Abstract
The construction of a trypsin column for rapid and efficient protein digestion in proteomics is described. Electrospun and alcohol-dispersed polymer nanofibers were used for the fabrication of highly stable trypsin coatings, which was prepared by a two-step process of covalent attachment and enzyme crosslinking. In a comparative study with the trypsin coatings on as-spun and non-dispersed nanofibers, it has been observed that a simple step of alcohol dispersion improved not only the enzyme loading but also the performance of protein digestion. In-column digestion of enolase was successfully performed in less than twenty minutes. By applying the alcohol dispersion of polymer nanofibers, the bypass of samples was reduced by filling up the column with well-dispersed nanofibers, and subsequently, interactions between the protein and the trypsin coatings were improved, yielding more complete and reproducible digestions. Regardless of alcohol-dispersion or not, trypsin coatings showed better digestion performance and improved performance stability under recycled uses than covalently-attached trypsin, in-solution digestion, and commercial trypsin beads. The combination of highly stable trypsin coatings and alcohol-dispersion of polymer nanofibers has opened up a new potential to develop a trypsin column for on-line and automated protein digestion.
INTRODUCTION
An efficient protein digestion with high reproducibility is of great importance for successful bottom-up proteomics where proteins are digested by proteolytic enzymes such as trypsin to produce peptides for mass spectrometric analysis. Protein digestion is commonly performed in gel or in-solution. However, these processes have several drawbacks, such as long digestion time on the order of 4–15 h, low trypsin-to-substrate ratios due to trypsin autolysis, and loss of peptides during the sample preparation.1 The use of free soluble trypsin not only results in poor trypsin stability due to autolysis, but also limits high-throughput peptide identification as well as automated protein digestion.1,2 Many studies have been reported to overcome these drawbacks and improve the protein digestion efficiency. For example, trypsin was immobilized in or on solid supports such as sol-gel silica,3 magnetic particles,4 polymeric materials,5 monolithic columns,6 syringe,7 and microchips.8 The use of appropriate supports could reduce the digestion time by increasing the ratio of trypsin to substrate proteins. Further acceleration of protein digestion process was realized by applying energy inputs such as high temperature,9 pressure,10,11 ultrasound,12 microwave radiation,13 and a combination of immobilized trypsin and irradiation.4 On-line coupling of protein digestion to LC/MS/MS is desirable as it would increase throughput of entire proteomics experiments and minimize the sample losses.3,11,14–22 The most common system involves the immobilized enzyme reactors, which enables in-column digestion coupled with on-line separation columns prior to mass spectrometry detection.22–24 Despite many attempts, the ultimate process of automated and on-line protein digestion has not been realized due to several limitations such as poor enzyme activity and stability that usually result from autolysis and proteolysis by other proteases in the samples.
Recently, we reported successful stabilization of the trypsin (TR) activity in the form of enzyme coatings (EC) on the surface of electrospun polystyrene-poly(styrene-co-maleic anhydride) nanofibers.25 Trypsin-coated nanofibers (EC-TR/NF) showed high trypsin activity due to high enzyme loading, maintained its initial activity under recycled uses and rigorous shaking for one year, and was highly resistant to proteolytic digestion. EC-TR/NF was also successfully used in digesting bovine serum albumin and S. oneidensis proteome extract.25 EC-TR/NF, prepared by the fabrication of crosslinked trypsin molecules onto as-spun polymer nanofibers, forms multi-point covalent linkages on the surface of trypsin molecules, which can effectively prevent both denaturation and autolysis of trypsin. Crosslinking enzymes is well known for its effective enzyme stabilization by preventing the enzyme denaturation in the same mechanism. However, most of crosslinked enzyme systems have been developed in a form of carrier-free systems,26 which are difficult to be employed in repeated protein digestions due to the fragile nature of crosslinked enzymes with no carriers. The use of polymer nanofibers can find its advantages as a carrier of trypsin coatings due to their high surface area and durability. Most of trypsin coatings on polymer nanofibers are of nanometer-scale thickness and attached onto a large area of nanofiber surface. This structural feature of trypsin coatings on polymer nanofibers enables a tight binding of trypsin coatings on nanofibers, which led to an unprecedented success in stabilizing the trypsin activity.25
When a trypsin digestion column was attempted by using EC-TR/NF as an extended application, however, as-spun nanofibers created the void volume in a column due to their entangled form, which caused the sample bypassing and resulted in inefficient protein digestion. In the present work, we propose the alcohol dispersion of polymer nanofibers to achieve a well-packed column with highly-dispersed nanofibers. First, the trypsin coating was fabricated on the alcohol-dispersed nanofibers (EC-TR/EtOH-NF), and investigated in its activity, morphology, protein digestion performance, and digestion performance stability in a comparative study with the trypsin coating on as-spun nanofibers (EC-TR/NF). Then, the digestion performance of EC-TR/EtOH-NF was compared with those of in-solution digestion and commercially available trypsin beads by digesting enolase at 50°C. Finally, a trypsin column with reduced void volume was prepared by using EC-TR/EtOH-NF, and in-column digestion was performed with improved digestion efficiency and in a short time such as less than 20 min.
EXPERIMENTAL SECTION
Materials
Trypsin, glutaraldehyde (GA), Nα-Benzoyl-L-arginine 4-nitroanilide hydrochloride (L-BAPNA), enolase, N,N-dimethylformamide (DMF) was purchased from Sigma (St. Louis, MO, USA). Polystyrene (PS, Mw = 860,000) and poly(styrene-co-maleic anhydride) (PSMA, Mw = 224,000) was purchased from Aldrich (Milwaukee, WI, USA). Tetrahydrofuran was purchased from J. T. Baker (Phillipsburg, NJ, USA). Chromatography column was purchased from Omnifit (Cambridge, England). Commercial trypsin-immobilized beads were purchased from Promega Corp. (Madison, WI, USA). All other reagents were purchased from Sigma and Aldrich in the highest grade commercially available.
Electrospinning of polymer nanofibers and alcohol dispersion
Polymer nanofibers were prepared via electrospinning, as previously described.27 Polymer solution was prepared at room temperature by dissolving and mixing the 2:1 weight ratio of PS:PSMA mixture in the mixture of tetrahydrofuran and acetone with the 4:1 volume ratio for several hours. The polymer solution was loaded into a syringe equipped with 30-gauge stainless steel syringe needle. The voltage of 7 kV was applied, and the solution was fed at a rate of 0.1 ml h−1 using a syringe pump (PHD-2000 Infusion, Harvard Apparatus, Holliston, MA, USA). The electrospun nanofibers were collected on grounded aluminum foil at a suitable distance (7 to 10 cm) from the tip of the needle (Figure S-1). Electrospun polymer nanofibers was added into an aqueous alcohol solution (50%, v/v) and shaken at 200 rpm for 10 min. After the polymer nanofibers were fully dispersed (Figure S-2), they were excessively washed with 10 mM sodium phosphate buffer (pH 7.9) and kept in the same buffer.
Trypsin coating on polymer nanofibers
Either dispersed or as-spun nanofibers were incubated in 5 mL of 10 mg mL−1 trypsin solution in 10 mM sodium phosphate buffer (pH 7.9). After being shaken at 200 rpm for 30 min, and the vial was placed on the rocker (50 rpm) in a refrigerator (4°C) for 2 h in order to prepare covalent-attached trypsin onto nanofibers (CA-TR). For the preparation of trypsin coatings (EC-TR), the GA stock solution was added into the vial with the final GA concentration to be 0.5% (w/v), and the mixture was incubated at room temperature for 150 min and shaken on the rocker (50 rpm) at 4°C overnight. For the case of CA-TR, a buffer was added instead of the GA stock solution. After an overnight incubation, the trypsin nanofibers (both CA-TR and EC-TR) were washed with 100 mM sodium phosphate (pH 7.9). The unreacted aldehyde groups were capped by incubating the trypsin nanofibers in 100 mM Tris-HCl (pH 7.9) for 30 min. Trypsin nanofibers were washed excessively with 10 mM sodium phosphate buffer (pH 7.9), and were stored in the same buffer at 4°C until used.
Activity measurement of trypsin nanofibers
The TR activity of trypsin nanofibers were measured by the hydrolysis of L-BAPNA in an aqueous buffer (10 mM sodium phosphate, pH 7.9) (Figure S-3). Trypsin nanofibers were added to a vial containing 10 ml of L-BAPNA solution, the vial was shaken at 200 rpm, and aliquots were taken time-dependently from the reaction mixture. The concentration of p-nitroaniline in each aliquot was measured by the absorbance of 410 nm (A410) using a UV-VIS spectrophotometer (Shimadzu, UV-2450, Kyoto, Japan), and the initial activity was calculated from the slope of A410 with time.
In-vial protein digestion
Trypsin nanofibers were used for the digestion of enolase. First, 1 mL of 2 mM enolase in 100 mM ammonium bicarbonate (pH 8.0) was added into a vial containing trypsin nanofibers in 1 mL of 100 mM ammonium bicarbonate buffer (pH 8.0). The digestion was carried out at room temperature under shaking (200 rpm) for overnight. Finally, 1.5 mL of solution was transferred to a new glass vial, and formic acid wad added to quench the reaction. The samples were stored at −70°C until analyzed by LC/MS/MS. Trypsin nanofibers were excessively washed and stored at 4°C until the next digestion. To compare the digestion performance of trypsin-coated nanofibers (EC-TR/EtOH-NF) with in-solution digestion using free trypsin and commercially-available trypsin beads, enolase was digested under shaking (250 rpm) at 50°C for 6 hrs. For in-solution digestion, trypsin was added to the enolase solution in a weight ratio of 1:50 while the enolase digestion using the commercial beads was performed by following a standard protocol provided by Promega. Repeated digestions using EC-TR/EtOH-NF and commercial beads were carried out after excessively washing and incubating them under shaking (250 rpm) at 50°C until the next digestion. In the case of in-solution digestion, the sample was incubated under shaking (250 rpm) at 50°C, and an appropriate amount of aliquot was removed for each protein digestion at each time point.
Trypsin column and in-column digestion
Trypsin digestion columns were prepared by fabricating EC-TR on as-spun or alcohol-dispersed nanofibers in a column (3 mm i.d. and 5 cm long). First, a column was filled with as-spun nanofibers, and then an aqueous alcohol solution (50%, v/v) was delivered into the column at a flow rate of 1 mL h−1 for 10 h. Instead of alcohol solution, an aqueous buffer with no alcohol was used in this step for the preparation of EC-TR on as-spun nanofibers as a control. A trypsin solution (10 mg mL−1 in 10 mM sodium phosphate, pH 7.9) was pumped into the column at a flow rate of 2.5 mL h−1 for 2 h. Then, the aqueous mixture of 2.5 mg mL−1 trypsin and 0.05% w/v GA was fed into the column at a flow rate of 1 mL h−1 for 10 min, and the trypsin crosslinking was performed for 50 min. After enzyme crosslinking twice, the column was washed with 100 mM sodium phosphate (pH 7.9) and incubated with 100 mM Tris-HCl (pH 7.9) for 1 h to cap the unreacted aldehyde groups. The trypsin column was washed extensively with 10 mM sodium phosphate buffer (pH 7.9) at a flow rate of 5 mL h−1, and stored in the same buffer at room temperature. In-column protein digestion was performed at room temperature by making a flow of enolase solution (100 ng µL−1) through the column at a rate of 2 µL min−1. The effluents were analyzed by using LC/MS/MS.
LC/MS/MS analysis
The tryptic digests were analyzed by a nanoACQUITY UPLC (Waters, Milford, MA, USA) and 7-tesla Fourier transform ion cyclotron resonance mass spectrometer (FTICR, LTQ-FT, ThermoFinnigan, San Jose, CA, USA). The nanoACQUITY UPLC system was equipped with an in-house capillary column (75 µm i.d., 360 µm o.d., and 70 cm long; Polymicro Technologies, Phoenix, AZ, USA) that was packed with C18-bonded particles (3 µm diameter, 300 Å pore size, Phenomenex, Torrance, California, USA). The LC system was also equipped with an online SPE (Solid Phase Extraction) column (250 µm i.d. packed with the same 3 µm C18 particles). The tryptic peptides were loaded onto the online SPE column for 3 min with solvent A (0.1% formic acid in water) and eluted from the capillary column with a 60-min linear gradient of 10−60% solvent B (0.1% formic acid in acetonitrile). The linear low rate was 0.35 µL min−1. The LTQ-FT mass spectrometer was operated in a data-dependent mode, in which one full MS (from m/z 450 to 1800) scan was followed by three MS/MS scans. Normalized collision energy of 35% was used for peptide fragmentation. The mass spectrometric data was processed by the SEQUEST search algorithm (Bioworks 3.3, Thermo Fisher Scientific, New York, USA) program for an identification of the enolase tryptic peptides.
RESULTS AND DISCUSSION
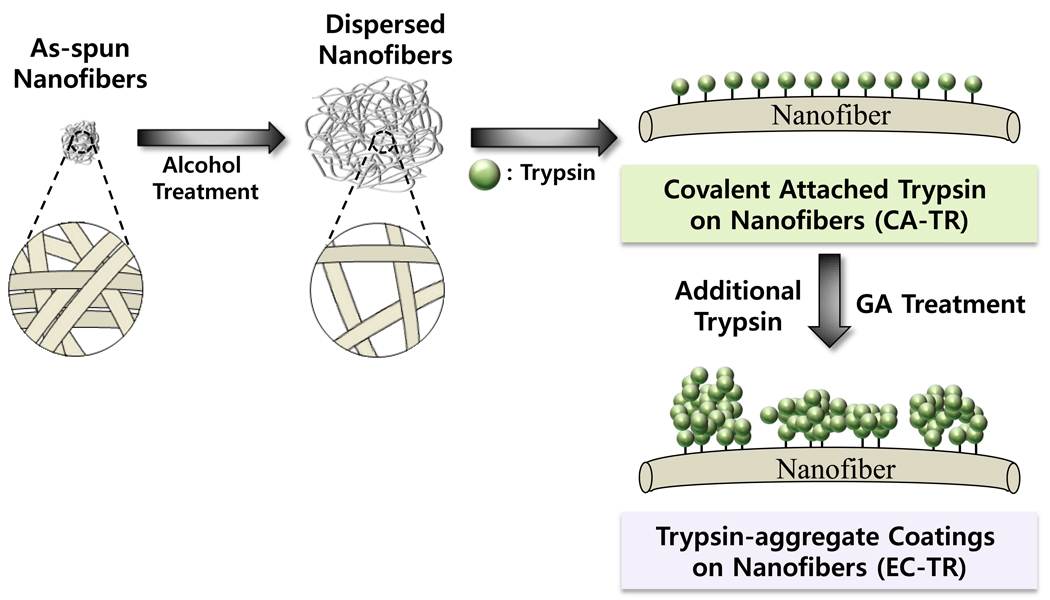
Polymer nanofibers were prepared by electrospinning the mixture of PS and PSMA in a mixture of tetrahydrofuran and acetone, and further dispersed in an aqueous solution of 50% (v/v) ethanol.28 Alcohol-dispersed polymer nanofibers were coated with trypsin aggregates by crosslinking additional trypsin molecules onto covalently-attached trypsin molecules on the nanofibers (Figure 1). The as-spun nanofibers floated on the water, but was observed to sink into the water upon treatment with an aqueous alcohol solution, which effectively dispersed the polymer nanofibers (Figure S-2).28 By considering the higher densities of polystyrene (1.05 g mL−1) and poly(styrene-co-maleic anhydride) (1.10 g mL−1) than that of water, the floating of as-spun nanofibers was explained by the hydrophobic nature of polymer nanofibers that allows tiny air-bubbles in a tightly-entangled form of nanofibers.28 Then, the treatment of as-spun nanofibers with the alcohol solution, which is more hydrophobic than water, could lead to the penetration of ethanol molecules into the matrix of polymer nanofibers, the removal of tiny air bubbles, and the dispersion of polymer nanofibers. Well-dispersed state of alcohol-treated nanofibers was maintained even after replacing the alcohol solution with water, supporting the mechanism of air-bubble removal during the alcohol dispersion. It was also observed that the formation of trypsin coating on nanofibers marginally helped to further immerse the nanofibers into the water (Figure S-2), which can be explained by the surface hydrophilization upon coating nanofibers with hydrophilic enzymes.
Figure 1.
Schematic diagram for the preparation of alcohol-dispersed nanofibers and trypsin coating (EC-TR).
The trypsin activity was measured by the hydrolysis of L-BAPNA in an aqueous buffer solution (10 mM sodium phosphate, pH 7.9) (Figure S-3). Table S-1 shows the kinetic constants of trypsin-coatings on as-spun and alcohol-dispersed nanofibers, which are represented by EC-TR/NF and EC-TR/EtOH-NF, respectively. Generally, the enzyme loading results are supposed to be obtained by calculating the disappeared protein amounts from the measurements of protein concentrations in solution before and after the enzyme immobilization. However, the enzyme crosslinking during the preparation of enzyme coating not only attaches the crosslinked enzyme coatings on nanofibers, but also generates insoluble form of crosslinked enzymes in an aqueous solution. This insoluble form of crosslinked enzymes interferes with the measurement of protein concentration that requires the soluble form of enzymes. As a result, the exact enzyme loading of trypsin-coated nanofibers could not be obtained. For this reason, the specific activity, defined as the activity per unit weight of enzyme, could not be calculated, and the comparative study was done by using the normalized activity per unit weight of polymer nanofibers. Table S-1 shows the kinetic values of Vmax, Km, and Vmax/Km of EC-TR/NF and EC-TR/EtOH-NF. The values of kcat (= Vmax / [E0]) and kcat/Km could not be obtained because the exact enzyme loading could not be measured. Instead, all the activity measurements for kinetic study were done by using trypsin-coatings on 1 mg of nanofibers. The Vmax/Km values of EC-TR/NF and EC-TR/EtOH-NF were 430 × 10−6 and 1380 × 10−6 s−1, respectively. The 3.2-fold increase of Vmax/Km by using dispersed nanofibers can be explained by the combination of 19-fold increased Vmax and 5.8-fold increased Km of EC-TR/EtOH-NF when compared to those of EC-TR/NF.
The increased Vmax of EC-TR/EtOH-NF can be attributed to the increased trypsin loading on dispersed nanofibers with more exposed surface area and inter-fiber space available for the immobilization of trypsin coating. To dissect the contributions of surface area and inter-fiber space to the improved trypsin loading on alcohol-dispersed nanofibers, the enzyme loadings of covalently-attached trypsin on as-spun (CA-TR/NF) and dispersed nanofibers (CA-TR/EtOH-NF) were obtained. Since the preparation of covalently-attached trypsin (CA-TR) does not require the step of enzyme crosslinking, the enzyme loading could be obtained via a conventional method that measures the disappeared enzyme amount from the solution during the enzyme immobilization. The trypsin loadings of CA-TR/NF and CA-TR/EtOH-NF were 200 µg and 640 µg per mg of nanofibers, respectively. This is a similar trend to the increased enzyme loading of covalently-attached lipase on alcohol-dispersed polymer nanofibers.28 Because covalently-attachment of enzymes results in a monolayer coverage of enzyme molecules on the surface of nanofibers, the 3.2-fold higher trypsin loading of CA-TR/EtOH-NF reveals that the dispersion of polymer nanofibers provided 3.2-fold increase of available surface area for the covalent attachment of enzyme. Then, the 19-fold increase of Vmax with EC-TR/EtOH-NF can be explained partly by 3.2-fold increase of accessible surface area on nanofibers, and the remaining 5.9-fold increase of Vmax (19/3.2 = 5.9) could be attributed to the other factors such as enlarged inter-fiber space that can also improve the enzyme loading in the form of multiple-layered enzyme coating. With an assumption that the kcat value of trypsin coating is not affected by using dispersed polymer nanofibers, it is estimated that the trypsin coating on dispersed nanofibers was 5.9 times thicker than that on as-spun nanofibers because the enlarged inter-fiber space enables the formation of thicker enzyme coating. The increased Km of EC-TR/EtOH-NF can be explained by more severe mass-transfer limitation through thicker trypsin coating on dispersed nanofibers. Interestingly, the 5.8-fold increase in Km of EC-TR/EtOH-NF well matched with the estimated 5.9-fold increase of coating thickness with EC-TR/EtOH-NF, which can be explained by the correlation between the thickness of enzyme coating and the mass transfer limitation.
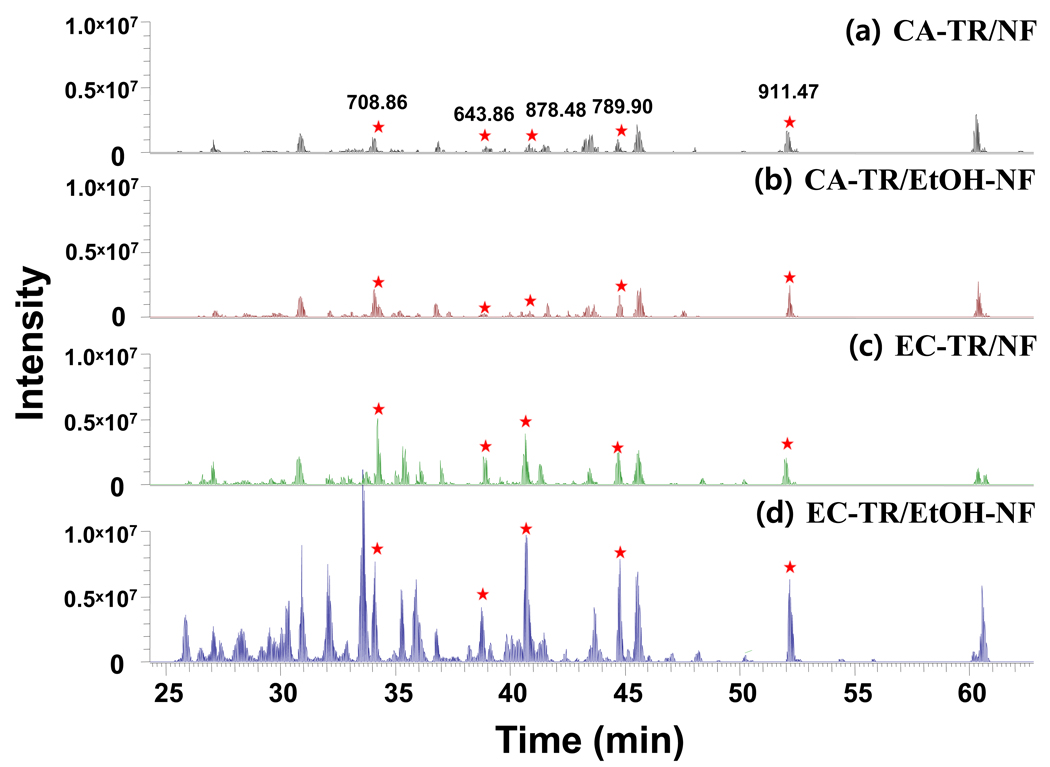
The protein digestions of enolase were carried out in vials by using four different trypsin nanofibers (CA-TR/NF, CA-TR/EtOH-NF, EC-TR/NF, and EC-TR/EtOH-NF) to compare their digestion performance under recycled uses for repeated protein digestions. The samples of CA-TR were added for a comparative study because the covalent attachment of enzymes is well known as one of conventional immobilization approaches that can stabilize the enzyme activity. Enolase was selected as a model protein because it results in tryptic peptides with a broad range of hydrophobicities that covers the entire LC gradient.29–33 After in-vial digestion of enolase by trypsin nanofibers, the digested peptides were analyzed by using LC/MS/MS. Figure 2 shows the LC-MS/MS spectra of enolase digests that were obtained from in-vial digestion by freshly-prepared trypsin nanofibers. The five tryptic peptides of enolase, marked with asterisks in Figure 2, were selected to quantitatively estimate the degree of tryptic digestion: GNPTVEVELTTEK, 708.8648; NVNDVIAPAFVK, 643.8589; TAGIQIVADDLTVTNPK, 878.4787; AVDDFLLSLDGTANK, 789.9040; and SGETEDTFIADLVVGLR, 911.4648. These five peptides are fully tryptic with no missed cleavages (Figure S-4) and cover from early to later gradients of LC separation. The summation of chromatographic peak areas of these five enolase-specific peptides is expected to better represent the degree of tryptic digestion (i.e. as the digestion proceeds, the MS intensities of these fully tryptic peptides increase barring degradation of these tryptic peptides) than measuring protein sequence coverage.34 As the current LC/MS/MS analysis platform was estimated to have sub-attomole detection sensitivity (Figure S-5), the simple estimation of protein sequence coverage might not be an accurate measure for the digestion efficiency. LC/MS/MS analysis of this sensitivity can result in a similar (if not the same) protein coverage for both low and high intensity chromatograms, resulting from low and high efficient digestions, respectively. As long as a peptide sequence is identified, it contributes the same to the protein sequence coverage regardless of its abundances. However, the chromatrographic areas of fully tryptic peptides represent the amount of the final products of tryptic digestion, which have a strong correlation to the digestion efficiency. With the increased digestion efficiency, it is expected to produce more fully tryptic peptides, leading to higher chromatrographic peak areas. The trypsin coatings generated higher intensity peaks of enolase-specific peptides than covalently-attached trypsin samples, and the use of alcohol-dispersed nanofibers further improved the performance of trypsin digestion over as-spun nanofibers as evidenced by increased peak intensities of the five peptides (Figure 2).
Figure 2.
LC/MS/MS spectra of the enolase digests obtained by using freshly-prepared trypsin nanofibers. (a) CA-TR/NF, (b) CA-TR/EtOH-NF, (c) EC-TR/NF, and (d) EC-TR/EtOH-NF.
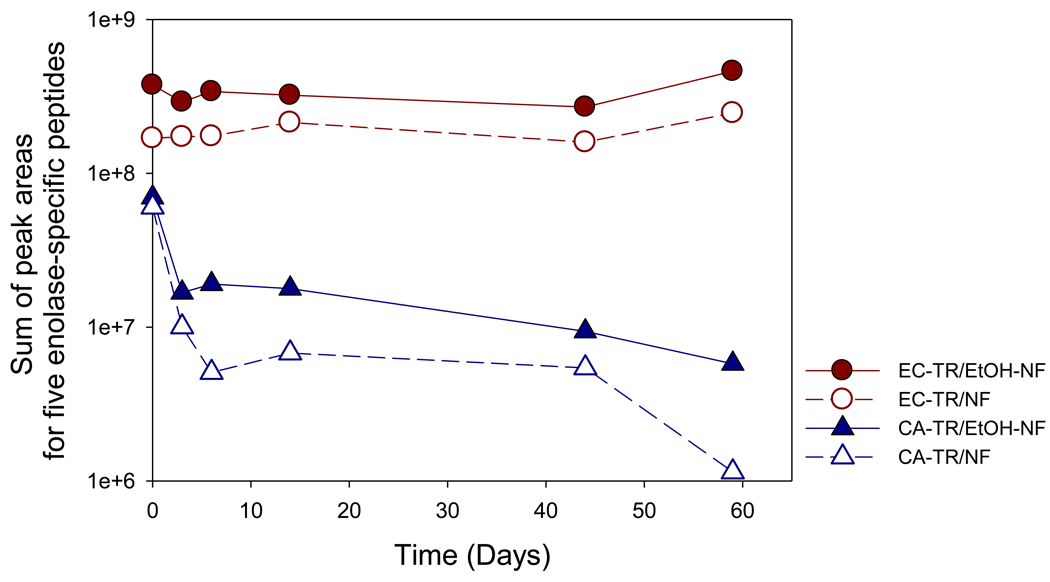
The stability of digestion performance was also investigated by recycling each of trypsin nanofibers and examining the resultant LC/MS/MS chromatograms. The digestion performance rapidly dropped when the samples of CA-TR were used for the enolase digestion while the trypsin coatings on both as-spun (EC-TR/NF) and dispersed nanofibers (EC-TR/EtOH-NF) maintained the initial digestion performance even after 59 days (Figure S-6). As expected, the summation of chromatographic peak areas of the five enolase-specific peptides better represented the changes of digestion performance of trypsin nanofibers than protein sequence coverage because the latter failed to show the decreasing digest performance of CA-TR (Figure S-7), which was evident in chromatographic peak areas (Figure S-6). Figure 3 compares time-dependent digestion performances of trypsin nanofibers, by calculating the total chromatographic peak areas of five tryptic enolase peptides. It is evident that the trypsin coatings were stable enough to maintain the initial peak area of five peptides, indicating reproducible digestions even after 59 days, while covalently-attached trypsin showed a rapid decrease of total peak area due to its poor stability. The poor stability of covalently-attached trypsin can be explained by both autolysis and denaturation of trypsin under shear stress of rigorous shaking for enolase digestion. On the other hand, the good performance stability of trypsin coatings can be explained by both the tight binding of enzyme coatings on nanofibers and the prevention of trypsin autolysis due to multi-point linkages on the trypsin molecules formed by chemical crosslinking.25,35
Figure 3.
In-vial enolase digestion catalyzed by trypsin nanofibers under recycled uses. The peak areas of five enolase-specific peptides were summed to check the performance of enolase digestion at each time point.
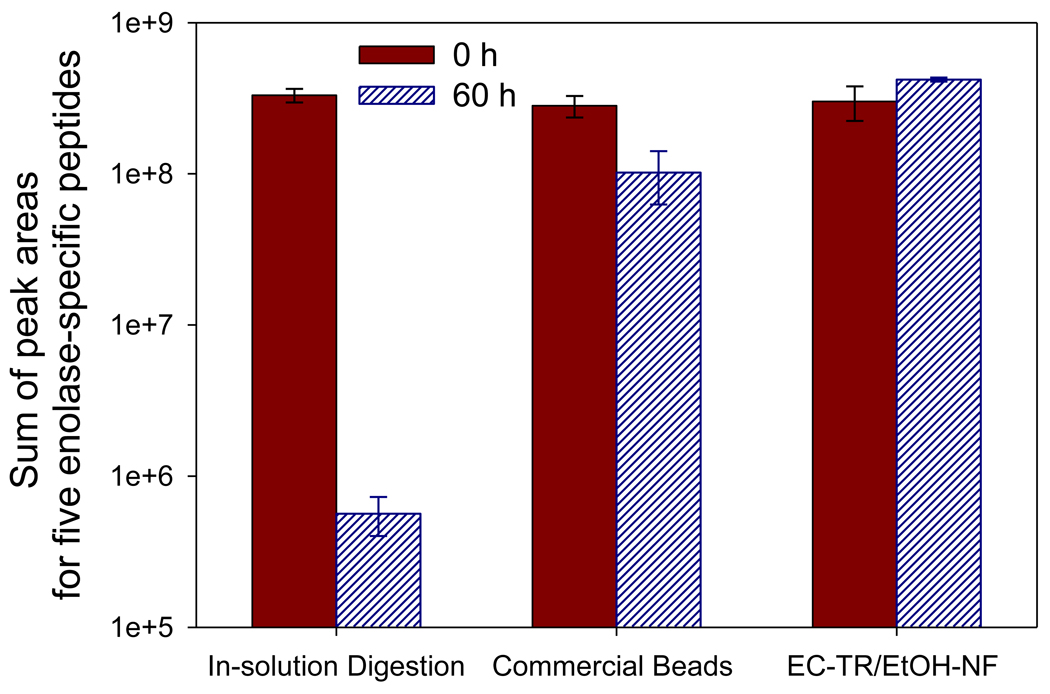
To compare the digestion performance of EC-TR/EtOH-NF with in-solution digestion and commercial trypsin beads, the enolase digestion was carried out in vials under shaking (250 rpm) at 50°C for 6 hrs. Figures 4 and S-8 show the stability of digestion performance of each sample after incubation at 50°C. At each time point, an aliquot of free trypsin solution was used for in-solution digestion while the samples of commercial trypsin beads and EC-TR/EtOH-NF were repeatedly used after excessive washings. The initial enolase digestion of EC-TR/EtOH-NF resulted in a slightly higher total peak area of five enolase-specific peptides than those of in-solution digestion and commercial trypsin beads. More importantly, the initial digestion performance of EC-TR/EtOH-NF was maintained after repeated digestions up to 60 hrs while both in-solution digestion and commercial trypsin beads showed a reduced digestion performance in the same condition (Figure 4). The digestion performance stability of each sample is more vividly shown in the extracted chromatogram of five enolase-specific peptides in a linear scale of peak intensity (Figure S-8). This result suggests that EC-TR/EtOH-NF is stable enough to be repeatedly used without losing the digestion performance at high temperature such as 50°C as well as at room temperature.
Figure 4.
In-vial enolase digestion at 50°C for the comparison of EC-TR/EtOH-NF with in-solution digestion and commercial trypsin beads in their digestion performance and stability. The peak areas of five enolase-specific peptides were summed to check the performance of enolase digestion at each time point.
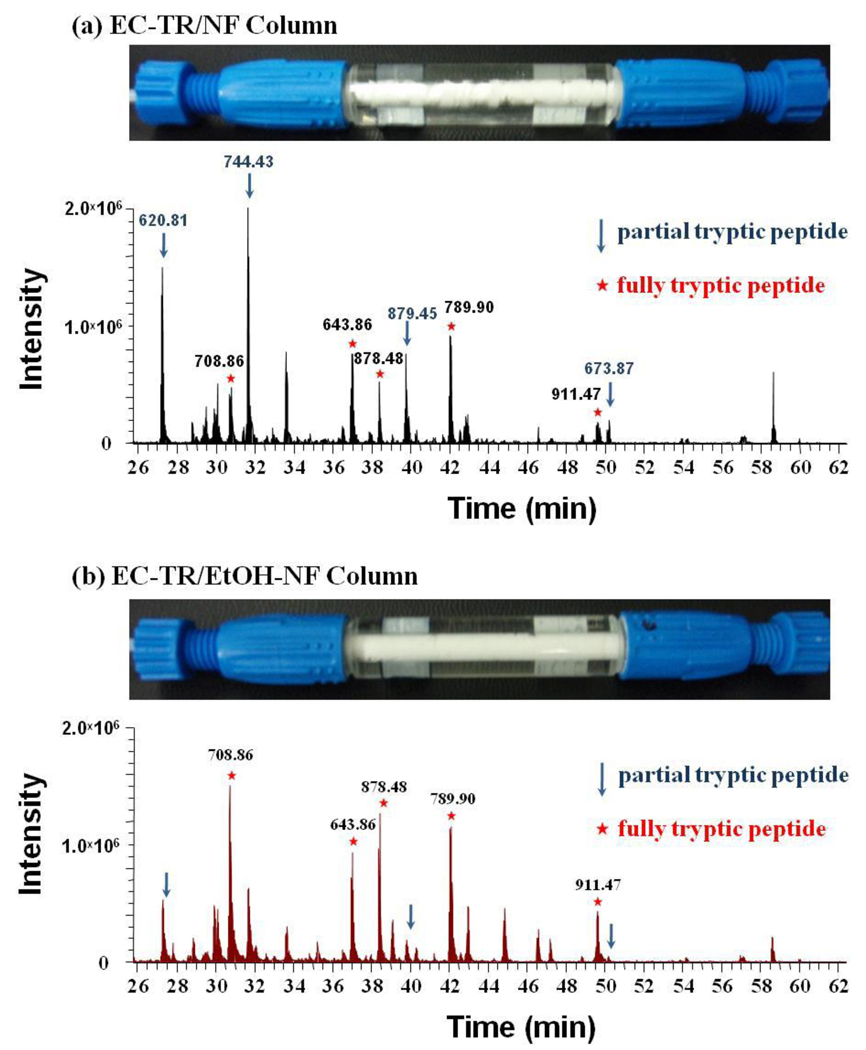
As an extension of successful in-vial digestion by using EC-TR/EtOH-NF, a trypsin column was prepared by filling up a column with as-spun nanofibers, dispersing polymer nanofibers via alcohol treatment, and fabricating the trypsin coating on dispersed nanofibers. As a control experiment, another trypsin column was prepared by omitting the second step of alcohol-dispersion. As shown in Figure 5, the alcohol dispersion was effective in filling up the column with fully-dispersed polymer nanofibers, which is critical in preventing the bypass of samples through the void volume of as-spun nanofibers in a column. The trypsin coating column with dispersed polymer nanofibers resulted in a better digestion performance by showing higher peaks of five enolase-specific peptides than the column with as-spun nanofibers under the same digestion condition (Figure 5). The total peak areas of five enolase-specific peptides were 2.5 × 107 and 4.2 × 107 for digestions in columns with as-spun and dispersed nanofibers, respectively. Better digestion performance of EC-TR/EtOH-NF column can be explained by the better contact of protein with trypsin coating on fully-dispersed nanofibers with less void volume. It should also be noted that in-column digestion was completed less than 20 min and can be further expedited by using higher flow rate. These promising results of EC-TR/EtOH-NF column have opened up a new potential to be employed for on-line and automated protein digestion at a much reduced time scale.
Figure 5.
Trypsin digestion columns, and LC/MS/MS chromatography of enolase digests from in-column digestion with (a) EC-TR/NF and (b) EC-TR/EtOH-NF at a flow rate of 2 µL/min.
CONCLUSIONS
Alcohol dispersion of electrospun polymer nanofibers, combined with the trypsin coating approach, has proven to be effective in improving enzyme loading, overall enzyme activity, and performance of protein digestion. The protein digestion column, filled with trypsin coating on alcohol-dispersed nanofibers, has demonstrated its potential to be used in on-line and automated protein digestion for facile proteomic analysis, which is being hampered mostly by poor trypsin stability. Although the current work is mainly focused on the development of trypsin digestion column, the combined approach of enzyme coating and alcohol-dispersion can be applied to the other enzymes for the development of various enzyme columns with high activity and performance, good stability and long lifetime, and reduced bypass of samples.
Supplementary Material
ACKNOWLEDGMENT
Portions of this work were supported by grants from the National Research Foundation (NRF) funded by the Korean Ministry of Education, Science & Technology (MEST) (No. 2009-0082314, No. 2009-0059861, and No. 2009-0075638). SL and MC acknowledge 21C Frontier Functional Proteomics Project (FPR08A1-010) from MEST, Converging Research Center for Mass Spectrometric Diagnosis, and the Fundamental R&D Program for Core Technology of Materials funded by the Ministry of Knowledge Economy. DLF and RDS also acknowledge the NIH National Center for Research Resources (NCRR, RR018522), NIH National Cancer Institute (R21 CA12619-01), and the Pacific Northwest National Laboratory’s (PNNL) Laboratory Directed Research and Development Program.
REFERENCES
- 1.Lopez-Ferrer D, Canas B, Vazquez J, Lodeiro C, Rial-Otero R, Moura I, Capelo JL. Trac-Trends Anal. Chem. 2006;25:996–1005. [Google Scholar]
- 2.Kim J, Kim B, Lopez-Ferrer D, Petritis K, Smith R. Proteomics. 2010;10:687–689. doi: 10.1002/pmic.200900519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakai-Kato K, Kato M, Toyo'oka T. Anal. Chem. 2002;74:2943–2949. doi: 10.1021/ac0200421. [DOI] [PubMed] [Google Scholar]
- 4.Chen WY, Chen YC. Anal. Chem. 2007;79:2394–2401. doi: 10.1021/ac0614893. [DOI] [PubMed] [Google Scholar]
- 5.Lin W, Skinner CD. J. Sep. Sci. 2009;32:2642–2652. doi: 10.1002/jssc.200900221. [DOI] [PubMed] [Google Scholar]
- 6.Peterson DS, Rohr T, Svec F, Frechet JMJ. Anal. Chem. 2002;74:4081–4088. doi: 10.1021/ac020180q. [DOI] [PubMed] [Google Scholar]
- 7.Wang S, Chen Z, Yang PY, Chen G. Proteomics. 2008;8:1785–1788. doi: 10.1002/pmic.200701042. [DOI] [PubMed] [Google Scholar]
- 8.Ji J, Zhang Y, Zhou X, Kong J, Tang Y, Liu B. Anal. Chem. 2008;80:2457–2463. doi: 10.1021/ac702218v. [DOI] [PubMed] [Google Scholar]
- 9.Havlis J, Thomas H, Sebela M, Shevchenko A. Anal. Chem. 2003;75:1300–1306. doi: 10.1021/ac026136s. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Ferrer D, Petritis K, Hixson KK, Heibeck TH, Moore RJ, Belov ME, Camp DG, Smith RD. Journal of Proteome Research. 2008;7:3276–3281. doi: 10.1021/pr7008077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Ferrer D, Petritis K, Lourette NM, Clowers B, Hixson KK, Heibeck T, Prior DC, Pasa-Tolic L, Camp DG, Belov ME, Smith RD. Analytical Chemistry. 2008;80:8930–8936. doi: 10.1021/ac800927v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Ferrer D, Capelo JL, Vazquez J. J. Proteome. Res. 2005;4:1569–1574. doi: 10.1021/pr050112v. [DOI] [PubMed] [Google Scholar]
- 13.Lin S, Yao GP, Qi DW, Li Y, Deng CH, Yang PY, Zhang XM. Anal. Chem. 2008;80:3655–3665. doi: 10.1021/ac800023r. [DOI] [PubMed] [Google Scholar]
- 14.Marie G, Serani L, Laprevote O. Anal. Chem. 2000;72:5423–5430. doi: 10.1021/ac000421z. [DOI] [PubMed] [Google Scholar]
- 15.Gao J, Xu JD, Locascio LE, Lee CS. Anal. Chem. 2001;73:2648–2655. doi: 10.1021/ac001126h. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Y, Lee CS. J. Chromatogr. A. 2001;924:315–322. doi: 10.1016/s0021-9673(01)00718-x. [DOI] [PubMed] [Google Scholar]
- 17.Lazar IM, Ramsey RS, Ramsey JM. Anal. Chem. 2001;73:1733–1739. doi: 10.1021/ac001420+. [DOI] [PubMed] [Google Scholar]
- 18.Kato M, Sakai-Kato K, Jin HM, Kubota K, Miyano H, Toyo'oka T, Dulay MT, Zare RN. Anal. Chem. 2004;76:1896–1902. doi: 10.1021/ac035107u. [DOI] [PubMed] [Google Scholar]
- 19.Massolini G, Calleri E. J. Sep. Sci. 2005;28:7–21. doi: 10.1002/jssc.200401941. [DOI] [PubMed] [Google Scholar]
- 20.Zhao C, Jiang HH, Smith DR, Bruckenstein S, Wood TD. Anal. Biochem. 2006;359:167–175. doi: 10.1016/j.ab.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Lee J, Soper SA, Murray KK. Anal. Chim. Acta. 2009;649:180–190. doi: 10.1016/j.aca.2009.07.037. [DOI] [PubMed] [Google Scholar]
- 22.Ma JF, Liu JX, Sun LL, Gao L, Liang Z, Zhang LH, Zhang YK. Anal. Chem. 2009;81:6534–6540. doi: 10.1021/ac900971w. [DOI] [PubMed] [Google Scholar]
- 23.Stigter ECA, de Jong GJ, van Bennekorn WP. Anal. Bioanal. Chem. 2007;389:1967–1977. doi: 10.1007/s00216-007-1584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye ML, Hu S, Schoenherr RM, Dovichi NJ. Electrophoresis. 2004;25:1319–1326. doi: 10.1002/elps.200305841. [DOI] [PubMed] [Google Scholar]
- 25.Kim BC, Lopez-Ferrer D, Lee SM, Ahn HK, Nair S, Kim SH, Kim BS, Petritis K, Camp DG, Grate JW, Smith RD, Koo YM, Gu MB, Kim J. Proteomics. 2009;9:1893–1900. doi: 10.1002/pmic.200800591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao LQ, van Langen L, Sheldon RA. Current Opinion in Biotechnology. 2003;14:387–394. doi: 10.1016/s0958-1669(03)00096-x. [DOI] [PubMed] [Google Scholar]
- 27.Kim BC, Nair S, Kim J, Kwak JH, Grate JW, Kim SH, Gu MB. Nanotechnology. 2005;16:S382–S388. doi: 10.1088/0957-4484/16/7/011. [DOI] [PubMed] [Google Scholar]
- 28.Nair S, Kim J, Crawford B, Kim SH. Biomacromolecules. 2007;8:1266–1270. doi: 10.1021/bm061004k. [DOI] [PubMed] [Google Scholar]
- 29.Chin CCQ, Brewer JM, Eckard E, Wold F. J. Biol. Chem. 1981;256:1370–1376. [PubMed] [Google Scholar]
- 30.Chin CCQ, Brewer JM, Wold F. J. Biol. Chem. 1981;256:1377–1384. [PubMed] [Google Scholar]
- 31.Holland MJ, Holland JP, Thill GP, Jackson KA. J. Biol. Chem. 1981;256:1385–1395. [PubMed] [Google Scholar]
- 32.Lebioda L, Stec B, Brewer JM. J. Biol. Chem. 1989;264:3685–3693. doi: 10.2210/pdb2enl/pdb. [DOI] [PubMed] [Google Scholar]
- 33.Brancia FL, Oliver SG, Gaskell SJ. Rapid Commun. Mass Spectrom. 2000;14:2070–2073. doi: 10.1002/1097-0231(20001115)14:21<2070::AID-RCM133>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 34.Olsen JV, Ong SE, Mann M. Mol. Cell. Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Mozhaev V, Melik-Nubarov N, Sergeeva M, ik nis V, Martinek K. Biocatal. Biotransformation. 1990;3:179–187. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.