Abstract
Two of the central issues in developing new strategies to interfere with viral infections concern the identification of cellular proteins involved in viral replication and/or antiviral measures and the dissection of the underlying molecular mechanisms. To gain initial insight into the role of host proteins in the life cycle of infectious bursal disease virus (IBDV), a double-stranded RNA virus, we examined the cellular nuclear factor 45 (NF45). NF45 was previously indicated to be involved in the replication process of other types of RNA viruses. Interestingly, by performing immunofluorescence studies, we found that in IBDV-infected cells the mainly nuclear NF45 accumulated at the sites of viral replication in the cytoplasm. NF45 was shown to specifically colocalize with the viral RNA-dependent RNA polymerase VP1, the capsid protein VP2, and the ribonucleoprotein VP3. Immunoprecipitation experiments indicated protein-protein associations between NF45 and VP1, VP2, and VP3. Expression of the individual VP3 or the combination of expression of VP1 and VP3 did not result in a cytoplasmic accumulation of NF45, which, among other data, showed that recruitment of the cellular protein in infected cells functionally correlates with the viral replication process. Since small interfering RNA(siRNA)-mediated downregulation of NF45 resulted in an approximately 5-fold increase of virus yield, our study suggests that NF45, by association with viral proteins, is part of a yet-uncharacterized cellular defense mechanism against IBDV infections.
The highly contagious infectious bursal disease virus (IBDV), first described by Cosgrove (5), is the causative agent of the immunosuppressive infectious bursa disease (IBD) in young chickens. Worldwide, IBDV infections cause significant economic losses in the poultry industry. The high mortality rate of the host animals may be directly related to the virus infection. Moreover, IBDV originates lytic infections of proliferating B lymphocytes in the bursa of Fabricius that lead to a general humoral immunosuppression and reduced response to vaccines, which in turn favor respiratory and enteric diseases.
IBDV is a nonenveloped, bisegmented (segment A and B) double-stranded RNA (dsRNA) virus belonging to the family Birnaviridae and the genus Avibirnavirus (7). Segment A contains two partially overlapping open reading frames (ORFs). The first ORF encodes the nonessential, nonstructural viral protein 5 (VP5) (33, 34, 51), which plays an important role in virulence and virus egress (28, 60). The second ORF encodes a polyprotein of 110 kDa that is autocatalytically cleaved by the viral protease VP4, yielding the pVP2 precursor (48 kDa) as well as VP4 (28 kDa) and VP3 (32 kDa) (3). Throughout virion maturation, pVP2 is further processed into the mature capsid protein VP2 (41 kDa) and four small peptides (6). While the largest peptide is autoproteolytically cleaved by VP2 (14), the other peptides are believed to be generated by trans-cleavage via the viral protease VP4 and/or cellular proteins (6). Segment B contains one ORF that encodes the viral RNA-dependent RNA polymerase VP1 (98 kDa) (55). VP2 and VP3 are the major structural proteins, constituting 51% and 40% of the virion, respectively (9). VP3 consists of 257 amino acids (aa), and it was shown that the protein's carboxy terminus exhibits several functions. A domain causing self-interaction is located between aa 224 and 247 (29). Moreover, VP3 was found to interact with VP1 via its 10 C-terminal amino acids (52) and to bind to the viral dsRNA, forming ribonucleoprotein complexes (29). During heterologous expression in insect cells, VP3 was found to colocalize with pVP2 but not with the mature form of VP2. VP3-pVP2 binding was observed to result in the formation of virus-like particles (37). VP3 is believed to act as a scaffolding protein for pVP2, and the protein is thought to be a key organizer in birnavirus morphogenesis (31).
This study aimed at examining a potential function of NF45 (also known as ILF2) in the replication process of IBDV. NF45 was originally shown to regulate interleukin-2 gene transcription (18). The protein forms a stable heterodimer with another nuclear factor, NF90 (also known as DRBP76, TCP80, ILF3, and NFAR-1) or a larger isoform of NF90, NF110 (also known as TCP110 and NFAR-2) (4, 18, 43, 53).
Numerous functions have been assigned to the NF45/NF90 heterodimeric protein complex, a complex that is predominantly nuclear but also present in the cytoplasm of many different cell types (61). Besides acting as a transcriptional cofactor (4, 38, 44-47), it has been found to be involved in RNA export and to mediate mRNA stability and translation (48, 54, 58, 59). In the nucleus, the NF45/NF90 complex was recently shown to function as a negative regulator in the microRNA processing pathway (42).
Recent observations suggest that NF45 and NF90 are significantly involved in the replication process of several different RNA viruses. For two members of the family Flaviviridae, bovine viral diarrhea virus (BVDV) and hepatitis C virus (HCV), both NF45 and NF90 were indicated to be part of viral replication machineries and suggested to be a part of the regulation of viral translation and RNA replication (15-17). Investigations of picornaviruses revealed that NF90 binds to the internal ribosomal entry site (IRES) of a chimeric poliovirus and that the association of NF45 to NF90 inhibits viral translation in neuron-derived cells (32). Data obtained by Shin et al. (49) suggest that both proteins may play a role in the priming activity of the HBV polymerase. Just recently, it was described that NF90 interacts with the nucleoprotein (NP) of influenza A virus, and suppression of NF90 expression stimulates viral replication (57).
So far, little is known about the function of cellular factors in the replication process of dsRNA viruses. By studying the role of NF45 in the replication of the birnavirus IBDV, our data indicate this cellular protein is a negative regulator of IBDV replication.
MATERIALS AND METHODS
Cells.
Cells of the chicken fibroblast cell line DF-1 (12), MDCK cells (CRL-2285; ATCC, Manassas, VA), chicken embryo cells (CEC) derived from 11-day-old embryonated specific-pathogen-free eggs (Sunrise Farms, Catskill, NY), and Vero cells (CRL-1587; ATCC) were grown in Dulbecco's modified Eagles's medium with 4.5 g/liter glucose (DMEM-4.5; Thermo Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; Mediatech, Manassas, VA). Quail muscle cells (QM-7; RIE 466; Collection of Cell Lines in Veterinary Medicine [CCLV], Insel Riems, Germany) and cells of the human embryonic kidney cell line HEK 293T (CRL-11268; ATCC) were grown in a mixture of equal parts of minimal essential medium (MEM; Invitrogen, Carlsbad, CA) with Earle's balanced salt solution and MEM with Hanks' balanced salt solution (Invitrogen, Carlsbad, CA) supplemented with 10% FBS. All cells were cultivated in a humidified atmosphere at 37°C with 5% CO2.
The insect cell lines of Spodoptera frugiperda (Sf9; Invitrogen, Carlsbad, CA) and ovarian cells of Trichoplusia ni (High Five; Invitrogen, Carlsbad, CA) were cultivated in serum-free SFX-Insect medium (Thermo Scientific, Waltham, MA) at 28°C.
RT-PCR and construction of plasmids.
RNA of CEC was isolated with the High Pure RNA isolation kit (Roche Diagnostics GmbH, Mannheim, Germany) following the manufacturer's instructions. Five microliters RNA was used for the SuperScript III One-Step reverse transcription-PCR (RT-PCR) system with Platinum Taq polymerase (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol with an RT step at 45°C for 30 min, a denaturation step of 94°C for 2 min, 5 cycles of 94°C for 15 s, 45°C for 30 s, and 68°C for 2 min, and 35 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 2 min. Amplification was performed using a chicken NF45 (ckNF45)-specific forward primer (NF45FP; CCGAATTCATGAGGGGCGACCGAGGCAGAGGCC) in combination with either NF45-RP-His (CCGCGGCCGCTCAATGGTGATGGTGATGGTGCTCCTGGGTCTCCATGCTTTCC) or NF45-RP-FLAG (CCGCGGCCGCTCACTTATCATCATCATCCTTGTAATCCTCCTGGGTCTCCATGCTTTCC). Primers were designed based on the published sequence of chicken NF45 (GenBank accesion number XP_423437) to add either a 6×His tag or a FLAG tag amino acid sequence (DYKDDDDK) at the C terminus of the NF45 protein. After amplification, both cDNA fragments were cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) following manufacturer's recommendations. Plasmids (pckNF45His and pckNF45FLAG) were sequenced using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). Sequence analysis was performed using the Lasergene software (DNAStar Inc., Madison, WI). After sequence verification, both plasmids (pckNF45His and pckNF45FLAG) were incubated with EcoRI and NotI. The appropriate fragments were gel eluted and ligated either into the appropriately cleaved pFastBac Dual (Invitrogen, Carlsbad, CA) to obtain pFAST-ckNF45Bac or the appropriately cleaved pcDNA3 (Invitrogen, Carlsbad, CA) to obtain pcDNA3-ckNF45FLAG.
Furthermore, a cDNA construct was generated by PCR where the VP3 coding region of pD78APD (amino acids 756 to 1012) (13), as determined by Lejal et al. (25), was amplified by PCR using an appropriate primer pair (VP3FP, GAATTCATGGCATCAGAGTTCAAAGAGACC CCCGAACTCG; VP3RP, GAGCGGCCGCTCACTCAAGGTCCTCATCAGAGACGGTCC). In addition, the coding region of VP1 was amplified by PCR using plasmid pD78BPD (13) and a primer pair (pcD78B-FP, GGGAATTCATGATTCTGCCACCATGAGTGAC; pcD78B-RP, GGGCGGCCGCTTAGCGGCTCTCCTTTTGGCGTTGC). Both PCR fragments were gel eluted and subsequently cleaved with appropriate restriction enzymes (EcoRI and NotI) and then ligated into the eukaryotic expression vector pcDNA3 (Invitrogen) to obtain pc3-VP3 and pc3-VP1. Plasmids were sequenced and used for subsequent transfection experiments.
Generation of recombinant baculovirus and purification of ckNF45-His.
Recombinant baculovirus DNA was generated using the Bac-to-Bac baculovirus expression system (Invitrogen, Carlsbad, CA) following the protocol provided by the manufacturer and using the plasmid pFAST-ckNF45Bac. Recombinant baculovirus DNA encoding ckNF45 was transfected into High Five cells by using Cellfectin (Invitrogen, Carlsbad, CA), and a recombinant baculovirus expressing ckNF45-His was rescued (BacNF45-His). Virus stocks were obtained by passaging the transfection supernatant on Sf9 cells as described previously (26). ckNF45-His was purified following a previously described protocol (26) with one exception: the recombinant protein was eluted with 150 mM imidazole-containing elution buffer. Samples containing ckNF45-His were pooled, and the protein amount was determined with the Micro BCA protein assay kit (Thermo Scientific, Waltham, MA). For the generation of ckNF45 antiserum, a rabbit was immunized at the Polyclonal Antibody Production Service facility (University of Georgia, Athens, GA) with the fraction containing the purified NF45-His proteins. The resulting serum was named r-anti-ckNF45 serum.
Generation of IBDV protein-specific antisera.
To perform these studies, it was necessary to generate sera from different species exhibiting specific reactivities against the IBDV proteins VP1 and VP3. To this end, VP1-His was expressed in Sf9 cells and purified as described previously (26). In addition, the VP3-encoding sequence (aa 735 to 1012) (25) of the IBDV strain D78 containing a 6×His sequence at its C terminus was cloned into pFastBac Dual (Invitrogen). A recombinant baculovirus encoding the VP3-6×His was generated using the Bac-to-Bac system (Invitrogen), and the protein was purified as described elsewhere (26). The purified proteins were used for the generation of antisera in two different species. The VP1-specific serum was generated in 3-week-old specific-pathogen-free chickens (Merial, Gainesville, GA) by repeated immunizations with a water in oil emulsion using the purified VP1-His and incomplete Freund's adjuvant (Sigma-Aldrich, St. Louis, MO) at the Poultry Diagnostic and Research Center (Athens, GA). For the generation of a rabbit VP3 antiserum, a New Zealand rabbit was immunized three times with a VP3/incomplete Freund's adjuvant emulsion at the Federal Research Institute for Animal Health (Insel Riems, Germany). To test the sera for specificity, DF-1 cells were infected with IBDV strain D78 or mock infected (see Fig. 1A). At 24 h postinfection (p.i.), cells were scraped into the medium and sedimented at 2,000 × g for 5 min. Cell pellets were lysed in 2× Laemmli buffer (21), heated, sonicated, and centrifuged at 15,000 × g for 5 min. The cellular lysates were separated on a 12% SDS-polyacrylamide gel and subjected to Western blot analysis (see below).
FIG. 1.
Generation and testing of viral protein and NF45-specific antisera. (A) To test the specificity of generated viral protein, DF-1 cells were either infected with the IBDV strain D78 (2) or left uninfected (1). The obtained cellular lysates were subjected to Western blot analysis. The membranes were incubated with either already-established rabbit sera (anti-VP1 R [3] anti-IBDV R [30]) or the newly established chicken anti-VP1 serum (anti-VP1 ch) and a rabbit anti-VP3 serum (anti-VP3 R). The locations of the IBDV proteins (VP) are indicated by arrows. (B) For monitoring the purification of ckNF45-His, samples were taken at different stages (L, lysate; S, supernatant after centrifugation; E, eluate) and subjected to SDS-PAGE. The gel was stained with Imperial protein stain. The eluate was monitored by using an HRP-conjugated anti-His monoclonal antibody and the rabbit ckNF45-His serum. The reactivity of the rabbit anti-ckNF45His serum was tested by Western blotting of cellular lysates from different species, namely, chicken (DF-1 and CEC), quail (QM-7), dog (MDCK), monkey (Vero), human (293T), and hamster (BHK 21). The molecular masses (in kDa) of protein markers (M) are indicated on the left. (C) BHK21 cells were transfected with a mammalian vector expressing ckNF45-FLAG (see Materials and Methods). Fixed cells were incubated with rabbit anti-ckNF45-His antiserum (NF45) and a mouse anti-FLAG MAb (FLAG) followed by incubation with appropriate conjugates (goat anti-mouse-Cy3 or goat anti-rabbit-FITC). The cellular nuclei were visualized by PI staining. An overlay of the confocal pictures is shown (merge).
Analysis of protein samples.
Electrophoreses were carried out on 12% SDS-polyacrylamide gels. The gels were either stained with Imperial protein stain (Thermo Scientific, Waltham, MA) or transferred to nitrocellulose and used for Western blot analysis. Several antibodies were used for the detection of antigens by Western blotting. The 6×His amino acid sequence was detected with a horseradish peroxidase (HRP)-conjugated mouse monoclonal antibody (MAb) directed against the 6×His amino acid sequence (MAb His-HRP; Sigma-Aldrich, St. Louis, MO). Chicken β-actin was detected with a mouse MAb directed against a β-actin peptide conjugated with HRP (MAb β-actin-HRP; Sigma-Aldrich, St. Louis, MO). The IBDV VP3 antigen and IBDV VP1 were detected using an anti-VP3 rabbit serum (described in this report) and an anti-VP1 rabbit serum (3), respectively. A polyclonal rabbit anti-IBDV serum (33) was used for the detection of IBDV VP2. The chicken NF45 was detected using r-anti-ckNF45 serum (see below). Binding of the diverse species antibodies was monitored by using HRP-conjugated goat antibodies against the appropriate species (Sigma-Aldrich, St. Louis, MO). The binding of the antibodies was visualized by using the chemiluminescence substrate Immobilon Western (Millipore, Billerica, MA) and Gel Logic 2200 (Carestream Health, New Haven, CT).
Infection of cell cultures.
For immunofluorescence analysis, various cell lines (DF-1, QM-7, and Vero) were infected with IBDV strains D78, GLS-05, and 8903 (kindly provided by Ruud Hein, Intervet Schering-Plough Animal Health, Millsboro, DE) or the low-pathogenicity avian influenza virus A/duck/NC/674964/07 (H5N2) (kindly provided by David Stallknecht, University of Georgia, Athens, GA). In each case a multiplicity of infection (MOI) of 1 was used. For the preparation of cellular lysates for immunoprecipitation, the cells were infected with an MOI of 100. Prior to infection, cells were washed with serum-free DMEM. Next, DMEM-diluted virus was added for 1 h at 37°C and the cells were washed with serum-free medium. Finally, medium containing 2% FCS and 1× penicillin-streptomycin (P/S; Sigma-Aldrich, St. Louis, MO) was added. Unless indicated otherwise, cells were fixed 12 h p.i. with ice-cold 96% ethanol.
Indirect immunofluorescence.
Cells were infected with the appropriate virus, fixed at the time points indicated with ice-cold 96% ethanol, and air dried. For indirect immunofluorescence (IIF), cells were rehydrated for 5 min with phosphate-buffered saline (PBS) and incubated with different combinations of the following antibodies: anti-FLAG MAb (anti-FLAG M2 antibody; Sigma-Aldrich, St. Louis, MO), MAb directed against IBDV VP3 (IBDV2) or IBDV VP4 (IBDV6) (10), MAb R63 directed against IBDV VP2 (50), rabbit anti-ckNF45His serum (described in this paper), chicken anti-VP1 serum (this study), rabbit anti-NP serum (19), and MAb anti-M1 (M2-1C6-4R3; ATCC, Manassas, VA) (62). The location of the cell nucleus was visualized by using either propidium iodide (PI; Invitrogen, Carlsbad, CA) or 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen, Carlsbad, CA). All fluorochrome-conjugated secondary antibodies (fluoroscein isothiocyanate [FITC]-conjugated goat anti-rabbit IgG, Cy5-conjugated goat anti-rabbit IgG, Cy5-conjugated goat anti-mouse IgG, Cy3-conjugated goat anti-mouse IgG, Cy3-conjugated goat anti-chicken IgY) were obtained from JacksonImmuno Research (West Grove, PA). Pictures were taken by confocal laser scan microscopy (CLSM) using the LSM 510 laser confocal microscopy system (Zeiss, Jena, Germany) at the AHRC Cell Imaging Core Facility, University of Georgia.
Transfection of cRNA.
As described earlier (35), cRNA of segment A was transcribed by using T7 polymerase. The cRNA was transfected into DF-1 cells grown in a four-well Lab-Tek II chamber slide system (Nunc, Roskilde, Denmark). Prior to transfection, 250 μl OPTI-MEM I (Invitrogen, Carlsbad, CA) was added to the QM-7 cells. A 100-μl volume OPTI-MEM was incubated for 45 min at room temperature with 6 μl Lipofectin (Invitrogen, Carlsbad, CA) in a 5-ml BD Falcon round-bottom tube (BD Biosciences, Franklin Lakes, NY). The cRNA was added to the mixture and incubated for 5 min on ice. Drop by drop, 50 μl of the OPTI-MEM I-RNA-Lipofectin mixture was added to the cells. The cells were incubated for 2 h at 37°C, and then the medium was removed and overlaid with the medium mixture containing 1× penicillin-streptomycin (Sigma Aldrich). At 24 h posttransfection (p.t.) cells were fixed with ice-cold 96% ethanol and processed for indirect immunofluorescence.
Transfection of DNA.
DF-1 cells were seeded in a six-well tissue culture plate or a four-well Lab-Tek II chamber slide system (Nunc) and allowed to grow to 90% confluence. Then, medium was removed and FBS-containing medium was added. In separate tubes, 100 μl of OPTI-MEM I was incubated with 4 μl Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). In another tube 100 μl of OPTI-MEM I was incubated with 3 μg DNA. After 5 min at room temperature both mixtures were combined, incubated for 30 min at room temperature, and added to the cells. For transfection of a chamber slide, 50 μl of the transfection mixture was used. At 3 h p.t. medium was removed and DMEM supplemented with 2% FCS and 1× penicillin-streptomycin (Sigma Aldrich) was added.
Immunoprecipitation.
For the analysis of protein-protein interactions, two sets of experiments were performed. The first set of experiments was performed using two wells each of DF-1 cells grown in a six-well tissue culture plate. Cells were transfected with pcDNA3-ckNF45FLAG or served as nontransfected controls. After 24 h, one well of nontransfected DF-1 cells as well as one well of transfected DF-1 cells were infected with IBDV strain D78 at an MOI of 100. For the second set of experiments DF-1 cells were either infected with IBDV strain D78 at an MOI of 1 or left noninfected. At 24 h (first set) or 12 h (second set) p.i., cells were prepared for immunoprecipitation. To this end cells were trypsinized, resuspended in 2 ml of 10% FBS-containing DMEM, and sedimented at 300 × g for 10 min at 4°C. The supernatant was discarded, and the cell pellet was resuspended in 200 μl lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.25% deoxycholic acid, 1% Igepal CA-630, 1 mM EDTA). The samples were incubated for 10 min on ice with three vortexing steps every 3 min. The lysates were cleared at 16,000 × g for 10 min at 4°C to remove the cellular nuclei from the lysate. The obtained supernatants were either used directly for immunoprecipitation or for RNase treatment followed by immunoprecipitation (see below). For immunoprecipitation, 5 μl of the anti-FLAG MAb, 100 μl of VP3 specific MAb IBDV2, or 20 μl of ascites fluid containing the VP2-specific MAb R63 was incubated overnight at 4°C with 20 μl protein G-Sepharose (Sigma-Aldrich, St. Louis, MO) on a rotor (Mini-Rotator; Glas-Col, Terre Haute, IN). Specifically for immunoprecipitation, supernatants from IBDV2-excreting hybridomas grown in protein-free medium (PFHM-II medium; Invitrogen) were concentrated with an Amicon Ultracel-30K apparatus (Millipore, Carrigtwohill, Ireland) following the manufacturer's instructions. After overnight incubation, the Sepharose-antibody matrix was washed twice with 1 ml wash buffer (20 mM Tris-HCl [pH 8.0], 150 mM KCl, 1% Igepal CA-630, 1 mM EDTA, 1 mM dithiothreitol [DTT]) and sedimented for 1 min at 16,000 × g. The cellular lysates were added to the sedimented Sepharose G-antibody matrix, mixed by pipetting, and incubated for 12 h at 4°C on the minirotator. After five wash steps as described above, samples were resuspended using 20 μl of 2× Laemmli buffer (23) and analyzed by Western blotting. For RNase treatment, prior immunoprecipitation lysates of the cytoplasmic fractions from both sets of experiments were used. To this end either, 1,000 units of RNase T1 (Fermentas, Glen Burnie, MD), 1.3 units of RNase III (New England Biolabs, Ipswich, MA), both RNases, or no RNase was added to the lyastes and incubated for 1 h at 37°C. The digestion of the RNA was analyzed after RNA isolation of one-half of the lysate using the High Pure RNA isolation kit (Roche Diagnostics GmbH, Mannheim, Germany) on a 1% agarose gel using 40 units of Optizyme RNase inhibitor (Fisher Scientific) in 10 mM DTT solution to protect the RNA from degrading. The second half of the lysate was used for immunoprecipitation with the anti-FLAG MAb as described above.
Transfection of siRNA.
The siRNA was designed using an available online program (RNAi Designer; Clontech). Two siRNAs were chosen and ordered as a duplex under desalted conditions (Sigma-Aldrich, St. Louis, MO). They were named 185 and 551, based on the starting nucleotide at which the siRNA binds (185, 5′-UGCAGAACAGGCUUCCAUUdTdT-3′; 551, 5′-GCUGCACUUGGACAUCAAAdTdT-3′). Numbering for the oligonucleotide was based on the number of nucleotides of the NF45 ORF as determined in this study. As a negative control, the Silencer Select negative control 2 siRNA (Invitrogen, Carlsbad, CA) was used.
During each experiment, for each treatment two wells in a six-well tissue culture plate were seeded with 2.75 × 106 DF-1 cells to obtain a double set of treated cells. Four hours later, cells were transfected with 150 pmol siRNA. To this end, the cells were overlaid with 1,250 μl 10% FBS containing DMEM. In parallel, 250 μl serum-free DMEM was mixed with 15 μl TransIT-siQUEST transfection reagent (Mirus, Madison, WI) and 150 pmol siRNA. The mixture was added to the cells after 5 min of incubation at room temperature and then incubated for 24 h at 37°C. At 24 h p.t. the transfection procedure was repeated. Cells transfected with the transfection reagent only and nontransfected cells were used as controls. At 24 h after the second transfection, one set of cells was trypsinized, resuspended in 10% FBS containing DMEM, sedimented at 700 × g for 5 min, and resuspended in 2× Laemmli buffer. The second set of treated cells was infected at an MOI of 10 with the IBDV strain D78 for viral growth studies. At 16 h p.i. cell culture supernatants were obtained and stored at −70°C. The infected cells were also trypsinized, resuspended in 10% FBS containing DMEM, sedimented at 700 × g for 5 min, and resuspended in 2× Laemmli buffer. The viral titers (50% tissue culture infective doses [TCID50]) were determined on DF-1. To this end, the virus was titrated by 10-fold dilutions in DMEM. A 100-μl aliquot of each virus dilution was pipetted into four wells each of a 96-well plate, and 100 μl of DF-1 cells with a density of 5 × 105 cells/ml was added in 10% FBS-containing DMEM. Cells were incubated for 5 days at 37°C in 5% CO2. Tissue culture wells showing cytopathic effect were counted as positive. The titer was calculated based on the method described by Reed and Munch (41).
RESULTS
Characterization of generated antisera.
Chicken anti-VP1 antiserum (ck-anti-VP1) and rabbit anti-VP3 antiserum (rab-anti-VP3) were characterized by Western blotting (Fig. 1 A). As expected the ck-anti-VP1 and r-anti-VP3 specifically reacted with the appropriate proteins, using an appropriate previously described antiserum as a control (3, 33).
For the generation of a rabbit anti-ckNF45 serum (r-anti-ckNF45), the ORF of ckNF45 protein was amplified by RT-PCR, cloned, and sequenced. The sequence revealed 100% identity of the in silico-translated amino acid sequence with the previously published amino acid sequence of interleukin enhancer binding factor 2 (XP_423437) or chicken NF45 (ckNF45). The purified recombinant ckNF45-His was analyzed by protein staining and via Western blotting with an anti-His tag monoclonal antibody (Fig. 1B). The analysis revealed two protein bands in the eluate with apparent molecular masses of approximately 48 kDa and a weak band at 42 kDa (Fig. 1B, left panel). The molecular masses of both larger bands were in the range of 45 kDa, which is in accordance with the determined theoretical molecular weight (http://www.expasy.ch/tools). The identity as ckNF45-His for both bands was confirmed by matrix-assisted laser desorption ionization-time of flight tandem mass spectrometry (Proteomics Resource Facility, University of Georgia, Athens, GA). Since Western blot analysis with anti-6×His antibody stained both large ckNF45-His bands, we assumed the larger band represents the full-length ckNF45 and the lower band represents a modified form and/or an N-terminal degradation product. The antiserum r-anti-ckNF45 was used in Western blotting and immunofluorescence assay for testing for its reactivity with cellular proteins and its specificity (Fig. 1B and C). The rabbit serum r-anti-ckNF45 recognized, in samples of purified ckNF45-His protein, a protein of approximately 26 kDa in addition to the 48-kDa/42-kDa proteins. For Western blot analysis, lysates of cells representing different species were tested, i.e., chicken (chicken embryo cells [DF-1 cells]), quail (QM-7 cells), monkey (Vero cells), dog (MDCK cells), hamster (BHK21 cells), and human (293 T cells) (Fig. 1B, right panel). In each case, the newly established r-anti-ckNF45 serum stained a protein band with a molecular mass of approximately 46 kDa, indicating a broad reactivity of the serum against the NF45 proteins from different species. In addition, a smaller protein of approximately 26 kDa was stained in some of the analyzed samples (CEC and MDCK). Lysates of QM7 and DF-1 cells also showed a reaction with a protein of ca. 42 kDa, while lysates of Vero cells detected a faint reactive protein band at approximately 35 kDa after prolonged exposure (data not shown). In the cell line of human origin (293T), a band of approximately 80 kDa was visible. As expected, IIF on Vero, DF-1, and QM7 cells revealed predominantly nuclear staining after incubation with the r-anti-ckNF45 serum (Fig. 2 A, C, and E), while in BHK21 cells the fluorescence was hardly visible and was not concentrated in the nucleus of the cell (Fig. 2G). We used BHK21 cells, the cells that revealed the least apparent IF signal, to test for the specificity of the generated r-anti-ckNF45 serum. To this end, BHK21 cells were transfected with pcDNA3-ckNF45FLAG for transient expression of ckNF45-FLAG. At 24 h p.t. the cells were fixed and incubated with both the r-anti-ckNF45 serum and a mouse anti-FLAG MAb (Fig. 1C). Notably, we observed an evident colocalization of NF45 and the FLAG peptide (Fig. 1C, merged image), providing additional evidence for the specificity of the r-anti-ckNF45 serum. Moreover, the heterologously expressed ckNF45 protein was shown to be functional; it was found to efficiently localize to the nucleus of the cell.
FIG. 2.
Cytoplasmic accumulation of NF45 during IBDV infection. DF-1 (A and B), QM-7 (C and D), Vero (E and F), and BHK21 (G and H) cells were infected with IBDV strain D78 or left uninfected to serve as controls (c). At 12 h p.i., the cells were ethanol fixed and incubated with anti-VP3 MAb IBDV2 (VP3), rabbit anti-ckNF45-His (NF45), and PI. The binding of the primary antibodies was visualized with species-specific conjugates (goat anti-mouse-Cy3 and goat anti-rabbit-FITC). An overlay of the confocal pictures is shown (merge).
In IBDV-infected cells NF45 accumulates in the cytoplasm.
To test for a potential contact between NF45 and the viral replication machinery, we infected chicken-derived DF-1 cells, QM-7 (quail) cells, and Vero (monkey) cells with IBDV strain D78. The cells were fixed at 12 h p.i. and incubated with r-anti-ckNF45 serum, PI to stain the nucleus, and the anti-IBDV VP3 MAb IBDV2. As shown in uninfected control cells of all three species, NF45 was found predominantly in the nucleus of the cells (Fig. 2A, C, and E). However, this phenotype dramatically changed when the cells were infected with IBDV as detected with the anti-VP3 MAb. In the infected cells, NF45 evidently accumulated in the cytoplasm of the cell. Most interestingly, the cytoplasmic fraction of NF45 accumulated in regions of the cell where accumulated VP3 was present, as indicated by the overlapping fluorescence (Fig. 2B, D, and F). This colocalization was species independent, as it was observed in cells derived from chicken, quail, and monkeys. Since Vero cells lack the ability to express type I (alpha/beta) interferons (IFNs) due to a chromosomal deletion in their genome (8), we concluded that the cytoplasmic accumulation of NF45 as a consequence of the viral infection occurred independently from the expression of IFN-regulated genes. To investigate if this reactivity of r-anti-ckNF45His serum was due to a nonspecific reaction with replication stages of IBDV, BHK21 cells were either IBDV infected (Fig. 2H) or left uninfected (Fig. 2G) and then stained with antibodies as described above. The result showed clearly that r-anti-ckNF45His serum did not react with replication stages of IBDV, since only VP3 fluorescence without accumulation of NF45 fluorescence was observed in the cytoplasm of infected cells.
In view of previously published data of Tacken et al. (52), who demonstrated an interaction of the viral RNA-dependent RNA polymerase VP1 with VP3, we next investigated whether VP1 was accumulating at places where NF45 was present. Accordingly, DF-1 cells were infected with IBDV strain D78, and the cells were fixed and incubated with r-anti-ckNF45-His serum, chicken anti-VP1, DAPI, and anti-VP3 MAb IBDV2. As expected, VP1 was found where VP3 and NF45 were accumulating in the cytoplasm of infected cells (Fig. 3 A). Interestingly, NF45 accumulated at the same spot as VP3 and VP1 in the infected cells, suggesting that NF45 accumulates in cell complexes where viral replication occurs. Next we tested possible colocalization of NF45 with VP2 and VP4 by using appropriate virus protein-specific MAb R63 and IBDV6, respectively. We observed that VP2 colocalized with NF45 (Fig. 3B, upper panel), whereas VP4 was detected in regions where NF45 was not visible (Fig. 3B, lower panel). Based on these results it can be concluded that NF45 accumulates in regions where IBDV proteins VP3, VP2, and VP1 are present.
FIG. 3.
NF45 colocalizes with several IBDV proteins, and its cytoplasmic accumulation is IBDV strain independent. DF-1 cells were infected with either the IBDV strain D78 (A and B) or the variant strains 8903 (E/Del subtype) and GLS-05 (GLS subtype) (C). At 12 h p.i., fixed cells were incubated with the anti-VP2 MAb R63 (VP2), the anti-VP4 MAb IBDV 6 (VP4), the chicken anti-VP1 serum (VP1), and the rabbit anti-ckNF45-His serum (NF45). The cellular nuclei were stained either with DAPI (A) or (B and C). Samples were incubated with appropriate conjugates: (A) goat anti-mouse-Cy5, goat anti-rabbit-FITC, or goat anti-chicken-Cy3; (B and C) goat anti-mouse-Cy5 or goat anti-rabbit-FITC. The overlay of the pictures taken by confocal laser scanning microscopy is shown (merge).
Further experiments were conducted to clarify whether this phenotype was dependent on the applied IBDV strain. That is, DF-1 cells were infected with IBDV strains 8903 and GLS-05, belonging to the variant strain subtypes E/Del and GLS, respectively. As observed earlier with the classical strain D78, infections with 8903 and GLS-05 showed essentially the same phenotype, i.e., an evident accumulation of NF45 at places in the cell where VP3 was present (Fig. 3C).
Cytoplasmic accumulation of NF45 correlates with the time course after viral infection.
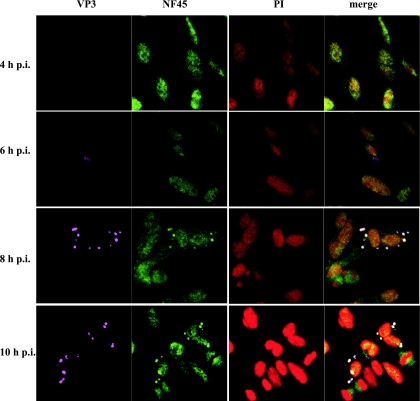
To investigate the dynamics of NF45 accumulation during infection, DF-1 cells were infected with IBDV strain D78, fixed at 4, 6, 8, and 10 h p.i., and analyzed by indirect immunofluorescence (Fig. 4). The detection of VP3 with the MAb IBDV2 was used as an indicator for viral replication. At 4 h p.i. VP3 was not detectable, and NF45 was monitored exclusively in the nucleus of the analyzed cells. At 6 h p.i., VP3 could be detected in the cytoplasm of the infected cells, while NF45 was still localized in the nucleus. Interestingly, this phenotype changed 2 h later. That is, at 8 h p.i., NF45 accumulated at places in the cell where VP3 was present. Accumulation of NF45 in the cytoplasm increased even more at 10 h p.i. These results led to the assumption that accumulation of NF45 in the cytoplasm corresponds to the time after viral infection.
FIG. 4.
NF45/VP3 colocalization correlates with the time course after viral infection. IBDV strain D78-infected DF-1 cells were ethanol fixed at different time points (4 h, 6 h, 8 h, and 10 h p.i.). The cells were subjected to indirect immunofluorescence analysis using the mouse anti-VP3 MAb IBDV 2 (VP3), anti-ckNF45-His serum (NF45), and PI followed by incubation with goat anti-rabbit-FITC and goat anti-mouse-Cy5. Colocalization of the antibody binding can be observed in the merged confocal picture.
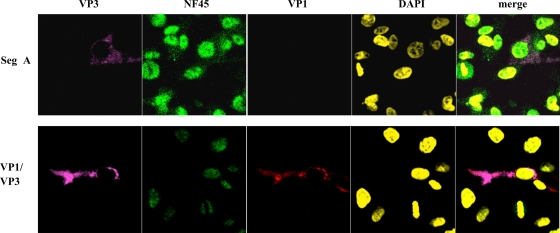
In an additional experiment, it was investigated whether cytoplasmic accumulation of NF45 was exclusively dependent on the presence of VP3. DF-1 cells were transfected with segment A cRNA as previously described (35), and 12 h p.t. the cells were analyzed for NF45 accumulation in the cytoplasm. As shown in Fig. 5 (upper panel), NF45 did not accumulate in the cytoplasm of the cell despite the fact that VP3 was present at high quantities. Furthermore, pc3-VP3 and pc3-VP1 were cotransfected to investigate if the presence of both proteins was sufficient to induce an accumulation of NF45 in the cytoplasm (Fig. 5, lower panel). At 24 h after transfection, it was observed that although VP1 and VP3 colocalized in the cell, NF45 did not accumulate in the cytoplasm. These results indicate that cytoplasmic accumulation of the cellular protein NF45 during viral infection is likely a cellular response to a certain but so-far-unknown stage of viral infection rather than being related to the dual expression of VP3 or VP1.
FIG. 5.
NF45 does not accumulate in the cytoplasm in the presence of VP3 and VP1. Either T7 polymerase in vitro-transcribed cRNA of segment A (Seg A) of IBDV strain D78 or recombinant plasmids encoding IBDV VP1 and VP3 (VP1/VP3) were transfected into DF-1 cells. At 12 h p.t., the cells were ethanol fixed and incubated with mouse anti-VP3 MAb IBDV2 (VP3), chicken anti-VP1 serum (VP1), or rabbit anti-ckNF45-His serum (NF45). The DNA in the cellular nuclei was stained with DAPI. The presence of the primary antibody was detected with goat anti-mouse-Cy5, goat anti-rabbit-FITC, and goat anti-chicken-Cy3. The pictures were taken by confocal laser scanning microscopy and were also merged.
Blocking of the Crm1-dependent export does not influence cytoplasmic accumulation of NF45 in IBDV-infected cells.
Next, we wanted to understand if the observed phenotype, namely, the accumulation of NF45 in the cytoplasm of IBDV-infected cells, was due to translocation of NF45 from the nucleus to the cytoplasm in a Crm1-dependent pathway. To address this question, we applied leptomycin B (LMB), a Streptomyces metabolite that is known to interact and inhibit Crm1 (20). It is a factor essential for the nuclear export of proteins containing nuclear export signals (NES). For example, it is known that LMB inhibits the translocation of the influenza A virus proteins NP and M1 from the nucleus to the cytoplasm (30). Therefore, we used influenza virus-infected cells as positive controls for the experiments. NF45 contains three potential NES (leu-187, leu-221, and leu-226 [http://www.cbs.dtu.dk/services/NetNES/]). Thus, DF-1 cells were infected with either IBDV strain D78 (Fig. 6, upper panel) or with an avian influenza virus subtype H5N2 as a positive control (Fig. 6, lower panel). At 3 h p.i. 2 ng/ml LMB was added, and at 12 h p.i. the cells were fixed and analyzed by IF as described above. The results showed that NP and M1 of H5N2 remained in the nucleus in the presence of LMB, indicating inhibition of the nuclear export of these two proteins as shown before (30). In contrast, NF45 accumulated in the cytoplasm of the IBDV-infected cells irrespective of the LMB treatment. We concluded that NF45 is not actively transported out of the nucleus via the Crm1 pathway, although other pathways for the protein export from the nucleus cannot be excluded. It is also possible that NF45 accumulates in the cytoplasm in response to IBDV infection before being translocated to the cellular nucleus.
FIG. 6.
Blocking of Crm1-dependent export does not influence cytoplasmic accumulation of NF45 in the cytoplasm of infected cells. DF-1 cells were infected with low-pathogenicity avian influenza virus H5N2 or IBDV strain D78. Cells were either treated with LMB (wLMB) or left untreated (w/oLMB). At 12 h p.i., cells were ethanol fixed. Indirect immunofluorescence of H5N2-infected cells was performed using a rabbit anti-NP antiserum (NP) and a mouse anti-M1 MAb (M1) (lower panels). IBDV-infected cells were incubated with the mouse anti-VP3 MAb IBDV2 and rabbit anti-ckNF45-His serum (upper panels). The binding of the antibodies was visualized by using goat anti-mouse-Cy5 and goat anti-rabbit-FITC conjugated antibodies. The nucleus was detected by PI staining. Colocalization was visualized by merging the pictures taken by confocal laser scanning microscopy.
NF45 association with viral proteins.
The following experiments were aimed at investigating NF45 for an association with viral proteins. For this purpose, DF-1 cells were transfected with the plasmid pcDNA3-ckNF45FLAG (see Materials and Methods) for transient expression of ckNF45-FLAG. At 24 h p.t., transfected and nontransfected cells were infected with IBDV strain D78 at an MOI of 100 for 24 h. Appropriate control experiments (nontransfected infected cells, transfected noninfected cells, and nontransfected noninfected cells) were performed in parallel. Due to the treatment with Igepal CA-630, the cellular nuclei were not destroyed and they were removed by centrifugation. Thus, only the cytoplasmic fraction of the cells was analyzed. The analysis of the lysates by Western blotting showed that the ectopic ckNF45-FLAG protein, due to the additional FLAG tag, migrated at a slightly higher molecular mass than the endogenous NF45 in the analytical SDS-PAGE. It was also always detected as a double band (Fig. 7 A, left panel; see also Fig. 1). After immunoprecipitation assays with anti-FLAG MAb (see Materials and Methods), NF45 was precipitated exclusively from the cytoplasmic lysates of the plasmid-transfected cells, again as a double band (Fig. 7A, right panel,). The viral protein VP3 was, as expected, found solely in the cytoplasmic lysates of infected cells (Fig. 7A, left panel). Interestingly, VP3 coimmunoprecipitated via the anti-FLAG antibody, indicating an evident association of NF45 and VP3 (Fig. 7A, right panel), although other viral or cellular proteins might be involved. Furthermore, in the cytoplasmic lysates of pVP2, VP2 and VP1 were also detected (Fig. 7A, left panel). Interestingly, VP1 also coimmunoprecipitated with the anti-FLAG antibody, although to a much lower extent than VP3. In contrast to the results obtained by immunofluorescence, VP2 was not detected after immunoprecipitation with the anti-FLAG MAb (Fig. 7A, right panel). This finding does not exclude an interaction between VP2 and NF45. In fact, in further experiments using infected but not transfected DF-1 cells, NF45 was immunoprecipitated by using the VP2-specific MAb R63 (Fig. 7B, right panel). Further immunoprecipitation experiments also showed that the anti-VP3 MAb IBDV2 was able to precipitate NF45 as well (Fig. 7C, right panel). In agreement with results from Tacken et al. (52), the VP3-specific MAb was able to immunoprecipitate VP1, confirming an association of VP3 with VP1. In contrast, the rabbit anti-VP1 serum was not able to immunoprecipitate NF45, but was able to immunoprecipitate VP3 (data not shown). Furthermore, the VP2-specific MAb R63 was able to precipitate VP3. The immunoprecipitation of NF45 confirmed the data obtained by immunofluorescence, in which colocalization of VP2 and VP3 with NF45 was observed (Fig. 3).
FIG. 7.
NF45 interacts with viral proteins VP1, VP2, and VP3. (A) Lysates of the cytoplasmic fractions of nontransfected DF-1 cells (DF1), DF-1 cells transfected with pcDNA3-ckNF45FLAG (DF1/NF45), nontransfected but IBDV-infected DF-1 cells (DF1/D78), and pcDNA3-ckNF45-FLAG-transfected and IBDV-infected Df1 cells (Df1/NF45/D78) were analyzed either directly in a Western blot assay (WB) or were immunoprecipitated (IP) using the anti-FLAG MAb followed by Western blot analysis of the IP products. (B and C) Lysates of the cytoplasmic fractions of noninfected DF-1 cells (DF1) and IBDV-infected DF-1 cells (DF1/D78) were either directly used for WB or for IP using the VP2-specific MAb R63 (B) or the VP3-specific MAb IBDV2 (C) followed by Western blot analysis. Western blots shown in all three panels were performed using rabbit anti-ckNF45-His serum or rabbit anti-VP1, -VP2, or -VP3 serum. The detected proteins are named and marked by arrows. The molecular masses (in kilodaltons) of protein markers are indicated to the left of each gel.
It was important to understand if the NF45-VP3 association was due to protein interactions or caused by nonspecific RNA bridging. Accordingly, we treated the cytoplasmic extracts of cells that were transfected with the ckNF45-FLAG plasmid and also infected with IBDV with RNase T1 (digesting single-stranded RNA), RNase III (digesting dsRNA), or both enzymes, respectively. As shown in Fig. 8 A (left panel), digestion with either RNase T1 or combinations of RNase T1 and III led to a complete digestion of internal RNA. Next, we performed immunoprecipitation assays with the RNase-treated extracts and a subsequent Western blot assay as described above. We found with all samples an evident coprecipitation of NF45 and VP3 via the anti-FLAG antibody (Fig. 8A, right panel). To investigate if a similar phenotype was observed after only IBDV infection of DF-1 cells, lysates of infected cells were RNase treated as described above and subjected to immunoprecipitation using either the VP2-specific (R63) or the VP3-specific (IBDV2) MAbs. We observed a very similar phenotype to what we observed in lysates that had been not treated with RNAses (Fig. 8B, right panel). These data suggest that the IBDV VP3 and NF45 association is not mediated by viral and/or cellular RNA.
FIG. 8.
Coimmunoprecipitation of NF45 and IBDV proteins after RNase treatment. Lysates of pcDNA3-ckNF45-FLAG-transfected and subsequent IBDV-infected DF-1 cells (A) or DF-1 cells only IBDV infected (B) were subjected to RNA digestion with either RNase T1 or RNase III or double-treated (T1+ R III) for 1 h at 37°C. Untreated cells were used as controls. One portion of each sample was subsequently analyzed for RNA content on a 1% agarose gel (left panel). The remaining portions of the samples were used for immunoprecipitations (IP). IP were performed with either the anti-FLAG MAb (A) or the VP2-specific MAb R63 and the VP3-specific MAb IBDV2 (B). The resulting precipitates were separated by SDS-PAGE and analyzed by Western blotting (WB) using either rabbit anti-ckNF45-His serum (anti-VP3 or -VP1) or IBDV protein-specific antiserum (rabbit anti-IBDV serum).
RNAi-mediated knockdown of NF45 increases IBDV replication.
Generally, two scenarios were conceivable to explain the observed viral/cellular protein associations linked with the viral replication process. On the one hand, NF45 association might support viral replication as observed earlier with the positive-strand RNA viruses BVDV and HCV (15, 16). Alternatively, it might inhibit viral replication, as found with the negative-strand RNA viruses vesicular stomatitis virus and influenza A virus (39, 57). To address this issue, DF-1 cells were transfected with siRNAs directed against the cNF45 mRNA (siRNA185, siRNA551, or a combination of both siRNAs; see Materials and Methods). At 24 h after the second transfection, one set of treated cells and the controls were harvested for assessment of NF45 expression by Western blotting. Subsequently, the second set of siRNA-transfected and control cells were infected with IBDV, and 16 h after infection the supernatants were harvested and the viral titers were determined. In addition, NF45 expression was also assessed by using cell lysates after infection. Nontransfected cells, mock-transfected cells, or cells transfected with a nonsilencer siRNA served as controls. Three independent experiments were performed and analyzed (Fig. 9). Western blot analysis of proteins of the whole cells showed that the amount of NF45 could be lowered by treatment with the specific siRNAs at both time points, before and after infection, to approximately 60% of the original endogenous level of cNF45. For normalization of the amount of analyzed proteins, the quantity of chicken β-actin was used. Analysis of the viral titers 16 h p.i. showed that the highest viral titers were observed in the nontransfected controls. Notably, the use of transfection reagent in the mock-transfected controls and the transfection with nonspecific siRNA resulted in viral titers of approximately 27% and 20% in comparison to the nontransfected controls (Fig. 9). This indicated that the transfection procedure alone influenced the cells and resulted in a decrease of viral titer. Based on these findings, which were obtained regardless of the different transfection reagents that were tested in the course of this study (data not shown), the virus titers obtained after transfection with the nonspecific siRNA were set as 100%. It was observed that transfection with NF45-specifc siRNAs and knockdown of endogenous NF45 was found to generally increase viral replication (Fig. 9). That is, the viral titers were on average 4.3-fold higher after transfection with each siRNA, whereas the combined transfection of the siRNA resulted in a 5.6-fold increase of the viral titer. In sum, these data revealed that lowering the amount of NF45 in the cytoplasm of the cell favors viral replication (see below).
FIG. 9.
Downregulation of NF45 expression increases viral IBDV titers. NF45 expression and virus titers in the supernatants of IBDV-infected DF-1 cells were investigated after transfection with the NF45-specific siRNAs 185 (si185) and 551 (si551), either alone or in combination (si185 si551). Three controls were applied in these experiments: a nonspecific siRNA (Con-siRNA), mock-transfected cells (Mock) and untransfected cells (Con). Protein expression of NF45 was analyzed by Western blotting with protein samples taken from parallel transfected cells before infection and at the time of harvest of the cell culture supernatant (after infection). The analyzed protein amount was normalized for chicken β-actin by using an HRP-conjugated mouse β-actin MAb. The presence of cellular NF45 was analyzed using the rabbit anti-ckNF45-His serum. The graphs show the average measured virus titers (in TCID50/ml) analyzed in DF-1 cells from three individual experiments. Error bars show the standard deviations. The average titer value is shown within the corresponding column.
DISCUSSION
In this report, we provide initial evidence on the involvement of a cellular factor, NF45, in the replication process of the dsRNA virus IBDV. NF45 has been indicated to act as an important regulatory factor involved in transcription, microRNA processing, nuclear export of RNAs, and posttranscriptional and translational regulation of gene expression (2, 11, 21, 22, 39, 40, 42, 56). Along this line, it is of particular interest that NF45 and its interaction partner, NF90, were found to be involved in various steps of the life cycles of different viruses (1, 15, 16, 27, 32, 39, 45, 48, 57). The exact functions of NF45 and the NF45/NF90 heterodimer in the diverse viruses life cycles are far from being understood. However, in sum, it appears that these proteins act as RNA binding components which modulate either viral RNA replication or the function of virus-encoded RNAs as in the case of adenovirus (27) (see below). Moreover, several studies have suggested that NF45 and NF90 are part of an as-yet-uncharacterized innate immune surveillance system and, perhaps, also host defense mechanism that protects against viral infections (references 39 and 57 and this study).
In our study with IBDV, we first generated the recombinant chicken ckNF45 and a specific antiserum that enabled specific investigations on infected cells. As reported earlier with human and bovine cells (15, 16), NF45 was found to be present in at least two forms in the cell. However, the exact natures of these forms were not further analyzed here. As with the closely related human and bovine counterparts, ckNF45 was found predominantly in the nucleus of noninfected cells (Fig. 2). However, possibly the most striking observation of this study was that this compartmentalization changed drastically as soon as the cells were infected with IBDV (Fig. 2). A considerable accumulation of ckNF45 in the cytoplasm of IBDV-infected cells was observed in different cell types and with different IBDV strains, and it was shown to occur independently of Crm1-dependent nuclear export of cellular protein (Fig. 2, 3, and 5). These data resembled earlier observations of Isken et al. (15), who conducted studies with bovine and human cells that were infected with BVDV and HCV. Moreover, cytoplasmic accumulation of NF45 was shown to be independent of an interferon response, as demonstrated in infection experiments with the interferon-negative Vero cells (Fig. 2).
Detailed immunofluorescence experiments further revealed an evident colocalization of NF45 with VP1, VP2, and VP3 at the sites of viral replication in the infected cell. They also showed a clear correlation of this cellular phenotype with the course of viral replication (Fig. 2, 3, and 4). The results of the immunoprecipitations suggest that proteins which are part of the replication complexes interact either directly or indirectly via another viral/cellular protein with NF45. The interaction of VP2 with VP3 was suggested before (37), as it was described for the VP3-VP1 interaction (52). Based on the obtained results, it seemed that VP3 stays in the center of the association with NF45, since VP1 immunoprecipitated in a very minor amount and VP2 was not detected after immunoprecipitation using NF45-FLAG as bait antibody. The immunoprecipitation with an anti-VP3 MAb resulted in a strong signal for VP1 and NF45, but no VP2 was detected. The VP3 center hypothesis was also supported by the finding that precipitation with an anti-VP2 MAb resulted in weak precipitation of both VP3 and NF45. Interestingly, immunoprecipitation assays that were performed in the presence of RNases demonstrated that the protein-protein association between NF45 and VP3 in infected cells was likely RNA independent. Furthermore, these protein associations, as well as NF45 accumulation in the cytoplasm, were undetectable if the IBDV polyprotein was heterologously expressed by only segment A or by coexpression of VP1 and VP3 (Fig. 5). Taken together, these data indicate a correlation between the formation of functional viral replication complexes and NF45 recruitment in the cytoplasm. It will be interesting to investigate whether NF90 and/or NF110 are also part of this viral recruitment.
Finally, we addressed the question of the functional relevance of this apparent recruitment of NF45 to the IBDV replication machinery. RNAi knockdown of NF45 and infection experiments resulted in an up-to-4.3-fold increase of the virus titer (Fig. 9). Moreover, after combined transfection of both NF45-specific siRNAs, the viral titers increased even more. This finding indicates that accumulation of NF45 is part of a host defense system. Such a scenario is congruent with data obtained by Wang et al. (57) and Pfeifer et al. (39), in which knockdown of the NF45 binding partner NF90 was found to increase the replication of influenza virus and vesicular stomatitis virus, respectively. It would also fit with earlier observations indicating that the heterodimeric complex of NF90 and NF45 was found associating with the PKR-modulating VA RNAII of adenovirus (27). Moreover, NF90/NF45 was also found to modulate the activity of PKR and ADAR, perhaps by direct interactions with these important players in cellular innate immunity (24, 36, 38). It will be important to further investigate this interesting aspect in the immediate future and to examine the role of VP3 within this context.
Acknowledgments
This study was supported by grants from the Georgia Research Alliance to E.M. and the Deutsche Forschungsgemeinschaft (DFG BE 1885/6) to S.-E.B.
We thank Jamie Barber (AHRC Cell Imaging Core Facility, University of Georgia) for help with the confocal laser scanning microscopy and Ruud Hein (Intervet-Schering/Plough) for providing the IBDV strains D78, 8903, and GLS-05.
Footnotes
Published ahead of print on 11 August 2010.
REFERENCES
- 1.Agbottah, E. T., C. Traviss, J. McArdle, S. Karki, G. C. St Laurent 3rd, and A. Kumar. 2007. Nuclear factor 90 (NF90) targeted to TAR RNA inhibits transcriptional activation of HIV. Retrovirology 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber, G. N. 2009. The NFAR's (nuclear factors associated with dsRNA): evolutionarily conserved members of the dsRNA binding protein family. RNA Biol. 6:35-39. [DOI] [PubMed] [Google Scholar]
- 3.Birghan, C., E. Mundt, and A. E. Gorbalenya. 2000. A non-canonical lon proteinase lacking the ATPase domain employs the ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J. 19:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corthésy, B., and P. N. Kao. 1994. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J. Biol. Chem. 269:20682-20690. [PubMed] [Google Scholar]
- 5.Cosgrove, A. S. 1962. An apparently new disease of chickens: avian nephrosis. Avian Dis. 6:385-389. [Google Scholar]
- 6.Da Costa, B., C. Chevalier, C. Henry, J. C. Huet, S. Petit, J. Lepault, H. Boot, and B. Delmas. 2002. The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2. J. Virol. 76:2393-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delmas, B., F. S. B. Kibenge, J. C. Leong, E. Mundt, V. N. Vakharia, and J. L. Wu. 2005. Birnaviridae, p. 561-569. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and A. L. Ball (ed.), Virus taxonomy, Academic Press, London, England.
- 8.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. U. S. A. 85:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobos, P., B. J. Hill, R. Hallett, D. T. Kells, H. Becht, and D. Teninges. 1979. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J. Virol. 32:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granzow, H., C. Birghan, T. C. Mettenleiter, J. Beyer, B. Köllner, and E. Mundt. 1997. A second form of infectious bursal disease virus-associated tubule contains VP4. J. Virol. 71:8879-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan, D., N. Altan-Bonnet, A. M. Parrott, C. J. Arrigo, Q. Li, M. Khaleduzzaman, H. Li, C. G. Lee, T. Pe'ery, and M. B. Mathews. 2008. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol. Cell. Biol. 28:4629-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 13.Icard, A. H., H. S. Sellers, and E. Mundt. 2008. Detection of infectious bursal disease virus isolates with unknown antigenic properties by reverse genetics. Avian Dis. 52:590-598. [DOI] [PubMed] [Google Scholar]
- 14.Irigoyen, N., D. Garriga, A. Navarro, N. Verdaguer, J. F. Rodríguez, and J. R. Castón. 2009. Autoproteolytic activity derived from the infectious bursal disease virus capsid protein. J. Biol. Chem. 284:8064-8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isken, O., M. Baroth, C. W. Grassmann, S. Weinlich, D. H. Ostareck, A. Ostareck-Lederer, and S. E. Behrens. 2007. Nuclear factors are involved in hepatitis C virus RNA replication. RNA 13:1675-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isken, O., C. W. Grassmann, R. T. Sarisky, M. Kann, S. Zhang, F. Grosse, P. N. Kao, and S. E. Behrens. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 22:5655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isken, O., C. W. Grassmann, H. Yu, and S. E. Behrens. 2004. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA 10:1637-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao, P. N., L. Chen, G. Brock, J. Ng, J. Kenny, A. J. Smith, and B. Corthésy. 1994. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J. Biol. Chem. 269:20691-20699. [PubMed] [Google Scholar]
- 19.Klopfleisch, R., O. Werner, E. Mundt, T. Harder, and J. P. Teifke. 2006. Neurotropism of highly pathogenic avian influenza virus /chicken/Indonesia/2003 (H5N1) in experimentally infected pigeons (Columbia livia f. domestica). Vet. Pathol. 43:463-470. [DOI] [PubMed] [Google Scholar]
- 20.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 21.Kuwano, Y., H. H. Kim, K. Abdelmohsen, R. Pullmann, Jr., J. L. Martindale, X. Yang, and M. Gorospe. 2008. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell. Biol. 28:4562-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuwano, Y., R. Pullmann, Jr., B. S. Marasa, K. Abdelmohsen, E. K. Lee, X. Yang, J. L. Martindale, M. Zhan, and M. Gorospe. 2010. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 38:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head for bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Langland, J. O., P. N. Kao, and B. L. Jacobs. 1999. Nuclear factor-90 of activated T-cells: a double-stranded RNA-binding protein and substrate for the double-stranded RNA-dependent protein kinase, PKR. Biochemistry 38:6361-6368. [DOI] [PubMed] [Google Scholar]
- 25.Lejal, N., B. Da Costa, J.-C. Huet, and B. Delmas. 2000. Role of Ser-652 and Lys-692 in the protease activity of infectious bursal disease virus VP4 and identification of its substrate cleavage sites. J. Gen. Virol. 81:983-992. [DOI] [PubMed] [Google Scholar]
- 26.Letzel, T., E. Mundt, and A. E. Gorbalenya. 2007. Evidence for functional significance of the permuted C motif in Co2+-stimulated RNA-dependent RNA polymerase of infectious bursal disease virus. J. Gen. Virol. 88:2824-2833. [DOI] [PubMed] [Google Scholar]
- 27.Liao, H. J., R. Kobayashi, and M. B. Mathews. 1998. Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc. Natl. Acad. Sci. U. S. A. 95:8514-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombardo, E., A. Maraver, I. Espinosa, A. Fernández-Arias, and J. F. Rodriguez. 2000. VP5, the nonstructural polypeptide of infectious bursal disease virus, accumulates within the host plasma membrane and induces cell lysis. Virology 277:345-357. [DOI] [PubMed] [Google Scholar]
- 29.Luque, D., I. Saugar, M. T. Rejas, J. L. Carrascosa, J. F. Rodríguez, and J. R. Castón. 2009. Infectious bursal disease virus: ribonucleoprotein complexes of a double-stranded RNA virus. J. Mol. Biol. 386:891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, K., A. M. Roy, and G. R. Whittaker. 2001. Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology 282:215-220. [DOI] [PubMed] [Google Scholar]
- 31.Maraver, A., A. Oña, F. Abaitua, D. González, R. Clemente, J. A. Ruiz-Díaz, J. R. Castón, F. Pazos, and J. F. Rodriguez. 2003. The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J. Virol. 77:6438-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merrill, M. K., and M. Gromeier. 2006. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J. Virol. 80:6936-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mundt, E., J. Beyer, and H. Müller. 1995. Identification of a novel viral protein in infectious bursal disease virus-infected cells. J. Gen. Virol. 76:437-443. [DOI] [PubMed] [Google Scholar]
- 34.Mundt, E., B. Köllner, and D. Kretzschmar. 1997. VP5 of infectious bursal disease virus is not essential for viral replication in cell culture. J. Virol. 71:5647-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mundt, E., and V. N. Vakharia. 1996. Synthetic transcripts of double-stranded Birnavirus genome are infectious. Proc. Natl. Acad. Sci. U. S. A. 93:11131-11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nie, Y., L. Ding, P. N. Kao, R. Braun, and J. H. Yang. 2005. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol. Cell. Biol. 25:6956-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oña, A., D. Luque, F. Abaitua, A. Maraver, J. R. Castón, and J. F. Rodríguez. 2004. The C-terminal domain of the pVP2 precursor is essential for the interaction between VP2 and VP3, the capsid polypeptides of infectious bursal disease virus. Virology 322:135-142. [DOI] [PubMed] [Google Scholar]
- 38.Parker, L. M., I. Fierro-Monti, and M. B. Mathews. 2001. Nuclear factor 90 is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded RNA-activated protein kinase. J. Biol. Chem. 276:32522-32530. [DOI] [PubMed] [Google Scholar]
- 39.Pfeifer, I., R. Elsby, M. Fernandez, P. A. Faria, D. R. Nussenzveig, I. S. Lossos, B. M. Fontoura, W. D. Martin, and G. N. Barber. 2008. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc. Natl. Acad. Sci. U. S. A. 105:4173-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranpura, S. A., U. Deshmukh, and P. P. Reddi. 2008. NF45 and NF90 in murine seminiferous epithelium: potential role in SP-10 gene transcription. J. Androl. 29:186-197. [DOI] [PubMed] [Google Scholar]
- 41.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 27:493-497. [Google Scholar]
- 42.Sakamoto, S., K. Aoki, T. Higuchi, H. Todaka, K. Morisawa, N. Tamaki, E. Hatano, A. Fukushima, T. Taniguchi, and Y. Agata. 2009. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol. Cell. Biol. 29:3754-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saunders, L. R., D. J. Perkins, S. Balachandran, R. Michaels, R. Ford, A. Mayeda, and G. N. Barber. 2001. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 276:32300-32312. [DOI] [PubMed] [Google Scholar]
- 44.Shaw, J. P., P. J. Utz, D. B. Durand, J. J. Toole, E. A. Emmel, and G. R. Crabtree. 1988. Identification of a putative regulator of early T cell activation genes. Science 241:202-205. [DOI] [PubMed] [Google Scholar]
- 45.Shi, L., W. R. Godfrey, J. Lin, G. Zhao, and P. N. Kao. 2007. NF90 regulates inducible IL-2 gene expression in T cells. J. Exp. Med. 204:971-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, L., D. Qiu, G. Zhao, B. Corthesy, S. Lees-Miller, W. H. Reeves, and P. N. Kao. 2007. Dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter in vivo in activated T-cells. Nucleic Acids Res. 35:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, L., G. Zhao, D. Qiu, W. R. Godfrey, H. Vogel, T. A. Rando, H. Hu, P. N. Kao. 2005. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J. Biol. Chem. 280:18981-18989. [DOI] [PubMed] [Google Scholar]
- 48.Shim, J., H. Lim, R. Yates, and M. Karin. 2002. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell 10:1331-1344. [DOI] [PubMed] [Google Scholar]
- 49.Shin, H. J., S. S. Kim, Y. H. Cho, S. G. Lee, and H. M. Rho. 2002. Host cell proteins binding to the encapsidation signal epsilon in hepatitis B virus RNA. Arch. Virol. 147:471-491. [DOI] [PubMed] [Google Scholar]
- 50.Snyder, D. B., D. P. Lana, P. K. Savage, F. S. Yancey, S. A. Mengel, and W. W. Marquardt. 1988. Differentiation of infectious bursal disease virus directly from infected tissues with neutralizing monoclonal antibodies: evidence of a major antigenic shift in recent field isolates. Avian Dis. 32:535-539. [PubMed] [Google Scholar]
- 51.Spies, U., H. Müller, and H. Becht. 1989. Nucleotide sequence of infectious bursal disease virus genome segment A delineates two major open reading frames. Nucleic Acids Res. 17:7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tacken, M. G., B. P. Peeters, A. A. Thomas, P. J. Rottier, and H. J. Boot. 2002. Infectious bursal disease virus capsid protein VP3 interacts both with VP1, the RNA-dependent RNA polymerase, and with viral double-stranded RNA. J. Virol. 76:11301-11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ting, N. S., P. N. Kao, D. W. Chan, L. G. Lintott, and S. P. Lees-Miller. 1998. DNA-dependent protein kinase interacts with antigen receptor response element binding proteins NF90 and NF45. J. Biol. Chem. 273:2136-2145. [DOI] [PubMed] [Google Scholar]
- 54.Tran, H., M. Schilling, C. Wirbelauer, D. Hess, and Y. Nagamine. 2004. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell 13:101-111. [DOI] [PubMed] [Google Scholar]
- 55.Von Einem, U. I., A. E. Gorbalenya, H. Schirrmeier, S. E. Behrens, T. Letzel, and E. Mundt. 2004. VP1 of infectious bursal disease virus is an RNA-dependent RNA polymerase. J. Gen. Virol. 85:2221-2229. [DOI] [PubMed] [Google Scholar]
- 56.Vumbaca, F., K. N. Phoenix, D. Rodriguez-Pinto, D. K. Han, and K. P. Claffey. 2008. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol. Cell. Biol. 28:772-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, P., W. Song, B. W. Mok, P. Zhao, K. Qin, A. Lai, G. J. Smith, J. Zhang, T. Lin, Y. Guan, and H. Chen. 2009. Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. J. Virol. 83:7850-7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, Y. H., and G. A. Grabowski. 1999. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid beta-glucosidase and other mRNAs. Mol. Genet. Metab. 68:441-454. [DOI] [PubMed] [Google Scholar]
- 59.Xu, Y. H., C. Busald, and G. A. Grabowski. 2000. Reconstitution of TCP80/NF90 translation inhibition activity in insect cells. Mol. Genet. Metab. 70:106-115. [DOI] [PubMed] [Google Scholar]
- 60.Yao, K., and V. N. Vakharia. 2001. Induction of apoptosis in vitro by the 17-kDa nonstructural protein of infectious bursal disease virus: possible role in viral pathogenesis. Virology 285:50-58. [DOI] [PubMed] [Google Scholar]
- 61.Yaseen, N. R., A. L. Maizel, F. Wang, and S. Sharma. 1993. Comparative analysis of NFAT (nuclear factor of activated T cells) complex in human T and B lymphocytes. J. Biol. Chem. 268:14285-14293. [PubMed] [Google Scholar]
- 62.Yewdell, J. W., E. Frank, and W. Gerhard. 1981. Expression of influenza A virus internal antigens on the surface of infected P815 cells. J. Immunol. 126:1814-1819. [PubMed] [Google Scholar]