Abstract
Adenosine deaminases acting on RNA (ADARs) are involved in editing of adenosine residues to inosine in double-stranded RNA (dsRNA). Although this editing recodes and alters functions of several mammalian genes, its most common targets are noncoding repeat sequences, indicating the involvement of this editing system in currently unknown functions other than recoding of protein sequences. Here we show that specific adenosine residues of certain microRNA (miRNA) precursors are edited by ADAR1 and ADAR2. Editing of pri–miR-142, the precursor of miRNA-142, expressed in hematopoietic tissues, resulted in suppression of its processing by Drosha. The edited pri–miR-142 was degraded by Tudor-SN, a component of RISC and also a ribonuclease specific to inosine-containing dsRNAs. Consequently, mature miRNA-142 expression levels increased substantially in ADAR1 null or ADAR2 null mice. Our results demonstrate a new function of RNA editing in the control of miRNA biogenesis.
ADARs bind dsRNAs and deaminate adenosine residues to inosine. The resulting A→I conversions replace A-U Watson-Crick pairs with I•U wobble pairs in the dsRNA1,2. Although I•U and isosteric G•U wobble base pairs, like Watson-Crick pairs, participate in forming helical regions in RNA folding, they seem to have unique conformational and biological features3. Three members of the ADAR gene family (ADAR1–3) have been identified in vertebrates4,5. In addition, two isoforms of ADAR1, an interferon-inducible, cytoplasmic 150-kDa protein (p150) and a constitutive, nuclear 110-kDa protein (p110) are synthesized by translation initiation at alternative methionine codons6. Members of the ADAR gene family contain multiple dsRNA-binding domains and a separate deaminase domain7–10. Although apparently functional domain features are conserved in ADAR3, its enzymatic activity has not yet been demonstrated9.
An inosine residue converted from adenosine in RNA is detected as an A→G change of the complementary DNA sequence, and the translation machinery reads inosine as guanosine, leading to alterations of codons. Relatively few editing sites located within the coding sequences of target genes have been identified by comparing individual cDNA sequences to their corresponding genomic sequences4,5. Involvement of both ADAR1 and ADAR2 in site-selective A→I RNA editing of these sites has been well established10,11. The RNA editing profoundly alters the functions of target genes such as glutamate receptor subunits and serotonin receptor 2C (5-HT2CR)11–14. Studies of in vitro RNA editing using purified recombinant proteins as well as analysis of in vivo editing-pattern alterations in ADAR1 null or ADAR2 null neurons indicate that ADAR1 and ADAR2 have substantial differences in site selectivity. The editing sites of ADAR1 and ADAR2 revealed by in vitro assays accurately predict their in vivo editing site selectivity. For instance, ADAR2 edits almost exclusively the ‘D’ site of 5-HT2CR precursor messenger RNA (pre-mRNA) both in vitro and in vivo, whereas ADAR1 selectively edits the ‘A’ and ‘B’ sites11,15,16. The editing site selectivity of different ADARs indicates a difference in their recognition of substrate RNA, possibly mediated through the functional interactions of the two monomers of the ADAR1 or ADAR2 homodimer17.
Many A→I RNA editing sites have been revealed recently through systematic computational analysis of human expressed sequence tag and genome databases18–20. Notably, almost all the new sites identified in the human transcriptome (over 10,000 sites mapped in ~2,000 different genes) reside in noncoding introns and 3′ untranslated regions that consist of inversely oriented repetitive elements18–20. These results implicate A→I RNA editing and ADARs in a new function, perhaps one very different from recoding of protein-coding genes, for example regulating noncoding RNAs and silencing of retrotransposons21.
Numerous small noncoding RNAs, miRNAs involved in RNA-mediated interference (RNAi), have been identified22–24. The miRNA precursors are transcribed from invertebrate and vertebrate genomes as well as certain DNA virus genomes. They are processed sequentially by two members of the RNase III superfamily, Drosha and Dicer, which recognize the foldback hairpin structure of the miRNA precursors25,26. First, nuclear Drosha together with its essential partner DGCR8 (refs. 27–30) cleaves long, primary transcripts (pri-miRNAs), releasing 60- to 70-nucleotide (nt) intermediates, pre-miRNAs25,26. Recognition of correctly processed pre-miRNAs and their nuclear export is carried out by exportin-5 and RanGTP31. Cytoplasmic Dicer, together with a dsRNA-binding protein, TRBP, then processes the pre-miRNAs into 20- to 22-nt short interfering RNA (siRNA)-like duplexes32,33. One or both strands of the duplex may serve as the mature miRNA. After integration into the RNA-induced silencing complex (RISC), miRNAs block translation of specific mRNAs containing partially complementary targets located in the 3′ untranslated regions or guide degradation of target mRNAs (as do siRNAs)34–36. The biological functions of miRNAs are largely unknown, except for a few examples34–36. It has recently been reported that miR-142 regulates promotion of T lymphoid–lineage cells37. Similar abundances of both sense (5p) and antisense (3p) miR-142 are expressed in hematopoietic tissues37. In the present study, we set out to investigate the interaction between the A→I RNA-editing and miRNA-biogenesis pathways. Both ADAR1 and ADAR2 edit specific adenosine residues of precursors of certain miRNAs including mouse miR-142. We demonstrate here that A→I editing alters the dsRNA structure of pri–miR-142, inhibits miR-142 processing by Drosha and, consequently, decreases mature miR-142-5p and miR-142-3p RNA levels. Thus, we reveal, for the first time, a new function of A→I RNA editing in the regulation of processing and expression of miRNAs.
RESULTS
Editing of miRNA precursors by ADAR1p110 and ADAR2
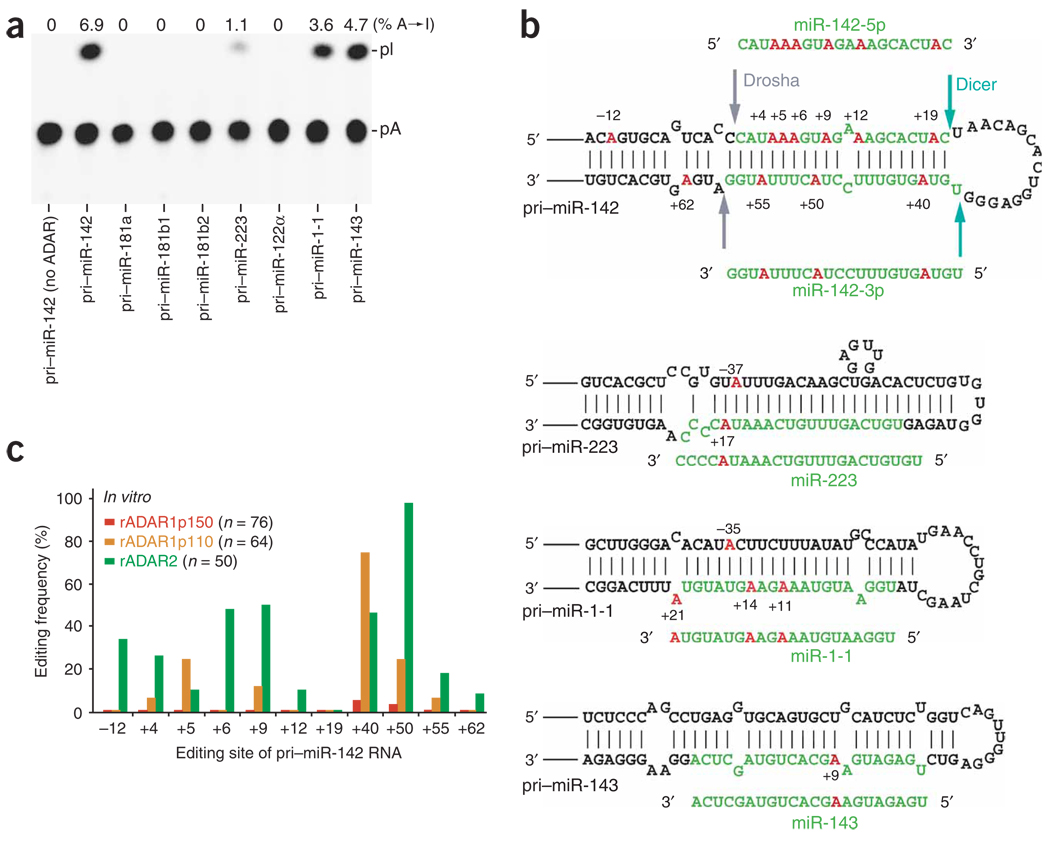
We chose eight miRNA precursors: pri–miR-142, pri–miR-181a, pri–miR-181b1, pri–miR-181b2, pri–miR-223 (expressed mainly in hematopoietic cells), pri-122α (liver), pri–miR-1-1 (heart) and pri– miR-143 (spleen, stomach, adipocyte), and determined whether they could serve as substrates for A→I RNA editing (Fig. 1a). The sequence covering 20–30 nt upstream and downstream of the pre-miRNA foldback dsRNA is sufficient to support Drosha and Dicer processing steps25,26. Therefore, the region surrounding the selected miRNA precursor sequence (~200–300 nt) was PCR-amplified using mouse chromosomal DNA and cloned into Bluescript KS vector. The synthetic pri-miRNAs labeled with 32P ATP were subjected to an in vitro RNA editing assay using one or a combination of recombinant ADAR proteins, and the reaction products were analyzed for base modification by thin-layer chromatography (TLC)7. A substantial fraction of adenosine residues of pri–miR-142, pri–miR-223, pri–miR-1-1 and pri–miR-143 RNAs were edited to inosines by ADAR1p110 and ADAR2. For instance, approximately 6.9% or an average of 6 out of 90 adenosine residues present in the pri–miR-142 RNA were edited. By contrast, no A→I modification of pri–miR-181a, pri–miR-181b1, pri–miR-181b2 or pri–miR-122α was detected (Fig. 1a).
Figure 1.
A→I RNA editing of pri-miR RNAs by ADAR. (a) TLC analysis of pri-miRNAs edited in vitro by a mixture of recombinant ADAR1p110 and ADAR2 proteins (rADAR1p110 and rADAR2, 20 ng each). (b) A→I editing sites of four pri-miRNAs. Red, edited adenosine residues (numbered by position with the 5′ end of the mature miRNA sequence counted as +1); green, the region to be processed into the mature miRNA; arrows in pri–miR-142 hairpin structure, cleavage sites for Drosha and Dicer. (c) A quantitative summary of the editing patterns revealed by sequencing of RT-PCR cDNA clones corresponding to pri–miR-142 RNAs edited in vitro by rADAR1p150, rADAR1p110 or rADAR2. Editing frequency is represented as a percentage (number of independent cDNA clones representing the edited pri–miR-142 sequence at that site divided by the total number of cDNA clones examined).
To identify adenosine residues that underwent editing, the reverse-transcription (RT)-PCR products of in vitro–edited pri-miRNAs were cloned and analyzed by sequencing individual cDNA isolates, revealing the specific sites of the pri-miRNAs edited in vitro by different ADARs. For instance, 11 specific adenosine residues of pri–miR-142 (Fig. 1b) edited frequently by ADAR1p110, ADAR2 or both were revealed as A→G changes within cDNA sequence inserts (Fig. 1c). Only a few DNA sequence alterations were detected among the 76 cDNA clones derived from pri–miR-142 RNAs treated in vitro with ADAR1p150 (Fig. 1c). The difference between ADAR1p150 and ADAR1p110 in editing pri–miR-142 is, to our knowledge, the first observed variation in enzymatic activity reported with these two ADAR1 isoforms. Possibly relevant to this, we have recently reported that ADAR1p150 binds siRNAs almost stoichiometrically and with extremely strong affinity, but without editing38. Similarly, we found that one specific adenosine residue (position +9) of pri–miR-143 was frequently edited by ADAR1p110 (60%) and ADAR2 (80%). ADAR2 also edited four specific adenosine residues (positions −35, +11, +14 and +21) of pri–miR-1-1 (up to 30%) and two sites (positions −37 and +17) of pri–miR-223 (10%) (Fig. 1b). Our findings suggest that A→I editing of miRNA precursors by ADAR1p110 and ADAR2 is selective, depending on the dsRNA foldback structure of each pri-miRNA.
Editing inhibits processing of pri–miR-142 by Drosha
Certain editing sites are located within the region corresponding to the sequence of mature miRNAs. For instance, six sites are located within the region corresponding to the mature miR-142-5p sequence, and three sites are located within the region corresponding to the mature miR-142-3p (Fig. 1b). If processed to mature miRNAs, the edited miR-142-5p and miR-142-3p might hybridize with and silence genes different than those targeted by the unedited miR-142-5p and miR-142-3p. Alternatively, A→I editing of miRNA precursors may affect their processing pathways. Editing of 11 adenosine residues identified within the pri–miR-142 sequence replaces A-U or U-A Watson-Crick pairs with I•U or U•I wobble pairs. Multiple I•U or U•I pairs reduce the overall stability and change dramatically the dsRNA structure, as detected by altered migration upon nondenaturing gel electrophoresis1,2.
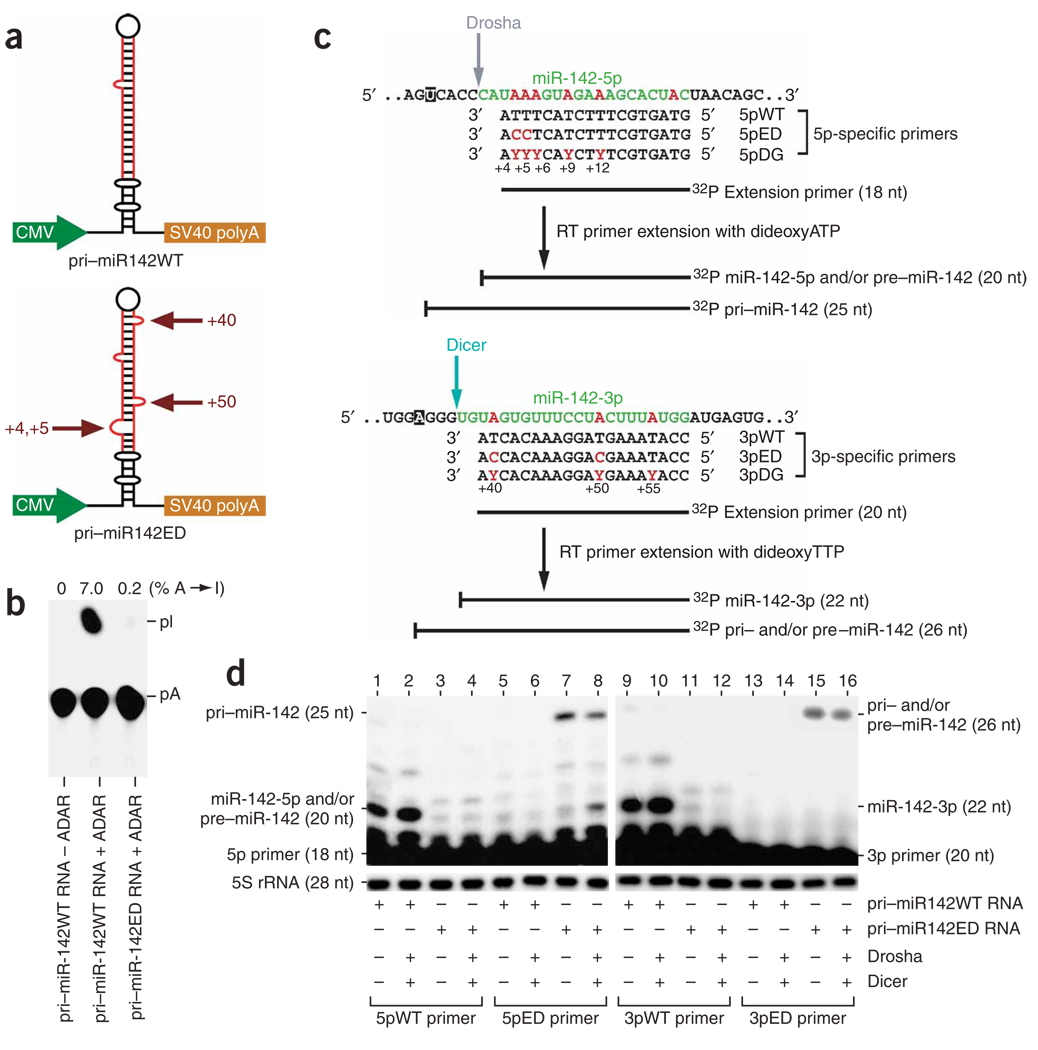
We investigated the effects of A→I RNA editing on pri–miR-142 RNA processing first in HEK293 cells transfected with miRNA precursor expression plasmids. We examined two pri–miR-142 RNA expression plasmids: one for synthesis of unedited, wild-type pri–miR-142 RNA (pri-miR142WT) and the other for synthesis of ‘pre-edited’ pri–miR-142 RNA carrying four guanosine (instead of adenosine) residues at the +4, +5, +40 and +50 sites (pri-miR142ED) (Fig. 2a). The structural alterations introduced in the pre-edited pri– miR-142 RNAs (four A-U or U-A pairs replaced with G•U or U•G pairs) are anticipated to be equivalent to those of the edited pri–miR-142 RNAs with four isosteric I•U or U•I pairs3. In contrast to the efficient editing of wild-type pri–miR-142 RNAs, essentially no additional A→I modification occurred for the pre-edited pri–miR-142 RNA when it was tested for in vitro editing with a combination of ADAR1p110 and ADAR2 (Fig. 2b).
Figure 2.
Analysis of pri–miR-142 processing in HEK293 cells. (a) Schematic of two pri–miR-142 RNA expression plasmids, one for wild-type, unedited (pri-miR142WT) and the other for pre-edited precursor RNA with A→G substitutions at the +4, +5, +40 and +50 positions (pri-miR142ED). (b) TLC analysis of unedited and pre-edited pri–miR-142 RNAs subjected to in vitro editing by a mixture of rADAR1p110 and rADAR2 proteins (20 ng each). (c) Primers used for differential primer-extension assay are shown, together with primer-extended DNA products of different sizes corresponding to pri–, pre– and mature miR-142 RNAs. A set of three primers complementary to the miR-142-5p sequence, 5pWT (wild-type and unedited), 5pED (pre-edited) and 5pDG (degenerate), were used to monitor the Drosha cleavage step and to quantitate simultaneously the expression levels of pri– and pre–miR-142 and mature miR-142-5p. The 25-nt products represent unprocessed pri–miR-142, whereas the 20-nt primer-extended products represent the pre–miR-142, mature miR-142-5p or both. The primer extension of both pre–miR-142 and mature miR-142-5p RNAs terminates at the same site generated by Drosha cleavage. The primer extension of pri– miR-142 extends five nucleotides upstream of the Drosha cleavage site in the presence of dideoxyATP. A separate set of three primers complementary to the miR-142-3p sequence, 3pWT, 3pED and 3pDG, were used to monitor the Dicer cleavage step and to quantitate simultaneously the expression levels of pri– and pre–miR-142 and mature miR-142-3p. The 26-nt products represent pri– and/or pre–miR-142, whereas the 22-nt primer-extended products represent mature miR-142-3p. The primer extension of both pri– and pre–miR-142 RNAs terminates at the same location in the presence of dideoxyTTP. The primer extension of mature miR-142-3p RNA terminates at the site generated by Dicer cleavage. Degenerate primers (5pDG and 3pDG) were used to monitor the total miR-142 RNAs (unedited and edited). Coloring is as in Figure 1b. The uracil and adenosine residues where the extension reaction is terminated by dideoxyATP and dideoxyTTP, respectively, are highlighted (filled box). Y in the DG primers denotes a random mix of T and C. (d) Analysis of pri– and pre–miR-142 and mature miR-142-5p and miR-142-3p RNAs processed in transfected HEK293 cells. Minor bands of primer-extended products with unexpected sizes (lanes 1 and 2 (22 nt) and lanes 9 and 10 (24 nt)) were also detected. At this time, we do not know how they are generated.
The expression levels of pri– and pre–miR-142 RNAs, as well as mature miR-142-5p and miR-142-3p, present in transfected HEK293 cells were quantitatively monitored by the dideoxynucleotide-quenched RT primer-extension assay using a set of primers corresponding to the miR-142-5p sequence and a separate set of primers corresponding to the miR-142-3p sequence (see Fig. 2c for details of these primers). Control experiments conducted separately by using in vitro–edited pri–miR-142 RNAs confirmed that the assay is differential and quantitative: unedited and edited pri–miR-142 RNAs are detected only with a specific, sequence-matching wild-type or pre-edited primer in proportion to their degree of editing. Additional experiments were conducted with degenerate primers (Fig. 2c) to monitor the total miR-142 RNA levels, independent of the editing extent (Supplementary Fig. 1 online). Accordingly, the two 5p-specific primers 5pWT (wild-type) and 5pED (pre-edited) distinguished unedited and pre-edited miR-142-5p RNAs synthesized in transfected HEK293 cells (Fig. 2d, lanes 1, 3, 5 and 7). Similarly, the two 3p primers, 3pWT and 3pED, distinguished unedited and pre-edited miR-142-3p RNAs (Fig. 2d, lanes 9, 11, 13 and 15).
Comparing the four different sizes of primer-extended products allowed us to quantitatively assess the effects of editing on the expression levels of pri– and pre–miR-142 as well as mature miR-142-5p and miR-142-3p. All primer-extended products were quantitated after normalizing their band intensities to those separately determined for 5S ribosomal RNA expression levels. Endogenous Drosha and Dicer present in HEK293 seem to efficiently process unedited miR-142 precursors. For instance, the primer-extension experiment conducted using the primer specific to the unedited miR-142-3p sequence (3pWT) generated very few 26-nt extended products, indicating that no substantial amount of pri– or pre–miR-142 RNAs was present (Fig. 2d, lane 9). Thus, almost all 20-nt 5pWT primer-extended products detected for analysis of RNAs derived from unedited pri–miR-142 RNA expression plasmid (pri-miR142WT) represented mature miR-142-5p and not pre–miR-142 (Fig. 2d, lane 1). Notably, expression levels of processed, mature miR-142-5p and miR-142-3p increased by ~1.5-fold (Fig. 2d, lanes 2 and 10) when expression plasmids driving synthesis of recombinant Drosha and Dicer were cotransfected (Supplementary Fig. 2 online). Analysis of RNAs derived from pre-edited pri–miR-142 RNA expression plasmid revealed that essentially no pre-edited mature miR-142-5p or pre–miR-142 (20-nt products extended with the 5pED primer) was present (Fig. 2d, lane 7), nor was pre-edited mature miR-142-3p (22-nt products extended with the 3pED primer; Fig. 2d, lane 15). However, we detected a substantial level of unprocessed pri–miR-142 (25-nt products extended with the 5pED primer; Fig. 2d, lane 7), indicating inefficient processing of pre-edited pri–miR-142 RNA (Fig. 2d, lanes 7 and 15) despite the presence of extra recombinant Drosha and Dicer enzymes (Fig. 2d, lanes 8 and 16). These results indicate that editing of the four sites examined interferes with the Drosha cleavage step.
Identification of sites inhibitory for Drosha cleavage
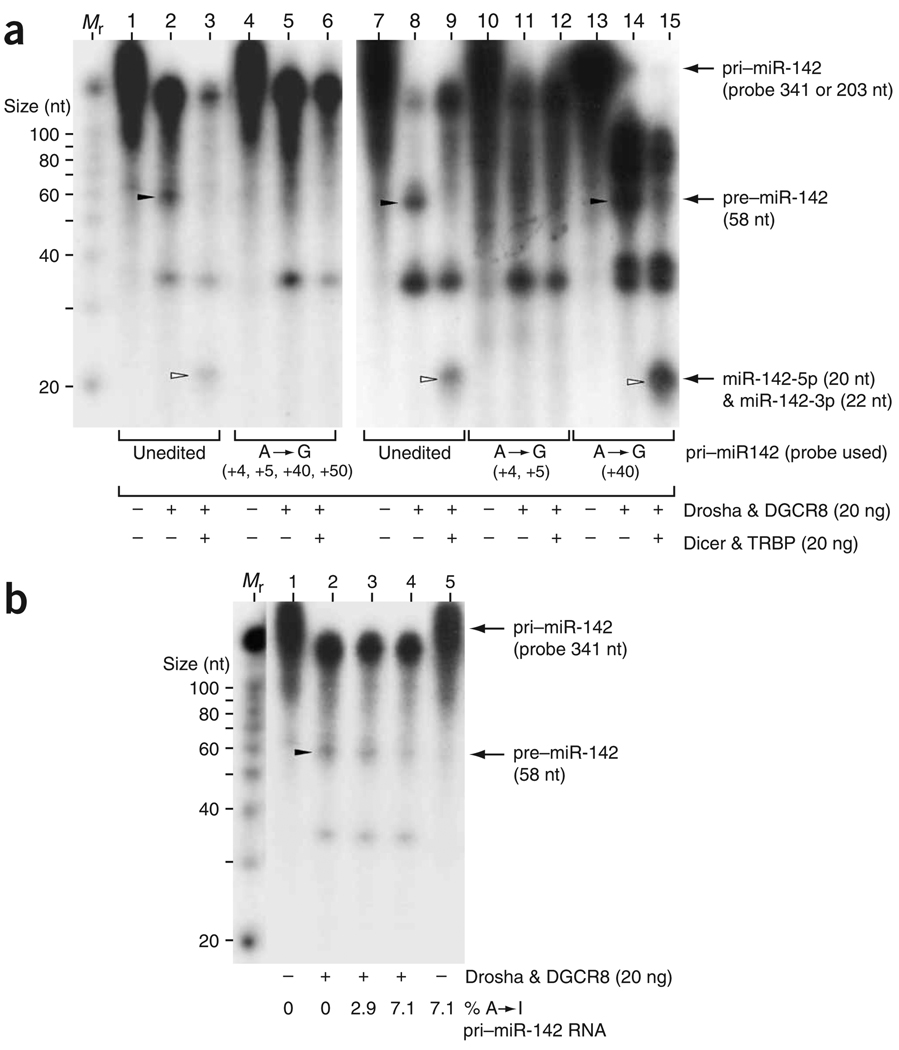
Among the four sites investigated in vivo, editing of particular adenosine residues may have disproportionately marked effects on processing of pri–miR-142 to pre–miR-142 by Drosha. As the pri– miR-142 RNA pre-edited at a single site was found to be further edited at additional sites by endogenous ADAR1p110 and ADAR2, it was not possible to identify the most crucial editing site that inhibits the processing of pri–miR-142 in transfected HEK293 cells (data not shown). Therefore, we next examined processing of pre-edited pri–miR-142 RNAs in vitro by using recombinant Drosha–DGCR8 and Dicer–TRBP miRNA processor complexes as described in refs. 28,32. As expected, unedited pri–miR-142 RNAs were efficiently cleaved to shorter pre–miR-142 RNAs (58 nt) by the Drosha–DGCR8 complex and then to mature miR-142-5p (20 nt) and miR-142-3p (22 nt) by the Dicer–TRBP complex (Fig. 3a, lanes 2, 3, 8 and 9). By contrast, processing of pre-edited pri–miR-142 RNAs (four adenosine residues substituted by guanosine at positions +4, +5, +40 and +50) to pre–miR-142 intermediates by the Drosha–DGCR8 complex was completely blocked (Fig. 3a, lane 5). Consequently, production of mature miR-142 RNAs by the Dicer–TRBP complex was not detected either (Fig. 3a, lane 6). Furthermore, analysis of a separate pre-edited pri–miR-142 RNA revealed that the editing sites inhibitory for processing of pri– to pre–miR-142 RNAs by the Drosha–DGCR8 complex are the +4 and +5 sites located in the foldback dsRNA stem near the Drosha cleavage site (Fig. 3a, lane 11). By contrast, editing of the +40 site seemed to have no noteworthy effects on either Drosha or Dicer cleavage (Fig. 3a, lanes 14 and 15).
Figure 3.
In vitro processing of pri–miR-142 RNAs by miRNA processor complexes. (a) Processing of pre-edited miR-142 precursor RNAs. Unedited pri–miR-142 RNAs as well as three pre-edited pri–miR-142 RNAs containing guanosine residues substituted for adenosine at the +4, +5, +40 and/or +50 sites were subjected to the Drosha cleavage reaction using the Drosha–DGCR complex and, in some experiments, then to the Dicer cleavage reaction using the Dicer–TRBP complex, as described previously28,32. Stability and structural changes introduced in pre-edited pri–miR-142 RNAs by G•U or U•G pairs are equivalent to those of isosteric I•U or U•I pairs. The total length of the pri–miR-142 RNA pre-edited only at the +40 site (203 nt) is shorter than that of the unedited and the two pre-edited RNAs (341 nt). The band positions of the pre–miR-142 RNA correctly cleaved by Drosha (58 nt) and mature miR-142 RNAs generated by Drosha (20 or 22 nt) are indicated. Nonspecific RNase activity of Drosha described previously28 is probably responsible for generation of other RNA products (~35 nt and longer). Mr indicates a lane containing molecular size markers. (b) Processing of pri–miR-142 RNAs that had been edited in vitro to different extents (0%, 2.9% or 7.1% A→I modifications); gel is labeled as in a.
Finally, we conducted in vitro Drosha–DGCR8 processing assays with pri–miR-142 RNAs that had been edited in vitro by ADAR1p110 and ADAR2. These experiments using in vitro–edited pri–miR-142 RNAs containing inosine residues (not substituted guanosine) clearly indicated that cleavage of pri–miR-142 to pre–miR-142 by Drosha– DGCR8 is progressively suppressed in proportion to the extent of A→I editing (Fig. 3b). The results confirm that Drosha recognizes both A→G and A→I changes.
Degradation of edited pri–miR-142 RNAs by Tudor-SN
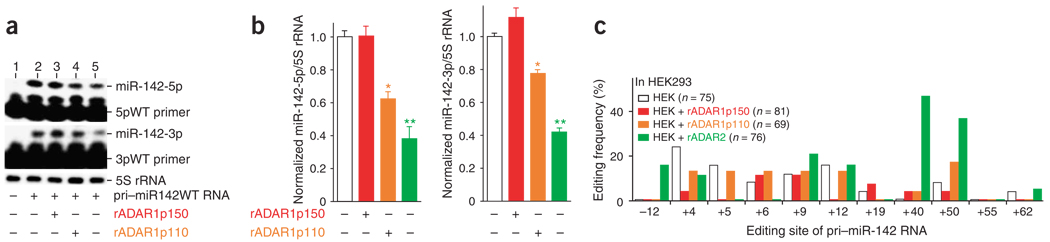
As expected, miR-142-5p and miR-142-3p expression levels were reduced substantially (Fig. 4a, lanes 4 and 5, and Fig. 4b) when an ADAR1p110 or ADAR2 expression plasmid was cotransfected (Supplementary Fig. 2), indicating that A→I editing of pri–miR-142 by these two ADARs inhibited their processing in transfected HEK293 cells. By contrast, when the pri–miR-181b1 RNA expression plasmid was transfected into HEK293 cells, no change in the expression level of miR-181b1 was detected, regardless of the presence or absence of coexpressed recombinant ADAR1p110 or ADAR2 proteins (data not shown). No editing sites were found in pri–miR-181b1 (Fig. 1a). The presence of extra recombinant ADAR1p150 protein (Supplementary Fig. 2) did not appreciably affect the overall expression levels of miR-142-5p and miR-142-3p (Fig. 4a, lane 3 and Fig. 4b), consistent with the fact that the p150 form of ADAR1 seldom edits pri–miR-142 RNAs in vitro (Fig. 1c).
Figure 4.
Suppression of pri–miR-142 processing in HEK293 cells transfected with ADAR expression plasmids. (a) Inhibitory effects of ADAR1p110 and ADAR2 on processing of pri–miR-142 in HEK293 cells. Primer-extension assays monitoring for expression levels of miR-142-5p (upper gel) and miR-142-3p (lower gel) as in Figure 2d are shown. (b) Histograms representing data like that in a from three independent experiments. The relative miR-142-5p and miR-142-3p levels in HEK293 cells cotransfected with the ADAR1p150, ADAR1p110 or ADAR2 expression plasmid were compared statistically by individual unpaired Student’s t-tests to those in HEK293 cells transfected only with pri-miR142WT plasmid. Significant differences are indicated as follows: one asterisk, P < 0.01; two asterisks, P < 0.001. Error bars, s.e.m. (c) A quantitative summary (as in Figure 1c) of the editing pattern revealed by sequencing of cDNA clones corresponding to pri–miR-142 RNAs edited in transfected HEK293 cells. White bars represent the results of experiments conducted with HEK 293 cells transfected solely with wild-type pri–miR-142 RNA expression plasmid.
Precursor RNAs of miR-142 were practically immeasurable by primer-extension assay in HEK293 cells cotransfected with either ADAR1p110 or ADAR2 expression plasmid. Therefore, the RT-PCR products corresponding to unprocessed miR-142 precursor RNAs present in HEK293 cells were cloned and the DNA sequences of cDNA isolates were examined (Fig. 4c). Notably, we found that several editing sites were also edited in vivo in HEK293 cells transfected with the pri-miR142WT plasmid alone, most likely by endogenous ADAR1p110 and ADAR2, at low frequencies (about 10%–20%; Fig. 4c, white bars). Editing of certain sites was increased by the presence of extra recombinant ADAR proteins; this could be seen, for example, at the +40 and +50 sites in HEK293 cells transfected with the ADAR2 expression plasmid (Fig. 4c). In view of the suppressive effects of A→I editing on the processing of pri–miR-142 and on the expression levels of mature miR-142-5p and miR-142-3p RNAs, however, we had anticipated detecting a substantial amount of the highly edited pri–miR-142 RNA sequences in HEK293 transfected with ADAR1p110 or ADAR2 expression plasmid (Fig. 4c). One possible explanation for their observed scarcity is that such highly edited precursor RNAs (prohibited from cleavage by Drosha) may be rapidly degraded, perhaps by a ribonuclease specific to inosine-containing, highly edited dsRNAs (I-dsRNAs)39. Thus, cDNA sequences derived from RT-PCR products may over-represent under-edited or unedited pri-miRNAs. In accordance with this hypothesis, accumulation of pre-edited pri–miR-142 RNAs derived from the pri-miR142ED plasmid was noted (Fig. 2d, lanes 7 and 15), probably because these pre-edited precursor RNAs contain guanosine residues in place of inosine and thus are not subject to the ribonuclease specific to I-dsRNAs. Detection of edited pri–miR-142 RNAs may depend on the overall balance of efficiency of the miRNA-processing machinery and degradation of the edited precursors by the ribonuclease specific to I-dsRNAs.
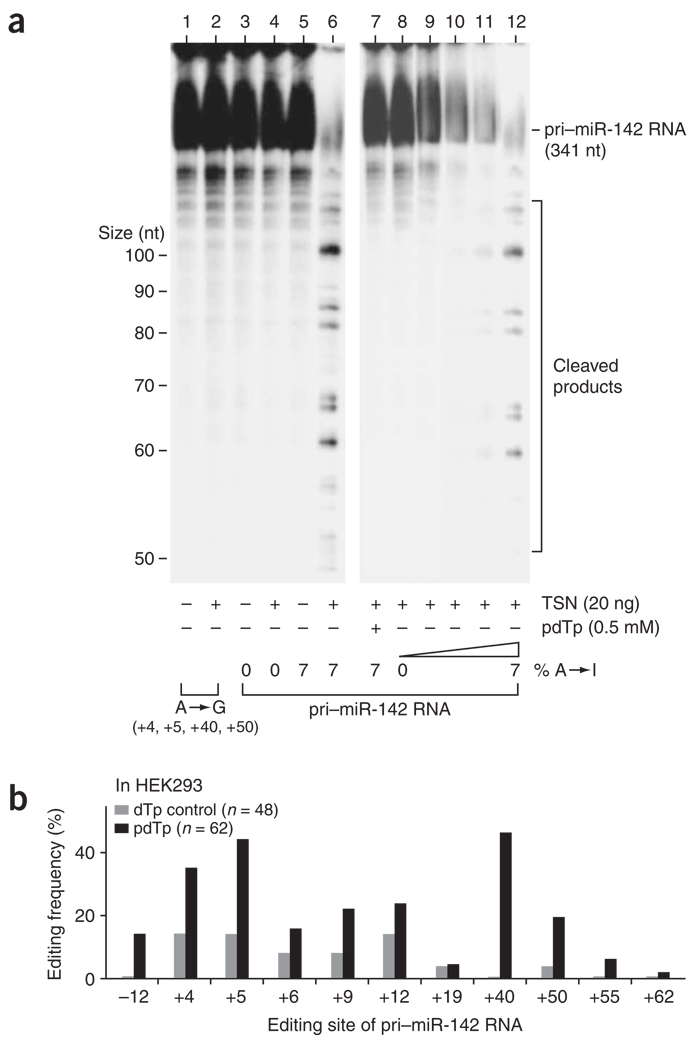
Notably, Tudor-SN (TSN), a member of the staphylococcal nuclease gene family and a known component of RISC40, has recently been identified as either the ribonuclease specific to I-dsRNA itself or an activator of the ribonuclease41. The ribonuclease activity has been tested so far only with synthetic, inter- and intramolecular dsRNA substrates containing U•I and I•U pairs in a specific sequence context39,41. No naturally occurring intramolecular duplexes containing multiple U•I and I•U pairs such as the edited pri–miR-142 RNA have been tested previously. Therefore, we ectopically expressed Flag epitope–tagged TSN proteins in HeLa cells and purified recombinant TSN proteins on anti-Flag affinity columns, which were then subjected to the ribonuclease assay for pri–miR-142 RNAs (Fig. 5a). The in vitro–edited pri–miR-142 RNAs became progressively sensitive to the TSN endonucleolytic cleavage in proportion to the number of A→I modifications (Fig. 5a, lanes 8–12), whereas the unedited pri–miR-142 RNA (Fig. 5a, lane 4) and the pre-edited pri–miR-142 RNA with four G•U and U•G pairs remained resistant (Fig. 5a, lane 2). A specific competitive inhibitor of staphylococcal nucleases, 2′-deoxythimidine 3′,5′-bisphosphate (pdTp), inhibits the activities of TSN and the I-dsRNA–specific ribonuclease40,41. Cleavage of highly edited pri–miR-142 RNAs by TSN was completely blocked in the presence of pdTp (Fig. 5a, lane 7). Notably, it has been reported that the recombinant TSN protein ectopically expressed in yeast requires supplementation of Xenopus laevis oocyte extracts for the I-dsRNA–specific ribonuclease activity41. When purified to a single band (Supplementary Fig. 3 online), our mammalian TSN preparation cleaved the edited pri–miR-142 RNAs, which indicates that TSN alone is responsible for the I-dsRNA–specific ribonuclease activity. However, we cannot completely rule out the possibility that other less abundant proteins (cofactors) were copurified with our mammalian TSN preparation.
Figure 5.
Degradation of highly edited pri–miR-142 RNAs by TSN. (a) In vitro assay for degradation of pri–miR-142 RNAs using purified TSN recombinant proteins. pri–miR-142 RNA edited to different extents (0%, 0.5%, 1.4%, 3.0% or 6.9% A→I modification) or pre-edited (four adenosine residues replaced by guanosine at the +4, +5, +40 and +50 sites) was subjected in vitro to endonucleolytic cleavage by TSN in the presence or absence of the inhibitor pdTp41. (b) Accumulation of highly edited pri–miR-142 RNAs in vivo in transfected HEK293 cells (quantitated as in Figure 1c) in the presence of the pdTp inhibitor.
We then tested our hypothesis that inhibition of the TSN activity by pdTp might increase the probability of detecting edited pri–miR-142 RNAs in vivo in transfected HEK293 cells. We examined the pri–miR-142 sequences of cDNA clones derived from RNAs extracted from HEK293 cells that had been transfected with pri-miR142WT plasmid in the presence of pdTp inhibitor or 2′-deoxythimidine 3′-monophosphate (dTp) control40. As anticipated, editing of several sites including the +4, +5, +40 and +50 sites dramatically increased in HEK293 cells treated with pdTp for 24 h (Fig. 5b). Together, our results support the hypothesis that the steady-state levels of edited pri-miRNAs are indeed affected by TSN. In addition, our results indicate that A→I editing of pri–miR-142 RNAs is a more widespread phenomenon than is predicted by the measured ratio of edited to unedited miRNA precursor cDNA sequences (see below).
Increased endogenous miR-142 levels in ADAR null mice
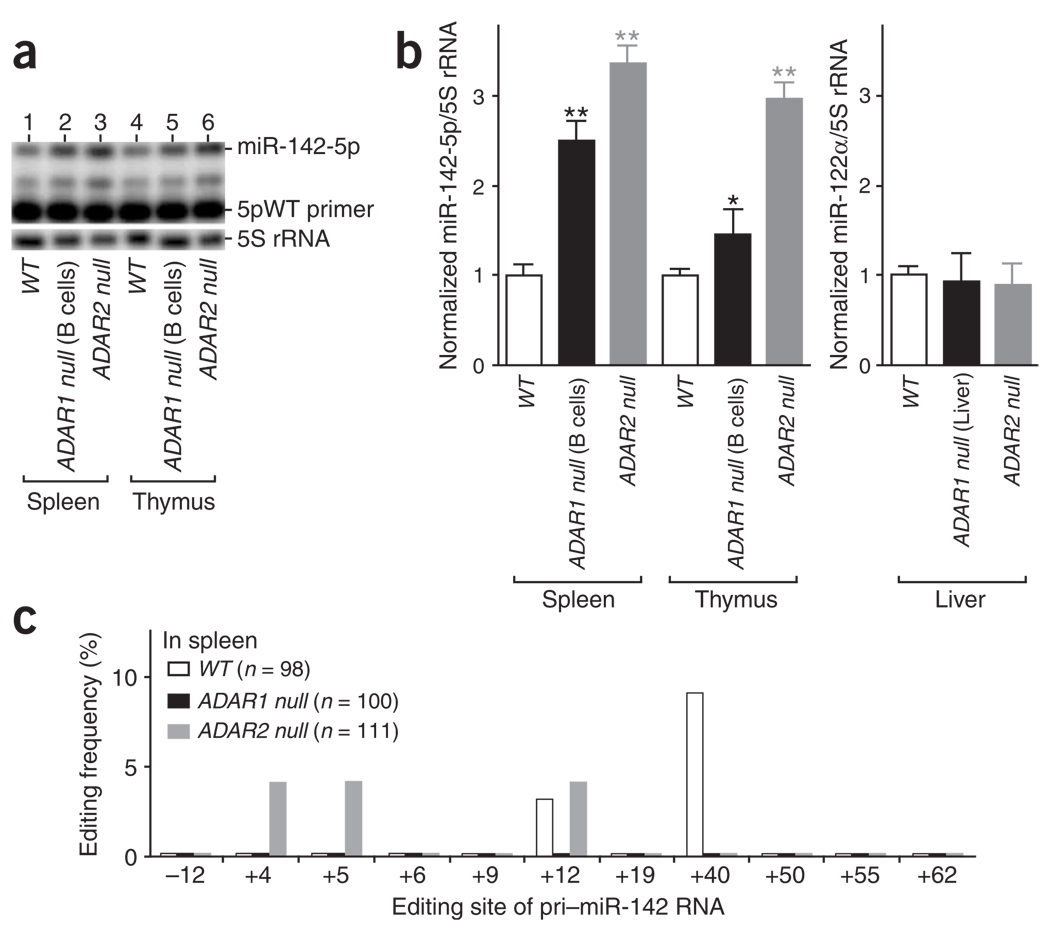
A major effect of A→I editing of pri–miR-142 seems to be suppression of pri– to pre–miR-142 processing by Drosha and, consequently, decreased expression of mature miR-142 RNAs. Accordingly, one may predict a substantial increase in the expression levels of miR-142-5p and miR-142-3p in the absence of ADARs. Therefore, we examined RNA samples extracted from spleens and thymuses of ADAR1 null mice (ADAR1flox/flox:CD19-Cre+) and ADAR2 null mice42. The B-lineage cell–specific ADAR1 null mutant mouse line, ADAR1flox/flox:CD19-Cre+, has been established recently in our laboratory. Endogenous miR-142-5p expression is substantially higher in the spleen of ADAR1 null mice (2.5-fold) and ADAR2 null mice (3.3-fold) and in the thymus of ADAR2 null mice (3.0-fold) compared to those of the wild-type control mice (Fig. 6a,b). We expected ADAR1 to be inactivated only in B-lineage cells of ADAR1flox/flox:CD19-Cre+ mice, and the thymus mainly contains T lymphocytes. Therefore, the reason for a slightly higher expression level (1.4-fold) of miR-142-5p detected also in the thymus of ADAR1flox/flox:CD19-Cre+ mice is currently unknown. We found identical results for miR-142-3p expression levels (data not shown). The changes detected in the endogenous miR-142-5p and miR-142-3p expression levels did not, however, result from a general effect on the level of every miRNA in ADAR null mice. For instance, we found no appreciable difference in the expression levels of miR-181b1 (B-lineage cell–promoting miRNAs) in ADAR1 null and ADAR2 null spleens (data not shown) and a liver-specific miR-122α in the RNA samples prepared from livers of ADAR1flox/flox:Alb-Cre+ (liver-specific ADAR1 null)16 and ADAR2 null mice (Fig. 6b). We found no editing sites in pri–miR-181b1 and pri–miR-122α RNAs (Fig. 1a). Analysis of cDNA clones derived from RT-PCR products corresponding to the pri–miR-142 RNAs revealed that, indeed, endogenous pri–miR-142 RNAs are edited at several sites in spleens of wild-type mice, although at low levels. This low-frequency editing was detected in spleens of even ADAR2 null mice but not of ADAR1 null mice (Fig. 6c). The steady-state abundances of edited endogenous pri–miR-142 RNAs are therefore certainly detectable but low, perhaps owing to their rapid degradation by TSN. Our results suggest that endogenous pri–miR-142 is also subject to A→I RNA editing by ADAR1p110, ADAR2 or both and that editing suppresses pri–miR-142 processing to mature miR-142-5p and miR-142-3p.
Figure 6.
Increased expression levels of miR-142 RNAs in spleen and thymus of ADAR null mice. (a) The wild-type sequence primer 5pWT was used for quantitation of mature miR-142-5p RNA expression levels as in Figure 2d. Identical results were obtained with a separate extension assay done by using the degenerate sequence primer 5pDG (data not shown). (b) The level of the mature miR-142-5p in wild-type and ADAR null mice. Results from three independent experiments are shown. The increase in the relative miR-142-5p levels in comparison to wild-type mouse tissue was examined statistically by individual unpaired Student’s t-tests. Significant differences are indicated as follows: one asterisk, P < 0.01; two asterisks, P < 0.001. Error bars, s.e.m. (c) The editing pattern (quantitated as in Figure 1c) revealed by sequencing of cDNA clones corresponding to the endogenous pri–miR-142 RNAs edited in vivo in spleens of wild-type, ADAR1 null and ADAR2 null mice.
DISCUSSION
In this study, we demonstrated that select miRNA precursors (four among eight randomly chosen) are edited by ADAR1p110 and ADAR2. Thus, A→I editing may occur for many miRNA precursors. In accordance with this proposition, editing of pri–miR-22 at very low levels but in many tissues has been reported recently43. However, the potential effects of editing on processing and expression of miRNA have never been investigated previously43. We found that A→I editing of pri–miR-142 results in a substantial reduction in mature miR-142-5p and miR-142-3p levels in transfected HEK293 cells. Furthermore, levels of endogenous miR-142-5p and miR-142-3p are substantially lower in wild-type mouse spleens than those in ADAR1−/− and ADAR2−/− spleens. Results of an in vitro miRNA processing assay identified the most crucial step affected by editing: processing of pri– to pre–miR-142 RNA by the Drosha–DGCR8 complex. Finally, we demonstrated that highly edited pri–miR-142 RNAs are degraded by TSN in vitro and in vivo. These results clearly indicate that A→I editing inhibits pri–miR-142 processing.
Modulation of miRNA biogenesis by RNA editing
Processing of primary miRNA transcripts by Drosha is a crucial step for generation of final mature miRNAs25,44. Previous studies using mutant miRNA precursors have indicated that Drosha may measure the junction of the terminal loop and the adjacent stem and then cleave approximately two helical turns into the stem26. The integrity of the precursor RNA stem seems to be essential for processing25,44. By contrast, the Drosha digestion step is less sensitive to alteration of the underlying sequence of the stem structure or terminal loop25,26,44. A→I editing of pri–miR-142 at eleven sites replaces an A-U or U-A Watson-Crick pair with a less stable I•U or U•I wobble pair, leading to substantial changes in the stem structure and stability. An in vitro miRNA processing assay with three pre-edited pri–miR-142 RNAs containing isosteric G•U or U•G pairs in place of I•U or U•I pairs revealed that editing at the +4 and +5 sites completely blocks the Drosha cleavage step, whereas editing at the +40 site (near the terminal loop) has no inhibitory effect on the Drosha or the Dicer cleavage step. However, sequence analysis of individual cDNA clones derived from pri–miR-142 edited in vitro or in vivo suggests that editing usually occurs at multiple sites, such as +4, +5 and +40 simultaneously. Therefore, most edited pri–miR-142 RNAs are likely to be blocked for processing by Drosha.
Finally, pri–miR-142 RNAs edited at multiple sites are degraded efficiently in vitro by TSN, a component of RISC and a recently identified ribonuclease specific to inosine-containing dsRNAs41. The results of in vivo experiments using a TSN inhibitor also indicate that TSN indeed degrades a substantial fraction of highly edited pri–miR-142 not promptly processed by Drosha. According to this hypothesis, however, highly edited pri–miR-142 RNAs, not having been processed by Drosha, must be exported to the cytoplasm, where TSN is localized39,41. Only properly processed pre-miR RNAs are exported efficiently to the cytoplasm by the RanGTP–exportin-5–dependent mechanism31. However, nuclear export of miRNA precursors longer than properly processed pre-miR RNAs (exportin-5–independent and perhaps nonspecific) has also been reported31. Because of the rapid degradation of highly edited pri-miR RNA molecules by TSN, such nonspecific export might have been unnoticed in previous studies.
Alternatively, A→I editing of miRNA precursors at a limited number of sites (for example, the +40 site of pri–miR-142) may have little effect on cleavage by Drosha and also may not trigger degradation by TSN. This would result in the processing and expression of mature miRNAs with edited sequences. Although we did not find the edited version of miR-142 in the miRNA Registry45, miR-142 RNAs with A→I edited sequences may not have been registered because they did not match any region of the genome. It has been reported that many miRNA-like molecules could not be assigned to a specific genomic sequence22,23. It may be worthwhile to re-examine certain miRNA-like molecules, identified through cloning but not assigned to a specific genomic sequence owing to few A→G mismatches, as ‘edited’ miRNAs. Notably, a recently reported viral miRNA, KSHV-miR-K12-10, with a single adenosine residue substituted by guanosine (miR-K12-10b) is frequently detected among cDNA isolates identified by the small-RNA cloning method. This indicates that editing of this particular site does not, in fact, inhibit pri–miR-K12-10 RNA processing, but leads to expression of mature miRNA with the edited sequence24.
Together, the structural changes of certain miRNA precursors caused by editing of multiple sites are significant for their interactions with the miRNA-processing machinery. Such changes inhibit processing, whereas editing of other miRNA precursors at few select sites may be tolerated. If editing sites are located in the mature miRNA sequence, as in the case of KSHV-miR-K12-10b, the edited miRNAs could be expressed and possibly target genes different from those silenced by the unedited miRNAs. In particular, the latter possibility should be considered when designing an miRNA expression vector aimed at silencing a specific target gene, as unexpected editing of the miRNA precursor RNA derived from the expression vector could silence unintended, wrong-target genes.
Ectopic expression of miR-142-5p in cultured hematopoietic progenitor cells from mouse bone marrow leads to a substantial increase (~40%) in the T lymphoid–lineage cells37. Accordingly, we expected that the number of T lymphoid–lineage cells might also be increased in the spleen of ADAR1flox/flox:CD19-Cre+ mice and in the spleen and thymus of ADAR2 null mice. However, we found no appreciable change in the T lymphoid lineage in these ADAR-mutant mice compared to wild-type control mice (Q.W. and K.N., unpublished data). The steady-state level of ectopic miR-142-5p achieved with the retroviral vector system used in the previous in vitro studies might be substantially higher than that resulting from ablation of ADAR1 or ADAR2. Alternatively, the increase of miR-142 may somehow be compensated to counterbalance the otherwise increased T lymphoid– lineage cells by a separate mechanism during development of ADAR null mutant mice.
Interaction of RNA editing and RNAi pathways
Double-stranded RNA–binding proteins, in general, have no apparent sequence specificity. Therefore, it has been speculated that ADARs may intersect with other cellular mechanisms, such as RNAi, that also act on dsRNA by competing for shared substrate dsRNAs46,47. Caenorhabditis elegans strains containing homozygous deletions of both the c.e.ADAR1 and c.e.ADAR2 genes show defective chemotaxis48. The phenotypic alteration of the mutant worms, however, can be reverted in RNAi-defective strains of C. elegans, indicating the dependence of ADAR null worm phenotypes on RNAi47. In addition, the cytoplasmic full-length isoform of ADAR1p150 sequesters siRNA and thereby limits the potency of siRNA in mammalian cells38. In the present studies, we have demonstrated the involvement of the A→I RNA editing system in the control of the miRNA biogenesis pathway and the expression levels of mature miRNAs. Finally, TSN, a known component of RISC, has now been identified as a ribonuclease that specifically cleaves dsRNAs containing I•U and U•I pairs, such as edited pri–miR-142. These findings all point to frequent interactions between the A→I editing mechanism and the RNAi mechanism as well as to a potential role for ADARs in regulating the RNAi pathway.
Inactivation of ADAR1 leads to an embryonic-lethal phenotype caused by deficiency in hematopoiesis and widespread apoptosis15,16. It seems that the editing of an unknown target dsRNA(s) protects developing embryos from massive apoptosis, which underlies the embryonic-lethal and apoptosis-prone phenotype of ADAR1 null mutant embryos. It is tempting to hypothesize that the phenotype of ADAR1 null mutant embryos may be due to the absence of interaction between A→I RNA editing and RNAi pathways during, for example, the biogenesis of particular miRNAs.
METHODS
Plasmids
Plasmid pBSmiR142 contains a 258-base-pair (bp) fragment of mouse chromosomal DNA encompassing the region that is transcribed to miR-142 precursor RNAs. The DNA fragment was PCR-amplified using mouse genomic DNA and PCR primers 142S1 and 142A1, and was cloned into pBluescript KS vector (Stratagene). Primer sequences were as follows: 142S1, 5′-CGGGATCCGAAGTTACACGGAGGGGAGGGGG-3′; 142A1, 5′-CGGAATTC GGCGTGTGAGAGATGCTCACCTGT-3′. Plasmids pCMV-pri-miR142WT and pCMV-pri-miR142ED contain a DNA fragment corresponding to the pri–miR-142 RNA (unedited and pre-edited at the +4, +5, +40 and +50 sites, respectively) cloned into pSilencer4.1-CMVpuro vector (Ambion).
In vitro RNA editing assay
The in vitro editing reaction mixture, containing 20 fmol of pri–miR-142 RNA prepared by in vitro transcription and 20 ng of Flag-tagged ADAR1p150, ADAR1p110 or ADAR2 protein, was incubated at 30 °C for 1 h as described previously49.
Processing of pri–miR-142 RNAs in HEK293 cells
A transfection mixture included the following: pCMV-pri-miR142WT or pCMV-pri-miR142ED (1 µg); pCMV-F-ADAR1p150, pCMV-F-ADAR1p110 or pCMV-F-ADAR2 (1 µg); pCMV-F-Drosha (1 µg) and/or pCMV-F-Dicer (1 µg); or vector-only control pRC/CMV (total DNA adjusted to 5 µg). The mixture was electroporated into HEK293 cells using Nucleofector (Amaxa Biosystems). The transient transfection efficiency was comparable between separate experiments (70% to 90%). The transfected HEK293 cells were harvested 36 h after electroporation for extraction of total RNA or protein.
Preparation of recombinant TSN proteins
A full-length human TSN cDNA–expressed sequence-tag clone (IMAGE 3345037) was used to construct a mammalian, Flag epitope–tagged TSN expression plasmid in p3XFLAGCMV-10 vector (Sigma). The resultant pCMV-3F-TSN plasmid was used to transform HeLa cells, which resulted in the establishment of a cell line overexpressing Flag-TSN proteins. TSN recombinant proteins were purified on an anti-Flag M2 affinity column as described previously8.
Effects of pdTp on edited pri–miR-142 RNA levels
Synthesis of pdTp was carried out using 10 mM dTp (Sigma), 10 mM ATP and 1 unit of T4 polynucleotide kinase (Roche) as described previously40. HEK293 cells transfected with pCMV-pri-miR142WTwere pretreated with 0.005% (v/v) digitonin (Sigma) for 6 h. After removing digitonin and replacing with fresh medium, pdTp or control dTp was added to 1 mM and the cells were cultured further for 24 h.
Preparation of ADAR1flox/flox:CD19-Cre+ mice
Establishment of ADAR1flox/flox mice, homozygous for ADAR1-2LoxP allele, was described previously16. Intercrossing ADAR1flox/flox mice with CD19-Cre mice50 resulted in B-lineage cell–specific ablation of ADAR1. All protocols adhered to the Institutional Animal Care and Use Committee of The Wistar Institute and were performed in accordance with the US National Institutes of Health Guidelines.
Dideoxyoligonucleotide/primer extension assay
Approximately 10 fmol of 32P-labeled extension primer was mixed with 10 µg of RNA derived from transfected HEK293 cells or 1 µg of mouse tissue RNA, heated to 70 °C for 10 min and annealed at 60 °C for 1 h. For analysis of miR-142-5p RNA expression levels, the primer-annealed RNA was extended with Superscript (Invitrogen) in the presence of 250 µM dideoxyATP at 60 °C for 1 h. For analysis of miR-142-3p and miR-122α expression levels, the extension reaction was carried out in the presence of dideoxyTTP. For analysis of 5S rRNA expression levels (normalization control), the extension reaction was performed in the presence of dideoxyGTP. Primers EX-122α (5′-CAAACACCATTGTCACACT-3′) and EX-5SR (5′-GCGGTCTCCCATCCAAGTACTAACC-3′) were used for the primer-extension assay to monitor miR-122α and 5S rRNA levels. All primer-extended DNA products were fractionated on a 15% (w/v) poly-acrylamide, 8 Murea gel. The ratio of the miRNA (miR-142-5p, miR-142-3p or miR-122α) to 5S rRNA expression levels was estimated by quantifying the radioactivity of the primer-extended product with a PhosphorImaging System (Molecular Dynamics).
Supplementary Material
ACKNOWLEDGMENTS
We thank J.M. Murray for critical reading of the manuscript and J.T. Lee for excellent technical assistance. This work was supported in part by grants from the US National Institutes of Health, the March of Dimes and the Commonwealth Universal Research Enhancement Program of the Pennsylvania Department of Health.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING INTERESTS STATEMENT: The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 2.Wagner RW, Smith JE, Cooperman BS, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. USA. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masquida B, Westhof E. On the wobble GoU and related pairs. RNA. 2000;6:9–15. doi: 10.1017/s1355838200992082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keegan LP, Gallo A, O’Connell MA. The many roles of an RNA editor. Nat. Rev. Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- 6.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai F, Drakas R, Nishikura K. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 9.Melcher T, et al. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 10.Melcher T, et al. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 11.Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi M, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intronexon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 13.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 14.Lomeli H, et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 15.Hartner JC, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 17.Cho DS, et al. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 2003;278:17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 18.Athanasiadis A, Rich A, Maas S. Widespread A→I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levanon EY, et al. Systematic identification of abundant A→I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 21.Nishikura K. Editing the message from A to I. Nat. Biotechnol. 2004;22:962–963. doi: 10.1038/nbt0804-962. [DOI] [PubMed] [Google Scholar]
- 22.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 24.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 26.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 28.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 29.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 32.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Förstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 35.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 36.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 37.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, et al. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J. Biol. Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scadden AD, O’Connell MA. Cleavage of dsRNAs hyper-edited by ADARs occurs at preferred editing sites. Nucleic Acids Res. 2005;22:5954–5964. doi: 10.1093/nar/gki909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caudy AA, et al. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- 41.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 42.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 43.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 47.Scadden AD, Smith CW. RNAi is antagonized by A→I hyper-editing. EMBO Rep. 2001;2:1107–1111. doi: 10.1093/embo-reports/kve244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dabiri GA, Lai F, Drakas RA, Nishikura K. Editing of the GLuR-B ion channel RNA in vitro by recombinant double-stranded RNA adenosine deaminase. EMBO J. 1996;15:34–45. [PMC free article] [PubMed] [Google Scholar]
- 50.Rickert RC, Rajewsky K, Roes J. Impairment of T-cell-dependent B-cell responses and B-1 cell development in CD19-deficient mice. Nature. 1995;376:352–355. doi: 10.1038/376352a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.