Abstract
In this report we describe insights into the function of the ribosome tunnel that were obtained through an analysis of an unusual 25 residue N-terminal motif (EspP1-25) associated with the signal peptide of the E. coli EspP protein. It was previously shown that EspP1-25 inhibits signal peptide recognition by the signal recognition particle (SRP), and we now show that fusion of EspP1-25 to a cytoplasmic protein causes it to aggregate. We obtained two lines of evidence that both of these effects are attributable to the conformation of EspP1-25 inside the ribosome tunnel. First, we found that mutations in EspP1-25 that abolished its effects on protein targeting and protein folding altered the crosslinking of short nascent chains to ribosomal components. Second, we found that a mutation in L22 that distorts the tunnel mimicked the effects of the EspP1-25 mutations on protein biogenesis. Our results provide evidence that the conformation of a polypeptide inside the ribosome tunnel can influence protein folding under physiological conditions and suggest that ribosomal mutations might increase the solubility of at least some aggregation-prone proteins produced in E. coli.
Keywords: protein folding, protein targeting, ribosome, signal peptide, translation
INTRODUCTION
One of the most prominent structural features of the ribosome is the long aqueous tunnel in the large subunit that nascent polypeptide chains traverse before emerging into the cytoplasm (Milligan and Unwin, 1986; Yonath et al., 1987). The ribosome tunnel extends ~100 Å from the peptidyltransferase center (PTC) to the exit point on the ribosome surface and has an average diameter of ~15 Å (Nissen et al., 2000). The walls of the tunnel are comprised primarily of 23S rRNA and the non-globular regions of three proteins. Two of the proteins (designated L4 and L22 in bacteria) are located near the tunnel entrance and the third protein, L23, resides near the exit site. The tunnel is irregular in shape and contains numerous grooves and cavities. It is especially narrow about 30 Å from the PTC at a point where a conserved b-hairpin loop of L22 comes into proximity to L4 (“constriction point”). In addition, the electrostatic potential of the tunnel varies considerably along its length (Lu et al., 2007).
As nascent polypeptides emerge from the tunnel they encounter factors that facilitate protein folding and localization. These factors bind to an exposed segment of L23 that is situated adjacent to the tunnel exit site. In bacteria, L23 serves as a docking site for trigger factor (TF), a molecular chaperone that functions cooperatively with DnaK to promote the folding of many different proteins (Deuerling et al., 1999; Teter et al., 1999; Kramer et al., 2002). Eukaryotic cells produce a distinct factor called the nascent chain-associated complex (NAC) that binds to the L23 homolog and that may have an analogous function (Wegryzn et al., 2006). The signal recognition particle (SRP) also binds to L23 (Pool et al., 2002; Ullers et al., 2003; Gu et al., 2003). SRP is a ribonucleoprotein that recognizes the signal peptides of nascent presecretory proteins and targets ribosome-nascent chain complexes (RNCs) to the Sec protein translocation channel in the bacterial inner membrane (IM) or endoplasmic reticulum (ER) cotranslationally (Keenan et al., 2001). Whereas mammalian SRP recognizes most signal peptides, E. coli SRP recognizes only especially hydrophobic signal peptides (Lee and Bernstein, 2001). Consequently, most E. coli presecretory proteins are targeted to the IM post-translationally. During cotranslational translocation, L23 also makes contact with the Sec complex following the dissociation of SRP (Beckmann et al., 2001).
Although the function of the ribosome tunnel has remained enigmatic, its predominantly hydrophilic nature originally led to the proposal that it creates a “Teflon-like” surface that prevents interactions with the diverse array of denatured polypeptides that it encounters(Nissen et al., 2000). In recent years, however, a growing body of evidence has challenged the idea that the tunnel is simply a passive conduit. The detection of specific peptide sequences inside the tunnel of bacterial, fungal and higher eukaryotic ribosomes has been shown to arrest translation at either the elongation or termination stage (Tenson and Ehrenberg, 2002). In addition, the recognition of hydrophobic transmembrane (TM) segments of integral membrane proteins inside the ribosome tunnel stimulates SRP binding in vitro and regulates the gating and structure of the protein translocation machinery in ER vesicles (Liao et al., 1997; Berndt et al., 2009; Pool, 2009). Both types of peptides appear to be detected by ribosomal components that line the tunnel, including the L4 and L22 proteins and segments of 23S RNA, which subsequently transmit a signal to distant segments of the ribosome (Nakatogowa and Ito, 2002; Woolhead et al., 2004; Cruz-Vera et al., 2005; Vasquez-Laslop et al., 2008). The interaction between the ribosome and external factors such as SRP and the Sec complex might be regulated by the transmission of a signal to L23 and possibly L24 and L29, two other proteins that are situated adjacent to the polypeptide exit site (Mitra and Frank, 2006). Molecular dynamics simulations have revealed binding crevices and free-energy barriers inside the tunnel that might be used in the detection of single amino acid side chains (Petrone et al., 2009). Early studies showed that the ribosome tunnel typically protects 30-40 amino acids and implied that these segments are in an extended conformation (Malkin and Rich, 1967; Blobel and Sabatini, 1970). More recent work, however, has provided evidence that at least some polypeptides fold partially in the tunnel and that the recognition of both translation arrest peptides and TM segments involves the formation of α-helical or other compacted structures (Hardesty and Kramer, 2001; Woolhead et al., 2004 and 2006; Lu and Deutsch, 2005; Tu and Deutsch, 2010). Cryo-EM studies have also shown that different polypeptides adopt distinct conformations inside the ribosome tunnel (Becker et al., 2009; Seidelt et al., 2009; Bhushan et al., 2010). Although most of the tunnel is too narrow to accommodate the acquisition of more than secondary structure, minimal tertiary structure can form near the exit site (Kosolapov and Deutsch, 2009).
To date the conformation of polypeptides inside the ribosome tunnel has only been investigated in vitro using static ribosome-nascent chain complexes, and beyond evidence that the conformation of regulatory peptides governs their detection inside the tunnel, it is unclear whether specific conformations adopted by other peptides are biologically significant. That is, it is unclear whether the conformations that have been observed in vitro actually form during active translation in vivo and, if so, whether they persist outside the tunnel long enough to affect the fate of a nascent chain once it enters the cytoplasm. The α-helical conformation of TM segments formed inside the tunnel, for example, has been shown to be unstable in an aqueous environment (Woolhead et al., 2004). In this report we describe an analysis of an unusual bacterial signal peptide that unexpectedly linked the conformation of a peptide inside the ribosome tunnel to events that occur in the cytoplasm in vivo. We previously showed that a 25 residue N-terminal motif (EspP1-25) that is part of the signal peptide of the E. coli O157:H7 autotransporter EspP exerts a dramatic effect on protein targeting by inhibiting SRP binding in vivo (Peterson et al., 2006), and here we show that the attachment of EspP1-25 to a cytoplasmic protein causes it to aggregate. Interestingly, chemical crosslinking experiments indicated that point mutations in EspP1-25 that suppress both of these effects alter the conformation of the peptide as it emerges from the ribosome tunnel. The observation that a mutation in L22 that alters the shape of the tunnel acts in trans to mimic the effect of the EspP1-25 point mutations corroborated the conclusion that the activity of the peptide is dictated by its conformation inside the tunnel. These results not only expand the range of biological activities associated with the ribosome tunnel, but also suggest that under physiological conditions the protein folding process can begin earlier than currently believed.
RESULTS
Mutations in an unusual signal peptide motif (EspP1-25) alter the targeting of a presecretory protein
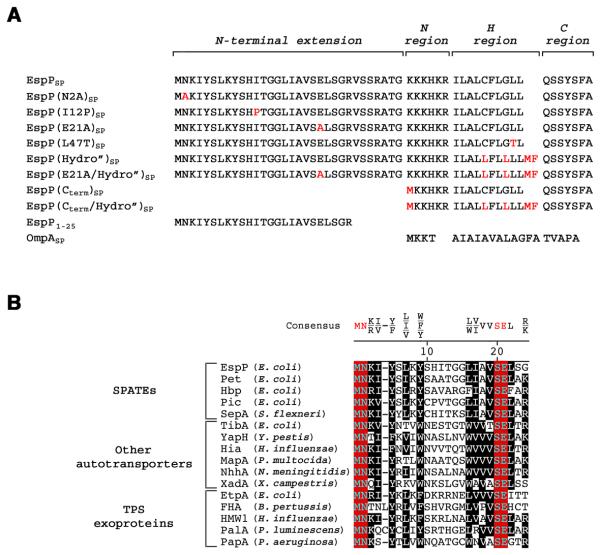
Previous studies have shown that the unusually long (>50 amino acid) signal peptide associated with EspP (EspPSP) and a subset of other bacterial virulence factors of the autotransporter and two-partner secretion (TPS) exoprotein superfamilies consist of two functional domains. The C-terminal ~25 residues contain characteristic N, H and C regions that are the hallmark of most signal peptides (von Heijne, 1985) and are sufficient to target model presecretory proteins to the E. coli Sec complex (Peterson et al., 2003; Desvaux et al., 2007). A signal peptide consisting of the C-terminal domain of EspP [EspP(Cterm)SP; Fig. 1A] routes OmpA, which is normally targeted to the IM post-translationally, into the SRP pathway (Peterson et al., 2003). Presumably because the hydrophobicity of the EspP(Cterm)SP H region is close to the threshold for SRP recognition, proteins containing this signal peptide can also be targeted effectively by SRP-independent mechanisms. Efficient secretion of proteins containing highly hydrophobic derivatives of EspP(Cterm)SP, however, requires SRP. The N-terminal 25 residues of EspPSP (designated EspP1-25) contain a unique sequence motif that is highly conserved among all of the autotransporters and TPS exoproteins that contain long signal peptides (Fig. 1B). Interestingly, the presence of this domain inhibits the interaction between SRP and the full-length signal peptide and even inhibits the interaction between SRP and highly hydrophobic EspPSP derivatives (Peterson et al., 2006). Unlike many other peptides that emerge from the ribosome tunnel, EspP1-25 does not appear to interact with TF (Peterson et al., 2006).
Fig. 1.
The signal peptides of a subset of autotransporters and TPS exoproteins contain a conserved sequence motif. A. The EspP signal peptide (EspPSP) consists of typical N, H and C regions plus a ~30 residue N-terminal extension. The derivatives of EspPSP that were used in this study are shown. The OmpA signal peptide (OmpASP) is shown for comparison. B. The first 24 residues of the N-terminal signal peptide extensions of selected serine protease autotransporters of Enterobacteriaceae (SPATEs), other autotransporters, and TPS exoproteins were aligned using Clustal V. The organism that produces each protein is indicated in parentheses. Invariant residues are shown in red and highly conserved residues are shown in black.
We used an in vivo protein targeting assay to gain insight into the mechanism by which EspP1-25 influences targeting pathway selection. In this assay E. coli were transformed with a plasmid that encodes HA-tagged OmpA containing its native signal peptide or a derivative of the EspP signal peptide. Cells were subjected to pulse-chase labeling, and the conversion of the precursor form of the protein to the mature form was monitored by conducting immunoprecipitations with an anti-HA antiserum. The precursor is not observed during SRP-mediated targeting because RNCs are targeted to the IM at an early stage of translation and signal peptide cleavage occurs before the completion of polypeptide synthesis (Lee and Bernstein, 2001). Conversely, derivatives that are targeted post-translationally are fully synthesized before they reach the IM. For this reason the presence of the precursor is diagnostic of post-translational targeting.
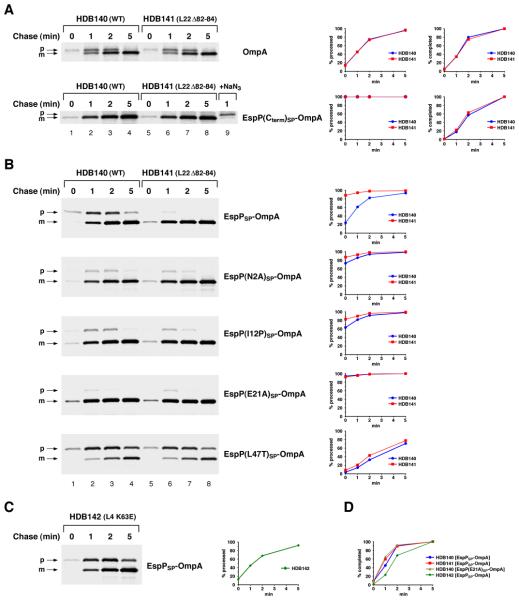
Because protein synthesis in E. coli is very rapid at 37°C and only relatively small amounts of the precursor form of post-translationally targeted proteins are observed, we incubated cells at 20°C to slow translation and to amplify the difference between post-translational and cotranslational targeting. Incubation at this temperature does not appear to affect the translocation of OmpA and its derivatives through the Sec channel (Peterson et al., 2003 and 2006). Translation times can be determined by measuring the time required for the completion of all of the polypeptide chains initiated during a short pulse labeling period (i.e., the time required for maximal incorporation of radioactivity into the protein) (Farewell and Neidhardt, 1998). When the wild-type strain HDB140 was pulse-labeled for 30 sec at 20°C, the translation time of the 346 residue OmpA precursor was slightly greater than 2 min (Fig. 2A, top panel, lanes 1-4). In agreement with a previous estimate, this time corresponds to an elongation rate of ~2.5-3 residues/sec (Farewell and Neidhardt, 1998). The observation that the conversion of the OmpA precursor to mature OmpA paralleled the completion of polypeptide synthesis confirmed that the protein was targeted to the IM post-translationally. In contrast, no EspP(Cterm)SP-OmpA precursor was observed at any time point (Fig. 2A, second panel, lanes 1-4), and therefore the protein must have reached the IM at an early stage of translation. Consistent with previous results, the presence of EspP1-25 radically slowed protein targeting, and EspPSP-OmpA was exported at essentially the same rate as wild-type OmpA (Fig. 2B, top panel, lanes 1-4 and blue circles). The finding that the EspPSP-OmpA precursor is located in the cytoplasm formally demonstrated that protein is targeted to the IM post-translationally (Peterson et al., 2006).
Fig. 2.
Mutations in EspP1-25 or L22 suppress the effect of EspP1-25 on protein targeting. A. HDB140 (wild-type) and HDB141 (L22 Δ82-84) cells transformed with a plasmid encoding HA-tagged OmpA or EspP(Cterm)SP-OmpA were grown in M9 at 37°C. Cultures were shifted to 20°C, IPTG was added to induce the expression of the plasmid-borne gene, and cells were subjected to pulse-chase labeling. Sodium azide (NaN3) was added to a portion of one culture to block secretion and to show the position of the precursor. The precursor (p) and mature (m) forms of the OmpA derivatives were immunoprecipitated with an anti-HA antiserum. The percent of the precursor processed to the mature form and the percent of polypeptide chains completed at each time point is shown. B. As in part A, except that HDB140 and HDB141 were transformed with a plasmid encoding EspPSP-OmpA or the indicated derivative. C. As in part A, except that HDB142 (L4 K63E) were transformed with a plasmid encoding EspPSP-OmpA. D. The fraction of EspPSP-OmpA or EspP(E21A)SP-OmpA completed in the indicated strain at each time point is shown.
The striking conservation of the N-terminal domain of autotransporter signal peptides suggests that the sequence is functionally significant. To test this idea, we mutated two of the three invariant residues (N2 and E21) to alanine. Based on the conjecture that the folding of EspP1-25 is important for its function, we also mutated a residue in the non-conserved middle region of the motif (I12) to proline, an amino acid that imposes conformational rigidity on the polypeptide backbone. Consistent with our hypothesis, all of the mutations accelerated the export of EspPSP-OmpA in HDB140 and largely negated the effect of EspP1-25 on targeting (Fig. 2B, panels 2-4, lanes 1-4 and blue circles). In contrast, a variant of EspPSP-OmpA containing a mutation in the C-terminal domain of the signal peptide that reduces hydrophobicity (L47T) and abolishes SRP recognition (Peterson et al., 2003) was still targeted to the IM post-translationally (Fig. 2B, bottom panel, lanes 1-4 and blue circles).
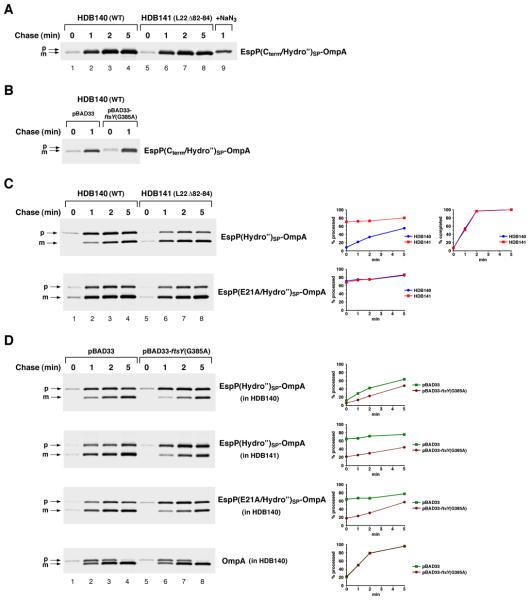
Although our results suggested that mutations that suppress the function of EspP1-25 restore SRP binding, it was necessary to increase the hydrophobicity of the signal peptide to test this hypothesis. EspPSP contains the same hydrophobic core as EspP(Cterm)SP, and EspPSP derivatives that are targeted cotranslationally are presumably likewise recognized by SRP. Like proteins that contain EspP(Cterm)SP, however, proteins such as EspP(E21A)SP-OmpA can be targeted effectively by SRP-independent mechanisms when the SRP pathway is inactivated (data not shown). As expected, OmpA containing the highly hydrophobic EspP(Cterm /Hydro”)SP signal peptide (Fig. 1A) was targeted to the IM cotranslationally in HDB140 (Fig. 3A, lanes 1-4). When cells were transformed with a plasmid that harbors a dominant-lethal SRP receptor mutation [ftsY(G385A); see Ulbrandt et al., 1997] under the control of the araBAD promoter and the SRP pathway was blocked by adding arabinose, the export of EspP(Cterm /Hydro”)SP-OmpA was impaired (Fig. 3B). In contrast, inactivation of ftsY did not affect the export of OmpA (Fig. 3D, bottom panel). Consistent with the observation that EspP1-25 prevents SRP from effectively recognizing even very hydrophobic signal peptides (Peterson et al., 2006), EspP(Hydro”)SP–OmpA was targeted to the IM predominantly post-translationally and inhibition of the SRP pathway only slightly impaired its export (Fig. 3C, top panel, lanes 1-4; Fig. 3D, top panel). Possibly as a result of aggregation, about half of the EspP(Hydro”)SP-OmpA protein remained in the cytoplasm even after a 5 min chase. Interestingly, the introduction of the E21A mutation into EspP(Hydro”)SP greatly accelerated OmpA export, and about 70% of the protein was exported after a 0 min chase (Fig. 3C, bottom panel, lanes 1-4 and blue circles). The observation that inactivation of ftsY caused EspP(E21A/Hydro”)SP-OmpA to be targeted with the same slow kinetics as EspP(Hydro”)SP-OmpA confirmed that the mutant signal peptide was recognized by SRP. Taken together, these results demonstrate the conserved elements of EspP1-25 create a functional unit that inhibits SRP binding.
Fig. 3.
Mutations in EspP1-25 or L22 restore SRP recognition of a highly hydrophobic signal peptide. A. HDB140 and HDB141 cells transformed with plasmid encoding HA-tagged EspP(Cterm/Hydro”)SP-OmpA were grown and treated as described in the legend to Fig. 2A. The precursor (p) and mature (m) forms of the OmpA derivative were immunoprecipitated with an anti-HA antiserum. B. As in part A, except that HDB140 harbored a second plasmid [either pBAD33 or pBAD33-ftsY(G385A)]. Arabinose was added to both cultures before they were shifted to 20°C. C. As in part A, except that HDB140 and HDB141 were transformed with a plasmid encoding EspP(Hydro”)SP-OmpA or EspP(E21A/Hydro”)SP-OmpA. The percent of the precursor processed to the mature form and the percent of EspP(Hydro”)SP-OmpA completed at each time point is shown. D. As in part B, except that HDB140 or HDB141 harbored a plasmid encoding the indicated OmpA derivative.
A specific mutation in the ribosome tunnel mimics the effect of EspP1-25 mutations on protein targeting
There are three possible mechanisms by which EspP1-25 might inhibit the binding of SRP to the EspPSP core. First, this segment might encode the binding site for a trans-acting factor that competes with SRP. To date, however, we have not been able to identify a cytoplasmic factor that recognizes EspP1-25 in crosslinking experiments (data not shown). In addition, we previously showed that the kinetics of EspPSP-OmpA targeting is independent of its level of expression (Peterson et al., 2006), and we have not been able to titrate out a hypothetical trans-acting factor that blocks the access of SRP to the EspPSP core in competition experiments (Fig. S1). Second, EspP1-25 might adopt a conformation either during or immediately after its transit through the ribosome tunnel that sterically hinders the binding of SRP to the signal peptide. Third, EspP1-25 might function as a signaling peptide. In this scenario the recognition of EspP1-25 by L23 and/or other components of the ribosome tunnel would transmit a signal to the surface of the ribosome that alters the interaction of SRP with L23.
An examination of the secretion kinetics of EspPSP-OmpA in strains that harbor ribosomal tunnel mutations provided strong evidence that EspP1-25 exerts its effect on protein targeting by adopting a specific conformation inside the tunnel. We hypothesized that if EspP1-25 begins to fold into a conformation that affects SRP binding before emerging into the cytoplasmic milieu, then ribosomal mutations that prevent folding by modifying the tunnel environment should restore cotranslational targeting. Consistent with our prediction, we found that EspPSP-OmpA [as well as EspP(N2A)SP-OmpA, EspP(I12P)SP-OmpA and EspP(E21A)SP-OmpA] was targeted to the IM in a predominantly cotranslational fashion in HDB141, a strain that harbors a deletion (Δ82-84) in the L22 β hairpin loop (Fig. 2B, panels 1-4, lanes 5-8, and red squares). The L22 Δ82-84 mutation has been shown to increase the diameter of the tunnel and to impair the recognition of translation arrest motifs (Gabashvili et al., 2001; Cruz-Vera et al., 2005; Woolhead et al., 2006; Vasquez-Laslop et al., 2008; Yap and Bernstein, 2009). In contrast, the L22 Δ82-84 mutation had no effect on the export of the post-translationally targeted proteins OmpA and EspP(L47T)SP-OmpA whose signal peptides are bypassed by SRP (Fig. 2A, lanes 5-8 and Fig. 2B, bottom panel, lanes 5-8). The observation that a mutation in L4 that narrows the tunnel (K63E) (Gabashvili et al., 2001) did not reroute EspPSP-OmpA into a cotranslational targeting pathway showed that the effect of the L22 Δ82-84 mutation was specific (Fig. 2C).
To show that the L22 Δ82-84 mutation restores the cotranslational targeting of proteins containing the EspP1-25 sequence motif by rerouting them into the SRP pathway, we examined the export of EspP(Hydro”)SP–OmpA in HDB141. Consistent with the results described above, EspP(Hydro”)SP–OmpA was processed much more rapidly in HDB141 than in wild-type cells (Fig. 3C, top panel, lanes 5-8 and red squares). As expected, inactivating the SRP pathway by inducing the expression of ftsY(G385A) slowed both the rate and efficiency of export considerably (Fig. 3D, second panel). Interestingly, the introduction of the L22 Δ82-84 mutation into the ribosome produced the same effect on targeting as the introduction of the E21A mutation into EspP(Hydro”)SP. While each mutation led to the export of ~70-80% of the presecretory protein at all time points, combining the two mutations did not produce a synergistic effect that increases secretion efficiency (Fig. 3C, compare the first panel, lanes 5-8 and the second panel). These results provided the first indication that the L22 mutation mimics the effect of mutations in EspP1-25 and might therefore exert the same effect on peptide folding.
Several control experiments confirmed that the L22 Δ82-84 mutation influences the targeting of EspPSP-OmpA by altering the tunnel environment rather than by altering ribosome structure or function. A previous study showed that unlike the L4 K63E mutation, which substantially impairs multiple ribosome functions, the L22 Δ82-84 mutation produces only minor effects on translation (O'Connor et al., 2004). Consistent with the results of this study, we found that only the L4 K63E mutation clearly slowed the elongation of OmpA derivatives (Fig. 2D, green diamonds; see also Figs. 2A and 3C). Furthermore, slowing translation elongation by treating cells with low to moderate amounts of chloramphenicol, spectinomycin or fusidic acid, drugs that act at different stages of the elongation cycle, did not alter the mode of EspPSP-OmpA targeting in either HDB140 or HDB141 (Fig. S2). We also obtained evidence that the L22 Δ82-84 mutation did not reroute EspP(Hydro”)SP–OmpA into the SRP pathway by enhancing the binding of SRP to the ribosome. On the contrary, experiments in which the amount of SRP associated with free ribosomes was analyzed indicated that the mutation slightly reduces the affinity of SRP for ribosomes (Fig. S3). Finally, the observation that the L22 Δ82-84 mutation produced an identical effect on targeting in an entirely different strain background (MC4100) ruled out the possibility that our results were due to a cryptic second mutation (data not shown).
To test the possibility that the conformation of EspP1-25 in the ribosome tunnel generates a signal that inhibits the binding of SRP to L23, we examined the effect of deleting the loop of L23 that is situated inside the tunnel on the targeting of EspPSP–OmpA and EspP(Hydro”)SP–OmpA. Indeed there is evidence that the presence of at least some nascent chains inside the tunnel can influence SRP binding by communicating through the L23 loop (Bornemann et al., 2008). If this hypothesis were correct, we would expect that deletion of the loop would restore cotranslational targeting. We found, however, that both proteins were targeted to the IM in a predominantly post-translational fashion in a strain containing the L23 Δ65-74 mutation and that, if anything, EspPSP–OmpA was targeted more slowly in the mutant strain than in the wild-type strain (Fig. S4). These results strongly suggest that EspP1-25 adopts a conformation inside the tunnel that directly affects the interaction of SRP with EspPSP once it emerges in the cytoplasm.
Mutations in both EspP1-25 and L22 suppress the aggregation of a cytoplasmic protein containing EspP1-25
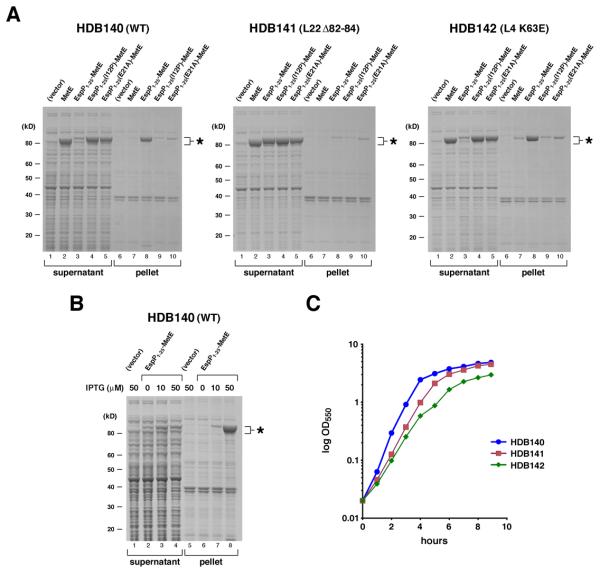
Because the experiments described above suggested that EspP1-25 adopts a conformation in the ribosome tunnel that affects the structure of at least the N-terminus of a presecretory protein, we conjectured that the presence of this segment might also affect the folding of a cytoplasmic protein. To test this idea we fused EspP1-25 to MetE, a protein that aggregates when it misfolds (Deuerling et al., 1999). HDB140 was transformed with a plasmid that encodes espP1-25-metE under the control of the trc promoter, and the synthesis of the fusion protein was induced by the addition of IPTG. Cells were lysed by sonication and aggregated proteins were isolated by high-speed centrifugation. Consistent with our hypothesis, almost all of the EspP1-25-MetE was found in the high-speed pellet when cultures were supplemented with 50 μM IPTG and the protein was produced at a substantial level (Fig. 4A, left panel, lane 8). Like typical aggregated proteins, the pelleted EspP1-25-MetE was insoluble in non-ionic detergents (data not shown). Interestingly, EspP1-25-MetE aggregation was concentration-dependent and was mitigated by the addition of lower concentrations of inducer (Fig. 4B). The results indicate that the presence of EspP1-25 does not inevitably cause MetE to misfold, but rather causes the protein to pass through an intermediate stage of folding in which it undergoes conditional self-association like proteins that form inclusion bodies (Speed et al., 1996).
Fig. 4.
Mutations in EspP1-25 or L22 suppress the effect of EspP1-25 on protein folding. A. HDB140, HDB141 and HDB142 transformed with pTRC99a (vector) or pTRC99a encoding MetE, EspP1-25-MetE, or a derivative of EspP1-25-MetE were grown in LB at 37°C. Expression of the plasmid-borne gene was induced by the addition of 50 μM IPTG, and cell lysates were subjected to high-speed centrifugation. Proteins present in the supernatant and pellet fractions were resolved by SDS-PAGE and visualized by Coomassie Blue staining. MetE-containing polypeptides are indicated with an asterisk. B. HDB140 transformed with pTRC99a or pTRC99a encoding EspP1-25-MetE were grown in LB, and expression of the plasmid-borne gene was induced by the addition of the indicated amount of IPTG. Cell lysates were processed as in part A. (C) The growth of HDB140, HDB141 and HDB142 harboring pTRC99a is shown.
Further analysis provided strong evidence that EspP1-25 adopted a sequence-dependent conformation in the ribosome tunnel that ultimately resulted in EspP1-25-MetE aggregation. Introduction of the I12P and E21A mutations into the fusion protein almost completely suppressed misfolding; only residual amounts of the protein were isolated in the high-speed pellets (Fig. 4A, left panel, lanes 4-5 and 9-10). Remarkably, the L22 Δ82-84 mutation had the same effect on EspP1-25-MetE folding as the point mutations in EspP1-25 (Fig. 4A, middle panel). Like the EspP1-25(I12P) and EspP1-25(E21A) mutations, the ribosomal mutation greatly increased the solubility of EspP1-25-MetE without altering the amount protein that was synthesized (Fig. S5). As in the protein targeting experiments, the mutations in EspP1-25-MetE and L22 did not act synergistically: the same amount of residual aggregated protein was observed when EspP1-25(I12P)-MetE and EspP1-25(E21A)-MetE were produced in either HDB140 or HDB141 (Fig. 4A, compare lanes 9-10 in the left and middle panels). These results provide additional evidence that the I12P and E21A mutations and the ribosomal mutation both affect the conformation of EspP1-25 in the same fashion. As in wild-type cells, almost all of the EspP1-25-MetE that was synthesized in a strain that harbors the L4 K63E mutation aggregated (Fig. 4A, right panel). The L4 mutation is more deleterious than the L22 mutation and causes a more profound growth defect (Fig. 4C), and it is therefore unlikely that the altered folding of EspP1-25-MetE in HDB141 was simply a secondary effect of a reduction in translational activity or growth rate.
Because TF is involved in the folding of MetE and the protein aggregates when TF and DnaK are depleted simultaneously (Deuerling et al., 1999), we considered the possibility that EspP1-25-MetE aggregation results from a failure of the protein to interact with TF. In one scenario, EspP1-25 would transmit a signal (that is disrupted by either cis-acting point mutations or the trans-acting L22 mutation) that leads to the dissociation of TF from ribosomes. In another scenario, EspP1-25 would direct the protein to one of the “alternate” ribosome tunnels whose existence was suggested by cryo-EM studies (Gabashvili et al., 2001) and the nascent chain would never encounter TF. In either case EspP1-25-MetE might aggregate when it is overproduced because the excess protein overwhelms the folding capacity of DnaK. A corollary of this hypothesis is that DnaK would become essential for EspP1-25-MetE folding even when the protein is produced at a low level. The overexpression of EspP1-25-MetE, however, did not cause a heat shock response and the concomitant increase in the concentration of DnaK that is typically observed when DnaK is overburdened (Fig. S6). Furthermore, the elimination of DnaK had no effect on EspP1-25-MetE solubility (Fig. S7A). Finally, although EspP1-25-MetE should aggregate in the absence of TF if the L22 Δ82-84 mutation suppresses TF dissociation, disruption of the tig gene in the L22 mutant strain did not significantly reduce the solubility of the protein (Fig. S7B). Taken together, these results provide additional evidence that EspP1-25 does not influence the interaction between the nascent chain and external factors by transmitting a signal or altering the path of the nascent chain.
The conformation of wild-type and mutant EspP1-25 peptides differs as they emerge from the ribosome tunnel
We next conducted chemical crosslinking experiments to obtain more direct information about the conformation of wild-type and mutant EspP1-25 peptides inside the ribosome tunnel. Chemical crosslinkers do not penetrate deep inside the E. coli ribosome tunnel and are useful primarily to probe interactions close to the polypeptide exit site and on the surface of the ribosome (Woolhead et al., 2006). In our experiments we synthesized nascent chains of varying lengths that had reactive lysine residues at fixed positions. Using this approach we were able to monitor polypeptide conformation not only by identifying ribosomal proteins that are in proximity to the nascent chain as it grows, but also by determining the number of residues that must be synthesized before the lysine residues become accessible to the crosslinking reagent.
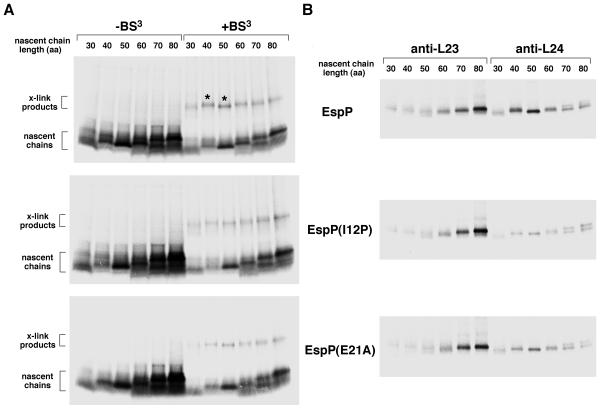
Initially we synthesized EspP, EspP(I12P), and EspP(E21A) 30-80 residue nascent chains in coupled in vitro transcription-translation reactions. The lysines in the N region of the signal peptide core (residues 33, 34, 35 and 37) were mutated to glutamine, so each nascent chain contained only two lysines at positions 3 and 8 (Fig. 1). Unlike mutations in EspP1-25, these mutations do not affect targeting pathway selection (Peterson et al., 2006). One portion of each reaction was untreated and an equal portion was subjected to crosslinking using the lysine-specific reagent BS3. The remainder of each sample was used for immunoprecipitations. Crosslinked products of ~16-20 kD were observed in all of the reactions and were particularly prominent in reactions in which EspP 40 and 50 residue nascent chains were synthesized (Fig. 5A; see asterisks in top panel). The products were only observed when a DNA template was added to the reactions (Fig. S8). The size of these products suggested that the nascent chains were crosslinked to a ~10-12 kD protein. Preliminary immunoprecipitations using antisera against L23, L24 and L29 and the heat shock protein hsp15, any of which might bind polypeptides as they exit the tunnel, revealed that EspP 50 residue nascent chains were crosslinked predominantly to L24 (data not shown). This result was surprising because only L23 and L29 were previously identified as nascent chain crosslinking partners (Ullers et al., 2003; Eisner et al., 2003 and 2006). Immunoprecipitations using portions of the reactions shown in Fig. 5A with anti-L23 and L24 antisera confirmed that the prominent bands resulted from the crosslinking of EspP 40 and 50 residue nascent chains to L24 (Fig. 5B, top panel). In contrast, we did not observe significant crosslinking between any of the EspP(I12P) nascent chains and L24, and EspP(E21A) 40 and 50 residue nascent chains were crosslinked to L24 with a much lower efficiency than the equivalent EspP nascent chains (Fig. 5B, middle and bottom panels). We also did not see significant crosslinking between EspP and L24 when we used an extract derived from an L22 Δ82-84 strain, but the data are difficult to evaluate partly because the translation efficiency was reduced (data not shown). Furthermore, we found that EspP, EspP(I12P), and EspP(E21A) 70 and 80 residue nascent chains were crosslinked to L23 with equal efficiency. This observation is consistent with previous results showing a persistent interaction between L23 and relatively long nascent chains (Eisner et al., 2003 and 2006). Taken together, the results indicate that when wild-type EspP reaches a length of ~40-50 residues, amino acids that are ~35-45 residues away from the PTC are uniquely situated close to L24.
Fig. 5.
Crosslinking of EspP, EspP(I12P) and EspP(E21A) 30-80 residue nascent chains to ribosomal proteins. A. N-terminal fragments containing the indicated number of amino acids (aa) of EspP, EspP(I12P) and EspP(E21A) were synthesized in coupled transcription-translation reactions and radiolabeled. Equal portions of each reaction that were untreated or treated with BS3 were resolved by SDS-PAGE. Prominent crosslinking products are denoted with an asterisk. B. Portions of the reactions shown in part A that were treated with BS3 were subjected to immunoprecipitation with anti-L23 and anti-L24 antisera.
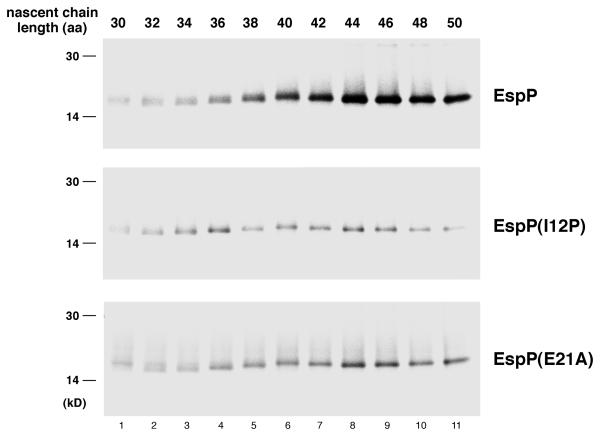
To obtain a more detailed picture of the disposition of the EspP polypeptide as it emerges from the ribosome tunnel, we repeated the crosslinking experiments described above except that we examined the nascent chain as it increased in size from 30 to 50 amino acids in two residue increments. Following the addition of BS3, translation products were subjected to immunoprecipitation with anti-L24 antiserum. Only a very low background signal was detected when either wild-type or mutant nascent chains were 30-34 residues in length and the lysine residues were protected inside the ribosome tunnel (Fig. 6, lanes 1-3). Crosslinking of wild-type EspP to L24 clearly increased when the nascent chain attained a length of 38 residues (at which point lysine 3 would be expected to just reach the surface of the ribosome if the nascent chain was in a relatively extended conformation) and ultimately peaked when the length reached 44-46 residues (Fig. 6, top panel). These results imply that at least one of the two lysine residues encounters L24 immediately upon exiting from the tunnel. In contrast, the crosslinking of EspP(I12P) nascent chains to L24 never significantly increased above the background level and only modest crosslinking of EspP(E21A) nascent chains was observed (Fig. 6, bottom two panels). These results strongly suggest that two mutations that compromise the effect of the EspP1-25 peptide on protein targeting and protein folding alter the conformation of the peptide as it emerges from the ribosome tunnel so that the lysines at positions 3 and 8 are oriented away from L24.
Fig. 6.
Differential crosslinking of EspP, EspP(I12P) and EspP(E21A) 30-50 residue nascent chains to L24. N-terminal fragments containing the indicated number of amino acids (aa) of EspP, EspP(I12P) and EspP(E21A) were synthesized in coupled transcription-translation reactions and radiolabeled. Equal portions of each reaction that were treated with BS3 were subjected to immunoprecipitation with an anti-L24 antisera.
Finally, we considered the possibility that the EspP nascent chain was not crosslinked to L24 because it adopted a unique conformation, but rather because a signaling process initiated by the recognition of EspP1-25 inside the ribosome tunnel led to the reorientation of the flexible L24 β hairpin loop. Indeed structural studies have suggested that the position of this loop could regulate the binding of external factors such as TF (Schlünzen et al., 2005). We found, however, that deletion of the loop had no effect on either the targeting of EspPSP-OmpA or the aggregation of EspP1-25-MetE (Fig. S9). While we cannot exclude the possibility that the L24 β hairpin loop reorients in response to the recognition of EspP1-25 inside the tunnel, these results imply that EspP1-25 does not influence protein biogenesis through the movement of the loop.
DISCUSSION
In this study we describe evidence that non-regulatory peptides can adopt specific conformations in the ribosome tunnel under physiological conditions. Furthermore, we demonstrate that those conformations are biologically significant in that they can affect the fate of a polypeptide after it leaves the tunnel. We originally wished to elucidate the mechanism by which an unusual signal peptide motif dramatically affects targeting pathway selection in E. coli. Mutations that alter the hydrophobicity of the H region of a bacterial signal peptide have been shown to reroute presecretory proteins from one targeting pathway to another, and the myristoylation of an ER targeting signal has been shown to inhibit SRP binding in mammalian cells (Lee and Bernstein, 2001; Colombo et al., 2005). The inhibition of SRP binding to even extremely hydrophobic H regions by an independent 25 residue signal peptide domain, however, is unprecedented. Because our results disfavored the possibility that EspP1-25 encodes a binding site for a cytoplasmic factor, we focused on the hypothesis that the activity of the peptide is attributable to its structure. The observation that the mutation of key residues in EspP1-25 suppressed its effects on the targeting of a presecretory protein as well as the folding a cytoplasmic protein to which it was fused was consistent with this hypothesis. The finding that an L22 mutation mimicked the point mutations (i.e., it suppressed the effects of EspP1-25 to the same degree and did not act synergistically with the point mutations) strongly suggested that folding of the peptide was initiated or at least influenced by its passage through the tunnel. The notion that EspP1-25 adopts a specific conformation inside the tunnel was corroborated by crosslinking experiments that provided direct evidence that wild-type and mutant peptides have distinct orientations as they emerge into the cytoplasm. Finally, several observations on the status of SRP and TF in wild-type and L22 mutant strains supported the argument that the effect of EspP1-25 on protein biogenesis was a direct consequence of its conformation rather than an indirect effect of a signaling process that leads to the dissociation of these factors from L23.
Our data challenge conventional wisdom by implying that the protein folding process can begin before a polypeptide emerges into the cytoplasm. It is important to emphasize, however, that the “folding” that occurs inside the ribosome tunnel may be extremely limited. Based solely on space constraints, it is difficult to imagine that EspP1-25 could acquire significant tertiary structure inside the tunnel. On the contrary, the observation that the crosslinking of EspP to L24 could be observed when the nascent chain reached a length of only ~38 residues (at which point the lysine at position 3 would be 35 residues from the PTC) suggests that the polypeptide is relatively extended. Consistent with this possibility, secondary structure predictions indicate that EspP1-25 has a high probability of forming two β strands connected by a loop and a very low probability of forming an α helix. Indeed EspP1-25 may have little or no secondary structure and may resemble the modestly kinked peptides observed inside the tunnel in recent cryo-EM studies (Becker et al., 2009; Seidelt et al., 2009). It is conceivable that EspP1-25 simply adopts a conformation inside the tunnel that biases the folding of the polypeptide once it emerges into the cytoplasm. This conformation might influence either short-range interactions that enable EspP1-25 to fold into a discrete domain or the interaction of EspP1-25 with more distant segments of the protein. Furthermore, the conformation of EspP1-25 inside the tunnel may not persist in the final folded state, but rather may be very transient and may only influence the range of folding intermediates a protein passes through. In any case, our results imply that as a consequence of passing through the ribosome tunnel EspP1-25 forms a structure that persists in the cytoplasm long enough to influence both the binding of external factors (like SRP) and protein folding inside a living cell.
It is intriguing that the L22 Δ82-84 mutation strongly influenced the biogenesis of proteins containing the EspP1-25 peptide whereas a second tunnel mutation that has a more deleterious effect on ribosome function did not. While the L22 mutation has not been reported to affect events outside the ribosome tunnel, it has been shown to perturb several different translation arrest phenomena and to alter the conformation of the SecM arrest peptide (Woolhead et al., 2006). One possible explanation of the results is that the L22 β hairpin loop plays a general and especially important role in determining the conformation of nascent chains inside the tunnel. L22 residues 83 and 84 are both basic and reside in a segment of the loop that extends below the constriction point, and it is conceivable that one of them forms a salt bridge with residue E21 in EspP1-25 that strongly affects the conformation of the peptide. Because the L22 Δ82-84 mutation impairs the function of a very short SecM arrest motif that does not reach the constriction point (Yap and Bernstein, 2009), however, it seems more likely that the mutation exerts a broad effect on the tunnel environment. Perhaps by widening the tunnel the mutation inhibits interactions between nascent chains and the tunnel walls that occur beyond the constriction point. Presumably the narrowing of the tunnel associated with the L4 K63E mutation, in contrast, is compatible with these interactions. It is also noteworthy that prolines are conspicuously absent from autotransporter and TPS exoprotein signal peptide extensions (Fig. 1B; Hiss and Schneider, 2009), and the introduction of a proline residue at a non-conserved position in the middle of EspP1-25 strongly affected both the function and conformation of the peptide inside the ribosome tunnel. Proline substitutions at non-critical positions in the middle of the E. coli SecM arrest motif likewise greatly reduce translation arrest activity, but conversely, prolines are essential for the activity of other SecM arrest motifs (Woolhead et al., 2006; Yap and Bernstein, 2009). The unique significance of proline residues in all of these contexts suggests that even a single constraint on the flexibility of the polypeptide backbone can strongly affect the formation of functional conformations inside the ribosome tunnel.
While our study expands the range of activities associated with the ribosome tunnel, the frequency with which the fate of a polypeptide is affected by its transit through the tunnel remains to be determined. Although the L22 Δ82-84 mutation profoundly affects the biogenesis of proteins that contain EspP1-25, E. coli strains that harbor the mutation are viable. Thus, any defects in protein folding or localization caused by the mutation are presumably restricted to a subset of cellular proteins. Beyond its effect on protein targeting, EspP1-25 appears to prevent the misfolding of EspP in the periplasm by mediating an unusual interaction with the SecYEG complex that delays signal peptide cleavage and transiently anchors the protein to the IM (Szabady et al., 2005). Thus the peptide has a highly specialized function that may impose constraints on its conformation at a very early stage of protein synthesis that do not apply to the vast majority of peptides that emerge from the ribosome tunnel. In any case, the observation that the L22 Δ82-84 mutation suppresses the aggregation of overexpressed EspP1-25-MetE raises the possibility that tunnel mutations may reduce the tendency of at least some recombinant proteins to form inclusion bodies. The overproduction of molecular chaperones has long been the most commonly used approach to increase the yield of recombinant proteins produced in E. coli (Georgiou and Valax, 1996), but has not always been successful. Our work suggests that modification of the ribosome tunnel might in some cases offer a useful alternative to this strategy.
EXPERIMENTAL PROCEDURES
Bacterial strains
The strains used for in vivo experiments are derivatives of N281 (Hfr relA1 spoT1 metB1 rplV281) and N282 (Hfr relA1 spoT1 metB1 rplD282) (Apirion, 1967) and include HDB140 [N281 rplV+ ΔlacZYA metB+ rpsL150(strR)] HDB141 [N281 ΔlacZYA metB+ rpsL150(strR)] and HDB142 [N282 ΔlacZYA metB+ rpsL150(strR)]. C41 (Miroux and Walker, 1996) was used for in vitro translations.
Plasmid construction
Plasmids pHL36 (pTRC99a-ompA-HA), pJH50 (pTRC99a-espPSP-ompA-HA), pJH51 [pTRC99a-espP(Cterm)SP-ompA-HA], pJH84 [pTRC99a-espP(Hydro”)SP-ompA-HA) and pJH88 [pTRC99a-espP(-6)SP-espP] have been described (Lee and Bernstein, 2002; Peterson et al., 2003 and 2006). pJH94 [pTRC99a-espP(Cterm/Hydro”)SP-ompA-HA] was made by site-directed mutagenesis of pJH51. pBAD33-ftsY(G385A) was constructed by subcloning an Eco RI-Hind III fragment from pTRC-ftsY(G385A) (Ulbrandt et al., 1997) into pBAD33 (ATCC). pJH96 (pTRC99a-metE) was constructed by first amplifying metE by PCR using the oligonucleotides 5′-CATATAATTAGAGGAAGAACATATGACAATATTGAATCAC-3′ and 5′-GCGTATGCTGGAAAGCTTTAAGCAGTATGG-3′ (MetERev) and E. coli genomic DNA from strain LE392 as a template. The resulting DNA product was then digested with Nde I and Hind III and cloned into pRLS6 (Szabady et al., 2005). To generate pJH99 (pTRC99a-espP1-25-metE), metE was amplified by PCR using the oligonucleotides 5′-GAGGAAGAAAAAACGGCCGTATTGAATCACACCC-3′ and MetERev, and a second Eag I site was introduced into pJH50 using the oligonucleotide 5′-CGCTGTTTCTGAATTATCCGGCAGAGCGGCCGCAAGAGCAACTGG-3′ and its complement to make pJH98. Both the metE PCR product and pJH98 were then digested with Eag I and Hind III and ligated together. All mutagenesis reactions (including the introduction of the EspP N2A, I12P, E21A and L47T mutations into the above plasmids) were performed using the QuikChange mutagenesis kit (Stratagene)
Protein targeting assays
Cells transformed with appropriate plasmids were grown at 37°C in M9 containing 0.2% glycerol and all of the L-amino acids except methionine and cysteine. Cultures were supplemented with ampicillin (100 μg/ml) and chloramphenicol (40 μg/ml) as needed. Overnight cultures were washed and diluted into fresh medium at OD550=0.025. In experiments that involved expression of ftsY (G385A), 0.2% arabinose was added when cultures reached OD550=0.15. All cultures were shifted to 20°C when they reached OD550=0.2, and IPTG (50 μM) was added 40 min later. After another 20 min cells were subjected to pulse-chase labeling and TCA precipitation, and immunoprecipitations were conducted using an anti-HA antiserum (Santa Cruz Biotechnology) essentially as described (Ulbrandt et al., 1997). Immunoprecipitated proteins were resolved by SDS-PAGE on 8-16% minigels (Invitrogen).
Protein aggregation assays
Cells transformed with appropriate plasmids were grown at 37°C in LB containing 100 μg/ml ampicillin. Overnight cultures were washed and diluted into fresh medium at OD550=0.02. When cultures reached OD550=0.4, IPTG (50 μM, unless otherwise noted) was added. After 30 min cells were poured over ice and pelleted (2500 × g, 10 min, 4°C). Cells were then resuspended in PBS at 5 OD/ml and sonicated. Unbroken cells were removed (2500 × g, 10 min, 4°C) and the cell lysates were centrifuged (100,000 × g, 30 min, 4°C) in a Beckman TLA-120.2 rotor. Proteins in both the pellet and supernatant were TCA precipitated and resolved by SDS-PAGE on 4-12% NuPage minigels (Invitrogen).
Chemical crosslinking
S-30 extracts were made from strain C41 essentially as described (Woolhead et al., 2006). Linear DNA fragments were amplified using the Ex Taq PCR kit (Takara) and pJH88 (or a derivative containing the EspP I12P or E21A mutation) as a template. In these reactions the 5′ primer (5′-CTGAAATGAGCTGTTGACAATTAATCATCCGG-3′) was situated upstream of the trc promoter. The 3′ primer was chosen to amplify an EspP fragment of a desired length, but the last two residues were changed to methionine to increase the radioactive signal. Coupled transcription-translation reactions (100 μl) were conducted essentially as described and chilled on ice (Woolhead et al., 2006). A portion of each reaction (7 μl) was overlayed onto 50 μl RNC buffer (20 mM Hepes pH 7.5/14 mM Mg(OAc)2/100 mM KOAc) containing 0.5 M sucrose (tube A) while the remainder was overlayed onto 100 μl of the same solution (tube B). Samples were then centrifuged in a Beckman TLA-100 rotor (436,000 × g, 6 min, 4°C). Pellet A was resuspended in 8 μl RNC buffer, treated with 100 μg/ml RNAse A/5 mM EDTA (26°C, 10 min), and heated to 85°C in Tricine SDS-PAGE sample buffer (“-BS3 sample”). Pellet B was resuspended in 88 μl RNC buffer containing 1 mM BS3 (Pierce) and incubated on ice for 2 h. The crosslinking reactions were quenched by the addition of 5 μl 1M Tris pH 8.0 and incubated at 20°C for 15 min. Subsequently 7 μl was removed and treated with RNAse A/EDTA and heated as described above (“+BS3 sample”). The remainder of each sample was TCA precipitated and used for immunoprecipitations with polyclonal rabbit antisera generated against the L23 and L24 peptides NH2-MIREERLLKVLRAPHVSEKAC-COOH and NH2-MAAKIRRDDEVIVLTGKDKGC-COOH. All samples were resolved by SDS-PAGE using 10-20% Tricine minigels (Invitrogen).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jim Bardwell, Bernd Bukau, Isabelle Iost, Rowena Matthews, Matthias Müller and Knud Nierhaus for gifts of strains and antisera. We also thank Manu Hegde and Frances Yap for helpful comments on the manuscript. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases and by BBSRC grant BB/G011281 to C.A.W.
REFERENCES
- Apirion D. Three genes that affect Escherichia coli ribosomes. J Mol Biol. 1967;30:255–275. [PubMed] [Google Scholar]
- Becker T, Bhushan S, Jarasch A, Armache J-P, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon E, Beckmann R. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- Berndt U, Oellerer S, Zhang Y, Johnson AE, Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc Natl Acad Sci USA. 2009;106:1398–1403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Gartmann M, Halic M, Armache J-P, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R. α-helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol. 2010;17:313–318. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- Blobel G, Sabatini DD. Controlled proteolysis of nascent polypeptides in rat liver cell fractions. J Cell Biol. 1970;45:130–145. doi: 10.1083/jcb.45.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornemann T, Jockel J, Rodnina MV, Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat Struct Mol Biol. 2008;15:494–499. doi: 10.1038/nsmb.1402. [DOI] [PubMed] [Google Scholar]
- Colombo S, Longhi R, Alcaro S, Ortuso F, Sprocati T, Flora A, Borgese N. N-myristoylation determine dual targeting of mammalian NADH-cytochrome b(5) reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J Cell Biol. 2005;168:735–745. doi: 10.1083/jcb.200407082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell. 2005;19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Desvaux M, Scott-Tucker A, Turner SM, Cooper LM, Huber D, Nataro JP, Henderson IR. A conserved extended signal peptide region directs posttranslational protein translocation via a novel mechanism. Microbiology. 2007;153:59–70. doi: 10.1099/mic.0.29091-0. [DOI] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- Eisner G, Koch H-G, Beck K, Brunner J, Müller M. Ligand crowding at a nascent signal sequence. J Cell Biol. 2003;163:35–44. doi: 10.1083/jcb.200306069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner G, Moser M, Schäfer U, Beck K, Müller M. Alternate recruitment of signal recognition particle and trigger factor to the signal sequence of a growing nascent polypeptide. J Biol Chem. 2006;281:7172–7179. doi: 10.1074/jbc.M511388200. [DOI] [PubMed] [Google Scholar]
- Farewell A, Neidhardt FC. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J Bacteriol. 1998;180:4704–4710. doi: 10.1128/jb.180.17.4704-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabashvili IS, Gregory ST, Valle M, Grassucci R, Worbs M, Wahl MC, Dahlberg AE, Frank J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol Cell. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- Georgiou G, Valax P. Expression of correctly folded proteins in Escherichia coli. Curr Opin Biotechnol. 1996;7:190–197. doi: 10.1016/s0958-1669(96)80012-7. [DOI] [PubMed] [Google Scholar]
- Gu SQ, Peske F, Wieden HJ, Rodnina MV, Wintermeyer W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 2003;9:566–573. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardesty B, Kramer G. Folding of a nascent peptide on the ribosome. Prog Nucleic Acid Res Mol Biol. 2001;66:41–66. doi: 10.1016/s0079-6603(00)66026-9. [DOI] [PubMed] [Google Scholar]
- Hiss JA, Schneider G. Domain organization of long autotransporter signal peptides. Bioinform Biol Insights. 2009;3:189–204. doi: 10.4137/bbi.s3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Kosolapov A, Deutsch C. Tertiary interactions within the ribosomal exit tunnel. Nat Struct Mol Biol. 2009;16:405–411. doi: 10.1038/nsmb.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Rauch T, Rist W, Vorderwülbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- Lee HC, Bernstein HD. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc Natl Acad Sci USA. 2001;98:3471–3476. doi: 10.1073/pnas.051484198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Bernstein HD. Trigger factor retards protein export in Escherichia coli. J Biol Chem. 2002;277:43527–43535. doi: 10.1074/jbc.M205950200. [DOI] [PubMed] [Google Scholar]
- Liao S, Lin J, Do H, Johnson AE. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- Lu J, Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005;44:8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- Lu J, Kobertz WR, Deutsch C. Mapping the electrostatic potential within the ribosomal exit tunnel. J Mol Biol. 2007;371:1378–1391. doi: 10.1016/j.jmb.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Malkin LI, Rich A. Partial resistance of nascent polypeptide chains to proteolytic digestion due to ribosomal shielding. J Mol Biol. 1967;26:329–346. doi: 10.1016/0022-2836(67)90301-4. [DOI] [PubMed] [Google Scholar]
- Milligan RA, Unwin PN. Location of exit channel for nascent protein in 80S ribosome. Nature. 1986;319:693–695. doi: 10.1038/319693a0. [DOI] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Mitra K, Frank J. A model for co-translational translocation: ribosome-regulated nascent polypeptide translocation at the protein-conducting channel. FEBS Lett. 2006;580:3353–3360. doi: 10.1016/j.febslet.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- O'Connor M, Gregory ST, Dahlberg AE. Multiple defects in translation associated with altered ribosomal protein L4. Nucl Acids Res. 2004;32:5750–5756. doi: 10.1093/nar/gkh913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JH, Szabady RL, Bernstein HD. An unusual signal peptide extension inhibits the binding of bacterial presecretory proteins to the signal recognition particle, trigger factor, and the SecYEG complex. J Biol Chem. 2006;281:9038–9048. doi: 10.1074/jbc.M508681200. [DOI] [PubMed] [Google Scholar]
- Peterson JH, Woolhead CA, Bernstein HD. Basic amino acids in a distinct subset of signal peptides promote interaction with the signal recognition particle. J Biol Chem. 2003;278:46155–46162. doi: 10.1074/jbc.M309082200. [DOI] [PubMed] [Google Scholar]
- Petrone PM, Snow CD, Lucent D, Pande VS. Side-chain recognition and gating in the ribosome exit tunnel. Proc Natl Acad Sci USA. 2008;105:16549–16554. doi: 10.1073/pnas.0801795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool MR. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J Cell Biol. 2009;185:889–902. doi: 10.1083/jcb.200807066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–1348. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- Schlünzen F, Wilson DN, Tian P, Harms JM, McInnes SJ, Hansen HAS, Albrecht R, Buerger J, Wilbanks SM, Fucini P. The binding mode of the trigger factor on the ribosome: implications for protein folding and SRP interaction. Structure. 2005;13:1685–1694. doi: 10.1016/j.str.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache J-P, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed MA, Wang DIC, King J. Specific aggregation of partially folded polypeptide chains: the molecular basis of inclusion body composition. Nat Biotechnol. 1996;14:1283–1287. doi: 10.1038/nbt1096-1283. [DOI] [PubMed] [Google Scholar]
- Szabady RL, Peterson JH, Skillman KM, Bernstein HD. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci USA. 2005;102:221–226. doi: 10.1073/pnas.0406055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T, Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell. 1999;97:755–65. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- Tu LW, Deutsch C. A folding zone in the ribosomal exit tunnel for Kv1.3 helix formation. J Mol Biol. 2010;396:346–1360. doi: 10.1016/j.jmb.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrandt ND, Newitt JA, Bernstein HD. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- Ullers RS, Houben EN, Raine A, ten Hagen-Jongman CM, Ehrenberg M, Brunner J, Oudega B, Harms N, Luirink J. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J Cell Biol. 2003;161:679–684. doi: 10.1083/jcb.200302130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Von Heijne G. Signal sequences: the limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- Wegrzyn RD, Hofmann D, Merz F, Nikolay R, Rauch T, Graf C, Deuerling E. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J Biol Chem. 2006;281:2847–2857. doi: 10.1074/jbc.M511420200. [DOI] [PubMed] [Google Scholar]
- Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- Woolhead CA, Johnson AE, Bernstein HD. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol Cell. 2006;22:587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Yap M-N, Bernstein HD. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol Cell. 2009;34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonath A, Leonard KR, Wittmann HG. A tunnel in the large ribosomal subunit revealed by three-dimensional image reconstruction. Science. 1987;236:813–816. doi: 10.1126/science.3576200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.