Abstract
The human papillomavirus type 16 E5 oncoprotein (16E5) enhances acute, ligand-dependent activation of the epidermal growth factor receptor (EGFR) and concomitantly alkalinizes endosomes, presumably by binding to the 16-kDa “c” subunit of the V-ATPase proton pump (16K) and inhibiting V-ATPase function. However, the relationship between 16K binding, endosome alkalinization, and altered EGFR signaling remains unclear. Using an antibody that we generated against 16K, we found that 16E5 associated with only a small fraction of endogenous 16K in keratinocytes, suggesting that it was unlikely that E5 could significantly affect V-ATPase function by direct inhibition. Nevertheless, E5 inhibited the acidification of endosomes, as determined by a new assay using a biologically active, pH-sensitive fluorescent EGF conjugate. Since we also found that 16E5 did not alter cell surface EGF binding, the number of EGFRs on the cell surface, or the endocytosis of prebound EGF, we postulated that it might be blocking the fusion of early endosomes with acidified vesicles. Our studies with pH-sensitive and -insensitive fluorescent EGF conjugates and fluorescent dextran confirmed that E5 prevented endosome maturation (acidification and enlargement) by inhibiting endosome fusion. The E5-dependent defect in vesicle fusion was not due to detectable disruption of actin, tubulin, vimentin, or cytokeratin filaments, suggesting that membrane fusion was being directly affected rather than vesicle transport. Perhaps most importantly, while bafilomycin A1 (like E5) binds to 16K and inhibits endosome acidification, it did not mimic the ability of E5 to inhibit endosome enlargement or the trafficking of EGF. Thus, 16E5 alters EGF endocytic trafficking via a pH-independent inhibition of vesicle fusion.
High-risk human papillomaviruses (HPVs) are the causative agent of cervical cancer (63) and HPV type 16 (HPV-16) is associated with a majority of cervical malignancies worldwide (13). HPV-16 encodes three oncoproteins: E5, E6, and E7. While the contributions of E6 and E7 to cellular immortalization and transformation have been characterized in detail (20), the role of HPV-16 E5 (16E5) is poorly understood (53). Nevertheless, a number of studies suggest that 16E5 does contribute to the development of cervical cancer. Most high-risk HPV types encode an E5 protein (48), and targeted expression of the three HPV-16 oncogenes in basal epithelial cells of transgenic mice (4) leads to a higher incidence of cervical cancer than does the expression of E6 and E7 alone (44). In addition, targeted epithelial expression of 16E5 (without E6 and E7) in transgenic mice induces skin tumors (21). It may be noteworthy that unlike high-risk HPV-18, which integrates into the host DNA and potentially disrupts E5 gene expression (20, 64), the HPV-16 genome often persists in episomal form in malignant lesions (12, 16, 24, 36, 42).
Biological activities of 16E5 that may facilitate carcinogenesis include evading host immune detection by interfering with the transport of antigen-presenting major histocompatibility complex (MHC) class I molecules to the cell surface (6), promoting anchorage-independent growth (33, 41, 52) and disrupting gap junctions responsible for cell-cell communication (37, 58). The 16E5 phenotype most frequently linked to the development of cancer is enhanced ligand-dependent activation of the epidermal growth factor receptor (EGFR) (15, 41, 46, 52). 16E5 stimulates EGF-dependent cell proliferation in vitro (7, 33, 40, 41, 52, 60) and in vivo (21), which might expand the population of basal or stemlike keratinocytes and thereby increase the probability that some of these cells would undergo malignant transformation. A number of studies indicate that 16E5 may enhance ligand-dependent EGFR activation by interfering with the acidification of early endosomes containing EGF bound to activated EGFRs (17, 51, 57). It has been hypothesized that 16E5 inhibits the H+ V-ATPase responsible for maintaining an acidic luminal pH in late endosomes and lysosomes (28) by associating with the V-ATPase 16-kDa “c” subunit (16K) (1, 5, 14, 22, 46) and disrupting assembly of the V-ATPase integral (Vo) and peripheral (Vi) subcomplexes (10). In contrast, Thomsen et al. (57) reported that 16E5 inhibits early endosome trafficking in fibroblasts by completely depolymerizing actin microfilaments.
Due to the unavailability of antibodies that recognize native 16E5 and 16K, direct association of 16E5 with 16K has only been observed by overexpressing epitope-tagged forms of both proteins in vitro (5, 46) or in vivo (1, 14, 22). It is uncertain, therefore, whether these associations occur when the proteins are expressed at “physiological” levels. In yeast, both wild-type 16E5 (10) and several 16E5 mutants that associate with 16K in COS cells (1) inhibit vacuolar acidification, although another study in yeast concludes the opposite (5). 16K is a component of the V-ATPase Vo subcomplex, which is assembled in the endoplasmic reticulum (ER) (28), and 16E5 localizes to the ER and nuclear envelope in epithelial cells (32, 54). Thus, the export of Vo from the ER could potentially be inhibited by a significant level of 16K binding to 16E5, although the differential alkalinization of endosomes rather than the Golgi apparatus (17) would require specificity for those proton pumps directed to those sites.
In the present study, we generated an antibody against native 16K and used it to determine whether 16K/16E5 complexes formed in primary keratinocytes. We also synthesized a new pH-sensitive fluorescent EGF conjugate to evaluate whether there was a correlation between E5-induced EGFR activation, trafficking and endosome alkalinization. Finally, we simultaneously monitored EGFR endocytic trafficking (using pH-insensitive fluorescent EGF), endosome fusion (using fluorescent EGF and dextran), and the status of cellular filaments and microtubules to evaluate whether E5 might disrupt some of these structures that mediate vesicle transport.
MATERIALS AND METHODS
Antibodies and fluorescent probes.
Following the protocol of Chen et al. (11), we generated a rabbit polyclonal antiserum that can immunoprecipitate 16K. The antiserum was produced by New England Peptide (Gardner, MA) using a 16K-derived peptide, C36KSGTGIAAMSVMRPEQ51, that was conjugated to keyhole limpet hemocyanin. A rabbit polyclonal antibody that recognizes the AU1 epitope tag (DTYRYI) (34) was purchased from Covance (Princeton, NJ). 12CA5 mouse ascites fluid recognizing the influenza virus hemagglutinin (HA) epitope tag (YPYDVPDYASL) (38) was a gift from J. Bolen (Millennium Pharmaceuticals, Cambridge, MA). Anti-EGFR rabbit polyclonal and anti-phosphotyrosine mouse monoclonal (clone 4G10) antibodies were obtained from Millipore (Temecula, CA).
Custom synthesis of the pH-sensitive EGF conjugate, pHrodo-EGF, was performed by Molecular Probes (Eugene, OR). Briefly, the amine-reactive succinimidyl ester form of pHrodo dye was covalently linked to streptavidin, which was then complexed with EGF that has a single biotin molecule attached to its N terminus. Alexa Fluor 488-EGF complex, Alexa Fluor 594-anionic fixable dextran (10,000 molecular weight), LysoTracker Red, LysoTracker Yellow, and Alexa Fluor 488-phalloidin are commercially available from the same supplier.
Cells and viruses.
Retroviruses encoding codon-optimized 16E5 with an N-terminal AU1 epitope tag in the vector pLXSN (18) were generated by using the Phoenix cell system (39).
Primary human foreskin keratinocytes (HFKs) were isolated from neonatal foreskins as described previously (49) and were grown as monolayer cultures at 37°C and 5% CO2 in keratinocyte growth medium (KGM; Invitrogen, Carlsbad, CA) containing gentamicin sulfate (10 μg/ml). HFKs expressing 16E5 (or harboring the empty pLXSN expression vector) were generated by retroviral infection (31) and selection in the presence of Geneticin G418 (100 μg/ml). For some experiments, cells were cultured in the absence of EGF by removing the KGM, washing twice with Dulbecco's phosphate-buffered saline (PBS), and adding keratinocyte serum-free medium without supplements (KSFM; Invitrogen) for 24 h.
COS cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum, 100 U of penicillin G/ml, and 100 μg of streptomycin sulfate (Invitrogen)/ml. Cells were transfected with AU1 epitope-tagged, codon-optimized 16E5 in the pJS55 expression vector (50) or with HA epitope-tagged 16K in the expression vector pSVL (2), using Lipofectamine 2000 (Invitrogen) as previously described (30).
Metabolic labeling, immunoprecipitation, and immunoblotting.
Ten-centimeter tissue culture dishes of HFKs (or COS cells 24 h after transfection) were washed with PBS (15 ml per dish) and then incubated for 2 h in DMEM without cysteine and methionine (4 ml per dish; Invitrogen). [35S]methionine/[35S]cysteine protein labeling mix (Easy Tag; Perkin-Elmer, Waltham, MA) was added (200 μCi per dish) for 3 h. Proteins were immunoprecipitated from cell lysates, separated on sodium dodecyl sulfate (SDS)-polyacrylamide mini-gels (Invitrogen), and transferred to Immobilon-P membranes (Millipore) as previously described (55). 35S-labeled proteins were detected by autoradiography using BioMax MS film and low-energy intensity intensifying screens (Eastman Kodak, Rochester, NY) at −70°C. Membranes were labeled with 12CA5 anti-HA ascites fluid (1:5,000 dilution) as described previously (55). Ten-centimeter tissue culture dishes of HFKs (in KSFM) were processed for analysis of EGFR tyrosine phosphorylation as described previously for the platelet-derived growth factor receptor (56). The relative intensities of polypeptides on exposed films were measured by analysis of scanned images using Kodak MI software.
Immunofluorescence microscopy.
Procedures for the transfection, paraformaldehyde fixation, saponin permeabilization, and immunolabeling of COS cells grown on 22-by-22-mm glass coverslips have been described in detail (32). The following primary antibodies were used: 1:1,500-diluted rabbit anti-AU1 polyclonal antibody (Covance), 1:300-diluted anti-tubulin mouse monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), 1:150-diluted anti-vimentin mouse monoclonal antibody (Santa Cruz Biotechnology), and 1:1,000-diluted anti-pan-keratin mouse monoclonal antibody (Cell Signaling Technology, Danvers, MA), which reacts with keratins 4, 5, 6, 8, 10, 13, and 18. To label actin filaments, cells on coverslips were fixed and washed as described above but were permeabilized with PBS containing 0.1% (wt/vol) Triton X-100 (5 min), blocked with PBS containing 1% (wt/vol) bovine serum albumin (BSA; 20 min), and labeled with PBS-BSA containing Alexa Fluor 488-phalloidin (8 U/ml). A Zeiss Axioskop microscope (Carl Zeiss Microimaging, Thornwood, NJ) equipped with a 63× oil immersion objective lens, Orca-ER charge-coupled device camera (Hamamatsu Corp., Bridgewater, NJ), and Openlab digital imaging software (Perkin-Elmer) was used for microphotography.
Alexa Fluor 488-EGF (0.5 μg/ml) and pHrodo-EGF (2 μg/ml) were prebound to EGF-starved HFKs on coverslips (60 min at 4°C) in KSFM containing 1% (wt/vol) BSA. Cells were then washed twice with PBS at 4°C before adding KSFM and transferring the cells to a 37°C tissue culture incubator. Where indicated, KSFM was supplemented with 0.33 μM bafilomycin A1 (Enzo Life Sciences, Plymouth Meeting, PA) by the addition of a 100 μM stock solution dissolved in (CH3)2SO. At intervals, coverslips were inverted over 18 μl of 37°C KSFM containing 20 μM Hoechst dye 33342 (Sigma, St. Louis, MO) on a microscope slide for fluorescence imaging of live cells (within 5 min) using a 40× objective lens. For high-resolution imaging using a 63× oil immersion objective lens (Alexa Fluor 488-EGF only), cells were washed with PBS, fixed with paraformaldehyde, and mounted on slides as described above. The size of EGF-containing endosomes was determined by using Kodak MI software. Cells on coverslips were labeled with 75 nM LysoTracker Red (in KSFM) for 60 min at 4°C, followed by 30 to 60 min at 37°C and paraformaldehyde fixation.
To measure endosome fusion, Alexa Fluor 488-EGF (1.0 μg/ml) was prebound to EGF-starved HFKs on coverslips at 4°C as described above. 10 min after warming to 37°C, the cells were pulsed with Alexa Fluor 594-fixable dextran (0.5 mg/ml) for 10 min. The coverslips were then placed on ice, washed twice with PBS at 4°C, and shifted back to 37°C for 10 to 25 min before paraformaldehyde fixation and mounting on slides.
Flow cytometry.
Cells harvested from 75-cm2 tissue culture flasks using standard trypsin-EDTA treatment were collected by centrifugation for 5 min at 250 × g and resuspended in KSFM (or KGM) at 4°C. Aliquots containing 2.5 × 105 cells were transferred to siliconized microcentrifuge tubes, centrifuged for 3 min at 250 × g, and resuspended in 1 ml of staining buffer (PBS containing 0.2% [wt/vol] BSA, 1 mM MgCl2, and 0.09% [wt/vol] NaN3) at 4°C. The cells were centrifuged again and resuspended in 0.4 ml of staining buffer containing 2 μg of Alexa Fluor 488-EGF/ml or in 0.2 ml of staining buffer containing 20 μl of R-phycoerythrin-conjugated anti-EGFR mouse monoclonal antibody (BD Biosciences, San Jose, CA) for 30 min at 4°C. After two washes with 1 ml of staining buffer (at 4°C), the cells were resuspended in 0.5 ml of staining buffer containing 0.5% (wt/vol) paraformaldehyde for analysis using a FACSStar Plus dual-laser system (Becton Dickinson, San Jose, CA). At least 40,000 events were recorded for each sample.
Adherent cultures of 16E5- and LXSN-HFKs (75-cm2 tissue culture flasks containing equal numbers of cells) were labeled with LysoTracker Yellow for 60 min at 37°C, harvested using trypsin-EDTA treatment (as described above), and resuspended in 0.75 ml per flask staining buffer containing 0.5% paraformaldehyde for flow cytometry analysis.
RESULTS
16E5 association with 16K.
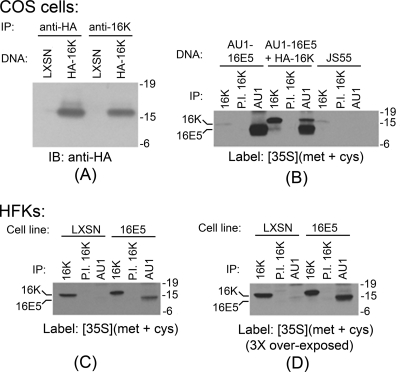
Our initial experiment was designed to verify the specificity of the 16K antibody. Lysates were prepared from COS cells transfected with DNA encoding HA epitope-tagged 16K or with the empty expression vector. As shown in Fig. 1 A, both anti-HA and anti-16K antibodies immunoprecipitated a 16-kDa polypeptide only from cells transfected with 16K DNA. Moreover, the polypeptide immunoprecipitated by anti-16K serum contained the HA epitope.
FIG. 1.
Significant 16E5 binding to 16K occurs only when both proteins are highly overexpressed. (A) Antibodies recognizing both the HA epitope tag and 16K immunoprecipitate a 16-kDa polypeptide, which is HA positive on immunoblots specifically from COS cells transfected with HA epitope-tagged 16K DNA. Molecular mass markers (in kilodaltons) are shown on the right. IP, immunoprecipitation; IB, immunoblotting. (B) Association of 16E5 and 16K in metabolically labeled COS cells transfected with AU1 epitope-tagged 16E5 (AU1-16E5), AU1-16E5 and HA epitope-tagged 16K (HA-16K), or the empty pJS55 expression vector (JS55). Immunoprecipitations (IP) were performed with anti-16K antiserum (16K), preimmune serum from the same rabbit (P.I. 16K), or anti-AU1 antibody (AU1). (C) Immunoprecipitates (IP) from lysates of metabolically labeled HFKs stably expressing AU1-tagged 16E5 or harboring the empty pLXSN expression vector. The autoradiography exposure time was identical to that in panel B. (D) Threefold longer exposure of panel C. In all cases, immunoprecipitations were performed on cell lysates containing equal amounts of protein.
Because the anti-16K antibody was not useful for immunoblotting or immunofluorescence microscopy, we used it to immunoprecipitate 16K from [35S]methionine-labeled COS cells to demonstrate that 16E5 associated with 16K when both proteins were highly overexpressed. This method detects endogenous 16K and shows that its expression increases 15-fold after transfection with the plasmid encoding 16K (Fig. 1B). In addition, 16E5 is easily visible in anti-16K immunoprecipitates from cells transfected with both 16K and AU1 epitope-tagged 16E5 (Fig. 1B). In the same cells, a majority (62%) of total 16K coprecipitates with 16E5 (Fig. 1B). This result confirms previous reports of 16K binding to 16E5 when both proteins were highly overexpressed (1, 5, 14, 22, 46, 55). In contrast, the association of 16K with 16E5 was markedly decreased when these proteins were immunoprecipitated from HFKs that stably express lower levels of 16E5. For example, there was no apparent interaction of 16E5 and 16K when gels were exposed to film for the same time as those for the COS cell experiments (Fig. 1C). However, if the gels were exposed for longer times, only a small fraction of endogenous 16K (ca. 5%) coprecipitated with 16E5 (Fig. 1D). This indicates that 16E5 binding to 16K is dependent upon its relative concentration.
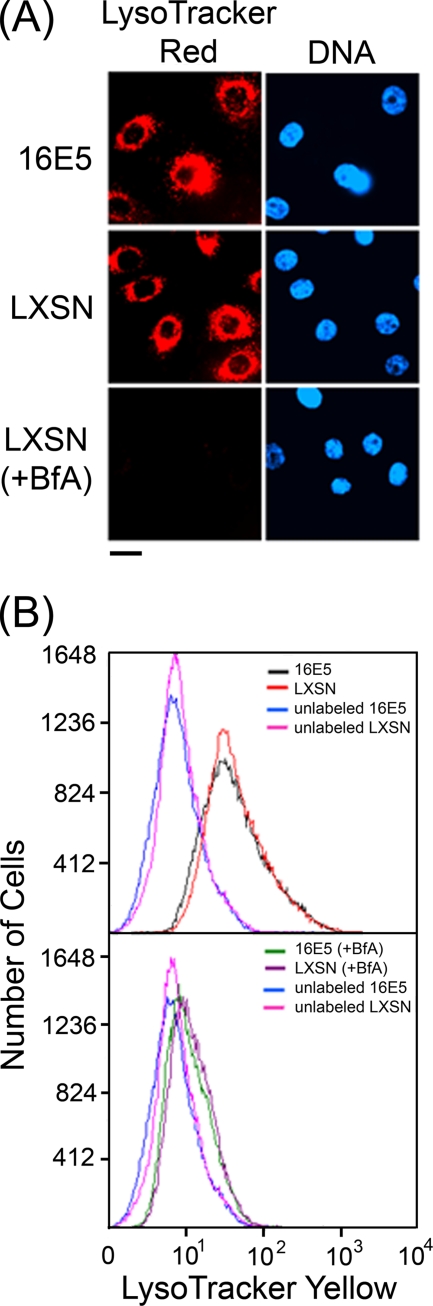
Although we do not know the “physiologic” level of E5 expression in cells, we speculated that the low level of E5/16K binding observed in the HFKs might not be sufficient to generally inhibit the acidification of intracellular compartments. This idea is supported by the observation that 16E5-expressing and control (LXSN) HFKs are labeled with the same pattern (Fig. 2 A) and to the same extent (Fig. 2B) by the acidophilic fluorescent probes LysoTracker Red and LysoTracker Yellow. In contrast, LysoTracker labeling is undetectable (Fig. 2) if the HFKs are treated with bafilomycin A1, a highly specific H+ V-ATPase inhibitor (9) that binds to 16K (8). Clearly, 16E5 is incapable of alkalinizing the major acidified cellular compartments.
FIG. 2.
16E5 does not generally inhibit organelle acidification. (A) Acidic compartments in EGF-starved HFKs stably expressing 16E5 or harboring the empty pLXSN expression vector were labeled with LysoTracker Red for 60 min at 37°C. Where indicated, LXSN-HFKs were treated with 0.33 μM bafilomycin A1 during labeling (+BfA). Nuclei were costained with Hoechst dye 33342 (DNA). Cells were imaged, using a fluorescence microscope. Scale bar, 20 μm. (B) Flow cytometry of EGF-starved 16E5- and LXSN-HFKs labeled with LysoTracker Yellow as described in Materials and Methods. Where indicated, cells were treated with 0.33 μM bafilomycin A1 during labeling (+BfA). The geometric mean fluorescence values were as follows: 37 (16E5-HFKs), 39 (LXSN-HFKs), 11 (16E5-HFKs + BfA), 12 (LXSN-HFKs + BfA), 7 (unlabeled 16E5-HFKs), and 8 (unlabeled LXSN-HFKs).
16E5 enhances acute EGFR activation.
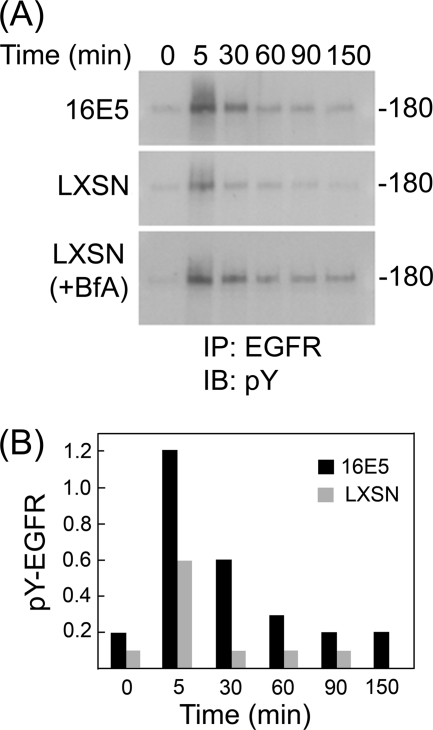
Enhanced ligand-dependent activation of the EGFR by 16E5 has been postulated to result from impaired endosome acidification due to the association of 16E5 with 16K. Since we found that 16E5 does not exhibit significant binding to 16K and does not generally inhibit organelle acidification in HFKs, we sought to determine whether 16E5 nevertheless enhances ligand-dependent activation of the EGFR in these cells. For this analysis, EGF was prebound to EGF-starved HFKs at 4°C. The cells were then washed to remove excess EGF and were warmed to 37°C for various periods of time prior to lysis in the presence of SDS and immunoprecipitation of EGFRs. Western blots of the immunoprecipitates were labeled with an anti-phosphotyrosine antibody to detect activated EGFRs. As shown (Fig. 3), initial activation of the EGFR (5 min at 37°C) is 2-fold higher in 16E5-expressing HFKs compared to control (LXSN) cells. Moreover, 16E5 greatly slows subsequent EGFR inactivation: levels of active EGFRs are elevated 6-fold after 30 min, 3-fold after 60 min and 2-fold after 150 min. Bafilomycin A1 similarly alters the kinetics of EGFR inactivation in control cells to an even greater extent (Fig. 3A). These results confirm that both 16E5 expression and bafilomycin-induced H+ V-ATPase inhibition enhance acute ligand-dependent EGFR activation in HFKs.
FIG. 3.
16E5 and bafilomycin A1 enhance ligand-dependent EGFR activation. (A) Recombinant human EGF (100 ng/ml; Invitrogen) was prebound to EGF-starved HFKs expressing 16E5 (or harboring the empty pLXSN expression vector) for 60 min at 4°C. The levels of EGFR tyrosine phosphorylation subsequently were determined before (0 min) and up to 150 min after warming to 37°C using anti-phosphotyrosine immunoblots (IB) of EGFR immunoprecipitates (IP). For LXSN (+BfA), 0.33 μM bafilomycin A1 was added to LXSN-HFKs as they were shifted to 37°C. Molecular mass marker (in kilodaltons) is shown on the right. (B) Quantitative analysis of panel A using densitometry.
Measurement of endosome acidification using pHrodo-EGF.
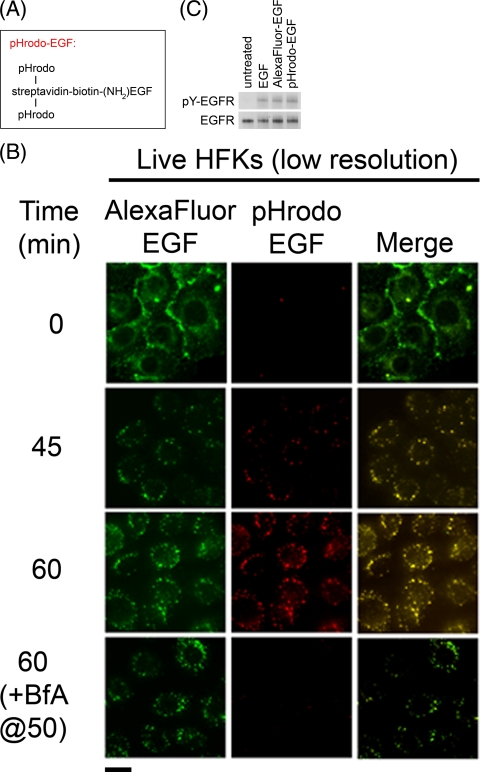
To determine whether prolonged EGFR activation correlates with impaired acidification of EGF-containing endosomes in HFKs, it was important to develop an EGF conjugate that was fluorescent only in acidic environments. We therefore covalently linked the pH-sensitive red fluorescent dye, pHrodo, to streptavidin and complexed the streptavidin with recombinant human EGF that is substituted with a single biotin molecule at its amino terminus (pHrodo-EGF; Fig. 4 A). When pHrodo-EGF and Alexa Fluor 488-EGF, which emits pH-independent green fluorescence, were bound to EGF-starved LXSN-HFKs at 4°C and visualized (in live cells) by fluorescence microscopy, only green fluorescence was detected, a finding consistent with the neutral pH of the culture medium (Fig. 4B, 0 min). After shifting the HFKs to 37°C for 45 min, a punctate pattern of intracellular red and green fluorescence was observed, consistent with endocytosis of the EGF conjugates and acidification of EGF-containing endosomes (Fig. 4B). pHrodo-EGF fluorescence was even stronger after 60 min but could be completely eliminated by adding bafilomycin A1 to the cells for 10 min (Fig. 4B), which is consistent with the rapid neutralization of acidic organelles that accompanies H+ V-ATPase inhibition (17, 47). The complete merging of red and green fluorescence patterns at 45 and 60 min indicates that both EGF conjugates localize to the same intracellular compartments (Fig. 4B). In addition, since both pHrodo-EGF and Alexa Fluor 488-EGF activate the EGFR to the same extent as unconjugated EGF (Fig. 4C), they both maintain biological activity. Thus, pHrodo-EGF is a useful probe for monitoring the pH of EGF-containing endosomes in live cells.
FIG. 4.
pHrodo-EGF measures the acidification of EGF-containing endosomes in live cells. (A) Design of pHrodo-EGF. (B) Trafficking and acidification of endosomes in unfixed EGF-starved LXSN-HFKs labeled with Alexa Fluor 488-EGF and pHrodo-EGF at 4°C before (0 min) and up to 60 min after warming to 37°C. Where indicated, 0.33 μM bafilomycin A1 (BfA) was added to cells after 50 min at 37°C, followed by imaging 10 min later. Scale bar, 20 μm. (C) Fluorescent EGF conjugates are biologically active. Anti-phosphotyrosine immunoblot of EGFR immunoprecipitates (pY-EGFR) from EGF-starved HFKs that were treated for 5 min at 37°C with recombinant human EGF, Alexa Fluor 488-EGF, or pHrodo-EGF (all at a concentration of 960 nM). The immunoblot subsequently was stripped and relabeled to detect total EGFR in the immunoprecipitates.
16E5 inhibits endosome trafficking and acidification.
pHrodo-EGF was used to monitor the acidification kinetics of EGF-containing endosomes in 16E5-expressing and control (LXSN) HFKs, whereas Alexa Fluor 488-EGF was used to simultaneously monitor the trafficking (transport and/or fusion) of the same endosomes. For this analysis, both EGF conjugates were bound to EGF-starved cells at 4°C, followed by the removal of any unbound conjugate, transfer of the cells to 37°C, and fluorescence imaging without fixation.
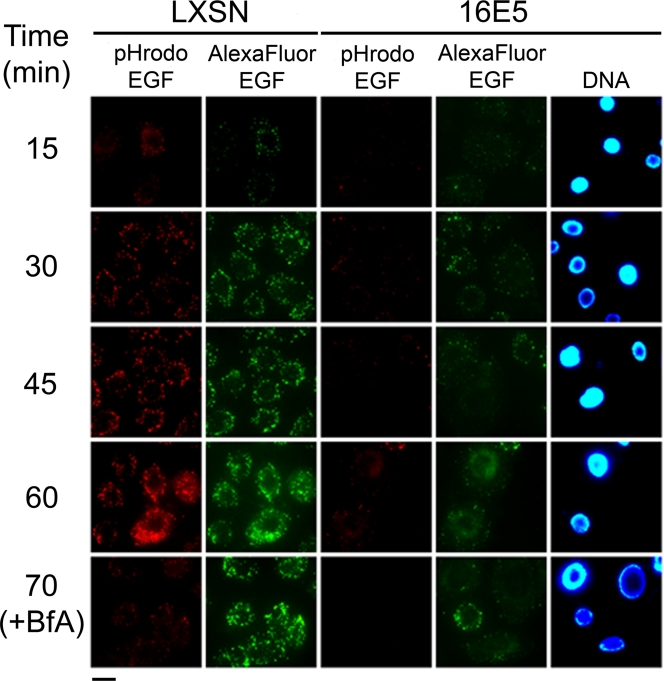
In LXSN-HFKs, weak acidification of some EGF-containing endosomes was observed after 15 min at 37°C. Endosomal acidification dramatically increased over the next 45 min and coincided with the formation of numerous larger EGF-containing vesicles (Fig. 5) due to the fusion of early endosomes with acidic intracellular compartments that represent later stages of the endocytic pathway (23, 26). Importantly, inhibition of the H+ V-ATPase by bafilomycin A1 did not interfere with the generation of these large, perinuclear vesicles containing Alexa Fluor 488-EGF (70 min at 37°C, Fig. 5), indicating that bafilomycin did not interfere with endosome maturation. Thus, H+ V-ATPase activity is not required for the trafficking of EGF-containing endosomes in HFKs, in contrast to some other systems where this proton pump is necessary for membrane trafficking (27, 62).
FIG. 5.
16E5 inhibits endosome acidification and trafficking. Trafficking and acidification of endosomes in unfixed EGF-starved HFKs expressing 16E5 (or empty pLXSN vector) after labeling with Alexa Fluor 488-EGF and pHrodo-EGF at 4°C and warming to 37°C for up to 70 min was examined. Where indicated, 0.33 μM bafilomycin A1 was added to cells as they were shifted to 37°C (+BfA). Scale bar, 20 μm.
In contrast to bafilomycin, 16E5 inhibited both the acidification and the trafficking of EGF-containing endosomes in HFKs. Prebound pHrodo-EGF did not become highly fluorescent after warming 16E5-HFKs to 37°C, indicating that endosome acidification did not occur, and early endosomes containing Alexa Fluor 488-EGF did not undergo fusion with later endocytic compartments to form larger fluorescent structures (Fig. 5). The most straightforward interpretation of these results is that 16E5 prevents the acidification of early endosomes containing EGF by inhibiting their subsequent fusion to acidic vesicles of the endocytic pathway. Moreover, 16E5 interferes with endocytic membrane trafficking through a mechanism that is independent of endosome acidification, since bafilomycin-induced endosome alkalinization does not perturb endocytic trafficking.
16E5 inhibits endosome fusion.
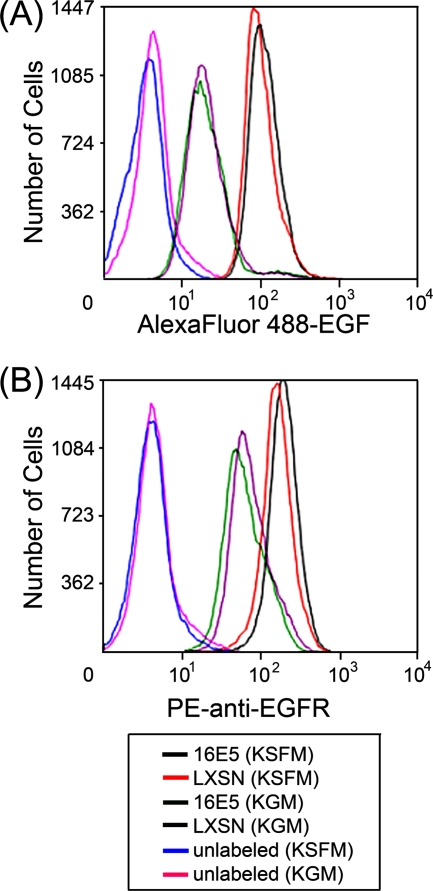
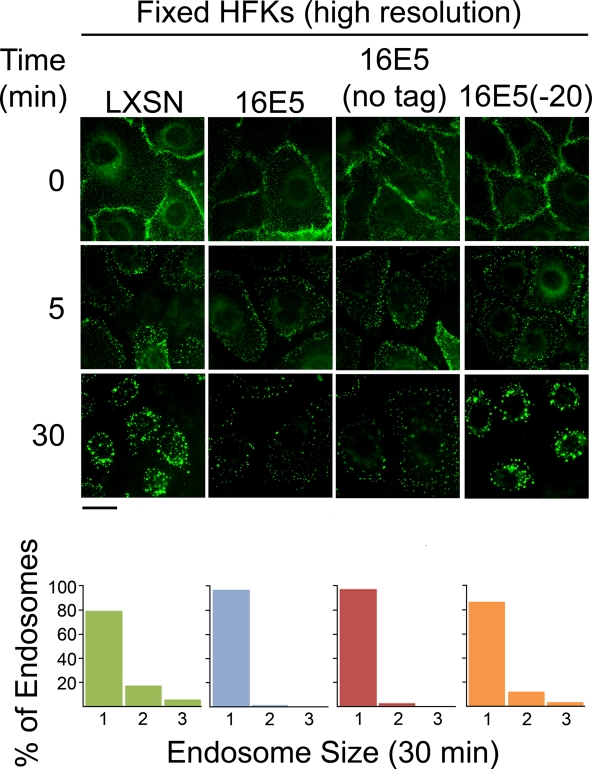
To rule out the possibility that EGFR trafficking appears to be inhibited in 16E5-expressing HFKs because the EGF binding capacity or surface expression of EGFRs is reduced, we directly measured these parameters in EGF-starved (and in proliferating) 16E5- and LXSN-HFKs by flow cytometry. Although EGF deprivation greatly increased the EGF binding capacity (Fig. 6 A) and number of EGFRs on the cell surface (Fig. 6B), no significant differences in these values were associated with 16E5 expression (Fig. 6). We also found that the initial endocytosis of prebound EGF occurred similarly in 16E5- and LXSN-HFKs. After binding Alexa Fluor 488-EGF to EGF-starved HFKs on coverslips at 4°C, the cells were fixed and mounted on microscope slides 5 min after warming them to 37°C, so that high-resolution optics could be used to visualize the very small fluorescent endosomes present at this early stage of the endocytic pathway. As shown (Fig. 7; 5 min), 16E5 did not alter the initial number of endocytic vesicles subjacent to the plasma membrane but did block subsequent fusion to form larger compartments (Fig. 7; 30 min). The latter observation is in agreement with our results using live (unfixed) cells (Fig. 5). Importantly, Fig. 7 also shows that the AU1 epitope tag is neither necessary nor inhibitory for the 16E5-induced membrane trafficking defect and extends our previous finding that AU1 epitope tagging does not affect the ability of 16E5 to induce koilocytosis in keratinocytes (31). In addition, a 20-amino-acid C-terminal 16E5 deletion mutant that is defective for enhanced ligand-dependent EGFR activation (46) and koilocyte formation (31) did not inhibit endosome fusion (Fig. 7).
FIG. 6.
16E5 does not alter surface EGFR expression or EGF binding capacity. (A) Flow cytometry of Alexa Fluor 488-EGF bound to the surface of growing (KGM) and EGF-starved (KSFM) 16E5- and LXSN-HFKs. The geometric mean fluorescence values were as follows: 21 (16E5-HFKs in KGM), 21 (LXSN-HFKs in KGM), 113 (16E5-HFKs in KSFM), and 102 (LXSN-HFKs in KSFM). (B) Flow cytometry of growing (KGM) and EGF-starved (KSFM) 16E5- and LXSN-HFKs labeled with an anti-EGFR antibody conjugated to R-phycoerythrin (PE). The geometric mean fluorescence values were as follows: 62 (16E5-HFKs in KGM), 74 (LXSN-HFKs in KGM), 195 (16E5-HFKs in KSFM), and 160 (LXSN-HFKs in KSFM).
FIG. 7.
16E5 does not slow the initial endocytosis of prebound EGF. Trafficking of Alexa Fluor 488-EGF bound to the surface of EGF-starved LXSN-HFKs, 16E5-HFKs (with or without the AU1 epitope tag), and 16E5(−20)-HFKs (which express a 20-amino-acid C-terminal 16E5 deletion mutant) at 4°C (0 min) and after warming to 37°C for 5 min or 30 min. Cells were fixed at the indicated times to allow high-resolution imaging using a 63× oil immersion objective lens. Scale bar, 20 μm. Images taken at 30 min were analyzed using Kodak MI software to determine the size of EGF-containing endosomes. Bar graphs indicate the percentages of endosomes that fall within a defined range of sizes: 0 to 165 pixels (group 1), 166 to 331 pixels (group 2), or >331 pixels (group 3).
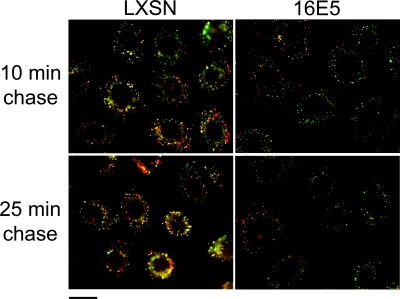
To confirm that 16E5 inhibits the fusion of EGF-containing endosomes with later stages of the endocytic pathway, 16E5- and control LXSN-HFKs were given a 10-min pulse of Alexa Fluor 594-dextran (red) 10 min after the cells had internalized prebound Alexa Fluor 488-EGF (green). In control cells, partial colocalization of the fluorescent probes 10 min later, and more extensive colocalization in larger vesicles 25 min later (Fig. 8; yellow), indicated that the two sets of endosomes had fused with the same sorting and/or late endocytic compartments. In contrast, endosome fusion was strongly inhibited in cells expressing 16E5, since almost no colocalization of green and red endosomes was observed (Fig. 8). As a consequence of the inhibition of vesicle fusion, the formation of larger EGF-containing vesicles was also greatly decreased by 16E5.
FIG. 8.
16E5 inhibits endosome fusion. Alexa Fluor 488-EGF (green) was prebound to EGF-starved LXSN-HFKs and 16E5-HFKs at 4°C and was internalized for 10 min at 37°C before fixable Alexa Fluor 594-dextran (red) was added to the culture medium for an additional 10 min. The cells were moved to ice, washed to remove noninternalized dextran, and shifted back to 37°C for 10 or 25 min before fixation. The fusion of endosomes containing EGF with endosomes containing dextran is evidenced by merging of the green and red fluorescence signals (yellow). Scale bar, 20 μm.
16E5 does not disrupt cytoskeletal filament networks.
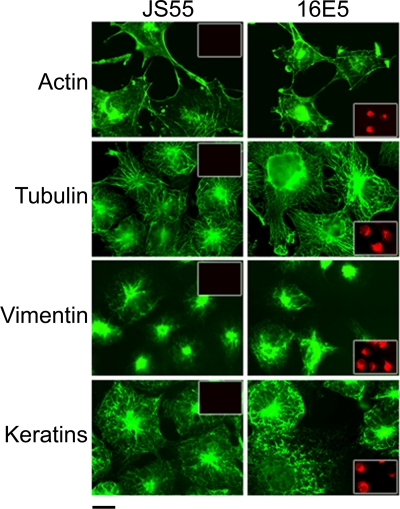
A previous study in fibroblasts proposed that 16E5 inhibits endosome acidification (and the inactivation of internalized EGFRs), without inhibiting V-ATPase-mediated H+ transport, by interfering with the fusion of early and late endosomes (57). This endocytic trafficking defect was attributed to the complete 16E5-dependent depolymerization of actin microfilaments (57). Our results, while consistent with the published data, suggest that the inhibition of endocytic trafficking occurs at the level of membrane fusion rather than interference with vesicle transit mediated by actin microfilaments. We have shown previously that 16E5 does not depolymerize microfilaments or microtubules in stable human ectocervical cell lines (54). However, to eliminate the possibility that the level of E5 in the ectocervical cells was insufficient to induce alterations in actin, we fluorescently labeled cytoskeletal filament networks in transfected COS cells that highly overexpressed 16E5. Compared to control COS cells transfected with the empty JS55 expression vector, 16E5 did not alter or disrupt actin microfilaments, microtubules, or intermediate filaments composed of vimentin or cytokeratins (Fig. 9). It appears highly unlikely, therefore, that the inhibition of vesicle fusion is due to defective vesicle transport.
FIG. 9.
16E5 does not disrupt cytoskeletal filament networks. Immunofluorescence microscopy showing actin, tubulin, vimentin, and cytokeratin filaments (green) in COS cells 24 h after transfection with AU1 epitope-tagged 16E5 or the empty pJS55 expression vector. Scale bar, 20 μm. The cells were colabeled with anti-AU1 antibody (red) to demonstrate 16E5 expression (insets).
DISCUSSION
A role for 16E5 in promoting cervical cancer has most frequently been linked to increased EGFR activation (53, 59), yet aside from the hypothesis that impaired endosome acidification is involved (17, 51, 57), the biology underlying this potentially important 16E5 phenotype remains ambiguous. The purpose of our study was to use several new reagents that we have generated to better understand the mechanism of 16E5-dependent EGFR activation in primary HFKs, a physiological host cell for HPVs.
First, we demonstrated that interfering with endosome acidification does augment acute ligand-dependent activation of the EGFR in HFKs, independently of 16E5. Both the initial extent and the duration of EGFR activation are increased in control cells treated with bafilomycin A1, a pharmacological inhibitor that is highly selective for the H+ V-ATPase responsible for organelle acidification. We found that 16E5 similarly enhances EGFR activation and have used a novel biologically active, pH-sensing EGF conjugate, pHrodo-EGF, to show that 16E5 specifically inhibits the acidification of EGF-containing endosomes. Previous studies have measured bulk endosomal pH using dextran conjugates (51) or organelle-specific fluorescent proteins (17).
With a few exceptions (5, 57), 16E5 was thought to interfere with endosome acidification in a manner similar to that of bafilomycin: by inhibiting the H+ V-ATPase via binding to its 16K subunit. Due to a lack of appropriate antibodies against the native proteins, studies of 16E5-16K association relied upon overexpressing epitope-tagged versions of both proteins in vitro (5, 46), in COS cells (1, 14), or in 293-T cells (22). We have used an antibody that immunoprecipitates wild-type 16K to show that 16K expression increases 15-fold in COS cells that are cotransfected with expression vectors encoding 16K and 16E5 and to confirm that 62% of the 16K coprecipitates with 16E5 under these conditions. However, we show that only 5% of endogenous 16K is bound to 16E5 in primary HFKs that have been selected for stable 16E5 expression. If E5 functions by binding and sequestering 16K in a 1:1 ratio, it is difficult to envision how the low level of 16E5-16K association prevents assembly of the H+ V-ATPase complex or otherwise inhibits its function. In support of this conclusion, we found that bafilomycin, but not 16E5, induced a generalized neutralization of acidic organelles that label with LysoTracker Red and LysoTracker Yellow.
The absence of significant 16E5-16K association in cells that stably express 16E5 argues against the hypothesis that the E5 protein affects organelle acidification directly. Indeed, 16E5 has been reported to both inhibit (10) and have no effect (5) on vacuolar acidification in yeast. The first study to demonstrate impaired endosomal acidification in 16E5-expressing HFKs (51) used endocytosis of a pH-sensitive dextran conjugate. As in the present study, the failure of these dextran-containing endosomes to acidify may indicate a 16E5-dependent defect in membrane trafficking. Disbrow et al. (17) used ratiometric single-cell imaging of 16E5- and LXSN-HFKs that expressed a fluorescent pH-sensitive cellubrevin construct to conclude that 16E5 inhibits endosome acidification. However, cellubrevin is an integral membrane protein that is synthesized in the ER and exported to endosomes in a BAP31-dependent manner (3). Since 16E5 binds to the BAP31 domain that is essential for cellubrevin export (3, 43), the E5 protein may have caused retention of the cellubrevin pH probe in the ER lumen, which is not acidic (29).
The progressive acidification and maturation of early endosomes requires that they undergo fusion with acidic sorting endosomes, late endosomes, and lysosomes (23, 28). This fusion gives rise to larger endocytic vesicles that can easily be observed using fluorescence microscopy (45, 61). An important finding of our study is that 16E5 inhibits the acidification of EGF-containing early endosomes by impeding their fusion with later endocytic compartments, so that larger fluorescent vesicles which arise in control cells do not form in 16E5-HFKs. We further show that the merging of EGF-containing endosomes with a distinct population of dextran-containing endosomes, which occurs in control cells as a result of both populations fusing with later endocytic compartments, is strongly inhibited in cells that express 16E5. Future work will focus on the identification of specific proteins that regulate endocytic trafficking that may be targeted by 16E5; however, several potential mechanisms for the 16E5-dependent trafficking defect can already be ruled out. In kidney proximal tubule epithelial cells, endosome acidification is necessary for recruitment of the small GTPase Arf6 (27, 35), which is a regulator of the receptor-mediated endocytic pathway (19). This pH-dependent recruitment explains why endosome acidification is required for endocytic trafficking in these cells (27). In contrast, we find that the trafficking of endosomes containing EGF is not disrupted in HFKs by bafilomycin treatment, which neutralizes all acidic intracellular compartments. Therefore, 16E5 inhibits endocytic trafficking of the EGFR through a mechanism that is independent of endosome acidification. A report by Thomsen et al. (57), which was the first to show that 16E5 inhibits endocytic trafficking, attributed the phenotype to defective endosomal transport caused by 16E5-dependent depolymerization of the actin cytoskeleton. The effect of 16E5 on actin filaments may be unique to the C127 rodent fibroblasts used in their study, since we previously have shown that 16E5 does not perturb actin filaments in human ectocervical cells (54), and the present study demonstrates that even a high level of 16E5 expression in COS cells does not alter the organization of actin, tubulin, vimentin, or cytokeratin filaments.
In epithelial cells, 16E5 localizes to membranes of the ER and nuclear envelope with its C terminus exposed to the cytoplasm (32). If the 16E5 C terminus associates with one or more proteins that regulate membrane fusion, these proteins might become tethered to the ER and nuclear envelope and therefore be unable to participate in endocytic processing. It may be relevant that 16E5 cooperates with HPV E6 proteins to induce koilocytosis, the formation of large perinuclear membrane vacuoles (31). While 16E5 inhibits endocytic trafficking in primary HFKs which do not express E6, an accumulation of membrane fusion regulatory protein(s) at the nuclear envelope may also be part of the mechanism underlying koilocytosis. This hypothesis is supported by the observation that a 20-amino-acid C-terminal 16E5 deletion mutant that is defective for inducing koilocytosis (31) also fails to inhibit endocytic trafficking (Fig. 7).
Two recent publications claim that overexpression of 16E5 in HaCaT cells by means of an inducible adenovirus promoter leads to its presence in the plasma membrane, where the extracellular 16E5 carboxyl terminus induces cell-cell fusion and the formation of binucleated cells (25, 26). However, these findings do not seem pertinent to our present study for several reasons. In stable keratinocyte lines, 16E5 is expressed at lower levels, is restricted to the ER membrane, and does not generate binucleated cells (32). Moreover, 16E5 does not promote intracellular membrane fusion in our system, even though its carboxyl terminus is located in the cytoplasm (32). Rather, 16E5 inhibits endosome fusion, and the 16E5 carboxyl terminus is required for the inhibitory activity. It is most likely that some of the differences in E5 biology and localization observed in these studies are attributable to differences in the level of expressed E5 protein.
In brief, 16E5 alters EGF endocytic trafficking and endosome maturation via a pH-independent, transport-independent mechanism, most likely by altering vesicle fusion events. This observation may explain the ability of E5 to not only interfere with EGF processing and augment EGFR signaling but also interfere with its ability to alter the transport of HLA proteins, cholesterol, gangliosides, and lipid raft proteins that function in controlling signal transduction pathways.
Acknowledgments
We are grateful to the Flow Cytometry and Cell Sorting Shared Resource Facility of The Lombardi Comprehensive Cancer Center for invaluable technical assistance.
This study was supported by grant R01-CA053371 from the National Cancer Institute (to R.S.).
Footnotes
Published ahead of print on 4 August 2010.
REFERENCES
- 1.Adam, J. L., M. W. Briggs, and D. J. McCance. 2000. A mutagenic analysis of the E5 protein of human papillomavirus type 16 reveals that E5 binding to the vacuolar H+-ATPase is not sufficient for biological activity, using mammalian and yeast expression systems. Virology 272:315-325. [DOI] [PubMed] [Google Scholar]
- 2.Andresson, T., J. Sparkowski, D. J. Goldstein, and R. Schlegel. 1995. Vacuolar H+-ATPase mutants transform cells and define a binding site for the papillomavirus E5 oncoprotein. J. Biol. Chem. 270:6830-6837. [DOI] [PubMed] [Google Scholar]
- 3.Annaert, W. G., B. Becker, U. Kistner, M. Reth, and R. Jahn. 1997. Export of cellubrevin from the endoplasmic reticulum is controlled by BAP31. J. Cell Biol. 139:1397-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arbeit, J. M., K. Munger, P. M. Howley, and D. Hanahan. 1994. Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J. Virol. 68:4358-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashby, A. D., L. Meagher, M. S. Campo, and M. E. Finbow. 2001. E5 transforming proteins of papillomaviruses do not disturb the activity of the vacuolar H+-ATPase. J. Gen. Virol. 82:2353-2362. [DOI] [PubMed] [Google Scholar]
- 6.Ashrafi, G. H., M. Haghshenas, B. Marchetti, and M. S. Campo. 2006. E5 protein of human papillomavirus 16 downregulates HLA class I and interacts with the heavy chain via its first hydrophobic domain. Int. J. Cancer 119:2105-2112. [DOI] [PubMed] [Google Scholar]
- 7.Bouvard, V., G. Matlashewski, Z. M. Gu, A. Storey, and L. Banks. 1994. The human papillomavirus type 16 E5 gene cooperates with the E7 gene to stimulate proliferation of primary cells and increases viral gene expression. Virology 203:73-80. [DOI] [PubMed] [Google Scholar]
- 8.Bowman, B. J., and E. J. Bowman. 2002. Mutations in subunit c of the vacuolar ATPase confer resistance to bafilomycin and identify a conserved antibiotic binding site. J. Biol. Chem. 277:3965-3972. [DOI] [PubMed] [Google Scholar]
- 9.Bowman, E. J., A. Siebers, and K. Altendorf. 1988. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. U. S. A. 85:7972-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs, M. W., J. L. Adam, and D. J. McCance. 2001. The human papillomavirus type 16 E5 protein alters vacuolar H+-ATPase function and stability in Saccharomyces cerevisiae. Virology 280:169-175. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J., M. A. Skinner, W. Shi, Q. C. Yu, A. G. Wildeman, and Y. M. Chan. 2007. The 16-kDa subunit of vacuolar H+-ATPase is a novel sarcoglycan-interacting protein. Biochim. Biophys. Acta 1772:570-579. [DOI] [PubMed] [Google Scholar]
- 12.Choo, K. B., C. C. Pan, M. S. Liu, H. T. Ng, C. P. Chen, Y. N. Lee, C. F. Chao, C. L. Meng, M. Y. Yeh, and S. H. Han. 1987. Presence of episomal and integrated human papillomavirus DNA sequences in cervical carcinoma. J. Med. Virol. 21:101-107. [DOI] [PubMed] [Google Scholar]
- 13.Clifford, G. M., J. S. Smith, M. Plummer, N. Munoz, and S. Franceschi. 2003. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br. J. Cancer 88:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad, M., V. J. Bubb, and R. Schlegel. 1993. The human papillomavirus type 6 and 16 E5 proteins are membrane-associated proteins which associate with the 16-kilodalton pore-forming protein. J. Virol. 67:6170-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crusius, K., E. Auvinen, B. Steuer, H. Gaissert, and A. Alonso. 1998. The human papillomavirus type 16 E5 protein modulates ligand-dependent activation of the EGF receptor family in the human epithelial cell line HaCaT. Exp. Cell Res. 241:76-83. [DOI] [PubMed] [Google Scholar]
- 16.Cullen, A. P., R. Reid, M. Campion, and A. T. Lorincz. 1991. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J. Virol. 65:606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Disbrow, G. L., J. A. Hanover, and R. Schlegel. 2005. Endoplasmic reticulum-localized human papillomavirus type 16 E5 protein alters endosomal pH but not trans-Golgi pH. J. Virol. 79:5839-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Disbrow, G. L., I. Sunitha, C. C. Baker, J. Hanover, and R. Schlegel. 2003. Codon optimization of the HPV-16 E5 gene enhances protein expression. Virology 311:105-114. [DOI] [PubMed] [Google Scholar]
- 19.D'Souza-Schorey, C., G. Li, M. I. Colombo, and P. D. Stahl. 1995. A regulatory role for ARF6 in receptor-mediated endocytosis. Science 267:1175-1178. [DOI] [PubMed] [Google Scholar]
- 20.Fehrmann, F., and L. A. Laimins. 2003. Human papillomaviruses: targeting differentiating cells for malignant transformation,. Oncogene 22:5201-5207. [DOI] [PubMed] [Google Scholar]
- 21.Genther-Williams, S. M., G. L. Disbrow, R. Schlegel, D. Lee, D. W. Threadgill, and P. F. Lambert. 2005. Requirement of epidermal growth factor receptor for hyperplasia induced by E5, a high-risk human papillomavirus oncogene. Cancer Res. 65:6534-6542. [DOI] [PubMed] [Google Scholar]
- 22.Gieswein, C. E., F. J. Sharom, and A. G. Wildeman. 2003. Oligomerization of the E5 protein of human papillomavirus type 16 occurs through multiple hydrophobic regions. Virology 313:415-426. [DOI] [PubMed] [Google Scholar]
- 23.Gruenberg, J., and F. R. Maxfield. 1995. Membrane transport in the endocytic pathway. Curr. Biol. 7:552-563. [DOI] [PubMed] [Google Scholar]
- 24.Hafner, N., C. Driesch, M. Gajda, L. Jansen, R. Kirchnayr, I. B. Runnebaum, and M. Durst. 2008. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene 27:1610-1617. [DOI] [PubMed] [Google Scholar]
- 25.Hu, L., and B. P. Ceresa. 2009. Characterization of the plasma membrane localization and orientation of HPV16 E5 for cell-cell fusion. Virology 393:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, L., K. Plafker, V. Vorozhko, R. E. Zuna, M. H. Hanigan, G. J. Gorbsky, S. M. Plafker, P. C. Angeletti, and B. P. Ceresa. 2009. Human papillomavirus 16 E5 induces bi-nucleated cell formation by cell-cell fusion. Virology 384:125-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurtado-Lorenzo, A., M. Skinner, J. El Annan, M. Futai, G. H. Sun-Wada, S. Bourgoin, J. Casanova, A. Wildeman, S. Bechoua, D. A. Ausiello, D. Brown, and V. Marshansky. 2006. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8:124-136. [DOI] [PubMed] [Google Scholar]
- 28.Jefferies, K. C., D. J. Cipriano, and M. Forgac. 2008. Function, structure and regulation of the vacuolar (H+)-ATPases. Arch. Biochem. Biophys. 476:33-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, J. H., L. Johannes, B. Goud, C. Antony, C. A. Lingwood, R. Daneman, and S. Grinstein. 1998. Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proc. Natl. Acad. Sci. U. S. A. 95:2997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krawczyk, E., J. A. Hanover, R. Schlegel, and F. A. Suprynowicz. 2008. Karyopherin β3: a new cellular target for the HPV-16 E5 oncoprotein. Biochem. Biophys. Res. Commun. 371:684-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krawczyk, E., F. A. Suprynowicz, X. Liu, Y. Dai, D. P. Hartmann, J. Hanover, and R. Schlegel. 2008. Koilocytosis: a cooperative interaction between the human papillomavirus E5 and E6 oncoproteins. Am. J. Pathol. 173:682-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krawczyk, E., F. A. Suprynowicz, S. R. Sudarshan, and R. Schlegel. 2010. Membrane orientation of the human papillomavirus type 16 E5 oncoprotein. J. Virol. 84:1696-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leechanachai, P., L. Banks, F. Moreau, and G. Matlashewski. 1992. The E5 gene from human papillomavirus type 16 is an oncogene which enhances growth factor-mediated signal transduction to the nucleus. Oncogene 7:19-25. [PubMed] [Google Scholar]
- 34.Lim, P. S., A. B. Jenson, L. Cowsert, Y. Nakai, L. Y. Lim, X. W. Jin, and J. P. Sundberg. 1990. Distribution and specific identification of papillomavirus major capsid protein epitopes by immunocytochemistry and epitope scanning of synthetic peptides. J. Infect. Dis. 162:1263-1269. [DOI] [PubMed] [Google Scholar]
- 35.Maranda, B., D. Brown, S. Bourgoin, J. E. Casanova, P. Vinay, D. A. Ausiello, and V. Marshansky. 2001. Intra-endosomal pH-sensitive recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from cytoplasm to proximal tubule endosomes. J. Biol. Chem. 276:18540-18550. [DOI] [PubMed] [Google Scholar]
- 36.Matsukura, T., S. Koi, and K. Sugase. 1989. Both episomal and integrated forms of human papillomavirus type 16 are involved in invasive cervical cancers. Virology 172:63-72. [DOI] [PubMed] [Google Scholar]
- 37.Oelze, I., J. Kartenbeck, K. Crusius, and A. Alonso. 1995. Human papillomavirus type 16 E5 protein affects cell-cell communication in an epithelial cell line. J. Virol. 69:4489-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pati, U. K. 1992. Novel vectors for expression of cDNA encoding epitope-tagged proteins in mammalian cells. Gene 114:285-288. [DOI] [PubMed] [Google Scholar]
- 39.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U. S. A. 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedroza-Saavedra, A., E. W. Lam, F. Esquivel-Guadarrama, and L. Gutierrez-Xicotencatl. 2010. The human papillomavirus type 16 E5 oncoprotein synergizes with EGF-receptor signaling to enhance cell cycle progression and the downregulation of p27Kip1. Virology 400:44-52. [DOI] [PubMed] [Google Scholar]
- 41.Pim, D., M. Collins, and L. Banks. 1992. Human papillomavirus type 16 E5 gene stimulates the transforming activity of the epidermal growth factor receptor. Oncogene 7:27-32. [PubMed] [Google Scholar]
- 42.Pirami, L., V. Giache, and A. Becciolini. 1997. Analysis of HPV16, 18, 31, and 35 DNA in pre-invasive and invasive lesions of the uterine cervix. J. Clin. Pathol. 50:600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regan, J. A., and L. A. Laimins. 2008. Bap31 is a novel target of the human papillomavirus E5 protein. J. Virol. 82:10042-10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riley, R. R., S. Duensing, T. Brake, K. Munger, P. F. Lambert, and J. M. Arbeit. 2003. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 63:4862-4871. [PubMed] [Google Scholar]
- 45.Roberts, R. L., M. A. Barbieri, J. Ullrich, and P. D. Stahl. 2000. Dynamics of rab5 activation in endocytosis and phagocytosis. J. Leukoc. Biol. 68:627-632. [PubMed] [Google Scholar]
- 46.Rodriguez, M. I., M. E. Finbow, and A. Alonso. 2000. Binding of human papillomavirus 16 E5 to the 16-kDa subunit c (proteolipid) of the vacuolar H+-ATPase can be dissociated from the E5-mediated epidermal growth factor receptor overactivation. Oncogene 19:3727-3732. [DOI] [PubMed] [Google Scholar]
- 47.Schapiro, F., J. Sparkowski, A. Adduci, F. Suprynowicz, R. Schlegel, and S. Grinstein. 2000. Golgi alkalinization by the papillomavirus E5 oncoprotein. J. Cell Biol. 148:305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiffman, M., R. Herrero, R. Desalle, A. Hildesheim, S. Wacholder, A. C. Rodriguez, M. C. Bratti, M. E. Sherman, J. Morales, D. Guillen, M. Alfaro, M. Hutchinson, T. C. Wright, D. Solomon, Z. Chen, J. Schussler, P. E. Castle, and R. D. Burk. 2005. The carcinogenicity of human papillomavirus types reflects viral evolution. Virology 337:76-84. [DOI] [PubMed] [Google Scholar]
- 49.Schlegel, R., W. C. Phelps, Y. L. Zhang, and M. Barbosa. 1988. Quantitative keratinocyte assay detects two biological activities of human papillomavirus DNA and identifies viral types associated with cervical carcinoma. EMBO J. 7:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sparkowski, J., J. Anders, and R. Schlegel. 1994. Mutation of the bovine papillomavirus E5 oncoprotein at amino acid 17 generates both high- and low-transforming variants. J. Virol. 68:6120-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Straight, S. W., B. Herman, and D. J. McCance. 1995. The E5 oncoprotein of human papillomavirus type 16 inhibits the acidification of endosomes in human keratinocytes. J. Virol. 69:3185-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Straight, S. W., P. M. Hinkle, R. J. Jewers, and D. J. McCance. 1993. The E5 oncoprotein of human papillomavirus type 16 transforms fibroblasts and effects the downregulation of the epidermal growth factor receptor in keratinocytes. J. Virol. 67:4521-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suprynowicz, F. A., M. S. Campo, and R. Schlegel. 2006. Biological activities of papillomavirus E5 proteins, p. 97-113. In M. S. Campo (ed.), Papillomavirus research: from natural history to vaccines and beyond. Caister Academic Press, Wymondham, United Kingdom.
- 54.Suprynowicz, F. A., G. L. Disbrow, E. Krawczyk, V. Simic, K. Lantzky, and R. Schlegel. 2008. HPV-16 E5 oncoprotein upregulates lipid raft components caveolin-1 and ganglioside GM1 at the plasma membrane of cervical cells. Oncogene 27:1071-1078. [DOI] [PubMed] [Google Scholar]
- 55.Suprynowicz, F. A., G. L. Disbrow, V. Simic, and R. Schlegel. 2005. Are transforming properties of the bovine papillomavirus E5 protein shared by E5 from high-risk human papillomavirus type 16? Virology 332:102-113. [DOI] [PubMed] [Google Scholar]
- 56.Suprynowicz, F. A., J. Sparkowski, A. Baege, and R. Schlegel. 2000. E5 oncoprotein mutants activate phosphoinositide 3-kinase independently of platelet-derived growth factor receptor activation. J. Biol. Chem. 275:5111-5119. [DOI] [PubMed] [Google Scholar]
- 57.Thomsen, P., B. van Deurs, B. Norrild, and L. Kayser. 2000. The HPV16 E5 oncogene inhibits endocytic trafficking. Oncogene 19:6023-6032. [DOI] [PubMed] [Google Scholar]
- 58.Tomakidi, P., H. Cheng, A. Kohl, G. Komposch, and A. Alonso. 2000. Connexin 43 expression is downregulated in raft cultures of human keratinocytes expressing the human papillomavirus type 16 E5 protein. Cell Tissue Res. 301:323-327. [DOI] [PubMed] [Google Scholar]
- 59.Tsai, T. C., and S. L. Chen. 2003. The biochemical and biological functions of human papillomavirus type 16 E5 protein. Arch. Virol. 148:1445-1453. [DOI] [PubMed] [Google Scholar]
- 60.Valle, G. F., and L. Banks. 1995. The human papillomavirus (HPV)-6 and HPV-16 E5 proteins cooperate with HPV-16 E7 in the transformation of primary rodent cells. J. Gen. Virol. 76:1239-1245. [DOI] [PubMed] [Google Scholar]
- 61.Visser Smit, G. D., T. L. Place, S. L. Cole, K. A. Clausen, S. Vemuganti, G. Zhang, J. G. Koland, and N. L. Lill. 2009. Cbl controls EGFR fate by regulating early endosome fusion. Sci. Signal. 2:ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wada, Y., G. H. Sun-Wada, H. Tabata, and N. Kawamura. 2008. Vacuolar-type proton ATPase as a regulator of membrane dynamics in multicellular organisms. J. Bioenerg. Biomembr. 40:53-57. [DOI] [PubMed] [Google Scholar]
- 63.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 64.zur Hausen, H., and E. M. de Villiers. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427-447. [DOI] [PubMed] [Google Scholar]