Abstract
Phosphodiesterase 4 catalyzes the hydrolysis of cyclic AMP and is a target for the development of anti-inflammatory agents. We have designed and synthesized a series of phenyl alkyl ketones as PDE4 inhibitors. Among them, 13 compounds were identified as having submicromolar IC50 values. The most potent compounds have IC50 values of in the mid- to low-nanomolar range. Compound 5v also showed preference for PDE4 with selectivity of >2000-fold over PDE7, PDE9, PDE2, and PDE5. Docking of 5v, 5zf, and 5za into the binding pocket of the PDE4 catalytic domain revealed a similar binding profile to PDE4 with rolipram except that the fluorine atoms of the difluoromethyl groups of 5v, 5za, and 5zf are within a reasonable range for hydrogen bond formation with the amide hydrogen of Thr 333 and the long alkyl chain bears additional van der Waals interactions with His 160, Asp 318, and Tyr 159.
Introduction
Phosphodiesterases (PDEs) are responsible for the hydrolysis of second messengers cyclic adenosine and guanosine monophosphates (cAMP and cGMP).a,1 Because of the important roles that cAMP and cGMP play, PDEs are excellent targets for the development of therapeutic agents. About 100 isoforms of PDEs are coded by 21 human genes and differentially expressed in various cell types.2–7 Such a large number of isoforms presents a unique challenge to the design and synthesis of PDE inhibitors that are specific for PDE isoforms of interest for pharmaceutical applications. We are especially interested in developing PDE4 inhibitors because of its implication in inflammatory diseases such as asthma and chronic obstructive pulmonary disease.3,8–13 PDE4 specifically catalyzes the hydrolysis of cAMP14 and has been found in inflammatory15,16 as well in immune cells.17 Several selective PDE4 inhibitors have entered into clinical trials, the results from which have validated PDE4 as a pharmacological target for the development of therapeutic agents for the treatment of inflammatory and chronic obstructive pulmonary diseases.18–22 Some PDE4 inhibitors, such as rolipram (Figure 1), have been dropped from clinical studies because of side effects, which presumably arise from nonspecific inhibition of other isoforms, while others including roflumilast and cilomilast (Figure 1) are still in clinical trials.3,23,24 Up until now no PDE4 inhibitors have been approved for clinical use.
Figure 1.
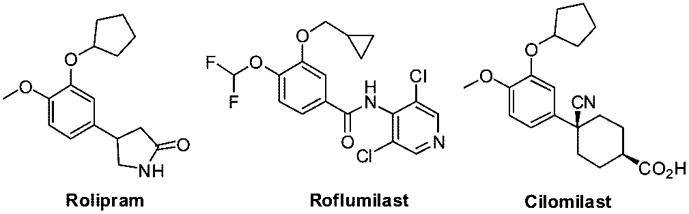
Structures of selected PDE4 inhibitors.
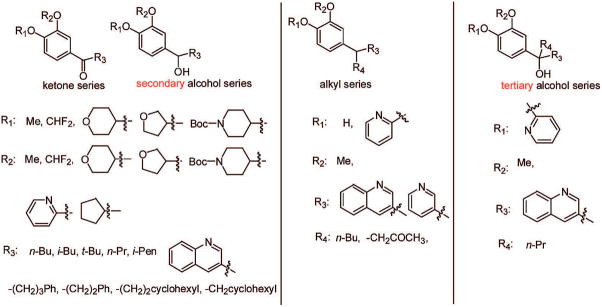
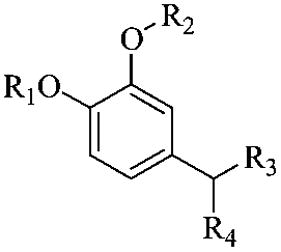
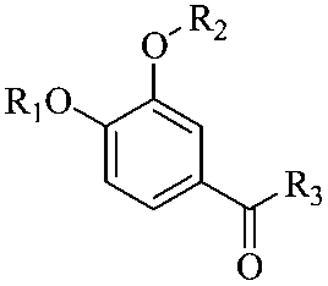
We are interested in the design, synthesis, and evaluation of selective PDE4 inhibitors for potential pharmaceutical applications. The design is based on published crystal structural information.25 Forty-one potential PDE4 inhibitors have been designed, synthesized, and evaluated. Figure 2 shows the core structure and different substituents in general terms.
Figure 2.
General structures of potential inhibitors designed.
Results and Discussion
Design
The design was based on published cocrystal structures of PDE4D2 and rolipram.25,26 From the crystal structure, it was clear that the side chain region of rolipram still has plenty of space for structural optimization and exploration of selectivity. Therefore, three series of compounds with ketone, alcohol, and alkyl side chains as point of modification were designed with the ketone series looking especially promising because of the planar structure of the side chain moiety at the aryl attachment point. However, the other compounds also seemed like reasonable candidates (Figure 2).
Chemical Synthesis
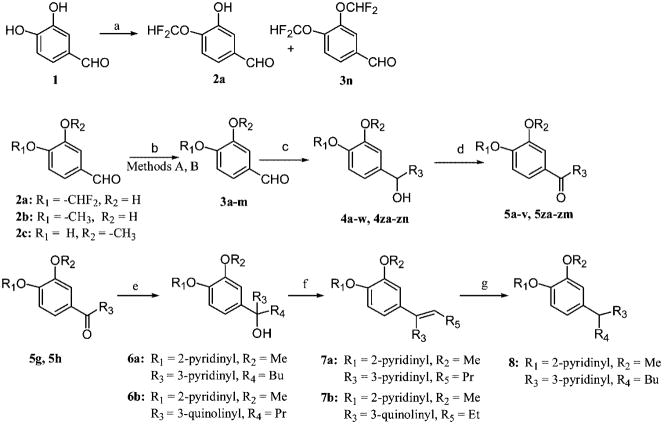
The synthesis of the designed analogues started with catechol, which required differential alkylation of the two hydroxyl groups of the catechol core. Therefore, the synthesis started with the alkylation of 1 at either the 3- or 4-position (Scheme 1). In one series, a difluoromethyl group needed to be attached to the 4-position, which was achieved through the alkylation of 3,4-dihydroxybenzaldehyde using chlorodifluoroacetate to give 4-difluoromethoxy-3-hydroxybenzaldehyde 2a in 45% yield and 3,4-bis(difluoromethoxy)benzaldehyde 3n in 20% yield.27,28 For the series with methyl substitution at either the 3- or 4-position of the catechol core, commercially available 3-hydroxy-4-methoxybenzaldehyde 2b or 4-hydroxy-3-methoxybenzaldehyde 2c was used as the starting material. The second alkyl group on the catechol moiety was attached by reacting 2a–c with racemic alcohols in THF via the Mitsunobu reaction (method A) to give racemic compounds 3a–j or by O-alkylation with bromides (method B) to give compounds 3k–m. Treatment of aldehydes 3a–m with n-butyllithium, tert-butyllithium, or a Grignard reagent gave the corresponding diastereomeric mixtures of alcohols 4a–w and 4za–zn. Oxidation of the alcohol with pyridium chlorochromate (PCC) afforded the corresponding ketones 5a–v and 5za–zm. Reaction of 5g,h with pyridinyllithium or quinolinyl-lithium gave the respective alcohols 6a,b. Dehydration of 6a afforded olefins 7a. Hydrogenation of 7a on Pd/C afforded 8.
Scheme 1.
a
a (a) CClF2COOMe, Cs2CO3, DMF; (b) (i) method A, R1OH or R2OH, PPh3, t-BuO2CN=NCO2-t-Bu, THF; method B, R1Br or R2Br, CsCO3, DMF; (c) R3Li or R3MgBr, THF, −78 °C; (d) PCC, DCM; (e) R4Br, n-BuLi, THF, −78 °C; (f) AcOH, H2SO4, 80 °C; (g) H2, Pd/C, room temp.
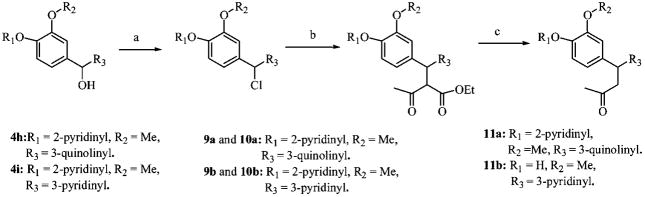
Compounds 4h and 4i were converted to their respective chlorides 9a and 9b upon reaction with thionyl chloride (Scheme 2). Reaction of 9a,b with the sodium salt of methyl acetoacetate afforded a diastereomeric mixture of esters 10a,b. Saponification followed by acidification led to spontaneous decarboxylation to give derivatives 11a,b (Scheme 2).
Scheme 2.
a
a(a) SOCl2, DCM; (b) ethyl acetoacetate, CH3ONa, THF; (c) LiOH, THF/MeOH/H2O.
SAR of Potential PDE4D Inhibitors
The enzyme inhibitory activities of the synthesized compounds were evaluated against PDE4 using human PDE4D2 (79–438) expressed in E. coli strain BL21 (Codonplus) as a model.25 The results are summarized in Tables 1–3.
Table 1.
IC50 (μM) Values of Phenyl Alkyl Alcohol PDE4 Inhibitorsa
 | ||||||
|---|---|---|---|---|---|---|
| Entry | Compd | R1 | R2 | R3 | IC50(μM) | |
| Rolipram | 0.55 | |||||
| 1 | 4b |
|
-CH3 | n-pr | >100 | |
| 2 | 4c |
|
-CH3 | n-Bu | >100 | |
| 3 | 4d |
|
-CH3 | n-pr | >100 | |
| 4 | 4e |
|
-CH3 | n-Bu | >100 | |
| 5 | 4l | -CH3 |
|
n-Bu | 10 | |
|
| ||||||
| ||||||
| Entry | Compd | R1 | R2 | R3 | R4 | IC50(μM) |
|
| ||||||
| 6 | 6b | 2-py | -CH3 | 3-quninolinyl | n-pr | 6 |
All potent compounds (low nanomolar) were assayed 2–3 times, and their inhibition results are expressed with standard deviations. All other results were from single runs.
Table 3.
IC50 (μM) Values of Phenylalkyl PDE4 Inhibitorsa
 | ||||||
|---|---|---|---|---|---|---|
| entry | compd | R1 | R2 | R3 | R4 | IC50 (μM) |
| 39 | 11a | 2-py | –CH3 | 3-quninolinyl | –CH2COCH3 | 6 |
| 40 | 11b | H | –CH3 | 3-py | –CH2COCH3 | 80 |
| 41 | 8 | 2-py | –CH3 | 3-py | n-Bu | 10 |
All potent compounds (low nanomolar) were assayed 2–3 times, and their inhibition results are expressed with standard deviations. All otsher results were from single runs.
The alcohol series of compounds (entries 1–5, Table 1) only exhibited weak inhibition activities with IC50 values in the range from 10 μM to over 100 μM, which were many fold less potent than the corresponding phenyl alkyl ketones (entries 9–12, 16, Table 2). Such results indicate that a planar geometry is desirable at this position for compounds with the catechol core. The inhibition results with the disubsituted alcohol (entry 6, Table 1) further suggested that the planar geometry was required. With the additional group on 6b the inhibition potency did not improve significantly compared to the ketone series. Within the alcohol series, there are two substitution patterns: one with a bulky alkyl group on the 3-position and the other with a bulky alkyl group on the 4-position of the catechol core. It is very clear that the compound (entry 5, IC50 = 10 μM) with a small methyl group on the 4-position and the bulky substitution on the 3-position has much higher potency than the analogues with a reverse substitution pattern (entries 1–4, IC50 > 100 μM). The same trend is reflected in the phenyl alkyl ketone series. For example, the 4-methoxy substituted phenyl compounds (entries 14–21, Table 2) are 20- to 500-fold more potent than the corresponding 3-methoxy substituted compounds (entries 7–12, Table 2). Such results are consistent with literature reports as well.19
Table 2.
IC50 (μM) Values of Phenyl Alkyl Ketone PDE4 Inhibitorsa
 | |||||
|---|---|---|---|---|---|
| Entry | Compd | R1 | R2 | R3 | IC50(μM) |
| 7 | 5a |
|
-CH3 | n-Bu | 93 |
| 8 | 5b |
|
-CH3 | n-Pr | 29 |
| 9 | 5c |
|
-CH3 | n-Pr | 60 |
| 10 | 5d |
|
-CH3 | n-Bu | 71 |
| 11 | 5e |
|
-CH3 | n-Pr | 24 |
| 12 | 5f |
|
-CH3 | n-Bu | 100 |
| 13 | 5j | -CH3 |
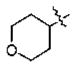
|
3-quinolinyl | 45 |
| 14 | 5k | -CH3 |
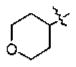
|
n-Pr | 3.5 |
| 15 | 5l | -CH3 |
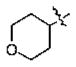
|
n-Bu | 0.72 |
| 16 | 5m | -CH3 |
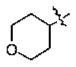
|
n-Bu | 0.22 |
| 17 | 5n | -CH3 |
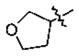
|
n-Pr | 0.43 ±0.11 |
| 18 | 5o | -CH3 |
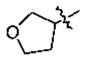
|
-(CH2)3Ph | 2.3 |
| 19 | 5p | -CH3 |
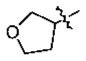
|
-(CH2)2Cyclohexyl | 1.9 |
| 20 | 5q | -CH3 |
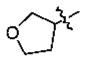
|
-(CH2)2Ph | 2.6 |
| 21 | 5r | -CH3 |
|
-CH2cyclohexyl | 2.6 |
| 22 | 5i | -CH3 |
|
n-Bu | >100 |
| 23 | 5zi | -CHF2 | 2-py | n-Pr | 40 |
| 24 | 5zh | -CHF2 |
|
n-Pr | 0.35 |
| 25 | 5v | -CHF2 |
|
i-Bu | 0.026 ± 0.004 |
| 26 | 5zb | -CHF2 |
|
-(CH2)2cyclohexyl | 1.1 |
| 27 | 5zd | -CHF2 |
|
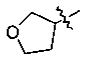
-(CH2)2Ph | 0.18 ± 0.03 |
| 28 | 5za | -CHF2 |
|
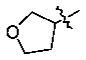
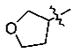
n-Bu | 0.061 ± 0.006 |
| 29 | 5zl | -CHF2 | -CHF2 | -(CH2)2Ph | 0.82 ± 0.03 |
| 30 | 5zm | -CHF2 | -CHF2 | -CH2-cyclohexyl | 5.2 |
| 31 | 5zj | -CHF2 | -CHF2 | -(CH2)3Ph | 44 |
| 32 | 5zf | -CHF2 |
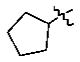
|
i-Bu | 0.028 ± 0.001 |
| 33 | 5zg | -CHF2 |
|
n-Bu | 2.2 |
| 34 | 5zc | -CHF2 |
|
-CH2-cyclohexyl | 0.667 |
| 35 | 5ze | -CHF2 |
|
-(CH2)3Ph | 0.636 |
| 36 | 5s | -CHF2 |
|
t-Bu | 0.41 |
| 37 | 5t | -CH3 |
|
i-Bu | 0.19 ± 0.02 |
| 38 | 5u | -CH3 |
|
i-Pen | 0.38 ± 0.05 |
All potent compounds (low nanomolar) were assayed 2–3 times, and their inhibition results are expressed with standard deviations. All other results were from single runs.
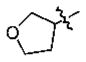
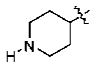
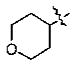
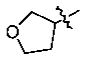
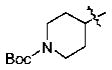
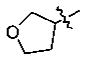
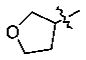
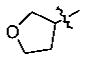
With the initial indication that the 4-methoxy ketones are more active, the subsequent effort was focused on optimizing this series of compounds. One approach adopted was to use a 4-difluoromethoxy group in place of the methoxy group with the idea of improved metabolic stability (entries 24–28 and 33–35, Table 2).27,28 It turned out that these 4-difluoromethoxy compounds were also 2- to 14-fold more potent than the corresponding 4-methoxy substituted compounds (entries 14, 16, 18–20, and 37, Table 2). Within this series, the substituent effect at the 3-position was also studied. The tetrahydrofuran-3-yloxy substituted phenyl compounds (entries 16, 17) have 3-to 8.5-fold higher potencies than the corresponding tetrahydropyran-3-yloxy substituted phenyl compounds (entries 14, 15). The replacement of the tetrahydrofuran ring by a cyclopentyl ring (entry 25) gave a compound (entry 32) with comparable potency (IC50 = 28 nM). The tetrahydrofuran-3-yloxy substituted phenyl compound (entry 28) also seems to be more active than the corresponding Boc protected piperidi-4-nyl substituted compound (entry 33) by more than 20- to 30-fold, and the 3,4-bis(difluoromethoxy)phenyl compounds (entry 31) are also less active than the corresponding 4-difluoromethyl-3- tetrahydrofuran-3-yloxy substituted phenyl compounds (entries 27, 28, 34, and 35) by 2.7- to 60-fold. Similarly, replacing the tetrahydrofuran ring with a deprotected tetrahydropyrrol ring (entries 25, 28) also decreased the potency of IC50 by over 100 μM (entry 22) from 26 and 61 nM.
In the series of the 4-methoxy-3-tetrahydrofuran-3-yloxy phenyl ketones (entries 16–21, 37, 38) and the series of the 4-difluoromethoxy-3-tetrahydrofuran-3-yloxy phenyl ketones (entries 25–28 and 34–36), both ketones (entries 25, 37) with an isobutyl side chain have the highest potency (IC50 of 26 and 176 nM, respectively) and the ketones (entries 28 and 16) with a butyl side chain have the second highest potency (IC50 of 61 and 220 nM, respectively). The ketones with the bulky cyclohexyl (entries 21 and 26) and phenylalkyl (entries 20 and 18) side chains (R3) constantly showed the lowest potency.
We have also tested the compounds with further transformations of the ketone group into a sp3 carbon (entries 39–41, Table 3). None of these transformations led to improved potency.
Some representatives of the PDE4 inhibitors were tested for their PDE4 selectivity using human PDE7A1, 9A2, 2A3, and 5A1 subtypes, which were cloned and expressed in E. coli strain BL21 (Codonplus) (Table 4).25 Most of the compounds that we have synthesized showed high PDE4 selectivity. For example, 5v showed preference for PDE4 with selectivity of at least about 2000-fold over PDE7A1, PDE9A2, PDE2A3, and PDE5A1, respectively; 5k had selectivity at least 14-fold over PDE7A1, PDE9A2, PDE2A3, and PDE5A1.
Table 4.
IC50 Values (μM) of Selected Lead Inhibitors on Various PDEs
| compd | PDE4D2 | PDE7A1 | PDE9A2 | PDE2A3 | PDE5A1 |
|---|---|---|---|---|---|
| 5v | 0.026 | >50 | >50 | >50 | >50 |
| 11a | 6 | >50 | >50 | >50 | >50 |
| 5k | 3.5 | >50 | >50 | >50 | >50 |
| 6b | 6 | 17 | >50 | >50 | 40 |
The inhibition of TNF-α production induced by lipopolysaccharide is an essential assay for evaluation of the pharmacological effect of PDE4 inhibitors in anti-inflammation and the treatment of COPD and asthma.29,30 Normal and nonstimulated human blood does not contain detectable levels of TNF-α. Upon stimulation with LPS, activated monocytes express and secrete TNF-α for up to 8 h and plasma levels remain stable for 24 h. Published studies have shown that inhibition of TNF-α production by increasing intracellular cAMP via PDE4 inhibition and/or enhanced adenylyl cyclase activity occurs at the transcriptional level. Therefore, we selected the two most active compounds and studied their ability to inhibit TNF-α production. Specifically, the effect of 5v and 5zf for their ability to inhibit LPS-mediated TNF-α production by human peripheral blood mononuclear cells (PBMCs) was studied (Figure 3). Rolipram (IC50 = 60 nM) was included as a reference. The results show that both 5v and 5zf were able to inhibit LPS-mediated TNF-α production with IC50 values of 40 and 100 nM, respectively. IC50 values for TNF-α release from the literature may not be directly comparable because of variations of assay systems and conditions. However, if roliprarn is taken as a relative reference, the potency of our 5v is higher than that of rolipram,30 a compound that went into clinical trials for treatment of COPD.
Figure 3.
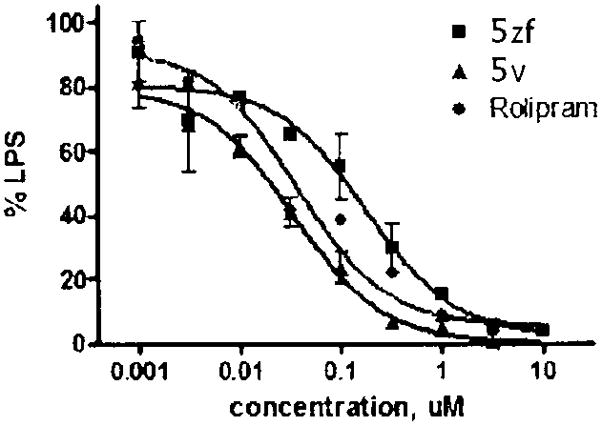
Inhibitory effect of 5v (▲) and 5zf (■) against LPS-mediated TNF-α production, compared with rolipram (●).
Molecular Modeling
In an effort to gain an understanding of the structural basis for the empirical structure–activity relationships observed, we also studied the binding of the synthesized inhibitors with PDE4D2 through computer modeling. The crystal structure of PDE4D2–rolipram complex25,26 was used as the starting point.
Molecular Docking
In this investigation, the 3D structure of PDE4D was taken from a Protein Data Bank entry (PDB entry 1OYN) having the ligand rolipram in place.26 The new inhibitors were built and optimized at the Hartree–Fock level with the 6-311 G basis set in the Gaussian 03 program.31 The optimized ligands were then embedded with Gastiger–Hückel partial charges using the SYBYL 7.1 package.30 DOCK 5.232 was employed to dock these ligands into the PDE4D active site. The resulting structures were then analyzed using HBPLUS 3.0633 and Ligplot 4.2234 programs to identify specific contacts between ligands and receptor.
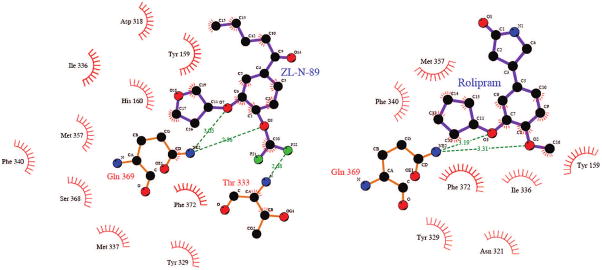
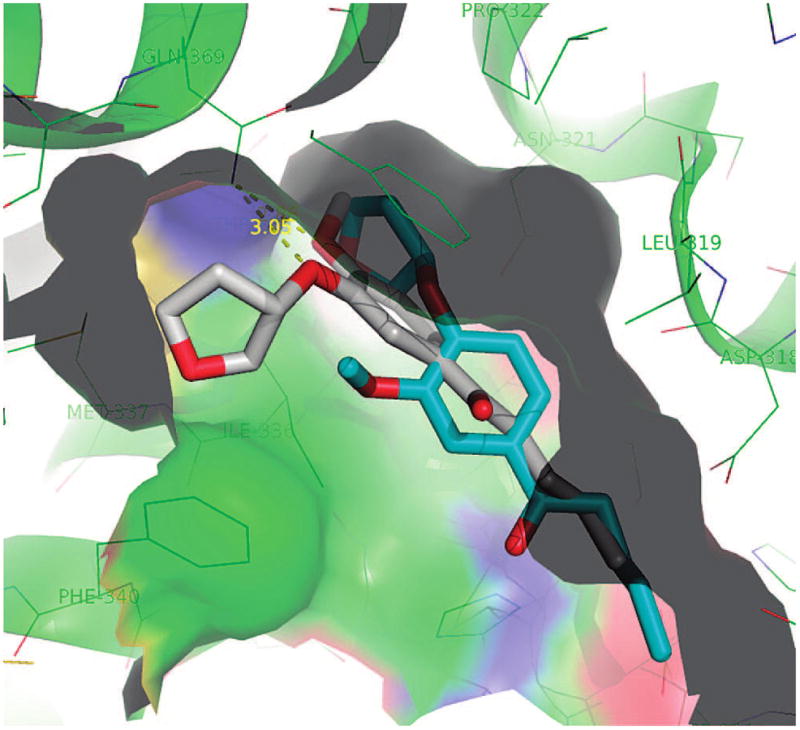
Initially, we selected the most potent compounds 5v, 5za, and 5zf for the docking experiments. These three compounds adopt the same alignments as rolipram in the active site. Such alignments have also been confirmed by a preliminary crystallographic study, the details of which will be published elsewhere. Figure 4 presents the docking conformations of rolipram (white), 5za (yellow), 5v (blue), and 5zf (red) in the active site of PDE4. Figure 5 depicts a schematic binding profile of 5za. Similar to the binding of rolipram (Figure 5), 5za, 5v, and 5zf have a hydrogen bond with Gln 369 and hydrophobic contact with Ile 336, Met 357, Phe 340, Ser 368, Met 337, Phe 372, and Tyr 3. Different from rolipram binding are the possible interactions of the difluoromethyl group of 5za, 5v, and 5zf with the Thr 333 N–H, which could engage in hydrogen bond interactions, and the additional van der Waals interactions of the long alkyl chain with His 160, Asp 318, and Tyr 159. In the case of hydrogen bond interactions, the F–O distance is 2.68 Å but with a nonideal C–F–H angle of 34.7°.
Figure 4.
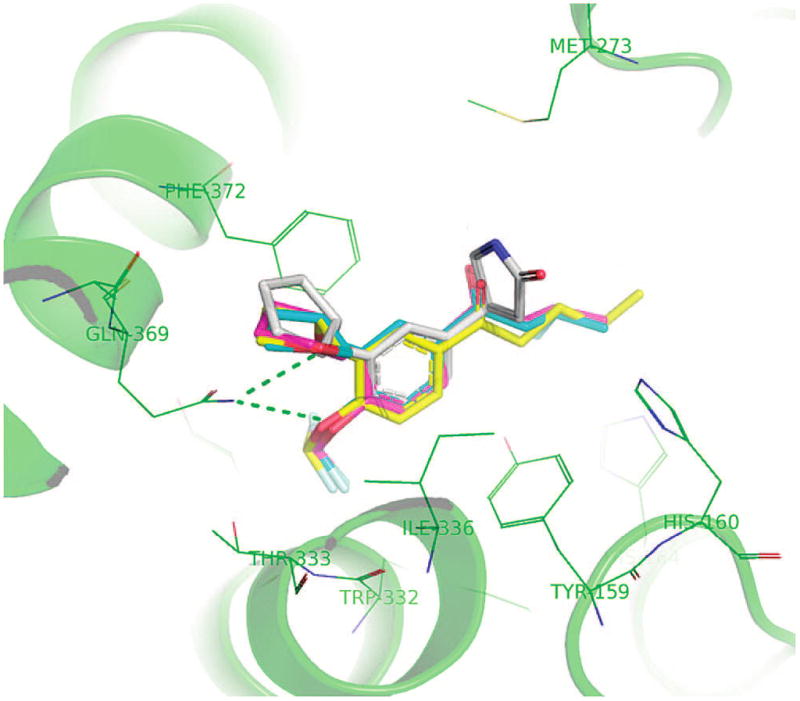
Key residues in the binding site of rolipram (white), 5za (yellow), 5v (blue), and 5zf (red) to PDE4.
Figure 5.
Binding profile of 5za and rolipram with PDE4D2 obtained from Ligplot analysis.
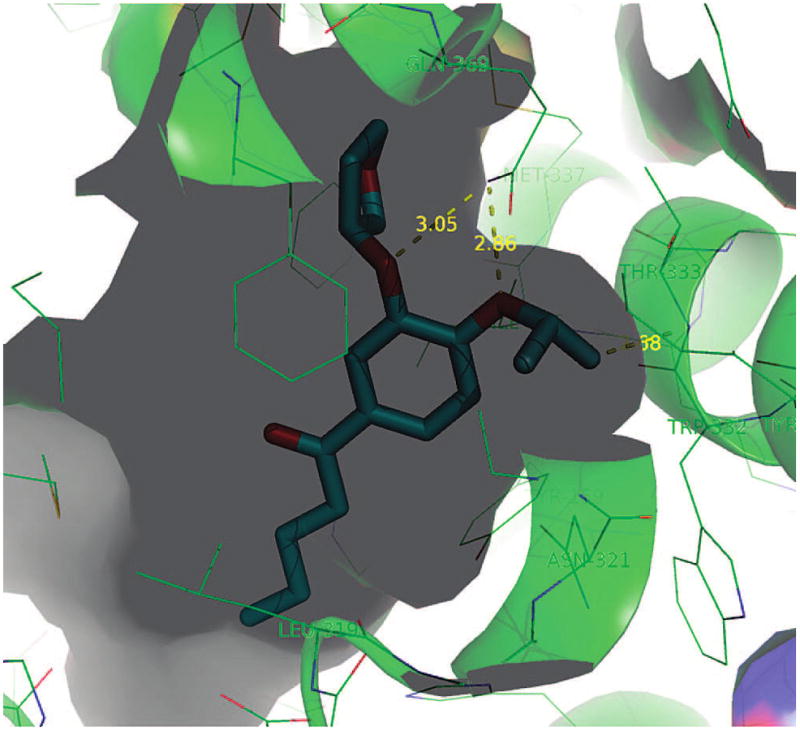
The computer modeling results are also consistent with the empirically observed structure–activity relationship. From the modeling results it is clear that the 4-methyl/difluoromethyl group occupies a small pocket. Anything larger at this position would result in steric hindrance and therefore unfavorable interactions (Figure 6). Such a finding is consistent with earlier studies with rolipram26 and the fact that 4-position substitution with a bulkier group leads to compounds with diminished potency (entries 1–6, Table 1; entries 7–12, Table 2). Figure 6 shows the unfavorable interactions of an inhibitor with large substituents at the 4-position. For 5za, the difluoromethyl group has an additional hydrogen bond with Thr 333; meanwhile, the long alkyl chain also bears additional van der Waals interactions with His 160, Asp 318, and Tyr 159. This difference could be the reason that these difluoremethyl compounds have higher potencies than others (Figures 5 and 7). The 3-position has a fairly large hydrophobic pocket, allowing favorable interactions with bulkier groups such as tetrahydropyran and tetrahydrofuran (entries 13–28, 32–38, Table 2). At this site, there is Lys 367, which under normal physiological conditions should be positively charged. This helps to explain why the deprotected amino group in compound 5i (entry 22, Table 2), which introduces a positively charged protonated amine, leads to reduced potency.
Figure 6.
Negative effect observed for a bulky substitution at 4-position: blue stick, 5f; gray stick, 5m.
Figure 7.
Docking conformation of 5za in the PDE4D active site.
Within the ketone series (entries 7–38, Table 2), it seems that compounds with a four-carbon side chain have the highest potency. This is consistent with the existence of a hydrophobic binding pocket accommodating the side chain, which is somewhat narrow but deep. Bulkier groups such as the phenyl and cyclohexyl side chains protrude away from this hydrophobic pocket, resulting in unfavorable interactions (Figures 4 and 7). The planar sp2-hybridized ketone group seems to have the flexibility to put the hydrophobic side chain in the right position and orientation for favorable hydrophobic interactions. In contrast, a tetrahedral sp3 carbon would fix the side chain in a position for less favorable hydrophobic contact with PDE4, which explains why the compounds with a sp3 carbon at the benzylic position showed low potency (Figure 8).
Figure 8.

Comparison of sp2-hybridized ketone (blue stick, 5m) and sp3 alcohol (gray stick, 4l).
Molecular Similarity Index Analysis (CoMSIA)
In order to achieve some semiquantitative understanding of the contribution of various parameters to binding affinity, we have also conducted a structure-based comparative molecular similarity index analysis (CoMSIA) implemented in SYBYL 7.1.30 CoMSIA is an extension of the CoMFA (comparative molecular field analysis) methodology.35,36 Both are forms of 3D-QSAR and are based on the assumption that changes in ligand binding affinities are related to changes in molecular properties, represented by fields. They differ only in the implementation of the fields.35,36 In this study the docking conformations of all compounds synthesized were used as the input of molecular alignments (Figure 9).
Figure 9.
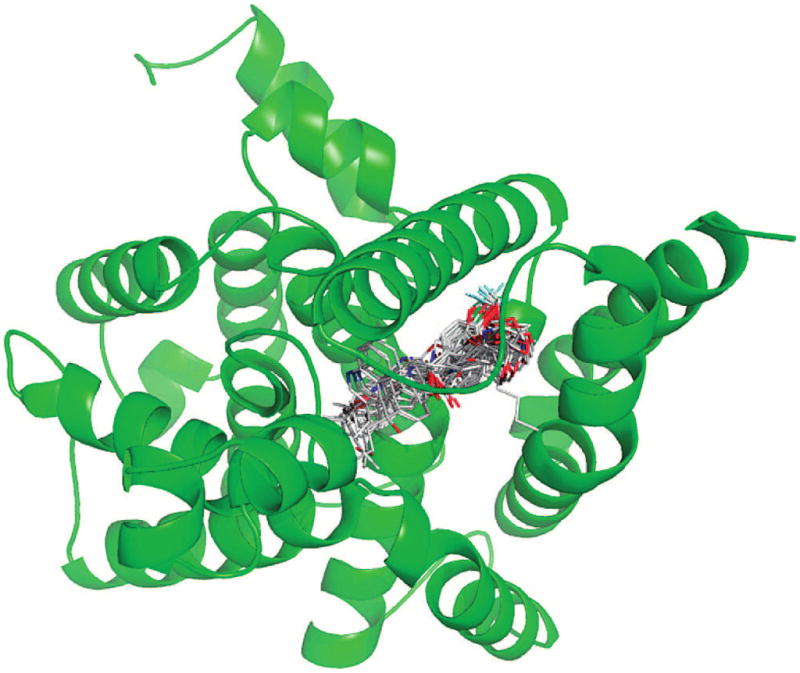
Docking conformations of synthesized inhibitors in the PDE4D active site.
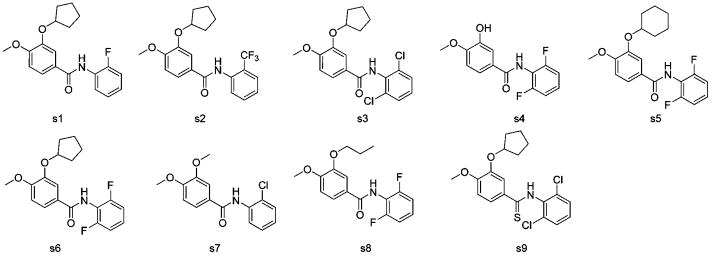
The CoMSIA model analysis indicates that the contribution of steric, electrostatic, hydrophobic, hydrogen bond donor, and hydrogen bond acceptor of this series of PDE4 inhibitors to binding affinity is 5.8%, 26.4%, 25.4%, 23.7%, and 18.7%, respectively. In PLS analysis, the standard error of estimate of 0.312, non-cross-validated correlation coefficient r2 of 0.939, cross-validated coefficient q2 of 0.832, and F value of 584.051 show that our CoMSIA model has good prediction. In order to validate this CoMSIA model again, we introduced an external test set consisting of rolipram plus nine compounds (s1–s9) from literature (Figure 10).19 After prediction by the CoMSIA model, an r2 of 0.803 and q2 of 0.706 were obtained. This external validation again suggests a reliable CoMSIA model we harvested. Table 5 shows a comparison of the calculated pIC50 and experimentally determined pIC50 values using the CoMSIA model developed, and Figure 11 shows the schematic correlation of such data. Such results will help future optimization efforts.
Figure 10.
Chemical structures for test set validation.
Table 5.
Actual pIC50, Estimated pIC50, and Residual Values of Molecules Used for CoMSIA
| compd | actual pIC50 | estimated pIC50 | residual |
|---|---|---|---|
| 4b | 2.82 | 3.15 | −0.33 |
| 4c | 3.27 | 3.15 | 0.12 |
| 4d | 3.28 | 3.39 | −0.11 |
| 4e | 3.40 | 3.23 | 0.17 |
| 5i | 3.99 | 3.85 | 0.14 |
| 5f | 4.00 | 3.92 | 0.08 |
| 5a | 4.03 | 4.15 | −0.12 |
| 11b | 4.10 | 4.02 | 0.08 |
| 5d | 4.15 | 4.10 | 0.05 |
| 5c | 4.22 | 4.30 | −0.08 |
| 5j | 4.35 | 4.09 | 0.26 |
| 5zj | 4.36 | 4.49 | 0.13 |
| 5zi | 4.40 | 4.14 | 0.26 |
| 5b | 4.54 | 4.27 | 0.27 |
| 5e | 4.62 | 4.45 | 0.17 |
| 4l | 5.00 | 5.35 | −0.35 |
| 8 | 5.00 | 4.99 | 0.01 |
| 11a | 5.22 | 5.24 | −0.02 |
| 5zm | 5.28 | 5.60 | −0.32 |
| 5k | 5.46 | 5.77 | −0.31 |
| 5q | 5.59 | 6.08 | −0.49 |
| 5r | 5.59 | 5.78 | −0.19 |
| 5o | 5.64 | 5.98 | −0.34 |
| 5zg | 5.66 | 5.51 | 0.11 |
| 5p | 5.72 | 5.54 | 0.18 |
| 5zb | 5.96 | 6.34 | −0.38 |
| 5l | 6.14 | 5.88 | 0.26 |
| 5zc | 6.18 | 6.37 | −0.19 |
| 5ze | 6.20 | 6.43 | −0.23 |
| 5zl | 6.30 | 6.07 | 0.23 |
| 5s | 6.39 | 6.90 | −0.01 |
| 5n | 6.40 | 6.25 | 0.15 |
| 5u | 6.42 | 6.21 | 0.21 |
| 5zh | 6.46 | 6.39 | 0.07 |
| 5m | 6.7 | 6.06 | 0.14 |
| 5zd | 6.74 | 6.31 | 0.43 |
| 5t | 6.75 | 6.99 | −0.24 |
| 5za | 7.21 | 7.20 | 0.01 |
| 5zf | 7.66 | 6.80 | −0.25 |
| 5v | 7.74 | 7.48 | 0.26 |
| Test set rolipram | 6.26 | 6.87 | −0.61 |
| s1 | 4.97 | 4.56 | 0.41 |
| s2 | 5.51 | 6.12 | −0.61 |
| s3 | 7.55 | 7.93 | −0.38 |
| s4 | 4.43 | 4.82 | −0.39 |
| s5 | 6.40 | 7.01 | −0.61 |
| s6 | 7.15 | 6.54 | 0.61 |
| s7 | 6.19 | 7.01 | −0.82 |
| s8 | 5.80 | 5.82 | −0.02 |
| s9 | 6.70 | 7.1 | −0.40 |
Figure 11.
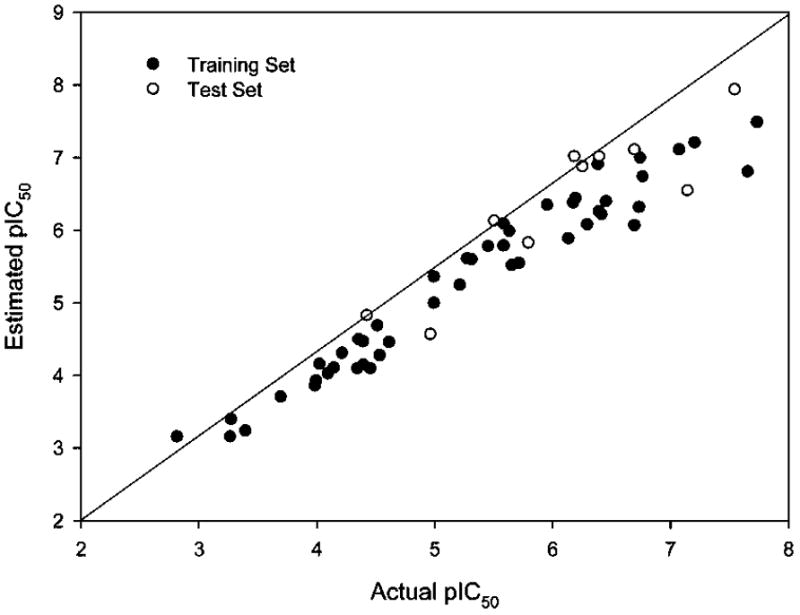
Actual versus predicted pIC50 for CoMSIA 3D-QSAR model.
NMR Studies of the Interactions of 5i with PDE4D2 Protein
In order to achieve some experimental verification of the modeled binding modes of the inhibitors synthesized, NMR experiments were conducted in solution. We initially selected two compounds, 5i (IC50 = 102 μM) and 5zi (IC50 = 40 μM), for the studies. These two compounds have moderate affinities, which allow high sensitivity detection of transferred effects through observation of spectra from ligands in 10- to 100-fold excess over protein.37 Under the conditions of rapid exchange, normally associated with low to moderate binding affinities, effects such as nuclear Overhauser effects (NOEs) and saturation transfer differences (STDs) originating with ligands bound to PDE4 are carried into solution. Because of enhanced efficiencies in magnetization transfer in large complexes, the bound-state properties dominate the average effects observed through the narrow lines and high intensities of the free ligand resonances. Unfortunately, 5zi had solubility and nonspecific binding problems that prevented the collection of meaningful data. Therefore, only 5i was used for the NMR studies. Both STD and transferred NOEs (trNOE) experiments were performed to study the inhibitor. STD provides information on the proximity of protons on the bound ligand to protons on the protein. The relative signal intensities in an STD spectrum are interpreted to produce an epitope map with protons giving the strongest signal closest to the protein surface.37 The trNOE experiment provides distance information between protons within the ligand, which can be used to determine the compound’s bound state conformation.37 The solubility of 5i (up to 800 μM in D2O) was confirmed by NMR. A reference 1H spectrum allowed assignment of resonances as indicated in Figure 12a.
Figure 12.
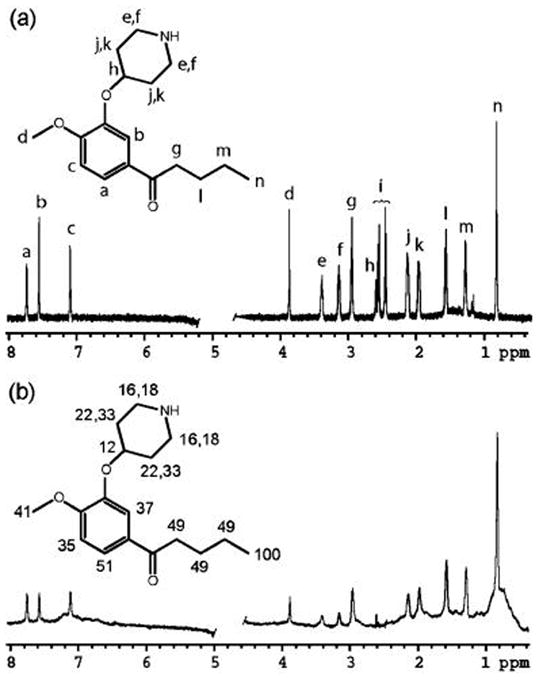
(a) Reference 1D 1H NMR spectrum of 400 μM 5i with assignments indicated by letters on the chemical structure. The peak for the proton in the 4-position of the piperidinyl group (labeled “h” on the chemical structure) is buried under the water peak. Peak “i” was not assigned and is believed to be a contaminant. (b) STD spectrum of 800 μM 5i with 10 μM PDE4D2 using a 50 Hz Gaussian saturation pulse train of 250 ms centered at 0.5 ppm. The protein background was not subtracted or suppressed. The normalized STD percentages calculated from the STD and reference spectra of 5i are superimposed on the chemical structure. Both spectra were collected using a Varian INOVA 800 MHz NMR instrument equipped with a cryogenic probe. The water peaks in the center of the spectra were removed for clarity.
The STD spectrum of 5i in the presence of PDE4D2 is presented in Figure 12b. In an effort to map the binding epitope, the peak integrals from the reference and STD spectra were compared and normalized to give a STD percentage for each proton. The STD percentages are shown on the chemical structure in Figure 12b. The STD percentages give an indication of which protons of bound 5i are close to the protein, i.e., an epitope map. For 5i, peak “i” was not observed in the STD spectrum, indicating that it does not bind PDE4D2 providing evidence that it is a contaminant. The n-butanyl, methyl, and catechol protons show stronger STD contacts than the piperidinyl protons. While it is possible to check these intensities for consistencies with specific models when available,38 it is also useful to seek a more qualitative interpretation. The results as presented are consistent with computer modeling studies indicating strong hydrophobic interactions of the alkyl chain and adjacent parts of the catechol ring. Close approach combined with the higher density of nonexchangeable protons on hydrophobic groups would lead to the observed effects.
A trNOEsy spectrum of 5i in the presence of PDE4D2 is presented in Figure 13. A short mixing time of 50 ms was used to avoid spin diffusion artifacts and provide a more accurate interpretation. The cross peaks show the same sign as the diagonal peaks indicating negative NOEs originating from the bound form of 5i. The reference NOESY spectrum of 5i (not shown) has positive NOEs typical of small molecules free in solution and confirms that the NOEs in the trNOEsy spectrum reflect the bound state structure. The appearance of a set of cross peaks similar to those seen in the reference NOESY spectrum is indicative of some similarity in the bound form compared to the solution form of the drug. Under an initial rate assumption, the relative intensities of the NOESY cross peaks are inversely related to the sixth power of the distance between proton pairs and qualitative information on a structure can readily be determined by comparing the cross peak intensities in Figure 13. Structurally restraining NOEs include contacts between the catechol protons and the methyl, piperidinyl, and n-butanyl protons. The catechol proton in the 6-position (peak “a”) has an NOE to the n-butanyl protons (peak “g”), and the catechol proton in the 5-position (peak “c”) has an NOE to the methyl protons (peak “d”). This NOE would suggest a conformation different from the structure used to illustrate the NMR spectrum (Figure 12) with the keto chain rotated 180° about the chain to catechol bond. This geometry is in fact consistent with the geometries shown for 5v, 5za, and 5zf (Table 2). NOEs are also seen from the catechol 2-position to the piperidinyl protons in the 4-position (peak “h”) and the NMR equivalent methylene protons in the 3- and 5-positions (peaks “j” and “k”). This NOE restrains the relative orientation of the piperidinyl and catechol rings and is consistent with the docking models of 5za in Figure 7.
Figure 13.
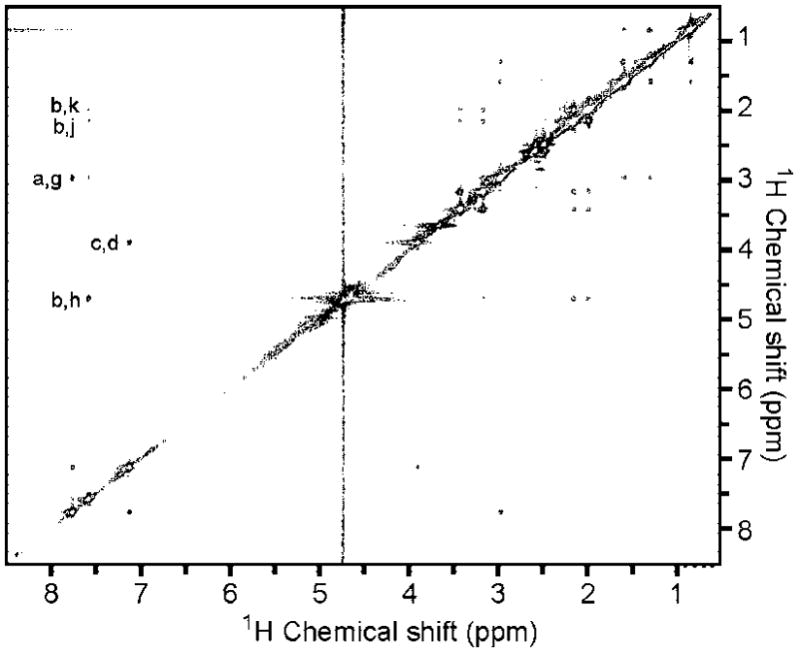
The 2D NOESY spectrum of 800 μM 5i and 10 μM PDE4D2 with a 50 ms mixing time is shown. The cross peaks have positive intensities (i.e., negative NOEs), which indicate that 5i binds PDE4D2.
Conclusions
A new series of phenyl alkyl ketones with variable side chains was investigated as potential PDE4D inhibitors. Thirteen alkyl ketones were identified to have submicromolar IC50 values. Among them, the most potent compounds 5v, 5zf, and 5za have IC50 values of 26, 28, and 61 nM, respectively. Compound 5v also showed a preference for PDE4D with selectivity of >2000 fold over PDE7A1, PDE9A2, PDE2A3, and PDE5A1, respectively. Docking of 5v, 5zf, and 5za into the binding pocket of the PDE4 catalytic domain revealed a similar binding profile with PDE4 as rolipram. trNOEsy and STD NMR experiments on 5i provided experimental support for many of the modeling projections. Compound 5i appears to adopt the same overall conformation in the free and enzyme bound form and to dock in a similar manner as 5v, 5za, and 5zf.
Experimental Section
General Chemistry
All reagents were purchased from Acros and Aldrich. N-(tert-Butoxycarbonyl)-4-piperidinol was synthesized according to literature procedures.1 Microwave heating was performed in the single-mode microwave cavity of a Discover Synthesis System (CEM Co.), and all microwave reactions were conducted in a heavy-walled glass vials sealed with Teflon septa. 1H NMR, 13C NMR, and DEPT spectra were recorded at 400 and 100 MHz, respectively, on a Bruker 400 NMR spectrometer. Combustion analyses and mass spectra were performed by the analytical and the mass spectrometry facilities at Georgia State University.
4-Difluoromethoxy-3-hydroxybenzaldehyde (2a) and 3,4-Bisdifluoromethoxybenzaldehyde (3n)
A mixture of 3,4-dihydroxybenzaldehyde (1.38 g, 0.01 mol), methyl chlorofluoroacetate (1.87 g, 0.013 mol), and cesium carbonate (4.24 g, 0.013 mol) in DMF (20 mL) was stirred at 65 °C for 3 h under nitrogen atmosphere. Then DMF was removed in vacuo and the residue was partitioned between aqueous 3 N HCl and ether. The aqueous layer was extracted three times with ether. The combined organic solution was washed with water and brine, dried over MgSO4, filtered, and evaporated. The residue was purified by flash chromatography to afford 2a as a white solid (0.85 g, 45%) and 3n as a yellowish liquid (0.48 g, 20%).
2a
1H NMR (CDCl3): 9.92 (1H, s), 7.55 (1H, d, J = 4.0 Hz), 7.45 (1H, dd, J = 4.0 and 8.4 Hz), 7.28 (1H, d, J = 8.4 Hz), 6.67 (1H, t, J = 72.8 Hz), 6.17 (1H, s, br). 13C NMR (CDCl3): 191.1 (d), 147.8 (s), 143.0 (s), 134.5 (s), 123.2 (d), 119.3 (d), 117.1 (d), 115.6 (triplet, J = 262 Hz, d). MS-EI: 188 (M+).
3n
1H NMR (CDCl3): 9.95 (1H, s), 7.79–7.77 (2H, m), 7.43 (1H, dd, J = 8.8 Hz), 6.68 (1H, t, J = 72.4 Hz), 6.63 (1H, t, J = 72.8 Hz). 13C NMR (CDCl3): 190.1 (d), 147.1 (s), 142.3 (s), 134.1 (s), 128.7 (d), 122.1 (d), 121.4 (d), 115.5 (triplet, J = 263 Hz, d), 115.3 (triplet, J = 262 Hz, d). MS-EI: 238 (M+).
General Procedure for the Synthesis of 3 by Mitsunobu Reaction of 2a and 2c with R2OH
To the mixture of compound 2 (1 mmol), R2OH (1.1 mmol), and di-tert-butyl azodicarboxylate (1.2 mmol) in THF (5 mL) was added dropwise triphenylphosphine (1.2 mmol) in THF (5 mL) at 0 °C. The resulting solution was stirred until compound 2 was consumed completely (usually within 24 h). Solvent was removed in vacuo and the residue was purified by flash chromatography to afford 3.
4-Difluoromethoxy-3-(tetrahydropyran-4-yloxy)benzaldehyde (3a)
Yield: 28%. 1H NMR (CDCl3): 9.93 (1H, s), 7.52 (1H, d, J = 2.0 Hz), 7.48 (1H, dd, J = 2.0 and 8.0 Hz), 7.33 (1H, d, J = 8.0 Hz), 6.70 (1H, t, J = 74.0 Hz), 4.65 (1H, m), 3.98 (2H, m), 3.61 (2H, m), 2.06 (2H, m), 1.85 (2H, m). 13C NMR (CDCl3): 190.6 (d), 149.2 (s), 146.0 (s), 134.4 (s), 125.0 (d), 122.3 (d), 115.6 (triplet, J = 259 Hz, d), 114.1 (d), 73.3 (d), 64.7 (t), 31.4 (t). MS-EI: 272 (M+)
4-Difluoromethoxy-3-(tetrahydrofuran-3-yloxy)benzaldehyde (3b)
Yield: 84%. 1H NMR (CDCl3): 9.93 (1H, s), 7.48 (1H, dd, J = 1.6 and 8.4 Hz), 7.44 (1H, d, J = 1.6 Hz), 7.33 (1H, d, J = 8.4 Hz), 6.67 (1H, t, J = 74.4 Hz), 5.07 (1H, m), 4.06–3.91 (4H, m), 2.30 (1H, m), 2.19 (1H, m). 13C NMR (CDCl3): 190.5 (d), 149.4 (s), 145.6 (s), 134.3 (s), 125.2 (d), 122.2 (d), 115.5 (triplet, J = 260 Hz, d), 112.9 (d), 78.8 (d), 72.5 (t), 67.0 (t), 32.7 (t). MS-EI: 258 (M+).
4-(2-Difluoromethoxy-5-formylphenoxy)piperidine-1-carboxylic Acid tert-Butyl Ester (3c)
Yield: 54%. 1H NMR (CDCl3): 9.93 (1H, s), 7.52 (1H, s), 7.48 (1H, dd, J = 1.2 and 8.4 Hz), 7.33 (1H, d, J = 8.4 Hz), 6.66 (1H, t, J = 74.0 Hz), 4.63 (1H, m), 3.69 (2H, m), 3.41 (2H, m), 1.96 (2H, m), 1.81 (2H, m), 1.48 (9H, s). 13C NMR (CDCl3): 190.6 (d), 154.6 (s), 149.3 (s), 146.0 (triplet, J = 3 Hz, s), 134.5 (s), 125.2 (d), 122.3 (d), 115.6 (triplet, J = 260 Hz, d), 114.1 (d), 79.7 (s), 74.0 (d), 40.4 (multiple, t), 30.2 (t), 28.3 (q).
3-Methoxy-4-(tetrahydropyran-4-yloxy)benzaldehyde (3d)
Yield: 65%. 1H NMR (CDCl3): 9.85 (1H, s), 7.43–7.42 (2H, m), 7.01 (1H, d, J = 8.8 Hz), 4.62 (1H, m), 4.03 (2H, m), 3.93 (3H, s), 3.59 (2H, m), 2.07 (2H, m), 1.89 (2H, m). 13C NMR (CDCl3): 190.7 (d), 152.2 (s), 150.7 (s), 130.4 (s), 126.1 (d), 114.0 (d), 110.0 (d), 73.2 (d), 65.1 (t), 56.0 (q), 31.7 (t). MS-EI: 236 (M+).
3-Methoxy-4-(tetrahydrofuran-3-yloxy)benzaldehyde (3e)
Yield: 71%. 1H NMR (CDCl3): 9.84 (1H, s), 7.44–7.41 (2H, m), 6.93 (1H, d, J = 8.4 Hz), 5.06 (1H, m), 4.02 (4H, m), 3.90 (3H, s), 2.06 (2H, m). 13C NMR (CDCl3): 190.4 (d), 152.1 (s), 150.0 (s), 130.0 (s), 125.8 (d), 112.8 (d), 109.5 (d), 78.4 (d), 72.4 (t), 66.8 (t), 55.5 (q), 32.6 (t). MS-EI: 222 (M+).
4-(4-Formyl-2-methoxyphenoxy)piperidine-1-carboxylic Acid tert-Butyl Ester (3f)
Yield: 76%. 1H NMR (CDCl3): 9.85 (1H, s), 7.44–7.41 (2H, m), 7.03 (1H, m), 4.61 (1H, m), 3.91 (3H, s), 3.77 (2H, m), 3.32 (2H, m), 1.97 (1H, m), 1.82 (1H, m), 1.46 (9H, s). 13C NMR (CDCl3): 190.7 (d), 154.6 (s), 152.1 (s), 150.7 (s), 130.3 (s), 126.1 (d), 114.1 (d), 110.0 (d), 79.5 (s), 73.7 (d), 56.0 (q), 40.5 (multiple, t), 30.3 (t), 28.3 (q).
4-Methoxy-3-(tetrahydropyran-4-yloxy)benzaldehyde (3g)
Yield: 52%. 1H NMR (400 MHz, CDCl3): 1.84–1.86 (2H, q, J = 4.4, 4.4 Hz), 2.06 (2H, d, J = 8.8 Hz), 3.57 (2H, t, J = 9.2 Hz), 3.95 (3H, s), 4.05 (2H, m), 4.51 (1H, m), 7.01 (1H, d, J = 8.4 Hz), 7.44 (1H, s), 7.47–7.49 (1H, q, J = 1.6, 6.4 Hz) 9.84 (1H, s) ppm. 13C NMR (100 MHz, CDCl3): 31.86, 56.16, 65.31, 73.49, 111.25, 113.92, 127.15, 1.99, 147.09, 156.06, 190.80 ppm. HRMS (ESI+): calcd for C13H16O4, 237.1127; found, 237.1121.
4-Methyoxy-3-(tetrahydrofuran-3-yloxy)benzaldehye (3h)
Yield: 40%. 1H NMR (400 MHz, CDCl3): 2.25 (2H, q, J = 6.0, 8.0 Hz), 3.94 (4H, d, J = 12 Hz), 4.04 (3H, d, J = 2.4 Hz), 5.02 (1H, s), 7.00 (1H, d, J = 8.0 Hz), 7.35 (1H, s), 7.48 (1H, q, J = 2.0, 6.4 Hz), 9.84 (1H, s) ppm. 13C NMR (100 MHz, CDCl3): 32.98, 56.16, 67.72, 72.91, 78.67, 111.07, 112.18, 127.21, 1.96, 147.58, 155.56, 190.73 ppm. HRMS (ESI+): calcd for C12H14O4, 223.0970; found, 223.0965.
4-(5-Formyl-2-methoxyphenoxy)piperidine-1-carboxylic Acid tert-Butyl Ester (3i)
Yield: 53%. 1H NMR (400 MHz, CDCl3): 1.47 (9H, s), 1.80 (2H, q, J = 3.2, 3.6 Hz), 1.95 (2H, q, J = 4.0, 4.8 Hz), 3.30 (2H, m), 3.80 (2H, s), 3.94 (3H, s), 4.52 (1H, s), 7.01 (1H, d, J = 8.0 Hz), 7.44 (1H, s), 7.49 (1H, d, J = 8.0 Hz), 9.84 (1H, s) ppm. 13C NMR (100 MHz, CDCl3): 28.45, 30.61, 56.17, 74.20, 79.62, 111.26, 114.11, 127.21, 130.04, 147.16, 154.78, 156.13, 190.79 ppm. HRMS (ESI+): calcd for C18H25NO5, 336.1811; found, 336.1814.
tert-Butyl 3-(2-(Difluromethoxy)-5-formylphenoxy)pyrrolidine-1-carboxylate (3j)
Yield: 26%. 1H NMR (400 MHz, CDCl3): 1.46 (9H, s), 2.20 (2H, d, J = 7.6 Hz), 3.65–3.49 (4H, m), 5.01 (1H, s), 6.59 (1H, (d) t, J = 74 Hz), 7.32 (1H, s), 7.45 (1H, s), 7.49 (1H, d, J = 8.4 Hz), 9.26 (1H, s) ppm. 13C NMR (100 MHz, CDCl3): 28.46, 31.53 (d), 44.00 (d), 51.47 (d), 79.72 (d), 113.54 (d), 122.53 (d), 125.64, 154.54, 190.55 ppm. HRMS (ESI+): calcd for C13H13F2NO5 (M+ – tert-butyl), 302.0840; found, 302.0823.
3-Cyclopentyloxy-4-difluoromethoxybenzaldehyde (3k)
A mixture of 4-difluoromethoxy-3-hydroxybenzaldehyde 2a (0.38 g, 2.0 mmol), cyclopentyl bromide (0.45 g, 3.0 mmol), and potassium carbonate (0.41 g, 3.0 mmol) in DMF (10 mL) was stirred overnight at 65 °C. After the mixture was filtered, solvent was removed in vacuo and the residue was purified by flash chromatography to afford 3-cyclopentyloxy-4-difluoromethoxybenzaldehyde (0.18 g, 35%). 1H NMR (CDCl3): 9.93 (1H, s), 7.48 (1H, d, J = 1.6 Hz), 7.43 (1H, dd, J = 1.6 and 8.0 Hz), 7.00 (1H, d, J = 8.0 Hz), 6.65 (1H, t, J = 74.8 Hz), 4.90 (1H, m), 1.98–1.80 (6H, m), 1.67 (2H, m). 13C NMR (CDCl3): 190.9 (d), 150.1 (s), 145.7 (s), 134.5 (s), 124.4 (d), 122.4 (d), 115.7 (triplet, J = 259 Hz, d), 113.3 (d), 80.9 (d), 32.7 (t), 23.9 (t).
General Procedure for the Preparation of 3- or 4-(Pyridine-2-yloxy)benzaldehydes (3l or 3m)
A mixture of 4-difluoromethoxy-3-hydroxybenzaldehyde 2a (2.0 mmol) or 2c, 2-bromopyridine (0.95 g, 6.0 mmol), and potassium carbonate (0.41 g, 3.0 mmol) in DMSO (1 mL) was heated under microwave irradiation for 10 min. After the solvent was removed in vacuo, the residue was purified by flash chromatography to afford the 3-(pyridine-2-yloxy)benzaldehyde 3l or 3m.
4-Difluoromethoxy-3-(pyridine-2-yloxy)benzaldehyde (3l)
Yield: 11%. 1H NMR (CDCl3): 9.94 (1H, s), 8.09 (1H, m), 7.78–7.72 (3H, m), 7.41 (1H, d, J = 8.8 Hz), 7.06–7.02 (2H, m), 6.52 (1H, t, J = 74.0 Hz). 13C NMR (CDCl3): 190.1 (d), 162.6 (s), 148.3 (s), 147.2 (d), 145.5 (s), 139.8 (d), 134.5 (s), 127.7 (d), 124.4 (d), 121.3 (d), 119.2 (d), 118.3 (d), 115.7 (triplet, J = 260 Hz, d), 117.4 (d). MS-EI: 264 (M+).
3-Methoxy-4-(pyridine-2-yloxy)benzaldehyde (3m)
Yield: 85%. 1H NMR (CDCl3): 9.95 (1H, s), 8.13 (1H, m), 7.71 (1H, m), 7.54–7.50 (2H, m), 7.00 (1H, d, J = 8.0 Hz), 7.02–6.99 (2H, m), 3.83 (3H, s). 13C NMR (CDCl3): 191.0 (d), 162.9 (s), 152.3 (s), 148.2 (s), 147.4 (d), 139.4 (d), 134.1 (s), 125.1 (d), 122.9 (d), 118.7 (d), 111.3 (d), 111.1 (d), 55.9 (q). MS-EI: 198 (M+).
General Procedure for the Synthesis of 4 by Nucleophilic Addition to Aldehydes 3
To aldehyde 3 (1 mmol) in THF (5 mL) was added dropwise n-butyllithium or Grignard reagent at −78 °C. The resulting mixture was stirred overnight, allowing the temperature too gradually warm from −78 °C to room temperature, and then quenched with water. The aqueous layer was extracted three times with ethyl acetate. The combined organic solution was dried over MgSO4, filtered, and evaporated. The residue was purified by flash chromatography to afford 4.
4-[4-(1-Hydroxypentyl)-2-methoxyphenoxy]piperidine-1-carboxylic Acid tert-Butyl Ester (4a)
Yield: 42%. 1H NMR (CDCl3): 6.91 (1H, d, J = 2.0 Hz), 6.87 (1H, d, J = 8.0 Hz), 6.81 (1H, dd, J = 2.0 and 8.0 Hz), 4.59 (1H, t, J = 6.8 Hz), 4.37 (1H, m), 3.84 (3H, s), 3.76 (2H, m), 3.23 (2H, m), 2.25 (1H, s, br), 1.89 (2H, m), 1.79–1.73 (4H, m), 1.46 (9H, s), 1.43–1.24 (4H, m), 0.89 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 154.7 (s), 150.9 (s), 145.6 (s), 139.3 (s), 118.1 (d), 117.5 (d), 109.9 (d), 79.4 (s), 74.5 (d), 74.3 (d), 55.8 (q), 40.6 (t), 38.7 (t), 30.7 (t), 28.3 (q), 27.8(t), 22.5 (t), 13.9 (q). MS-EI: 393 (M+).
1-[3-Methoxy-4-(tetrahydropyran-4-yloxy)phenyl]butan-1-ol (4b)
Yield: 70%. 1H NMR (CDCl3): 6.91 (1H, s), 6.87 (1H, d, J = 8.0 Hz), 6.81 (1H, d, J = 7.6 Hz), 4.59 (1H, m), 4.36 (1H, m), 3.98 (1H, m), 3.84 (3H, s), 3.50 (2H, m), 2.32 (1H, s, br), 1.97 (2H, m), 1.82–1.76 (3H, m), 1.64 (1H, m), 1.42 (1H, m), 1.31 (1H, m), 0.93 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 150.9 (s), 145.5 (s), 139.3 (s), 118.0 (d), 117.4 (d), 109.9 (d), 74.0 (d), 73.8 (d), 65.3 (t), 55.8 (q), 41.1 (t), 32.0 (t), 19.0 (t), 13.9 (q). MS-EI: 280 (M+).
1-[3-Methoxy-4-(tetrahydropyran-4-yloxy)phenyl]pentan-1-ol (4c)
Yield: 68%. 1H NMR(CDCl3): 6.92 (1H, s), 6.88 (1H, d, J = 8.0 Hz), 6.82 (1H, d, J = 8.0 Hz), 4.59 (1H, m), 4.38 (1H, m), 4.00 (2H, m), 3.86 (3H, s), 3.53 (2H, m), 1.99 (3H, m), 1.83–1.67 (4H, m), 1.36–1.25 (4H, m), 0.89 (3H, t, J = 6.8 Hz). 13C NMR (CDCl3): 151.0 (s), 145.7 (s), 139.2 (s), 118.1 (d), 117.4 (d), 110.0 (d), 74.4 (d), 73.9 (d), 65.4 (t), 55.9 (q), 38.7 (t), 32.1 (t), 28.1 (t), 22.6 (t), 13.9 (q). MS-EI: 4 (M+).
1-[3-Methoxy-4-(tetrahydrofuran-3-yloxy)phenyl]butan-1-ol (4d)
Yield: 77%. 1H NMR (CDCl3): 6.90 (1H, s), 6.80 (1H, d, J = 8.0 Hz), 6.75 (1H, d, J = 8.0 Hz), 4.90 (1H, m), 4.57 (1H, m), 3.99–3.82 (7H, m), 2.66 (1H, s, br), 2.14 (2H, m), 1.74 (1H, m), 1.63 (1H, m), 1.41 (1H, m), 1.00 (1H, m), 0.92 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 150.6 (s), 145.8 (s), 139.0 (s), 117.9 (d), 115.4 (d), 109.8 (d), 78.7 (d), 73.8 (d), 72.7 (t), 66.9 (t), 55.7 (q), 41.1 (t), 32.8 (t), 18.9 (t), 13.8 (q). MS-EI: 266 (M+).
1-[3-Methoxy-4-(tetrahydrofuran-3-yloxy)phenyl]pentan-1-ol (4e)
Yield: 66%. 1H NMR (CDCl3): 6.90 (1H, s), 6.81 (1H, d, J = 8.0 Hz), 6.76 (1H, d, J = 8.0 Hz), 4.90 (1H, m), 4.56 (1H, m), 4.00–3.83 (7H, m), 2.54 (1H, s, br), 2.14 (2H, m), 1.76 (1H, m), 1.67 (1H, m), 1.35–1.23 (4H, m), 0.88 (3H, t, J = 6.8 Hz). 13C NMR (CDCl3): 150.1 (s), 145.8 (s), 139.0 (s), 118.0 (d), 115.4 (d), 109.8 (d), 78.7 (d), 74.1 (d), 72.8 (t), 67.0 (t), 55.7 (q), 38.7 (t), 32.9 (t), 27.9 (t), 22.4 (t), 13.9 (q). MS-EI: 280 (M+).
1-[3-Methoxy-4-(pyridin-2-yloxy)phenyl]pentan-1-ol (4f)
Yield: 45%. 1H NMR (CDCl3): 8.06 (1H, dd, J = 1.2 and 4.8 Hz), 7.57 (1H, m), 7.03 (1H, d, J = 8.0 Hz), 7.01 (1H, d, J = 2.0 Hz), 6.90–6.85 (2H, m), 6.81 (1H, d, J = 8.0 Hz), 4.60 (1H, m), 3.69 (3H, s), 2.89 (1H, s, br), 1.76 (2H, m), 1.43–1.31 (4H, m), 0.88 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 163.6 (s), 151.5 (s), 147.2 (d), 143.1 (s), 141.7 (s), 138.8 (d), 122.4 (d), 118.3 (d), 117.7 (d), 110.8 (d), 110.3 (d), 73.9 (d), 55.7 (q), 38.6 (t), 27.8 (t), 22.3 (t), 13.7 (q).
1-[3-Methoxy-4-(pyridin-2-yloxy)phenyl]butan-1-ol (4g)
Yield: 80%. 1H NMR (CDCl3): 8.11 (1H, dd, J = 2.0 and 5.2 Hz), 7.63 (1H, m), 7.07 (1H, d, J = 8.0 Hz), 7.03 (1H, s), 6.94–6.87 (3H, m), 4.65 (1H, t, J = 6.0 Hz), 3.75 (3H, s), 2.40 (1H, s, br), 1.78 (1H, m), 1.69 (1H, m), 1.46 (1H, m), 1.36 (1H, m), 0.94 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 163.6 (s), 151.6 (s), 147.4 (d), 143.0 (s), 141.5 (s), 139.1 (d), 122.6 (d), 118.4 (d), 117.9 (d), 110.5 (d), 110.3 (d), 74.0 (d), 55.8 (q), 41.1 (t), 19.0 (t), 13.9 (q). HRMS-ESI (+): calcd for C16H20NO3, 274.1443; found, 274.1431 [M +H]+.
[3-Methoxy-4-(pyridin-2-yloxy)phenyl]quinolin-3-ylmethanol (4h)
Yield: 68%. 1H NMR (CDCl3): 8.75 (1H, d, J = 1.2 Hz), 8.16 (1H, s), 8.03–8.00 (2H, m), 7.74 (1H, d, J = 8.0 Hz), 7.65–7.59 (2H, m), 7.48 (1H, m), 7.09 (1H, d, J = 1.6 Hz), 7.04 (1H, d, J = 8.0 Hz), 6.94–6.84 (3H, m), 5.95 (1H, s), 5.76 (1H, s, br), 3.63 (3H, s). 13C NMR (CDCl3): 163.4, 151.7, 149.9, 147.2, 146.8, 141.8, 141.2, 139.3, 137.0, 133.1, 1.3, 128.5, 127.9, 127.7, 126.7, 122.7, 119.3, 118.1, 111.0, 110.6, 73.4, 55.7. MS-ESI (+): 359.58 [M + H]+.
[3-Methoxy-4-(pyridin-2-yloxy)phenyl]pyridin-3-ylmethanol (4i)
Yellow oil. Yield: 40%. 1H NMR (CDCl3): 8.51 (1H, s), 8.33 (1H, d, J = 3.6 Hz), 8.03 (1H, m), 7.72 (1H, m), 7.62 (1H, m), 7.22 (1H, m), 7.07–7.05 (2H, m), 6.93–6.85 (3H, m), 5.78 (1H, s), 5.26 (1H, s, br), 3.67 (3H, s). 13C NMR (CDCl3): 163.4 (s), 151.7 (s), 147.9 (d), 147.8 (d), 147.1 (d), 141.7 (s), 141.4 (s), 139.9 (s), 139.2 (d), 134.5 (d), 123.4 (d), 122.6 (d), 119.0 (d), 118.0 (d), 110.9 (d), 110.6 (d), 73.1 (d), 55.7 (q).
1-[4-Methoxy-3-(tetrahydropyran-4-yloxy)phenyl]butan-1-ol (4j)
Yellow oil. Yield: 60%. 1H NMR (400 MHz, CDCl3): 0.92 (3H, t, J = 7.6 Hz), 1.44 (3H, m), 1.68 (1H, m), 1.87 (4H, m), 2.01 (2H, m), 3.56–3.51 (2H, dd, J = 2.4, 8.8 Hz), 3.84 (3H, s), 4.03 (2H, m), 4.44 (1H, m), 4.59 (1H, s), 6.86 (1H, d, J = 8.4 Hz), 6.92 (1H, d, J = 8.4 Hz), 6.95(1H, s) ppm. 13C NMR (100 MHz, CDCl3): 13.97, 19.10, 32.12, 41.24, 56.05, 65.38, 73.80, 73.96, 112.10, 115.34, 119.66, 137.75, 146.40, 150.28 ppm. HRMS (ESI+): calcd for C16H22O3-H2O, 263.1647; found, 263.1645.
1-[4-Methoxy-3-(tetrahydropyran-4-yloxy)phenyl]pentan-1-ol (4k)
Yellow oil. Yield: 72%. 1H NMR (400 MHz, CDCl3): 0.88 (3H, t, J = 6.8 Hz), 1.25 (1.32 (3H, m), 1.65 (1H, m), 1.85 (4H, m), 2.01 (2H, s), 3.53 (2H, t, J = 8.8 Hz), 3.84 (3H, s), 4.01 (2H, t, J = 5.6 Hz), 4.43 (1H, d, J = 4.0 Hz), 4.57 (1H, s), 6.86 (1H, d, J = 8.4 Hz), 6.92 (1H, d, J = 8.4 Hz), 6.95 (1H, s) ppm. 13C NMR (100 MHz, CDCl3): 14.03, 22.58, 28.05, 32.10, 38.81, 56.02, 65.34, 73.76, 74.14, 112.09, 115.40, 119.69, 137.83, 146.35, 150.24 ppm. HRMS (ESI+): calcd for C17H27O4 – H2O, 277.1804; found, 277.1795.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]pentan-1-ol (4l)
Yellow oil. Yield: 41%. 1H NMR (400 MHz, CDCl3): 0.88 (3H, t, J = 6.4 Hz), 1.22–1.35 (4H, m), 1.65–1.77 (2H, m), 1.86 (1H, s), 2.18 (2H, d, J = 5.6 Hz), 3.84 (2H, s), 3.89–3.92 (1H, m), 4.00 (3H, s), 4.58 (1H, s), 4.96 (1H, s), 6.84–6.91 (3H, m) ppm. 13C NMR (100 MHz, CDCl3): 14.03, 22.59, 28.03, 33.04, 38.80, 56.05, 67.19, 72.98, 74.26, 78.83, 112.02, 113.65, 119.35, 137.76, 146.83, 149.62 ppm. HRMS (ESI+): calcd for C16H25O4 – H2O, 263.1647; found, 263.1643.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]butan-1-ol (4m)
Yield: 40%. 1H NMR (400 MHz, CDCl3): 0.92 (3H, t, J = 7.2 Hz), 1.30 (1H, dd, J = 6.4, 9.6 Hz), 1.42 (1H, d, J = 7.2 Hz), 1.63 (1H, s), 1.81 (2H, bs), 2.19 (2H, m), 3.84 (2H, s), 3.90 (2H, s), 4.00 (3H, s), 4.60 (1H, s), 4.97 (1H, s), 6.89 (3H, m) ppm. 13C NMR (100 MHz, CDCl3): 13.97, 19.08, 33.07, 41.24, 56.08, 67.22, 73.01, 74.07, 78.88, 112.05, 113.66, 119.33, 137.68, 146.89, 149.70 ppm. HRMS (ESI+): calcd for C15H23O4 – H2O, 249.1491; found, 249.1480.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-4-phenylbutan-1-ol (4n)
Yield: 20%. 1H NMR (400 MHz, CDCl3): 1.60 (1H, m), 1.74 (2H, m), 1.85 (2H, m), 2.18 (2H, m), 2.64 (2H,t, J = 6.4 Hz), 3.83 (3H, s), 3.90 (2H, m), 4.04 (3H, s), 4.60 (1H, m), 4.95 (1H, m), 6.83 (1H, d, J = 2 Hz), 6.84 (1H, s), 6.87–6.89 (1H, dd, J = 1.6, 1.6 Hz), 7.15 (3H, m), 7.25 (2H, d, J = 6.8 Hz) ppm. 13C NMR (100 MHz, CDCl3): 27.61, 33.07, 35.72, 38.51, 56.06, 67.22, 73.02, 74.17, 78.84, 112.01, 113.49, 119.36, 125.77, 128.30, 128.42, 137.39, 142.22, 146.92,149.73 ppm. HRMS (ESI+): calcd for C21H27O4 – H2O, 325.1804; found, 325.1792.
3-Cyclohexyl-1-[4-methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]propan-1-ol (4o)
Yield: 70%. 1H NMR (400 MHz, CDCl3): 0.86 (2H, t, J = 12 Hz), 1.09–1.30 (7H, m), 1.61–1.69 (6H, m), 1.74–1.77 (2H, m), 2.16–2.20 (2H, m), 3.84 (3H, d, J = 2.8 Hz), 3.88–3.91 (1H, m), 4.54 (1H, d, J = 6.8 Hz), 4.96 (1H, s), 6.83–6.85 (2H, dd, J = 2.8,5.2 Hz), 6.70 (1H, d, J = 7.6 Hz) ppm. 13C NMR (100 MHz, CDCl3): 26.36, 26.65, 33.06, 33.35, 33.53, 36.43, 37.64, 56.06, 67.22, 73.01, 74.67, 78.84, 112.00, 113.60, 119.37, 137.67, 146.85, 149.66 ppm. HRMS (ESI+): calcd for C20H31O4 – H2O, 317.2117; found, 317.2112. Anal. Calcd for C20H30O4: C, 71.82; H, 9.04. Found: C, 71.92; H, 9.10.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]--phenylpropan-1-ol (4p)
Yield: 99%. 1H NMR (400 MHz, CDCl3): 1.96–2.00 (1H, m), 2.09 (1H, s), 2.11–2.18 (2H, m), 2.63–2.72 (2H, m), 3.83 (3H, s), 3.82 (3H, m), 3.85–3.90 (1H, m), 3.94–4.03 (3H, m), 4.59 (1H, t, J = 7.2 Hz), 4.93–4.95 (1H, m), 6.83 (1H, s), 6.85 (1H,s), 6.91 (1H, d, J = 8.4 Hz), 7.17 (3H, t, J = 4.4 Hz), 7.25–7.00 (2H, dd, J = 7.2, 7.6 Hz) ppm. 13C NMR (100 MHz, CDCl3): 32.14, 33.07, 40.49, 56.07, 67.22, 73.00, 73.49, 78.86, 112.08, 113.63, 119.41, 125.89, 128.42, 128.44, 137.35, 141.79, 146.92, 149.76 ppm. HRMS (ESI+): calcd for C20H25O4 – H2O, 311.1647; found, 311.1639. Anal. Calcd for C20H24O4, C, 73.15; H, 7.37; found, C, 73.25; H, 7.56.
2-Cyclohexyl-1-[4-methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]ethanol (4q)
Yield: 55%. 1H NMR (400 MHz, CDCl3): 0.94 (2H, m), 1.20 (2H, m), 1.36 (1H, m), 1.48 (1H, m), 1.69 (7H, m), 2.18 (2H, m), 3.84 (3H, s), 3.90 (1H, m), 4.00 (2H, m), 4.71 (1H, s), 4.96 (1H, s), 6.83 (1H, s), 6.85 (1H, s), 6.92–6.89 (1H, q, J = 2.0, 6.0 Hz) ppm. 13C NMR (100 MHz, CDCl3): 26.17, 26., 26.56, 33.03, 33.07, 33.91, 34., 46.97, 56.07, 67.23, 71.79, 73.01, 78.86, 112.06, 113.59, 113.63, 119.27, 138.03, 146.88, 149.68 ppm. HRMS (ESI+): calcd for C19H28O4 – H2O, 303.1960; found, 303.1975.
1-[4-Difluoromethoxy-3-(tetrahydrofuran-3-ylxoy)phenyl]-2,2-dimethylpropan-1-ol (4r)
Yield: 8.6%. 1H NMR (400 MHz, CDCl3): 0.92 (9H, d, J = 0.8 Hz), 1.95 (1H, s), 2.16–2.20 (2H, m), 3.90–3.92 (1H, m), 3.98–4.00 (3H, m), 4.36 (1H, s), 4.96 (1H, d, J = 2.0 Hz), 6.52 (1H, (d) t, J = 75.2 Hz), 6.85–6.87 (1H, dd, J = 2.0, 6.0 Hz), 6.89–6.91 (1H, dd, J = 2.0, 2.4 Hz), 7.11 (1H, d, J = 8.0 Hz) ppm. 13C NMR (100 MHz, CDCl3): 25.88, 0.71, 32.98, 33.02, 35.71, 67.16, 72.80, 72.83, 78.72, 81.73, 114.69 (t, J = 160 Hz), 120.96, 121.93, 140.05, 140.88, 148.17 ppm. MS (EI+): calcd for C16H22F2O4, 316; found, 316.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-3-methyl-butan-1-ol (4s)
Yield: 34%. 1H NMR (400 MHz, CDCl3): 0.91–0.94 (6H, q, J = 4.4, 2.0 Hz), 1.24 (1H, s), 1.44–1.49 (1H, m), 1.64–1.72 (2H, m), 1.92 (1H, bs, O–H), 2.16–2.19 (2H, m), 3.83 (3H, s), 3.86–3.91 (1H, m), 3.97–4.02 (3H, m), 4.64–4.67 (1H, q, J = 5.6, 2.0 Hz), 4.94–4.97 (1H, m), 6.84 (1H, d, J = 8.4 Hz), 6.86 (1H, J = 2 Hz), 6.91–6.89 (1H, dd, J = 2, 6.4 Hz) ppm. 13C NMR (100 MHz, CDCl3): 22.34, 23.09, 24.84, 33.04, 48.28, 56.06, 67.21, 72.41, 72.99, 78.85, 112.04, 113.62, 119.32, 137.96, 146.89, 149.68 ppm. HRMS (ESI+): calcd for C16H22O3 – H2O, 263.1647; found, 263.1648.
3-[2-Difluoromethoxy-5-(1-hydroxypentyl)phenoxy]pyrrolidine-1-carboxylic Acid tert-Butyl Ester (4t)
Yield: 25%. 1H NMR (400 MHz, CDCl3): 0.89 (3H, t, J = 6.8 Hz), 1.36 (4H, m), 1.46 (9H, s), 1.73 (2H, m), 1.95 (1H, d, J = 2.8 Hz), 2.20 (2H, m), 3.67 (4H, m), 4.63 (1H, s), 4.93 (1H, s), 6.65 (1H, t, J = 75.2 Hz), 6.91 (1H, s), 6.96 (1H, d, J = 1.6 Hz), 7.12 (1H, d, J = 6.8 Hz) ppm. 13C NMR (100 MHz, CDCl3): 14.0, 22.55, 27.9, 28.47, 0.72, 30.82, 31.67, 39.02, 44.07, 51.08, 51.57, 74.02, 78.11, 79.61, 113.4, 119.4, 122.89 ppm. HRMS (ESI+): calcd for C21H31F2NO5, 416.2248; found, 416.2247.
3[2-Difluoromethoxy-5-(1-hydroxy-3-methylbutyl)phenoxy]-pyrrolidine-1-carboxylic Acid tert-Butyl Ester (4u)
Yield: 14%. 1H NMR (400 MHz, CDCl3): 0.96 (6H, d, J = 6.4 Hz), 1.46 (9H, s), 1.69 (2H, m), 1.90 (1H, d, J = 2.8 Hz), 2.20 (2H, m), 3.57 (4H, m), 4.71 (1H, s), 4.94 (1H, s), 6.65 (1H, d (t), J = 75.2 Hz), 6.92 (1H, d, J = 6.8 Hz), 6.97 (1H, d, J = 1.2 Hz), 7.12 (1H, d, J = 6.4 Hz) ppm. 13C NMR (100 MHz, CDCl3): 14.1 (d), 22.6, 24.7, 28.4, 31.5, 43.6 (d), 48.5, 51.5 (d), 60.3, 72.1, 79.60, 113.5 (d, J = 14 Hz), 119.4, 122.9, 140.3 ppm. HRMS (ESI+): calcd for C17H21F2NO4 – (H2O + tert-butyl), 342.1515; found, 342.1159.
4-[5-(1-Hydroxypentyl)-2-methoxyphenoxy]piperidine-1-carboxylic Acid tert-Butyl Ester (4v)
Yield: 84%. 1H NMR (400 MHz, CDCl3): 0.95 (3H, t, J = 7.2 Hz), 1.44 (2H, q, J = 7.2, 7.6 Hz), 1.47 (9H, s), 1.69 (4H, m), 1.73 (2H, s), 1.92–1.96 (2H, m), 2.91 (2H, t, J = 7.2 Hz), 3.23–3.00 (2H, m), 3.80 (2H, m), 4.63 (1H, s), 3.91 (3H, s), 4.50–4.51 (1H, m), 4.70–4.72 (1H, s), 6.92 (1H, d, J = 8.4 Hz), 7.57 (1H, d, J = 2 Hz), 7.60–7.63 (1H, dd, J = 2, 6.4 Hz) ppm. 13C NMR (100 MHz, CDCl3): 14.03, 22.59, 28.06, 28.45, 30.83, 38.79, 56.04, 74.28, 74.53, 79.52, 112.13, 115.49, 119.75, 137.72, 146.47, 150.37, 154.84 ppm. HRMS (ESI+): calcd for C18H25NO4 – (H2O + tert-butyl): 320.1862; found, 320.1882.
1-[4-Difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-3-methylbutan-1-ol (4w)
Yield: 15%. 1H NMR (CDCl3): 7.13 (1H, d, J = 8.4 Hz), 6.95 (1H, s), 6.90 (1H, d, J = 8.4 Hz), 6.51 (1H, t, J = 75.2 Hz), 5.00 (1H, m), 4.72 (1H, m), 4.03–3.90 (4H, m), 2.20 (2H, m), 1.94 (1H, s, br), 1.70 (2H, m), 1.47 (1H, m), 0.96 (6H, d, J = 6.0 Hz). 13C NMR (CDCl3): 149.0, 144.3, 140.1, 122.9, 118.9, 116.3 (triplet, J = 258 Hz), 112.8, 78.8, 72.8, 72.2, 67.1, 48.5, 33.0, 24.8, 23.1, 22.2. MS-EI: 316 (M+).
1-[4-Difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-pentan-1-ol (4za)
Yield: %. 1H NMR (CDCl3): 7.13 (1H, d, J = 8.0 Hz), 6.94 (1H, s), 6.90 (1H, dd, J = 2.0 and 8.4 Hz), 6.52 (1H, t, J = 75.2 Hz), 5.00 (1H, m), 4.64 (1H, m), 4.03–3.91 (4H, m), 2.18 (2H, m), 1.98 (1H, s, br), 1.75 (1H, m), 1.67 (1H, m), 1.42–1.24 (4H, m), 0.90 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 149.0 (s), 143.8 (s), 140.1 (s), 122.9 (d), 119.0 (d), 116.3 (triplet, J = 258 Hz, d), 112.8 (d), 78.7 (d), 74.0 (d), 72.8 (t), 67.1 (t), 39.0 (t), 33.0 (t), 27.8 (t), 22.5 (t), 14.0 (q). MS-EI: 316 (M+).
3-Cyclohexyl-1-[4-difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]propan-1-ol (4zb)
Yield: 51%. 1H NMR (CDCl3): 7.11 (1H, d, J = 8.0 Hz), 6.94 (1H, s), 6.89 (1H, d, J = 8.4 Hz), 6.50 (1H, t, J = 74.8 Hz), 4.97 (1H, m), 4.59 (1H, m), 4.01–3.87 (4H, m), 2.18 (2H, m), 1.94 (1H, s, br), 1.80–1.67 (7H, m), 1.34–1.12 (6H, m), 0.88 (2H, m). 13C NMR (CDCl3): 149.1, 143.9, 140.4, 122.8, 119.2, 116.4 (triplet, J = 258 Hz), 113.4, 79.1, 74.4, 72.8, 67.1, 37.7, 36.7, 33.4, 33.3, 33.1, 26.6, 26.3.
2-Cyclohexyl-1-[4-difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]ethanol (4zc)
Yield: 35%. 1H NMR (CDCl3): 7.14 (1H, d, J = 8.4 Hz), 6.95 (1H, d, J = 2.0 Hz), 6.91 (1H, dd, J = 2.0 and 8.0 Hz), 6.53 (1H, t, J = 75.2 Hz), 5.01 (1H, m), 4.77 (1H, m), 4.03–3.91 (4H, m), 2.20 (2H, m), 2.00 (1H, s, br), 1.83–1.66 (6H, m), 1.52–1.39 (2H, m), 1.31–1.15 (3H, m), 0.98 (2H, m). 13C NMR (CDCl3): 149.0 (s), 144.3 (s), 140.0 (s), 122.9 (d), 118.91 (d), 118.89 (d), 116.3 (triplet, J = 258 Hz, d), 112.8 (d), 78.7 (d), 72.8 (t), 71.5 (d), 67.1 (t), 47.2 (t), 34.2 (d), 33.9 (t), 33.0 (t), 32.8 (t), 26.5 (t), 26.2 (t), 26.1 (t).
1-[4-Difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-3-phenylpropan-1-ol (4zd)
Yield: 46%. 1H NMR (CDCl3): 7.30–7.25 (2H, m), 7.20–7.11 (4H, m), 6.92–6.88 (2H, m), 6.50 (1H, t, J = 74.8 Hz), 4.96 (1H, m), 4.64 (1H, m), 3.99–3.89 (4H, m), 2.72 (2H, m), 2.18–1.97 (5H, m). 13C NMR (CDCl3): 149.0 (s), 143.5 (s), 141.4 (s), 140.1 (s), 128.42 (d), 128.36 (d), 126.0 (d), 122.9 (d), 119.0 (d), 116.2 (triplet, J = 258 Hz, d), 112.81 (d), 112.78 (d), 78.7 (d), 73.2 (d), 72.7 (t), 67.1 (t), 40.6 (t), 32.9 (t), 31.9 (t).
1-[4-Difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-4-phenylbutan-1-ol (4ze)
Yield: 56%. 1H NMR (CDCl3): 7.28–7.25 (2H, m), 7.19–7.10 (4H, m), 6.89–6.85 (2H, m), 6.50 (1H, t, J = 75.2 Hz), 4.95 (1H, m), 4.64 (1H, m), 3.99–3.89 (4H, m), 2.63 (2H, t, J = 6.8 Hz), 2.15 (2H, m), 2.01 (1H, s, br), 1.78–1.61 (4H, m). 13C NMR (CDCl3): 149.0 (s), 143.6 (s), 142.0 (s), 140.1 (s), 128.37 (d), 128.31 (d), 125.8(d), 122.9 (d), 119.0 (d), 116.2 (triplet, J = 258 Hz, d), 112.7 (d), 78.7 (d), 73.8 (d), 72.8 (t), 67.1 (t), 38.6 (t), 35.6 (t), 33.0 (t), 27.4 (t).
1-(3-Cyclopentyloxy-4-difluoromethoxyphenyl)-3-methyl-butan-1-ol (4zf)
Yield: 34%. 1H NMR (CDCl3): 7.09 (1H, d, J = 8.4 Hz), 6.97 (1H, d, J = 1.6 Hz), 6.83 (1H, dd, J = 1.6 and 8.4 Hz), 6.51 (1H, t, J = 75.6 Hz), 4.82 (1H, m), 4.70 (1H, m), 1.92–1.79 (7H, m), 1.72–1.62 (4H, m), 1.46 (1H, m), 0.95 (6H, d, J = 6.0 Hz). 13C NMR (CDCl3): 149.6 (s), 144.0 (s), 140.0 (triplet, J = 3.0 Hz, s), 122.7 (d), 117.9 (d), 116.4 (triplet, J = 257 Hz, d), 112.6 (d), 80.5 (d), 72.4 (d), 48.4 (t), 32.74 (t), 32.73 (t), 24.8 (d), 23.8 (t), 23.1 (q), 22.2 (q).
4-[2-Difluoromethoxy-5-(1-hydroxypentyl)phenoxy]piperidine-1-carboxylic Acid tert-Butyl Ester (4zg)
Yield: 57%. 1H NMR (CDCl3): 7.12 (1H, d, J = 8.0 Hz), 7.02 (1H, d, J = 1.2 Hz), 6.90 (1H, dd, J = 1.6 and 8.0 Hz), 6.51 (1H, t, J = 74.8 Hz), 4.63 (1H, m), 4.52 (1H, m), 3.67 (2H, m), 3.41 (2H, m), 2.11 (1H, s, br), 1.90 (2H, m), 1.80–1.63 (4H, m), 1.47 (9H, s), 1.42–1.28 (4H, m), 0.88 (3H, t, J = 7.2 Hz).
1-[4-Difluoromethoxy-3-(tetrahydropyran-4-yloxy)phenyl]-butan-1-ol (4zh)
Yield: 35%. 1H NMR (CDCl3): 7.13 (1H, d, J = 8.4 Hz), 7.02 (1H, s), 6.90 (1H, d, J = 8.0 Hz), 6.54 (1H, t, J = 75.2 Hz), 4.64 (1H, m), 4.52 (1H, m), 3.97 (2H, m), 3.56 (2H, m), 2.02 (3H, m), 1.82–1.61 (4H, m), 1.44 (1H, m), 1.33 (1H, m), 0.94 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 148.8 (s), 143.9 (s), 140.7 (s), 122.8 (d), 119.2 (d), 116.4 (triplet, J = 258 Hz, d), 114.2 (d), 73.8 (d), 73.3 (d), 64.8 (t), 41.4 (t), 31.7 (t), 18.9 (t), 13.9 (q). MS-EI: 316 (M+).
1-[4-Difluoromethoxy-3-(pyridin-2-yloxy)phenyl]>butan-1-ol (4zi)
Yield: 52%. 1H NMR (CDCl3): 8.07 (1H, m), 7.70 (1H, m), 7.26–7.15 (3H, m), 7.00–6.96 (2H, m), 6.40 (1H, t, J = 74.4 Hz), 4.64 (1H, m), 2.33 (1H, s, br), 1.70–1.60 (2H, m), 1.48–1.26 (2H, m), 0.92 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 163.0 (s), 147.3 (d), 145.0 (s), 144.1 (s), 142.4 (s), 139.6 (d), 123.2 (d), 121.9 (d), 121.3 (d), 118.7 (d), 116.3 (triplet, J = 258 Hz, d), 111.2 (d), 73.3 (d), 41.1 (t), 18.9 (t), 13.9 (q).
1-(3,4-Bisdifluoromethoxyphenyl]-4-phenylbutan-1-ol (4zj)
Yield: 43%. 1H NMR (CDCl3): 7.00–7.14 (8H, m), 6.52 (1H, t, J = 73.6 Hz), 6.50 (1H, t, J = 73.6 Hz), 4.68 (1H, m), 2.64 (2H, t, J = 7.2 Hz), 2.11 (1H, s, br), 1.92–1.58 (4H, m).
1-(3,4-Bisdifluoromethoxyphenyl]pentan-1-ol (4zk)
Yield: 25%. 1H NMR (CDCl3): 7.24–7.18 (3H, m), 6.53 (1H, t, J = 73.6 Hz), 6.51 (1H, t, J = 73.6 Hz), 4.67 (1H, m), 1.92 (1H, s, br), 1.77–1.67 (2H, m), 1.40–1.23 (4H, m), 0.89 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 144.1 (s), 142.3 (s), 141.2 (s), 123.9 (d), 122.3 (d), 119.8 (d), 115.8 (triplet, J = 261 Hz, d), 73.5 (d), 38.9 (t), 27.8 (t), 22.5 (t), 13.9 (q).
1-(3,4-Bisdifluoromethoxyphenyl]butan-1-ol (4zl)
Yield: 80%. 1H NMR (CDCl3): 7.24–7.17 (3H, m), 6.53 (1H, t, J = 73.6 Hz), 6.51 (1H, t, J = 73.6 Hz), 4.66 (1H, t, J = 6.4 Hz), 2.19 (1H, s, br), 1.74–1.60 (2H, m), 1.43–1.30 (2H, m), 0.93 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 144.1 (s), 142.3 (s), 141.2 (s), 123.9 (d), 122.2 (d), 119.7 (d), 115.8 (triplet, J = 260 Hz, d), 73.2 (d), 41.3 (t), 18.8 (t), 13.7 (q). MS-EI: 282 (M+).
1-(3,4-Bisdifluoromethoxyphenyl]-3-phenylpropan-1-ol (4zm)
Yield: 73%. 1H NMR (CDCl3): 7.30–7.17 (8H, m), 6.51 (1H, t, J = 74.0 Hz), 6.50 (1H, t, J = 73.6 Hz), 4.67 (1H, m), 2.71 (2H, m), 2.18–1.98 (3H, m). 13C NMR (CDCl3): 143.8, 142.3, 141.2, 128.9, 128.5, 128.4, 126.0, 123.9, 122.3, 119.8, 115.8 (triplet, J = 260 Hz), 72.6, 40.5, 31.8.
1-(3,4-Bisdifluoromethoxyphenyl]-2-cyclohexylethanol (4zn)
1H NMR (CDCl3): 7.24–7.17 (3H, m), 6.53 (1H, triplet, J = 74.0 Hz), 6.51 (1H, triplet, J = 74.0 Hz), 4.76 (1H, m), 2.01 (1H, s, br), 1.82–1.62 (6H, m), 1.44 (2H, m), 1.26–1.14 (3H, m), 0.94 (2H, m). 13C NMR (CDCl3): 144.6, 142.3, 141.2, 123.8, 122.3, 119.7, 115.8 (triplet, J = 260 Hz), 71.0, 47.2, 34.1, 33.9, 32.7, 26.5, 26.2, 26.1.
General Procedure for the Synthesis of 5 by Oxidization of the Alcohols with PCC
To a solution of the alcohol 4 (1 mmol) in dichloromethane (5 mL) was added PCC in portions. The resulting mixture was stirred at room temperature for 2 h. The solvent was removed in vacuo and the residue was purified by flash chromatography to afford 5.
4-(2-Methoxy-4-pentanoylphenoxy)piperidine-1-carboxylic Acid tert-Butyl Ester (5a)
Yellow solid, mp 41–43 °C. Yield: 18%. 1H NMR (CDCl3): 7.55–7.53 (2H, m), 6.92 (1H, d, J = 8.8 Hz), 4.56 (1H, m), 3.90 (3H, s), 3.78 (2H, m), 3.30 (2H, m), 2.92 (2H, t, J = 7.2 Hz), 1.95 (2H, m), 1.81 (2H, m), 1.70 (2H, m), 1.47 (9H, s), 1.40 (2H, m), 0.95 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 199.2 (s), 154.7 (s), 150.8 (s), 150.4 (s), 130.9 (s), 122.2 (d), 114.4 (d), 111.2 (d), 79.6 (s), 73.9 (d), 56.0 (q), 40.6 (t), 37.8 (t), 30.5 (t), 28.4 (q), 26.8(t), 22.5 (t), 14.0 (q). MS-EI: 391 (M+).
4-(4-Butyryl-2-methoxyphenoxy)piperidine-1-carboxylic Acid tert-Butyl Ester (5b)
Yield: 40%. 1H NMR (CDCl3): 7.55–7.53 (2H, m), 6.93 (1H, d, J = 8.8 Hz), 4.56 (1H, m), 3.90 (3H, s), 3.77 (2H, m), 3.30 (2H, m), 2.92 (2H, t, J = 7.6 Hz), 1.94 (2H, m), 1.85–1.74 (4H, m), 1.47 (9H, s), 1.00 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 198.8 (s), 154.6 (s), 150.7 (s), 150.3 (s), 130.9 (s), 122.2 (d), 114.4 (d), 111.1 (d), 79.5 (s), 73.8 (d), 55.9 (q), 40.6 (t), 39.9 (t), 30.4 (t), 28.3 (q), 17.9 (t), 13.8 (q). MS-EI: 377 (M+).
1-[3-Methoxy-4-(tetrahydropyran-4-yloxy)phenyl]butan-1-one (5c)
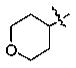
Yellow solid, mp 60–62 °C. Yield: 93%. 1H NMR (CDCl3): 7.55–7.53 (2H, m), 6.93 (1H, d, J = 8.0 Hz), 4.57 (1H, m), 4.02 (2H, m), 3.92 (3H, s), 3.58 (2H, m), 2.90 (2H, t, J = 7.2 Hz), 2.04 (2H, m), 1.88 (2H, m), 1.77 (2H, m), 1.00 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 199.0 (s), 150.7 (s), 150.3 (s), 130.8 (s), 122.2 (d), 114.2 (d), 111.2 (d), 73.2 (d), 65.1 (t), 56.0 (q), 40.0 (t), 31.9 (t), 18.0 (t), 13.9 (q). MS-EI: 278 (M+).
1-[3-Methoxy-4-(tetrahydropyran-4-yloxy)phenyl]pentan-1-one (5d)
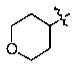
Yield: 99%. 1H NMR (CDCl3): 7.55–7.54 (2H, m), 6.93 (1H, d, J = 8.0 Hz), 4.58 (1H, m), 4.02 (2H, m), 3.91 (3H, s), 3.57 (2H, m), 2.92 (2H, t, J = 7.2 Hz), 2.05 (2H, m), 1.87 (2H, m), 1.71 (2H, m), 1.42 (2H, m), 0.95 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 199.0 (s), 150.7 (s), 150.2 (s), 130.8 (s), 122.2 (d), 114.2 (d), 111.1 (d), 73.2 (d), 65.1 (t), 55.9 (q), 37.8 (t), 31.7 (t), 26.7 (t), 22.4 (t), 13.8 (q). MS-EI: 2 (M+).
1-[3-Methoxy-4-(tetrahydrofuran-3-yloxy)phenyl]butan-1-one (5e)
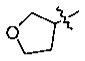
Yield: 75%. 1H NMR (CDCl3): 7.56–7.54 (2H, m), 6.83 (1H, d, J = 8.0 Hz), 5.02 (1H, m), 4.03–3.90 (7H, m), 2.90 (2H, t, J = 7.2 Hz), 2.22 (2H, m), 1.76 (2H, m), 1.00 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 198.7 (s), 150.9 (s), 149.6 (s), 130.6 (s), 122.1 (d), 112.9 (d), 110.7 (d), 78.4 (d), 72.6 (t), 66.9 (t), 55.7 (q), 39.8 (t), 32.8 (t), 17.8 (t), 13.7 (q). MS-EI: 264 (M+).
1-[3-Methoxy-4-(tetrahydrofuran-3-yloxy)phenyl]pentan-1-one (5f)
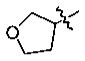
Yield: 55%. 1H NMR (CDCl3): 7.56–7.54 (2H, m), 6.83 (1H, d, J = 8.8 Hz), 5.02 (1H, m), 4.03–3.90 (7H, m), 2.92 (2H, t, J = 7.2 Hz), 2.22 (2H, m), 1.71 (2H, m), 1.41 (2H, m), 0.95 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 199.1 (s), 151.0 (s), 149.8 (s), 130.7 (s), 122.2 (d), 113.0 (d), 111.0 (d), 78.5 (d), 72.8 (t), 67.1 (t), 55.9 (q), 37.8 (t), 33.0 (t), 26.7 (t), 22.4 (t), 13.8 (q). MS-EI: 278 (M+).
1-[3-Methoxy-4-(pyridine-2-yloxy)phenyl]pentan-1-one (5g)
Yield: 60%. 1H NMR (CDCl3): 8.13 (1H, dd, J = 2.0 and 5.2 Hz), 7.70 (1H, m), 7.66 (1H, d, J = 2.0 Hz), 7.61 (1H, dd, J = 2.0 and 8.0 Hz), 7.20 (1H, d, J = 8.0 Hz), 7.00–6.98 (2H, m), 3.82 (3H, s), 2.96 (2H, t, J = 7.2 Hz), 1.77 (2H, m), 1.42 (2H, m), 0.96 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 199.3 (s), 163.1 (s), 151.8 (s), 147.4 (d), 146.7 (s), 139.4 (d), 134.7 (s), 122.4 (d), 121.8 (d), 118.5 (d), 111.8 (d), 111.1 (d), 55.9 (q), 38.1 (t), 26.5 (t), 22.4 (t), 13.9 (q). MS-ESI (+): 286.24 [M + H]+.
1-[3-Methoxy-4-(pyridine-2-yloxy)phenyl]butan-1-one (5h)
Yield: 88%. 1H NMR (CDCl3): 8.13 (1H, dd, J = 2.0 and 5.2 Hz), 7.70 (1H, m), 7.66 (1H, d, J = 2.0 Hz), 7.61 (1H, dd, J = 2.0 and 8.0 Hz), 7.20 (1H, d, J = 8.0 Hz), 7.00–6.98 (2H, m), 3.82 (3H, s), 2.94 (2H, t, J = 7.2 Hz), 1.78 (2H, m), 1.01 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 199.1 (s), 163.1 (s), 151.8 (s), 147.4 (d), 146.7 (s), 139.4 (d), 134.7 (s), 122.4 (d), 121.8 (d), 118.5 (d), 111.8 (d), 111.1 (d), 55.9 (q), 40.3 (t), 17.8 (t), 13.9 (q). HRMS-ESI (+): calcd for C16H18NO3, 272.1287; found, 272.1282 (M+).
1-[4-Methoxy-3-(piperidin-4-yloxy)phenyl]pentan-1-one (5i)
Yellow oil. Yield: 96%. 1H NMR (400 MHz, CDCl3): 0.93 (3H, t, J = 7.2 Hz), 1.36–1.43 (2H, m), 2.15 (4H, d, J = 3.6 Hz), 2.91 (2H, t, J = 7.6 Hz), 3.33 (2H, d, J = 12 Hz), 3.53 (2H, s), 3.91 (3H, s), 4.66 (1H, t, J = 3.2 Hz), 6.95 (1H, d, J = 8.4 Hz), 7.56 (1H, d, J = 2 Hz), 7.66–7.69 (1H, dd, J = 2, 6.4 Hz) ppm. 13C NMR (100 MHz, CDCl3): 13.82, 22.43, 26.97 (d), 37.96, 40.48, 55.91, 70.00, 111.28, 113.98, 116.83, 117.24, 125.08, 1.99, 145.59, 155.60, 161.00 (d), 200.65 ppm. HRMS (ESI+): calcd for C17H26NO3, 2.1913; found, 2.1900.
4-Methoxy-3-(tetrahydropyran-4-yloxyphenyl]quinolin-3-ylmethanone (5j)
White solid, mp 72–74 °C. Yield: 88%. 1H NMR (400 MHz, CDCl3): 1.85–1.92 (2H, m), 2.04–2.10 (2H, m), 3.53–3.59 (2H, m), 3.97 (3H, s), 4.02 (2H, t, J = 6.0 Hz), 4.00–4.05 (2H, m), 4.56 (1H, m), 6.97 (2H, d, J = 8.4 Hz), 7.45–7.47 (1H, dd, J = 2.0, 6.4 Hz), 7.58 (1H, d, J = 1.6 Hz), 7.65 (1H, t, J = 7.2 Hz), 7.86 (1H, t, J = 6.8 Hz), 8.21 (1H, d, J = 8.4 Hz), 8.54 (1H, d, J = 2.0 Hz), 9.27 (1H, d, J = 2.0 Hz) ppm. 13C NMR (100 MHz, CDCl3): 0.72, 31.95, 56.19, 65.34, 73.75, 110.89, 117.15, 126.71, 127.60, 1.02, 1.51, 131.65, 138.27, 146.75, 150.27 ppm. MS (ESI+): calcd for C22H21NO4, 363.1; found, 363.
1-[4-Methoxy-3-(tetrahydropyran-4-yloxy)phenyl]butan-1-one (5k)
Yellow oil. Yield: 63%. 1H NMR (400 MHz, CDCl3): 1.02 (3H, t, J = 7.2 Hz), 1.78–1.73 (2H, q, J = 7.6, 6.4 Hz), 1.85 (2H, m), 2.05 (2H, d, J = 10.8 Hz), 2.89 (2H, t, J = 7.2 Hz), 3.55 (2H, t, J = 10 Hz), 3.92 (3H, s), 4.02 (2H, t, J = 6.0 Hz), 4.51 (1H, s), 6.91 (1H, d, J = 8.0 Hz), 7.57 (1H, s), 7.62 (1H, d, J = 8.4 Hz) ppm. 13C NMR (100 MHz, CDCl3): 13.95, 18.10, 32.00, 40.08, 56.07, 65.41, 73.70, 110.87, 115.63, 123.46, 130.33, 146.44, 154.84, 199.14 ppm. HRMS (ESI+): calcd for C16H22O4, 279.1596; found, 279.1606.
1-[4-Methoxy-3-(tetrahydropyran-4-yloxy)phenyl]pentan-1-one (5l)
Yield: 75%. 1H NMR (400 MHz, CDCl3): 0.95 (3H, t, J = 7.2 Hz), 1.41 (2H, q, J = 7.2, 7.2 Hz), 1.72 (2H, q, J = 7.2, 7.2 Hz), 1.84 (2H, t, J = 4.4 Hz), 2.05 (2H, d, J = 10.0 Hz), 2.90 (2H, t, J = 7.2 Hz), 3.55 (2H, t, J = 9.2 Hz), 3.92 (3H, s), 4.03 (2H, d, J = 11.2 Hz), 4.52 (1H, s), 6.91 (1H, d, J = 8.0 Hz), 7.57 (1H, s), 7.62 (1H, d, J = 8.4 Hz) ppm. 13C NMR (100 MHz, CDCl3): 13.95, 22.52, 26.83, 31.99, 37.86, 56.04, 65.37, 73.68, 110.87, 115.67, 123.48, 130.27, 146.42, 154.83, 199.14 ppm. HRMS (ESI+): calcd for C17H25O4, 3.1753; found, 3.1754. Anal. Calcd for C17H24O4 · 1/8H2O: C, 69.24; H, 8.23. Found: C, 69.27; H, 8.61.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]pentan-1-one (5m)
Yield: 92%. 1H NMR (400 MHz, CDCl3): 0.95 (3H, t, J = 7.2 Hz), 1.43 (2H, q, J = 6.8, 7.2 Hz), 1.72 (2H, t, J = 6.8 Hz), 2.24 (2H, m), 2.91 (2H, t, J = 6.8 Hz), 3.92 (4H, s), 4.03 (3H, s), 5.03 (1H, s), 6.90 (1H, d, J = 7.6 Hz), 7.48 (1H, s), 7.61 (1H, d, J = 8 Hz) ppm. 13C NMR (100 MHz, CDCl3): 13.96, 22.54, 26.84, 33.01, 37.86, 56.07, 67.23, 72.97, 78.74, 110.72, 113.90, 123.30, 130.22, 146.90, 154.24, 199.10 ppm. HRMS (ESI+): calcd for C16H23O4, 279.1596; found, 279.1598.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]butan-1-one (5n)
Yield: 64%. 1H NMR (400 MHz, CDCl3): 1.00 (3H, t, J = 7.2 Hz), 1.75 (2H, m), 2.24 (2H, m), 2.91 (2H, t, J = 7.2 Hz), 3.91 (4H, s), 4.06 (3H, s), 5.02 (1H, s), 6.91 (1H, d, J = 8.4 Hz), 7.48 (1H, s), 7.61 (1H, d, J = 8.4 Hz) ppm. 13C NMR (100 MHz, CDCl3): 13.98, 18.12, 33.03 (d), 40.08, 56.07, 67.25 (d), 72.99 (d), 78.77, 110.76 (d), 113.92, 123.31, 130.27 (d), 146.92, 154.28 (d), 198.95 ppm. HRMS (ESI+): calcd for C15H21O4, 265.1440; found, 265.1436.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-4-phenyl-butan-1-one (5o)
Yield: 80%. 1H NMR (400 MHz, CDCl3): 2.11(2H, d, J = 7.6 Hz), 2.24 (2H, d, J = 5.2 Hz), 2.76 (2H, t, J = 7.6 Hz), 2.96 (2H, t, J = 7.6 Hz), 3.94 (4H, s), 4.06 (3H, s), 6.92 (1H, d, J = 8.4 Hz), 7.26 (3H, d, J = 7.2 Hz), 7.30–7.35 (2H, q, J = 7.6, 4.8 Hz), 7.50 (1H, s), 7.60 (1H, d, J = 8 Hz) ppm. 13C NMR (100 MHz, CDCl3): 26.08, 33.00, 35.27, 37.22, 56.08, 67.24, 72.97, 78.71, 110.72, 113.77, 123.27, 125.95, 128.40, 128.54, 130.11, 141.75, 146.90, 154.30, 198.63 ppm. HRMS (ESI+): calcd for C21H25O4, 341.1753; found, 341.1748.
3-Cyclohexyl-1-[4-methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]propan-1-one (5p)
Yield: 95%. 1H NMR (400 MHz, CDCl3): 0.88–0.97 (2H, q, J = 11.6, 10 Hz), 1.31 (4H, m), 1.76 (7H, m), 2.20 (2H, m), 2.90 (2H, t, J = 7.6 Hz), 3.90 (4H, s), 4.02 (3H, s), 5.01 (1H, s), 6.89 (1H, d, J = 8.4 Hz), 7.47 (1H, s), 7.60 (1H, d, J = 7.6 Hz) ppm. 13C NMR (100 MHz, CDCl3): 26., 26.58, 32.18, 33.01, 33.23, 35.69, 37.48, 56.07, 67.24, 72.98, 78.72, 110.71, 113.89, 123., 130.16, 146.89, 154.22, 199.38 ppm. HRMS (ESI+): calcd for C20HO4, 333.2066; found, 333.2067. Anal. Calcd for C20H28O4: C, 72.26; H, 8.49. Found: C, 71.95; H, 8.76.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-3-phenyl-propan-1-one (5q)
Yield: 98%. 1H NMR (400 MHz, CDCl3): 2.21 (2H, s), 3.06 (2H, s), 3.25 (2H, s), 3.91 (4H, s), 4.02 (3H, s), 5.01 (1H, s), 6.89 (1H, d, J = 7.6 Hz), 7.21–7.30 (5H, m), 7.48 (1H, s), 7.60 (1H, d, J = 6.8 Hz) ppm. 13C NMR (100 MHz, CDCl3): 30.42, 33.00, 39.99, 56.08, 67.24, 72.96, 78.73, 110.74, 113.80, 123.27, 126.14, 128.45, 128.54, 1.98, 141.41, 146.93, 154.40, 197.72 ppm. HRMS (ESI+): calcd for C20H23O4, 327.1596; found, 327.1609. Anal. Calcd for C20H22O4: C, 73.60; H, 6.79. Found: C, 73.57; H, 7.09.
2-Cyclohexyl-1-[4-methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]ethanone (5r)
Yellow oil. Yield: 78%. 1H NMR (400 MHz, CDCl3): 1.02 (2H, m), 1. (3H, m), 1.76 (5H, m), 1.96 (1H, m), 2.22 (2H, m), 2.76 (2H, d, J = 6.8 Hz), 3.91 (1H, s), 4.03 (3H, s), 5.02 (1H, s), 6.90 (1H, d, J = 8.4 Hz), 7.47 (1H, d, J = 2.0 Hz), 7.60 (1H, q, J = 2.0, 6.4 Hz) ppm. 13C NMR (100 MHz, CDCl3): 26.18, 26.27, 33.01, 33.50, 34.96, 45.73, 56.08, 67.24, 72.99, 78.72, 110.68, 113.89, 123.45, 130.62, 146.88, 154.24, 198.76 ppm. HRMS (ESI+): calcd for C19H26O4, 319.1909; found, 319.1911
1-[4-Difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-2,2-dimethylpropan-1-one (5s)
Yellow oil. Yield: 81%. 1H NMR (400 MHz, CDCl3): 1.36 (9H, s), 2.19–2.25 (2H, m), 3.91–3.95 (1H, m), 3.99–4.03 (3H, m), 5.00 (1H, s), 6.59 (1H, t, J = 74.8 Hz), 7.18 (1H, d, J = 8.4 Hz), 7.30 (1H, d, J = 2.0 Hz), 7.40–7.42 (1H, dd, J = 2.0, 1.6 Hz) ppm. 13C NMR (100 MHz, CDCl3): 28.15, 28.71, 32.96, 44.22, 67.16, 72.81, 78.94, 115.84 (d, J = 36 Hz), 121.77 (d), 136.04, 148.72, 206.85 ppm. MS (EI+): calcd for C16H20F2O4, 314; found, 314.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-3-methyl-butan-1-one (5t)
Yield: 93%. 1H NMR (400 MHz, CDCl3): 0.99 (6H, d, J = 6.4 Hz), 2.19–2.28 (3H, m), 2.77 (3H, d, J = 6.8 Hz), 3.90 (4H, d, J = 4.8 Hz), 4.02 (3H, t, J = 3.6 Hz), 5.02 (1H, d, J = 3.2 Hz), 6.89 (1H, d, J = 8.4 Hz), 7.47 (1H, d, J = 1.6 Hz), 7.57–7.59 (1H, dd, J = 2.0, 1.6 Hz) ppm. 13C NMR (100 MHz, CDCl3): 22.82, 25.48, 0.70, 33.00, 47.01, 56.05, 67.23, 72.97, 78.72, 110.67, 113.87, 123.39, 130.54, 146.90, 154.24, 198.73 ppm. MS (EI+): calcd for C16H22O4, 278; found, 278.
1-[4-Methoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-4-methyl-pentan-1-one (5u)
Yield: 93%. 1H NMR (400 MHz, CDCl3): 0.95 (6H, d, J = 6.0 Hz), 1.59–1.64 (3H, m), 2.19–2.25 (2H, m), 2.90 (2H, t, J = 7.6 Hz), 3.88–3.94 (4H, m), 4.02–4.06 (3H, m), 5.01–5.03 (1H, q, J = 2.4, 2.8 Hz), 6.91 (1H, d, J = 8.4 Hz), 7.48 (1H, d, J = 1.6 Hz), 7.59–7.62 (1H, dd, J = 6.8, 1.6 Hz) ppm. 13C NMR (100 MHz, CDCl3): 22.46, 27.90, 29.70, 33.00, 33.64, 36.16, 56.05, 67.22, 72.97, 78.72, 110.71, 113.88, 123., 130.15, 146.90, 154.23, 199.28 ppm. MS (EI+): calcd for C17H24O4, 292; found, 292.
1-[4-Difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-3-methylbutan-1-one (5v)
Yield: 65%. 1H NMR (CDCl3): 7.56–7.53 (2H, m), 7.23 (1H, d, J = 8.8 Hz), 6.62 (1H, t, J = 74.4 Hz), 5.06 (1H, m), 4.06–3.93 (4H, m), 2.80 (2H, d, J = 6.8 Hz), 2.32–2.18 (3H, m), 1.01 (6H, d, J = 6.8 Hz). 13C NMR (CDCl3): 198.6 (s), 149.0 (s), 144.5 (s), 135.5 (s), 122.2 (d), 122.0 (d), 115.7 (triplet, J = 259 Hz, d), 114.0 (d), 78.9 (d), 72.8 (t), 67.1 (t), 47.3 (t), 32.9 (t), 25.2 (s), 22.7 (q). MS-EI: 314 (M+).
1-[4-Difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-3-pentan-1-one (5za)
Yield: 43%. 1H NMR (CDCl3): 7.57–7.55 (2H, m), 7.23 (1H, d, J = 8.0 Hz), 6.62 (1H, t, J = 74.4 Hz), 5.06 (1H, m), 4.03–3.92 (4H, m), 2.93 (2H, t, J = 7.2 Hz), 2.32–2.15 (2H, m), 1.71 (2H, m), 1.41 (2H, m), 0.96 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 199.0 (s), 149.0 (s), 144.5 (s), 135.2 (s), 122.08 (d), 122.05 (d), 115.7 (triplet, J = 260 Hz, d), 114.0 (d), 78.9 (d), 72.7 (t), 67.1 (t), 38.2 (t), 32.9 (t), 26.5 (t), 22.4 (t), 13.9 (q). MS-EI: 314 (M+).
Cyclohexyl-1-[4-difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]propan-1-one (5zb)
Yield: 83%. 1H NMR (CDCl3): 7.57–7.54 (2H, m), 7.23 (1H, d, J = 8.0 Hz), 6.62 (1H, t, J = 74.4 Hz), 5.06 (1H, m), 4.03–3.92 (4H, m), 2.94 (2H, t, J = 7.6 Hz), 2.28–2.18 (2H, m), 1.76–1.59 (7H, m), 1.31–1.16 (4H, m), 0.97 (2H, m). 13C NMR (CDCl3): 199.3 (s), 149.0 (s), 144.5 (s), 135.2 (s), 122.1 (d), 115.7 (triplet, J = 258 Hz, d), 114.0 (d), 78.9 (d), 72.7 (t), 67.1 (t), 37.4 (d), 36.0 (t), 33.2 (t), 32.9 (t), 31.8 (t), 26.5 (t), 26.3 (t). MS-EI: 368 (M+).
2-Cyclohexyl-1-[4-difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]ethanone (5zc)
Yield: 89%. 1H NMR (CDCl3): 7.56–7.53 (2H, m), 7.23 (1H, d, J = 8.8 Hz), 6.62 (1H, t, J = 74.4 Hz), 5.06 (1H, m), 4.06–3.90 (4H, m), 2.79 (2H, d, J = 6.8 Hz), 2.30–2.18 (2H, m), 1.96 (1H, m), 1.77–1.65 (5H, m), 1.31–1.15 (4H, m), 1.03 (2H, m). 13C NMR (CDCl3): 198.7 (s), 149.0 (s), 144.5 (s), 135.6 (s), 122.2 (d), 122.0 (d), 115.7 (triplet, J = 260 Hz, d), 114.0 (d), 78.9 (d), 72.8 (t), 67.1 (t), 46.0 (t), 34.7 (d), 33.4 (t), 32.9 (t), 26.2 (t), 26.1 (t). MS-EI: 354 (M+).
1-[4-Difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-3-phenylpropan-1-one (5zd)
Yellow oil. Yield: 88%. 1H NMR (CDCl3): 7.55–7.52 (2H, m), 7.31–7.19 (6H, m), 6.61 (1H, t, J = 74.4 Hz), 5.03 (1H, m), 4.03–3.89 (4H, m), 3.27 (2H, t, J = 7.6 Hz), 3.05 (2H, t, J = 7.6 Hz), 2.27–2.14 (2H, m). 13C NMR (CDCl3): 197.6 (s), 149.0 (s), 144.6 (s), 141.0 (s), 134.9 (s), 128.51 (d), 128.35 (d), 126.2 (d), 122.00 (d), 121.99 (d), 115.7 (triplet, J = 260 Hz, d), 113.9 (d), 78.8 (d), 72.7 (t), 67.1 (t), 40.2 (t), 32.9 (t), 30.1 (t). MS-EI: 362 (M+).
1-[4-Difluoromethoxy-3-(tetrahydrofuran-3-yloxy)phenyl]-4-phenylbutan-1-one (5ze)
Yellow oil. Yield: 91%. 1H NMR (CDCl3): 7.51–7.47 (2H, m), 7.31–7.26 (2H, m), 7.21–7.19 (4H, m), 6.61 (1H, t, J = 74.4 Hz), 5.03 (1H, m), 4.04–3.90 (4H, m), 2.94 (2H, t, J = 7.2 Hz), 2.72 (2H, t, J = 7.2 Hz), 2.28–2.14 (2H, m), 2.08 (2H, m). 13C NMR (CDCl3): 198.5 (s), 149.0 (s), 144.6 (s), 141.5 (s), 135.1 (s), 128.48 (d), 128.42 (d), 126.0 (d), 122.05 (d), 122.04 (d), 115.7 (triplet, J = 259 Hz, d), 113.9 (d), 78.8 (d), 72.7 (t), 67.1 (t), 37.5 (t), 35.1 (t), 32.9 (t), 25.7 (t). MS-EI: 376 (M+).
1-(3-Cyclopentyloxy-4-difluoromethoxyphenyl]-3-methyl-butan-1-one (5zf)
Yield: 74%. 1H NMR (CDCl3): 7.58 (1H, d, J = 4.0 Hz), 7.49 (1H, dd, J = 4.0 and 8.4 Hz), 7.19 (1H, d, J = 8.4 Hz), 6.63 (1H, t, J = 75.2 Hz), 4.90 (1H, m), 2.80 (2H, d, J = 6.8 Hz), 2.28 (1H, m), 1.97–1.79 (7H, m), 1.69–1.64 (2H, m), 1.00 (6H, d, J = 6.8 Hz). 13C NMR (CDCl3): 198.9 (s), 149.6 (s), 144.4 (s), 135.5 (s), 122.0 (d), 121.3 (d), 115.8 (triplet, J = 259 Hz, d), 114.1 (d), 80.8 (d), 47.3 (t), 32.7 (t), 25.3 (d), 23.7 (t), 22.7 (q). MS-EI: 312 (M+).
4-(2-Difluoromethoxy-5-pentanoylphenoxy)piperidine-1-carboxylic Acid tert-Butyl Ester (5zg)
Yield: 10%. 1H NMR (CDCl3): 7.62 (1H, d, J = 1.6 Hz), 7.55 (1H, dd, J = 1.6 and 8.4 Hz), 7.23 (1H, d, J = 8.4 Hz), 6.61 (1H, t, J = 74.4 Hz), 4.60 (1H, m), 3.67 (2H, m), 3.40 (2H, m), 2.93 (2H, t, J = 6.8 Hz), 1.93 (2H, m), 1.80 (2H, m), 1.71 (2H, m), 1.47 (9H, s), 1.40 (2H, m), 0.96 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 199.1 (s), 154.7 (s), 148.9 (s), 145.0 (s), 135.3 (s), 122.1 (d), 122.0 (d), 115.8 (triplet, J = 259 Hz, d), 115.2 (d), 79.7 (s), 74.1 (d), 40.5 (t), 38.2 (t), 30.4 (t), 28.4 (q), 26.5 (t), 22.4 (t), 13.9 (q). MS-EI: 371 [M – C4H9 (t-Bu) + H]+, 354 [M – C4H9O (t-BuO)]+, 327 [M – C5H9O2 (Boc) + H]+.
1-[4-Difluoromethoxy-3-(tetrahydropyran-4-yloxy)phenyl]-butan-1-one (5zh)
Colorless oil. Yield: 59%. 1H NMR (CDCl3): 7.62 (1H, s), 7.55 (1H, d, J = 8.4 Hz), 7.23 (1H, d, J = 8.4 Hz), 6.64 (1H, t, J = 74.4 Hz), 4.62 (1H, m), 3.98 (2H, m), 3.60 (2H, m), 2.92 (1H, t, J = 7.2 Hz), 2.03 (2H, m), 1.84–1.74 (4H, m), 1.01 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 198.9 (s), 148.8 (s), 145.0 (s), 135.4 (s), 122.14 (d), 122.07 (d), 115.9 (triplet, J = 259 Hz, d), 115.3 (d), 73.5 (d), 64.9 (t), 40.4 (t), 31.6 (t), 17.8 (t), 13.8 (q). MS-EI: 314 (M+).
1-[4-Difluoromethoxy-3-(pyridin-2-yloxy)phenyl]butan-1-one (5zi)
Yield: 99%. 1H NMR (CDCl3): 8.08 (1H, m), 7.85–7.83 (2H, m), 7.71 (1H, m), 7.32 (1H, d, J = 8.8 Hz), 7.03–6.99 (2H, m), 6.48 (1H, t, J = 73.6 Hz), 2.91 (2H, t, J = 7.2 Hz), 1.76 (2H, m), 0.99 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 198.1 (s), 162.7 (s), 147.2 (d), 144.8 (s), 139.7 (d), 135.3 (s), 125.9 (d), 123.7 (d), 120.8 (d), 118.9 (d), 115.8 (triplet, J = 260 Hz, d), 111.2 (d), 40.4 (t), 17.6 (t), 13.8 (q). HRMS-ESI (+): calcd for C19H15NO2F, 308.1087; found, 308.1085.
1-(3,4-Bisdifluoromethoxyphenyl)-4-phenylbutan-1-one (5zj)
Yield: 63%. 1H NMR (CDCl3): 7.81 (1H, s), 7.77 (1H, dd, J = 2.0 and 8.4 Hz), 7.32–7.27 (3H, m), 7.22–7.19 (3H, m), 6.59 (1H, t, J = 72.8 Hz), 6.56 (1H, t, J = 73.2 Hz), 2.93 (2H, t, J = 7.2 Hz), 2.72 (2H, t, J = 7.2 Hz), 2.08 (2H, m). 13C NMR (CDCl3): 197.6 (s), 145.9 (s), 141.9 (s), 141.4 (s), 135.1 (s), 128.48 (d), 128.45 (d), 126.5 (d), 126.1 (d), 121.9 (d), 121.2 (d), 115.52 (triplet, J = 262 Hz, d), 115.33 (triplet, J = 263 Hz, d), 37.5 (t), 35.0 (t), 25.5 (t). MS-EI: 356 (M+).
1-(3,4-Bisdifluoromethoxyphenyl)butan-1-one (5zk)
Yield: 72%. 1H NMR (CDCl3): 7.85–7.82 (2H, m), 7.33 (1H, d, J = 8.0 Hz), 6.63 (1H, t, J = 72.8 Hz), 6.60 (1H, t, J = 73.2 Hz), 2.92 (2H, t, J = 7.2 Hz), 1.76 (2H, m), 1.01 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 197.9 (s), 145.8 (s), 141.9 (s), 135.1 (s), 126.5 (d), 121.8 (d), 121.1 (d), 115.6 (triplet, J = 261 Hz, d), 115.4 (triplet, J = 263 Hz, d), 40.3 (t), 17.5 (t), 13.6 (q). MS-EI: 280 (M+).
1-(3,4-Bisdifluoromethoxyphenyl)-3-phenylpropan-1-on (5zl)
Yield: 46%. 1H NMR (CDCl3): 7.83–7.80 (2H, m), 7.30–7.24 (6H, m), 6.59 (1H, t, J = 74.4 Hz), 6.55 (1H, t, J = 72.8 Hz), 3.26 (2H, s), 3.06 (2H, s). 13C NMR (CDCl3): 196.8, 146.0, 141.9, 140.8, 134.9, 128.6, 128.4, 126.5, 126.3, 121.3, 121.2, 115.5 (triplet, J = 262 Hz), 115.3 (triplet, J = 263 Hz), 40.3, 0.9. MS-EI: 342 (M+).
1-(3,4-Bisdifluoromethoxyphenyl)-2-cyclohexylethanone (5zm)
Yield: 92%. 1H NMR (CDCl3): 7.84–7.81 (2H, m), 7.33 (1H, d, J = 7.6 Hz), 6.62 (1H, triplet, J = 72.8 Hz), 6.60 (1H, triplet, J = 72.8 Hz), 2.79 (2H, d, J = 5.2 Hz), 1.96 (1H, m), 1.76–1.68 (5H, m), 1.31–1.15 (3H, m), 1.03 (2H, m). 13C NMR (CDCl3): 197.7 (s), 145.8 (s), 141.9 (s), 135.5 (s), 126.6 (d), 121.9 (d), 121.1 (d), 115.6 (triplet, J = 262 Hz, d), 115.4 (triplet, J = 263 Hz, d), 46.0 (t), 34.4 (d), 33.3 (t), 26.1(t), 26.0 (t). MS-EI: 334 (M+).
General Procedure for the Synthesis of 6 by Nucleophilic Addition to the Ketones 5
To ketones 5g or 5h (1 mmol) in THF (5 mL) was added dropwise 3-pyridyllithium or 3-quinolyllithium at −78 °C. The resulting mixture was stirred overnight, during which time the mixture was allowed to gradually warm from −78 °C to room temperature and quenched with water. The aqueous layer was extracted three times with ethyl acetate. The combined organic solution was dried over MgSO4, filtered, and evaporated. The residue was purified by flash chromatography to afford 6.
1-[3-Methoxy-4-(pyridin-2-yloxy)phenyl]-1-pyridin-3-yl-pentan-1-ol (6a)
Yield: 36%. 1H NMR (CDCl3): 8.61 (1H, s), 8.32 (1H, s), 8.08 (1H, dd, J = 1.6 and 5.2 Hz), 7.79 (1H, d, J = 8.0 Hz), 7.63 (1H, m), 7.21 (1H, m), 7.15 (1H, d, J = 1.6 Hz), 7.05 (1H, d, J = 8.4 Hz), 6.98–6.91 (2H, m), 6.86 (1H, d, J = 8.0 Hz), 4.27 (1H, s, br), 3.67 (3H, s), 2.27 (2H, t, J = 7.2 Hz), 1.40–1.21 (4H, m), 0.86 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 163.4 (s), 151.4 (s), 147.31 (d), 147.26 (d), 144.4 (s), 142.8 (s), 141.4 (s), 139.1 (d), 134.1 (d), 122.9 (d), 122.2 (d), 118.5 (d), 118.0 (d), 111.2 (d), 110.6 (d), 76.5 (s), 55.8 (q), 41.6 (t), 25.7 (t), 22.9 (t), 13.9 (q). MS-EI: 364 (M+).
1-[3-Methoxy-4-(pyridin-2-yloxy)phenyl]-1-quinolin-3-ylbutan-1-ol (6b)
Yield: 53%. 1H NMR (CDCl3): 8.90 (1H, d, J = 1.6 Hz), 8.27 (1H, d, J = 1.6 Hz), 8.13 (1H, dd, J = 1.2 and 4.4 Hz), 8.07 (1H, d, J = 8.0 Hz), 7.83 (1H, d, J = 8.0 Hz), 7.71–7.62 (2H, m), 7.55 (1H, m), 7.16 (1H, s), 7.10–7.03 (2H, m), 6.94 (1H, m), 7.89 (1H, d, J = 8.4 Hz), 3.70 (3H, s), 2.53 (1H, s, br), 2.38 (2H, m), 1.47 (1H, m), 1.00 (1H, m), 0.97 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 163.5 (s), 151.7 (s), 149.9 (d), 147.5 (d), 146.8 (s), 144.0 (s), 141.9 (s), 139.3 (s), 139.2 (d), 132.1 (d), 1.3 (d), 1.0 (d), 128.1 (d), 127.5 (s), 126.8 (d), 122.5 (d), 121.8 (d), 118.7 (d), 118.1 (d), 111.3 (d), 110.8 (d), 77.3 (s), 56.0 (q), 44.2 (t), 17.1 (t), 14.4 (q). HRMS-ESI (+): calcd for C25H25N2O3, 401.1865; found, 401.1853 [M + H]+.
Synthesis of 3-{1-[3-Methoxy-4-(pyridin-2-yloxy)phenyl]-but-1-enyl}quinoline (7a)
To a solution of 6a (0.33 g, 0.8 mmol) in acetic acid (5 mL) was added dropwise concentrated H2SO4 (0.3 mL). The mixture was heated at 80 °C for 1 h and then cooled and neutralized with saturated NaHCO3. The aqueous solution was extracted with ethyl acetate three times. The combined organics were washed with brine, dried over MgSO4, filtered, concentrated. The residue was purified by flash chromatography to afford 0.12 g (37%) of 7a as a mixture of Z- and E-isomers. 1H NMR (CDCl3): 8.97 and 8.79 (1H, s), 8.20–7.49 (6H, m), 7.19–6.82 (5H, m), 6.00–6.25 (1H, m), 3.71 and 3.68 (3H, s), 2.30 (1H, m), 2.17 (1H, m), 1.11 (3H, m). 13C NMR (CDCl3): 163.6 (s), 163.5 (s), 150.1 (d0, 151.6 (s), 151.45 (s), 150.0 (d), 147.53 (d), 147.46 9d), 147.1 (s), 147.0 (s), 142.0 (s), 139.9 (s), 139.24 (d), 139.15 (d), 137.9 (s), 137.2 (s), 136.6 (s), 136.3 (d), 135.4 (s), 134.3 (d), 133.9 (d), 133.5 (d), 133.1 (s), 1.4 (d), 1.2 (d), 128.99 (d), 128.96 (d), 127.9 (d), 127.8 (d), 127.8 (s), 126.8 (d), 126.7 (d), 122.63 (d), 122.60 (d), 120.3 (d), 118.2 (d), 118.1 (d), 114.4 (d), 111.8 (d), 110.9 (d), 110.7 (d), 55.9 (q), 23.5 (t), 23.3 (t), 14.4 (q). HRMS-ESI (+): calcd for C25H23N2O2, 383.1760; found, 383.1742 [M + H]+.
Synthesis of 8
To a solution of 6a (0.30 g, 0.80 mmol) in acetic acid (5 mL) was added dropwise concentrated H2SO4 (0.3 mL). The mixture was heated at 80 °C for 1 h. After 6a was converted to 7a completely, without workup the product was hydrogenated at room temperature overnight using 10% Pd/C (0.03 g) as catalyst. The reaction mixture was neutralized with saturated NaHCO3. The aqueous solution was extracted with ethyl acetate three times. The combined organic solution was washed with brine, dried over MgSO4, filtered, and concentrated. The residue was purified by flash chromatography to afford 0.11 g (38%) of 8 as a white solid, mp 56–58 °C. 1H NMR (CDCl3): 8.57 (1H, s), 8.44 (1H, d, J = 4.4 Hz), 8.11 (1H, dd, J = 2.0 and 5.2 Hz), 7.62–7.54 (2H, m), 7.20 (1H, m), 7.70 (1H, d, J = 8.0 Hz), 6.91–6.84 (4H, m), 3.91 (1H, t, J = 8.0 Hz), 3.70 (3H, s), 2.06 (2H, m), 1.38–1.24 (4H, m), 0.87 (3H, t, J = 7.2 Hz). 13C NMR (CDCl3): 163.4 (s), 151.4 (s), 149.34 (d), 147.37 (d), 147.20 (d), 141.6 (s), 140.8 (s), 140.1 (s), 138.9 (d), 134.9 (d), 123.2 (d), 122.6 (d), 119.8 (d), 117.8 (d), 111.4 (d), 110.4 (d), 55.7 (q), 48.5 (d), 35.0 (t), 0.8 (t), 22.3 (t), 13.7 (q).
Synthesis of 4-[3-Methoxy-4-(pyridin-2-yloxy)phenyl]-4-quinolin-3-ylbutan-2-one (11a) and 4-(4-Hydroxy-3-methoxyphenyl]-4-pyridin-3-ylbutan-2-one (11b)
To compound 4h or 4i (0.50 mmol) in CH2Cl2 (20 mL) at room temperature was added SOCl2 (0.75 mmol). After 45 min, the mixture was carefully poured into saturated NaHCO3 (5 mL). The layers were separated, and the aqueous phase was extracted with CH2Cl2. The combined organic solution was washed with saturated NaHCO3 and brine, dried over MgSO4, filtered, and concentrated. The thick yellow oil (9a or 9b) was used immediately in the next reaction without further purification. To a solution of ethyl acetoacetate (0.60 mmol) in THF at 0 °C was added sodium methoxide in portions. After the mixture was stirred for 30 min, a solution of 9a or 9b in THF was added dropwise. The mixture was stirred at room temperature overnight and then poured into saturated NH4Cl. The layers were separated, and the aqueous phase was extracted twice with ethyl acetate. The combined organic solution was washed with 25% NH4OAc buffer and brine, dried over MgSO4, filtered, and concentrated to provide crude 10a or 10b. 10a or 10b was dissolved in a mixture of THF/MeOH/H2O (3:1:1, 10 mL), and then 2 N LiOH (1 mL, 2 mmol) was added. The mixture was heated at 70 °C for 2 h. Volatile components were removed on the rotovap, and the residue was partitioned between saturated NaHCO3 and ethyl acetate. The aqueous phase was extracted twice with ethyl acetate. The combined organic solution was washed with brine, dried over MgSO4, filtered, and concentrated. The residue was purified by flash chromatography to afford 11a or 11b.
11a
Yellow oil. Yield: 28%. 1H NMR (CDCl3): 8.86 (1H, d, J = 2.0 Hz), 8.11 (1H, dd, J = 1.2 and 4.4 Hz), 8.06 (1H, d, J = 8.4 Hz), 7.96 (1H, d, J = 2.4 Hz), 7.76 (1H, d, J = 7.2 Hz), 7.68–7.59 (2H, m), 7.50 (1H, m), 7.07 (1H, d, J = 8.0 Hz), 6.93–6.84 (4H, m), 4.82 (1H, t, J = 7.2 Hz), 3.70 (3H, s), 3.31 (2H, d, J = 7.2 Hz), 2.14 (3H, s). 13C NMR (CDCl3): 205.5, 163.7, 152.1, 151.3, 147.6, 147.2, 142.0, 140.4, 139.1, 136.5, 133.7, 1.3, 1.1, 128.0, 127.7, 126.8, 13.2, 120.1, 118.1, 113.5, 110.8, 56.2, 49.6, 43.6, 30.5.
11b
Yellow oil. Yield: 24%. 1H NMR (CDCl3): 8.52 (1H, d, J = 2.0 Hz), 8.43 (1H, dd, J = 1.6 and 4.8 Hz), 7.52 (1H, d, J = 8.0 Hz), 7.20 (1H, m), 6.84 (1H, d, J = 8.0 Hz), 6.72–6.67 (2H, m), 6.15 (1H, s, br), 4.54 (1H, t, J = 7.6 Hz), 3.81 (3H, s), 3.17 (2H, d, J = 7.6 Hz), 2.11 (3H, s). 13C NMR (CDCl3): 206.1 (s), 145.0 (d), 147.7 (d), 146.8 (s), 144.6 (s), 139.7 (s), 135.3 (d), 134.5 (s), 123.5 (d), 119.8 (d), 114.6 (d), 110.7 (d), 55.9 (q), 49.4 (t), 43.2 (d), 30.6 (q).
PDE Inhibition Assays
The enzymatic activities of PDEs were assayed using 3H-cAMP and 3H-cGMP as substrates.39,40 The expression and purification of PDE has been described earlier.41 First, PDEs were incubated with the reaction mixture of 50 mM Tris-HCl, pH 7.8, 10 mM MgCl2, and 3H-cAMP or 3H-cGMP (40 000 cpm/assay) at 24 °C for 30 min. Then the reaction was terminated by addition of ZnSO4 and Ba(OH)2. The reaction product 3H-AMP or 3H-GMP was precipitated by BaSO4, while unreacted 3H-cAMP or 3H-cGMP remained in the supernatant. Radioactivity in the supernatant was measured by liquid scintillation. The activity was measured at eight concentrations of cAMP and cGMP, and each measurement was repeated three times. Vmax and Km values were calculated by linear plots of Lineweaver–Burk (1/V vs 1/[S]) and Eadie–Hofstee (V/[S] vs V)42 and also by nonlinear regression analysis as implemented in the program DynaFit.43 For measurement of IC50, 10 concentrations of inhibitors were used under the substrate concentration of one-tenth of KM and the suitable enzyme concentration. For the potent inhibitors, a double or triple measurement was performed, while a single measurement was carried out for the poor inhibitors
NMR Study of the Interaction of Inhibitors with PDE4D2 Protein
A reference 1H spectrum and a 2D NOESY (mixing time of 500 ms) spectrum were taken on a Varian 800 MHz NMR using a 400 μM sample in 20 mM potassium phosphate (pH 7.1) D2O buffer. A sample with a 1:80 molar ratio of the inhibitor to protein was made in 20 mM potassium phosphate (pH 7.1) D2O buffer with concentrations of 800 μM 5i and 10 μM PDE4D2. STD and trNOEsy (mixing time of 50 ms) experiments were performed. The sample was subsequently diluted, and another STD spectrum was collected to test for nonspecific binding.
Supplementary Material
Acknowledgments
We gratefully acknowledge the financial support by the Georgia Research Alliance, Georgia Cancer Coalition, and the Molecular Basis of Disease Program at Georgia State University. We also thank Dr. Lupei Du for technical assistance.
Footnotes
Abbreviations: PDE4, phosphodiesterase 4; cAMP, cyclic adenosine monophosphate; cGMP, cyclic guanosine monophosphate; PCC, pydrinium chlorochromate; Boc, di-tert-butyl dicarbonate; COPD, cardiovascular obstructive pulmonary disease; PBMCs, peripheral blood mononuclear cells; TNF, tumor necrosis factor; QSAR, quantitative structure–activity relationship; CoMSIA, comparative molecular similarity index analysis; NOE, nuclear Overhauser effect; STD, saturation transfer difference.
Supporting Information Available: Purity and NMR results for final compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Antoni FA. Molecular diversity of cyclic AMP signalling. Front Neuroendocrinol. 2000;21:103–132. doi: 10.1006/frne.1999.0193. [DOI] [PubMed] [Google Scholar]
- 2.Beavo JA. Cyclic nucleotide phosphodiesterases: functional implications of multiple isoforms. Physicol Rev. 1995;75:725–748. doi: 10.1152/physrev.1995.75.4.725. [DOI] [PubMed] [Google Scholar]
- 3.Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006;58:488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- 4.Ke H, Wang H. Crystal structures of phosphodiesterases and implications on substrate specificity and inhibitor selectivity. Curr Top Med Chem. 2007;7:391–403. doi: 10.2174/156802607779941242. [DOI] [PubMed] [Google Scholar]
- 5.Lugnier C. Cyclic nucleotide phosphodiesterase (PDE) superfamily: a new target for the development of specific therapeutic agents. Pharmacol Ther. 2006;109:366–398. doi: 10.1016/j.pharmthera.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Omori K, Kotera J. Overview of PDEs and their regulation. Circ Res. 2007;100:309–327. doi: 10.1161/01.RES.0000256354.95791.f1. [DOI] [PubMed] [Google Scholar]
- 7.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 8.Houslay MD, Schafer P, Zhang KYJ. Phosphodiesterase-4 as a therapeutic target. Drug Discovery Today. 2005;10:1503–1519. doi: 10.1016/S1359-6446(05)03622-6. [DOI] [PubMed] [Google Scholar]
- 9.Kanes SJ, Tokarczyk J, Siegel SJ, Bilker W, Abel T, Kelly MP. Rolipram: a specific phosphodiesterase 4 inhibitor with potential antipsychotic activity. Neuroscience. 2007;144:239–246. doi: 10.1016/j.neuroscience.2006.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Donnell JM, Zhang HT. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4) Trends Pharmacol Sci. 2004;25:158–163. doi: 10.1016/j.tips.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Huang Z, Mancini JA. Phosphodiesterase 4 inhibitors for the treatment of asthma and COPD. Curr Med Chem. 2006;13:3253–3262. doi: 10.2174/092986706778773040. [DOI] [PubMed] [Google Scholar]
- 12.Lerner A, Epstein PM. Cyclic nucleotide phosphodiesterases as targets for treatment of haematological malignancies. Biochem J. 2006;393:21–41. doi: 10.1042/BJ20051368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipworth BJ. Phosphodiesterase-4 inhibitors for asthma and chronic obstructive pulmonary disease. Lancet. 2005;365:167–175. doi: 10.1016/S0140-6736(05)17708-3. [DOI] [PubMed] [Google Scholar]
- 14.Torphy TJ. Phosphodiesterase isozymes: molecular targets for the novel antiasthma agents. Am J Respir Crit Care Med. 1998;157:351–370. doi: 10.1164/ajrccm.157.2.9708012. [DOI] [PubMed] [Google Scholar]
- 15.Lowe JA, Cheng JB. The PDE IV family of calcium-independent phosphodiesterase enzymes. Drugs Future. 1992;17:799–807. [Google Scholar]
- 16.Nicholson D, Challiss R, Shahid M. Differential modulation of tissue function and therapeutic potential of selective inhibitors of cyclic nucleotide phospohodiesterase isozymes. Trends Pharmacol Sci. 1991;12:19–27. doi: 10.1016/0165-6147(91)90484-a. [DOI] [PubMed] [Google Scholar]
- 17.Fleming YM, Frame MC, Houslay MD. PDE4-regulated cAMP degradation controls the assembly of integrin-dependent actin adhesion structures and REF52 cell migration. J Cell Sci. 2004;117:2377–2388. doi: 10.1242/jcs.01096. [DOI] [PubMed] [Google Scholar]
- 18.Marivet M, Bourguignon J, Lugnier C, Mann A, Stoclet J, Wermuth C. Inhibition of cyclic adenosine-3′,5′-monophophate phosphodiesterase from vasular smooth muscle by rolipram analogues. J Med Chem. 1989;32:1450–1457. doi: 10.1021/jm00127a009. [DOI] [PubMed] [Google Scholar]
- 19.Ashton MJ, Cook DC, Fenton G, Karlsson JA, Palfreyman MN, Raeburn D, Ratcliffe AJ, Souness JE, Thurairatnam S, Vicker N. Selective type IV phosphodiesterase inhibitors as antiasthmatic agents. The syntheses and biological activities of 3-(cyclopentyloxy)-4-methoxybenzamides and analogues. J Med Chem. 1994;37:1696–1703. doi: 10.1021/jm00037a021. [DOI] [PubMed] [Google Scholar]
- 20.Stafford JA, Veal JM, Feldman PL, Valvano NL, Baer PG, Brackeen MF, Brawley ES, Connolly KM, Domanico PL, Han B, Rose D, Rutkowske R, Sekut L, Stimpson S, Strickland A, Verghese M. Introduction of a conformational switching element on a pyrrolidine ring. J Med Chem. 1995;38:4972–4975. doi: 10.1021/jm00026a003. [DOI] [PubMed] [Google Scholar]
- 21.Christensen SB, Guider A, Forster CJ, Gleason JG, Bender PE, Karpinski JM, DeWolf WE, Jr, Barnette MS, Underwood DC, Griswold DE, Cieslinski LB, Burman M, Bochnowicz S, Osborn RR, Manning CD, Grous M, Hillegas LM, Bartus JO, Ryan MD, Eggleston DS, Haltiwanger RC, Torphy TJ. 1,4-Cyclohexanecarboxylates: potent and selective inhibitors of phosophodiesterase 4 for the treatment of asthma. J Med Chem. 1998;41:821–835. doi: 10.1021/jm970090r. [DOI] [PubMed] [Google Scholar]
- 22.Norman P. PDE4 inhibitors 1999. Expert Opin Ther Pat. 1999;9:1101–1118. [Google Scholar]
- 23.Jeon YH, Heo YS, Kim CM, Hyun YL, Lee TG, Ro S, Cho JM. Phosphodiesterase: overview of protein structures, potential therapeutic applications and recent progress in drug development. Cell Mol Life Sci. 2005;62:1198–1220. doi: 10.1007/s00018-005-4533-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatzelmann A, Schudt C. Anti-inflammatory and immunomodulatory potential of the novel PDE4 inhibitor roflumilast in vitro. J Pharmacol Exp Ther. 2001;297:267–279. [PubMed] [Google Scholar]
- 25.Huai Q, Liu Y, Francis SH, Corbin JD, Ke H. Crystal structures of phosphodiesterases 4 and 5 in complex with inhibitor 3-isobutyl-1-methylxanthine suggest a conformation determinant of inhibitor selectivity. J Biol Chem. 2004;279:13095–13101. doi: 10.1074/jbc.M311556200. [DOI] [PubMed] [Google Scholar]
- 26.Huai Q, Wang H, Sun Y, Kim HY, Liu Y, Ke H. Three-dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity. Structure. 2003;11:865–873. doi: 10.1016/s0969-2126(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 27.Ochiaia H, Ohtania T, Ishidaa A, Kusumia K, Katoa M, Kohnoa H, Kishikawab K, Obataa T, Nakai H, Todaa M. Highly potent PDE4 inhibitors with therapeutic potential. Bioorg Med Chem Lett. 2004;14:207–210. doi: 10.1016/j.bmcl.2003.09.087. [DOI] [PubMed] [Google Scholar]
- 28.Guay D, Hamel P, Blouin M, Brideau C, Chan CC, Chauret N, Ducharme Y, Huang Z, Girard M, Jones TR, Laliberte F, Masson P, McAuliffe M, Piechuta H, Silva J, Young RN, Girard Y. Discovery of L-791,943: a potent, selective, non emetic and orally active phophodiesteras-4 inhibitors. Bioorg Med Chem Lett. 2002;12:1457–1461. doi: 10.1016/s0960-894x(02)00190-7. [DOI] [PubMed] [Google Scholar]
- 29.Muise ES, Chute IC, Claveau D, Masson P, Boulet L, Tkalec L, Pon DJ, Girard Y, Frenette R, Mancini JA. Comparison of inhibition of ovalbumin-induced bronchoconstriction in guinea pigs and in vitro inhibition of tumor necrosis factor-alpha formation with phosphodiesterase 4 (PDE4) selective inhibitors. Biochem Pharmacol. 2002;63:1527–1535. doi: 10.1016/s0006-2952(02)00903-6. [DOI] [PubMed] [Google Scholar]
- 30.Ouagued M, Martin-Chouly CA, Brinchault G, Leportier-Comoy C, Depince A, Bertrand C, Lagente V, Belleguic C, Pruniaux MP. The novel phosphodiesterase 4 inhibitor, CI-1044, inhibits LPS-induced TNF-alpha production in whole blood from COPD patients. Pulm Pharmacol Ther. 2005;18:49–54. doi: 10.1016/j.pupt.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 31.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, revision C.02. Gaussian, Inc; Wallingford, CT: 2004. [Google Scholar]
- 32.Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput-Aided Mol Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 33.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 34.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 35.Klebe G, Abraham U, Mietzner T. Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem. 1994;37:4130–4146. doi: 10.1021/jm00050a010. [DOI] [PubMed] [Google Scholar]
- 36.Klebe G, Abraham U. Comparative molecular similarity index analysis (CoMSIA) to study hydrogen-bonding properties and to score combinatorial libraries. J Comput-Aided Mol Des. 1999;13:1–10. doi: 10.1023/a:1008047919606. [DOI] [PubMed] [Google Scholar]
- 37.Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem. 2003;42:864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 38.Jayalakshmi V, Krishna NR. CORCEMA refinement of the bound ligand conformation within the protein binding pocket in reversibly forming weak complexes using STD-NMR intensities. J Magn Reson. 2004;168:36–45. doi: 10.1016/j.jmr.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 39.Turko IV, Francis SH, Corbin JD. Hydropathic analysis and mutagenesis of the catalytic domain of the cGMP-binding cGMP-specific phosphodiesterase (PDE5). cGMP versus cAMP substrate selectivity. Biochemistry. 1998;37:4200–4205. doi: 10.1021/bi972448r. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, Ke H, Tretiakova AP, Jameson B, Colman RW. Identification of overlapping but distinct cAMP and cGMP interaction sites with cyclic nucleotide phosphodiesterase 3A by site-directed mutagenesis and molecular modeling based on crystalline PDE4B. Protein Sci. 2001;10:1481–1489. doi: 10.1110/ps.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Liu Y, Chen Y, Robinson H, Ke H. Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7. J Biol Chem. 2005;280:30949–30955. doi: 10.1074/jbc.M504398200. [DOI] [PubMed] [Google Scholar]
- 42.Cornish-Bowden A. Fundamentals of Enzymatic Kinetics. Butterworth & Co. Ltd; London: 1979. [Google Scholar]
- 43.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.