daudi cells for 24 h with higher concentrations of IFN led to overlapping AV and AP activities or to AP activity only (data not shown).
FIG. 2.
Kinetics of IFN AV activity on Daudi cells using anti-IFNAR1 MAb 64.10. Daudi cells were treated with IFN-α2c, HY-2, or IFN-α2a standard for 6, 15, or 24 h prior to the addition of neutralizing anti-IFNAR1 MAb 64.10 (1 μg/ml). At 24 h, cells were infected with VSV. Plates were developed 24 h postinfection.
Identification of genes associated with AV activity of IFN-α and confirmation of AV gene expression by qRT-PCR and Western blotting.
The conditions from the AV/MTT assay in conjunction with gene expression analysis were used to identify genes responsible for the AV properties of IFNs. Initially, 35 genes were identified by using concentrations of IFN-α2c or HY-2 associated with AV protection but not AP activity. To further narrow down the number of IFN-induced genes associated with AV activity, samples showing an AP phenotype (0.36 ng of IFN-α2c/ml, 36 ng of HY-2/ml) were compared to samples treated with the same concentration of IFNs but neutralized with anti-IFNAR1 MAb 64.10 to the level which showed AV protection only. Based on these studies, 25 genes represented a common AV gene signature for both IFN-α2c and HY-2 (Table 3). Analysis of the gene expression profile after treatment of AP phenotype cells and IFN-α2a standard (2.5 IU/ml) at 6 h identified 24 of these genes as early IFN-upregulated genes in Daudi cells (Table 4). FLJ11286 was not detected at 6 h.
TABLE 3.
IFN-α-induced genes associated with AV activity at 24 h
| Gene | Entrez Gene ID | Mean gene expression fold changea |
|||
|---|---|---|---|---|---|
| IFN-α2c (0.0036 ng/ml) | IFN-α2c (0.00036 ng/ml) | HY-2 (0.036 ng/ml) | HY-2 (0.0036 ng/ml) | ||
| BST2 | 684 | 1.72 | 1.15 | 2.30 | 1.17 |
| EIF2AK2 | 5610 | 2.27 | 1.42 | 3.08 | 1.69 |
| FLJ11286 | 55337 | 1.59 | 0.87 | 2.37 | 0.98 |
| GOT1 | 2805 | 2.18 | 0.66 | 4.32 | 0.85 |
| HSH2D | 84941 | 1.38 | 1.07 | 2.06 | 1.25 |
| IFI27 | 3429 | 9.84 | 1.62 | 11.72 | 1.57 |
| IFI35 | 3430 | 2.76 | 1.22 | 4.01 | 1.40 |
| IFI44 | 10561 | 6.47 | 1.18 | 11.62 | 1.74 |
| IFI44L | 10964 | 11.32 | 1.79 | 13.95 | 3.09 |
| IFI6 | 2537 | 9.00 | 2.00 | 6.99 | 1.82 |
| IFIT3 | 3437 | 3.81 | 1.07 | 7.85 | 1.11 |
| IFITM1 | 8519 | 5.12 | 0.77 | 13.02 | 0.86 |
| IRF7 | 3665 | 3.09 | 1.24 | 5.92 | 1.63 |
| IRF9 | 10379 | 2.77 | 1.20 | 3.93 | 1.89 |
| KCNJ1 | 3758 | 10.06 | 1.84 | 11.16 | 2.51 |
| LY6E | 4061 | 1.44 | 1.14 | 1.88 | 1.07 |
| MX1 | 4599 | 3.16 | 0.72 | 7.58 | 0.86 |
| OAS1 | 4938 | 2.68 | 1.14 | 5.48 | 1.48 |
| OAS2 | 4939 | 2.86 | 1.05 | 5.71 | 1.24 |
| PARP12 | 64761 | 2.02 | 1.18 | 3.25 | 1.34 |
| RSAD2 | 91543 | 2.26 | 0.63 | 6.98 | 0.45 |
| SP100 | 6672 | 1.61 | 0.90 | 2.10 | 0.97 |
| STAT1 | 6772 | 2.37 | 1.01 | 4.51 | 1.51 |
| TREX1 | 11277 | 1.53 | 1.20 | 2.41 | 1.20 |
| UBE2L6 | 9246 | 2.08 | 1.21 | 3.47 | 1.28 |
Gene expression levels in Daudi cells from microarray experiments after 24 h of IFN treatment. The numbers indicate the mean expression values of genes. In each experiment, a comparison of treated versus untreated cells was performed. Data are from three independent experiments.
TABLE 4.
IFN-α-induced genes associated with AV activity at 6 h
| Gene | Entrez Gene ID | Mean gene expression fold changea |
||
|---|---|---|---|---|
| IFN-α2c (0.36 ng/ml) | HY-2 (36 ng/ml) | IFN Std (2.5 IU/ml) | ||
| BST2 | 684 | 1.90 | 2.80 | 2.59 |
| EIF2AK2 | 5610 | 4.19 | 4.87 | 5.21 |
| GOT1 | 2805 | 9.42 | 8.23 | 4.05 |
| HSH2D | 84941 | 3.46 | 2.55 | 3.23 |
| IFI27 | 3429 | 3.40 | 4.57 | 1.57 |
| IFI35 | 3430 | 12.23 | 11.05 | 7.29 |
| IFI44 | 10561 | 35.77 | 33.39 | 24.65 |
| IFI44L | 10964 | 16.00 | 13.84 | 16.49 |
| IFI6 | 2537 | 5.05 | 4.55 | 7.64 |
| IFIT3 | 3437 | 16.88 | 7.36 | 9.99 |
| IFITM1 | 8519 | 28.87 | 21.53 | 8.93 |
| IRF7 | 3665 | 5.13 | 4.88 | 2.38 |
| IRF9 | 10379 | 4.26 | 4.44 | 4.41 |
| KCNJ1 | 3758 | 7.22 | 6.63 | 5.11 |
| LY6E | 4061 | 1.92 | 2.17 | 1.62 |
| MX1 | 4599 | 13.22 | 19.54 | 6.36 |
| OAS1 | 4938 | 9.25 | 6.78 | 6.28 |
| OAS2 | 4939 | 18.48 | 19.36 | 17.94 |
| PARP12 | 64761 | 8.80 | 6.63 | 6.10 |
| RSAD2 | 91543 | 18.91 | 18.72 | 13.01 |
| SP100 | 6672 | 4.71 | 4.97 | 2.93 |
| STAT1 | 6772 | 7.24 | 7.26 | 6.58 |
| TREX1 | 11277 | 2.82 | 2.64 | 1.58 |
| UBE2L6 | 9246 | 4.58 | 5.60 | 4.78 |
Gene expression levels in Daudi cells from microarray experiments after 6 h of IFN treatment. The numbers indicate the mean expression values of genes. In each experiment, a comparison of treated versus untreated cells was performed. Data are from three independent experiments.
qRT-PCR confirmed the upregulation for 23 of the 25 genes selected by microarray at 6 and 24 h (data not shown). Upregulation of 2 of 25 genes, GOT1 (glutamic-oxaloacetic transaminase 1) and KCNJ1 (potassium inwardly rectifying channel, subfamily J, member 1) were not confirmed. These results were obtained with three different primer sets. The discrepancy between microarray and qRT-PCR can be explained by nonspecific annealing of target sequences to the array (since dissociation curve analysis showed only a single peak for each primer set). Neutralizing anti-IFNAR1 MAb 64.10 reduced expression of all 23 genes at 24 h (data not shown). FLJ11286 was not detected by microarray at 6 h but was detected in qRT-PCR. This discrepancy may be explained by differences in the sensitivity of the methods.
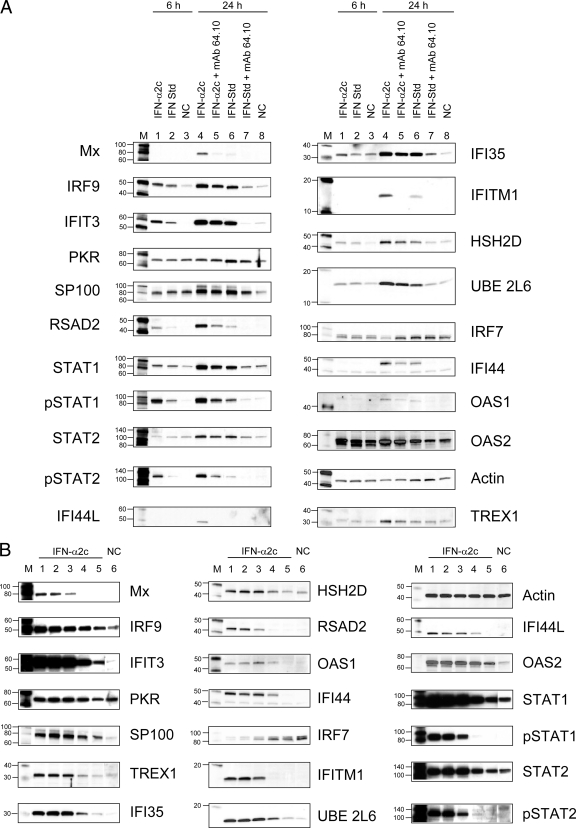
The WB analysis of samples treated with the selected concentrations of IFN-α2c at 6 h identified IRF9, IFIT3, RSAD2, STAT1, IFI35, HSH2D, and UBE2L6 as early IFN-induced proteins (Fig. 3A). Seventeen proteins, including MxA, IRF9, IFIT3, PKR, IFI35, SP100, IFITM1, HSH2D, RSAD2, UBE2L6, IFI44, IRF7, STAT1, OAS1, OAS2, IFI44L, and TREX-1, were detected by WB at 24 h (Fig. 3B).
FIG. 3.
(A) Detection of IFN-α induced AV proteins after 6 h of treatment and the influence of anti-IFNAR1 MAb 64.10 on the expression levels of these proteins at 24 h. To study early IFN-α-induced proteins, Daudi cells were treated for 6 h with concentrations of IFN-α2c that cause an AP effect, or a concentration of IFN-α2a standard, causing AV protection only (2.5 IU/ml). To compare the levels of IFN-induced proteins under conditions of AP activity versus conditions causing AV protection only and to confirm the association of upregulated proteins with IFN treatment, the same samples were incubated with or without neutralizing anti-IFNAR1 MAb 64.10 for 24 h. Lanes: M, molecular weight marker; 1, IFN-α2c (0.36 ng/ml) AP; 2, IFN-α2a standard (2.5 IU/ml) AV; 3, negative control (NC), untreated cells; 4, IFN-α2c (0.36 ng/ml) AP; 5, IFN-α2c (0.36 ng/ml) + MAb 64.10 AV; 6, IFN-α2a standard (2.5 IU/ml) AV; 7, IFN-α2a standard (2.5 IU/ml) + MAb 64.10, no activity; 8, negative control (NC), untreated cells. AP, concentrations of IFN causing antiproliferative effect on Daudi cells. AV, concentrations of IFN causing antiviral effect on Daudi cells. No effect, concentrations of IFN not having any effect on Daudi cells. (B) Detection of IFN-α-induced AV proteins after 24 h of treatment. Daudi cells were treated for 24 h with different concentrations of IFNs. Lanes: M, molecular weight marker; 1, IFN-α2c (3.6 ng/ml) AP; 2, IFN-α2c (0.36 ng/ml) AP; 3, IFN-α2c (0.036 ng/ml) AP; 4, IFN-α2c (0.0036 ng/ml) AV; 5, IFN-α2c (0.00036 ng/ml) no activity; 6, negative control (NC), untreated cells.
Analysis of samples showing an AP phenotype and identically prepared samples transformed to the AV phenotype via additional treatment with anti-IFNAR1 MAb 64.10 (1 μg/ml) by WB showed reduced expression of MxA, RSAD2, IFI44L, IFITM1, IFI44, OAS1, and TREX1 (Fig. 3A, lanes 4 and 5). The same concentration of anti-IFNAR1 MAb completely neutralized 2.5 IU of IFN-α2a standard/ml, which further decreased the levels of IRF9, IFIT3, SP100, RSAD2, STAT1, STAT2, IFI35, IFITM1, HSH2D, UBE2L6, IFI44, OAS1, and OAS2 (Fig. 3A, lanes 6 and 7). Similar results were obtained for HY-2 (data not shown). Thus, results of neutralizing experiments provide compelling evidence of a correlation between expression of these proteins and IFN AV activity.
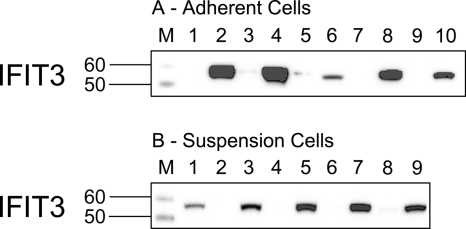
Further analysis of IFIT3 by Western blotting confirmed the presence of IFIT3 in all tested cell lines (Fig. 4A and B). HSH2D was detected in different suspension cell lines (Daudi, monocyte, B-JAB, and Jurkat cells) and in OVCAR-3 and HeLa cells (data not shown).
FIG. 4.
Detection of IFIT3 by Western blotting (24 h). (A) Adherent cell lines. Lanes: M, molecular weight marker; 1, A549, negative control, untreated cells (NC); 2, A549 IFN-α2c (0.036 ng/ml); 3, Huh7 (NC); 4, Huh7 IFN-α2c (3.6 ng/ml); 5, OVCAR-3 (NC); 6, OVCAR-3 (0.36 ng/ml); 7, 2fTGH (NC); 8, 2fTGH (0.36 ng/ml); 9, HeLa (NC); 10, HeLa (0.36 ng/ml). (B) Suspension cell lines. Lanes: M, molecular weight marker; 1, Daudi IFN-α2c (3.6 ng/ml), positive control; 2, monocyte (NC); 3, monocyte IFN-α2c (0.36 ng/ml); 4, B-JAB (NC); 5, B-JAB (0.36 ng/ml); 6, U-937 (NC); 7, U-937 (0.36 ng/ml); 8, Jurkat (NC); 9, Jurkat (0.36 ng/ml).
AV protein expression after viral infection.
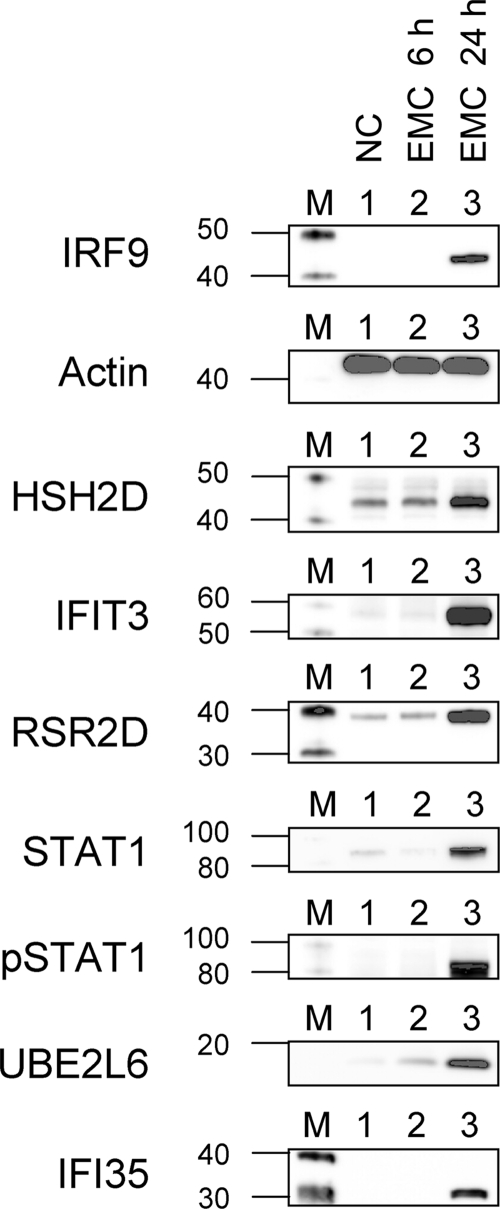
Treatment of Daudi cells with EMCV leads to induction of type I IFN, followed by induction of IFN-stimulated genes. All early gene products previously detected after treatment of Daudi cells with IFN-α2c and HY-2 were detected 24 h after EMCV treatment (Fig. 5). Interestingly, EMCV did not replicate in Daudi cells and did not cause cell death. EMCV infection did, however, induce high levels of type I IFN. The presence of induced IFN inhibited cell proliferation at 48 h after EMCV treatment, as confirmed by using anti-IFNAR2 neutralizing antibody (data not shown).
FIG. 5.
Detection of AV proteins at 6 and 24 h after EMCV infection. Lanes: M, molecular weight marker (MWM); 1, negative control (NC), no virus present; 2, cells 6 h after EMCV treatment; 3, cells 24 h after EMCV infection.
WB analysis of Daudi cells incubated with VSV and 2.5 IU of IFN-α2a/ml (the concentration allowing maximal AV protection without an AP effect) showed that VSV had no effect on the levels of proteins in 6 h after IFN and virus treatment. Interestingly, a decreased level of Ube2L6 was observed in cells treated with VSV only. In addition, a significant decrease in levels of all examined genes was observed 12 h after IFN and VSV treatment (data not shown).
RNA interference and overexpression of IFIT3.
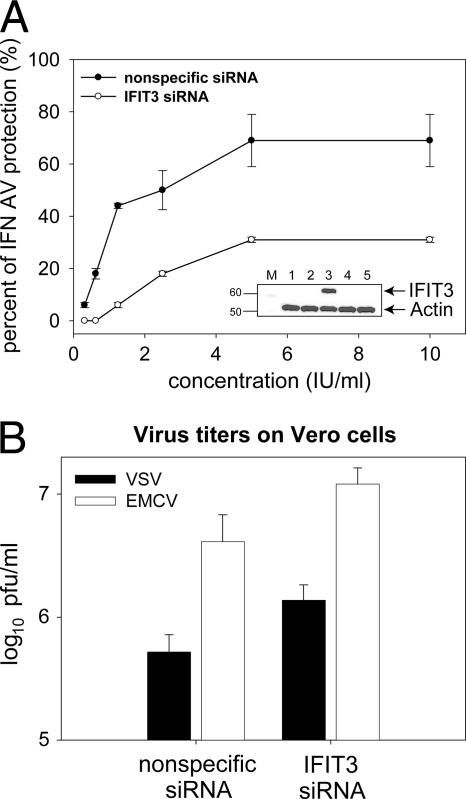
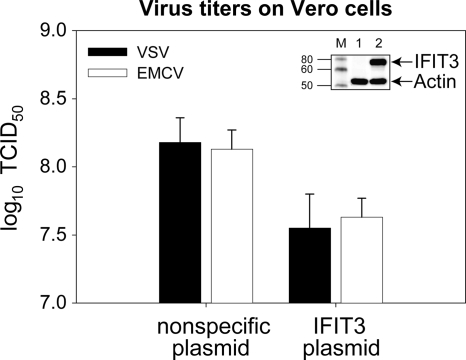
To further elucidate the role of IFIT3 in AV activity of IFN, siRNA knock-down experiments were performed. Because it is not possible to transfect Daudi cells with siRNA, A549 cells, which are similarly sensitive to the AV activity of type I IFN and have similar gene products (data not shown), were substituted. The addition of a specific siRNA for IFIT3 prior to IFN treatment resulted in full inhibition of IFN AV activity against VSV and partial inhibition against EMCV compared to nonspecific (NS) siRNA (Fig. 6A). A concentration of 10 IU of IFN-α2a/ml and 2-fold dilutions from that concentration were selected to avoid masking the IFIT3 siRNA inhibition induced by higher concentrations of IFN. The IFIT3 knock-down efficiency was confirmed by WB (Fig. 6A). Finally, a comparison of virus titers derived from selected samples treated with nonspecific versus specific siRNA for IFIT3 confirmed the decreased efficiency of IFN AV effect (Fig. 6B). Overexpression of IFIT3 in Vero cells resulted in a decrease in the virus titers of both VSV and EMCV (Fig. 7).
FIG. 6.
IFIT3 RNA interference decreases efficiency of IFN AV effect. (A) Influence of IFIT3 siRNA on IFN AV protection against EMCV (expressed as percentage of cells protected by IFN from EMCV) and analysis of the efficiency of IFIT3 RNA interference by WB (inset). Lanes: M, molecular weight marker; 1, untreated cells; 2, nonspecific siRNA (20 nM); 3, nonspecific siRNA + IFN-α2a (2.5 IU/ml); 4, IFIT3 siRNA (20 nM); 5, IFIT3 siRNA + IFN-α2a (2.5 IU/ml). (B) Analysis of efficiency of IFIT3 RNA interference by plaque assay. To analyze effect of IFIT3-siRNA transfection on virus titers, cell supernatants were harvested after treatment with 5 IU of IFN-α2a/ml 24 h postinfection. The virus titer was examined by plaque assay on Vero cells.
FIG. 7.
Overexpression of IFIT3 decreases virus titers. Influence of IFIT3 on VSV and EMCV virus titers in Vero cells, and analysis of IFIT3 overexpression by WB (inset). Lanes: M, molecular weight marker; 1, cells transfected with plasmid pIRESpuro3; 2, cells transfected with plasmid pIRESpuro3-IFIT3.
DISCUSSION
Gene expression microarray analysis is a widely used method of identifying genes that are induced by IFN (5, 13, 16, 23, 28, 34, 40). However, most of these studies use high concentrations of IFN for treatment of cells. Since IFNs are produced in mammalian cells at picomolar concentrations, and these concentrations are sufficient for AV protection, it is important to identify genes and proteins responsible for the AV activities of IFNs under these conditions. Separation of IFN antiviral genes from genes with other functions has been challenging. Moreover, identification of IFN-induced genes under conditions for antiviral protection but no growth inhibition has not previously been reported. The MTT assay was modified to identify conditions of AV protection, but not growth inhibition, on Daudi cells. Gene expression profiles were then examined using these conditions at 6 and 24 h. The use of two different IFNs (HY-2 and IFN-α2c) helped to narrow down the list of genes and resulted in the identification of 25 gene candidates associated with AV protection in Daudi cells.
Some of the upregulated genes we identified, including PKR, MxA, OAS1, OAS2, UBE2L6, IRF7, IRF9, STAT1, RSAD2, SP100, IFI 35, BST2, and IFITM1, have previously been reported as associated with IFN AV function (2, 4, 14, 20, 24-26, 29, 33). Further, a connection between IFN and the genes IFI6, IFI44, TREX-1, IFIT3, PARP12, FLJ11286, IFI44L, IFI27, and LY6E has been previously reported (5, 13, 16, 23, 28, 34, 39, 40). However, the role of these genes in type I IFN AV activity was not described. HSH2D is reported for the first time as IFN-stimulated gene.
IFIT3 was originally described as an all-trans-retinoic acid (ATRA)- and IFN-induced gene in NB4 cells, which were derived from a patient with acute promyelocytic leukemia (38). An association between ATRA and IFIT3 could be explained based on the fact that ATRA induces IFN-α (22). As our results showed, IFIT3 was the most upregulated among all genes at 6 and 24 h in Daudi cells after IFN-α treatment. In addition, the presence of this protein after IFN-α treatment in numerous cell lines (A549, Huh7, OVCAR-3, 2fTGH, HeLa, Daudi, monocyte, B-JAB, U937, and Jurkat cells) suggested the importance of IFIT3 in the mechanism of IFN actions (36). IFIT3 belongs to the IFI54/IFIT2 family (6). Subcellular distribution of IFIT3 showed a diffuse distribution pattern in cytoplasm (38). Analogues of IFIT3 were identified in different species (6). The ubiquity of this protein during the presence of IFN suggests IFIT3 may be an excellent biomarker of IFN action.
HSH2D was not previously reported as being upregulated by type I IFN. It is a protein that was found to protect the WEHI-231 B-cell line from undergoing apoptosis in response to B-cell antigen receptor complex (BCR) ligation through its ability to directly or indirectly promote mitochondrial stability (9). Our results, together with those of several earlier studies, suggest that HSH2D is expressed in lymphoid, as well as myeloid, lineage cells (7, 21). It has been hypothesized that HSH2D may be involved in cytokine-induced signaling (7). Expression of human HSH2D was shown to inhibit interleukin-2 and promote activation in Jurkat T cells (7). Similar to STAT1, HSH2D contains an SH2 domain (19). It has been shown that the carboxyl-terminal segment of HSH2D is critical for the trafficking of HSH2D between cytoplasm and nucleus. It has also been reported that nuclear export is a CRM1-dependent process (27). However, the role of HSH2D in IFN AV function remains to be elucidated.
In conclusion, our novel MTT AV/AP assay is the first to allow examination of IFN AP and AV activities simultaneously on a suspension cell line. This assay is compatible with a broad range of IFNs and is applicable to different cell lines (e.g., OVCAR-3 [data not shown]). The microarray analysis of samples developed by using the MTT AV/AP assay allowed identification of 25 genes associated with IFN AV activity; 10 of these 25 genes have not been previously reported as linked to AV activity of IFN. HSH2D is reported for the first time as a gene being upregulated in response to IFN treatment. IFIT3 was the most upregulated gene. The siRNA knockdown of IFIT3 results in increased sensitivity of A549 to two different viruses. Overexpression of IFIT3 in VERO cells led to a decrease in virus titer after infection. Thus, the results of the present study suggested IFIT3 as a key element of IFN-α AV activity. To better understand the IFN AV signaling pathway, future work will be required to shed light on the function of these genes. Finally, uncoupling of AV and AP effects may be important for maximizing efficacy and minimizing adverse effects with IFN treatment for a variety of chronic viral infections and cancers.
Acknowledgments
We thank Huiqin Nie (National Institutes of Health/National Institute of Allergy and Infectious Disease [NIH/NIAID]) for technical help in pilot MTT/AV assay on Daudi cells; J. Hartley, D. Esposito, and W. Gillette (SAIC) for expression and purification of IFN-α2c and HY-2; H. Young (National Cancer Institute), S. Kottilil (NIAID), F. Schmeisser (U.S. Food and Drug Administration), J. Bekisz (NIAID), K. Miller (NIAID), and the NIH Fellows Editorial Board for reviewing the manuscript and valuable discussions. Biodefense and Emerging Infections Research Resource Repository (BEI) supplied the IFN international standard.
This research was supported by the Intramural Research Program of the NIH (NIAID).
Footnotes
Published ahead of print on 4 August 2010.
REFERENCES
- 1.Armstrong, J. A. 1981. Cytopathic effect inhibition assay for interferon: microculture plate assay. Methods Enzymol. 78:381-387. [DOI] [PubMed] [Google Scholar]
- 2.Brass, A. L., I.-C. Huang, Y. Benita, S. P. John, M. N. Krishnan, E. M. Feeley, B. J. Ryan, J. L. Weyer, L. van der Weyden, E. Fikrig, D. J. Adams, R. J. Xavier, M. Farzan, and S. J. Elledge. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chieux, V., W. Chehadeh, P. Hautecoeur, J. Harvey, P. Wattre, and D. Hober. 2001. Increased levels of antiviral MxA protein in peripheral blood of patients with a chronic disease of unknown etiology. J. Med. Virol. 65:301-308. [DOI] [PubMed] [Google Scholar]
- 4.Deblandre, G. A., O. P. Marinx, S. S. Evans, S. Majjaj, O. Leo, D. Caput, G. A. Huez, and M. G. Wathelet. 1995. Expression cloning of an interferon inducible 17-kDa membrane protein implicated in the control of cell growth. J. Biol. Chem. 270:23860-23866. [DOI] [PubMed] [Google Scholar]
- 5.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Veer, M. J., H. Sim, J. C. Whisstock, R. J. Devenish, and S. J. Ralph. 1998. IFI60/ISG60/IFIT4, a new member of the human IFI54/IFIT2 family of interferon-stimulated genes. Genomics 54:267-277. [DOI] [PubMed] [Google Scholar]
- 7.Greene, T. A., P. Powell, C. Nzerem, M. J. Shapiro, and V. S. Shapiro. 2003. Cloning and characterization of ALX, an adaptor downstream of CD28. J. Biol. Chem. 278:45128-45134. [DOI] [PubMed] [Google Scholar]
- 8.Han, J., H. Lee, N. Y. Nguyen, S. L. Beaucage, and R. K. Puri. 2005. Novel multiple 5′-amino-modified primer for DNA microarrays. Genomics 86:252-258. [DOI] [PubMed] [Google Scholar]
- 9.Herrin, B. R., A. L. Groeger, and L. B. Justement. 2005. The adaptor protein HSH2 attenuates apoptosis in response to ligation of the B-cell antigen receptor complex on the B lymphoma cell line, WEHI-231. J. Biol. Chem. 280:3507-3515. [DOI] [PubMed] [Google Scholar]
- 10.Hu, R., Y. Gan, J. Liu, D. Miller, and K. C. Zoon. 1993. Evidence for multiple binding sites for several components of human lymphoblastoid interferon-α. J. Biol. Chem. 268:12591-12595. [PubMed] [Google Scholar]
- 11.Hu, R., J. Bekisz, M. Hayes, S. Audet, J. Beeler, E. Petricoin, and K. Zoon. 1999. Divergence of binding, signaling, and biological responses to recombinant human hybrid IFN. J. Immunol. 163:854-860. [PubMed] [Google Scholar]
- 12.Hu, R., J. Bekisz, H. Schmeisser, P. McPhie, and K. Zoon. 2001. Human IFN-α protein engineering: the amino acid residues at positions 86 and 90 are important for antiproliferative activity. J. Immunol. 167:1482-1489. [DOI] [PubMed] [Google Scholar]
- 13.Jaitin, D. A., and G. Schreiber. 2007. Upregulation of a small subset of genes drives type I interferon-induced antiviral memory. J. Interferon Cytokine Res. 27:653-664. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, D., H. Guo, Ch. Xu, J. Chang, B. Gu, L. Wang, T. M. Block, and J. T. Guo. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82:1665-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kessler, D. S., D. E. Levy, and J. E. Darnell. 1988. Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc. Natl. Acad. Sci. U. S. A. 85:8521-8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khabar, K. S. A., L. Al-Haj, F. Al-Zoghaibi, M. Marie, M. Dhalla, S. J. Polyak, and B. R. G. Williams. 2004. Expressed gene clusters associated with cellular sensitivity and resistance toward antiviral and anti-proliferative action of interferon. J. Mol. Biol. 342:833-846. [DOI] [PubMed] [Google Scholar]
- 17.Levy, D. E., D. S. Kessler, R. Pine, N. Reich, and J. E. Darnell. 1988. Interferon-induced nuclear factors that bind a shared promoter element correlated with positive and negative transcriptional control. Genes Dev. 2:383-393. [DOI] [PubMed] [Google Scholar]
- 18.Lou, Y.-J., X.-R. Pan, P.-M. Jia, D. Li, S. Xiao, Z.-L. Zhang, S.-J. Chen, Z. Chen, and J.-H. Tong. 2008. IRF-9/STAT2 functional interaction drives retinoic acid-induced gene G expression independently of STAT1. Cancer Res. 8:3673-3680. [DOI] [PubMed] [Google Scholar]
- 19.Machida, K., and B. J. Mayer. 2004. The SH2 domain: versatile signaling module and pharmaceutical target. Biochim. Biophys. Acta 1747:1-25. [DOI] [PubMed] [Google Scholar]
- 20.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425-430. [DOI] [PubMed] [Google Scholar]
- 21.Oda, T., M. A. Muramatsu, T. Isogai, Y. Masuho, S. Asano, and T. Yamashita. 2001. HSH2: a novel SH2 domain-containing adaptor protein involved in tyrosine kinase signaling in hematopoietic cells. Biochem. Biophys. Res. Commun. 288:1078-1086. [DOI] [PubMed] [Google Scholar]
- 22.Pelicano, O., C. Brumpt, P. M. Pitha, and M. K. Chelbi-Alix. 1999. Retinoic acid resistance in NB4 APL cells is associated with lack of interferon a synthesis STST1 and p48 induction. Oncogene 18:3944-3953. [DOI] [PubMed] [Google Scholar]
- 23.Rani, M. R. S., J. Shrock, S. Appachi, R. A. Rudick, B. R. G. Williams, and R. M. Ransohoff. 2007. Novel interferon-β-induced gene expression in peripheral blood cells. J. Leukoc. Biol. 82:1353-1360. [DOI] [PubMed] [Google Scholar]
- 24.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infection. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 25.Sabile, A., A. Rhodes-Feuillette, F. Z. Jaoui, J. Tobaly-Tapiero, M. L. Giron, J. Lasneret, J. Peries, and M. Canivet. 1996. In vitro studies on interferon-inducing capacity and sensitivity to IFN of human foamy virus. Res. Virol. 147:29-37. [DOI] [PubMed] [Google Scholar]
- 26.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro, M. J., Y.-Y. Chen., and V. S. Shapiro. 2005. The carboxyl-terminal segment of the adaptor protein ALX directs its nuclear export during T cell activation. J. Biol. Chem. 280:38242-38246. [DOI] [PubMed] [Google Scholar]
- 28.Sanda, C., P. Weitzel, T. Tsukahara, J. Schaley, H. J. Edenberg, M. A. Stephens, J. N. McClintick, L. M. Blatt, L. Li, L. Brodsky, and M. W. Taylor. 2006. Differential gene induction by type I and type II interferons and their combination. J. Interferon Cytokine Res. 26:462-472. [DOI] [PubMed] [Google Scholar]
- 29.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN inducible transcription factor IRF7. FEBS Lett. 441:106-110. [DOI] [PubMed] [Google Scholar]
- 30.Schanen, C., V. Chieux, P. E. Lobert, J. Harvey, and D. Hober. 2006. Correlation between the anti-virus-induced cytopathic effect activity of interferon-α subtypes and induction of MxA protein in vitro. Microbiol. Immunol. 50:19-24. [DOI] [PubMed] [Google Scholar]
- 31.Schmeisser, H., I. Gorshkova, P. H. Brown, P. Kontsek, P. Schuck, and K. C. Zoon. 2007. Two interferons alpha influence each other during their interaction with the extracellular domain of human type interferon receptor subunit 2. Biochemistry 46:14638-14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmeisser, H., P. Kontsek, D. Esposito, W. Gillette, G. Schreiber, and K. C. Zoon. 2006. Binding characteristids of interferon-α subvariants to IFNAR2-EC and influence of the 6-histidine tag. J. Interferon Cytokine Res. 26:866-876. [DOI] [PubMed] [Google Scholar]
- 33.Tan, J., W. Qiao, J. Wang, F. Xu, Y. Li, J. Zhou, Q. Chen, and Y. Geng. 2008. IFP35 is involved in the antiviral function of interferon by association with the viral tas transactivator of bovine foamy virus. J. Virol. 82:4275-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, W. M., T. Tsukahara, J. N. McClintick, H. J. Edenberg, and P. Kwo. 2008. Cyclic changes in gene expression induced by Peg-interferon α2b plus ribavirin in peripheral blood monocytes (PBMC) of hepatitis C patient during the first 10 weeks of treatment. J. Transl. Med. 6:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuno, T., J. Mejido, T. Zhao, H. Schmeisser, A. Morrow, and K. C. Zoon. 2009. IRF-9 is a key factor for eliciting the antiproliferative activity of IFN-α. J. Immunother. 8:803-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao S., D. Li, H.-Q. Zhu, M.-G. Song, X.-R. Pan, P.-M. Jia, L.-L. Peng, A.-X. Dou, G.-Q. Chen, S.-J. Chen, Z. Chen, and J.-H. Tong. 2006. RIG-G as a key mediator of the antiproliferative activity of interferon-related pathways through enhancing p21 and p27 proteins. Proc. Natl. Acad. Sci. U. S. A. 103:16448-16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeh, T. J., P. T. McBridge, J. C. Overall, Jr., and J. A. Green. 1982. Automated, quantitative cytopathic effect reduction assay for interferon. J. Clin. Microbiol. 16:413-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, M., J.-H. Tong, M. Mao, L.-X. Kan, M.-M. Liu, Y.-W. Sun, G. Fu., Y.-K. Jing, L. Yu., D. Lepaslier, M. Lanotte, Z.-Y. Wang, Z. Chen, S. Waxman, Y.-X. Wang, J.-Z. Tan, and S.-J. Chen. 1997. Cloning of a gene (RIG-G) associated with retinoic acid-induced differentiation of acute promyelocytic leukemia cells and representing a new member of a family of interferon-stimulated genes. Proc. Natl. Acad. Sci. U. S. A. 94:7406-7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao, D., D. Peng, L. Li, Q. Zhang, and C. Zhang. 2008. Inhibition of G1P3 expression found in the differential display study on respiratory syncytial virus replication. Virol. J. 5:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmerer, J. M., G. B. Lesinski, A. S. Ruppert, M. D. Radmacher, C. Noble, K. Kendra, M. J., and W. E. Carson. 2008. Gene expression profiling reveals similarities between the in vitro and in vivo responses of immune effectors cells to IFN-α. Cancer Ther. 14:5900-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoon, K. C., D. L. zur Nedden, J. C. Enterline, J. F. Manischewits, D. R. Dyer, R. A. Boykins, J. Bekisz, and T. L. Gerrard. 1986. Chemical and biological characterisation of natural human lymphoblastoid interferon alphas, p. 567-569. In K. Cantell and H. Schellekens (ed.), The biology of the interferon system. Martinus Nijhoff Publishers, Dordrecht, Netherlands.